Abstract
Accumulation of Plasmodium falciparum-infected erythrocytes in the placenta is a key feature of maternal malaria. This process is mediated in part by the parasite ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1) at the surface of the infected erythrocyte interacting with the host receptor chondroitin sulfate A (CSA) on the placental lining. We have localized CSA binding activity to two adjacent domains in PfEMP1 of an adherent parasite line and shown the presence of at least three active glycosaminoglycan binding sites. A putative CSA binding sequence was identified in one domain, but nonlinear binding motifs are also likely to be present, since binding activity in the region was shown to be dependent on conformation. Characterization of this binding region provides an opportunity to investigate further its potential as a target for antiadhesion therapy.
In Africa alone, over 20 million women each year are exposed to the risk of maternal malaria, an infection that leads to intrauterine growth retardation, low-birth-weight babies, and high levels of infant mortality (16). The key feature of this infection is the accumulation of Plasmodium falciparum- infected erythrocytes in the placenta (17), which is associated with parasite adhesion to the glycosaminoglycans (GAGs) chondroitin sulfate A (CSA) (2, 8) and hyaluronic acid (3) on the placental lining. Blocking the interaction between the infected erythrocyte and the GAG receptors, for example by an antiadhesive vaccine or small molecule, is therefore a rational therapeutic strategy for the disease, but elucidation of the mechanism of adhesion is a prerequisite.
The parasite protein P. falciparum erythrocyte membrane protein 1 (PfEMP1) has been identified recently as the ligand mediating CSA binding (5, 12), but the highly variable nature of the protein (4, 13) would appear to limit its suitability as a therapeutic target. Development of natural immunity requires the acquisition of a wide range of variant-specific antibodies directed against PfEMP1, following exposure to many different parasite variants (6). In contrast, antibodies which block the adhesion of infected erythrocytes to CSA may develop after a limited number of infections in pregnant women. Such antibodies may be strain independent and are associated with reduced placental malaria (M. Fried, F. Nosten, A. Brockman, B. J. Brabin, and P. E. Duffy, Letter, Nature 395:851–852, 1998). Given the large potential for generation of diversity in PfEMP1 (15), it is possible that this represents the presence of a functionally conserved binding site with a restricted variant antigenic type, which is the antibody target. Thus, characterization of the CSA binding region of PfEMP1 is critical for understanding pathogenesis and immunity.
Here we use a competitive enzyme inhibition assay to identify sites within a CSA binding region of PfEMP1 that interact directly with GAGs and to assess the importance of protein conformation to this activity. This information is essential for the wider investigation of binding-site conservation and future assessment of the feasibility of antiadhesive therapeutic strategies.
MATERIALS AND METHODS
Expression of fusion proteins.
The glutathione S-transferase fusion proteins were expressed from PCR products of the CS2 var gene in Escherichia coli by using pGeX expression vectors (Amersham Pharmacia) and affinity purified as previously described (12). The locations of the six proteins described are as follows: CIDRa, amino acids (aa) 404 to 736; CIDRb, aa 716 to 905; DBL3, aa 911 to 1204; DBL3-C, aa 979 to 1123; DBL3-5′, aa 911 to 1076; and DBL3-3′, aa 1063 to 1204. The full gene sequence is available from GenBank under accession number AF134154.
Reduction and alkylation of fusion proteins.
The CIDRb and DBL3 fusion proteins were reduced at 45°C for 1 h in 2-nmol amounts in the presence of 6 M guanidine–0.02 M Tris (pH 8) buffer containing 20 mM dithiothreitol (7). The proteins were then alkylated by the addition of 0.1 M iodoacetic acid and incubation for 1 h at room temperature in the dark. After this time, the reduced and alkylated protein was immediately buffer exchanged into 62 mM sodium acetate (pH 4.8) buffer using a Nap-5 column (Amersham Pharmacia). A nonreduced and nonalkylated sample of each protein was buffer exchanged down a Nap-5 column in a similar manner to control for protein loss.
Homogeneous enzyme-based binding assay.
