Abstract
Introduction
People with HIV (PWH) of African ancestry have faster decline of kidney function and faster progression to end-stage renal disease than PWH of European ancestry. DNA methylation have been associated with kidney function in the general population, however, their relationships are unclear for PWH of African ancestry.
Methods
We performed epigenome-wide association studies (EWAS) of estimated glomerular filtration rate (eGFR) among PWH of African ancestry in 2 subsets of the Veterans Aging Cohort Study cohort (N = 885), followed by a meta-analysis to combine the results. Replication was conducted among independent African American samples without HIV.
Results
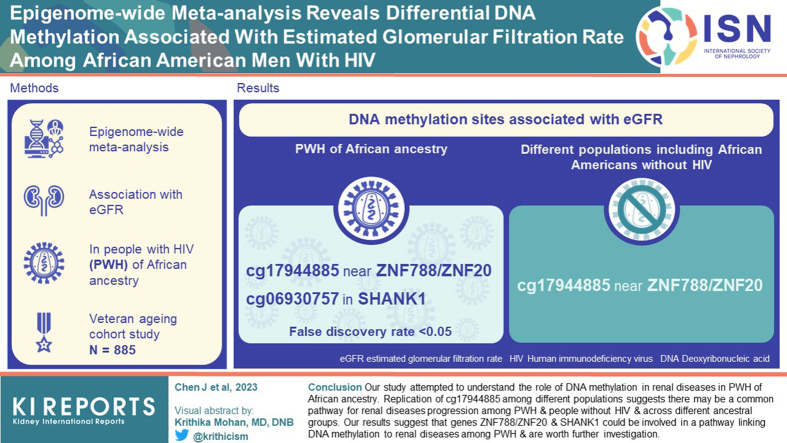
DNA methylation sites cg17944885 near Zinc Finger Family Member 788 (ZNF788) and Zinc Finger Protein 20 (ZNF20), and cg06930757 in SHANK1 were significantly associated with eGFR among PWH of African ancestry (false discovery rate < 0.05). DNA methylation site cg17944885 was also associated with eGFR among different populations including African Americans without HIV.
Conclusions
Our study attempted to address an important gap in the literature and to understand the role of DNA methylation in renal diseases in PWH of African ancestry. Replication of cg17944885 among different populations suggests there may be a common pathway for renal diseases progression among PWH and people without HIV, and across different ancestral groups. Our results suggest that genes ZNF788/ZNF20 and SHANK1 could be involved in a pathway linking DNA methylation to renal diseases among PWH and are worth further investigation.
Keywords: African ancestry, DNA methylation, eGFR, EWAS, HIV
Graphical abstract
Introduction
Chronic kidney disease (CKD), a common complication of both HIV infection and its treatment, continues to be a critical cause of shortened life span among PWH.1 Although the biological mechanisms leading to CKD among PWH remain incompletely understood, multiple factors, including direct exposure to HIV; superinfections; the systemic immune response to infection;1,2 as well as traditional CKD risk factors such as body mass index, hypertension, diabetes, and cigarette smoking have been linked to a higher burden of kidney dysfunction among PWH.3, 4, 5 PWH of African ancestry have a markedly faster decline of kidney function and faster progression to end-stage renal disease than PWH of European ancestry.6 Apart from these observations, our understanding of the underlying molecular and pathophysiologic pathways of developing HIV-induced kidney disease remains limited, particularly among PWH of African ancestry.
Multiancestral genome-wide association studies have successfully identified common genetic variants associated with kidney function and CKD susceptibility.7 However, only a small proportion of CKD risk can be explained by genetics alone.8 Integration of regulatory annotations with the leading genetic variants supports altered transcriptional regulation as an essential mechanism for CKD.9 DNA methylation, the most studied epigenetic modification in humans, can precipitate both transient and stable changes in gene expression. Investigation of DNA methylation related to CKD and kidney function is of particular interest.10, 11, 12
Early EWAS of eGFR, a measure of renal filtration function and CKD in the general population had small sample sizes or were limited to European ancestry.13, 14, 15 Later on, EWAS of multiethnic participants, including both non-Hispanic White and African Americans, identified several significant cytosine-phosphate-guanine dinucleotide (CpG, a specific form of DNA methylation) sites associated with eGFR.10,16 A meta-analysis of 2878 African Americans revealed 4 associations between DNA methylation and eGFR.11 An EWAS in the Genetic Epidemiology Network of Arteriopathy (GENOA) study discovered 7 CpG sites associated with damage of target organs, which included kidney function measured by eGFR, among African Americans.17 Furthermore, epigenetic associations with eGFR were also observed in native Americans.18
To date, only 1 EWAS of eGFR has been conducted among PWH, in which 3 hypermethylated CpG sites were associated with low eGFR.12 However, this study included only 567 PWH of multiple ancestries and the epigenome-wide coverage was limited in comparison with newer array platforms, such as the EPIC 850K BeadChip. The epigenetic associations from this study have not been replicated in an independent sample of PWH and the primary outcome eGFR was calculated based on an outdated creatinine-based equation that considered Black ancestry.12,19 There is now a newer equation called Chronic Kidney Disease Epidemiology Collaboration 2021 Creatinine-based Equation, which excludes ancestry in the calculation of eGFR and is considered more accurate for clinical usage.20 To identify epigenetic associations with kidney function in African Americans with HIV, we conducted an EWAS of eGFR calculated using the Chronic Kidney Disease Epidemiology Collaboration 2021 Creatinine-based Equation among 885 African American PWH and pursued external replications in independent African American samples without HIV.
Methods
Study Population and Outcome
We utilized phenotypic and epigenetic data from the Veterans Aging Cohort Study (VACS), which is a prospective study of veterans in care at Department of Veterans Affairs medical centers across the United States.21 Between 2005 and 2007, 1525 PWH and 843 participants without HIV from VACS consented to provide blood samples for future biomarker and genetic studies, forming the VACS Biomarker Cohort as previously described.22 Whole blood samples collected from each VACS-Biomarker Cohort participant were used for profiling of DNA methylation.22 This study was approved by the human research protection program of Yale University and the institutional research committee at the Connecticut Veterans Healthcare System. The current analysis included VACS-Biomarker Cohort PWH with available DNA methylation data.
Demographic and clinical information closest to the date of blood sample collection, including age, sex, ancestry, chronic health conditions, use of medications, creatinine, and HIV viral load were obtained from the electronic health record. VACS surveys completed by participants closest to the date of blood sample collection were used to obtain information on cigarette smoking.
