Abstract
BACKGROUND:
Schizophrenia (SCZ) is a debilitating psychiatric disorder with a large genetic contribution; however, its neurodevelopmental substrates remain largely unknown. Modeling pathogenic processes in SCZ using human induced pluripotent stem cell–derived neurons (iNs) has emerged as a promising strategy. Copy number variants confer high genetic risk for SCZ, with duplication of the 16p11.2 locus increasing the risk 14.5-fold.
METHODS:
To dissect the contribution of induced excitatory neurons (iENs) versus GABAergic (gamma-aminobutyric acidergic) neurons (iGNs) to SCZ pathophysiology, we induced iNs from CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 isogenic and SCZ patient–derived induced pluripotent stem cells and analyzed SCZ-related phenotypes in iEN monocultures and iEN/iGN cocultures.
RESULTS:
In iEN/iGN cocultures, neuronal firing and synchrony were reduced at later, but not earlier, stages of in vitro development. These were fully recapitulated in iEN monocultures, indicating a primary role for iENs. Moreover, isogenic iENs showed reduced dendrite length and deficits in calcium handling. iENs from 16p11.2 duplication-carrying patients with SCZ displayed overlapping deficits in network synchrony, dendrite outgrowth, and calcium handling. Transcriptomic analysis of both iEN cohorts revealed molecular markers of disease related to the glutamatergic synapse, neuroarchitecture, and calcium regulation.
CONCLUSIONS:
Our results indicate the presence of 16p11.2 duplication-dependent alterations in SCZ patient–derived iENs. Transcriptomics and cellular phenotyping reveal overlap between isogenic and patient-derived iENs, suggesting a central role of glutamatergic, morphological, and calcium dysregulation in 16p11.2 duplication-mediated pathogenesis. Moreover, excitatory dysfunction during early neurodevelopment is implicated as the basis of SCZ pathogenesis in 16p11.2 duplication carriers. Our results support network synchrony and calcium handling as outcomes directly linked to this genetic risk variant.
Schizophrenia (SCZ) is a debilitating psychiatric disorder that is highly heritable, with 60% to 90% of risk attributed to genetic background (1–3). Despite extensive genome-wide association studies (GWASs) and exome-wide association studies (4–8) and an expansive array of SCZ animal model paradigms [reviewed in (9)], the etiological basis of SCZ in early neurodevelopment remains largely unknown.
The clearest insights into SCZ pathogenesis arise from patient studies, which have revealed neuronal deficits likely implicated in disease etiology and pathophysiology. GWASs have identified synaptic plasticity as a key molecular pathway disrupted in SCZ, with strong enrichment of variants within synaptic proteins and voltage-gated calcium channels (6,10). Postmortem studies in patients with SCZ have also identified consistent alterations in cortical neuron morphology; SCZ neurons display a dramatic reduction in dendritic length and complexity (11,12) and a reduction in dendritic spine density [reviewed in (13)]. These findings are consistent with alterations in neuronal network activity observed within the cortex of patients with SCZ (14,15) that have been linked to working memory deficits (16). Together, these human studies support the presence of pervasive deficits in the neurodevelopment of synaptic circuits in individuals with SCZ.
Despite these alterations to neuronal circuitry in SCZ, little is known about the early neurodevelopmental deficits that may contribute to SCZ pathogenesis. This is important because SCZ is largely viewed as a neurodevelopmental disorder (NDD) despite the typical SCZ age of onset at 15 to 30 years (17). In support of this, there is a high degree of genetic overlap between SCZ and early-onset disorders such as autism spectrum disorder (ASD), bipolar disorder, and intellectual disability (ID) (18–20). In addition, intellectual impairment (21) and altered gamma oscillations within the cortex (22) are observed before the onset of psychoses and formal SCZ diagnoses. Together, these observations support the hypothesis that the etiological roots of SCZ lie in early neurodevelopment. Therefore, appropriate paradigms are required to address the etiology of SCZ during this developmental period. Human induced pluripotent stem cells (iPSCs) have emerged as a powerful tool in the study of complex genetic NDDs, including SCZ [reviewed in (23,24)]. iPSC-derived neurons (iNs) provide a faithful representation of early prenatal neurodevelopment and allow the impact of SCZ risk genes to be evaluated at this critical time point. Therefore, iNs provide an unrivaled approach to dissecting early neurodevelopmental dysfunction (23,25–27).
To accurately model early SCZ, appropriate iPSC-derived models are required. The recently developed ability to direct the differentiation of iPSCs into pure subpopulations of glutamatergic excitatory neurons (iENs) (28) and GABAergic (gamma-aminobutyric acidergic) neurons (iGNs) (29) provides a unique means to isolate neuronal subtype-specific deficits that may contribute to excitatory/inhibitory imbalance and SCZ susceptibility (30). However, the heterogenetic nature of SCZ necessitates large patient cohorts to identify conserved markers of neuronal impairment—a major hurdle in identifying pathogenic processes in SCZ. One powerful approach is the study of genetically defined cohorts (31) to allow for the assessment of shared large effect size genetic variants that confer strong risk for SCZ and of the manner in which they alter neurodevelopment and contribute to SCZ pathogenesis.
Copy number variants (CNVs) have the largest effect size in SCZ (32), and CNVs at the 16p11.2 locus contribute considerable risk for NDDs (32–35). Whereas both deletion and duplication confer risk for ASD and ID (36–38), only duplications are associated with SCZ and psychosis (36,39,40) and bipolar disorder (41). Isogenic iPSC and animal models cannot present the full, complex interplay of common low-effect-size genetic factors that have been observed to be significant contributing factors in patients with SCZ (6). For these reasons, studying the effect of 16p11.2 duplication in SCZ patient–derived iPSCs may be required to reveal mechanisms that contribute to SCZ progression. The relatively common incidence of 16p11.2 duplication [~1:4000 live births (42)] facilitates the generation of SCZ-16p11.2 duplication patient-derived cohorts. Combined with the large contribution to SCZ risk, 16p11.2 duplication SCZ patient–derived iPSCs provide an attractive model to assess the etiological roots of SCZ. However, to date, no SCZ patient–derived iN studies have been performed on the 16p11.2 duplication risk factor.
We therefore set out to investigate the role of 16p11.2 duplication in SCZ pathogenesis using iNs. Due to phenotypic noise associated with diverse patient genetic backgrounds, these analyses were first performed using CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9-modified isogenic lines in which the 16p11.2 duplication was introduced into an iPSC line derived from a healthy individual. To determine the contribution of excitatory versus inhibitory populations, we assessed phenotypic outcomes in iEN/iGN cocultures and in iEN moncultures. We observed deficits in morphology, calcium handling, and network activity, alongside altered molecular signatures. These observations were recapitulated in iENs from SCZ patients harboring the 16p11.2 duplication, indicating the presence of strong markers of 16p11.2 duplication-mediated dysfunction. These results indicate the presence of strong markers of 16p11.2 duplication-mediated dysfunction, in line with observed cellular phenotypes.
Taken together, our comparative analysis supports excitatory neuronal dysfunction early in neurodevelopment as a basis for SCZ pathogenesis and identifies synaptic circuit and calcium handling deficits directly linked to this genetic variant. Moreover, these observations are closely related to phenotypes reported in idiopathic SCZ iNs (25,43) and have strong correlates to patient intermediate phenotypes (14). Our studies also identified phenotypic differences between isogenic and patient-derived neurons potentially linked to sex or genetic background.
METHODS AND MATERIALS
Cell Lines
Isogenic CRISPR-Cas9-modified 16p11.2 duplication cells were generated from GM08330 iPSCs (Control, Coriell Institute) and provided for this study by Dr. Talkowski. The generation and characterization of these CRISPR-modified and control lines has been outlined previously (44). SCZ patient (02C12626, 04C27671, 06C58821) and age/ethnicity-matched cryopreserved lymphocytes (06C52982, 05C51897, 05C44627) were sourced from the MSG1/2 SCZ clinical cohorts (45–47), study accessions phs000021.v2.p1 and phs000167.v1.p1. Derivation, culture, and polygenic risk scoring are described in Supplemental Methods in Supplement 1.
iN Differentiation
iEN (28) and iGN (29) were generated by Ngn2 and Ascl1/Dlx2 induction, respectively. The full protocol is available in Supplemental Methods in Supplement 1.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism v.8.0.1 (GraphPad Software Inc.). For details of clone and patient replication, see Supplemental Methods in Supplement 1. All data were assessed for Gaussian distribution by the Shapiro-Wilk test; based on the results, parametric or nonparametric two-tailed unpaired t tests were performed with p-value thresholds as noted in the figure legends. A single round of outlier detection (ROUT Q = 1%) was performed for each complete dataset, and figures represent the cleaned data.
