Distinct genomic structural variations are found in pediatric high-grade glioma, affecting tumor gene expression.
Abstract
Pediatric high-grade gliomas (pHGGs), including glioblastoma multiforme (GBM) and diffuse intrinsic pontine glioma (DIPG), are morbid brain tumors. Even with treatment survival is poor, making pHGG the number one cause of cancer death in children. Up to 80% of DIPGs harbor a somatic missense mutation in genes encoding histone H3. To investigate whether H3K27M is associated with distinct chromatin structure that alters transcription regulation, we generated the first high-resolution Hi-C maps of pHGG cell lines and tumor tissue. By integrating transcriptome (RNA-seq), enhancer landscape (ChIP-seq), genome structure (Hi-C), and chromatin accessibility (ATAC-seq) datasets from H3K27M and wild-type specimens, we identified tumor-specific enhancers and regulatory networks for known oncogenes. We identified genomic structural variations that lead to potential enhancer hijacking and gene coamplification, including A2M, JAG2, and FLRT1. Together, our results imply three-dimensional genome alterations may play a critical role in the pHGG epigenetic landscape and contribute to tumorigenesis.
INTRODUCTION
Pediatric high-grade glioma (pHGG) is a highly morbid brain tumor, with a 5-year survival of less than 20% despite treatment. pHGG is the most common malignant brain tumor in children and therefore represents the greatest cause of pediatric cancer–related death. pHGG may arise in the cerebral hemispheres, where it is known as glioblastoma multiforme (GBM), or in the midline thalamus or brainstem [diffuse intrinsic pontine glioma (DIPG)]. Recent molecular profiling of pHGG reveals genetic and epigenetic characteristics distinct from adult glioma. Notably, a high rate of somatic missense mutations in genes encoding histone H3 isoforms, including H3F3A and HIST1H3B, occurs in pediatric GBM and DIPG but are rarely detected in adult gliomas. Up to 80% of DIPGs harbor the mutant histone protein H3K27M, which results from when a methionine is encoded rather than lysine as the 27th amino acid on the histone H3.1 and H3.3 N-terminal tail. The H3K27M mutation is associated with a more aggressive clinical course and a poorer overall response to therapy, as well as distinct patterns of DNA methylation, chromatin structure, gene transcription, and protein expression (1). As a result, the H3K27M mutant protein is thought to play a role in gliomagenesis by altering epigenetic control of gene expression (2, 3).
For example, in DIPG, the H3K27M mutation is accompanied by a reduction in global genomic H3K27 trimethylation (H3K27me3), a transcriptionally repressive mark rendered by the polycomb-repressive complex 2 (PRC2). In turn, this loss of H3K27me3 has been shown to be associated with a reciprocal increase in genomic H3K27ac, a mark recognized by bromodomain (BRD) and extraterminal (BET)/BRD proteins to recruit RNA polymerase II and activate transcription. In addition, there are specific regions of increased H3K27me3 enrichment in H3K27M cells relative to wild type, further altering the epigenetic landscape of DIPG. Therefore, the effect of the H3K27M protein on chromatin structure and function may be regionally specific, dependent on the local genetic environment. Recent studies revealed variant-specific enhancer architecture in DIPG (4), and a global increase in the level of other activating histone marks such as H3K36 di- and trimethylation (H3K36me2/3) (5), in the setting in H3K27M mutant glioma, suggesting a more complex effect of this oncohistone on the tumor epigenome.
The mechanism by which the H3K27M protein affects chromatin structure and function, and thereby gliomagenesis, is still not completely understood. While multiple recent studies evaluate the epigenetic factors contributing to pHGG biology, characterization of the tridimensional (3D) structure of the pHGG genome has yet to be reported. Disruption of genome architecture is increasingly understood to contribute to cancer biology via dysregulation of gene expression programs (6). In adult glioma, 3D genome studies revealed hypermethylation of a CCCTC-binding factor (CTCF) binding motif in some IDH1 mutant gliomas, thereby eliminating a domain boundary and resulting in sustained expression of PDGFRA, a known glioma oncogene. Further, through integration of genomics and 3D structural genomics datasets, CD276 targeting was recently identified as a novel therapeutic strategy for GBM via suppression the tumor stem cell self-renewal. Thus, characterizing the 3D genome of pHGG could provide valuable insight structural variations (SVs) and enhancer-promoter interactions that lead to tumor formation, particularly in the setting of histone H3 mutations.
Here, we integrated transcriptome analysis [RNA sequencing (RNA-seq)], chromatin immunoprecipitation sequencing (ChIP-seq), high-throughput chromosome conformation capture (Hi-C), and chromatin accessibility [assay for transposase-accessible chromatin using sequencing (ATAC-seq)] datasets for H3 mutant and wild-type pHGG specimens, including tumor cell lines and tissues. We identified tumor-specific enhancers and regulatory networks for known oncogenes in DIPG and GBM. In addition, we saw genome distinct SVs between DIPG and GBM that lead to enhancer hijacking events, resulting in aberrant oncogenic gene expressions. This is, to our best knowledge, the first comprehensive genomic characterization of its kind for pHGG, lending new insight into mechanisms of transcription regulation in these tumors and revealing previously unknown therapeutic vulnerabilities.
RESULTS
To better understand how SVs in genome structure and enhancer-promoter interactions affect gene expression in pHGG in the presence and absence of the H3K27M mutation, we integrated high-throughput multiomics data in patient-derived cell lines (DIPG, n = 6; GBM, n = 3; normal, n = 2; table S1A) and frozen tissue specimens (DIPG, n = 1; normal brainstem, n = 1; table S1B). Analyses included cell line RNA-seq; ChIP-seq for H3K27ac, H3K27me3, and H3K27M; and genome-wide chromatin conformation capture (Hi-C), as well as tissue Hi-C and ATAC-seq (Fig. 1A and fig. S1). All data generated are available in Gene Expression Omnibus (GEO) series GSE162976.
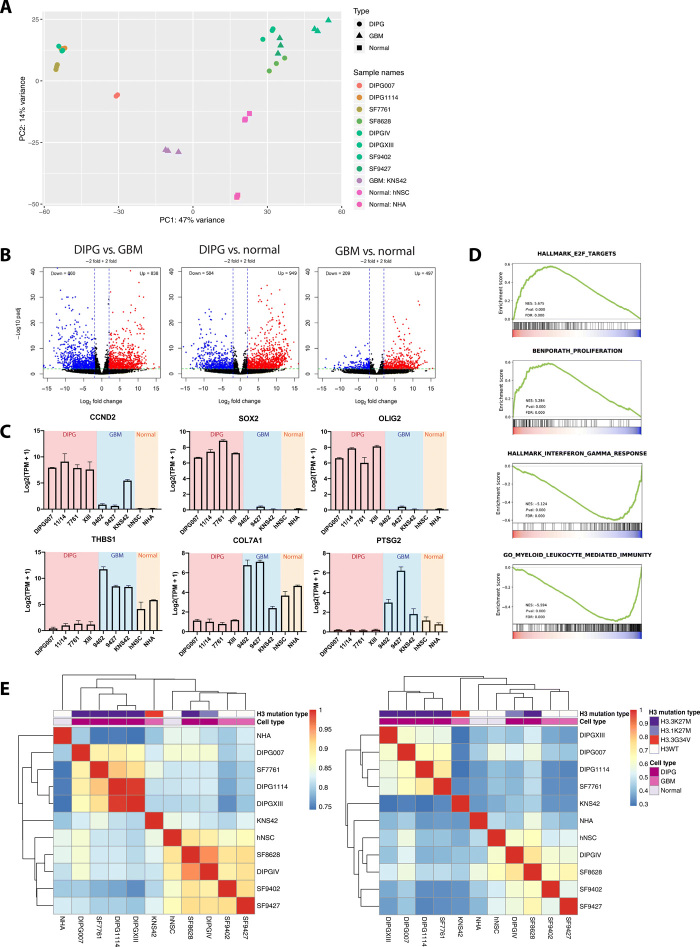
Fig. 1. DIPG and GBM cells harbor distinct transcriptomes corresponding with H3K27ac enhancer enrichment.
(A) PCA of cell transcriptome data: Unsupervised clustering of technical replicates (n = 3) and diagnosis (DIPG, n = 6; GBM, n = 3; normal, n = 2). PC, principal component. (B) Differentially expressed genes (DEGs) between DIPG, GBM, and normal cell lines. Red, up-regulated relative to normal cell lines; blue, down-regulated (P < 0.01, |log2FC| ≥ 2). (C) Differentially expressed genes between DIPG and GBM cell lines. Y axis = expression level for each gene [log2(TPM + 1)]. TPM, transcripts per million. (D) Gene Set Enrichment Analysis (GSEA) of cell transcriptomes: Increased E2F signaling and repression of cell proliferation and immune response pathways in DIPG versus GBM. Signal-to-noise ratio was used to rank the genes per correlation with DIPG (red) or GBM (blue). Green curve = enrichment score. Normalized enrichment score (NES), corresponding P value (Pval), and false discovery rate (FDR) are reported. Enriched pathways: Cell cycle targets of E2F transcription factors (TFs), genes expressed in human breast tumors, genes up-regulated in response to interferon gamma, and genes involved in immune response by a myeloid leukocyte. (E) Unsupervised hierarchical clustering of whole transcriptome RNA-seq data (left) and genome-wide H3K27ac signals (right), demonstrating similar transcriptome and H3K27ac landscapes. Correlations for RNA-seq data were calculated using the Pearson correlation coefficient from the log-transformed raw read counts, and correlations for K27ac ChIP-seq data were calculated on the basis of genome-wide bigwig signals at 10-kb resolution using deeptools: DIPG tissue (n = 8), normal pons tissue (n = 2), DIPG cell lines (n = 6), GBM cell lines (n = 3), and nontumor cell lines (n = 2), including NHAs and human normal stem cells (hNSCs). Tissue data were obtained from published results (table S1). WT, wild type.
Transcriptome analysis reveals tumor-specific patterns of gene expression
To evaluate the tumor transcriptomic landscape, RNA-seq was performed on all cell lines studied. Principal components analysis (PCA) was conducted for the top 500 variable genes across all specimens. In all samples, replicates clustered tightly together (Fig. 1A). Spearman correlation was calculated for uniquely mapped reads among replicates, with an average of 0.968, demonstrating the technical reproducibility of our RNA-seq data (table S2A). Of the six H3K27M mutant DIPG cell lines analyzed, two distinct clusters were identified on PCA: one group with SF7761, DIPGXIII, and DIPG1114 and a second with SF8628 and DIPGIV (the latter being the only H3.1K27M mutant line analyzed). Therefore, to obtain uniform differential expression gene sets, we excluded DIPGIV and SF8628 from subsequent analyses characterized below. Two H3 wild-type normal cell lines, normal human astrocytes (NHAs) and human neural stem cells, clustered independently from the cancer cell lines. As expected, PCA results also indicated that the two different pHGG types, DIPG and GBM, are more distinct from normal cells than from each other. There is a clear distinction between DIPG and GBM clusters. This observation is recapitulated in the heatmap depicting unsupervised hierarchical analysis of the top 500 most variable genes across all cell lines studied (fig. S2A), which separates transcriptome profiles into respective diagnostic categories (DIPG, GBM, and normal) by technical replicate. Of the GBM cell lines analyzed, the H3 wild-type lines SF9402 and SF9427 clustered together, distinctly from KNS42, which harbors an H3.3G34V mutation. Because of this unique mutation status, we also excluded KNS42 from subsequent analyses.
