Abstract
Cells produce multiple mRNAs through alternative splicing, which ensures proteome diversity. Because most human genes undergo alternative splicing, key components of signal transduction pathways are no exception. Cells regulate various signal transduction pathways, including those associated with cell proliferation, development, differentiation, migration, and apoptosis. Since proteins produced through alternative splicing can exhibit diverse biological functions, splicing regulatory mechanisms affect all signal transduction pathways. Studies have demonstrated that proteins generated by the selective combination of exons encoding important domains can enhance or attenuate signal transduction and can stably and precisely regulate various signal transduction pathways. However, aberrant splicing regulation via genetic mutation or abnormal expression of splicing factors negatively affects signal transduction pathways and is associated with the onset and progression of various diseases, including cancer. In this review, we describe the effects of alternative splicing regulation on major signal transduction pathways and highlight the significance of alternative splicing.
Subject terms: Alternative splicing, Extracellular signalling molecules
Molecular genetics: RNA splicing in health and disease
Cell signaling processes are affected by the varying ways that sections of messenger RNA (mRNA), the molecule that carries genetic instructions copied from a gene, are spliced together to generate several different proteins from a single gene. Kee K. Kim and colleagues at Chungnam National University in Daejon, South Korea, review the significance for cell signaling of the alternative splicing patterns of mRNAs. Mutations that lead to the splicing processes going awry are implicated in a range of diseases, including cancer. The authors examine the most recent research insights gained by applying emerging methods of genetic analysis to the role of mRNA splicing in several specific cell signaling pathways vital for normal development and health. They suggest that increasing understanding of faulty splicing in disease could open avenues towards new forms of treatment.
Introduction
Splicing is a process in which introns, noncoding regions, are removed from a precursor messenger RNA transcript (pre-mRNA) and exons, coding regions, are joined to generate mature mRNA. The process of selective recombination of exons by site-selective splicing of pre-mRNA is called alternative splicing. Since alternative splicing allows a single gene to produce multiple mRNAs, it is a key mechanism conferring intracellular proteome diversity. Since 95% of human genes with multiple exons undergo alternative splicing, most genes encoding major signal transduction pathway components undergo alternative splicing regulation1. Signal transduction is the process by which a specific environmental stimulus is converted into a biochemical signal that is transferred inside a cell and the subsequent translation of the signal that leads to a change in gene expression2. Cells rely on various signal transduction pathways to perform functions, such as proper growth, development, differentiation, migration, and apoptosis. Because proteins produced through alternative splicing can be expressed at different levels and be involved in various biological functions, splicing regulation generally affects signal transduction pathways. For example, through the selective combination of exons encoding important domains, the protein products may interact with various other proteins with differing binding affinities3. Splicing-generated isoforms may change a post-translational modification (PTM) or the protein localization, thereby exhibiting unique functions4,5. Additionally, these isoforms may affect transcriptional or enzymatic activity6. Signal transduction pathway components interact with each other to form a network and transmit signals downstream. Therefore, changes in protein localization, activity, interaction, or PTM can change the cellular effect mediated by the signal transduction pathway. As a result of alternative splicing regulation, unstable proteins with reduced expression levels can be generated, and prematurely terminated truncated proteins can be generated due to frameshifting7. Signal transduction can be activated or inhibited by various splicing isoforms. Additionally, aberrant splicing caused by mutations or abnormal expression of splicing factors may change the behavior of a signal transduction pathway and is thus associated with the onset and progression of various diseases, including cancer. In this review, we describe the effects of various alternative splicing outcomes on key signal transduction pathways. Therefore, this review highlights the importance of alternative splicing in fine-tuning intracellular signal transduction pathway effects.
Regulatory mechanism of alternative splicing
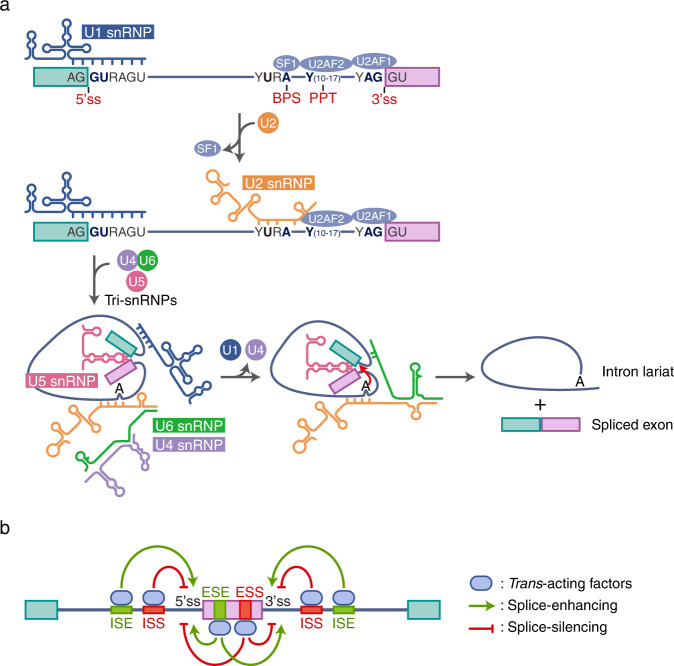
Splicing is regulated by the spliceosome, a large ribonucleic acid protein complex. The spliceosome components are U-enriched small nuclear RNA (snRNA) and small nuclear ribonucleoprotein particles (snRNPs), which are composed of several nuclear proteins8,9. snRNPs are classified according to their association with snRNAs; U1, U2, U4, U5, and U6 are representative snRNPs. Splicing is controlled by regulatory consensus sequences on pre-mRNA that define exon/intron boundaries10; these sequences include a 5′ splice donor site (5′ss), branch point site (BPS), polypyrimidine tract (PPT), and a 3′ splice acceptor site (3′ss) (Fig. 1a). U1 snRNA binds to the GU dinucleotide of the 5’ss of a pre-mRNA via a base-pair interaction. The splicing factor SF1 recognizes and binds to a BPS. In contrast, U2AF is a heterodimer composed of U2AF2 and U2AF1, which promotes the replacement of SF1 with a U2 snRNP in the BPS after binding to the PPT and an AG dinucleotide sequences in the 3’s, respectively. Then, the tri-snRNP complex containing the U4, U5, and U6 complexes is recruited. The binding of U4/U5/U6 tri-snRNPs to mRNA induces a conformational rearrangement, and U1 and U4 are then released. This sequence change induces the activation of two transesterification reaction steps, triggering 5’ss cleavage and catalyzing 3’ss and exon ligation. Finally, the mRNA is generated with introns cleaved, and the snRNPs are released from the spliceosome complex and recycled for consumption in additional splicing rounds11–13.