RESULTS
Deficits in Neuronal Network Development in 16p11.2 Duplication iEN/iGN Cocultures
The study of large CNVs, such as the 16p11.2 duplication, has been hindered by the inability to generate faithful genetic models to accurately determine molecular and neuronal dysfunction. However, the recent generation of a CRISPR-Cas9-edited isogenic 16p11.2 duplication iPSC model (44) provides the means to identify phenotypes arising directly from the 16p11.2 duplication CNV, independent of variability from diverse genetic backgrounds. Thus, we used isogenic 16p11.2 duplication (DUP) and corresponding control (Ctrl) iPSC lines for the generation of iNs. These isogenic cell lines were previously subject to extensive quality control, including RNA sequencing, chromosomal microarray, and Western blot analysis (44), as well as pluripotency, karyotype analysis, and iPSC expression of 16p11.2 genes (48) confirming the presence of the 16p11.2 duplication and the absence of additional chromosomal abnormalities. Because both excitatory and inhibitory neurons have been implicated in SCZ, we differentiated iPSCs (44) into excitatory [iEN (28)] (Figure 1A) and GABAergic [iGN (29)] (Figure 1B) lineages. iEN/iGN cocultures were generated by combining iENs and iGNs at an 80%:20% ratio (Figure 1C), reflecting the neuronal content of the cortex, and matured over a 7-week in vitro (7WIV) period on multielectrode arrays (MEAs) (Figure 1D). Cocultured iEN/iGN (Figure S1A in Supplement 1), iEN (Figure S1B in Supplement 1), and iGN (Figure 1C) cultures were observed to express neuronal markers MAP2 and Synapsin (Syn1), and iGNs expressed the GABAergic marker, GABA, confirming the presence of mature neuronal cultures suitable for the assessment of 16p11.2 duplication phenotypes. Green fluorescent protein–labeled iENs (Figure S1B in Supplement 1), iGNs (Figure S1C in Supplement 1), and cocultured green fluorescent protein/red fluorescent protein iENs/iGNs (Figure S1A in Supplement 1) were assessed in terms of neuronal induction efficiency. Greater than 98% of green fluorescent protein/red fluorescent protein iNs in mono- or cocultures expressed the neuronal marker MAP2 (Figure S1D–F in Supplement 1), indicating successful and highly homogeneous neuronal induction with no altered differentiation efficiency between genotypes. Within cocultures, assessment of the iEN/iGN ratio confirmed that there were no differences in seeding density or iEN/iGN viability between genotypes at 7WIV (Figure S1G in Supplement 1).
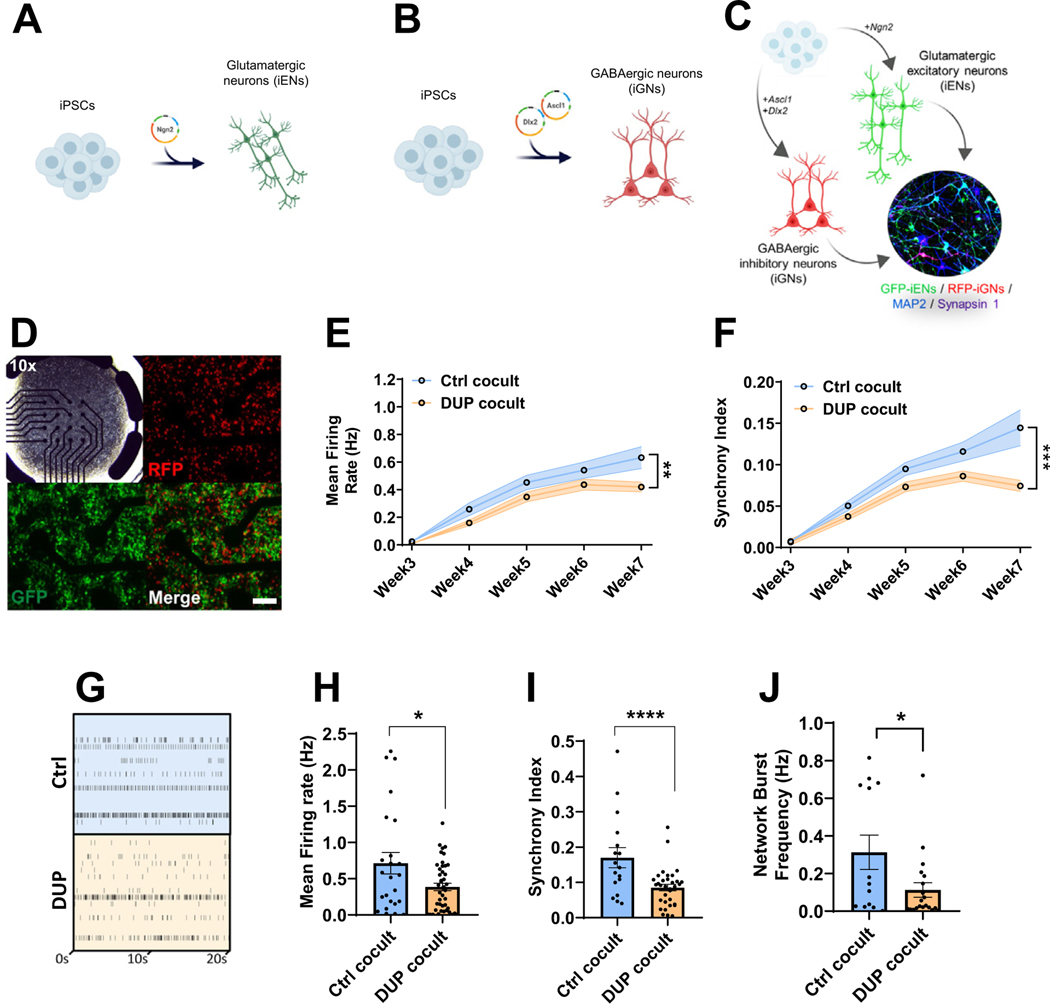
Figure 1.
Modeling 16p11.2 duplication using isogenic 16p11.2 duplication iPSC-derived neurons. (A) Schematic overview of iEN induction. (B) Schematic overview of iGN induction. (C) Schematic overview of iEN/iGN coculture model. (D) Representative multielectrode array plate well of cocultured iENs (GFP) and iGNs (RFP, at 20×, transmission at 10×). Developmental time course of activity (E) and network synchrony (F). (G) Representative raster plot of cocultured iENs/iGNs electrical activity at 7 weeks in vitro. Each well electrode is shown (rows) vs. time (x-axis). Mean firing rate (H), synchrony index (I), and network burst frequency (J) at 7 weeks in vitro (N = 6, 2 clones per condition from 3 independent differentiations, n = 24 wells per condition). *p < .05, ****p < .001. Ctrl cocult: control iENs/iGNs; DUP cocult: 16p11.2 duplication isogenic iENs/iGNs. Cocult, coculture; Ctrl, control lines; DUP, 16p11.2 duplication; GABAergic, gamma-aminobutyric acidergic; GFP, green fluorescent protein; iENs, induced excitatory neurons; iGNs, induced GABAergic neurons; iPSC, induced pluripotent stem cell; RFP, red fluorescent protein.
In both Ctrl and DUP, iEN/iGN network activity developed over 7WIV in terms of firing rate (Figure 1E) and synchronous (simultaneous events across multiple electrodes, Figure S2A in Supplement 1) firing (Figure 1F), indicating the presence of a mature and functional network. 7WIV DUP cocultures displayed a significant decrease in mean firing rate (Figure 1E) and synchrony index (Figure 1F) compared with Ctrl lines. Firing rate and network properties were assessed in depth at this time point (Figure 1G). In addition to reduced mean firing rate (Figure 1H), DUP neurons exhibited reduced synchrony (Figure 1I) and network burst frequency (Figure 1J), bursts occurring over multiple electrodes simultaneously (see Figure S2B in Supplement 1), and low clonal variability was observed (Figure S2C–E in Supplement 1). Together, these results reveal highly dysregulated electrical activity and neuronal network formation as a direct result of 16p11.2 duplication.