The primary outcome is eGFR2021, which denotes eGFR calculated from standardized serum creatinine levels (an endogenous filtration marker), sex, and age using the Chronic Kidney Disease Epidemiology Collaboration 2021 Creatinine-based Equation (eGFR = 142 × [minimum of standardized creatinine/K or 1]α × [maximum of standardized creatinine/K or 1]−1.2 × 0.9938Age × [1.012 for female], where α = −0.241 for females or −0.302 for males and K = 0.7 for females and 0.9 for males).20 Iin addition, for comparison with previous research, we considered a secondary outcome, eGFR2006, which denotes eGFR calculated using the Modification of Diet in Renal Disease Study Equation (eGFR = 175 × [standardized creatinine]−1.154 × [age]−0.203 × [0.742 for females] × [1.212 if Black]).19
Within VACS-Biomarker Cohort, blood samples of a subset of participants were selected for epigenome-wide CpG sites profiling using the Illumina Infinium Human Methylation 450K BeadChip (VACS-450K).23 Subsequently, because of the availability of a newer generation of DNA methylation array that has a higher genomic coverage, another subset of participants was profiled using the Illumina Methylation EPIC (850K) BeadChip (VACS-850K).24 VACS-450K and VACS-850K were mutually exclusive, and the assignment of VACS participants to the 2 DNA methylation platforms was arbitrary.
DNA Methylation Measures and Quality Control
Genomic DNA was extracted from whole blood samples using PAXgene collection tubes and FlexiGene DNA extraction kits. DNA methylation was measured using either Illumina 450K or 850K BeadChips following the standard protocol. Intensity files in IDAT format were imported using minfi R package,25 followed by quality control. Probes with detection P-values greater than or equal to 0.001 were considered not successfully detected and set to missing, and probes that failed detection in at least 95% of the samples were removed. Probes that were within 10 base pairs from a single-nucleotide polymorphism or mapped to multiple genomic locations were removed from further analysis.26 After quality control, there were 412,583 CpG sites in VACS-450K data set, 797,676 CpG sites in the VACS-850K data set, and 374,642 CpG sites common to both datasets. All samples had a detection rate of at least 95%. The CpG sites measured by the EPIC and 450K chips were mapped to Genome Research Consortium human build 37 (GRCh37).
All raw intensity values of each probe were quantile-normalized using minfi package in R. Normalized intensity values were then used to generate a methylation β value for each CpG site; β values ranged from 0 to 1, with 0 representing complete unmethylation and 1 for complete methylation. Proportions of 6 blood cell types (CD4+ T cells, CD8+ T cells, natural killer T cells, B cells, monocytes, and granulocytes) were calculated using cell-type specific CpG sites from a reference panel implemented in R minfi package.27
Statistical Model
The outline of the study design and main analyses are shown in Figure 1. We restricted our analyses to 885 self-reported non-Hispanic African American PWH. To examine the difference in demographic and clinical characteristics between VACS450K and VACS850K, standard 2-sample t-tests were used for numeric variables and chi-squared tests were used for categorical variables. Linear mixed effect regression models were utilized to assess the association between DNA methylation at individual CpG sites and each eGFR separately in VACS-450K and VACS-850K. The regression models were adjusted for age, body mass index, systolic blood pressure, hepatitis C virus infection, hepatitis B virus infection, smoking status, antiretroviral therapy status, diabetes status, antihypertensive medication usage, HIV viral load, top 10 principal components of epigenome-wide DNA methylation, and calculated cell-type proportions as fixed effects. We also included chip ID as the random effect to account for potential batch effect. To account for additional potential unmeasured bias and cryptic relatedness, we applied a Bayesian method based on an empirical null distribution, bacon to the EWAS results.28 Briefly, bacon constructs an empirical null distribution using a Gibbs Sampling algorithm by fitting a 3-component normal mixture on z-scores, with 1 forced component using prior knowledge and the other 2 components capturing the assumed-to-be-small number of true associations present in the data.
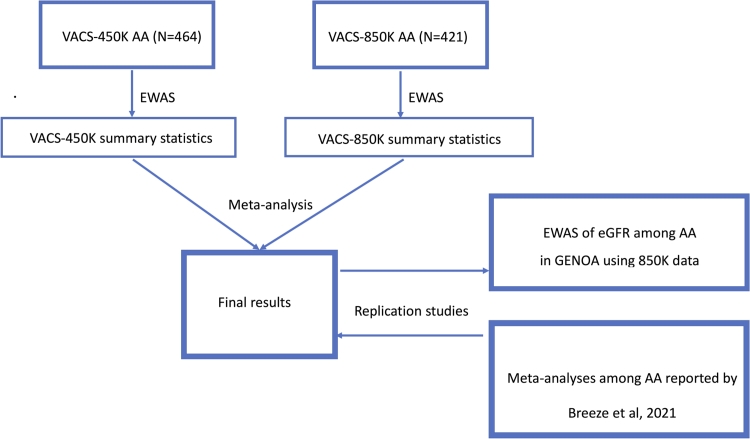
Figure 1.
Outline of the study design and main analyses among African Americans in the Veteran Aging Cohort Study (VACS). Meta-analyses were used to combine EWAS summary statistics (beta-coefficients, standard errors and P-values) from VACS-450K and VACS-850K. Replication of findings in our study were sought in African Americans in the GENOA study. Results from meta-analyses in our study were used to replicate findings from previous epigenetic association studies of eGFR in people without HIV.
Meta-analysis for 374,642 CpG sites common to VACS-450K and VACS-850K was conducted using the inverse variance-based approach in METAL software.29 We defined epigenome-wide significance for meta-analysis as Benjamini-Hochberg false discovery rate (FDR-q) <0.05. We also considered a more stringent threshold, Bonferroni corrected P-value threshold (0.05/374,642 = 1.3 ×10−7). To evaluate the surrounding signals of the identified eGFR-associated CpG sites, coMet R package was used to create regional plots, which included all tested CpG sites within the neighboring region of each significant association (± 10,000 base pairs).30
Replication Studies
Because of a lack of data for African American PWH for an independent eGFR EWAS, we pursued replication of epigenetic association with eGFR in 961 African American participants from the GENOA study (Figure 1).17 The outcome eGFR in the GENOA study was calculated using the CKD-EPI 2009 Creatinine equation (eGFR=141 × [minimum of standardized creatinine/K or 1]α × [maximum of standardized creatinine/K or 1]−1.209 × 0.993Age × [1.018 for female] × [1.159 for Black], where α = −0.329 for females or −0.411 for males, and K = 0.7 for females and 0.9 for males),31 denoted by eGFR2009.