DIPG transcriptomes are enriched for cell proliferation and immune suppression genes
To identify differentially expressed genes between subgroups, we compared gene expression levels in four H3.3K27M mutant DIPG cell lines (DIPG007, DIPGXIII, SF7761, and DIPG1114) and two H3K27 wild-type GBM cell lines (SF9427 and SF9402). A total of 838 genes were identified as up-regulated in DIPG compared to GBM cells (log2fold change > 2 and adjusted P < 0.01), while 860 genes were down-regulated (log2fold change < −2 and adjusted P < 0.01). When gene expression profiles of glioma cell lines were compared to normal cell lines, we observed a higher number of cancer-specific differentially expressed genes in DIPG compared to normal cell lines. A total of 949 and 497 genes were up-regulated in DIPG and GBM compared to normal cell lines, respectively (Fig. 1B). These results indicate differential transcription regulation, especially activation, of distinct subsets of genes in DIPG and GBM. For example, three markers of stem and glial progenitors, CCND2, SOX2, and OLIG2, are significantly up-regulated in DIPG cell lines: SOX2 and OLIG2 have been previously shown to be highly expressed in DIPGs (Fig. 1C) (7). In contrast, THBS1, COL7A1, and PTSG2 are significantly up-regulated in GBM relative to DIPG cell lines.
To identify functional annotations that are differentially overrepresented in the two cancer types, we performed the Gene set enrichment analysis (GSEA) for DIPG- and GBM-specific differentially expressed genes (Fig. 1D). In DIPG, we detected the enrichment of gene sets associated with immune response, including targets of E2F, and cell proliferation pathways. Gene sets differentially enriched in GBM cell lines were also involved in immune system responses, such as interferon-γ response and myeloid leukocyte–mediated immunity. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses of DIPG-specific up-regulated genes revealed similar results and also implicated pathways involved in synaptic assembly and axonal guidance (fig. S3).
Additional functional pathways analysis of differentially expressed genes between DIPG and normal cell lines implicated SOX2 and SUZ12 as top activated upstream regulators in DIPG (fig. S4, A and B), with cancer as the top represented disease process. Synaptogenesis signaling was identified as the top activated canonical pathway in DIPG relative to normal cells (fig. S4C). The top implicated upstream regulators between DIPG and GBM cell lines were SOX2 and TRIM24 activation (fig. S4D), with cancer as the top disease process. When comparing DIPG versus GBM gene expressions, axonal guidance signaling and synaptogenesis (fig. S4C) emerged as the top canonical pathways in DIPG.
DIPG-specific active enhancer landscapes
To ensure that our cell line data accurately represent the tumor genomic landscape, we performed ChIP followed by deep sequencing (ChIP-seq) of our cell lines (table S2B) using the ENCODE pipeline. Spearman correlation was calculated for genome-wide bigwig signals among cell line ChIP-seq replicates, with average within-sample correlation of >0.9, demonstrating the technical reproducibility of our ChIP-seq data (fig. S2B). As promoter H3K27ac enrichment is associated with transcription activation, we expected gene expression (RNA-seq data) to share similar clustering patterns with H3K27ac enrichment patterns (ChIP-seq data). To confirm this, we performed unsupervised hierarchical clustering of mRNA expression patterns and genomic H3K27ac enrichment separately (Fig. 1E). We saw distinct clusters by diagnosis (DIPG, GBM, and normal cell lines). Furthermore, H3.3K27M DIPG cell lines (DIPG007, SF7761, DIPG1114, and DIPGXIII) cluster together, away from H3 wild-type specimens. Distinct clusters by mutation status were also observed in tissue, although to a lesser degree. As with RNA-seq data, genomic H3K27Ac enrichment patterns in SF8628 and DIPGIV show higher correlation with enrichment in SF9427 and SF9402 compared to the other four DIPG cell lines. This is possibly due to the fact that the DIPGIV is H3.1K27M mutant and that SF8628 could senesce over time (8). More focused comparison between histone H3.3 mutant (DIPG) and histone H3.3 wild type (GBM) demonstrated consistency of the clustering from gene expression and genome-wide H3K27Ac enrichment, indicating that H3K27Ac-marked enhancers could potentially contribute to the tumor subtype differential gene transcription.
Tumor-specific enhancer landscapes lead to differential oncogenic signaling
Aberrant enhancer activity is a key driver of gene expression that contributes to tumor formation, maintenance, and progression (9). Therefore, we hypothesized that DIPG and GBM show distinct, locus-specific gains and losses of enhancer activity across the epigenome, thereby resulting in distinct mechanisms of transcription regulation by tumor type and H3K27M mutation status. Distal H3K27ac peaks that are 2.5 kb away from gene promoter regions have been used as markers for active enhancers. To identify tumor type–specific enhancers, we first merged all the filtered H3K27ac peaks (P < 1 × 10−5 and q < 0.01) and obtained a total enhancer peak set of 328,242 peaks. H3K27ac signals from each cell line were calculated across this peak set, with 2883 peaks identified as enriching DIPG-specific enhancers, 4690 peaks as enriching H3 wild-type GBM-specific enhancers, and 4289 peaks as enriching KNS42-specific enhancers (Fig. 2A, left). Since KNS42 harbors a unique H3G34V mutation that sets it apart from the other two GBM cell lines analyzed, we excluded it from subsequent analyses to ensure a more homogenous GBM genomic landscape for comparison to DIPG. We then plotted H3.3K27M signals at the identified enhancer regions and observed colocalization of H3.3K27M and H3K27ac at DIPG-specific enhancer regions across all DIPG cell lines. Tumor-specific distal H3K27me3 signals were also identified using the same strategy. Consistent with previous studies, H3K27me3 signals were globally decreased in DIPG compared to GBM (fig. S2C) (10), and H3.3K27M colocalized with H3K27ac (Fig. 2A, right). Representative genome browser tracks are shown to depict H3K27ac-enriched DIPG-specific enhancer regions, including ASCL1, JAK3, and PHLDA1, and H3K27ac-enriched GBM-specific enhancer regions, including IMP3, SMAD3, and ATAD1 (Fig. 2B).
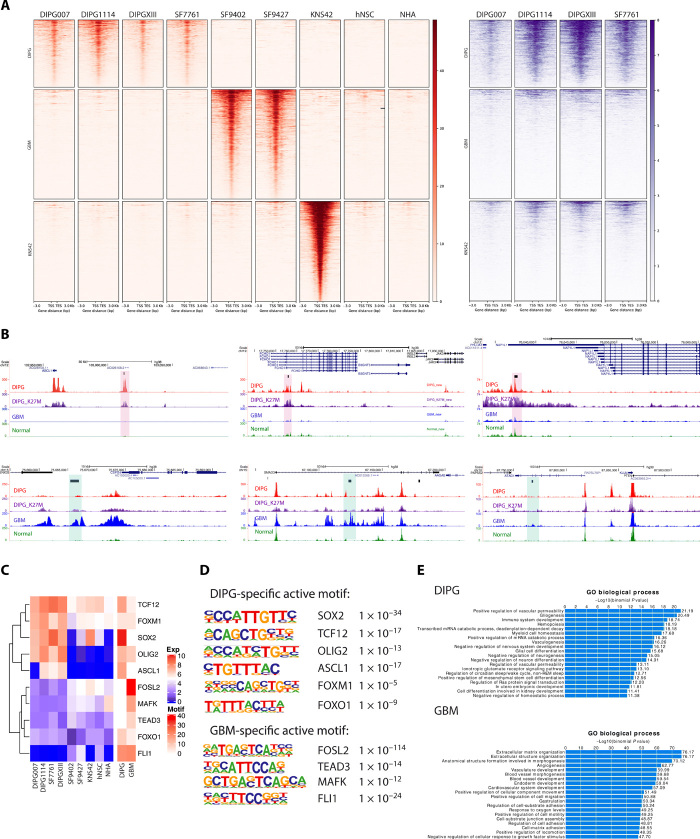
Fig. 2. ChIP-seq data revealed tumor-specific enhancer landscapes that result in specific motif and TF enrichment.
(A) H3K27ac ChIP-seq reveals tumor-specific distal enhancers (red). DIPG-, GBM-, and G34V-specific distal enhancers are shown, with normal cell lines as controls. H3.3K27M signals were shown for DIPG cell lines at the same regions of H3K27ac (purple). The GBM cell line KNS42 (H3G34V mutant) is excluded from subsequent motif enrichment analysis due to its distinct molecular profile relative to the H3 wild-type GBM cell lines (n = 2). (B) Representative genome browser view of DIPG- and GBM-specific enhancers. Tracks are superimposed H3K27ac and H3.3K27M ChIP-seq tracks from DIPG (n = 4), GBM (n = 2), and normal (n = 2) cell lines. Tumor-type specific enhancer peak calls are represented as black bars above tracks. DIPG-specific enhancers are highlighted in red and GBM-specific enhancers in blue. (C) Motif enrichment with differentially expressed genes between DIPG and GBM cell lines. Expression levels of the motif genes are shown (left); heatmap shows the motif enrichment level (−log10 P value; right). Tumor-specific active motifs, defined as highly enriched motifs with high differential gene expression in a given tumor type, are shown (table, right). (D) TF motif enrichment is distinct in DIPG versus GBM cell lines. (E) GREAT analysis for DIPG- and GBM-specific distal enhancer regions TF motif enrichment implicates distinct biological processes in DIPG versus GBM.
We then performed motif enrichment analysis at the tumor-specific, H3K27ac-enriched enhancer regions. In DIPG, FOX, SOX, STAT, and SMAD families were among the top motifs enriched with H3K27ac. Similarly, BACH, ETS, bZIP, TEAD, and FOS families were enriched in H3K27ac at GBM-specific enhancers. Transcription factor (TF) members inside identified that families usually share similar motifs, which makes it difficult to identify the real functional TF. We identified motif enrichment of genes that are significantly differentially expressed between DIPG and GBM (Fig. 2C) and of additional genes that were not differentially expressed these two groups (fig. S2D). To identify candidate functional TFs, we took the expression level of TFs into consideration as well. For example, in DIPGs, SOX2, OLIG2, TCF12, ASCL1, FOXM1, and FOXO1 were all highly expressed and showed significant motif enrichment (Fig. 2, C and D). These motifs were therefore identified as DIPG-specific active motifs. In contrast, FOSL2, TEAD3, MAFK, and FLI1 were identified as GBM-specific active motifs (Fig. 2, C and D). Last, to determine how the differences in enhancer architecture between DIPG and GBM translate to oncogenic signaling, Genomic Regions Enrichment of Annotations Tool (GREAT) analysis was performed for tumor-specific enhancers. In accordance with GSEA analysis for RNA-seq data, DIPG-specific enhancers also showed enrichment in proliferation-related pathways, whereas GBM-specific enhancers are more enriched for cell migration, hypoxia, and inflammation-related pathways (Fig. 2E).