Fig. 1. Mechanism of pre-mRNA splicing.
a The spliceosome machinery recognizes and binds the 5’ splice site (5’ss; GU), 3’ splice site (3’ss; AG), branch point site (BPS), and polypyrimidine tract (PPT). U1 snRNP generates base pairs with the 5’ss, and U2 snRNP base pairs with the BPS. Then, tri-snRNPs consisting of U4, U5, and U6 are recruited, and U1 and U4 snRNPs are released. This step induces conformational changes in the pre-mRNA and catalyzes two successive transesterification reactions to generate ligated exon and intron lariats. b Exonic and intronic splicing enhancers (ESEs and ISEs, respectively) are motif sequences that enhance splicing, and exonic and intronic splicing silencers (ESSs and ISSs) are motif sequences that silence splicing. Trans-acting factors bind to these cis-regulatory sequences to regulate alternative splicing.
Additionally, cis-regulatory sequences in exons and introns increase the fidelity of the splicing process (Fig. 1b). Cis-regulatory sequences include exonic and intronic splicing enhancers (ESEs and ISEs, respectively) or exonic and intronic splicing silencers (ESSs and ISSs, respectively)14. Exons and introns are recognized based on the presence and interaction of cis-regulatory sequences and trans-acting factors, such as RNA-binding proteins. Cis-regulatory sequences modulate alternative splicing by either activating or inhibiting the occupation of nearby splice sites by recruiting trans-acting factors to enhancer or silencer sites. Arginine/serine dipeptide-rich (SR) proteins, trans-acting factors, recruit U1 snRNP, or U2AF to a 5’ss and BPS at an RNA recognition motif, forming the nascent spliceosome E complex and promoting bound exon incorporation15–18. In contrast to SR proteins, which are splicing enhancers, nuclear heterogeneous RNPs (hnRNPs) bind to an ISS and inhibit exon inclusion19,20. However, various trans-acting factors that induce exon inclusion and exclusion may exert opposite effects depending on specific site to which it binds21,22. These trans-acting factors block snRNP recruitment or access to consensus sequences and induce structural changes in RNA, thereby affecting splice site selection. They further fine-tune alternative splicing through strong cooperative and competitive effects23–25.
Systematic analysis using next-generation sequencing and computational techniques has thus far revealed seven basic types of alternative splicing. Among alternative splicing events, exon skipping is the most frequent alternative splicing type in higher eukaryotes, and the exon that is skipped is called a cassette exon (Fig. 2a). The next most common alternative splicing type is called an alternative 5’ss or 3’ss event, in which the length of an exon with two or more splice sites is changed (Fig. 2b, c). Introns can be retained in mature mRNA, which is a common alternative splicing outcome in plants and lower metazoans (Fig. 2d)26. A mutually exclusive exon is generated when one exon of two consecutive alternative exons is included when the other exon is excluded (Fig. 2e). Finally, alternatively spliced transcripts are generated by alternative promoters or alternative polyadenylation involving more than one initiator or terminator exon (Fig. 2f, g). Key signal transduction pathway components are subjected to these different alternative splicing regulatory mechanisms. The effects of alternative splicing on each of the major signal transduction pathways are described below.
Fig. 2. Types of alternative splicing.
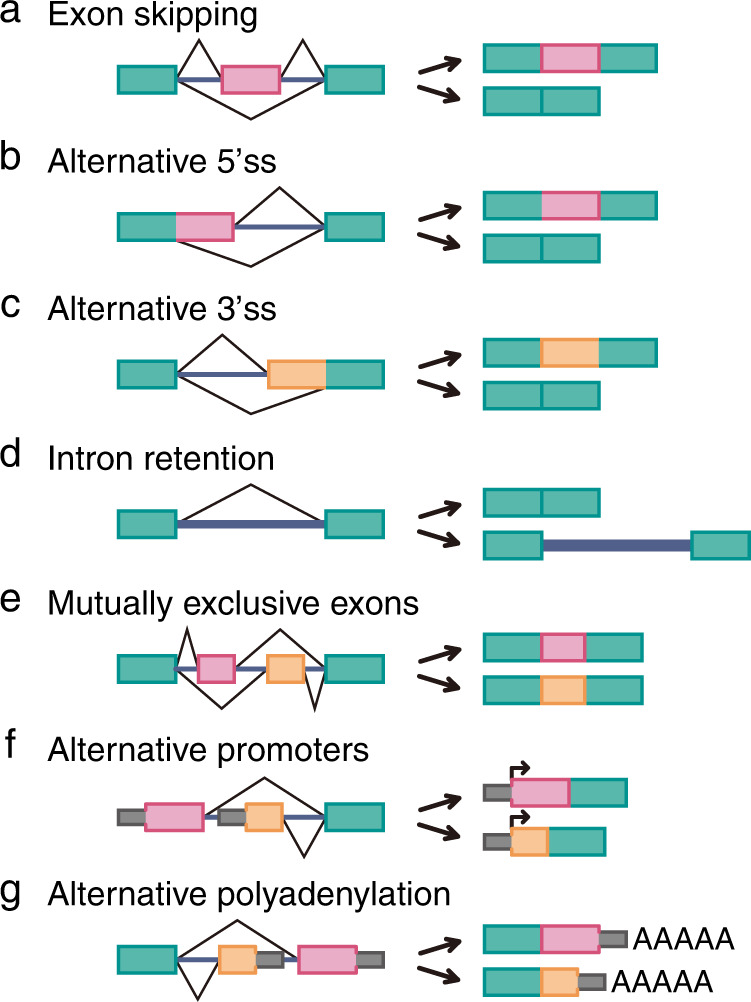
Seven main types of alternative splicing events are depicted. Exons are represented by boxes, and introns are represented by lines. Constitutive exons are green, and alternatively spliced exons are pink and orange. Bent lines indicate an alternative splicing event. Promoters (P) are indicated with arrows and polyadenylation sites with AAAAA. Exon skipping (a), alternative 5’ss (b), alternative 3’ss (c), intron retention (d), mutually exclusive exons (e), alternative promoter (f), and alternative polyadenylation (g) are shown.