DNA methylation in GENOA was measured using the Infinium MethylationEPIC BeadChip. Probes with detection P-value < 10−16 were considered successful detection. Samples and probes with a failed detection rate of at least 10% were excluded. Data for cross-hybridizing probes and those on sex chromosomes, sex mismatched samples, and outliers were also excluded (details of the quality control procedure has been published elsewhere).17 M-values were calculated from methylation beta values using logit transformation (log2[β/(1-β)]), followed by adjustment for white blood cell type counts and batch effects (sample plate, chip ID, and sample row) using linear mixed models. Values of eGFR2009 were rank-based inverse normalized. EWAS testing the association between M-values for each CpG site and normalized eGFR2009 were performed using linear mixed models implemented in the Genome-wide Efficient Mixed Model Association software,32 controlling for age, sex, smoking status, systolic and diastolic blood pressures, antihypertensive medicine use, body mass index, and top principal components. In the linear mixed model, genetic relatedness matrix was used as random effect to adjust for familial relationship.
We examined 4 eGFR2009-associated CpG sites from a recent meta-analysis of 1820 African Americans followed-up with a successful replication among 203 African Americans in their own study.11 We also attempted to replicate significant findings among multiancestry populations reported by Breeze et al.11 (25 CpG sites) and Chu et al.10 (18 CpG sites). Successful replication of an associated CpG in the VACS meta-analysis results was defined as having a consistent direction of association and a multiple testing corrected P-value less than 0.05. If the previously established CpG sites were not among the 374,642 common CpG sites used in our meta-analysis, epigenetic associations from either VACS-450K or VACS-850K subsets were used for replication.
Results
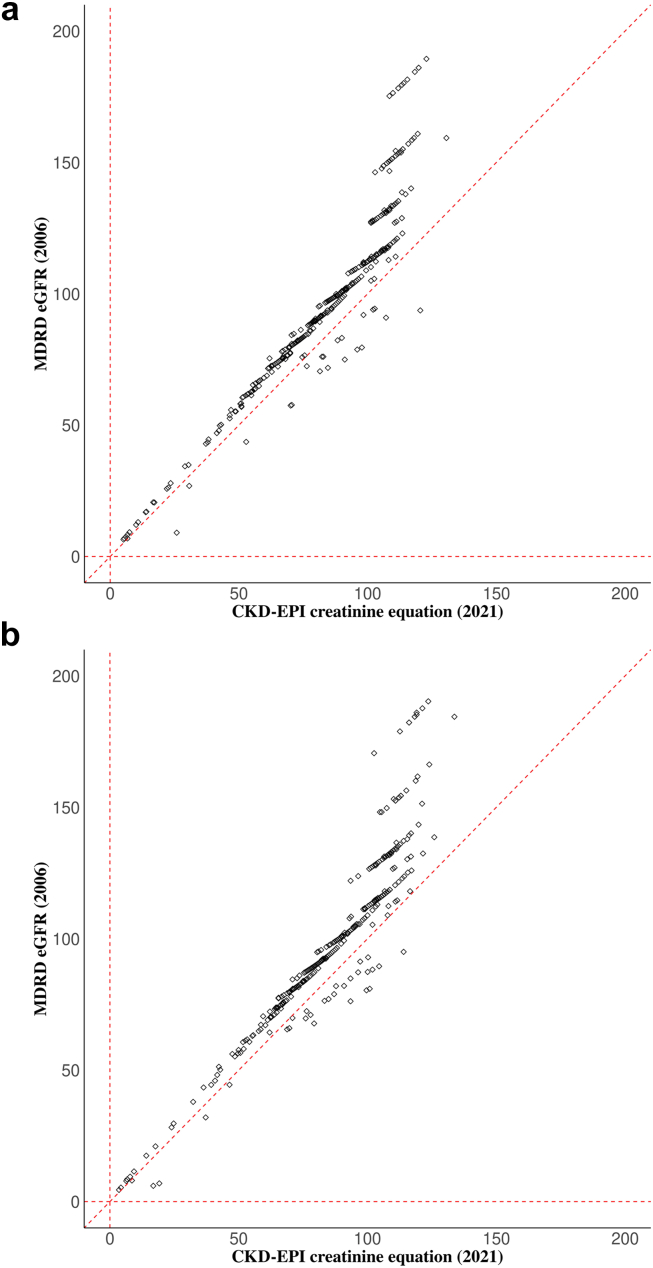
Demographic and clinical characteristics were generally similar in the 2 subpopulations (Table 1). Participants from both VACS-450K (N = 464) and VACS-850K (N = 421) were all male, African Americans, with an average age of 52.2 ± 7.5 and 50.8 ± 8.0 years respectively. The proportion with hepatitis C virus infection was higher in VACS-450K (55.0%) than in VACS-850K (32.5%). Average eGFR (in ml/min per 1.73 m2) was 85.7 and 86.7, using eGFR2021; and 99.9 and 100.3, using eGFR2006. Although the 2 eGFR measures were highly correlated (r = 0.92) in both samples, deviations from the regression lines (eGFR2021 vs. eGFR2006) were seen for participants with higher eGFR (Figure 2).
Table 1.
Demographic and clinical characteristics of the African Americans in the 2 subsets of VACS
| Demographic and clinical characteristics | VACS-450K (N = 464) | VACS-850K (N = 421) | P-value |
|---|---|---|---|
| eGFR2021 (ml/min per 1.73 m2), mean ± SD | 85.7 ± 23.7 | 86.7 ± 23.2 | 0.505 |
| eGFR2006 (ml/min per 1.73 m2), mean ± SD | 99.9 ± 35.0 | 100.3 ± 33.3 | 0.865 |
| Age (yr), mean ± SD | 52.2 ± 7.5 | 50.8 ± 8.0 | 0.005 |
| Antiretroviral therapy, n (%) | 387 (83.4%) | 348 (82.7%) | 0.837 |
| Diabetes, n (%) | 89 (19.2%) | 79 (18.8%) | 0.943 |
| Hepatitis B virus infection, n (%) | 49 (10.6%) | 37 (8.8%) | 0.438 |
| Hepatitis C virus infection, n (%) | 255 (55.0%) | 137 (32.5%) | <0.001 |
| Body mass index (kg/m2), mean ± SD | 25.6 ± 4.7 | 26.2 ± 5.2 | 0.059 |
| Systolic blood pressure (mm Hg), mean ± SD | 129.6 ± 14.7 | 129.2 ± 14.7 | 0.695 |
| Smoking status | 0.146 | ||
| Nonsmoker, n (%) | 92 (19.8%) | 106 (25.1%) | |
| Past smoker, n (%) | 97 (20.9%) | 87 (20.7%) | |
| Current smoker, n (%) | 275 (59.3%) | 228 (54.2%) | |
| Anti-hypertensive medication use, n (%) | 331 (71.3%) | 288 (68.4%) | 0.381 |
| Viral load (copies/ml), median (Q1,Q3) | 75 (50,941000) | 75 (50,750000) | 0.659 |
eGFR2021, estimated glomerular filtration rate calculated using Chronic Kidney Disease Epidemiology Collaboration 2021 equation; eGFR2006, estimated glomerular filtration rate calculated using Modification of Diet in Renal Disease equation; (Q1,Q3), (Quantile 1, Quantile 3); VACS, Veteran Aging Cohort Study.