DUP iEN Monocultures Recapitulate iEN/iGN Coculture Network Deficits
To delineate iGN-mediated versus intrinsic iEN deficits, we took advantage of the ability to culture iENs independently of GABAergic input and compared phenotypes in DUP and Ctrl iEN monocultures. We seeded iENs on MEAs (Figure 2A) and matured them over 7WIV. DUP iENs were observed to have significantly lower firing rate (Figure 2B) and synchrony (Figure 2C). At 7WIV, iENs recapitulated all activity and network (Figure 2D–H) deficits observed in iEN/iGN, and these effects were not due to clonal variation (Figure S2F–H in Supplement 1). As expected, tetrodotoxin totally abolished all events detected by MEA (Figure S3 in Supplement 1). Application of ion channel antagonists at 4WIV indicated that ~40% of activity was due to synaptic AMPA receptor and 20% was due to NMDA receptor activity (Figure S3A, C in Supplement 1). However, at 7WIV, NBQX inhibited 90% and APV blocked ~50% of activity (Figure S3B, D in Supplement 1), suggesting maturation of synaptic connectivity between 4WIV and 7WIV coinciding with the presentation of network synchrony and of DUP phenotypes. Moreover, these results confirm a pronounced effect of 16p11.2 duplication on excitatory neuronal function and network formation independent of iGNs. These results were not due to the lack of integrity of iGNs within the iENs/iGNs cocultures because transcriptomic analyses indicated the presence of interneuron markers, and MEA analyses demonstrated the functionality of iGNs in altering the activity, network formation, and longitudinal development of cultures (Figure S4 in Supplement 1).
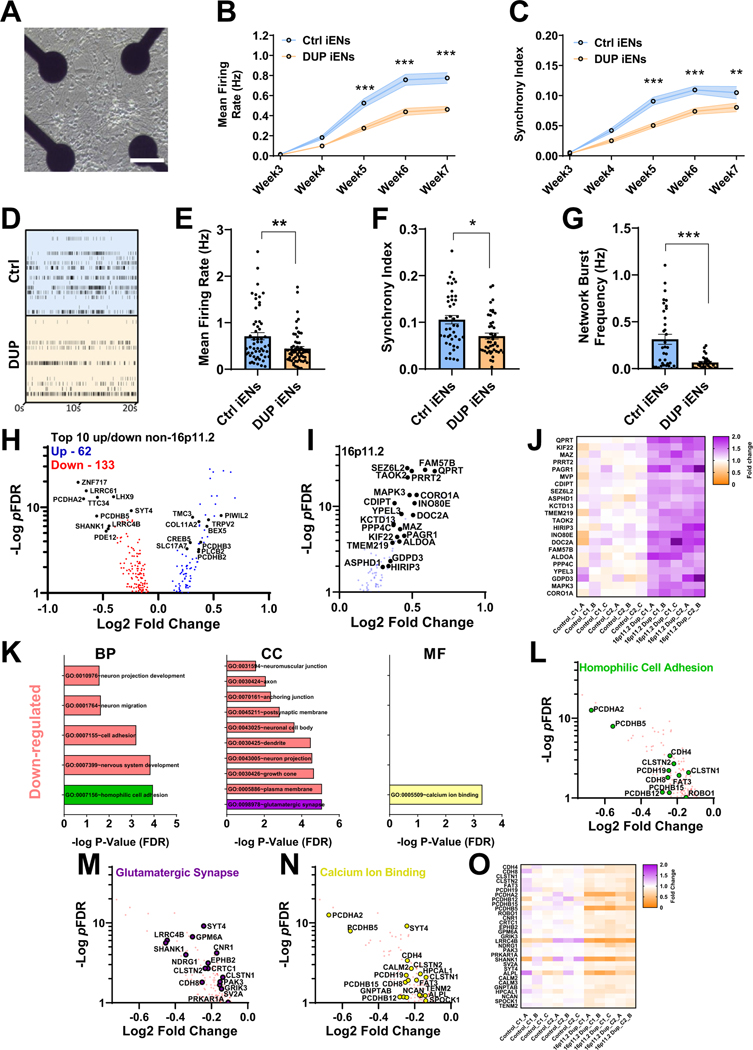
Figure 2.
Analysis of neuronal network activity and transcriptome of isogenic 16p11.2 duplication iEN monoculture. (A) Representative multielectrode array well of monocultured iENs. Scale bar = 50 μm. Developmental time course of iEN activity (B) and network synchrony (C). (D) Representative raster plot of iEN activity at 7 weeks in vitro. Well and plate averages of firing rate (E), synchrony index (F), and network burst frequency (G) at 7 weeks in vitro (N = 6, 2 clones from 3 independent differentiations, n = 30 wells per condition). (H) Volcano plot of DEGs identified in isogenic DUP lines (pFDR < .1, blue: upregulated, red: downregulated; 16,698 expressed with mean read counts > 5) with top 10 non-16p11.2 genes highlighted (black). (I) Volcano plot and (J) heatmap of 16p11.2 gene expression (fold change over mean control). C1_A indicates clone1, sample A, C1_B indicates clone1, sample B, etc. (K) Significantly enriched GO terms within DUP iENs. Volcano plot of top GO terms; homophilic cell adhesion, green (I), glutamatergic synapse, purple (M), and calcium ion binding, yellow (N) and heatmap of aggregated DEGs from these terms (O). *p < .05, **p < .01, ***p < .005. BP, biological process; CC, cellular component; Ctrl, control lines; DEGs, differentially expressed genes; DUP, 16p11.2 duplication; FDR, false discovery rate; GO, gene ontology; iENs, induced excitatory neurons; MF, molecular function.
To guide further analyses of cellular phenotypes, we used an unbiased approach to determine altered molecular pathways that may contribute to iEN dysfunction. Ctrl and DUP iENs were matured to 7WIV before RNA extraction and sequencing. Sixty-two genes were found to be upregulated and 133 downregulated within DUP iEN (Figure 2H; Table S1 in Supplement 2). As expected, expressed 16p11.2 genes were significantly upregulated 1.2- to 1.7-fold, consistent with the presence of a third copy of each gene (Figure 2I, J). Gene ontology (GO) analyses were performed on significantly altered upregulated and downregulated gene sets independently (excluding 16p11.2 genes, false discovery rate (FDR)-normalized p value < .1) to identify molecular pathways that could direct cellular phenotyping (Table S2 in Supplement 2). GO terms associated with significantly downregulated differentially expressed genes (DEGs) included homophilic cell adhesion, neuron projection development, glutamatergic synapse, and calcium ion binding (Figure 2K). Few GO terms were associated with upregulated DEGs, with the strongest term being regulation of cell shape (pFDR = .44). 16p11.2 duplication is a risk factor for numerous NDDs, such as ASD, ID, and epilepsy, as well as SCZ (36). To assess the DUP transcriptome in terms of disease profile, enrichment of risk genes was ascertained from exome sequencing and GWAS in SCZ, epilepsy, ASD, and ID (4,5,7,10,49). We found significant enrichment exclusively in SCZ exome-wide de novo variants (Figure S5A in Supplement 1) and SCZ GWAS loci (Figure S5B in Supplement 1), as well as the presence of multiple high confidence SCZ risk genes (Figure S5C in Supplement 1) (50). These results support the suitability of isogenic DUP iENs to model molecular pathways relevant to SCZ. Moreover, SCZ-associated risk genes (Figure S5D in Supplement 1) were observed within the observed GO terms, such as PCDHA2 (Figure 2L), SHANK1 (Figure 2M), and NCAN (Figure 2N), and DEGs were highly consistent between clones/replicates (Figure 2O).