Hedgehog pathway
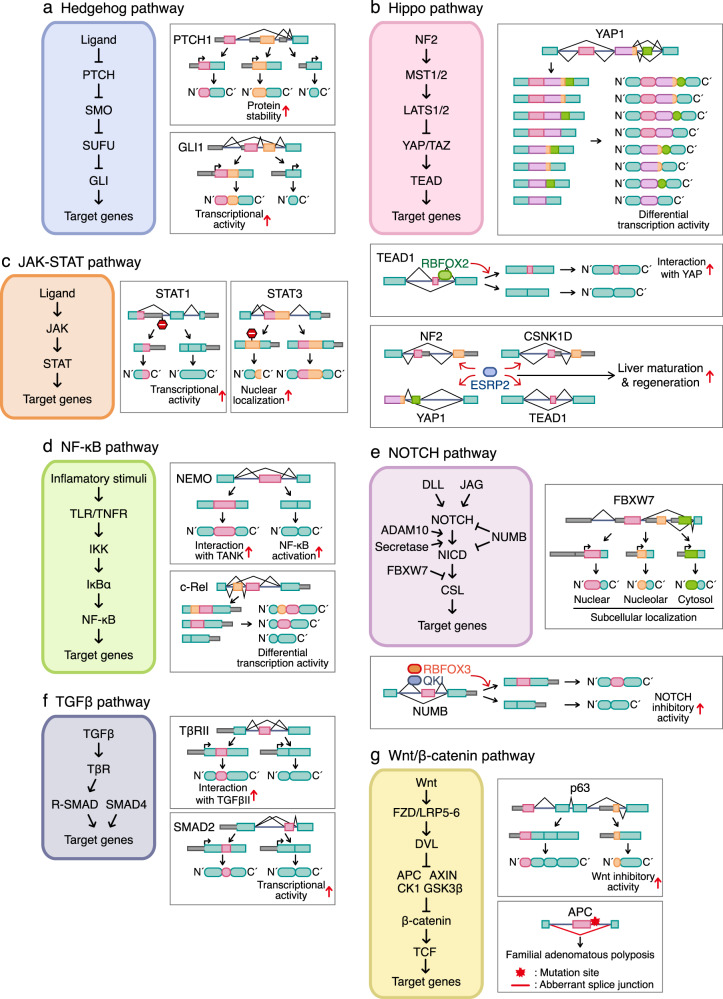
The hedgehog pathway is essential for normal embryonic patterning and development and plays an important role in the maintenance, regeneration, and homeostasis of adult tissues27,28. In the absence of the hedgehog ligand, Ptch (protein patched homolog) is located in primary cilia, sequestering the downstream transducer SMO (smoothened homolog) into intracellular compartments and inhibiting SMO activity27–34. The transcription factor GLI is sequestered in the cytoplasm by SUFU and phosphorylated by protein kinase A, CK1 (casein kinase 1), and GSK3β (glycogen synthase kinase 3β). Phosphorylated GLI is processed by the proteasome into its repressor forms (GLI2R and GLI3R). Therefore, Hh pathway target gene expression is turned off. When the hedgehog ligand is present, Ptch leaves cilia and does not inhibit SMO, allowing it to accumulate in the primary ciliary membrane. SMO is then activated, mitigating the inhibitory effect of the cytoplasmic sequestrated factor SUFU and leading to GLI activation. Activated GLI migrates to the nucleus to activate the transcription of Hedgehog pathway target genes (Fig. 3a).
Fig. 3. Differential activities of splicing isoforms in cell signal transduction.
Several genes that are involved in signal transduction produce alternative splicing-derived proteins with differential activities in regulating signaling pathways. A brief mechanism of intracellular signal transduction and representative alternative splicing events that regulate them are presented. The seven major signal transduction pathways include the Hedgehog pathway (a), Hippo pathway (b), JAK-STAT pathway (c), NF-κB pathway (d), NOTCH pathway (e), TGF-β pathway (f), and Wnt/β-catenin pathway (g).
Because of their alternative promoter, PTCH1 isoforms with different first exons show higher protein stability than wild-type PTCH1 and thus inhibited SMO, GLI1, and GLI2 to a greater extent35–39. Thus, isoforms with different N-terminal tails induce various levels of Hedgehog signaling inhibition. Additionally, tissue-specific alternative splicing and nevoid basal cell carcinoma syndrome (NBCCS)-related abnormal PTCH1 splicing, which includes exons 1-5, exon 10, and the novel exon 12b, were detected using exon junction microarrays40. A PTCH2 variant lacking exons 9 and 10, which are in the conserved sterol-sensing domain, and a PTCH2 variant lacking the final exon, exon 22, have been reported41. Of these two variants, only the isoform in which PTCH2 exon 22 was skipped inhibited hedgehog activity. Both isoforms failed to inhibit the activated form of SMO but they translocated SMO that had been dispersed in the cytoplasm to the juxtanuclear region.
Longer transcripts (α and β in mice and β in humans) and shorter transcripts (γ) are generated according to the number of noncoding exons in the GLI1 5’ UTR35,42. The longest variants were expressed in normal tissues but not in BCC (basal cell carcinoma) or human keratinocyte cell lines, whereas GLI-γ was expressed in BCC and HaCaT cells. Additionally, GLI-γ expression was induced by TPA (12-O-tetradecanoylphorbol 13-acetate) treatment in mouse skin, but the levels of the longer variants were reduced. These observations suggest an association between GLI-γ with actively proliferating keratinocytes and GLI-α and GLI-β expression and quiescent cells, demonstrating a clear correlation with proliferative status. Moreover, an increased 5’-UTR length has been confirmed to reduce the translation efficiency of GLI1 mRNA, and differential inclusion of the first exon has been shown to depend on SNPs (rs10783826, rs10783827, and rs118093490)43. GLI1-γ expression after 5’ noncoding exon skipping may be an important oncogenic contributor. GLI1 variants encoded a truncated GLI1 (GLI1ΔN) protein, which lacked the N-terminal SUFU-binding domain because exons 2 and 3 were skipped44. In adult human tissues, GLI1ΔN mRNA was expressed with wild-type GLI1 at an approximate 1:1 ratio, but a generally lower and variable expression pattern of GLI1ΔN mRNA was observed in tumor cell lines. GLI1ΔN weakly activated transcription; however, in specific cellular contexts, GLI1ΔN exerted a more potent transcription-inducing effect than full-length GLI1. A truncated GLI1 splice variant (tGLI1) in which all of exon 3 and part of exon 4 were skipped, was not expressed in normal cells but was highly expressed in glioblastoma multiforme (GBM) and other cancer cells45. In contrast to wild-type GLI1, tGLI1 specifically targeted CD24, an invasion-associated gene, inducing its transcription and was involved in increased tumor aggressiveness by promoting GBM cell motility and invasiveness.