DIPG and GBM exhibit distinct 3D genome structures
Previous studies have shown that 3D chromatin organization is associated with epigenetic activation or silencing of gene transcription (11–13). Therefore, to better understand the impact of chromatin structure on regulation of DIPG cancer genome, we performed high-resolution Hi-C experiments (>700 M reads each) for DIPG (DIPGXIII and DIPG007), GBM (SF9427), and NHA cell lines, as well as a fresh DIPG tumor tissue (3810) (fig. S5, A to E, and table S2C). Normalized Hi-C maps revealed differences in topologically associating domains (TADs) and DNA looping between H3 mutant (DIPG) and wild-type (GBM and NHA) cell lines at biologically relevant genomic regions. For example, at MYCN, we observed sub-TAD structures and more frequent interactions within this TAD region in DIPGs compared to GBM and NHA. In line with this finding, we also observed heightened within-TAD interaction at MYCN in DIPG tissue (Fig. 3A). The expression of MYCN gene is significantly higher in the DIPG cell lines, except for the DIPG007, compared to the H3.3 wild-type GBM cell lines (fig. S7A). Differences in TAD structure between tumor types were observed in multiple genomic regions, including GAS7 (figs. S6 and S7A), which was highly expressed in all the four DIPG cell lines. This indicates that the 3D genomic feature change may be one of the contributing factors that regulate the gene expression, although it might not be the only one.
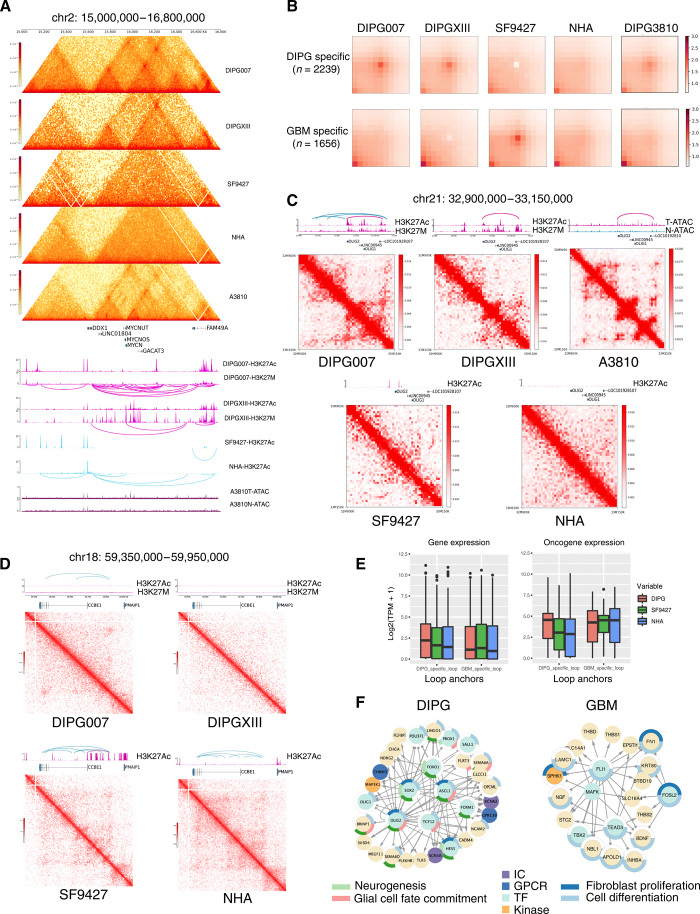
Fig. 3. DIPG and GBM showed distinct 3D genome organization affecting multiple promotor and enhancer regions in each tumor type.
(A) Hi-C contact maps and H3K27ac ChIP-seq tracks at MYCN (chr2: 15,000,000 to 16,750,000 bp). Loops are shown under H3K27ac and K27M tracks. DIPG cell lines (DIPG007 and DIPGXIII) and tissue (3810 T) have higher within-TAD interaction frequency and intensities compared to GBM (SF9427) and normal (NHA) cells. Sub-TADs are formed near MYCN. ATAC-seq shows DIPG tissue (3810 T) with greater chromatin accessibility compared to normal brain stem tissue from the same patient (3810 N). (B) APA plot shows aggregated signals from tumor type-specific loops. (C) DIPG-specific loops link OLIG2 with a distal enhancer located within the gene body of lncRNA LOC101928107. Loops are shown above each H3K27ac track. Hi-C maps show genomic regions chr21: 32,900,000 to 33,260,000 at 5-kb resolution. DIPG-specific loops were not observed in SF9426 or NHA. ATAC-seq of tissue samples shows DIPG (3810 T) to have greater chromatin accessibility in the OLIG2 enhancer region compared to normal brainstem (3810 N). (D) GBM-specific loops link the CCBE1 promoter with an upstream enhancer highlighted in purple. Hi-C maps show genomic regions chr18: 59,350,000 to 59,950,000 at 5-kb resolution. Loops were not observed in DIPG or normal cell lines in this region. (E) Gene expression in DIPG, GBM, and NHA at DIPG-specific and GBM-specific loop anchors (left). Oncogene expression in cell lines at these loop anchors (right). (F) Gene regulatory networks in DIPG and GBM. Each link represents tumor type–specific loops linking the tumor type–specific H3K27Ac enriched enhancers (shown in Fig 2A) and gene promoters. The circle surrounding the nodes represents functional pathways enrichment. IC, ion channel protein; GPCR, G protein–coupled receptors.
To systematically study the impact of 3D genomic features on transcription, we identified ~2877 TADs across cell lines at 40-kb resolution, according to the insulation scores (range: 2794 to 2963). When interrogating the differences of TADs between specimen types, we observed that most of the TAD boundaries remained stable across specimens (fig. S7B). In addition, we observed a slight gain in the number of TAD boundaries in DIPGs compared to GBM and normal cells, as well as in GBM compared to normal cells (fig. S7B). We next explored the consistency between the DIPG cell lines and patient tissue sample of the gained and loss TAD boundaries comparing to the GBM cell lines. We observed 333 (~26%) gained and 519 (35 to 49%) loss of overlapped boundary regions among the three DIPG samples (fig. S7C). The consistency was further confirmed by the lower insulation scores among DIPG samples at the 333 DIPG gained of TAD boundaries, and the valley of insulations score at the loss of TAD boundaries only occurred in the GBM sample (fig. S7, D and E). We further evaluated the relationship between TAD boundaries and gene expressions. Genes surrounding TAD boundries gained in DIPG had lower expression in DIPG samples, while genes with loss of TAD boundaries had higher espression in DIPG samples (fig. S7F). We further examined specifically for oncogenes within the boundary regions, and we observed similar patterns (fig. S7F). We further interrogated the H3K27M and H3K27Ac bindings within the TAD boundaries. We observed colocalization of H3K27ac and H3.3K27M peaks within TAD boundaries, consistent with previous observations (fig. S8, A and B) (10). Almost half of the TAD boundaries contain the H3K27M binding (fig. S8, C and D). In addition, the TAD boundaries that contain H3K27M or H3K27Ac peaks also had notable overlap. The H3K27M- and H3K27Ac-bound TAD boundaries also showed consistency between cell lines (fig. S8, C and D).
Next, examined DNA interaction loops for each cell line. An average of 17,141 loops were identified across cell lines (range: 8091 to 29,124) (table S3). To identify distinct looping structures in DIPG versus GBM, we first compared each DIPG cell line separately with the GBM line, SF9427, using diffPeakachu. We identified 2236 DIPG007-specific loops and 704 SF9427-specific loops, comparing DIPG007 with SF9427 (fig. S9B). Likewise, upon comparing DIPGXIII to SF9427, we identified 1231 DIPGXIII-specific loops and 1190 SF9427-specific loops (fig S9B). Differential loops identified via these two comparisons were then merged into a union set, which yielded 2239 DIPG-specific loops and 1656 GBM-specific loops (Fig. 3B). Aggregated peak analysis plots at these loop regions demonstrated tumor type–specific enrichment, as expected (Fig. 3B). It further showed the consistency from the DIPG 3810 tumor tissue sample (Fig. 3B). We also observed that average loop distances in DIPG-specific loops are longer than those specific to GBM (fig. S9C). This indicated a potentially more condensed 3D structure, which brought together longer distance regulatory elements. We further investigated the impact of differential loops on gene expression. We observed that for genes within the DIPG-specific loops, the expressions were higher in the two DIPG samples, while the expressions of genes within the GBM-specific loops were higher in the GBM sample (Fig. 3E). We observed similar patterns for the oncogenes (Fig. 3E). In conclusion, the 3D genomic plasticity between the two subtypes of brain tumor indicated that the rearranged enhancer promoter circuitry might be important for brain tumor development.
Tumor-specific enhancer-promoter loops and regulatory networks identified for known oncogenes
Physical contact or interaction between regulatory elements and gene promoters facilitates gene transcription regulation over long regions of the genome. Therefore, we next interrogated the roles of specific epigenetic regulatory features identified within differential loops regions in DIPG versus GBM. We observed a DIPG-specific loop between the OLIG2 promoter region and a downstream enhancer region located within the intron of LOC101928107 in our two DIPG cell lines and in DIPG tumor tissue (Fig. 3C). Comparing ATAC-seq data from DIPG tumor tissue and normal brainstem sample from the same patient, we found that DIPG chromatin has greater accessibility in the OLIG2 enhancer region compared to normal brainstem tissue (Fig. 3C). We also identified GBM-specific loops at CCBE1 and LNC01303 regions (Fig. 3D and fig S9A). These interactions were not observed in DIPG or NHA. To obtain an overview of the loop anchor distributions within each cell line, we queried for loop anchor regions that overlapped with gene promoter and enhancer regions. More than 20% of loops in DIPG, and 30% in GBM, can be categorized as enhancer-promoter loops (fig. S9D).
The significant distinctions in gene expression, enhancer landscape, and enhancer-promoter interactions between DIPG and GBM specimens implicate distinct regulatory networks of gene transcription. Therefore, we sought to predict the gene regulatory networks specific to DIPG and GBM. We first performed a motif scan using MEME-FIMO at tumor type–specific enhancer regions, revealing tumor specific enhancer-promoter interactions upstream of tumor-specific up-regulated genes. Focusing on DIPG-specific active motifs, we observed a network of interaction between OLIG2, TCF12, SOX2, and ASCL1. OLIG2 was also identified upstream of a number of neurogenesis related genes, including HES5, SEMA6D, and OLIG1. These findings imply that OLIG2 plays a central role in this DIPG-specific signaling network (Fig. 3F, left). In addition, we performed gene ontology enrichment analysis of this gene network, identifying neurogenesis and glial cell fate commitment as the top enriched functional pathways. Similarly, inspection of GBM-specific genes with enhancer-promoter interactions revealed a number of genes previously implicated in tumorigenesis, including TBX2, THBS1, and BDNF (Fig. 3F, right). Gene Ontology analysis of this gene network showed enrichment in regulation of fibroblast proliferations and cell differentiation pathways.