To assess synaptic DEG function relating to the glutamatergic synapse, synapse-specific GO analyses (Table S3 in Supplement 2) were performed using combined up/down gene sets and including 16p11.2 genes. An enrichment in synaptic (Figure S5D, E in Supplement 1) and presynaptic vesicle release genes (Figure S5E in Supplement 1, blue) was observed, supporting previous observations of deregulated synaptic transmission in SCZ (51,52). Protein-protein interaction analysis (Figure S5F in Supplement 1) revealed that DEG products formed into a network with 121 nodes and 211 edges associated with neuroarchitecture terms (Figure S5G, H in Supplement 1; Table S4 in Supplement 2). These findings suggest the presence of numerous molecular alterations that may contribute to activity and network phenotypes.
Deficits in Dendritic Length and Calcium Homeostasis in DUP iENs
Downregulation of neuron projection development, dendrite, and growth cone DEGs suggest potential deficits in DUP iEN architecture. Therefore, we analyzed dendritic morphology in 7WIV iENs after staining for MAP2. Sholl (Figure 3A) and branch point analysis (Figure S6A in Supplement 1) revealed no alteration in dendritic arbor complexity. However, we observed a significant decrease in total neurite length (Figure 3B, C) and the length of the longest neurite (Figure S6B in Supplement 1) consistent with previous observations (53,54). As MAP2 stains only the dendritic arbor but not axons (Figure S6D in Supplement 1), this alteration in neurite length can be ascribed to a reduction in dendritic length, consistent with SCZ postmortem observations (11,12). We employed the presynaptic marker Syn1 to assess gross alterations to presynaptic number. DUP iENs showed no alterations in the size (Figure S6C in Supplement 1) or number of presynaptic puncta (Figure 3D, E) as previously reported (55). Results were highly consistent between clones (Figure S6E–I in Supplement 1).
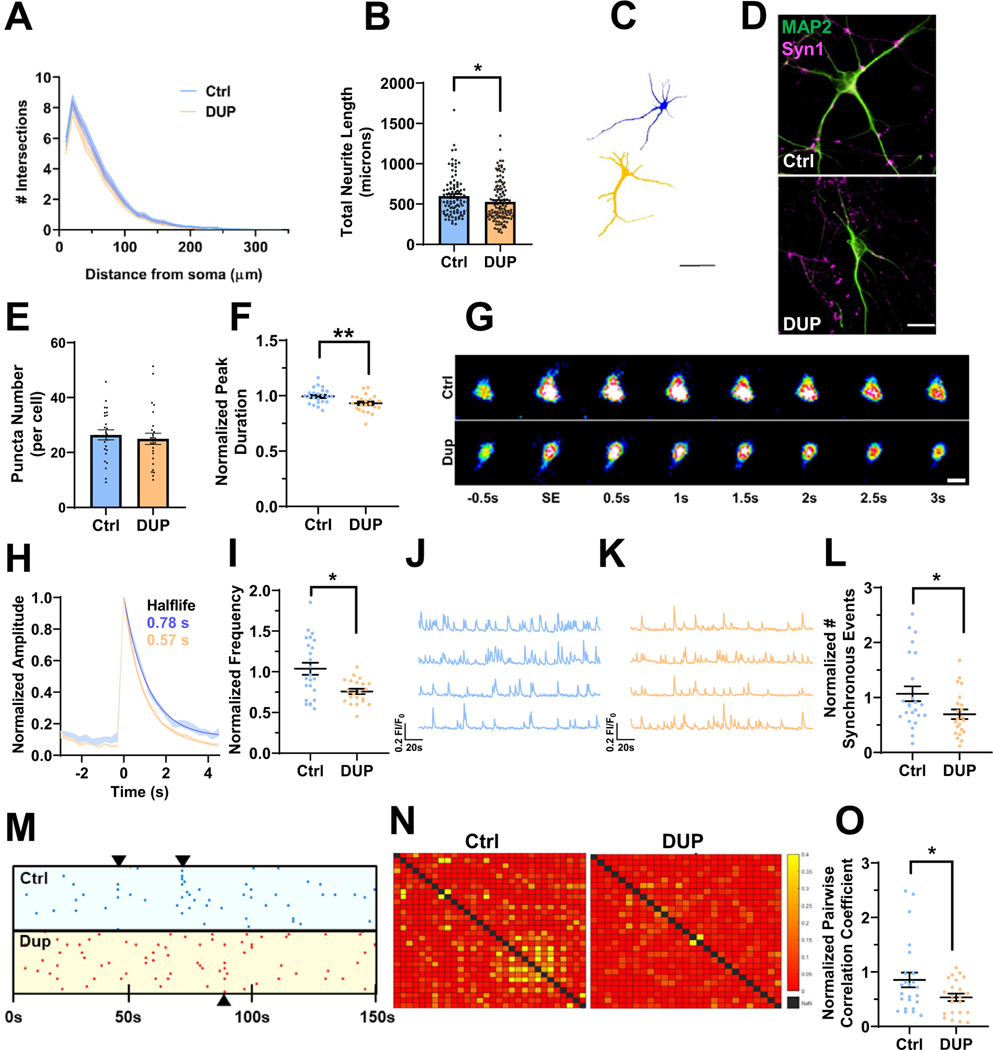
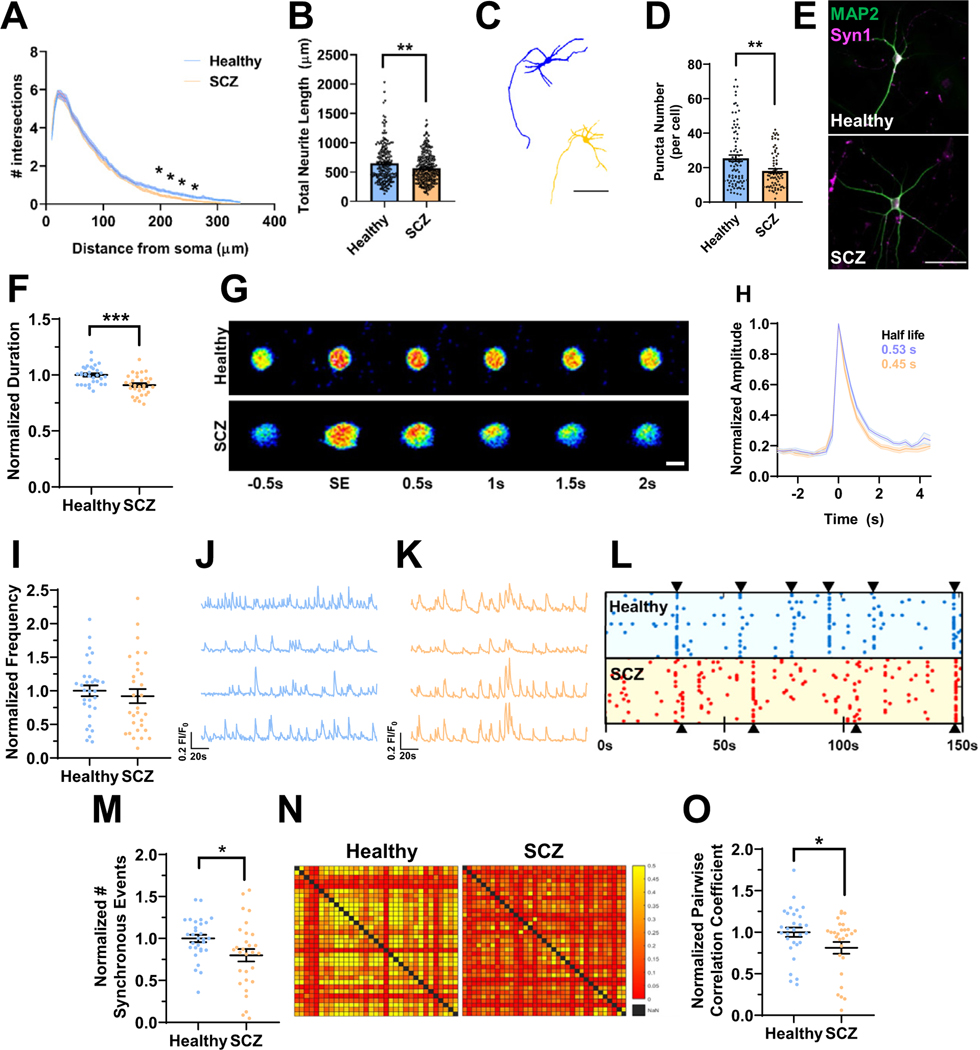
Figure 3.