Various GLI2 variants have been reported, and the isoform in which GLI2 exon 3 is skipped induced reporter gene activation similar to the full-length isoform46,47. In contrast, the isoform in which GLI2 exons 4 and 5 were skipped was expressed at relatively low protein levels but induced 10-fold greater reporter gene activation. The expression levels of GLI2∆N (GLI2 with the N-terminus truncated) and GLI2∆C (with the C-terminus truncated) were increased by TGF-β, and these isoforms were expressed mainly in cells with a highly migratory and invasive phenotype48. Mutations (c.473 + 5 G > A and c.473 + 3 A > T) leading to GLI3 exon 4 skipping and lack the most functionally important domains and have been identified in patients with polydactyly49,50.
Hippo pathway
The Hippo pathway is an evolutionarily conserved signaling pathway that regulates many biological processes, including cell growth, survival, metastasis, and organ size and regeneration51,52. Thus, Hippo pathway dysregulation leads to abnormal cell growth and the development of tumors53. This pathway is turned on and off by various internal and external signals, such as those related to mechanotransduction, cell‒cell contact inhibition, cell adhesion, cell polarity, hormone signaling, and extracellular matrix (ECM) stiffness52,53. In soft-ECM, small-surface, and high-cell-density conditions, the Hippo pathway phosphorylated MST1/2 and activated LATS1/2 to phosphorylate YAP/TAZ. Phosphorylated YAP/TAZ interact with 14-3-3 proteins and cause their cytoplasmic retention54,55. Further YAP/TAZ phosphorylation by casein kinase 1 induced β-TrCP-mediated ubiquitination and proteasomal degradation56–58. In contrast, under conditions such as a stiff ECM, large surface areas, sparse cells, and stretched cells, the Hippo pathway is turned off, and YAP/TAZ was not phosphorylated and was translocated to the nucleus. YAP/TAZ bound to TEAD, a transcription factor, and induced the transcription of genes important for cell proliferation, survival, and migration (Fig. 3b).
The human YAP1 gene comprises nine exons, including exons 4 and 6, which are cassette exons, and exon 5, which carries a 5’ss. Using these exon combinations, researchers identified eight transcripts encoding an alternatively spliced YAP1 isoform59. The YAP1 isoform with exon 5 extended, exon 6, or both was predicted to disrupt the leucine zipper region in terms of protein structure and to affect various protein‒protein interactions. The second YAP WW domain is encoded by exon 4, and TEAD-mediated transcriptional activity is reduced when exon 4 is skipped60. In particular, the leucine zipper disruption by the extended exon 5 led to a greater decrease in transcriptional activity than the absence of the second WW domain.
The SR protein splicing factor SRSF1, a direct target of transcription factor and oncoprotein MYC, promotes TEAD1 exon 5 inclusion61. The TEAD1 full-length isoform showed greater transcriptional activity and oncogenic properties than the TEAD1 isoform without exon 63. Exon 6 is the region coding for the DNA binding domain of TEAD1, but there was no difference in the DNA binding activity of the two isoforms, and rather, it was confirmed that the interaction with YAP was affected. Through a TCGA database analysis, the Hippo target gene expression level did not correlate with the expression level of TEAD1 itself but showed a positive correlation with the percent spliced in (PSI) value of exon 6, which comprises only 12 nucleotides (nt). In addition, patients with high TEAD1 exon 6 levels exhibited a lower survival rate than those not carrying TEAD1 exon 6. TEAD1 exon 6 is harbored by RBFOX2, and it has been confirmed that RBFOX2 affects Hippo target gene expression because of TEAD1 splicing changes.
TEAD4 variants with exon 3 skipped by RBM4 are translated starting at exon 6, yielding a short TEAD4 protein isoform (TEAD4-S)62. TEAD4-S does not carry an N-terminal DNA-binding domain but harbors a C-terminal YAP-binding motif. In contrast to full-length TEAD4 (TEAD4-FL), which predominantly localizes to the nucleus, TEAD4-S is located in both the nucleus and cytoplasm and exerts a dominant-negative effect on YAP activity by antagonizing TEAD4-FL. TEAD4-S expression was decreased in cancer cells, and TEAD4-S inhibited cancer cell proliferation and the epithelial-mesenchymal transition (EMT). Consistent with this, patients with high TEAD4-S scores showed longer survival.
ESRP2 (epithelial splicing regulatory protein 2) downregulation reactivated the neonatal splicing program and induced the exclusion of alternative Nf2, Csnk1d, Yap1, and Tead1 exons, which are involved in the Hippo pathway63. ESRP2 has been shown to modulate hepatocyte proliferation in response to chronic liver injury by attenuating Hippo pathway activation and activating gene expression associated with cell proliferation. Similarly, TNF-α (tumor necrosis factor-α) and IL-1β (interleukin-1β), proinflammatory cytokines that accumulate in alcohol-damaged livers, have been shown to inhibit ESRP2 in adult hepatocytes64. ESRP2 loss increased YAP/TAZ activity by regulating the splicing of CSNK1D and NF2, thereby upregulating Hippo target genes64,65. ESRP2 inhibition increased the viability and regenerative capacity of hepatocytes, increasing their survival. These results revealed the cause of liver failure in patients with SAH (severe alcoholic hepatitis) from the perspective of an ESRP2-dependent RNA-splicing program.