Assays were performed essentially as described previously (9). To determine the working concentration of CSA, 250 μl of a serial dilution of porcine rib CSA (Sigma Aldrich), 250 μl of 80 mM p-nitrophenyl phenylphosphonate (Sigma Aldrich), 250 μl of 100-U/ml deoxyRNase II (Sigma Aldrich), and 250 μl of 62 mM sodium acetate (pH 4.8) buffer were mixed in glass tubes and incubated at 37°C for 15 min. All components were diluted and reconstituted in sodium acetate (pH 4.8) buffer. The reaction was then stopped with 100 μl of 2 N NH3H2O, and the absorbance was read at 405 nm. A “negative” control value (substrate and buffer) was subtracted from all readings, and a “positive” control value (substrate, enzyme, and buffer) was used to calculate enzyme inhibition as (1 − test absorbance/positive-control absorbance) × 100. The plot comparing inhibition with CSA concentration was a steep exponential curve (data not shown), and 1 μg of CSA per ml was seen to inhibit enzyme activity by approximately 80% and correspond to a phase of the reaction at which a small change in concentration profoundly affected the degree of inhibition observed.
For assay of the fusion proteins, a similar protocol was used with the CSA concentration constant at 1 μg/ml, and the aliquot of buffer exchanged for a serial dilution of the protein in the range 0.5 to 0.004 nmol. Arbitrary enzyme activity was calculated relative to the “positive” control tube by subtracting the value calculated by the formula above from 100.
Binding of peptides to CSA.
A biotinylated peptide was synthesized (Chiron) corresponding to the putative CS2-CSA binding motif KKKTIMDKLI. As a negative control, the homologous region (GKRTIMDELI) of a var gene expressed by a CD36 binding, non-CSA binding 3D7 line 5B1 (Genebank accession number AC005140) was also synthesized. Microtiter plates (Falcon 3077) were coated for 1 h at 37°C with alternate rows of 50-μg/ml porcine rib CSA (Sigma Aldrich) or 0.1% bovine serum albumin (BSA). The plates were washed three times in 0.1% BSA, blocked for 2 h at 37°C with 0.1% BSA, and then washed twice more in 0.1% BSA. Serial dilutions of the two peptides (400 to 50 nmol) were made, and 25-μl aliquots added to adjacent rows containing CSA and BSA before the mixture was incubated for 1 h at room temperature. The plates were again washed twice in 0.1% BSA, and adherent peptide was detected by incubating for 15 min at room temperature with horseradish peroxidase-streptavidin (Silenus; 1/1,000 dilution), washing four times with 0.1% BSA, and developing with a chromogenic substrate (2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid [ABST]). All components were diluted in TBS (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2). The developed color was read spectrophotometrically at 405 nm, and values for each test (CSA) well had the corresponding background control (BSA) values subtracted before plotting absorbance against peptide concentration.
RESULTS AND DISCUSSION
We previously described the cloning and sequencing of a dominant var gene expressed by the P. falciparum line CS2, which had been selected for its high-level binding to CSA (12). The major domains of the gene were expressed as recombinant fusion proteins, and antibodies raised against them were tested for their ability to inhibit CSA binding. These data suggested that the binding activity lay within the cysteine-rich interdomain region (CIDR) and Duffy binding-like region 3 (DBL3) (Fig. 1A). The DBL3 domain was also implicated in subsequent work by others using a different CSA-binding isolate, although no specific homology was found between the DBL domains (5).
FIG. 1.
(A) Schematic representation of the location of the CIDR and DBL3 domains of the CS2 var gene. (B) Amino acid sequence of the CIDRb protein, showing a putative CSA binding motif (boxed) and an overlap with a region associated with CD36 binding in other isolates (underlined). (C) Amino acid sequence of the DBL3 protein and the smaller DBL3-5′ (underlined), DBL3-C (highlighted), and DBL-3′ (dashed underline) polypeptides.