Deficits in neuroarchitecture and calcium handling in isogenic 16p11.2 duplication iENs at 7 weeks in vitro. (A) Sholl analysis of Ctrl and DUP lines. (B) Total neurite length (2 clones, 2 independent differentiations, n = ~100 cells). (C) Representative traces of Ctrl (blue) and DUP (tan) iENs. Scale bar = 50 μm. (D) Representative image of MAP2 (green) and Syn1 (magenta) stained iENs. (E) Syn1 puncta density (total normalized to cell number per field of view; 2 clones, 2 independent differentiations, n = 20 images per condition). (F) Average calcium peak half width maximum (duration of event normalized to control per independent experiment). (G) Representative time course of a single spontaneous calcium event. Scale bar = 10 μm. (H) Representative trace of Ctrl (blue) and DUP (tan) calcium event (average trace of 5 independent calcium events from 10 cells from 3 coverslips per condition). (I) Calcium event frequency (normalized to control condition within experiment). Representative calcium trace of Ctrl (blue) (J) and DUP (tan) (K). (L) Synchronous calcium network event frequency (normalized to control condition within each independent experiment). (M) Raster plot of representative Ctrl (blue) and DUP (red) coverslips (points indicate the time [x-axis] of calcium events for each cell within the coverslip [y-axis]). (N) Representative heatmap of pairwise correlation of each cell to each other cell within a field of view (no correlation between a pair = red, 50% of spontaneous event activity correlated in a pairwise fashion = yellow). (O) Pairwise correlation coefficient (normalized to control condition within each independent experiment). N = 6; 2 clones, 3 independent differentiations, n = 20–24 coverslips. *p < .05, **p < .01. Ctrl, control lines; DUP, 16p11.2 duplication; iENs, induced excitatory neurons; SE, spontaneous event.
Calcium signaling is a major driver of synaptic plasticity and calcium ion binding DEGs were observed within DUP iENs. Therefore, dysregulated calcium homeostasis may contribute to electrical and network phenotypes in DUP iENs. To assess this, calcium handling and activity was observed by incubating 7WIV iENs with Cal520, a calcium-sensitive fluorescent probe. Observed calcium transients were dependent on neuronal activity, which was confirmed by their responsiveness to synaptic and sodium channel blockers (Figure S7 in Supplement 1). At the single-cell level, we observed a reduction in the duration of calcium events within DUP iENs (Figure 3F, G). Analysis of the average calcium peak profile revealed a pronounced enhancement in the half-maximal recovery time to baseline (Figure 3H), but no difference in event amplitude (Figure S8A in Supplement 1) or resting calcium levels (Figure S8B in Supplement 1). At the network level, we observed a decrease in the frequency of spontaneous (Figure 3I–K) and synchronous network calcium events (Figure 3L, M) in DUP iENs. Moreover, the correlation in activity of each cell to each other cell within the network (pairwise correlation) (Figure 3N, O) was impaired, further supporting a deficit in the formation of functional networks. Observed phenotypes were not driven by clonal differences (Figure S8C–K in Supplement 1). These results confirm alterations in activity and network properties observed by MEAs and reveal a pronounced disruption to calcium handling in DUP iENs.
Abnormalities in Network Activity and Transcriptomic Profile of iENs Derived From SCZ Patients With the 16p11.2 Duplication
To date, no studies have been performed on iNs derived from SCZ-patient iPSCs harboring the 16p11.2 duplication. Comparison of phenotypes shared between patient-derived and isogenic lines would allow the causal assignment of such phenotypes to the duplication versus genetic background. Therefore, we assembled a 16p11.2 duplication SCZ patient–derived iPSC cohort to assess early neurodevelopmental alterations that may contribute to SCZ risk. iPSCs were derived from cryopreserved lymphocytes of 3 SCZ patients carrying the 16p11.2 duplication alongside age/ethnicity-matched controls from a well-characterized SCZ cohort (45–47) (Figure S9A in Supplement 1). iPSCs were characterized based on morphology, alkaline phosphatase uptake, and the presence of pluripotency markers (Nanog, Oct-4, Sox2) (Figure S9B, C in Supplement 1). CNV microarrays confirmed the presence and size of 16p11.2 duplications in each patient with SCZ (Figure S9D in Supplement 1); 16p11.2 CNVs closely matched the region disrupted in the isogenic DUP lines employed [740 kbp (44)].
To assess the role of 16p11.2 duplication in altered neurodevelopment, iENs were derived from SCZ-patient iPSCs harboring 16p11.2 duplications (SCZ) alongside healthy control lines. MEA (Figure 4A) analyses of SCZ iENs showed progressive deficits in network synchrony throughout development (Figure 4C); however, mean spontaneous firing rate (Figure 4B) was unchanged. At 7WIV (Figure 4D), SCZ iEN showed unaltered firing rate (Figure 4E) and significant reductions in synchronous firing (Figure 4F) and network burst frequency (Figure 4G). As expected, there was high variability between patients with SCZ, potentially linked to background/sex (Figure S10 and Supplemental Discussion in Supplement 1). These data reveal network synchrony as a significantly altered phenotype induced by 16p11.2 duplication.
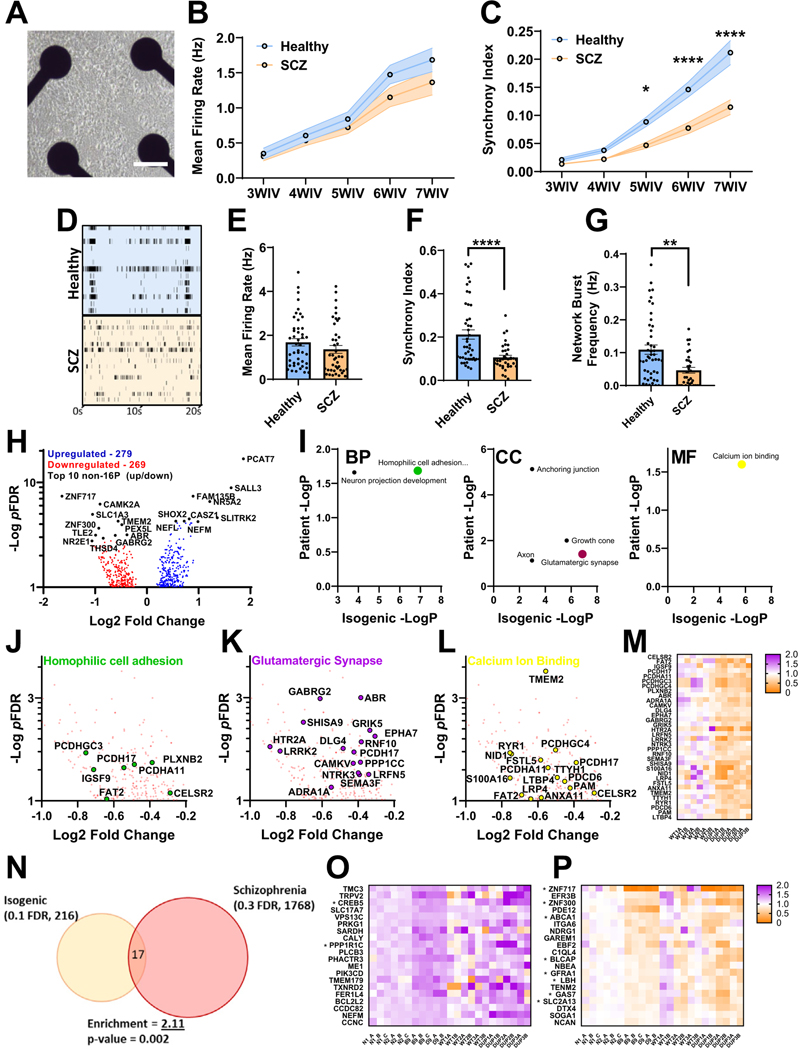
Figure 4.
