Abstract
The leukotoxin of Pasteurella (Mannheimia) haemolytica is believed to play a significant role in pathogenesis, causing cell lysis and apoptosis that lead to the lung pathology characteristic of bovine shipping fever. Using a system for Cre-lox recombination, a nonpolar mutation within the lktC transacylase gene of the leukotoxin operon was created. The lktC locus was insertionally inactivated using a loxP-aph3-loxP cassette, and then the aph3 marker was excised from the chromosome by Cre recombinase expressed from a P. haemolytica plasmid. The resulting lktC strain (SH2099) secretes inactive leukotoxin and carries no known antibiotic resistance genes. Strain SH2099 was tested for virulence in a calf challenge model. We inoculated 3 × 108 or 3 × 109 CFU of wild-type or mutant bacteria into the lungs of healthy, colostrum-deprived calves via transthoracic injection. Animals were observed for clinical signs and for nasal colonization for 4 days, after which they were euthanized and necropsied. The lower inoculum (3 × 108 CFU) caused significantly fewer deaths and allowed lung pathology to be scored and compared, while the 3 × 109 CFU dose of either the wild-type or mutant was lethal to ≥50% of the calves. The estimated 50% lethal dose of SH2099 was four times higher than that of the wild-type strain. Lung lesion scores were reduced twofold in animals inoculated with the mutant, while clinical scores were nearly equivalent for both strains. The wild-type and mutant strains were equally capable of colonizing the upper respiratory tracts of the calves. In this study, the P. haemolytica lktC mutant was shown to be less virulent than the parent strain.
Pasteurella (Mannheimia) haemolytica serotype A1 is the primary bacterial agent of bovine pneumonic pasteurellosis, or shipping fever. This multifactorial fibrinonecrotizing pneumonia may be triggered by viral infection, overcrowding, stress, or immunosuppression, which allows the normally commensal Pasteurella bacterium to gain access to the lower respiratory tract, where it becomes pathogenic (13). Leukotoxin has long been thought to be the primary virulence factor of P. haemolytica, and vaccine development studies have focused on the leukotoxin as an important protective antigen (6).
The P. haemolytica leukotoxin (LktA) is a calcium-dependent cytotoxin that is a member of the RTX (repeats in toxin) family. This family includes the Escherichia coli hemolysin (HlyA), the Bordetella pertussis adenylate cyclase/hemolysin (CyaA), and the Actinobacillus pleuropneumoniae Apx toxins (44). P. haemolytica leukotoxin is species specific and has cytolytic activity against ruminant lymphoid cells (18, 34). LktA is also weakly hemolytic (25). Though the leukotoxin can bind to cells from a variety of species (39), cytolysis requires a specific interaction with the lymphocyte function-associated antigen 1, or β2-integrin, on the target (1, 21). At high concentrations, the toxin creates pores in the cell membrane that lead to cell swelling and lysis (5). At sublytic concentrations, the toxin activates neutrophils (8), induces inflammatory cytokine production (48), invokes cytoskeletal changes, and causes apoptosis (37, 38). Like other members of the RTX family, the leukotoxin is expressed from an operon that encodes the protoxin and a transacylase (LktC) that is required to convert the protoxin to an active form (16). Inactive leukotoxin has neither cytolytic nor apoptotic activity in vitro (10, 38, 40), though the inactive toxin can be secreted from bacterial cells (10), and inactive pro-LktA binds with high affinity to target cells (39).
In addition to the in vitro activities described above, the role of leukotoxin in pneumonic disease has been examined in vivo. When instilled into the bovine lung, partially purified leukotoxin was shown to cause cytopathic changes in bovine alveolar macrophages (47), and leukotoxin has been found physically associated with membranes of degenerating macrophages and neutrophils in the alveoli (46). In an attempt to further clarify the role of leukotoxin in bovine pasteurellosis, leukotoxin-negative strains have been constructed and tested for virulence in calves. In one study, a genetically uncharacterized leukotoxin mutant, created by chemical mutagenesis (4), caused reduced mortality and smaller lung lesions than the wild type when inoculated intratracheally (29). In another study, a lktA deletion mutant was tested using bacteria delivered endobronchially (41). A similar reduction of mortality and lung lesions was observed. In the latter study, the authors concluded that the mutant “revealed significant reduction in virulence” (41). Based on these studies, we hypothesized that a P. haemolytica strain that lacked the lktC gene would express and secrete inactive leukotoxin and that such a mutation should cause significant attenuation of the organism. Unlike previous mutants, a lktC strain should express all protective epitopes, including those carried on the leukotoxin, and such a strain could be useful as an attenuated live vaccine.
In a prior report, we developed a method for site-specific chromosomal mutagenesis and created a lktC strain by inserting a nonpolar promoterless chloramphenicol acetyltransferase gene into lktC on the P. haemolytica chromosome (10). Since a chloramphenicol-resistant strain is not acceptable as a live vaccine candidate, we sought to develop a lktC P. haemolytica strain that lacked a resistance marker. Though we had attempted to create unmarked mutations in P. haemolytica using counterselections such as sacB/sucrose and tetracycline sensitivity, we were unsuccessful. We turned, then, to the use of site-specific recombination systems that promote precise and efficient excision between two directly repeated sequences that flank a selectable marker. Systems such as Cre/lox (36), FLP/FRT (2), and λInt/att (22) have been used to create unmarked mutations in bacteria, yeast, and higher eukaryotes (20). Among systems for site-specific recombination, the phage P1 Cre/lox system seemed to be the most suitable for genetic manipulation of P. haemolytica since it requires the activity of only one enzyme, Cre recombinase. In addition, Cre-mediated excision leaves behind a single 34-bp loxP site, which creates a nonpolar, stable mutation in the target gene (36). We have succeeded in developing the Cre-loxP system for site-specific recombination in P. haemolytica and have used it to create an unmarked lktC strain. This was tested for its ability to cause disease in a calf challenge model of Pasteurella pneumonia.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
E. coli strain XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10]; Stratagene, La Jolla, Calif.) was used for plasmid propagation and cloning. E. coli was grown at 37°C in liquid or on solid Luria-Bertani medium. P. haemolytica strain SH789, isolated from the pneumonic lung of a calf, was kindly provided by Glynn Frank (National Animal Disease Center, U.S. Department of Agriculture, Ames, Iowa). Strain SH1217, a plasmid-cured derivative of SH789, was used as the host for genetic manipulations (11). P. haemolytica was grown at 37°C in liquid or solid brain heart infusion (BHI; Difco, Detroit, Mich.) or on 5% sheep blood agar plates (Remel, Lenexa, Kans.). The following plasmids were used: pUC4K (Pharmacia, Piscataway, N.J.), pBCKS+ (Stratagene), pHSG-cre (3), pBS30 (33), pNF2176 (11), and pNF2232 (10). Antibiotics were used at the following concentrations for E. coli and P. haemolytica, respectively: ampicillin, 50 and 25 μg/ml; kanamycin, 50 and 25 μg/ml; streptomycin, 20 and 70 μg/ml.