Last, on functional pathways analysis of the 35 genes with increased expression, H3K27ac enhancer enrichment, and loops within 50 kb of the transcription start site (TSS) in DIPG007 versus GBM cells, γ-aminobutyric acid (GABA) receptor signaling was identified as a top canonical pathway (P = 8.9 × 10−3). In addition, this gene set is mapped to cellular movement, nervous system development, and cell-to-cell signaling as top networks of molecular interaction (fig. S10A). Tamzemetostat, an inhibitor of the PRC2 subunit EZH2 (enhancer of zeste homolog 2), was identified as the top upstream regulator for this gene set (P = 1.23 × 10−3; fig. S10B).
Effects of pharmacologic alteration of BRD on DIPG genome structure
BET proteins, including BRD-containing protein 2 and 4 (BRD2 and BRD4, respectively), bind to acetylated lysine on histones on chromatin to subsequently facilitate transcriptional activation of RNA polymerase II. The role of BRD proteins in DIPG has been previously described (9, 10). Specifically, strong co-occupancy between BRD2 and BRD4 proteins and H3.3K27M-K27ac heterotypic nucleosomes was observed, suggesting a potential role of BRD proteins in DIPG pathogenesis (14). Abrogating the antidifferentiation effect of H3.3K27M-K27ac heterotypic nucleosomes through the use of BRD/BET inhibitors has been proven to decrease DIPG cell proliferation and promote terminal neuronal differentiation in vitro, as well as prolong DIPG mouse xenograft animal survival in vivo (10, 14).
Therefore, to explore the effect of BRD/BET inhibition on DIPG genome structures, we performed Hi-C analyses of DIPG007 cells treated with a BET BRD inhibitor (BBi), BRD4i, and a BRD degrader, dBET6. We found the BRD degrader to exert more pronounced antitumor effect compared to BRD inhibitor. Protein analyses showed greater BRD4 degradation and H3K27ac reduction with dBET6 at both 24 and 48 hours of drug treatment (Fig. 4A). Since BRD4 and H3K27ac levels were lowest at 24 hours of drug treatment, we selected this pharmacological treatment duration for both drugs for all subsequent analyses. In addition, dBET6 had strong antiproliferative effect on our DIPG cell line at 24 hours of treatment, consistent with the findings in a previous study using adult GBM cell lines (fig. S11) (15).
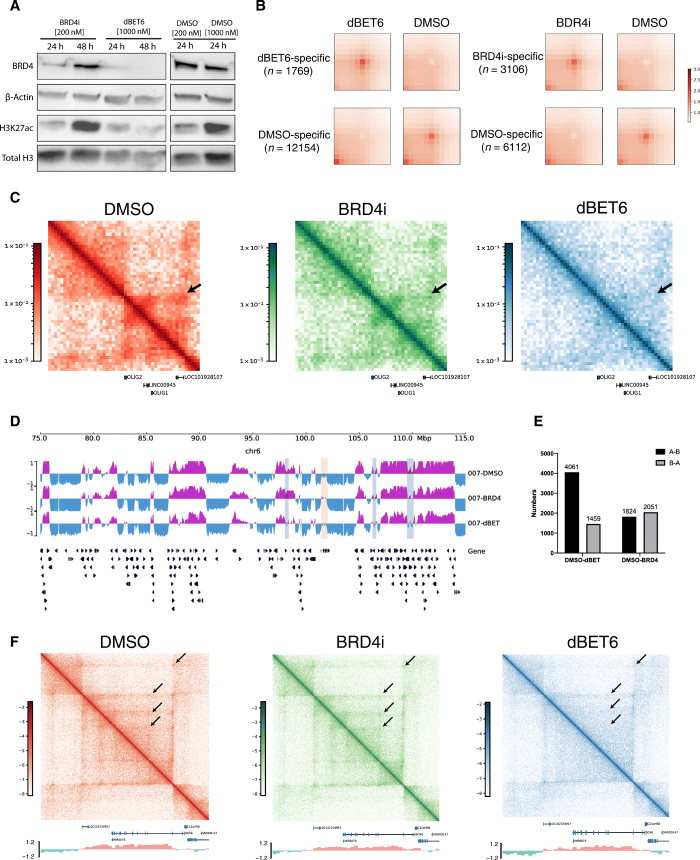
Fig. 4. Bromodomain-targeted treatment results in changes in DIPG 3D genome structure.
(A) Western blot analysis of BRD4i, β-actin, H3K27ac, and total H3 levels upon BRD4i or dBET6 treatment for 24 and 48 hours. The effect of both drugs on BRD4 levels was strongest at 24 hours, with greatest decrease after dBET6 treatment. (B) APA plot depicting aggregated signals from drug treatment–specific loops versus DMSO-specific loops (control). (C) Hi-C maps depicting genomic region chr21: 32,900,000 to 33,260,000, encompassing OLIG2, at 5-kb resolution in DMSO-, BRD4i-, and dBET6-treated cells. Tumor-specific `interaction (arrow) is disrupted 24 hours after BRD4i and dBET6 treatment, with dBET6 exerting a stronger effect. (D) A/B compartment designations at genomic region chr6: 75,000,000 to 115,000,000 at 40-kb resolution in DMSO-, BRD4i-, and dBET6-treated cells. A compartment, purple; B, blue. Regions of A to B switch between DMSO and drug treatments, light blue; regions of B to A switch, light yellow. Both A to B and B to A compartmental switches occur with drug treatment. (E) Number of genome-wide A/B compartment switches between DMSO and dBET6- or BRD4i-treated cells. A ➔ B and B ➔ A switches were observed with dBET6 and BRD4i treatment, compared to DMSO control. dBET6 exhibits a greater effect on compartmental switching. (F) Hi-C maps showing genomic region chr11: 15,200,000 to 17,000,000 bp, encompassing SOX6, at 5-kb resolution in DMSO-, BRD4i-, and dBET6-treated cells. A/B compartment signals are shown below Hi-C maps. Weakened interactions between the promoter region of SOX6 and surrounding regions are observed in dBET6-treated conditions, compared to DMSO control.
Next, we determined the effects of BRD/BET inhibition on DNA looping structures. We identified 36,996 loops in dimethyl sulfoxide (DMSO)–treated cells, 32,147 loops in BRD4i-treated cells, and 21,293 loops in dBET6-treated cells using Peakachu. To assess the genome-wide effects of drug treatment on chromatin interactions, we systematically identified the treatment-specific loops using diffPeakchu. We found that a large number of loops (12,154) were specific to the DMSO treatment compared to the dBET6 treatment, indicating significant loss of loops after the dBET6 treatment. This is similar in the BRD4i treatment but to a lesser extent (6112 loops loss), indicating stronger effects of dBET6 on 3D chromatin interactions (Fig. 4B).
As we previously identified OLIG2 as a core TF in the DIPG gene regulatory network with strong enhancer-promoter interactions in both DIPG cell lines and tumor tissue, we questioned whether H3K27Ac-targeted drug treatment could disrupt these enhancer-promoter interactions. The enhancer-promoter interaction at OLIG2 was disrupted after both BRD4i and dBET6 treatment (Fig. 4C), with dBET6 having a stronger effect. Given that BET/BRD inhibition reduces effect of H3K27ac, an active histone modification, we suspected this treatment would also be associated with the A/B compartment switches. We observed both A to B and B to A switches between drug treatments and DMSO control cell lines (Fig. 4, D and E). In general, there were 4488 A to B and 2005 B to A compartment switches from DMSO to dBET6 treatment. The number of A to B (2303) and B to A (2711) compartment switches from DMSO to BRD4i were not significantly different. These further indicated the stronger inhibitory effects of dBET6 treatment on active gene transcription relative to BRD4i. Last, the interactions surrounding the SOX6 regions were subsequently weakened with dBET6 treatment relative to DMSO controls (Fig. 4F). These results indicated that both BRD4i and dBET6 have the potential to change the 3D genomic structures, but dBET6 demonstrated a stronger effect, which might contribute to the stronger phenotypic effect.
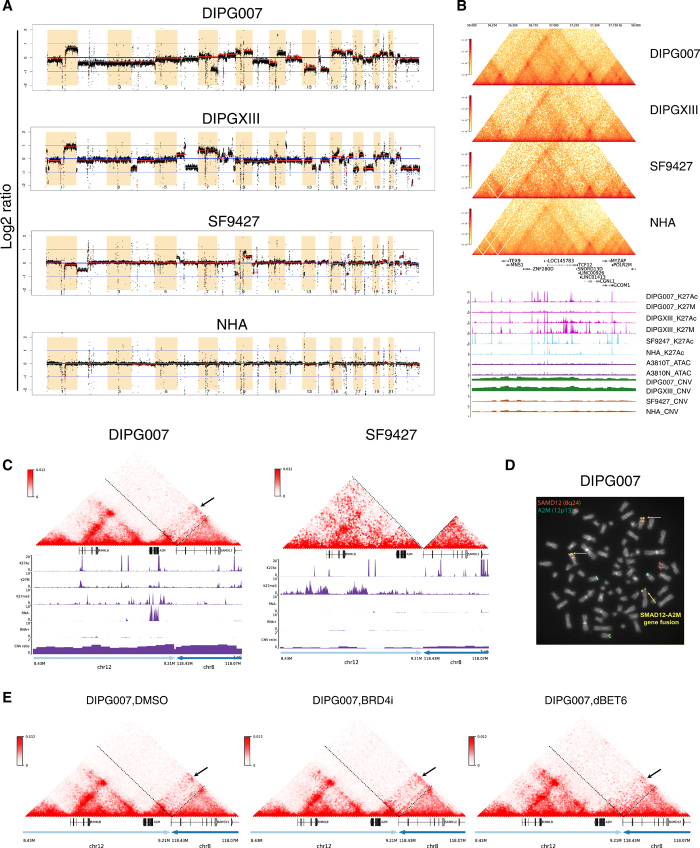
SVs lead to enhancer hijacking and coamplification events in pHGG
SV is of great relevance in cancer genetics. Studies have shown that SVs not only affect gene expression but also alter the copy number of regulatory elements, such as promoters or enhancers, and modify the 3D genome by disrupting TADs. The result of this so-called “position effect” is the expression of genes distant from SV breakpoints, thereby causing a disease-specific genotype (16, 17). Thus, to evaluate the effects of SVs on regulation of gene expression, we first used our cell line Hi-C data to infer copy number variation (CNV) profiles for the cells analyzed (Fig. 5A). As expected, we observed more regions of CNV in our glioma cell lines relative to NHAs, with the DIPG lines exhibiting the greatest number of CNVs. These genome-wide CNV profiles from our Hi-C data are consistent with previous comparative genomic hybridization studies in DIPG (18, 19), where 1q, 7p, and 7q gain was observed. 10q loss, which is commonly identified in DIPGs, was not observed in our data. We observed CNV gain in DIPG cell lines for 16.5 to 19% of DIPG-specific differentially up-regulated genes and CNV loss in 13.8 to 18.3% of DIPG-specific differentially down-regulated genes, depending on the DIPG cell line analyzed (fig. S12, A and B). Enhancer coamplification was also previously shown to contribute to gene up-regulation through extrachromosomal DNA in glioblastoma (9). Here, we observed coamplification of TCF12 gene and the enhancer within the TCF12 intron region, indicating that TCF12 up-regulation in DIPG is likely due to both copy number gain and enhancer coamplification (Fig. 5B).
Fig. 5. SVs lead to enhancer hijacking and gene coamplification in DIPG.