To further characterize the GAG binding activity of these regions, we investigated the direct interaction of recombinant proteins with CSA by using a fluid-phase homogeneous enzyme-based binding assay. The assay is based on the principle that at an acidic pH, the activity of deoxyRNase II, which converts a substrate to a colored product, can be nonspecifically inhibited by GAGs. When a GAG binding protein is introduced, it competes the GAG off the enzyme and restores activity, giving a reliable indication of the relative binding affinity of the protein (9).
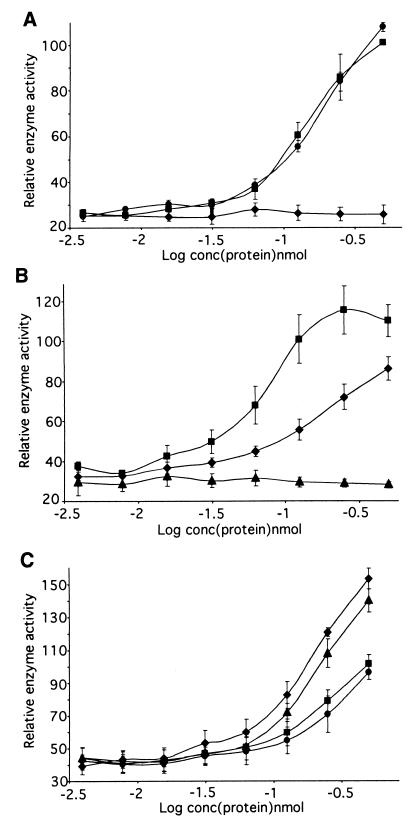
Recombinant proteins corresponding to defined regions of CS2-PfEMP1 (Fig. 1A) interacted with CSA in a specific manner in this assay. The presence of either DBL3 or CIDRb caused a steep recovery of enzyme activity, whereas the adjacent CIDRa had no effect (Fig. 2A). A full-length DBL4 fusion protein showed no activity (data not shown), consistent with the antibody inhibition data, in which anti-CIDR and DBL3 inhibited CSA binding but anti-DBL4 did not (12), and reinforcing the specificity of the assay. This identifies the end of the CIDR and the abutting DBL3 domain (Fig. 1B and C) as the GAG-CSA binding region of CS2-PfEMP1. Whether the binding sites in these two regions act independently or cooperatively in the native protein remains to be determined. Of particular importance for potential antiadhesive therapy is that antibodies directed to either individual site (CIDRb or DBL3) or, indeed, an adjacent noninteracting site (CIDRa) have been shown to inhibit adhesion (12). This suggests that targeting a single motif in the binding region can be an effective strategy, even in the presence of multiple binding sites.
FIG. 2.
Fusion proteins derived from the CS2 var gene sequence were assayed for direct interaction with CSA. (A) The CIDRb and DBL3 fusion proteins interacted with CSA, whereas the CIDRa protein showed no activity. ⧫, CIDRa; ■, CIDRb; ●, DBL3. (B) The end regions of DBL3 had independent CSA binding activity, but the center region did not bind. ⧫, DBL3-5′; ▴, DBL3-C; ■, DBL3-3′. (C) Reduction and alkylation of CIDRb and DBL3 proteins substantially reduces their affinity for CSA. ⧫, CIDRb; ■, CIDRb reduced and alkylated; ▴, DBL3; ●, DBL3 reduced and alkylated. All results are mean and standard error of the mean for three independent assays.
To map the binding activity of DBL3 more closely, recombinant proteins corresponding to the 5′ (DBL3-5′), 3′ (DBL3-3′), and central (DBL3-C) regions of the domain were tested in the assay system (Fig. 1C). The two end regions were seen to interact with CSA independently, indicating the presence of at least two sites with GAG binding ability within DBL3, but the central region (DBL3-C) showed no activity (Fig. 2B). Activity of the DBL3 proteins appeared to be restricted to those which contain the conserved cysteine-rich end blocks and raised the possibility that binding may be dependent on conformation. Reduction and alkylation of CIDRb and DBL3, which disrupts disulfide bonding and destabilizes the tertiary structure, reduced the CSA binding activity of the proteins by 30 to 40% in the assay (Fig. 2C). This effect suggests that some conformational restrictions do apply to the binding sites. However, residual activity demonstrates the presence of a proportion of motifs which do not require folding to interact with GAGs.