To create the inoculum for the calf virulence trial, P. haemolytica strains were grown in Trypticase soy broth (Difco) to an optical density at 600 nm (OD600) of 0.6; then 0.1-ml aliquots were plated on sheep blood agar plates and incubated overnight at 37°C. Cells were recovered from the plates by washing with RPMI 1640 medium (Sigma, St. Louis, Mo.) and were resuspended to a final OD600 per milliliter of 3.0 or 0.3 in 0.5× RPMI 1640. Aliquots of the inocula were serially diluted in phosphate-buffered saline, and aliquots were plated on sheep blood agar plates to determine the number of CFU per milliliter.
Genetic and recombinant DNA manipulations.
Standard recombinant DNA techniques were used (32). Plasmid DNAs were isolated from P. haemolytica or from E. coli using a Plasmid Midi kit (Qiagen, Valencia, Calif.). Chromosomal DNAs were isolated using a PUREGENE DNA isolation kit (Gentra, Minneapolis, Minn.). Both E. coli and P. haemolytica cells were transformed by electroporation as previously described (7). To transfer genes between E. coli and P. haemolytica, we used an ampicillin-resistant (Apr) P. haemolytica-E. coli shuttle vector, pNF2176, derived from a native streptomycin-resistant (Smr) plasmid, pYFC1 (11). All shuttle plasmids used in this work are derivatives of pYFC1 and are therefore incompatible. This property allows selective heteroplasmid segregation. Oligonucleotides SH117 (TTTTGTTTAATTTCCCTACATTTTGTATAAC, lktC 5′) and SH151 (GCGTCTGTCACCAGACTGCC, lktC 3′) were used as primers to amplify DNA flanking the mutagenesis target.
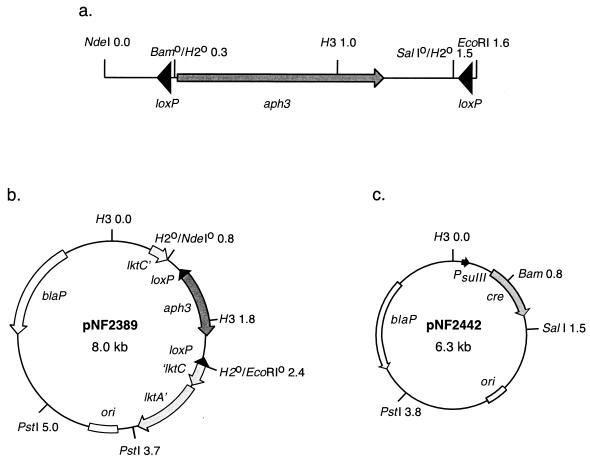
To construct a loxP-aph3-loxP cassette, the aph3 gene from pUC4K was subcloned onto pBS30, which carries two head-to-tail loxP sites separated by the yeast LEU2 gene (33). The 2.5-kb BamHI-SalI LEU2 fragment of pBS30 was replaced with the 1.3-kb HindII fragment carrying the aph3 gene. The resulting loxP-aph3-loxP cassette (Fig. 1a), on a 1.85-kb EcoRI-NdeI fragment, was inserted into the HindII site within the lktC gene on plasmid pNF2232 (10), to create the mutagenic plasmid pNF2389 (Fig. 1b). Plasmid pNF2389 was electroporated into P. haemolytica SH1217, and a double-crossover recombinant, carrying the loxP-aph3-loxP cassette at the lktC locus, was created as previously described (10).
FIG. 1.
(a) Abbreviated restriction map of the loxP-aph3-loxP cassette. (b) Restriction map of plasmid pNF2389 used for allelic exchange at the lktC locus. (c) Restriction map of the P. haemolytica plasmid that expresses Cre recombinase under control of the sulfonamide promoter, PsulII. Abbreviations: Bam, BamHI; H2, HindII; H3, HindIII.
Plasmid pNF2442 (Fig. 1c) was constructed to express Cre recombinase in P. haemolytica. The 1.3-kb EcoRI-SalI cre fragment of pHSG-cre was subcloned onto pBCKS+, which was linearized with the same enzymes. An EcoRI-KpnI cre fragment was then subcloned onto pNF2176, linearized with EcoRI and KpnI, to create pNF2442. On pNF2442, the cre gene is expressed under control of a P. haemolytica PsulII promoter located 5′ of the EcoRI site. Following excision of the loxP-aph3-loxP cassette from the chromosome of SH2040, the Cre plasmid was cured using novobiocin as previously described (11).
Western blotting and leukotoxin ELISA.