JAK-STAT pathway
More than 50 cytokines and growth factors, such as hormones, interferons (IFNs), and interleukins (ILs), transmit signals downstream through the JAK (Janus kinase)-STAT (signal transducer of activators of transcription) pathway to regulate hematopoiesis, immune responses, pathogen resistance, cell growth and survival, tissue repair, apoptosis, adipogenesis, and cell differentiation66–69. When a cytokine binds to a specific receptor, the receptor multimerizes, bringing JAK into close physical proximity. JAK then mediates transphosphorylation of tyrosine residues to create a STAT-docking site that binds to the cytoplasmic domain of the receptor. STATs are in turn phosphorylated and activated, driving their dimerization. STAT-STAT dimers are translocated to the nucleus, where they bind directly to DNA and upregulate the expression of related genes (Fig. 3c).
JAK2 exon 14 skipping has been found in patients with primary myelofibrosis (PMF) and myeloproliferative neoplasms (MPNs)70,71. A somatic guanine-to-thymine substitution (c.1849G > T) located in the terminal part of JAK2 exon 14 was observed in MPN patients, resulting in changes in amino acids (V617F), altering the pseudokinase domain structure. JAK2-V617F has been observed in most patients with polycythemia vera and approximately 60% of patients with PMF. JAK2 exon 14 skipping was a more frequent event in patients with the JAK2-V617F mutation. The truncated JAK2 isoform generated by JAK2 exon 14 skipping dimerized with wild-type JAK2 to activate the kinase domain and possibly triggered JAK2-STAT pathway activation, suggesting that it may play an important role in MPN pathophysiology70. However, the hypothesis suggesting that the truncated protein isoform of JAK2 may exert an antiproliferative effect has also been proposed71; therefore, further research is needed to determine truncated JAK2 function. Additionally, a JAK3 splice variant with a different C-terminus was isolated from breast (B) spleen (S) tissue and activated monocytes (M)72. JAK3-S and M encode amino acids 1068 (alanine) and 1069 (glutamine) as a result of differential splicing in which GCTGAG is the 5’ss, whereas JAK3-B generates a read-through transcript without accessing the 5’ss. JAK3-S has been detected mainly in hematopoietic cell lines, whereas JAK3-B and M have been detected in cell lines derived from a broad array of hematopoietic and epithelial tissues and showed a wider expression profile. JAK3-B did not show kinase activity because subdomain XI in the tyrosine kinase core was absent. Competition between kinase-active JAK3 and defective JAK3B attenuated downstream responses, suggesting that JAK3B is a dominant-negative isoform that transactivates JAK3 signaling. Additionally, these three JAK3 isoforms recruit different proteins and substrates within a cell, conferring signal transduction complexity. The mutation (c.2652 C > T; pV884V) in JAK3 exon 19, which has been identified in patients with severe combined immunodeficiency (SCID), did not encode an altered the amino acid sequence but created a new 5’ss73. As a result, 29 nt were deleted at the 3’ end of exon 19, resulting in a premature stop codon. A protein encoded by this variant was not detected, indicating that it was either not expressed or was rapidly degraded. Taken together, these studies suggest that JAK3 exons 17-23 encode a kinase domain and that splicing events involving these exons regulate the related C-terminal region and are very important for altering JAK3 function.
In addition to the full-length α-isoforms of STAT1, 3, 4, and 5, evolutionarily conserved β-isoforms carry truncated C-terminal transactivation domains (TADs)74,75. Initially, β-isoforms were considered to be dominant-negative variants, but some studies have reported overlapping or unique functions of α-isoforms76–78. STAT1β was found to be transcriptionally active at lower levels than STAT1α in response to IFN-γ signaling, and STAT1α and STAT1β affected the transcription of distinct sets of genes79,80. In experiments with knock-in mice, NK cells from Stat1β/β mice showed impaired maturation and effector functions, although the effects were less severe than those in NK cells from mice lacking STAT1 (Stat1 −/−)81. This study confirmed that NK cell function was more efficiently affected by STAT1α. Additionally, normal phosphorylation and DNA binding did not occur after STAT1 exon 3 skipping induced by the G372C mutation, suggesting that abnormal signal transduction was induced in response to IFN-γ and IFN-α, which are related to the susceptibility to intracellular pathogens and viruses82. An alternative 3’ss with a difference in STAT3 exon 23 at position 50 nt produced STAT3-α and a frame-shifted prematurely terminated STAT3-β isoform83. STAT3-α regulates cellular responses to IL-6 and mediates IL-10 function in macrophages. In vivo, STAT3-β rescued the embryonic lethality induced by STAT3-null mutations and led to the expression of specific STAT3 target genes. STAT3-β is more efficiently transported to the nucleus, can stay in the nucleus longer than STAT3α, and regulates a greater number of genes84. A splicing variant of the GTAGTT tandem 5’ss in STAT3 exon 21 has been identified85. These isoforms, with or without Ser-701 inclusion, were required for optimal STAT3 function in ABC DLBCL (activated B-cell-like diffuse large B-cell lymphoma) cells86. Stat4α was required for the maximal induction of IL-12-induced IFN-γ production, whereas Stat4β was required for IL-12-stimulated proliferative responses87. Additionally, certain genes were regulated in common, but other target genes were specifically regulated by the STAT4 isoform.
NF-κB pathway
The nuclear factor kappa B (NF-κB) family of transcription factors regulates the expression of various genes that mediate many aspects of innate and adaptive immunity, inflammation, proliferation, cell death, angiogenesis, EMT, and tumor cell invasion and metastasis88–94. In the NF-κB signaling pathway, TNF-α, LPS (lipopolysaccharides), and IL-1 activate TNFRs (TNF receptor), TLRs (Toll-like receptors), and IL-1R (IL-1 receptor), respectively. Integral membrane receptors activate the IKK (IκB kinase) complex. The IKK complex is composed of a heterodimer comprising catalytic IKKα and IKKβ subunits and NEMO (NF-κB essential modulator). Activated IKKβ in the IKK complex phosphorylates serine residues 32 and 36 in human IκBα, leading to the ubiquitination and proteasomal degradation of IκBα. When IκBα is degraded, NF-κB homo or heterodimers are activated by PTM and translocated to the nucleus, where they bind to DNA consensus sequences and activate the transcription of target genes (Fig. 3d).
A variant of TNFRSF1B, encoding TNFR type 2, icp75TNFR carries a 5’-UTR and an alternative first exon95. Wild-type TNFRSF1B was found on the cell surface, but due to the alternative first exon not carrying a signal peptide, the icp75TNFR isoform was diffused throughout the cell and colocalized with some mitochondria. icp75TNFR bound to TNF, which resulted in NF-κB activation in TNF-stimulated cells. A variant in which human TLR1 exon 2 was skipped and five variants generated by two alternative 3’ss and three 5’ss of TLR2 exon 2 produced identical proteins96,97. However, the 5’-UTR difference may have affected mRNA stability and translation.