(A) Genome-wide CNV profiles generated from Hi-C data using HiNT. Red lines, CNV segmentations for each cell line. Y axis, log2 copy number ratio. Greater CNVs are observed in glioma relative to normal cell lines, with the greatest number of CNVs identified in DIPGs. (B) Hi-C, H3K27ac, and H3K27M ChIP-seq; CNV; and ATAC-seq tracks at TCF12 (chr15: 56,000,000 to 58,000,000). In DIPG cell lines, coamplification is observed at TCF12 and the enhancer within the TCF12 intron region. Copy number gain is observed in DIPG but not GBM or normal cell lines, indicating that TCF12 up-regulation is due to both copy number gain and enhancer coamplification in DIPG. There is no difference in chromatin accessibility between DIPG and normal tissue at this region. (C) Enhancer hijacking events identified in DIPG007 based on SV analysis. Interchromosomal translocation is observed between chr12: 8,430,000 to 9,210,000 and chr8: 118,070,000 to 118,430,000. Copy number gain is also observed in this region, indicating simultaneous enhancer hijacking and coamplification upstream of the A2M gene. (D) Fluorescence in situ hybridization (FISH) using dual fusion translocation probes were hybridized to metaphases for detection of translocation t(8;12)(q24;p13) of SAMD12 and A2M genes. FISH analysis of the SAMD12 gene was identified by fluorescent spectrum orange signals and A2M gene by spectrum green signals. A fusion event was identified by fused orange and green signals = yellow arrows. Additional concurrent copy number gains of the SAMD12 and A2M loci were observed. (E) Hi-C maps after DMSO, BRD4i, and dBET6 treatment for the same translocation region as shown in (C).
We also identified additional SVs in our cell lines (table S3) using Hi-C breakfinder. By reassembling the genomic locations of breaks based on SV data, we identified several potential enhancer hijacking events that exerted influence on observed gene expression levels. For example, we identified an interchromosomal translocation event for chr12: 8,430,000 to 9,210,000 and chr8: 118,070,000 to 118,430,000 regions from DIPG007 Hi-C data (Fig. 5C). Neo-loops were identified between the A2M promoter and candidate enhancers on chr8. Copy number gain was also observed at these regions, indicating simultaneous enhancer hijacking and coamplification events in DIPG007 upstream of the A2M gene. The enhancer hijacking and coamplification might contribute to the highly expression of this gene in the DIPG007 cell line (Fig. 5C). To verify our finding of a previously unidentified SAMD12-A2M interchromosomal translocation event, t(8;12)(q24;p13), we performed fluorescence in situ hybridization (FISH) in DIPG007 cells and observed the SAMD12-A2M fusions with concurrent copy number gains of the SAMD12 and A2M loci in metaphase from the DIPG007 cells (Fig. 5D), confirming the complex inversional translocations and copy number alterations observed by Hi-C data. Furthermore, we also observed weakened interactions with the dBET6 treatment, indicating this hijacked enhancer as a potential drug target (Fig. 5E). We observed similar such events for WNT3 (inversion, interchromosomal translocation, and amplification) and JAG2 (inversion and deletion) in DIPG007 (fig. S13, A and B).
DISCUSSION
pHGGs, including GBM and DIPG, are highly malignant brain tumors characterized by a high rate of somatic missense mutations in genes encoding histone H3 proteins. Greater understanding of the tumor structural and epigenetic landscape can provide insight into the mechanism of gliomagenesis, with the potential to reveal therapeutic vulnerabilities. The transcriptional programs of pediatric brain tumors have been well characterized in recent years. Here, we report that DIPG and GBM harbor distinct 3D genome alterations that, in addition to previously identified oncogenic driver mutations and dysregulated pathways, may play critical roles in downstream gene regulations and, hence, tumorigenesis. By comparing H3 mutant and wild-type cell lines, we also identified structural differences that may potentially be attributed to the H3.3K27M mutation. Our integrated genomic and epigenomic analyses of pHGG specimens allowed identification of previously unknown regulatory networks and gene targets for further study.
Among the genes we identified as up-regulated and H3K27ac-enriched in H3K27M mutant DIPG relative to H3 wild-type GBM and nontumor cell lines, OLIG2, SOX2, and TCF12 are all well studied in the context of glioma. OLIG2 is known to be highly expressed in DIPG and coexpressed with SOX2 and NES by multipotent neuroprogenitor cells thought to act as glioma stem cells (4, 7, 20). In addition, the coordinated activity of core TFs, including POU3F2, SOX2, and OLIG2, is capable of reprogramming differentiated GBM cells to restore tumorigenic capacity (21). Furthermore, reduction of H3K27me3 at the OLIG2 locus and OLIG2 overexpression is observed in DIPG cell lines harboring the H3K27M mutation (7). An OLIG2-dependent transcription program is thought to be critical for DIPG initiation and pathogenesis (7). Recent studies also found that SOX2- and OLIG1- positive progenitor cells give rise to both H3K27M DIPG and adult GBM (7, 22). In addition, OLIG2 and RUNX2 were found to robustly segregate two distinct glioma cell populations with differential response to drug therapy (23), suggesting that these factors might contribute uniquely to therapeutic resistance (24).
We also found TCF12, ASCL1, FOSL2, and FOX genes to be highly expressed in DIPG. Mutation of TCF12 has previously been reported in anaplastic oligodendroglioma (25). ASCL1 was shown to regulate other neurodevelopmental TFs and cell cycle genes in glioma mouse models, and its overexpression is associated with DIPG cell fitness and tumorigenicity (26, 27). FOSL2 was validated as a master regulator of mesenchymal state (28). Two TFs from the FOX family were also previously identified as highly differentially expressed in DIPG: Although prior studies on FOX function in DIPG are limited, these genes are reported to play a proproliferative role in glioma stem–like cells (29). In contrast, we found COL7A1, PTSG2, and THBS1 to be significantly up-regulated in GBM relative to DIPG cell lines. COL7A1 is reported to contribute to the invasive nature of GBM (30). Inducing THBS1 expression via SMAD3 contributes to the invasive behavior during GBM expansion (31). PTSG2 up-regulation is also related to GBM gliomagenesis and progression (32). Hence, our RNA-seq results are in line with prior findings of differential gene expressions in these tumors.
Given our findings of DIPG- and GBM-specific H3K27ac enhancer enrichments in DIPG cell lines that correlate with observed levels of gene expressions, we investigated the effect of H3K27ac-targeted therapy on the 3D genome structure in DIPG. BBIs block the acetyl-lysine binding activity of BET proteins and have broad effects against a wide range of cancer types (33–35). However, the efficacy of BBIs is counteracted by the emergence of both primary and acquired resistance in various types of cancers. In brain tumors, a growing dependency of adult GBM cells on BET proteins was observed regardless of their sensitivities to BBIs (15, 36). Superior anti-GBM activities were instead observed following treatment with dBET6, a CRBN-dependent BET protein degrader, where dBET6-mediated depletion of BET proteins showed distinct transcriptional and cellular responses compared to BRD inhibition (15). Furthermore, dBET6 treatment does not result in BBI resistance (15). Here, we observed that both BRD4i and dBET6 led to the disruption of tumor-specific enhancer-promoter interactions, A/B compartment switching, and reduction of genome-wide chromatin interactions in DIPG cell lines. All of these observations were more pronounced with dBET6 treatment, highlighting the advantage of BRD degraders over inhibitors in this context. These findings reveal a potential new therapeutic vulnerability in DIPG that warrants further exploration.
Next, because of the long-range activity of regulatory genomic regions, associating the activities of regulatory regions of the genome, including specific superenhancers, to a particular gene can be challenging. However, high-resolution Hi-C maps can provide valuable insight on individual, physical contacts between specific regulatory regions and specific genes. Therefore, we applied Hi-C analysis to our cell lines and a DIPG tumor tissue and observed distinct looping interactions in both DIPG and GBM that affect tumor-specific chromatin landscapes and transcription regulation. First, we found that the total number of looping interactions within a specific gene did not correlate with increased gene expression, but specific loops throughout the genome did have a strong effect on transcriptional output. For example, we observed DIPG-specific looping between the promoter region of OLIG2 and a downstream enhancer located within an intron of LOC101928107, which may contribute to the observed increased OLIG2 expression in DIPG. We also found enhanced expression of some genes corresponding to loops lacking overlap with CTCF binding site or enhancers. Overall, these observed tumor-specific looping events could underlie tumorigenic transcriptional programs in pHGG, such as the observed differential expression of OLIG2, SOX2, and FOXO1 in these tumors. Forced chromatin looping via CRISPR-dCas9 has been shown to induce gene activation in a reversible manner and, hence, may have therapeutic implications (37). This technique enables redirection of enhancers away from genes harboring disease-causing mutations. By identifying unique enhancer-promoter interactions at OLIG2 and SOX2 and the heightened chromatin accessibility in the OLIG2 region, our work reveals these genomic regions as potential therapeutic vulnerabilities in DIPG and GBM.
Our study also provided evidence of a greater number of TAD boundaries and longer loop distances in H3K27M mutant DIPG compared to H3 wild-type GBM cell lines. These differential chromatin structures led to distinct up-regulation of several genes in DIPG via oncogenic rearrangement of enhancers due to translocation/inversion, also known as “enhancer hijacking” events. This SV-dependent redistribution of genes from regions of transcriptionally silent chromatin to regions populated with active enhancers highlights the diverse and complex interplay between the cancer genome and epigenome (38). For example, we identified DIPG-specific enhancer hijacking events at A2M, JAG2, PTK2, and FLRT1, including an inversion/translocation event involving SAMD12 and A2M, confirmed by metaphase FISH. These findings are in line with reports of structural genome alterations in adult glioma, where boundary element disruption via hypomethylation leads to enhancer hijacking and oncogene activation (39). Enhancer hijacking was also recently identified as a driver of oncogene activation in medulloblastoma (39). Enhancer hijacking events present a therapeutic vulnerability that can be exploited for clinical cancer treatment. Drugs that target key components of superenhancers, such as BET protein inhibitors that target BRD4 and CDK inhibitors that target CDK7, have been studied for DIPG treatment (10, 40). Our newly identified enhancer hijacking events in DIPG may therefore provide insight to the mechanism of tumorigenesis and targeted treatment susceptibility or resistance.
Last, our integrated sequencing studies lend important insight into potential pathways of pediatric gliomagenesis. For example, of those differentially expressed genes found to be expressed in DIPG, a subset (n = 35) was also observed to harbor CNVs, H3K27ac distal peaks, and loops within 50 kb of the TSS in DIPG007. The functional pathway analysis of these genes implicated GABA receptor signaling as the top canonical pathway, which is in line with prior observations that GABA type A receptors are down-regulated in GBM (41), reducing the growth inhibitory effects of GABA signaling. In addition, the top disease implicated by differentially expressed genes in DIPG was cancer, while top implicated molecular and cellular functions included neuronal migration, cellular signaling, and neuron/oligodendrocyte development. These findings correspond with recent single-cell RNA-seq analysis of H3K27M DIPG specimens, which revealed that an oligodendrocyte precursor cell tumor subpopulation exhibited greater self-renewal, tumor propagating potential, and transcriptional programs (27). Functional pathway analyses of our sequencing data also implicated EZH2, a long studied target for DIPG treatment (40), as well as SOX2 and TRIM24 as top upstream regulators for differentially expressed genes in DIPG. Hence, our findings provide new insight for therapeutic strategies to treat DIPG that are in line with current understanding of DIPG biology and tumor heterogeneity. These data support further investigation into how the 3D genome structure contributes to tumor subclonal diversity and developmental hierarchies to identify more effective targeted therapies.