Figure 2.
Scatter plots showing linear relationship between estimated glomerular filtration rate (eGFR) calculated using Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) 2021 equation and Modification of Diet in Renal Disease equation (MDRD) among (a) VACS-450K and (b) VACS-850K participants. For each participant, the eGFR values from CKD-2021 and MDRD equations were plotted on the X-axis and Y-axis, respectively. The Pearson's correlation coefficient between 2 eGFR measures were 0.92 in both VACS-450K and VACS-850K.
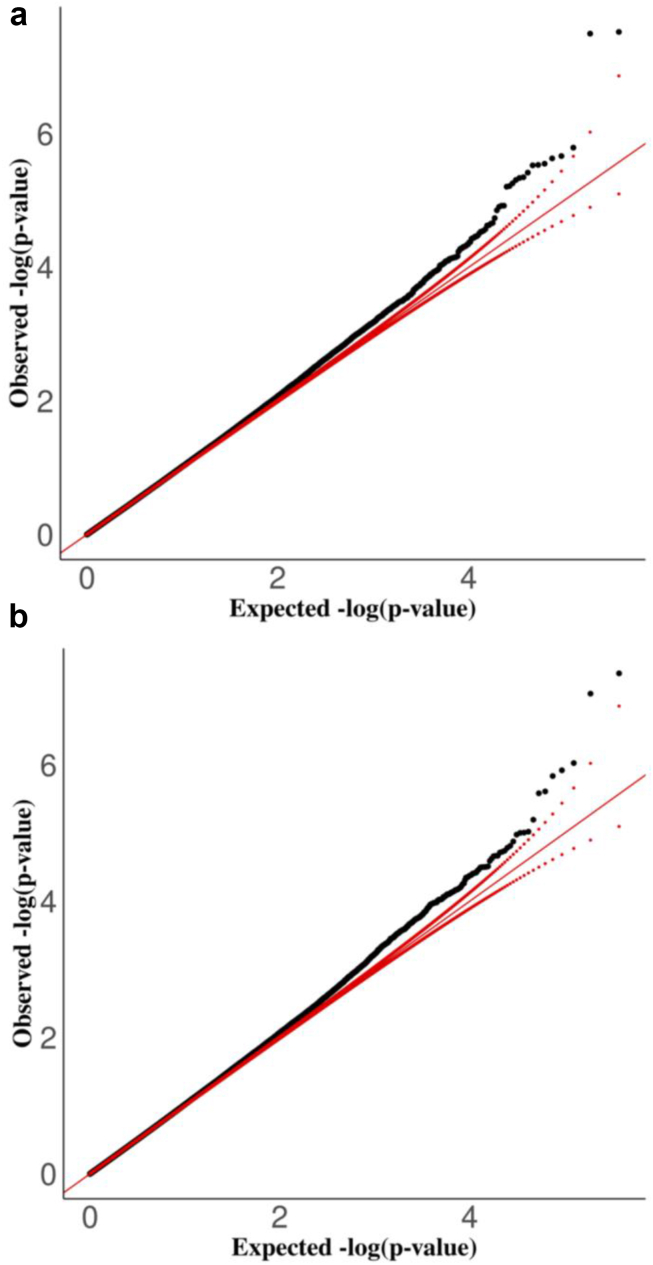
There were no significant CpG sites associated with eGFR2021 or eGFR2006 in VACS-450K or VACS-850K. Our meta-analyses focused on 375,014 common CpG sites available in both datasets. The quantile-quantile plot (Figure 3) comparing the observed P-values to the expected P-values in meta-analyses for eGFR2021 and eGFR2006 showed trivial global inflations (inflation factor = 1.01 and 1.03, respectively).
Figure 3.
Quantile-Quantile plots for the associations between DNA methylation and estimated glomerular filtration rate calculated using (a) Chronic Kidney Disease Epidemiology Collaboration 2021 equation and (b) Modification of Diet in Renal Disease equation from meta-analyses among African Americans in the Veteran Aging Cohort Study. These quantile-quantile plots compare the observed P-values to the expected P-values in meta-analyses for eGFR2021 and eGFR2006. X-axis represents -log10(expected P-values) and Y-axis represents -log10(observed P-values). The red lines indicate the distribution of expected P-values (solid diagonal line) and their 95% confidence interval (dotted lines). The inflation factors, which estimate the amount of inflation by comparing observed P-values to expected P-values under the hypothesis of no effect, equal 1.03 and 1.01 respectively in (a) and (b).
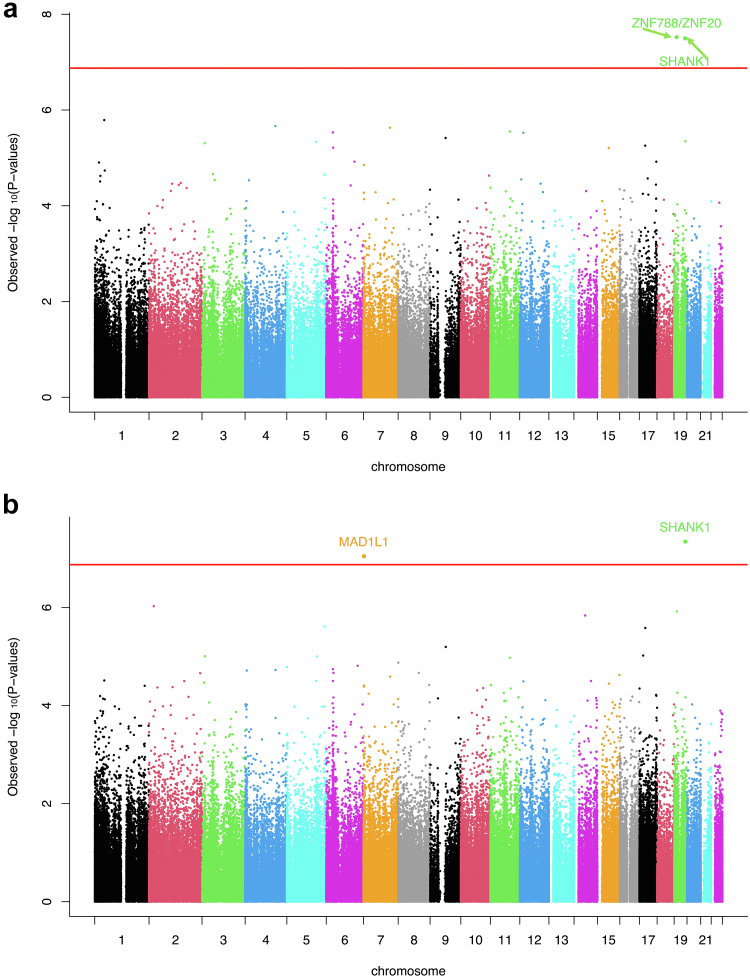
Meta-analysis for eGFR2021 identified 2 CpG sites significantly associated with eGFR2021 among African American PWH with FDR-q < 0.05 (Table 2). Both CpG sites remained significant using the Bonferroni-corrected P-value threshold (Figure 4a). We found that a 1% increase in the β value of CpG site cg17944885, located between genes ZNF788 and ZNF20, was associated with a 1.30-unit decrease in eGFR2021 (95% confidence interval, −1.77 to −0.83, P-value = 3.01 × 10−8). A 1% increase in value of CpG site cg06930757, located on gene SH3 And Multiple Ankyrin Repeat Domains 1 (SHANK1) was associated with a 1.28-unit increase in eGFR2021 (95% confidence interval, 0.83–1.73, P-value = 3.19 × 10−8). There were no additional CpG sites associated with eGFR2021 with nominal P-value < 0.05 in the ZNF788/ZNF20-cg17944885 and SHANK1-cg06930757 neighboring regions (Supplementary Figure S1).