The isoform caused by exon 5 skipping in NEMO, the noncatalytic subunit of IKK, efficiently mediated IKK and NF-κB activation but did not mediate IKK activation induced by HTLV-1 (T-cell leukemia virus type-1) Tax98. A mutation in the 5’ss of exon 6 (IVS6 + 5 G → A(1027 + 5 G → A)) identified in a family with severe immunodeficiency induced skipping of NEMO exons 4, 5, and 6, resulting in a small protein lacking part of the coiled-coil motif (CC) 1 and the linker between CC1 and CC299. This mutation resulted in low levels of IκBα degradation and impaired NF-κB signaling.
NFKBIZ, the gene encoding IκBζ, generates 15 alternative splicing variants, but only 3 of these mRNAs encode proteins100. A long IκBζ(L) variant consisting of 14 exons was induced by LPS stimulation101. The short IκBζ(S) variant lacks exon 3, where the initiation codon is located, and therefore lacks 99 amino acids in the N-terminus. As a result of alternative splicing at exon 7, the central part of the transactivating domain (TAD), the IκB-ζ(D) variant with the shortened exon 7, was induced upon LPS stimulation, similar to IκBζ(L)102. IκB-ζ(D) showed NF-κB inhibitory activity but no transactivation activity due to the TAD deletion.
The NF-κB family comprises five genes: NF-κB1, NF-κB2, relA, c-rel, and relB. Multiple splicing variants of relA, relB, and NF-κB2 have been identified in the lungs of CD14 knock-out mice after burn injury103. Most variants encode proteins that include frameshifts, are prematurely terminated or lack domains essential for NF-κB transcription factor function. Thus, these splicing changes could serve as one of the mechanisms regulating excess NF-κB activity in response to stress signals, such as burns.
Two NFKB1 SNPs (rs230511 and rs230504), which induced exon 4 and 5 skipping and produced an out-of-frame NFKB1 mRNA, have been identified in the Aymara people from the Andean highlands104. These two SNPs correlated with higher hemoglobin levels and lower white blood cell counts. Since truncated nonfunctional NF-κB inhibits inflammation, the NFKB1 haplotype variant is thought to be the main explanation for the adaptation of the Aymara population to high altitudes. Additionally, numerous NFKB1 mutations in patients with common variable immunodeficiency (CVID) or related diseases are located at the splice site and produce abnormally spliced mRNA variant105. c-Rel with an Alu sequence inserted between exons 8 and 9 generates a 32 amino acid longer variant and a variant with exon 9 skipping106. Both c-Rel isoforms showed enhanced DNA binding and transactivation compared with these functions in wild-type c-Rel.
NOTCH pathway
The Notch pathway is an evolutionarily conserved signal transduction pathway that regulates embryonic development, cell proliferation, differentiation, and death; determines cell fate; and maintains adult tissue homeostasis. Mammals carry Notch family members (Notch-1, -2, -3, -4), which act as receptors, and Delta-like ligands (DLL1, DLL3, and DLL4) and Jagged ligands (JAG1, JAG2), which are ligands in the Notch pathway107. Notch receptors are produced in the endoplasmic reticulum and transported to the plasma membrane. The interaction of a Notch extracellular domain with a cognate ligand result in a cascade of receptor cleavage reactions mediated by ADAM10 and secretase. After cleavage, the Notch intracellular domain is released into the cytoplasm, migrates to the nucleus, and binds to the NOTCH pathway transcription factor CSL to activate the transcription of Notch target genes, such as Myc, p21, HES, and HEY family genes (Fig. 3e)108.
Aberrant splicing variants have been identified in patients with myeloid leukemia, in which exon 12 and exons 17 and 18, which encode parts of the NOTCH2 extracellular EGF-like domains, are skipped109. These NOTCH2 splice variants impair the function of full-length NOTCH2 and act as dominant-negative variants, blocking the signaling pathway in AML cells. Additionally, exon 12 in NOTCH2 has been suggested to be related to apoptosis110. A mutation in which a part of the NOTCH3 intron 3 branch point site was deleted, causing abnormal splicing, has been found in a family with late-onset cerebral autosomal dominant arteriopathy with subcortical infarcts and a leukoencephalopathy (CADASIL) phenotype, suggesting an association between this variant and the pathologically confirmed CADASIL phenotype111. Significantly lower expression of NOTCH3 exon 16 mRNA encoding three EGF-like domains has been observed in patients with the GCB (B-cell-like) subtype of DLBCL (diffuse large B-cell lymphoma), classified as a vincristine-resistant variant112. These patients exhibited low survival rates, demonstrating the potential of NOTCH3 exon 16 transcript levels to be used as prognostic and predictive biomarkers. In addition, the A-to-G transition mutation in the 3’ ss of NOTCH3 exon 4 found in CADASIL patients resulted in an in-frame deletion of 7 amino acids, including the 6th cysteine residue of the second EGF domain113. The unpaired cysteine residue promoted abnormal NOTCH3 oligomerization, possibly playing a key role in the prognosis of patients with CADASIL. Taken together, these studies suggest that the splicing of exons encoding EGF-like domains in extracellular domains can affect receptor-ligand interactions that play important roles in the function of NOTCH.
A JAG2 variant that lacks exon 24 has been identified in breast cancer114. Exon 24 encodes 984-994 amino acids included in the Von Willebrand Factor C domain, but further research is needed to determine the function of this isoform. NOTCH is ubiquitinated and degraded by FBXW7 (F-Box and WD repeat domain-containing 7). FBXW7 encodes three isoforms, α, β, and γ, which differ in the N-terminus as a result of an alternative promoter. FBXW7α is located mainly in the nucleus, β is located in the cytoplasm, and γ is located in the nucleolar region115. The FBXW7α mRNA expression level in cells was the highest, approximately 100-fold that of γ116. Although FBXW7α is predominantly expressed in most human tissues, FBXW7β mRNA is abundantly expressed in the brain and thymus and is absent or minimally expressed in other tissues, and the expression of FBXW7γ mRNA appears to be restricted to the heart and skeletal muscle117. There are seven FBXW7α 5′-UTR variants that produce the same protein but demonstrate differential translational efficiencies. In addition to FBXW7β reducing the stability of the NOTCH intracellular domain, Notch-regulated HES5 has been shown directly repressing FBXW7β transcription, forming a Notch/Hes5/FBXW7β positive feedback loop118. FBXW7β expression was increased by genotoxic stress stimuli, and p53 upregulated FBXW7β. In contrast, the expression of FBXW7α and γ was independent of p53 activity and showed a limited response to most stress stimuli.