While our study provides insight into the genomic and epigenetic profiles of pHGGs, a notable limitation is that our integrated analyses were performed largely using pHGG cell lines. Previous studies have identified tumor cell subpopulations with distinct gene expressions and developmental hierarchy in DIPG tumor tissue (4, 27). In particular, Nagaraja and colleagues (4) found H3.3K27M and H3.1K27M DIPG tumors to be driven by distinct profiles of active regulatory elements, with some shared, but mostly distinctive oncogenic signaling pathways. While our studies overlap on the characterization of DIPG enhancer landscapes, Nagaraja and colleagues (4) further investigated the lineage of the cells and found that DIPG-specific superenhancers were enriched at oligodendrocyte lineage TFs. On the other hand, we focused on the difference in chromatin architecture that could contribute to the enhancer profiles observed in DIPG. It would therefore be of great interest to see whether the H3.3K27M and H3.1K27M DIPGs harbor analogous SVs via Hi-C analyses of additional DIPG tissue specimens. Further, as H3.3K27M and H3.1K27M mutant DIPG exhibit distinct transcriptomes and active enhancer profiles (4), determining differences in 3D genome structure between them may provide additional valuable insight. Given the data presented here, additional, similar analyses of rare DIPG tissue specimens are warranted.
In summary, we present the first integrated transcriptome, enhancer landscape, and 3D genome structure characterization of pHGG specimens, including DIPG and GBM cell lines, and DIPG tissue. By integrating transcriptome and epigenome data, we identified DIPG- and GBM-specific regulatory networks and potential therapeutic vulnerabilities including unique structure variations leading to enhancer hijacking and coamplification events that influence gene expression levels. The aberrant, DIPG-specific 3D genome structural characteristics presented here suggest a potential role for targeted CRISPR-dCas9 or pharmacologic BET inhibition as a strategy for DIPG treatment. Hence, these data provide a valuable new perspective on pHGG biology, with the potential to improve our understanding and clinical management of this devastating disease.
MATERIALS AND METHODS
Experimental design
To better understand how SVs in genome structure and enhancer-promoter interactions affect gene expression in pHGG, we integrated high-throughput multiomics data in patient-derived cell lines (DIPG, n = 6; GBM, n = 3; normal, n = 2; table S1A) and frozen tissue specimens (DIPG, n = 1; normal brainstem, n = 1; table S1B). We performed RNA-seq; ChIP-seq for H3K27ac, H3K27me3, and H3K27M; and genome-wide chromatin conformation capture (Hi-C), as well as tissue Hi-C and ATAC-seq (Fig. 1A and fig S1). Publicly available pediatric glioma tissue ChIP-seq data were integrated with our cell line ChIP-seq data (table S1C).
Cell lines analyzed
A total of 11 patient-derived cell lines were analyzed: H3K27M DIPG (n = 6), H3K27 wild-type GBM (n = 3), neural stem cells (n = 1), and NHA (n = 1). The source and culture media of each cell line are listed in table S1A. All cell lines were acquired and analyzed in accordance with institutionally approved protocols at Northwestern University (STU00202063). Cell lines were maintained in culturing media recommended by cell line providers. All cells were grown in an incubator with 5% CO2 at 37°C.
Tissue specimens analyzed
A total of 50 mg of DIPG tumor tissue and 50 mg of normal frontal cortex tissue from the same patient were used for ATAC-seq. Fifty milligram of each of the two DIPG tumor tissue samples was used for Hi-C. Tissue specimens were obtained in accordance with institutionally approved protocols at Ann and Robert H. Lurie Children’s Hospital of Chicago (Institutional Review Board no. 2012-14877) and Northwestern University (STU00202063) (table S1B).
RNA extraction
Cells were harvested at 80% confluency and washed with phosphate-buffered saline (PBS) twice. Cells were homogenized with QIAshredder (79654, QIAGEN). Homogenized whole-cell lysate then were used for RNA isolation via the manufacturer’s protocol (74104, QIAGEN). Extracted RNA was stored in −80°C or proceeds directly to library construction.
RNA library preparation and sequencing
RNA-seq libraries were prepared using the Illumina TruSeq Stranded Total RNA Preparation Kit (RS-122-2201) with ribodepletion. Input RNA quality was validated using the Agilent RNA 6000 Nano Kit (5067-1511). A total of 1 mg of total RNA was used as a starting material. One microgram of RNA was used as a starting material. Libraries were validated using the Agilent DNA 1000 Kit (5067-1504) before proceeding to single-read sequencing at 50 base pairs (bp) on the Illumina NextSeq 500 Sequencing System.
Chromatin immunoprecipitation
ChIP was performed on cell lines in triplicate, following previously described protocols (10). Briefly, 30 to 50 million cells were cross-linked using 1% formaldehyde for 10 min and quenched with 0.125 M glycine for 5 min at room temperature. Cells were then rinsed, harvested, resuspended in extraction buffer for 10 min at 4°C, and then centrifuged at 1350g for 5 min at 4°C. Cell pellet was resuspended and transferred to a 1-ml milliTUBE (520135, Covaris). After sonication (Covaris E220 ultrasonicator), samples were spun down, and the supernatant containing chromatin was collected. Chromatin was decross-linked for 3 hours at 65°C, and DNA was extracted (28104, QIAGEN). The sample (10%) was saved as input. Chromatin was incubated with primary antibody overnight at 4°C (table S1D). The following day, protein A/G agarose beads (sc-2003, Santa Cruz Biotechnology) were added to the samples and incubated for 3 hours. Beads were centrifuged and washed with 1 ml of radioimmunoprecipitation assay (RIPA) buffer four times. The beads and the 10 ml of input were eluted for 30 min at 65°C and then spun down. Supernatant was decross-linked overnight at 65°C. DNA was extracted and submitted for library preparation.
ChIP library preparation and sequencing
ChIP-seq libraries were prepared with the KAPA Library Preparation Kit (KK8234, Kapa Biosystems) and NETflex DNA barcodes (514104, Bioo Scientific). A total of 10 mg of DNA was used as starting material for input and immunoprecipitation samples. Libraries were amplified with a thermocycler for 13 cycles. Postamplification libraries were size-selected at 250 to 45 bp in length using the Agencourt AMPure XP beads (A63881, Beckman Coulter). Libraries were validated using the Agilent High Sensitivity DNA Analysis Kit (5067-4626). Libraries were sequenced at 30 million reads on the Illumina NextSeq 500 Sequencing System.
High-throughput chromosome conformation capture
Hi-C in cell and tissue was performed following the manufacturer’s protocol (A510008, Arima Genomics). Cells were cultured and cross-linked using 1% formaldehyde for 10 min and quenched with 0.125 M glycine for 5 min at room temperature. A total of 3 million cells were used for each Hi-C experiment. Tissue samples were frozen with liquid nitrogen, pulverized using mortar and pestle until the sample resembles a fine powder, and then cross-linked using the same condition as cell lines. Hi-C libraries were generated and quality-checked per manufacturer’s protocol. A total of 300 to 600 million reads per each sample were generated on the basis of total usable reads above 20 kb.
Assay for transposase-accessible chromatin (ATAC-seq)
ATAC-seq in tissue was performed following the previously published protocol with minor modifications. Approximately 5 mg of tissue was crushed into a fine powder in liquid nitrogen, then suspended in 1 ml of cold PBS, and spun down. The tissue pellet was resuspended in 1 ml of lysis buffer, followed by isolation of 50,000 nuclei. Then, the 50 μl of transposition reaction mix was added to the isolated nuclei and incubated at 37°C for 1 hour. The transposal DNA was purified with the QIAGEN MinElute Polymerase Chain Reaction (PCR) Purification Kit (QIAGEN, 28006) and then amplified with appropriate number of cycles. Libraries were sequenced on the Illumina HiSeq X Ten PE150 platform.
Pharmacological treatment of cells
DIPG007 cells were treated with 200 nM BRD4i (BMS986158, Bristol Myers Squibb) and 1000 nM dBET6 (S8762, Selleckchem) for 24 hours. DMSO treatment at 200 and 1000 nM for 24 hours was also included as vehicle controls. For all treatment conditions, 2 million cells were plated in six-well plates 24 hours before drug dosing. Twenty-four hours after administration of drugs, cells were cross-linked with 1% formaldehyde for 10 min and quenched with 0.125 M glycine for 5 min. Cells were then rinsed, harvested, and stored in −80°C before use.
Western blotting for pharmacologically treated cells
Protein (whole-cell extract) was extracted using RIPA buffer (89900, Thermo Fisher Scientific). Concentration of extracted protein was determined with the Pierce BCA Protein Assay Kit (23225, Thermo Fisher Scientific). Thirty micrograms of protein was separated by electrophoresis in a 4 to 15% precast protein gel (4561086, Bio-Rad) and transferred to polyvinylidene difluoride membrane. Blocking was subsequently done with 5% nonfat milk in Tris Buffered Saline Tween (TBST), followed by overnight incubation with anti-H3K27Ac or anti-BRD4 primary antibody (8173S and 13440, Cell Signaling Technology). After five washes with TBST, membrane was incubated with horseradish peroxidase (HRP)–conjugated anti-rabbit immunoglobulin G (IgG) antibody (7074, Cell Signaling Technology) for 1 hour. Pierce ECL Plus (32132, Thermo Fisher Scientific) was used to detect protein bands. Blot was then stripped (46430, Thermo Fisher Scientific) and reprobed with anti–total H3 primary antibody (14269S, Cell Signaling Technology) as our loading control. HRP-conjugated anti-mouse IgG antibody (7076, Cell Signaling Technology) was used to detect total H3 signal. Densitometry analysis was performed using ImageJ.
FISH for DIPG cells
FISH probes for the detection of A2M-SAMD12 fusion was customized (Empire Genomics LLC (Buffalo, NY). The SAMD12 gene locus (8q24.12) was labeled with spectrum orange, and the A2M gene locus (12p13.31) was labeled with spectrum green. The normal signal pattern shows two green and two orange signals, whereas the typical dual color dual fusion shows one green, one orange, and two fusion signals, indicating a balanced translocation. FISH was performed on the tumor cell line following the standard laboratory procedures in the clinical Cytogenetics laboratory.
RNA isolation and real-time quantitative PCR
RNA isolation and complementary DNA preparation were using the RNeasy Mini Kit (163037260, QIAGEN) and the SuperScript III First-Strand Synthesis System Kit (Invitrogen) following the manufacturer’s protocol, respectively. The target genes expression was verified by quantitative PCR (qPCR) with KAPA SYBR FAST qPCR Master Mix (KK4600), and glyceraldehyde-3-phosphate dehydrogenase was used as a control.
RNA-seq data processing
The FASTQ files were first trimmed for adapter and then mapped against hg38 human reference genome using STAR. Gene-level abundance was quantitated using RSEM program. The same procedure was applied to the downloaded FASTQ files from public resources. The data reproducibility for replicates was calculated for all genes using Pearson correlation.