Leukotoxin production by P. haemolytica strains was assessed by Western blotting as previously described (15). Antigens were detected using polyclonal bovine convalescent serum. Antileukotoxin antibodies were quantitated by an endpoint enzyme-linked immunosorbent assay (ELISA) using leukotoxin prepared by the method of Vega et al. (42). Fifty micrograms of leukotoxin, suspended in 50 μl of blocking buffer (10 mM Tris [pH 7.6], 0.9% NaCl, 2% milk powder, 0.05% Tween), was bound to each well of an Immulon IV plate (Dynex, Chantilly, Va.) by incubation overnight at 4°C. The plate was washed three times with Tris-buffered saline (TBS; 10 mM Tris [pH 7.6], 0.9% NaCl); then bovine serum, diluted in 50 μl of blocking buffer, was added to the plates. Following a 1-h incubation at 37°C, the plate was washed five times with TBS and then incubated with 50 μl of horseradish peroxidase-conjugated goat anti-bovine immunoglobulin G (IgG; 1/200 dilution in TBS; Kirkegaard & Perry, Gaithersburg, Md.) for an additional hour at 37°C. The plate was washed five times more with TBS and then developed for 10 min at room temperature with 50 μl of 2,2′-azino-di-3-ethyl-benzthiazoline sulfonate (400 μg/ml; Roche, Indianapolis, Ind.)–3% H2O2. The reaction was stopped by the addition of 20 μl of 2% sodium dodecyl sulfate per well, and the absorbance of each well was read at 405 nm.
Virulence trial.
Sixty-two 100- to 500-lb, mixed-gender, colostrum-deprived Holstein calves were screened for the presence of antileukotoxin antibodies. Thirty calves, selected as having titers less than 400 (reciprocal of serum dilution at which no reactivity was observed), were transported to the test site 5 days before the beginning of the trial (day −4). Serum samples were collected on days −5, 0, and 4, and antileukotoxin IgG titers were determined by ELISA (Table 1). Beginning on day −2, animals were examined daily for clinical signs, as noted in Table 2, and nasal cultures were collected. Calves were assigned to groups in random fashion. Because some of the animals had been used 1 month earlier for a bovine respiratory synctial virus (BRSV) trial, they were first sorted into three groups based on BRSV status: nonvaccinated but challenged (8 calves; group A), vaccinated and challenged (12 calves; group B), and nonvaccinated and nonchallenged (10 calves; group C). Animals in each BRSV group were then assigned a number using a random number generator, and group assignment (1, 2, 3, or 4) was then determined by consecutive numbering. On day 0, each calf received a 5-ml suspension of P. haemolytica bacteria (mutant or wild type) delivered transthoracically into each lung as described by Panciera and Corstvet (27). Calves that died before the end of the trial (day 4) were necropsied, and lungs and trachea were removed for subsequent analyses. On day 4 (96 h postinoculation), surviving calves were euthanized and necropsied. The lungs were removed, and the focal injection site lesions, edema, pleuritis, and inflammatory spread were scored by the method of Panciera et al. (28). Mean clinical and lesion scores were analyzed by the Student's t test, and 50% lethal doses (LD50s) were calculated by the method of Reed and Muench (30).
TABLE 1.
Groups, anti-LktA IgG titers, and survival statistics
| Group | Strain | Inoculum (CFU/lung) | No. in group | No. of deaths | Anti-LktA IgG titera (mean ± SD) |
|---|---|---|---|---|---|
| 1 | SH2099 | 3 × 108 | 8 | 1 | 240 ± 110 |
| 2 | SH2099 | 3 × 109 | 8 | 4 | 210 ± 98 |
| 3 | SH789 | 3 × 108 | 7 | 2 | 200 ± 110 |
| 4 | SH789 | 3 × 109 | 7 | 6 | 250 ± 95 |
Mean of titers from samples collected on days −5 and 0, reported as the reciprocal of the greatest serum dilution at which a positive reaction was observed by ELISA.
TABLE 2.
Clinical scores
| Group | Strain | Inoculum (CFU/lung) | No. of survivors | Clinical scorea (mean ± SD)
|
|
|---|---|---|---|---|---|
| All calves | Surviving calves | ||||
| 1 | SH2099 | 3 × 108 | 7b | 3.4 ± 4.5 | 1.9 ± 1.6 |
| 2 | SH2099 | 3 × 109 | 4c | 9.0 ± 5.2 | 5.8 ± 5.4 |
| 3 | SH789 | 3 × 108 | 5 | 3.4 ± 4.6 | 0.8 ± 1.1 |
| 4 | SH789 | 3 × 109 | 1 | 12 ± 2.0 | 13 |
Assigned as follows: Depressed and/or off feed, 1; sneezing and/or coughing, 2; purulent nasal discharge, 1; purulent ocular discharge, 1; respiratory distress, 3; oral and/or nasal mucosal lesions, 4; diarrhea (loose, watery, or bloody stools), 3; moderate rectal temperature elevation (103.5 to 105°C), 3; severe rectal temperature elevation (>105°C), 4 (maximum score, 22).
P = 0.03, group 1 versus group 3.
P = 0.005, group 2 versus group 4.
Lung tissue was cultured for bacteria by sampling the cut surface of a lesion with a sterile cotton swab and then by culturing the inoculum on sheep blood agar plates overnight at 37°C. Nasal swabs were similarly cultured on blood agar. Presumptive P. haemolytica isolates were identified and single-colony purified. Bacteria were characterized with respect to oxidase activity, mannitol and sucrose fermentation, and hemolytic phenotype and by PCR analysis of leukotoxin locus, using primers SH117 and SH151. Bacterial serotyping was performed using type-specific antisera provided by Glynn Frank by the slide agglutination method (14).
RESULTS
Construction of antibiotic-sensitive lktC strain SH2099.
To create an unmarked lktC strain, we first disrupted the lktC gene by allelic exchange in P. haemolytica, using a plasmid incompatibility system to enrich for the isolation of double crossovers (10). Briefly, the mutagenic plasmid pNF2389 (Fig. 1b), carrying lktC insertionally inactivated with the loxP-aph3-loxP cassette, was introduced into strain SH1217. The resulting strain was then electroporated with the incompatible Smr plasmid, pYFC1. Transformants were pooled and propagated overnight in BHI broth containing streptomycin to permit plasmid segregation. To identify isolates where the aph3 gene had been rescued by allelic exchange at the leukotoxin locus, an aliquot of the overnight culture was spread onto sheep blood agar plates containing kanamycin and streptomycin. Double recombinants (Smr Kmr Aps) were detected by replica plating onto ampicillin plates. Three nonhemolytic Aps Kmr strains were identified out of 400 Smr Kmr colonies screened, and one, strain SH2040, was chosen for further analysis.