NUMB prevents the translocation of the NOTCH intracellular domain to the nucleus. RBFOX3 plays an important role in neuronal differentiation progression during vertebrate development by inhibiting exon 12 inclusion through two UGCAUG elements present in NUMB intron 11119. QKI also inhibits NUMB exon 12 inclusion by recognizing two QKI-binding sequences adjacent to the 3’ splice site of intron 12 and selectively competing with SF1 for binding to the branch point site120. The exclusion of exon 12, which encodes 48 amino acids in the protein-protein interaction domain PRR, inhibits cell proliferation and prevents Notch pathway activation. NOVA1 excludes SORBS2 exon 3, and SORBS2 containing exon 3 inhibits NOVA1 and NOTCH1, promoting colorectal cancer cell migration mediated through the NOTCH pathway121.
TGF-β pathway
The TGF-β (transforming growth factor beta) superfamily includes ligands such as TGF-β, activin/inhibins/Nodal, BMPs (bone morphogenetic proteins), GDFs (growth and differentiation factors), and AMH (anti-Müllerian hormone)122–125. The TGF-β pathway is involved in tissue homeostasis and plays an important role in disease development by regulating various cellular processes, including cell proliferation, differentiation, apoptosis, angiogenesis, immune responses, cell invasion, migration, EMT, and ECM production. TGF-β signaling is initiated by the binding of TGF-β to a specific set of TβRII (serine and threonine kinase receptors, type II) and TβRI (serine and threonine kinase receptors, type I) receptors on the cell membrane. TβRII binds to TGF-β and induces the formation of a heterotetrameric receptor complex that activates TβRI and trans-phosphorylates it. This phosphorylated receptor, in turn, phosphorylates TGF-β receptor-specific SMADs (R-SMAD: SMAD2 and SMAD3) inducing TGF-β and activin signaling and Smad1/5/8 for BMP signaling. Carboxy-terminal phosphorylation of Smads by activated receptors leads to the formation of a heterocomplex with the co-Smad Smad4 and is translocated to the nucleus, where it cooperates with transcription factors and transcriptional coactivators to regulate target gene transcription. SMAD6 and SMAD7 inhibit signaling by antagonizing R-SMAD activation. In addition to activating SMAD proteins, TGF-β signaling can transmit ERK, MAPK, SAPK/JNK, PI3K-AKT, and NF-κB signals. This canonical and noncanonical TGF-β signaling is also affected by other signaling pathways, such as the Hedgehog, Notch, RAS, TNF, interferon, and Wnt pathways (Fig. 3f).
The TGFBR1 mutation c.973 + 1 G > A has been found in patients with LDS (Loeys–Dietz syndrome), and c.806-2 A > C has been found in patients with MSSE (multiple self-healing squamous epitheliomas) 126. In the LDS variant, activation of a cryptic 5’ss located 9 nt upstream of the 5’ splice site in intron 5 was induced, or exon 5 was skipped, generating two functionally inactive proteins. The MSSE variant is a 3’splice site mutation in intron 4 that activates a cryptic 3’ss located 76 bp downstream to generate an out-of-frame transcript. The frequency of a G → A mutation located 24 nt downstream of TGFBR1 intron 7 was significantly increased in RCC (renal cell carcinoma) and TCC (transitional cell carcinoma) patients compared to individual controls without cancer127. Abnormal splicing of the exon encoding the kinase domain leads to functional alterations in the TGF-β signaling pathway and shows the potential to induce various diseases, such as LDS, MSSE, RCC, and TCC.
Insertion of a 75 nt intron between exons 1 and 2 of TGFBR2 produces the membrane-anchor isoform TβRII-B128. TβRII binds only to isoforms TGF-β1 and TGF-β3, whereas TβRII-B binds to TGF-β1, 2, and 3 and forms a complex with TβRI, TβRII, and TβRIII. TβRII-B transinduces TGF-β2 signaling mediated through Smad2, independent of TβRIII, and TβRII-B signaling mediated through the Smad2/3 pathway was induced mainly through TβRI activation129.
The exon 3-skipped isoform of Smad2, which encodes a part of MH1 (Mothers against decapentaplegic (Mad) homology domain-1) that is critical for direct DNA binding, binds to DNA containing the AP-1 (activating protein-1) site, and its transcriptional activity is higher than that of an exon 3 inclusion isoform130,131. Additionally, the level of the alternatively spliced variant of Smad2 lacking exon 3 was increased during early postnatal brain maturation in mice and was expressed at higher levels than the exon 3-included variant at all stages, suggesting that it has a special function during brain differentiation132. Smad3 exon 3 skipping generates an isoform with a partially truncated linker region that shows TGF-β-dependent transcriptional activity133. Smad8B is generated by the deletion of 47 amino acids, including the SSXS motif that is phosphorylated by the BMP type I receptor ALK2 in the C-terminal MH2 region of Smad8134. This isoform is located in the cytoplasm and is a dominant BMP signaling inhibitor.
Wnt/β-catenin pathway
The Wnt/β-catenin pathway modulates embryonic development and adult tissue homeostasis by influencing physiological activities, such as cell proliferation, division, differentiation, migration, polarity, and apoptosis135–139. In the absence of the Wnt ligand, β-catenin is phosphorylated by the destruction complex, which is composed of Axin, APC (adenomatous polyposis coli), CK1α (casein kinase 1α), and GSK3β. Then, β-catenin phosphorylation leads to subsequent ubiquitin-proteasome degradation. In the Wnt-activation state, the Wnt ligand binds to a coreceptor composed of Frizzled and LRP5/6 (low-density lipoprotein receptor-related protein 5/6). The receptor LRP5/6 is phosphorylated by CK1 and GSK3β, and DVL and AXIN are then recruited, leading to GSK3β inhibition. This process induces the disassembly of the destruction complex, and β-catenin accumulates in the cytoplasm, is translocated to the nucleus, and then interacts with TCF/LEF transcription factors to induce target gene transcription (Fig. 3g).