Table 2.
Meta-analyses of differential DNA methylation at CpG sites associated with eGFR2021 or eGFR2006 among African Americans in 2 subsets of the Veteran Aging Cohort Study
| eGFR | CpG sites | Chr | Position (bp) | Gene | Meta-analysis |
VACS450K (N = 476) |
VACS850K (N = 369) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P-value | FDR-q | Beta | SE | P-value | Beta | SE | P-value | |||||
| 2021 | cg17944885 | 19 | 12225735 | ZNF788/ZNF20 | −1.30 | 0.24 | 3.01 × 10−8 | 5.98 × 10−3 | −1.22 | 0.37 | 8.95 × 10−4 | −1.37 | 0.32 | 2.69 × 10−5 |
| 2021 | cg06930757 | 19 | 51216389 | SHANK1 | 1.28 | 0.23 | 3.19 × 10−8 | 5.98 × 10−3 | 1.19 | 0.33 | 3.14 × 10−4 | 1.37 | 0.34 | 9.07 × 10−5 |
| 2006 | cg06930757 | 19 | 51216389 | SHANK1 | 1.88 | 0.34 | 4.53 × 10−8 | 0.02 | 1.73 | 0.50 | 5.42 × 10−4 | 2.01 | 0.47 | 2.34 × 10−5 |
| 2006 | cg06329547 | 7 | 1937739 | MAD1L1 | −2.27 | 0.42 | 9.01 × 10−8 | 0.03 | −2.70 | 0.47 | 1.67 × 10−8 | −0.28 | 1.01 | 0.78 |
bp, base-pair; Beta, Beta-coefficient; Chr, chromosome; CpG sites, cytosine-phosphate-guanine dinucleotide sites; eGFR2021, estimated glomerular filtration rate calculated using Chronic Kidney Disease Epidemiology Collaboration 2021 equation; eGFR2006, estimated glomerular filtration rate calculated using Modification of Diet in Renal Disease equation; SE, Standard error.
Linear mixed models were performed separately in VACS450K and VACS850K, followed by a meta-analysis for 374,642 CpG sites common to VACS-450K and VACS-850K. Beta refers to the beta-coefficient estimates of eGFR change per 1% changes in DNA methylation β-value, estimated by meta-analysis and linear mixed models in VACS450K and VACS850K respectively. Base-pair positions of CpG sites were mapped to Genome Research Consortium human build 37 (GRCh37).
Figure 4.
Manhattan plots of epigenome-wide CpG sites exhibiting differential associations for estimated glomerular filtration rate calculated using (a) Chronic Kidney Disease Epidemiology Collaboration 2021 equation and (b) Modification of Diet in Renal Disease equation from meta-analyses among African Americans in the Veteran Aging Cohort Study. Y-axis is showing -log10 (observed P-values) grouped by each chromosome in different colors (X axis). Red lines indicate genome-wide significance after adjusting for multiple testing. Gene annotations for significant CpG sites are shown.
Meta-analysis for eGFR2006 also detected a significant cg06930757-eGFR2006 association, where a 1% increase in value was associated with a 1.88-unit increase in eGFR2006 (95% confidence interval, 1.21–2.55, P-value = 4.53 × 10−8) (Table 2). CpG site cg06329547 resides in Mitotic Arrest Deficient Like 1 (MAD1L1) and was negatively associated with eGFR2006 (Beta-coefficient: −2.27, 95% confidence interval, −3.09 to −1.45, P-value = 9.01 × 10−8) (Table 2). Both CpG sites remained significant using the Bonferroni-corrected P-value threshold (Figure 4b).
We attempted to replicate our findings in the GENOA study (Table 3). CpG site cg06930757 was not included in the GENOA EWAS after quality control. CpG sites cg17944885 and cg06329547 showed consistent directionality in the GENOA results but only the association with cg17944885 was statistically significant (P-value = 1.62 × 10−4) using Bonferroni corrected P-value threshold (0.05/2 = 0.025). This CpG site was also reported as the most significant signal with eGFR in other EWAS.10,11 None of the recently reported eGFR-associated CpG sites among African Americans in a large meta-analysis study by Breeze et al.11 were replicated in our meta-analysis results in PWH (Table 4).11 However, 6 out of 15 (40%) eGFR-associated CpG sites among multiancestry populations in the same study11 were successfully replicated in our eGFR2021 meta-analysis results with FDR-q less than 0.05 (Supplementary Table S1). In addition, 9 out of 18 (50%) significant CpG sites in multiancestry populations reported by Chu et al.10 were successfully replicated in our eGFR2021 meta-analysis results (Supplementary Table S2). We found similar replications of these previously identified CpG sites using eGFR2006 meta-analysis results (Supplementary Tables 1 and 2).
Table 3.