To excise the Kmr marker from the chromosome, we expressed Cre recombinase in P. haemolytica on plasmid pNF2442 (Fig. 1c). Plasmid pNF2442 was electroporated into strain SH2040 with selection for Apr. Transformed cells were propagated overnight and screened for the loss of Kmr. Approximately 1% of the colonies were Kms, presumably resulting from Cre-mediated excision of the aph3 gene. One Kms isolate, strain SH2099, was cured of pNF2442 and was characterized further.
Verification of the lktC′-loxP-′lktC mutation in the chromosome of SH2099.
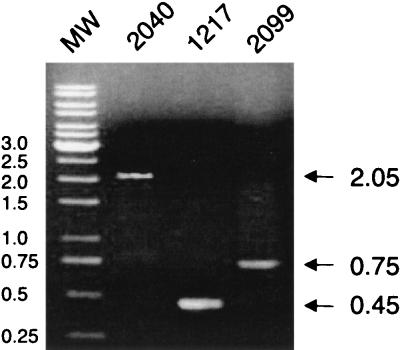
The presence of loxP at the leukotoxin locus on the SH2099 chromosome was demonstrated by PCR analysis (Fig. 2) and verified by Southern blotting (data not shown). A pair of lktC-specific primers (SH117 and SH151) was used to amplify chromosomal DNA from the SH2040, SH2099, and wild-type SH1217 strains. As illustrated in Fig. 1, the 0.45-kb wild-type amplimer from SH1217 was replaced by a 2.05-kb fragment in SH2040. Excision of the cassette from SH2040 to create SH2099 resulted in amplification of a 0.75-kb fragment, consistent with expectations. The DNA sequence of the 0.75-kb fragment from SH2099 was determined to verify the precise location of insertion (data not shown). The sequence confirmed that the insert contains the loxP site plus 300 bp of flanking DNA that was present on the fragment excised from the pBS30 vector (Fig. 1a). The insertion caused a frameshift at codon 76 of the lktC gene.
FIG. 2.
PCR analysis of the lktC locus in wild-type strain SH1217, mutant strain SH2040 (carries the loxP-aph3-loxP insertion within lktC), and mutant strain SH2099 (contains a single loxP site within lktC). Positions of DNA molecular weight markers (MW) are reported in kilobases.
Strain SH2099 secretes inactive leukotoxin.
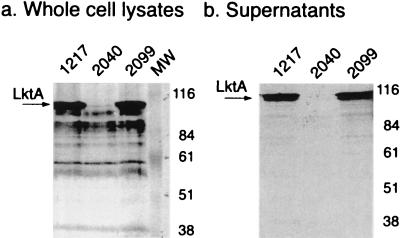
The mutant strains were tested for cytolytic activity against bovine erythrocytes. Both SH2040 and SH2099 were nonhemolytic on blood agar plates, indicating a loss of cytotoxic activity. trans complementation with the lktC plasmid restored hemolysis to SH2099 but not to SH2040 (data not shown). Cytotoxicity of SH2099 was not quantitated because a nonhemolytic, inactivated leukotoxin, created by a similar nonpolar insertion in lktC, had no leukotoxic activity (10). Loss of hemolytic phenotype has been 100% correlated with loss of leukotoxicity both in P. haemolytica mutants (10, 25, 41) and in complementation studies in E. coli (12, 16). To examine leukotoxin expression and secretion, Western blot analysis of P. haemolytica cell lysates and supernatants was performed using bovine polyclonal convalescent serum (Fig. 3). As expected, both SH1217 and SH2099 expressed and secreted LktA at approximately equivalent levels. Strain SH2040, which carries the polar loxP-aph3-loxP cassette, does not produce leukotoxin. Thus, the single loxP insertion is nonpolar and does not significantly affect downstream leukotoxin expression or secretion in SH2099. The stability of the loxP insertion in SH2099 was verified by passaging the strain in BHI broth and plating cells on sheep blood agar plates in an effort to detect hemolytic revertants. No reversion was observed following approximately 100 generations of growth in BHI broth.
FIG. 3.
Western blot of whole cell lysates (a) and cell-free supernatants (b) of wild-type strain SH1217, mutant strain SH2040, and mutant strain SH2099. Positions of protein molecular weight markers (MW) are reported in kilodaltons.
Virulence properties of SH2099.
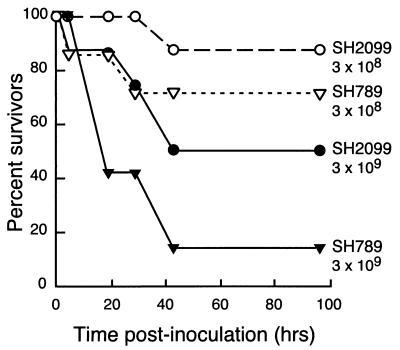
The virulence of SH2099 was compared to that of the wild-type parent, SH789, using a transthoracic intrapulmonic challenge-exposure protocol (27). Animals were assigned to one of four groups containing seven or eight animals each, and each lung was injected with either 3 × 108 or 3 × 109 CFU of the wild type or mutant in a 5-ml suspension (Table 1). Following the injections, the calves exhibited depression and labored breathing that persisted for about 8 h. By 4 h postinoculation, two of the calves (one each in groups 2 and 3) had died. Four calves were dead 18 h postinoculation (Fig. 4). Additional animals were found dead at 28 and 42 h; totals are summarized in Table 1. The difference in survival for both the high and low inocula was significant (Fig. 4). At the 3 × 108 CFU inoculum, final survival was 87% for the mutant and 71% for the wild-type strain (P = 0.03). At 3 × 109 CFU, 50% of the calves injected with the mutant bacteria survived, while only 14% (one calf) injected with the wild-type survived (P = 0.005). LD50s were calculated based on the numbers of deaths in each group. The LD50 of SH789 was 7.1 × 108 CFU/lung and the LD50 for the SH2099 mutant was 2.9 × 109 CFU/lung. Thus, the mutant appears to be about one-fourth as lethal as the wild type when tested by transthoracic challenge in susceptible calves.
FIG. 4.
Calf mortality of wild-type (SH789) and mutant (SH2099) P. haemolytica strains when injected transthoracically with 3 × 108 or 3 × 109 CFU/lung.