RNA-seq data clustering
We first combined the raw counts of 32 RNA-seq data from 11 cell lines and 9 patient RNA-seq data from public resources. Genes with overall read counts, from all the 41 RNA-seq data, less than 20 were filtered out. The raw counts were then normalized using variance-stabilizing transformation function from DEseq2 package. Top 500 variable genes within the 11 cell lines were used to perform PCA analysis. Further, Pearson correlations were calculated between all the replicates from cell lines. After confirming the reproducibility of the replicates, we calculated the mean-normalized expression value for each cell line and calculated the Pearson correlations between all cell lines and patient samples. These samples were then clustered according to correlation matrix.
Differentially expressed genes and pathway enrichment analysis
We used DEseq2 to identify the differential expressed genes between tumor and normal cell lines. We calculated the mean expressions from replicates for the same cell line and considered the cell lines from the same tumor type as replicates when identifying DIPG versus GBM, DIPG versus normal, and GBM versus normal differential genes. For DIPG versus GBM differential genes, we performed GSEA analysis for the curated gene sets (c2.all.v7.1.symbols.gmt) using pyGSEA, the prerank function. Top enriched pathways were selected out.
ChIP-seq and ATAC-seq data process
We used the standard ENCODE ChIP-seq pipeline for mapping and peak calling of histone ChIP-seq enrichment. Specifically, the raw ChIP-seq FASTQ files were first trimmed using Cutadapt and mapped back to hg38 human reference genome using Bowtie2. Duplicated aligned reads were marked and removed. MACS2 was used to call peaks from H3K27ac, H3.3K27M, and H3K27me3 ChIP-seq data. The public ChIP-seq data were processed using the same pipeline and parameters. ATAC-seq data were processed using the standard ENCODE ATAC-seq pipeline. The overall procedures are similar to ChIP-seq analysis. We used the bigwig files that contains −log10(P value) generated by MACS2 for genome browser visualization, calculating correlations between samples and generating heatmap of tumor-type special peak signal enrichment.
Tumor type–specific enhancers identification
We first extended the H3K27ac peaks 250 bp up/downstream from peak summit, with each peak extending 500 bp, and then merged the peaks together using mergeBed function from BEDTools. The original H3K27ac peak regions were kept if the peaks exist in only one sample. If several peaks were merged into one, then the peak that gives the best MACS2 scores was kept. Peaks that were located within 2.5 kb of RefSeq gene TSS regions were removed from the ensemble peak set. Next, we calculated the bigwig signals for each sample over filtered ensemble peaks, and the signals were then scaled across the nine cell lines analyzed for each peak. This allows identification of tumor-specific enhancers based on this matrix.
Motif analysis
Motif enrichment analysis was performed for both DIPG- and GBM-specific enhancer regions using HOMER findMotifsGenome function with -size 1000 -mask settings. TFs with high-expression level in DIPG cell lines and significant H3K27Ac enrichment in DIPG-specific enhancers were identified as DIPG-specific active motifs. Similar criteria apply for the GBM-specific active motifs.
Hi-C data processing
We first trimmed the adapters of the Hi-C raw FASTQ files and then mapped the trimmed files against hg38 human reference genome using runHi-C pipeline, which is based on the 4DN consortium. Specifically, Burrows-Wheeler Aligner was used for the FASTQ file alignment and aligned reads with low quality, and PCR duplicates were filtered. Aligned reads were then paired on the basis of read pairs and filtered for fragments that contain ligations of at least two different restriction fragments. These reads were then binned at 5-kb resolution. To generate the contact matrix at multiple resolutions (5, 10, 25, 40, 50, 100, 250, 500 kb, 1, 2.5, 5, and 10 Mb), we used the run-cool2multirescool script from 4DN consortium, which performed the ICE normalization at the same time. We used coolbox to visualize ICE normalized genomic Hi-C data. Juicer tool was also used to generate multiresolution .hic files, which can be visualized using juicebox.
Tumor type–specific loops identification
We used Peakachu to call loops from both DIPG and GBM cell line Hi-C data. We then used diffPeakachu to compare each of the two DIPG cell lines with the GBM cell line. Tumor type–specific loops were then merged using BEDTools pairtopair function with a negative slop of 20 kb.
Gene regulatory network construction
For DIPG gene regulatory network construction, we first overlapped the DIPG-specific loops with previously identified tumor type–specific enhancers using BEDTools pairToBed function with 20-kb extension of the loops. We then further used pairToBed overlapping the DIPG-specific loops with one anchor located in enhancer regions with the promoter of DIPG-specific highly expressed genes. MEME-FIMO was then used to scan the locations of the DIPG-specific active TFs at enhancers that have DIPG-specific loops linked between DIPG-specific highly expressed genes. The existence of the motif sites within the enhancer region indicated the regulatory relationship between the TF and the gene. The regulatory network was then constructed with each node as a DIPG-specific highly expressed gene and each edge as a TF-gene regulation pair. We used the same strategy to construct the GBM-specific gene regulatory network. The final networks were visualized using Cytoscape 3.7.2. Node annotation and pathway enrichment analysis of nodes were accomplished by using String app plugin in Cytoscape.
SV analysis
To infer CNV from Hi-C data, we used the CNV function of HiNT program (42). Here, we directly used the previously generated .hic files as input and predicted CNV events at 50-kb resolution. In addition, we also used Hi-C breakfinder (43) to identify large structural translocations, deletions, and inversions. Bam files were used as input, with low-quality reads filtered out. To identify neo-loops or enhancer hijacking events, we used our in-house pipeline. Briefly, we reassembled the genomic locations based on the breakpoints identified from Hi-C breakfinder, and then we called loops using Peakachu (44) at the reassembled regions.
Functional pathway analysis
Functional pathways analysis and upstream regulator predictions were performed on differentially expressed gene sets using Ingenuity Pathways Analysis software (Qiagen).
Statistical analysis
Data reproducibility for replicates was calculated for all genes and ChIP-seq peaks using Pearson correlation. After confirming the reproducibility of the replicates, the mean-normalized expression value for each cell line was calculated. The Pearson correlation between all cell lines and patient samples was then calculated.
Acknowledgments
We would like to thank D. M. Scholtens and K. L. Bell Burdett for the assistance with statistical analyses. Funding: Research reported in this publication was supported by the NIH’s Natural Institute of Neurological Disease and Stroke, grant number K08NS097264 (to A.S.). F.Y. is supported by NIH grants R35GM124820 and R01HG009906. E.B. is supported by the NIH grant R50CA221848. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. Author contributions: T.Y.-T.H.: Conceptualization, methodology, data collection, validation, data processing and analysis, writing (original draft), writing (review and editing), and visualization. J.W.: Data processing and analysis, writing (original draft), writing (review and editing), and visualization. Y.H.: Data collection and validation. E.B.: Data processing and analysis. X.L.: Data collection and validation. A.S.: Writing (review and editing) and supervision. F.Y.: Conceptualization, methodology, writing (review and editing), supervision, and funding acquisition. A.S.: Conceptualization, methodology, writing (original draft), writing (review and editing), supervision, and funding acquisition. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All raw data generated (RNA-seq, ChIP-seq, Hi-C, and ATAC-seq) are available in the GEO database (GSE162976). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabg4126/DC1
REFERENCES AND NOTES
- 1.Mackay A., Burford A., Carvalho D., Izquierdo E., Fazal-Salom J., Taylor K. R., Bjerke L., Clarke M., Vinci M., Nandhabalan M., Temelso S., Popov S., Molinari V., Raman P., Waanders A. J., Han H. J., Gupta S., Marshall L., Zacharoulis S., Vaidya S., Mandeville H. C., Bridges L. R., Martin A. J., Al-Sarraj S., Chandler C., Ng H.-K., Li X., Mu K., Trabelsi S., Brahim D. H.-B., Kisljakov A. N., Konovalov D. M., Moore A. S., Carcaboso A. M., Sunol M., de Torres C., Cruz O., Mora J., Shats L. I., Stavale J. N., Bidinotto L. T., Reis R. M., Entz-Werle N., Farrell M., Cryan J., Crimmins D., Caird J., Pears J., Monje M., Debily M.-A., Castel D., Grill J., Hawkins C., Nikbakht H., Jabado N., Baker S. J., Pfister S. M., Jones D. T. W., Fouladi M., von Bueren A. O., Baudis M., Resnick A., Jones C., Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32, 520–537.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones C., Karajannis M. A., Jones D. T. W., Kieran M. W., Monje M., Baker S. J., Becher O. J., Cho Y.-J., Gupta N., Hawkins C., Hargrave D., Haas-Kogan D. A., Jabado N., Li X.-N., Mueller S., Nicolaides T., Packer R. J., Persson A. I., Phillips J. J., Simonds E. F., Stafford J. M., Tang Y., Pfister S. M., Weiss W. A., Pediatric high-grade glioma: Biologically and clinically in need of new thinking. Neuro Oncol. 19, 153–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lulla R. R., Saratsis A. M., Hashizume R., Mutations in chromatin machinery and pediatric high-grade glioma. Sci. Adv. 2, e1501354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagaraja S., Quezada M. A., Gillespie S. M., Arzt M., Lennon J. J., Woo P. J., Hovestadt V., Kambhampati M., Filbin M. G., Suva M. L., Nazarian J., Monje M., Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol. Cell 76, 965–980.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stafford J. M., Lee C.-H., Voigt P., Descostes N., Saldaña-Meyer R., Yu J.-R., Leroy G., Oksuz O., Chapman J. R., Suarez F., Modrek A. S., Bayin N. S., Placantonakis D. G., Karajannis M. A., Snuderl M., Ueberheide B., Reinberg D., Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci. Adv. 4, eaau5935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flavahan W. A., Drier Y., Liau B. B., Gillespie S. M., Venteicher A. S., Stemmer-Rachamimov A. O., Suvà M. L., Bernstein B. E., Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson J. L., Muraleedharan R., Oatman N., Klotter A., Sengupta S., Waclaw R. R., Wu J., Drissi R., Miles L., Raabe E. H., Weirauch M. L., Fouladi M., Chow L. M., Hoffman L., De Wire M., Dasgupta B., The transcription factor Olig2 is important for the biology of diffuse intrinsic pontine gliomas. Neuro Oncol. 