Analysis of neuronal network activity and transcriptome in SCZ patient–derived iENs. (A) Representative well of patient iENs on multielectrode arrays. (B) Developmental time course of healthy and SCZ-patient iEN mean firing rate. (C) Developmental time course of healthy and SCZ-patient iEN synchrony index. (D) Representative raster plot of healthy and SCZ-patient line activity. Well and plate averages of firing rate (E), synchrony index (F), and network burst frequency (G) at 7WIV (N = 6, 3 patients per condition from 2 independent differentiations, n = 30 wells per condition). (H) Volcano plot of dysregulated gene expression in patients with SCZ with top 10 DEGs highlighted (black, 14,315 expressed with mean read counts > 5). (I) Significantly enriched gene ontology terms within SCZ iENs overlapping with isogenic lines. Volcano plot of top overlapping gene ontology terms; homophilic cell adhesion (J), glutamatergic synapse (K), and calcium ion binding (L) and heatmap of aggregated DEGs from these terms (M). (N) Venn diagram of overlapping DEGs from isogenic (FDR ˂ 0.1) and patient (FDR ˂ 0.3) lines showing shared direction. An enrichment of 2.11, p = .002, was observed. (O) Heat plot of significantly upregulated (FDR < 0.1) isogenic DEGs with shared direction in patient lines. (P) Heat plot of significantly downregulated (FDR ˂ 0.1) isogenic DEGs with shared direction in patient lines. *p < .05, **p < .01, ****p < .001. BP, biological process; CC, cellular component; DEG, differentially expressed gene; FDR, false discovery rate; iENs, induced excitatory neurons; MF, molecular function; SCZ, schizophrenia; WIV, week in vitro.
To assess transcriptomic changes, messenger RNA was harvested from healthy and SCZ iENs at 7WIV, and RNA sequencing was performed. A total of 568 DEGs (pFDR < .1) (Figure 4H; Table S5 in Supplement 2) were observed, with a significant increase in the expression of genes within the 16p11.2 locus (Figure S11A, B in Supplement 1) with the exception of MAZ and QPRT, which were unchanged with regard to healthy control iEN. An enrichment in genes affected by de novo risk variants in SCZ and ASD, but no SCZ GWAS loci, were identified (Figure S11C–F in Supplement 1). Interestingly, a number of GO terms were shared between isogenic and patient lines (Figure 4I); homophilic cell adhesion (Figure 4J), glutamatergic synapse (Figure 4K), and calcium ion binding (Figure 4L) and DEGs arising from these GO terms were well conserved within patient lines (Figure 4M). Synapse-specific GO analysis revealed a shared dysregulation of synaptic vesicle exocytosis in both isogenic and patient-derived lines (Figure S11G, H in Supplement 1). We also observed an enrichment in bipolar disorder variants in both isogenic and patient samples when compared with postmortem cross-disorder brain transcriptomic datasets (Figure S12 in Supplement 1) (56,57). Overlapping DEGs were observed between isogenic and patient DEGs (Figure 4N) (11 up, 6 down). Assessment of DEGs revealed low consistency in upregulated genes (Figure 4O), but downregulated genes showed high consistency between isogenic and patient lines (Figure 4P). We analyzed recent DUP transcriptomic datasets from a range of neuronal models/tissues (Figures S13–S16 in Supplement 1) (48,53,55,58). Notably, GO terms identified in isogenic and SCZ iEN were identified in multiple previous studies (Figure S17 in Supplement 1). The presence of shared gene ontologies between isogenic, patient, and previous 16p11.2 duplication RNA sequencing experiments suggests the presence of conserved molecular markers of 16p11.2 duplication-dependent dysfunction.
We set out to assess the neuroarchitecture of SCZ versus healthy lines. Sholl analysis revealed a deficit in distal dendrites (Figure 5A); however, no alterations in branch point analyses were observed (Figure S18A in Supplement 1), suggesting shorter dendritic length, which was confirmed by a reduction in total and maximum dendrite length (Figure 5B; Figure S18B in Supplement 1). iENs of patients with SCZ showed a reduction in the number of Syn1 puncta (Figure 5D, E), but no change in puncta size (Figure S18C in Supplement 1), and morphological/presynaptic deficits were highly consistent between patient lines (Figure S18D–J in Supplement 1).
Figure 5.
Deficits in neuroarchitecture and calcium handling in SCZ patient–derived 16p11.2 duplication iENs. (A) Sholl analysis of healthy control and 16p11.2 duplication SCZ lines. (B) Total neurite length (n = ~200 cells). (C) Representative traces of healthy (blue) and SCZ (tan) iENs. Scale bar = 50 μM. (D) Syn1 puncta density (total normalized to cell number per field of view; n = 20 images per condition). (E) Representative image of MAP2 (green) and Syn1 (magenta) stained healthy and SCZ iENs. Scale bar = 100 μM. (F) Average calcium peak duration (duration of event normalized to control condition per experiment). (G) Time course of a representative spontaneous event. Scale bar = 10 μM. (H) Representative trace of healthy (blue) and SCZ (tan) calcium events (average trace of 5 independent calcium events from 10 cells from 3 coverslips). (I) Calcium event frequency (normalized per experiment to control condition). Representative calcium trace of healthy (blue) (J) and SCZ (tan) (K) lines. (L) Raster plot of representative healthy (blue) and SCZ (red) coverslips (points indicate the time [x-axis] of calcium events for each cell within the coverslip [y-axis]). (M) Synchronous calcium event frequency (normalized to control condition within each independent experiment). (N) Representative heatmap of pairwise correlation of each cell to each other cell within a field of view (no correlation between a pair = red, 50% of spontaneous event activity correlated in a pairwise fashion = yellow). (O) Pairwise correlation coefficient (normalized to control condition within each independent experiment). N = 6; 2 clones, 3 independent differentiations, n = 20–24 coverslips. *p < .05, **p < .01, ***p < .005. iENs, induced excitatory neurons; SCZ, schizophrenia; SE, spontaneous event.
Calcium ion-binding DEGs were observed in both isogenic and patient transcriptomic profiles. Calcium imaging revealed alterations to calcium event duration in SCZ iENs (Figure 5F–H), as observed in isogenic DUP lines. However, SCZ lines also showed a significant reduction in resting calcium levels (Figure S19A in Supplement 1) and an increase in calcium event amplitude (Figure S19B in Supplement 1), phenotypes which are absent in isogenic DUP lines. These deficits were also evident following KCl-induced depolarization (Figure S20 in Supplement 1). Patients with SCZ showed no significant alteration in the mean frequency of calcium events (Figure 5I–K) consistent with MEA activity (Figure 4E), but the strength of these results was likely impacted by patient line variability (Figure S19C–J and Supplemental Discussion in Supplement 1). SCZ patient–derived lines showed a reduction in the number of synchronous events (Figure 5L, M) and in the pairwise correlation of neuronal firing within the network (Figure 5N, O).
DISCUSSION
The 16p11.2 duplication is one of the most penetrant risk factors for SCZ, unlike deletions of this region (36). However, identifying the role of 16p11.2 duplication in the etiology of SCZ has been challenging due to the inability to access appropriate neural tissue, the limited availability of 16p11.2 duplication carriers with SCZ, and the heterogenetic nature of SCZ. To address these issues, we used SCZ patient–derived iNs to study the 16p11.2 duplication. To complement this, we assessed 16p11.2 duplication using an isogenic line, whereby the 16p11.2 duplication was introduced into iPSCs derived from a healthy subject (44). This complementary patient/isogenic approach has been proposed as a powerful manner with which to assess the role of CNVs in genetically defined NDDs by reducing phenotypic noise from diverse patient backgrounds (31). Using this method, we identified novel 16p11.2 duplication-dependent alterations that likely contribute to the molecular and neuronal etiology of SCZ during early neurodevelopment.
Neuronal activity was dysfunctional in isogenic iEN, with a strong reduction in activity at more mature time points (7WIV; Figures 2, 3). Interestingly, assessment of MEA activity in the presence of AMPA receptor and NMDA receptor inhibitors at 4WIV and 7WIV indicate a strong enhancement of synaptic activity during this period (Figure S3 in Supplement 1), coinciding with the development of network activity. Moreover, it is at these time points that 16p11.2 activity-dependent iEN phenotypes emerged, supporting reductions in glutamatergic synapse gene expression, observed in both isogenic and patient lines, as being key to 16p11.2 duplication activity phenotypes. This is particularly compelling considering the enrichment in synaptic genes in SCZ GWAS, exome, and RNA sequencing studies (6,18,59). Thus, alterations in synaptic gene expression are likely to contribute to neuronal activity deficits in isogenic and patient lines, as supported by results of previous studies (Figure S21 and Supplemental Discussion in Supplement 1).