Clinical signs were not significantly different for animals receiving wild-type or mutant bacteria, though scores did correlate with bacterial dose (Table 2). Mean rectal temperatures were slightly elevated in group 3 animals (wild type, 3 × 108 CFU). In contrast, temperatures for calves receiving the larger inoculum demonstrated a significant dip on day 1 postinoculation (data not shown). This reflected a high proportion of animals that were near death following the inoculation on day 0.
Gross lung pathology provided the most rigorous means of distinguishing between the wild-type and mutant strains. A comparison of lung lesion scores for all animals (Table 3) showed lower scores for animals receiving 3 × 108 CFU of mutant versus the wild-type organism (P < 0.10). Specific scoring criteria were also compared for calves in groups 1 and 3 that survived the challenge and were sacrificed on day 4 (Table 3). For these surviving animals, focal necrotic lesions were always observed at the injection site, but calves receiving the wild-type bacteria had larger lesions. The edema (P < 0.05) and pleuritis (P < 0.01) scores were significantly greater in calves receiving wild-type bacteria, and trans- and interlobular extension scores also were higher in these calves than in calves receiving the mutant strain.
TABLE 3.
Mean lung lesion criteria and total scores for animals receiving 3 × 108 CFU/lung
| Group (strain) | Scorea (mean ± SD)
|
||||||
|---|---|---|---|---|---|---|---|
| Total lung (all calves) | Surviving calves
|
||||||
| Focus | Edema | Interlobular extension | Translobular extension | Pleuritis | Total lung | ||
| 1 (SH2099) | 4.0 ± 6.7b | 0.4 ± 1.2 | 0c | 0.9 ± 1.5c | 0.3 ± 0.3b | 0.1 ± 0.4d | 1.7 ± 1.7d |
| 3 (SH789) | 9.5 ± 7.6 | 1.0 ± 1.9 | 0.6 ± 1.3 | 2.2 ± 1.8 | 0.7 ± 0.8 | 0.9 ± 0.6 | 5.4 ± 3.7 |
Determined as described by Panciera et al. (28). Lungs from calves that died before the end of the experiment were not scored for specific lung criteria and were assigned a score of 20.
P < 0.10.
P < 0.05.
P ≤ 0.01.
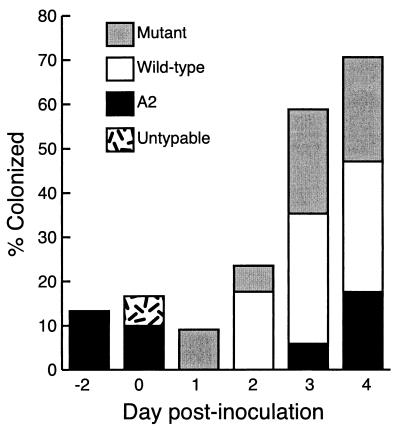
All blood cultures were negative, indicating the inoculum was confined to the respiratory tract. Bacteria isolated from nasal swabs and lung tissue were identified as P. haemolytica, first by morphology and then by biotyping and serotyping. Serotype A1 isolates were further characterized by antibiotic resistance profile and by PCR to detect the presence of the mutated lktC gene. Nasal carriage was monitored to provide information on upper respiratory tract colonization (Fig. 5). Overall, animals inoculated with SH2099 (low dose) had the fewest positive cultures, supporting its reduced virulence. Nasal swabs of group 3 animals were culture positive twice as frequently as the group 1 animals, but a number of atypical P. haemolytica isolates were also obtained. For example, strain SH789 was recovered from two group 1 nasal swabs, and nontypeable P. haemolytica isolates were recovered from calves in groups 2 and 4, but only on day 0. Serotype A2 was found sporadically in nasal swabs of all groups but was not observed in lung lesions. Since P. haemolytica A2 is a common pathogen of sheep, it is possible that its source was a lamb barn on the farm where the calf trial was held. Nevertheless, A2 is also a normal part of the bovine upper respiratory flora, and the animals may have been colonized before transport to the trial location. Lung lesions yielded positive cultures for P. haemolytica, with the exception of one right and four left lungs in group 1 and two right lungs in group 3. All lung cultures were pure bacterial cultures. Unexpectedly, SH2099 was recovered from the right lung lesions of two group 3 animals. We suspect that these samples were contaminated during sampling or on subculture of postnecropsy samples and believe that the error did not occur during transthoracic inoculation because a single injection was prepared per animal. Finally, two Tcs Smr isolates of SH789 were cultured from group 3 and 4 lung samples. These may represent plasmid-cured derivatives of the original inoculum.
FIG. 5.
Nasal colonization of calves with P. haemolytica.
DISCUSSION
This is the first report of creation of a defined, unmarked, nonpolar mutation within the genome of P. haemolytica. The insertion of a single copy of the bacteriophage P1 loxP site within the lktC open reading frame caused a nonpolar frameshift mutation that had no effect on downstream expression of the lktA, lktB, and lktD genes. This created strain SH2099, which produces and secretes inactive but antigenic leukotoxin. The loxP-aph3-loxP cassette described here should be useful for construction of other such mutations in the members of the family Pasteurellaceae. Similar loxP cassettes carrying other resistance markers have been constructed (N. D. Fedorova and S. K. Highlander, unpublished data). Placement of a loxP site in the P. haemolytica chromosome should permit us to create targeted insertions at the lktC locus or at other sites where allelic exchange can be accomplished. The system can also be used to create large deletions in the P. haemolytica chromosome.