19, 1068–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asby D. J., Killick-Cole C. L., Boulter L. J., Singleton W. G., Asby C. A., Wyatt M. J., Barua N. U., Bienemann A. S., Gill S. S., Combined use of CDK4/6 and mTOR inhibitors induce synergistic growth arrest of diffuse intrinsic pontine glioma cells via mutual downregulation of mTORC1 activity. Cancer Manag. Res. 10, 3483–3500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morton A. R., Dogan-Artun N., Faber Z. J., MacLeod G., Bartels C. F., Piazza M. S., Allan K. C., Mack S. C., Wang X., Gimple R. C., Wu Q., Rubin B. P., Shetty S., Angers S., Dirks P. B., Sallari R. C., Lupien M., Rich J. N., Scacheri P. C., Functional enhancers shape extrachromosomal oncogene amplifications. Cell 179, 1330–1341.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piunti A., Hashizume R., Morgan M. A., Bartom E. T., Horbinski C. M., Marshall S. A., Rendleman E. J., Ma Q., Takahashi Y.-h., Woodfin A. R., Misharin A. V., Abshiru N. A., Lulla R. R., Saratsis A. M., Kelleher N. L., James C. D., Shilatifard A., Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 23, 493–500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowley M. J., Nichols M. H., Lyu X., Ando-Kuri M., Rivera I. S. M., Hermetz K., Wang P., Ruan Y., Corces V. G., Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haddad N., Jost D., Vaillant C., Perspectives: Using polymer modeling to understand the formation and function of nuclear compartments. Chromosome Res. 25, 35–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J., Marco E., Pinello L., Yuan G.-C., Predicting chromatin organization using histone marks. Genome Biol. 16, 162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen N., Guo B., Zheng H., Xu L., Liang H., Wang Q., Wang D., Chen X., Zhang S., Li Y., Zhang L., Bromodomain inhibitor jq1 induces cell cycle arrest and apoptosis of glioma stem cells through the VEGF/PI3K/AKT signaling pathway. Int. J. Oncol. 55, 879–895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L., Chen Y., Mayakonda A., Koh L., Chong Y. K., Buckley D. L., Sandanaraj E., Lim S. W., Lin R. Y.-T., Ke X.-Y., Huang M.-L., Chen J., Sun W., Wang L.-Z., Goh B. C., Dinh H. Q., Kappei D., Winter G. E., Ding L.-W., Ang B. T., Berman B. P., Bradner J. E., Tang C., Koeffler H. P., Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc. Natl. Acad. Sci. U.S.A. 115, E5086–E5095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spielmann M., Lupiáñez D. G., Mundlos S., Structural variation in the 3D genome. Nat. Rev. Genet. 19, 453–467 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Buchanan J. A., Scherer S. W., Contemplating effects of genomic structural variation. Genet. Med. 10, 639–647 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Barrow J., Adamowicz-Brice M., Cartmill M., MacArthur D., Lowe J., Robson K., Brundler M.-A., Walker D. A., Coyle B., Grundy R., Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro Oncol. 13, 212–222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren K. E., Killian K., Suuriniemi M., Wang Y., Quezado M., Meltzer P. S., Genomic aberrations in pediatric diffuse intrinsic pontine gliomas. Neuro Oncol. 14, 326–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funato K., Major T., Lewis P. W., Allis C. D., Tabar V., Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346, 1529–1533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suvà M. L., Rheinbay E., Gillespie S. M., Patel A. P., Wakimoto H., Rabkin S. D., Riggi N., Chi A. S., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L., Rivera M. N., Rossetti N., Kasif S., Beik S., Kadri S., Tirosh I., Wortman I., Shalek A. K., Rozenblatt-Rosen O., Regev A., Louis D. N., Bernstein B. E., Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosty J., Lu F., Kupp R., Mehta S., Lu Q. R., Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas. Cell Cycle 16, 1654–1660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arabzade A., Stuckert A. J., Bertrand K. C., Mack S. C., Invited Review: The role and contribution of transcriptional enhancers in brain cancer. Neuropathol. Appl. Neurobiol. 46, 48–56 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Mack S. C., Singh I., Wang X., Hirsch R., Wu Q., Villagomez R., Bernatchez J. A., Zhu Z., Gimple R. C., Kim L. J. Y., Morton A., Lai S., Qiu Z., Prager B. C., Bertrand K. C., Mah C., Zhou W., Lee C., Barnett G. H., Vogelbaum M. A., Sloan A. E., Chavez L., Bao S., Scacheri P. C., Siqueira-Neto J. L., Lin C. Y., Rich J. N., Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J. Exp. Med. 216, 1071–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labreche K., Simeonova I., Kamoun A., Gleize V., Chubb D., Letouzé E., Riazalhosseini Y., Dobbins S. E., Elarouci N., Ducray F., de Reyniès A., Zelenika D., Wardell C. P., Frampton M., Saulnier O., Pastinen T., Hallout S., Figarella-Branger D., Dehais C., Idbaih A., Mokhtari K., Delattre J.-Y., Huillard E., Lathrop G. M., Sanson M., Houlston R. S.; POLA Network , TCF12 is mutated in anaplastic oligodendroglioma. Nat. Commun. 6, 7207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortin J., Tian R., Zarrabi I., Hill G., Williams E., Sanchez-Duffhues G., Thorikay M., Ramachandran P., Siddaway R., Wong J. F., Wu A., Apuzzo L. N., Haight J., You-Ten A., Snow B. E., Wakeham A., Goldhamer D. J., Schramek D., Bullock A. N., ten Dijke P., Hawkins C., Mak T. W., Mutant ACVR1 arrests glial cell differentiation to drive tumorigenesis in pediatric gliomas. Cancer Cell 37, 308–323.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filbin M. G., Tirosh I., Hovestadt V., Shaw M. L., Escalante L. E., Mathewson N. D., Neftel C., Frank N., Pelton K., Hebert C. M., Haberler C., Yizhak K., Gojo J., Egervari K., Mount C., van Galen P., Bonal D. M., Nguyen Q.-D., Beck A., Sinai C., Czech T., Dorfer C., Goumnerova L., Lavarino C., Carcaboso A. M., Mora J., Mylvaganam R., Luo C. C., Peyrl A., Popović M., Azizi A., Batchelor T. T., Frosch M. P., Martinez-Lage M., Kieran M. W., Bandopadhayay P., Beroukhim R., Fritsch G., Getz G., Rozenblatt-Rosen O., Wucherpfennig K. W., Louis D. N., Monje M., Slavc I., Ligon K. L., Golub T. R., Regev A., Bernstein B. E., Suvà M. L., Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carro M. S., Lim W. K., Alvarez M. J., Bollo R. J., Zhao X., Snyder E. Y., Sulman E. P., Anne S. L., Doetsch F., Colman H., Lasorella A., Aldape K., Califano A., Iavarone A., The transcriptional network for mesenchymal transformation of brain tumours. Nature 463, 318–325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S., Zhao B. S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E. P., Xie K., Bögler O., Majumder S., He C., Huang S., m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Towner R. A., Jensen R. L., Vaillant B., Colman H., Saunders D., Giles C. B., Wren J. D., Experimental validation of 5 in-silico predicted glioma biomarkers. Neuro Oncol. 15, 1625–1634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daubon T., Léon C., Clarke K., Andrique L., Salabert L., Darbo E., Pineau R., Guérit S., Maitre M., Dedieu S., Jeanne A., Bailly S., Feige J.-J., Miletic H., Rossi M., Bello L., Falciani F., Bjerkvig R., Bikfalvi A., Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat. Commun. 10, 1146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu J., Shi Z., Jiang J., Cyclooxygenase-2 in glioblastoma multiforme. Drug Discov. Today 22, 148–156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai L., Zhou B., Yang C.-Y., Ji J., McEachern D., Przybranowski S., Jiang H., Hu J., Xu F., Zhao Y., Liu L., Fernandez-Salas E., Xu J., Dou Y., Wen B., Sun D., Meagher J., Stuckey J., Hayes D. F., Li S., Ellis M. J., Wang S., Targeted degradation of BET proteins in triple-negative breast cancer. Cancer Res. 77, 2476–2487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter G. E., Buckley D. L., Paulk J., Roberts J. M., Souza A., Dhe-Paganon S., Bradner J. E., Drug Development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J., Qian Y., Altieri M., Dong H., Wang J., Raina K., Hines J., Winkler J. D., Crew A. P., Coleman K., Crews C. M., Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22, 755–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng Z., Gong Y., Ma Y., Lu K., Lu X., Pierce L. A., Thompson R. C., Muller S., Knapp S., Wang J., Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res. 19, 1748–1759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan S. L., Mariano N. C., Bermudez A., Arruda N. L., Wu F., Luo Y., Shankar G., Jia L., Chen H., Hu J.-F., Hoffman A. R., Huang C.-C., Pitteri S. J., Wang K. C., Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 8, 15993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plass C., Pfister S. M., Lindroth A. M., Bogatyrova O., Claus R., Lichter P., Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14, 765–780 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Northcott P. A., Lee C., Zichner T., Stütz A. M., Erkek S., Kawauchi D., Shih D. J. H., Hovestadt V., Zapatka M., Sturm D., Jones D. T. W., Kool M., Remke M., Cavalli F. M. G., Zuyderduyn S., Bader G. D., Berg S. V., Esparza L. A., Ryzhova M., Wang W., Wittmann A., Stark S., Sieber L., Seker-Cin H., Linke L., Kratochwil F., Jäger N., Buchhalter I., Imbusch C. D., Zipprich G., Raeder B., Schmidt S., Diessl N., Wolf S., Wiemann S., Brors B., Lawerenz C., Eils J., Warnatz H.-J., Risch T., Yaspo M.-L., Weber U. D., Bartholomae C. C., von Kalle C., Turányi E., Hauser P., Sanden E., Darabi A., Siesjö P., Sterba J., Zitterbart K., Sumerauer D., van Sluis P., Versteeg R., Volckmann R., Koster J., Schuhmann M. U., Ebinger M., Grimes H. L., Robinson G. W., Gajjar A., Mynarek M., von Hoff K., Rutkowski S., Pietsch T., Scheurlen W., Felsberg J., Reifenberger G., Kulozik A. E., von Deimling A., Witt O., Eils R., Gilbertson R. J., Korshunov A., Taylor M. D., Lichter P., Korbel J. O., Wechsler-Reya R. J., Pfister S. M., Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammad F., Weissmann S., Leblanc B., Pandey D. P., Højfeldt J. W., Comet I., Zheng C., Johansen J. V., Rapin N., Porse B. T., Tvardovskiy A., Jensen O. N., Olaciregui N. G., Lavarino C., Suñol M., de Torres C., Mora J., Carcaboso A. M., Helin K., EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 23, 483–492 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Jung E., Alfonso J., Osswald M., Monyer H., Wick W., Winkler F., Emerging intersections between neuroscience and glioma biology. Nat. Neurosci. 22, 1951–1960 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Wang S., Lee S., Chu C., Jain D., Kerpedjiev P., Nelson G. M., Walsh J. M., Alver B. H., Park P. J., HiNT: A computational method for detecting copy number variations and translocations from Hi-C data. Genome Biol. 21, 73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon J. R., Xu J., Dileep V., Zhan Y., Song F., Le V. T., Yardımcı G. G., Chakraborty A., Bann D. V., Wang Y., Clark R., Zhang L., Yang H., Liu T., Iyyanki S., An L., Pool C., Sasaki T., Rivera-Mulia J. C., Ozadam H., Lajoie B. R., Kaul R., Buckley M., Lee K., Diegel M., Pezic D., Ernst C., Hadjur S., Odom D. T., Stamatoyannopoulos J. A., Broach J. R., Hardison R. C., Ay F., Noble W. S., Dekker J., Gilbert D. M., Yue F., Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 50, 1388–1398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salameh T. J., Wang X., Song F., Zhang B., Wright S. M., Khunsriraksakul C., Ruan Y., Yue F., A supervised learning framework for chromatin loop detection in genome-wide contact maps. Nat. Commun. 11, 3428 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/23/eabg4126/DC1