Replication in the GENOA study
| CpG sites | Chr | Position (bp) | Gene | GENOAa |
|||
|---|---|---|---|---|---|---|---|
| Beta-coefficient | SE | P-value (Wald test) | FDR-q | ||||
| cg17944885 | 19 | 12225735 | ZNF788/ZNF20 | −0.22 | 0.06 | 1.62 x 10−4 | 3.32 x 10-4 |
| cg06930757 | 19 | 51216389 | SHANK1 | - | - | - | - |
| cg06329547 | 7 | 1937739 | MAD1L1 | −0.01 | 0.14 | 0.92 | 0.92 |
bp, base-pair; Chr, chromosome; CpG sites, cytosine-phosphate-guanine dinucleotide sites; eGFR2021, estimated glomerular filtration rate calculated using Chronic Kidney Disease Epidemiology Collaboration 2021 equation; eGFR2006, estimated glomerular filtration rate calculated using Modification of Diet in Renal Disease equation; GENOA, Genetic Epidemiology Network of Arteriopathy; SE, standard error.
GENOA study consisted of 2878 African Americans without HIV. Beta-coefficients, SE and P-value were estimated by linear mixed models used to replicate the 3 identified CpG associations with eGFR2021 or eGFR2006. In the GENOA study, estimated glomerular filtration rate was calculated using the Chronic Kidney Disease Epidemiology Collaboration 2009 equation, and was rank-based inverse normalized before performing EWAS, thus the Beta-coefficients and standard errors (SE) are in different scales from our study. Base-pair positions of CpG sites were mapped to Genome Research Consortium human build 37 (GRCh37). CpG site cg06930757 is not included in the EWAS in GENOA study after quality control.
Table 4.
Replication in Veteran Aging Cohort Study (VACS) participants of CpG associations identified by Breeze et al.11 (AA META)
| CpG sites | Chr | Position (bp) | Gene | AA META |
EWAS of eGFR2021 in VACS-850K |
EWAS of eGFR2006 in VACS-850K |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta-coefficient | SE | P-value | Beta | SE | P-value | FDR-q | Beta | SE | P-value | FDR-q | ||||
| cg14871770 | 10 | 96658622 | CYP2C9/CYP2C19 | −0.86 | 0.14 | 9.81 × 10−10 | −0.53 | 0.39 | 0.17 | 0.34 | −0.38 | 0.57 | 0.51 | 0.76 |
| cg13734658 | 17 | 73482882 | SDK2 | −0.61 | 0.11 | 9.70 × 10−9 | −0.45 | 0.31 | 0.14 | 0.34 | −0.49 | 0.46 | 0.28 | 0.76 |
| cg24097496 | 1 | 26288853 | UBXN11 | −0.72 | 0.13 | 4.12 × 10−8 | −0.23 | 0.34 | 0.50 | 0.66 | −0.28 | 0.50 | 0.57 | 0.76 |
| cg01065078 | 1 | 209768301 | TRAF3IP3 | −0.71 | 0.14 | 1.45 × 10−7 | 0.19 | 0.46 | 0.67 | 0.67 | −0.05 | 0.67 | 0.94 | 0.94 |
bp, base-pair; Beta, Beta-coefficients; CpG sites, cytosine-phosphate-guanine dinucleotide sites; Chr, Chromosome;SE, Standard error; eGFR, estimated glomerular filtration rate; eGFR2021, estimated glomerular filtration rate calculated using Chronic Kidney Disease Epidemiology Collaboration 2021 equation; EWAS, epigenome-wide association study.
In AA META, 23 CpG sites were found to be associated with eGFR in their discovery data set and 4 were successfully replicated in their replication data set. In AA META, eGFR was calculated using the Chronic Kidney Disease Epidemiology Collaboration 2009 equation. Beta-coefficients, standard error and P-values reported by AA META were obtained from a final meta-analysis of EWAS results in the discovery and replication data set and were located in Additional file 3: Supplementary Tables, Table S3b in their paper. Because these 4 CpG sites were not measured by 450K array, Beta, SE and P-values estimated from linear mixed models in VACS850K were used for replication. Beta refers to the changes in eGFR with 1% changes in DNA methylation β value.
Discussion
Our study investigated the DNA methylation association with eGFR2021 among PWH of African ancestry, a high-risk population for CKD and end-stage renal disease, and identified 2 significant CpG sites, cg17944885 and cg06930757. These eGFR-associated CpG sites mapped to genes ZNF788/ZNF20 and SHANK1 that may be related to eGFR among PWH of African ancestry in diverse pathways. We also examined DNA methylation association with eGFR2006, which detected a significant association with cg06930757 but not cg17944885, and revealed an additional CpG site cg06329547 that mapped to MAD1L1. There was a large difference between the values of eGFR2021 and eGFR2006, and the difference was larger among participants with higher eGFR. This was expected among African Americans because of the exclusion of ancestry in the calculation of eGFR2021.20,33 However, the 2 eGFR measures are highly correlated, which led to the identification of cg06930757-eGFR associations in both meta-analyses.
The leading CpG site cg17944885 showed a significant association with eGFR2021 in meta-analyses among PWH of African ancestry, which was replicated among people of African ancestry without HIV in the GENOA study. It was also reported in a meta-analysis performed among participants of the Women’s Health Initiative, the Multiethnic Atherosclerosis Study and the Jackson Heart Study.11 This CpG site is located between ZNF788 and ZNF20, whose function remains unknown. This association has also been observed across different racial groups, for example, in a subset of the Chronic Renal Insufficiency Cohort (473 participants, including 24% of non-Hispanic white ancestry and 54% of non-Hispanic Black ancestry); an EWAS of eGFR was performed in this cohort using linear regression with batch effect, age, sex, genetic background, hypertension, and cell heterogeneity as covariates and cytosine methylation (M values) as outcome.16 An EWAS among 181 diabetic Pima Indians also showed an association between M values at cg17944885 and baseline eGFR (P-value = 3.01 × 10−4).18 EWAS within the Atherosclerosis Risk in Communities study (2264 African Americans) and the Framingham Heart Study (2395 white participants) showed a significant, direction-consistent association between cg17944885 and eGFR with P-value = 1.61 × 10−7 in the Atherosclerosis Risk in Communities study, 2.03 × 10−17 in the Framingham Heart Study, and a pooled P-value = 1.20 × 10−23 from their meta-analysis.10 Moreover, the methylation level of cg17944885 was reported to show significant association with fibrosis in microdissected human kidney tubule samples.34 However, the methylation of cg17944885 did not show an observable association with albuminuria in the Chronic Renal Insufficiency Cohort.16 These findings indicate that this epigenetic association with eGFR is consistent across multiple ancestries, different tissue types, and populations with or without HIV.
Another significant CpG cg06930757 is located within SHANK1, which belongs to the SHANK family of proteins that encode scaffold proteins that interact with a variety of membrane and cytoplasmic proteins.35 Variants of the SHANK1 gene have been associated with a spectrum of neurodevelopmental disorders, and neurologic complications are reported to be highly prevalent among PWH and in CKD, irrespective of the cause.36, 37, 38, 39 This identified signal could be related to the development of neurologic diseases that are common among PWH and in CKD.