In addition, we observed DEGs associated with calcium ion binding and neuron projection development alongside dysregulation of calcium homeostasis and neuroarchitecture. Deregulated neuronal calcium signaling is an emerging finding among patient and genetic models of SCZ (6,60–62), and calcium ion binding was found to be the most significantly altered gene set within the SCZ brain (60). In addition, patient postmortem studies have consistently found impaired dendritic architecture in the SCZ cortex (63) suggesting that 16p11.2 duplication may impair dendritic morphology early in neuronal development, and this is maintained throughout SCZ progression. Therefore, these conserved phenotypes may constitute early 16p11.2 duplication-dependent neurodevelopmental alterations contributing to SCZ risk (see Supplemental Discussion in Supplement 1).
Perhaps the most compelling deficit was in iEN network synchrony, which was observed in isogenic and patient lines, by both calcium network imaging and MEAs. Neuronal network activity plays key roles during brain development, sensory processing, and cognition (14), and neuronal synchrony has been hypothesized to play a major role in the coding and propagation of information, with higher synchrony thought to confer faster responses with greater robustness of the information carried (64). Importantly, neuronal network synchrony alterations occur in SCZ and other NDDs (65), supporting observed reductions in network activity and synchrony as potentially relevant to SCZ pathophysiology. Of note, the strength of this phenotype in SCZ lines appeared to be influenced by additional genetic risk or resilience factors, or by patient sex (see Supplemental Discussion in Supplement 1).
Taken together, our findings showed the presence of early neuronal deficits in isogenic and SCZ patient–derived DUP neurons representing neurodevelopmental alterations that may contribute to the etiology of SCZ. Moreover, these deficits were dependent on impaired glutamatergic neuronal activity, supporting previous assertions of excitatory dysfunction as a driver of deficits in SCZ (66,67). Finally, these deficits coincided with impaired neuroarchitecture, the glutamatergic synapse, and calcium signaling, consistent with GWASs (4), postmortem studies (60), and genetic models of SCZ (43,61,62). These findings strongly implicate deregulated calcium homeostasis, neuronal network properties, and morphological development as 16p11.2 duplication-dependent mechanisms that impair neurodevelopment and contribute to SCZ pathophysiology.
Supplementary Material
Key Resource Table
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | Guinea pig MAP2 | Synaptic Systems | 188 004 | |
| Antibody | Rabbit synapsin1 | Cell signalling | D12G5 | |
| Antibody | Anti-GFP chicken | Abcam | ab13970 | |
| Antibody | Anti-NeuN | Cell Signalling | D3S3I | |
| Antibody | Alexafluor488 Goat anti-Guinnea pig | ThermoFisher | A-11073 | |
| Antibody | AlexaFluor 647 anti-rabbit | ThermoFisher | A32733 | |
| Antibody | Rabbit Anti-GABA | Sigma | A2052 | |
| Antibody | Mouse Anti-GABAARdelta1 | Neuromab | AB_2877385 | |
| Antibody | Guinea Pig Anti-GABAARGamma2 | Synaptic Systems | 224 004 | |
| Antibody | Nanog | Abcam Embryonic Stem Cell Marker Panel | ab109884 | |
| Antibody | 04-Oct | |||
| Antibody | SOX2 | |||
| Viral Strain | Ngn2 lentivrus | generated at NU GET-IN core | ||
| Viral Strain | Ascl3 lentivrus | generated at NU GET-IN core | ||
| Viral Strain | Dlx2 lentivrus | generated at NU GET-IN core | ||
| Viral Strain | GFP lentivrus | generated at NU GET-IN core | ||
| Viral Strain | RFP lentivrus | generated at NU GET-IN core | ||
| Isogenic Ctrl Clone N1 | iPSC line, M | PMID:26829649 | ||
| Isogenic Ctrl Clone N2 | iPSC line, M | PMID:26829649 | ||
| Isogenic CRISPR-Cas9 modified 16p11.2 duplication Clone B9 | iPSC line, M | PMID:26829649 | ||
| Isogenic CRISPR-Cas9 modified 16p11.2 duplication Clone D9 | iPSC line, M | PMID:26829649 | ||
| Schizophrenia line, 16p11.2 carrier | iPSC line, M | This paper, patients sourced from MSG1/2 schizophrenia cohorts (study accesions;phs000021.v2.p1 and phs000167.v1.p1) | 02C12626 | |
| Schizophrenia line, 16p11.2 carrier | iPSC line, F | 04C27671 | ||
| Schizophrenia line, 16p11.2 carrier | iPSC line, M | 06C58821 | ||
| Healthy control line | iPSC line, M | 06C52982 | ||
| Healthy control line | iPSC line, M | 05C51897 | ||
| Healthy control line | iPSC line, M | 05C44627 | ||
| Dye | Cal520-AM calcium-sensitive fluorogenic dye | AAT Bioquest | 21130 | |
| Commercial Assay Or Kit | TruSeq Stranded mRNA Sample Prep Kit v2 | Illumina, Inc. | Cat. No. RS-122-2101 | |
| Recombinant DNA | rtTA | Addgene | 20342 | |
| Recombinant DNA | FUW-TetO-Ngn2-P2A-puromycin | Addgene | 52047 | |
| Recombinant DNA | FUW-TetO-GFP | Addgene | 84041 | |
| Recombinant DNA | FUW-TetO-Ascl1-T2A-puromycin | Addgene | 27150 | |
| Recombinant DNA | FUW-TetO-Dlx2-IRES-hygromycin | Addgene | 97330 | |
| Cell Line | Primary Rat cortical astrocytes | Gibco | N7745100 | |
| Chemical Compound or Drug | DL-APV | Abcam | ab1200004 | |
| Chemical Compound or Drug | TTX | R & D Systems | 1078 | |
| Chemical Compound or Drug | NBQX | Tocris | 1044 | |
| Software; Algorithm | MATLAB v9.1 | Mathworks | RRID:SCR_001622 |
ACKNOWLEDGMENTS AND DISCLOSURES
This work was funded by the National Institute of Mental Health (Grant No. R01NS114977 [to PP]).
We thank the Northwestern Sequencing Core and Gene Editing and Transduction and Nanotechnology Core for their services.
Patient samples were sourced from the database of Genotypes and Phenotypes study (phs000021.v2.p1 and phs000167.v1.p1). Transcriptomic datasets are available on the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE215183, accession #GSE215183).
Schematics were created with BioRender.com.
Footnotes
The authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2022.11.005.