Although only specific PfEMP1 domains were seen to interact with GAGs, in this assay system it was not possible to demonstrate a preference of the proteins for CSA above other sulfated GAGs, such as CSC and heparin. This probably results from the use of relatively short recombinant proteins, which fail to achieve the correctly folded tertiary structure that appears to be important in determining the GAG specificity (10). We note, however, that the CS2 parasite line does not bind to heparin, CSC, or other polysulfated sugars (14) and that the DBL3 and CIDR region tested has been shown previously by antibody inhibition data to be linked to CSA adhesion (12). These findings make clear the importance of further studies on the native structure of this region in order to understand the molecular relationship of CSA affinity and specificity.
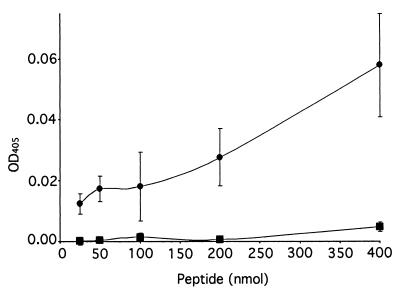
We examined the amino acid sequences of the DBL3 and CIDRb regions (Fig. 1B and C) for potential binding motifs, but sequence searches revealed no known glycoconjugate binding consensus sequences (10) in this region (or in any other part of the CS2 PfEMP1). However, a literature search revealed a description of a chondroitin sulfate binding motif, implicated in α4β1-integrin-mediated melanoma cell adhesion, ascribed to the 11-aa stretch KKEKDIMKKTI (11). A search for this motif revealed a sequence of high identity, KKKTIMDKLI, within the CIDRb sequence (Fig. 1B). This was clearly a prime candidate for the binding site in the CIDR, and a biotinylated peptide was synthesized with this sequence and shown in a solid-phase assay to bind to CSA in a dose-dependent manner (Fig. 3). A similarly synthesized peptide bearing the sequence of a homologous region from the CIDR from a var gene expressed by a non-CSA-adherent parasite did not bind in this assay, but once more the CS2 peptide was found to bind to other sulfated GAGs. The motif was not seen in the other reported var gene associated with CSA binding (5), and extensive sequence analysis of var genes from field isolates will be required to establish its general importance. A further point of interest is the location of the motif at the end of a region previously implicated in the binding of parasite-infected erythrocytes to CD36 (1) (Fig. 1B). This region appears well conserved in the numerous CD36 binding lines described, but the 10-aa CS2-PfEMP1 motif is absent from them. CSA-adherent placental parasites do not usually bind to CD36 (2, 8), and we speculate that a motif required for CSA binding which in some way disrupts the CD36 binding region could explain this observation.
FIG. 3.
A peptide corresponding to a putative CSA binding motif KKKTIMDKLI (●) in the CS2 CIDR region adhered to CSA in a solid-phase assay, whereas the homologous region GKRTIMDELI (■) from a non-CSA binding var gene did not. Results are mean and standard error of the mean for three independent assays. OD, optical density.
Characterization of the CSA binding region of CS2-PfEMP1 reveals fundamental characteristics of the adhesive interaction between P. falciparum and GAG/CSA. Particularly, it involves at least three potential binding sites, and protein conformation influences activity and GAG specificity. Additionally, we have identified a putative CSA binding motif that will be the focus of further investigation. Of major significance is the location of specific areas that can now be examined in a wide variety of CSA and placental binding isolates, enabling the identification of conserved elements and facilitation of the rational design of antiadhesive therapies.
ACKNOWLEDGMENTS
We thank Kathy Davern, Anne Thaus, and Tim Byrne for excellent technical assistance and Robert Flegg for assistance with the sequence searches.