MAD1L1 codes mitotic-arrest deficient 1 (MAD1) protein, which is targeted by human T-cell leukemia virus type 1 during its virus transformation.40 The role of MAD1L1 in the mechanisms of CKD among PWH might be related to modulation of specific immune responses to HIV. Genetic variants in MAD1L1 were reported to be associated with multiple mosaic chromosomal alterations (gains on chromosome 15 and mosaic Y chromosome loss),41,42 and some age-related mosaic chromosomal alterations could predispose to diverse types of infections.43 The association of MAD1L1 methylation with eGFR was found only among PWH but not in the replication cohort of persons without HIV, suggesting that the methylation changes associated with eGFR could be specific to PWH.
Many eGFR-associated CpG sites previously reported from multiancestry cohorts other than cg17944885 were successfully replicated in Black PWH in our study (40%−50%).10,11 This suggested that different ancestries may share some of the same eGFR-DNA methylation associations. At the same time, previously identified African ancestry-specific eGFR-DNA methylation associations were not replicated in our study, suggesting that multiancestry EWAS have better power than single-ancestry EWAS in identification of eGFR-DNA methylation associations.
To the best of our knowledge, this is the first study to assess the association between DNA methylation and eGFR among PWH of African ancestry. We used eGFR2021 calculated without ancestry as our primary outcome, which provided results that are more accurate and easier for translation into clinical usage. We also utilized the 850K array for 421 study participants for improved genomic coverage, however, meta-analysis could only be conducted on common CpG sites between the 450K array and the 850K array platform, which can limit the power for potential discovery in the entire epigenome. A key limitation of our study is that it included only men and the findings will need to be validated in women. Another limitation is the use of peripheral blood cells to measure DNA methylation. Although peripheral leukocytes are key cell types for research on immune functions in HIV research, their epigenetic associations may not reflect the functional effects in the kidney. The cross-sectional design also limits inference on the causal relationship between DNA methylation and decline in kidney function.
In conclusion, our study identified significant epigenetic associations with kidney function in PWH of African ancestry, an important gap in understanding the elevated risk for development of kidney disease in this high-risk population. Results of our study could potentially further the understanding of biological mechanisms and target early detection and intervention to help lift some of the burden of CKD among PWH. The association of cg17944885 with eGFR was replicated across different populations, suggesting a potential common epigenetic pathway for kidney function among PWH and people without HIV, and across different ancestral groups. On the other hand, we also identified unique eGFR-associated CpG sites among PWH, which could be promising targets for understanding HIV-associated kidney dysfunction. Large epigenetic epidemiologic studies of diverse groups representing at-risk populations may further reveal the risk profile of kidney disease among PWH.44, 45, 46
Disclosure
All the authors declared no competing interests.
Acknowledgments
This work was funded by the following grants: JC, ZW, QH, VCM, and YVS received support from the National Institute of Health (R01DK125187); VCM and YVS received support from Emory CFAR (P30AI050409) and I01BX006008; KX received support from the National Institute on Drug Abuse (R01DA042691, R01DA047820, R01DA047063); KS received support from NIH K01HL134147, VACS - NIH NIAAA U24 AA020794, U01 AA020790, U10 AA013566 completed. The Genetic Epidemiology Network of Arteriopathy is funded by R01 DK073537, R01 HL119443, R01 HL133221. The views and opinions expressed in this manuscript are those of the authors and do not necessarily represent those of the Department of Veterans Affairs or the United States government.
Data Sharing Statement
Because of US Department of Veterans Affairs (VA) regulations and our ethics agreements, the analytic data sets used for this study are not permitted to leave the VA firewall without a Data Use Agreement. This limitation is consistent with other studies based on VA data. However, VA data are made freely available to researchers with an approved VA study protocol. For more information, please visit https://www.virec.research.va.gov or contact the VA Information Resource Center at VIReC@va.gov.
Footnotes
Figure S1. Regional plots of significant CpG sites associated with estimated glomerular filtration rate calculated using Chronic Kidney Disease Epidemiology Collaboration 2021 equation from meta-analysis among African Americans in Veteran Aging Cohort Study. (a) Regional plot of CpG site cg17944885; (b) Regional plot of CpG site cg069307575.
Table S1. Replication in VACS participants of CpG associations with eGFR discovered and successfully replicated by meta-analysis among multiancestry population in Breeze et al.11 (MA META).
Table S2. Replication in VACS participants of CpG associations with eGFR discovered and successfully replicated by EWAS among multiancestry population in Chu et al.10
Supplementary Material
Figure S1. Regional plots of significant CpG sites associated with estimated glomerular filtration rate calculated using Chronic Kidney Disease Epidemiology Collaboration 2021 equation from meta-analysis among African Americans in Veteran Aging Cohort Study. (a) Regional plot of CpG site cg17944885; (b) Regional plot of CpG site cg069307575. (PDF)
Table S1. Replication in Veteran Aging Cohort Study participants of CpG associations with eGFR discovered and successfully replicated by meta-analysis among multiancestry population in Breeze et al.11 (MA META)
Table S2. Replication in VACS participants of CpG associations with eGFR discovered and successfully replicated by EWAS among multiancestry population in Chu et al.10
References
- 1.Cohen S.D., Kopp J.B., Kimmel P.L. Kidney diseases associated with human immunodeficiency virus infection. N Engl J Med. 2017;377:2363–2374. doi: 10.1056/NEJMra1508467. [DOI] [PubMed] [Google Scholar]
- 2.Nobakht E., Cohen S.D., Rosenberg A.Z., Kimmel P.L. HIV-associated immune complex kidney disease. Nat Rev Nephrol. 2016;12:291–300. doi: 10.1038/nrneph.2015.216. [DOI] [PubMed] [Google Scholar]
- 3.Miguez-Burbano M.J., Wyatt C., Lewis J.E., Rodriguez A., Duncan R. Ignoring the obvious missing piece of chronic kidney disease in HIV: cigarette smoking. J Assoc Nurs AIDS Care. 2010;21:16–24. doi: 10.1016/j.jana.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Wit S., Sabin C.A., Weber R., et al. Incidence and risk factors for new-onset diabetes in HIV-infected patients: the Data Collection on Adverse Events of anti-HIV Drugs (D:A:D) study. Diabetes Care. 2008;31:1224–1229. doi: 10.2337/dc07-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seaberg E.C., Munoz A., Lu M., et al. Association between highly active antiretroviral therapy and hypertension in a large cohort of men followed from 1984 to 2003. Aids. 2005;19:953–960. doi: 10.1097/01.aids.0000171410.76607.f8. [DOI] [PubMed] [Google Scholar]