REFERENCES
- 1.Cannon TD, Kaprio J, Lönnqvist J, Huttunen M, Koskenvuo M (1998): The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch Gen Psychiatry 55:67–74. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM (2009): Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: A population-based study. Lancet 373:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan PF, Kendler KS, Neale MC (2003): Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 60:1187–1192. [DOI] [PubMed] [Google Scholar]
- 4.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. (2014): De novo mutations in schizophrenia implicate synaptic networks. Nature 506:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howrigan DP, Rose SA, Samocha KE, Fromer M, Cerrato F, Chen WJ, et al. (2020): Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nat Neurosci 23:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, et al. (2013): Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, et al. (2016): Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia.Nat Neurosci 19:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. (2014): A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winship IR, Dursun SM, Baker GB, Balista PA, Kandratavicius L, Maiade-Oliveira JP, et al. (2019): An overview of animal models related to schizophrenia. Can J Psychiatry 64:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014): Biological insights from 108 schizophrenia-associated genetic loci. Nature 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalus P, Müller TJ, Zuschratter W, Senitz D (2000): The dendritic architecture of prefrontal pyramidal neurons in schizophrenic patients. Neuroreport 11:3621–3625. [DOI] [PubMed] [Google Scholar]
- 12.Black JE, Kodish IM, Grossman AW, Klintsova AY, Orlovskaya D, Vostrikov V, et al. (2004): Pathology of layer V pyramidal neurons in the prefrontal cortex of patients with schizophrenia. Am J Psychiatry 161:742–744. [DOI] [PubMed] [Google Scholar]
- 13.Moyer CE, Shelton MA, Sweet RA (2015): Dendritic spine alterations in schizophrenia. Neurosci Lett 601:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlhaas PJ, Singer W (2010): Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci 11:100–113. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Hu N, Zhang W, Tao B, Dai J, Gong Y, et al. (2019): Dysconnectivity of multiple brain networks in schizophrenia: A meta-analysis of resting-state functional connectivity. Front Psychiatry 10:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka-Koshiyama K, Koshiyama D, Miyakoshi M, Joshi YB, Molina JL, Sprock J, et al. (2020): Abnormal spontaneous gamma power is associated with verbal learning and memory dysfunction in schizophrenia. Front Psychiatry 11:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birnbaum R, Weinberger DR (2017): Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci 18:727–740. [DOI] [PubMed] [Google Scholar]
- 18.Forstner AJ, Hecker J, Hofmann A, Maaser A, Reinbold CS, Mühleisen TW, et al. (2017): Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS One 12:e0171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross-Disorder Group of the Psychiatric Genomics Consortium (2019): Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179:1469–1482e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. (2019): Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Focus (Am Psychiatr Publ) 17:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodberry KA, Giuliano AJ, Seidman LJ (2008): Premorbid IQ in schizophrenia: A meta-analytic review. Am J Psychiatry 165:579–587. [DOI] [PubMed] [Google Scholar]
- 22.Grent-’t-Jong T, Gross J, Goense J, Wibral M, Gajwani R, Gumley AI, et al. (2018): Resting-state gamma-band power alterations in schizophrenia reveal E/I-balance abnormalities across illness-stages. eLife 7: e37799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ardhanareeswaran K, Mariani J, Coppola G, Abyzov A, Vaccarino FM (2017): Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat Rev Neurol 13:265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennand KJ, Gage FH (2011): Concise review: The promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem Cells 29:1915–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. (2011): Modelling schizophrenia using human induced pluripotent stem cells. Nature 473:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad R, Sportelli V, Ziller M, Spengler D, Hoffmann A (2018): Tracing early neurodevelopment in schizophrenia with induced pluripotent stem cells. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann A, Ziller M, Spengler D (2018): Childhood-onset schizophrenia: Insights from induced pluripotent stem cells. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. (2013): Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang N, Chanda S, Marro S, Ng YH, Janas JA, Haag D, et al. (2017): Generation of pure GABAergic neurons by transcription factor programming. Nat Methods 14:621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao R, Penzes P (2015): Common mechanisms of excitatory and inhibitory imbalance in schizophrenia and autism spectrum disorders. Curr Mol Med 15:146–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennand KJ, Landek-Salgado MA, Sawa A (2014): Modeling heterogeneous patients with a clinical diagnosis of schizophrenia with induced pluripotent stem cells. Biol Psychiatry 75:936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. (2017): Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szatkiewicz JP, O’Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM, et al. (2014): Copy number variation in schizophrenia in Sweden. Mol Psychiatry 19:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. (2014): CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505:361–366. [DOI] [PubMed] [Google Scholar]
- 35.D’Angelo D, Lebon S, Chen Q, Martin-Brevet S, Snyder LG, Hippolyte L, et al. (2016): Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry 73:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niarchou M, Chawner SJRA, Doherty JL, Maillard AM, Jacquemont S, Chung WK, et al. (2019): Psychiatric disorders in children with 16p11.2 deletion and duplication [published correction appears in Transl Psychiatry 9:107]. Transl Psychiatry 9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. (2008): Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358:667–675. [DOI] [PubMed] [Google Scholar]
- 38.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. (2011): A copy number variation morbidity map of developmental delay. Nat Genet 43:838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, et al. (2014): Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry 204:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. (2009): Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 41:1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green EK, Rees E, Walters JT, Smith KG, Forty L, Grozeva D, et al. (2016): Copy number variation in bipolar disorder. Mol Psychiatry 21:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillentine MA, Lupo PJ, Stankiewicz P, Schaaf CP (2018): An estimation of the prevalence of genomic disorders using chromosomal microarray data. J Hum Genet 63:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naujock M, Speidel A, Fischer S, Kizner V, Dorner-Ciossek C, Gillardon F (2020): Neuronal differentiation of induced pluripotent stem cells from schizophrenia patients in two-dimensional and in three-dimensional cultures reveals increased expression of the Kv4.2 sub-unit DPP6 that contributes to decreased neuronal activity. Stem Cells Dev 29:1577–1587. [DOI] [PubMed] [Google Scholar]
- 44.Tai DJ, Ragavendran A, Manavalan P, Stortchevoi A, Seabra CM, Erdin S, et al. (2016): Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat Neurosci 19:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. (2009): Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460:753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. (2011): Copy number variants in schizophrenia: Confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry 168:302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanders AR, Levinson DF, Duan J, Dennis JM, Li R, Kendler KS, et al. (2010): The Internet-based MGS2 control sample: Self report of mental illness. Am J Psychiatry 167:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundberg M, Pinson H, Smith RS, Winden KD, Venugopal P, Tai DJC, et al. (2021): 16p11.2 deletion is associated with hyperactivation of human iPSC-derived dopaminergic neuron networks and is rescued by RHOA inhibition in vitro. Nat Commun 12:2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rees E, Han J, Morgan J, Carrera N, Escott-Price V, Pocklington AJ, et al. (2020): De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat Neurosci 23:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. (2018): Comprehensive functional genomic resource and integrative model for the human brain. Science 362:eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, et al. (2008): Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in postmortem cerebellum. PLoS One 3:e3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu JQ, Wang X, Beveridge NJ, Tooney PA, Scott RJ, Carr VJ, Cairns MJ (2012): Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS One 7:e36351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tai DJC, Razaz P, Erdin S, Gao D, Wang J, Nuttle X, et al. (2022): Tissue- and cell-type-specific molecular and functional signatures of 16p11.2 reciprocal genomic disorder across mouse brain and human neuronal models. Am J Hum Genet 109:1789–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deshpande A, Yadav S, Dao DQ, Wu ZY, Hokanson KC, Cahill MK, et al. (2017): Cellular phenotypes in human iPSC-derived neurons from a genetic model of autism spectrum disorder. Cell Rep 21:2678–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urresti J, Zhang P, Moran-Losada P, Yu NK, Negraes PD, Trujillo CA, et al. (2021): Cortical organoids model early brain development disrupted by 16p11.2 copy number variants in autism. Mol Psychiatry 26:7560–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. (2018): Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359:693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. (2018): Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rein B, Tan T, Yang F, Wang W, Williams J, Zhang F, et al. (2021): Reversal of synaptic and behavioral deficits in a 16p11.2 duplication mouse model via restoration of the GABA synapse regulator Npas4. Mol Psychiatry 26:1967–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall LS, Medway CW, Pain O, Pardiñas AF, Rees EG, Escott-Price V, et al. (2020): A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum Mol Genet 29:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vidal-Domènech F, Riquelme G, Pinacho R, Rodriguez-Mias R, Vera A, Monje A, et al. (2020): Calcium-binding proteins are altered in the cerebellum in schizophrenia. PLoS One 15:e0230400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan TA, Revah O, Gordon A, Yoon SJ, Krawisz AK, Goold C, et al. (2020): Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat Med 26:1888–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park SJ, Jeong J, Park YU, Park KS, Lee H, Lee N, et al. (2015): Disrupted-in-schizophrenia-1 (DISC1) regulates endoplasmic reticulum calcium dynamics. Sci Rep 5:8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forrest MP, Parnell E, Penzes P (2018): Dendritic structural plasticity and neuropsychiatric disease. Nat Rev Neurosci 19:215–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Panzeri S, Macke JH, Gross J, Kayser C (2015): Neural population coding: Combining insights from microscopic and mass signals. Trends Cogn Sci 19:162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mathalon DH, Sohal VS (2015): Neural oscillations and synchrony in brain dysfunction and neuropsychiatric disorders: It’s about time. JAMA Psychiatry 72:840–844. [DOI] [PubMed] [Google Scholar]
- 66.Lewis DA, Curley AA, Glausier JR, Volk DW (2012): Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci 35:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung DW, Fish KN, Lewis DA (2016): Pathological basis for deficient excitatory drive to cortical parvalbumin interneurons in schizophrenia. Am J Psychiatry 173:1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.