This work was supported by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Baruch D I, Ma X C, Singh H B, Bi X, Pasloske B L, Howard R J. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- 2.Beeson J G, Brown G V, Molyneux M E, Mhango C, Dzinjalamala F, Rogerson S J. Plasmodium falciparum isolates from infected pregnant women and children are associated with distinct adhesive and antigenic properties. J Infect Dis. 1999;180:464–472. doi: 10.1086/314899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beeson J G, Rogerson S J, Cooke B M, Reeder J C, Chai W, Lawson A M, Molyneux M E, Brown G V. Adhesion of Plasmodium falciparum-infected erythrocytes to hyaluronic acid in placental malaria. Nat Med. 2000;6:86–90. doi: 10.1038/71582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggs B A, Goozé L, Wycherley K, Wollish W, Southwell B, Leech J H, Brown G V. Antigenic variation in Plasmodium falciparum. Proc Natl Acad Sci USA. 1991;88:9171–9174. doi: 10.1073/pnas.88.20.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buffet P A, Gamain B, Scheidig C, Baruch D, Smith J D, Hernandez-Rivas R, Pouvelle B, Oishi S, Fujii N, Fusai T, Parzy D, Miller L H, Gysin J, Scherf A. Plasmodium falciparum domain mediating adhesion to chondroitin sulfate A: a receptor for human placental infection. Proc Natl Acad Sci USA. 1999;96:12743–12748. doi: 10.1073/pnas.96.22.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull P C, Lowe B S, Kortok M, Molyneux C S, Newbold C I, Marsh K. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med. 1998;4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creighton T E. Disulphide bonds between cysteine residues. In: Creighton T E, editor. Protein structure. A practical approach. Oxford, United Kingdom: IRL Press; 1990. pp. 155–168. [Google Scholar]
- 8.Fried M, Duffy P E. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 9.Guo X, Han I S, Yang V C, Meyerhoff M E. Homogeneous enzyme-based binding assay for studying glycosaminoglycan interactions with macromolecules and peptides. Anal Biochem. 1996;235:153–160. doi: 10.1006/abio.1996.0107. [DOI] [PubMed] [Google Scholar]
- 10.Holt G D. Identifying glyconjugate-binding domains. Building on the past. Glycobiology. 1991;1:329–336. doi: 10.1093/glycob/1.4.329. [DOI] [PubMed] [Google Scholar]
- 11.Iida J, Meijne A M L, Oegema T R J, Yednock T A, Kovach N L, Furcht L T, McCarthy J B. A role of chondroitin sulfate glycosaminoglycan binding sites in α4β1 integrin-mediated melanoma cell adhesion. J Biol Chem. 1998;273:5955–5962. doi: 10.1074/jbc.273.10.5955. [DOI] [PubMed] [Google Scholar]
- 12.Reeder J C, Cowman A F, Davern K M, Beeson J G, Thompson J K, Rogerson S J, Brown G V. The adhesion of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate A is mediated by PfEMP1. Proc Natl Acad Sci USA. 1999;96:5198–5202. doi: 10.1073/pnas.96.9.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts D J, Craig A G, Berendt A R, Pinches R, Nash G, Marsh G, Newbold C I. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogerson S J, Chaiyaroj S C, Ng K, Reeder J C, Brown G V. Chondroitin sulfate A is a cell surface receptor for Plasmodium falciparum-infected erythrocytes. J Exp Med. 1995;182:15–20. doi: 10.1084/jem.182.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubio J P, Thompson J K, Cowman A F. The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. EMBO J. 1996;15:4069–4077. [PMC free article] [PubMed] [Google Scholar]
- 16.Steketee R W, Wirima J J, Slutsker L, Heymann D L, Breman J G. The problem of malaria and malaria control in pregnancy in Sub-Saharan Africa. Am J Trop Med Hyg. 1996;55:2–7. doi: 10.4269/ajtmh.1996.55.2. [DOI] [PubMed] [Google Scholar]
- 17.Walter P R, Garin Y, Blot P. Placental pathologic changes in malaria. Am J Pathol. 1982;109:330–342. [PMC free article] [PubMed] [Google Scholar]