Since leukotoxin is believed to be a critical factor in P. haemolytica pathogenesis, we were surprised that inactivation of the toxin caused only a minor reduction in virulence in our calf challenge model. By comparing the strains at different bacterial doses, we established an LD50 for the wild-type strain of about 7 × 108 organisms/lung. This reinforces prior studies that showed that 109 CFU was required to reproducibly produce disease when inoculated into the lung (27, 35). The LD50 of the mutant strain was about four times higher than the wild-type value (3 × 109 CFU/lung), indicating that the mutant retained significant virulence. Survival curves were most illustrative of the differences between the strains. These curves revealed that a narrow dosage range exists for establishment of P. haemolytica pneumonia. In this study, 109 CFU/lung generally caused severe morbidity and mortality, while 108 CFU/lung failed to induce clinical signs and caused reduced pathological changes. Because of the toxicity of the higher dose, significant differences between the wild type and lktC mutant were obscured; at the 3 × 108 CFU dose, however, differences in gross lung pathology were apparent. For calves that survived the challenge, the total lung lesion score was significantly reduced (P < 0.01), as were pleuritis and edema scores (P < 0.05). Trans- and interlobular extension scores were also reduced, but to a lesser degree. By scoring specific criteria, it appears that presence of active leukotoxin is highly correlated with edema, pleuritis, and inflammation but not with lesion formation.
Our results are similar, but not equivalent, to those reported by Tatum et al., where the virulence of a lktA deletion mutant was examined in an endobronchial calf challenge (41). In contrast to our results, 5 × 109 CFU of the ΔlktA mutant caused no clinical signs of disease. Regardless, the mutant still produced lung lesions, but they were reduced 80% relative to the wild-type parent. Only four calves per group were compared, and an LD50 was not calculated. Petras et al. tested the virulence of a chemically induced, leukotoxin-minus mutant in calves and goats (29). Lung lesions created by the mutant strain were reduced 45 to 75% with respect to the wild type; clinical signs were not scored. Since statistical analyses were not applied to the data collected, and because of differences in animal immune status, inoculum preparation, and inoculation route, our study cannot be directly compared to these prior studies. We believe that the 60% reduction in lesion scores that we observed using SH2099 is in line with the level of reduction reported for the leukotoxin-deficient strains and suggests that this represents only partial attenuation of virulence.
The role of other RTX toxins in the pathogenesis of infectious disease has been studied but remains controversial. The bifunctional adenylate cyclase-hemolysin of B. pertussis is absolutely required for virulence in infant mice: the LD50 of a Cya− Hly− Tn5 insertion mutant was reduced 10,000-fold, though a mutant that was phenotypically Cya+ Hly− was only 200 times less virulent (43). This suggests that the adenylate cyclase portion of the molecule is key to B. pertussis virulence and that the hemolysin is less critical. E. coli hemolysin's role in disease continues to be elusive. Addition of a hemolysin plasmid to a clinical isolate of E. coli increased its virulence in a mouse ascending urinary tract model (26). Increased virulence was also observed when HlyA+ E. coli were inoculated intraperitoneally into mice (45). Nevertheless, a chromosomal hlyA deletion in an enterotoxigenic E. coli had no effect on virulence when administered orally to gnotobiotic piglets (24). Thus, HlyA may be most important to extraintestinal disease. The requirement for ApxI expression in porcine pleuropneumonia is also unreconciled. Nonhemolytic mutants of A. pleuropneumoniae serotype A5 had an LD50 in swine that was 10 times greater than the wild-type value (17). In contrast, a nonhemolytic A2 mutant produced fewer clinical signs than did the wild type but still produced significant lung lesion scores (31). All of these studies suggest that the RTX toxins contribute to the pathogenesis of the disease process but are not the sole, or primary, virulence factors.
Our calf challenge indicates that additional virulence factors in P. haemolytica play important roles in lung tissue inflammation and necrosis. It is probable that the early clinical signs that we observed (depression, wheezing) were the due to endotoxemia. Lipopolysaccharide (LPS) has been reported to represent 10 to 25% of the dry weight of P. haemolytica bacteria (19), and LPS forms high-molecular-weight aggregates with leukotoxin (23). Since LPS stimulates production of tumor necrosis factor alpha and interleukin-8, leading to inflammation, it is likely that some of the effects that we observed were LPS related. We presume that in the lktC strain, unlike leukotoxin knockout strains, leukotoxin-LPS complexes are still formed and secreted. If these complexes potentiate the action of LPS, just as LPS is thought to potentiate the action of RTX toxins (9), then a strain secreting an inactive toxin might well maintain significant virulence. Our findings underscore the need for continued studies of Pasteurella pathogenesis. Other factors (i.e., capsule, glycoprotease, and outer membrane proteins) may be found to be critical, and a search for additional virulence factors is in order.
Since P. haemolytica is an opportunistic pathogen, it continues to be of importance to examine the roles of stress and viral predisposition in the etiology of shipping fever pneumonia. In the absence of such factors, it can be difficult to produce bovine respiratory disease in experimental animals (35). By using the transthoracic model in colostrum-deprived calves, we were able to recreate P. haemolytica bacterial pneumonia without introducing additional factors that would complicate analysis. Nevertheless, the transthoracic inoculation method does not represent the natural mode of infection and fails to allow a test of upper respiratory tract colonization and subsequent descent of the bacteria into the lung.
The potential for the use of strain SH2099 as a live vaccine candidate is diminished by its minimal attenuation. In an earlier trial, using intranasal inoculation of young, colostrum-deprived calves, we were unable to create disease, even with the wild-type organism (D. M. Dusek, N. D. Fedorova, C. Rinehart, and S. K. Highlander, unpublished data). Since high titers of bacteria are required to cause disease (13), strain SH2099 could be tested for its protective antigenicity at lower doses using different routes of inoculation. Smaller doses of the mutant strain, introduced orally or intramuscularly, may yet be protective against shipping fever pneumonia.
ACKNOWLEDGMENTS
We thank Dave Carter and Lyle Kesl, Veterinary Resources, Inc., Ames, Iowa, for their expert assistance with the animal trial. We also thank NOBL Labs (Ames, Iowa) for assistance with bacteriological characterization.
This study was funded in part by Texas Higher Education Coordinating Board Technology, Transfer and Development grant 004949-037 and by USDA grant 96-35204-3825.