- 6.Lucas G.M., Lau B., Atta M.G., Fine D.M., Keruly J., Moore R.D. Chronic kidney disease incidence, and progression to end-stage renal disease, in HIV-infected individuals: a tale of two races. J Infect Dis. 2008;197:1548–1557. doi: 10.1086/587994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanzick K.J., Li Y., Schlosser P., et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat Commun. 2021;12:4350. doi: 10.1038/s41467-021-24491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köttgen A., Pattaro C., Böger C.A., et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattaro C., Teumer A., Gorski M., et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7 doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu A.Y., Tin A., Schlosser P., et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8:1286. doi: 10.1038/s41467-017-01297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breeze C.E., Batorsky A., Lee M.K., et al. Epigenome-wide association study of kidney function identifies trans-ethnic and ethnic-specific loci. Genome Med. 2021;13 doi: 10.1186/s13073-021-00877-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Huang Y., Hui Q., et al. Epigenetic associations with estimated glomerular filtration rate among men with human immunodeficiency virus infection. Clin Infect Dis. 2019;70:667–673. doi: 10.1093/cid/ciz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wing M.R., Devaney J.M., Joffe M.M., et al. DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC study. Nephrol Dial Transplant. 2014;29:864–872. doi: 10.1093/ndt/gft537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyth L.J., McKay G.J., Maxwell A.P., McKnight A.J., McKnight A.J. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9:366–376. doi: 10.4161/epi.27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sapienza C., Lee J., Powell J., et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2014;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 16.Sheng X., Qiu C., Liu H., et al. Systematic integrated analysis of genetic and epigenetic variation in diabetic kidney disease. Proc Natl Acad Sci U S A. 2020;117:29013–29024. doi: 10.1073/pnas.2005905117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ammous F., Zhao W., Ratliff S.M., et al. Epigenome-wide association study identifies DNA methylation sites associated with target organ damage in older African Americans. Epigenetics. 2021;16:862–875. doi: 10.1080/15592294.2020.1827717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu C., Hanson R.L., Fufaa G., et al. Cytosine methylation predicts renal function decline in American Indians. Kidney Int. 2018;93:1417–1431. doi: 10.1016/j.kint.2018.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levey A.S., Coresh J., Greene T., et al. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med. 2006;145:247–254. doi: 10.7326/0003-4819-145-4-200608150-00004. [DOI] [PubMed] [Google Scholar]
- 20.Inker L.A., Eneanya N.D., Coresh J., et al. New creatinine- and cystatin C–based equations to estimate GFR without race. N Engl J Med. 2021;385:1737–1749. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Justice A.C., Modur S.P., Tate J.P., et al. Predictive accuracy of the Veterans Aging Cohort Study index for mortality with HIV infection: a North American cross cohort analysis. J Acquir Immune Defic Syndr. 2013;62:149–163. doi: 10.1097/QAI.0b013e31827df36c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armah K.A., McGinnis K., Baker J., et al. HIV status, burden of comorbid disease, and biomarkers of inflammation, altered coagulation, and monocyte activation. Clin Infect Dis. 2012;55:126–136. doi: 10.1093/cid/cis406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Justice A.C., Hu Y., et al. Epigenome-wide differential DNA methylation between HIV-infected and uninfected individuals. Epigenetics. 2016;11:1–11. doi: 10.1080/15592294.2016.1221569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu C., Justice A.C., Zhang X., et al. DNA methylation biomarker selected by an ensemble machine learning approach predicts mortality risk in an HIV-positive veteran population. Epigenetics. 2021;16:741–753. doi: 10.1080/15592294.2020.1824097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aryee M.J., Jaffe A.E., Corrada-Bravo H., et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y.A., Lemire M., Choufani S., et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houseman E.A., Kelsey K.T., Wiencke J.K., Marsit C.J. Cell-composition effects in the analysis of DNA methylation array data: a mathematical perspective. BMC Bioinformatics. 2015;16:95. doi: 10.1186/s12859-015-0527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Iterson M., van Zwet E.W., BIOS Consortium. Heijmans B.T. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 2017;18:19. doi: 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin T.C., Yet I., Tsai P.C., Bell J.T. coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics. 2015;16:131. doi: 10.1186/s12859-015-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levey A.S., Stevens L.A., Schmid C.H., et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X., Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchkremer F., Segerer S. The 2009 and 2021 CKD-EPI equations: A graphical analysis of the effect of refitting GFR estimating equations without a race coefficient. Kidney Med. 2022;4 doi: 10.1016/j.xkme.2022.100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko Y.A., Mohtat D., Suzuki M., et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14:R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng M., Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 36.May H.J., Jeong J., Revah-Politi A., et al. Truncating variants in the SHANK1 gene are associated with a spectrum of neurodevelopmental disorders. Genet Med. 2021;23:1912–1921. doi: 10.1038/s41436-021-01222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M., Wang W., Song W., Qian W., Lin G.N. Integrative analysis identified key schizophrenia risk factors from an abnormal behavior mouse gene set. Life (Basel) 2021;11 doi: 10.3390/life11020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold R., Issar T., Krishnan A.V., Pussell B.A. Neurological complications in chronic kidney disease. JRSM Cardiovasc Dis. 2016;5 doi: 10.1177/2048004016677687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howlett W.P. Neurological disorders in HIV in Africa: a review. Afr Health Sci. 2019;19:1953–1977. doi: 10.4314/ahs.v19i2.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin D.Y., Spencer F., Jeang K.T. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 41.Wright D.J., Day F.R., Kerrison N.D., et al. Genetic variants associated with mosaic Y chromosome loss highlight cell cycle genes and overlap with cancer susceptibility. Nat Genet. 2017;49:674–679. doi: 10.1038/ng.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terao C., Suzuki A., Momozawa Y., et al. Chromosomal alterations among age-related haematopoietic clones in Japan. Nature. 2020;584:130–135. doi: 10.1038/s41586-020-2426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zekavat S.M., Lin S.-H., Bick A.G., et al. Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection. Nat Med. 2021;27:1012–1024. doi: 10.1038/s41591-021-01371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato M., Natarajan R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol. 2019;15:327–345. doi: 10.1038/s41581-019-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu L., Natarajan R., Chen Z. Epigenetic risk profile of diabetic kidney disease in high-risk populations. Curr Diab Rep. 2019;19:9. doi: 10.1007/s11892-019-1129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiels P.G., McGuinness D., Eriksson M., Kooman J.P., Stenvinkel P. The role of epigenetics in renal ageing. Nat Rev Nephrol. 2017;13:471–482. doi: 10.1038/nrneph.2017.78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.