REFERENCES
- 1.Ambagala T C, Ambagala P N, Srikumaran S. The leukotoxin of Pasteurella haemolytica binds to β2 integrins on bovine leukocytes. FEMS Microbiol Lett. 1999;179:161–167. doi: 10.1111/j.1574-6968.1999.tb08722.x. [DOI] [PubMed] [Google Scholar]
- 2.Babineau D, Vetter D, Andrews B J, Gronostajski R M, Proteau G A, Beatty L G, Sadowski P D. The FLP protein of the 1-micron plasmid of yeast. Purification of the protein from Escherichia coli cells expressing the cloned FLP gene. J Biol Chem. 1985;260:12313–12319. [PubMed] [Google Scholar]
- 3.Brunelli J P, Pall M L. Lambda/plasmid vector construction by in vivo cre/lox-mediated recombination. Bio/Technology. 1994;16:1061–1064. [PubMed] [Google Scholar]
- 4.Chidambaram M, Sharma B, Petras S F, Reese C P, Froshauer S, Weinstock G M. Isolation of Pasteurella haemolytica leukotoxin mutants. Infect Immun. 1995;63:1027–1032. doi: 10.1128/iai.63.3.1027-1032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinkenbeard K D, Mosier D A, Confer A W. Transmembrane pore size and role of cell swelling in cytotoxicity caused by Pasteurella haemolytica leukotoxin. Infect Immun. 1989;57:420–425. doi: 10.1128/iai.57.2.420-425.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlon J A, Shewen P E, Donnelly S F, Burger J P. Effects of Pasteurella haemolytica A1 culture supernatant on mechanisms controlling bovine alveolar macrophage oxygen radical production. Can J Vet Res. 1990;54:232–237. [PMC free article] [PubMed] [Google Scholar]
- 7.Craig F F, Coote J G, Parton R, Freer J H, Gilmour N J. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J Gen Microbiol. 1989;135:2885–2890. doi: 10.1099/00221287-135-11-2885. [DOI] [PubMed] [Google Scholar]
- 8.Czuprynski C J, Noel E J, Ortiz-Carranza O, Srikumaran S. Activation of bovine neutrophils by partially purified Pasteurella haemolytica leukotoxin. Infect Immun. 1991;59:3126–3133. doi: 10.1128/iai.59.9.3126-3133.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czuprynski C J, Welch R A. Biological effects of RTX toxins: the possible role of lipopolysaccharide. Trends Microbiol. 1995;3:480–483. doi: 10.1016/s0966-842x(00)89016-2. [DOI] [PubMed] [Google Scholar]
- 10.Fedorova N D, Highlander S K. Generation of targeted nonpolar gene insertions and operon fusions in Pasteurella haemolytica and creation of a strain that produces and secretes inactive leukotoxin. Infect Immun. 1997;65:2593–2598. doi: 10.1128/iai.65.7.2593-2598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fedorova N D, Highlander S K. Plasmids for heterologous expression in Pasteurella haemolytica. Gene. 1997;186:207–211. doi: 10.1016/s0378-1119(96)00704-4. [DOI] [PubMed] [Google Scholar]
- 12.Forestier C, Welch R A. Nonreciprocal complementation of the hlyC and lktC genes of the Escherichia coli hemolysin and Pasteurella haemolytica leukotoxin determinants. Infect Immun. 1990;58:828–832. doi: 10.1128/iai.58.3.828-832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank G H. Pasteurellosis of cattle. In: Adlam C, Rutter J M, editors. Pasteurella and pasteurellosis. New York, N.Y: Academic Press; 1989. pp. 197–222. [Google Scholar]
- 14.Frank G H, Wessman G E. Rapid plate agglutination procedure for serotyping Pasteurella haemolytica. J Clin Microbiol. 1978;7:142–145. doi: 10.1128/jcm.7.2.142-145.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Highlander S K, Chidambaram M, Engler M J, Weinstock G M. DNA sequence of the Pasteurella haemolytica leukotoxin gene cluster. DNA Cell Biol. 1989;8:15–28. doi: 10.1089/dna.1.1989.8.15. [DOI] [PubMed] [Google Scholar]
- 16.Highlander S K, Engler M J, Weinstock G M. Secretion and expression of the Pasteurella haemolytica leukotoxin. J Bacteriol. 1990;172:2343–2350. doi: 10.1128/jb.172.5.2343-2350.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inzana T J, Todd J, Ma J N, Veit H. Characterization of a non-hemolytic mutant of Actinobacillus pleuropneumoniae serotype 5: role of the 110 kilodalton hemolysin in virulence and immunoprotection. Microb Pathog. 1991;10:281–296. doi: 10.1016/0882-4010(91)90012-y. [DOI] [PubMed] [Google Scholar]
- 18.Kaehler K L, Markham R J F, Muscoplat C C, Johnson D W. Evidence for cytocidal effects of Pasteurella haemolytica on bovine peripheral blood mononuclear leukocytes. Am J Vet Res. 1980;41:1690–1693. [PubMed] [Google Scholar]
- 19.Keiss R E, Will D H, Collier J R. Skin toxicity and hemodynamic properties of endotoxin derived from Pasteurella haemolytica. Am J Vet Res. 1964;25:935–941. [PubMed] [Google Scholar]
- 20.Kilby N J, Snaith M R, Murray J A. Site-specific recombinases: tools for genome engineering. Trends Genet. 1993;9:413–421. doi: 10.1016/0168-9525(93)90104-p. [DOI] [PubMed] [Google Scholar]
- 21.Lally E T, Kieba I R, Sato A, Green C L, Rosenbloom J, Korostoff J, Wang J F, Shenker B J, Ortlepp S, Robinson M K, Billings P C. RTX toxins recognize a beta2 integrin on the surface of human target cells. J Biol Chem. 1997;272:30463–30469. doi: 10.1074/jbc.272.48.30463. [DOI] [PubMed] [Google Scholar]
- 22.Landy A. Dynamic structural and regulatory aspects of λ site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Clinkenbeard K D. Lipopolysaccharide complexes with Pasteurella haemolytica leukotoxin. Infect Immun. 1999;67:2920–2927. doi: 10.1128/iai.67.6.2920-2927.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moxley R A, Berberov E M, Francis D H, Xing J, Moayeri M, Welch R A, Baker D R, Barletta R G. Pathogenicity of an enterotoxigenic Escherichia coli hemolysin (hlyA) mutant in gnotobiotic piglets. Infect Immun. 1998;66:5031–5035. doi: 10.1128/iai.66.10.5031-5035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy G L, Whitworth L C, Clinkenbeard K D, Clinkenbeard P A. Hemolytic activity of the Pasteurella haemolytica leukotoxin. Infect Immun. 1995;63:3209–3212. doi: 10.1128/iai.63.8.3209-3212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Hanley P, Lalonde G, Ji G. Alpha-hemolysin contributes to the pathogenicity of piliated digalactoside-binding Escherichia coli in the kidney: efficacy of an alpha-hemolysin vaccine in preventing renal injury in the BALB/c mouse model of pyelonephritis. Infect Immun. 1991;59:1153–1161. doi: 10.1128/iai.59.3.1153-1161.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panciera R J, Corstvet R E. Bovine pneumonic pasteurellosis: model for Pasteurella haemolytica- and Pasteurella multocida-induced pneumonia in cattle. Am J Vet Res. 1984;45:2532–2537. [PubMed] [Google Scholar]
- 28.Panciera R J, Corstvet R E, Confer A W, Gresham C N. Bovine pneumonic pasteurellosis: effect of vaccination with live Pasteurella species. Am J Vet Res. 1984;45:2538–2542. [PubMed] [Google Scholar]
- 29.Petras S F, Chidambaram M, Illyes E F, Froshauer S, Weinstock G M, Reese C P. Antigenic and virulence properties of Pasteurella haemolytica leukotoxin mutants. Infect Immun. 1995;63:1033–1039. doi: 10.1128/iai.63.3.1033-1039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 31.Rycroft A N, Williams D, McCandlish I A, Taylor D J. Experimental reproduction of acute lesions of porcine pleuropneumonia with a haemolysin-deficient mutant of Actinobacillus pleuropneumoniae. Vet Rec. 1991;129:441–443. doi: 10.1136/vr.129.20.441. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shewen P E, Wilkie B N. Cytotoxin of Pasteurella haemolytica acting on bovine leukocytes. Infect Immun. 1982;35:91–94. doi: 10.1128/iai.35.1.91-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soo M K. Experimental bovine pneumonic pasteurellosis: a review. Vet Rec. 1989;124:141–144. doi: 10.1136/vr.124.6.141. [DOI] [PubMed] [Google Scholar]
- 36.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 37.Stevens P K, Czuprynski C J. Pasteurella haemolytica leukotoxin induces bovine leukocytes to undergo morphologic changes consistent with apoptosis in vitro. Infect Immun. 1996;64:110–117. doi: 10.1128/iai.64.7.2687-2694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Clinkenbeard K D, Clarke C R, Cudd L, Highlander S, Dabo M. Pasteurella haemolytica leukotoxin induced apoptosis of bovine lymphocytes involves DNA fragmentation. Vet Microbiol. 1998;1618:1–14. doi: 10.1016/s0378-1135(98)00286-7. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Clinkenbeard K D, Cudd L A, Clarke C R, Clinkenbeard P A. Correlation of Pasteurella haemolytica leukotoxin binding with susceptibility to intoxication of lymphoid cells from various species. Infect Immun. 1999;67:6264–6269. doi: 10.1128/iai.67.12.6264-6269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, Clinkenbeard K D, Ownby C L, Cudd L, Clarke C R, Highlander S K. Ultrastructural characterization of apoptosis in bovine lymphocytes exposed to Pasteurella haemolytica leukotoxin. Am J Vet Res. 2000;61:51–56. doi: 10.2460/ajvr.2000.61.51. [DOI] [PubMed] [Google Scholar]
- 41.Tatum F M, Briggs R E, Sreevatsan S S, Zehr E S, Hsuan S L, Whiteley L O, Ames T R, Maheswaran S K. Construction of an isogenic leukotoxin deletion mutant of Pasteurella haemolytica serotype 1: characterization and virulence. Microb Pathog. 1998;24:37–46. doi: 10.1006/mpat.1997.0181. [DOI] [PubMed] [Google Scholar]
- 42.Vega M V, Maheswaran S K, Leininger J R, Ames T R. Adaptation of a colorimetric microtitration assay for quantifying Pasteurella haemolytica A1 leukotoxin and antileukotoxin. Am J Vet Res. 1987;48:1559–1564. [PubMed] [Google Scholar]
- 43.Weiss A A, Hewlett E L, Myers G A, Falkow S. Pertussis toxin and extracytoplasmic adenylate cyclase as virulence factors of Bordetella pertussis. J Infect Dis. 1984;150:219–222. doi: 10.1093/infdis/150.2.219. [DOI] [PubMed] [Google Scholar]
- 44.Welch R A, Bauer M E, Kent A D, Leeds J A, Moayeri M, Regassa L B, Swenson D L. Battling against host phagocytes: the wherefore of the RTX family of toxins? Infect Agents Dis. 1995;4:254–272. [PubMed] [Google Scholar]
- 45.Welch R A, Dellinger E P, Minshew B, Falkow S. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature. 1981;294:665–667. doi: 10.1038/294665a0. [DOI] [PubMed] [Google Scholar]
- 46.Whiteley L O, Maheswaran S K, Weiss D J, Ames T R. Immunohistochemical localization of Pasteurella haemolytica A1-derived endotoxin, leukotoxin, and capsular polysaccharide in experimental bovine Pasteurella pneumonia. Vet Pathol. 1990;27:150–161. doi: 10.1177/030098589002700302. [DOI] [PubMed] [Google Scholar]
- 47.Whiteley L O, Maheswaran S K, Weiss D J, Ames T R. Morphological and morphometrical analysis of the acute response of the bovine alveolar wall to Pasteurella haemolytica A1-derived endotoxin and leucotoxin. J Comp Pathol. 1991;104:23–32. doi: 10.1016/s0021-9975(08)80085-0. [DOI] [PubMed] [Google Scholar]
- 48.Yoo H S, Rajagopal S, Maheswaran S K, Ames T R. Purified Pasteurella haemolytica leukotoxin induces expression of inflammatory cytokines from bovine alveolar macrophages. Microb Pathog. 1995;18:237–252. doi: 10.1016/s0882-4010(05)80001-4. [DOI] [PubMed] [Google Scholar]