Multiple APC splice site mutations have been identified in FAP (familial adenomatous polyposis) patients140. Exon 10 is skipped because of a distinct heterozygous 16 bp deletion (APC: c.1312 + 4_1312 + 19del) at the 5’ss of intron 10, and exon 9 is skipped because of a de novo indel (c.1226-1229delTTTTinsAAA) at the 5’ end of exon 9141,142. Exon skipping induces a frameshift, leading to the formation of premature stop codons. The resulting truncated protein lacks all functional domains and possesses only the homodimerization domain of wild-type APC, preventing the interaction of the mutant with other proteins such as β-catenin or axin. Therefore, β-catenin levels are increased, and the expression of growth-promoting genes can be increased through downstream T-cell transcription factor (Tcf) pathways. Mutations inducing exons 12 and 13 skipping resulted in the loss of armadillo functional domains without disrupting the open reading frame, and patients carrying this variant showed a less severe FAP phenotype than patients with truncation mutations143.
The LRP5 splice site mutation (NM_002335.4: c.686 + 1 G > T) identified in patients with familial exudative vitreoretinopathy (FEVR) causes LRP5 gene downregulation, resulting in complete retinal detachment and bilateral blindness and decreased BMD (bone mineral density)144. Several mutations that interfere with normal LRP5 splicing have been reported in OPPG (osteoporosis–pseudoglioma syndrome) patients, who also present with low BMD145. Gain-of-function mutations in the LRP5 gene leads to the acquisition of the high bone mass (HBM) phenotype in humans, whereas homozygous loss-of-function mutations can reduce the signaling activity of the Wnt signaling pathway, thereby reducing bone formation146–148. Mutations causing aberrant LRP5 splicing have also been shown to alter subcellular localization or disrupt intracellular transport compared to that of wild-type LRP5 and severely impair the Wnt pathway145.
DKK3 contains an alternative promoter that selectively carries exon 1a or 1b, which produces the same protein149,150. The exon 1a promoter showed stronger activity than the exon 1b promoter, and a 250 bp region further upstream of the promoter in exon 1a contained many CpG sequences. DKK3 expression downregulation has been associated with promoter methylation.
GSK3B shows an increased frequency with which exons 9 and 11 are skipped in the rs6438552 allele, which has been identified in PD (Parkinson’s disease) patients, and shows higher activity151. Additionally, five GSK3B splicing variants were identified in pig tissues, which showed differential expression patterns in fetal and adult tissues and were differentially regulated by insulin152. According to the GSK3β isoforms in pigs, the degree of influence on the mRNA expression of GY1 and GY2, encoding glycogen synthase, was different, and their roles in regulating phosphorylation and glycogen synthase activity were different. These findings suggest that GSK3β isoforms may play different roles in the insulin signaling pathway.
The homolog of the p53, p63, generates TAp63 and ΔNp63 isoforms due to an alternative promoter153. ΔNp63 lacking the N-terminal transactivation domain inhibits β-catenin phosphorylation and degradation mediated by GSK3β, resulting in β-catenin accumulation in the nucleus and increased transcriptional activity. Additionally, the TAp63 isoform exerts no effect, but the ΔNp63 isoform promotes normal mammary stem cell activation by enhancing Wnt signaling through increased Wnt receptor Fzd7 expression154. These findings suggest that ΔNp63 functions as a positive Wnt/β-catenin signaling regulator.
Summary and conclusions
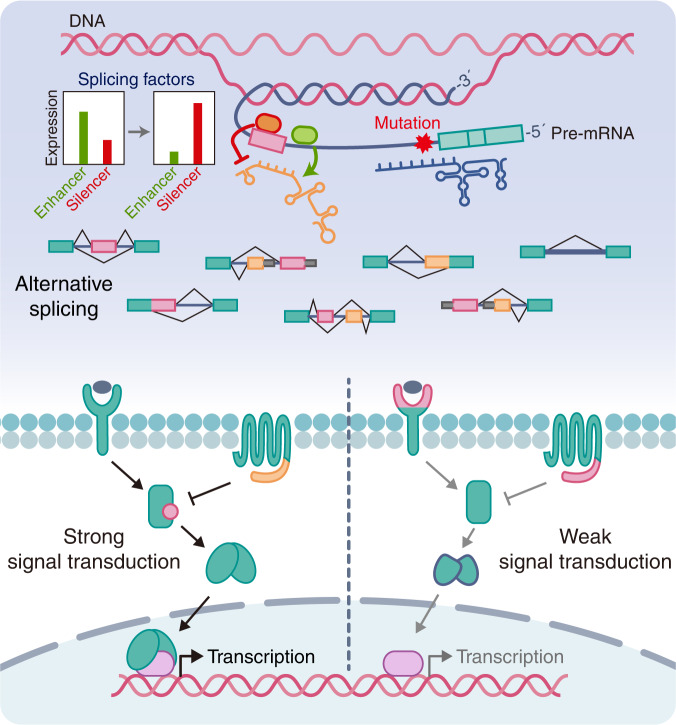
Taken together, the studies presented herein show that alternative splicing is a very important mechanism that can enhance or attenuate signal transduction and has been shown to stably and precisely regulate various signal transduction pathways in living organisms (Fig. 4). PTMs, such as phosphorylation, are essential in regulating signal transduction, but because PTMs can also be affected by alternative splicing, the importance of alternative splicing needs to be further emphasized. Signal transduction can exert a synergistic effect while reinforcing signaling mediated through crosstalk among pathways. Examples include the effect of STAT3 activation on the Ras and PI3K/Akt pathways and the connections of JAK2 with the PI3K and ERK pathways155–160. Thus, the effects of splice variants may be broader than those identified in a single pathway.
Fig. 4. Roles of alternative splicing in cell signal transduction.
Splicing factors systemically activate and regulate the alternative splicing of target gene pre-mRNA. A subset of genetic mutations induces aberrant alternative splicing. Alternative splicing produces multiple mRNAs, which increases proteome diversity. Splicing isoforms with differential functions induce the expression of specific gene sets by fine-tuning various signal transduction pathways. Therefore, dysregulation of alternative splicing leads to numerous human diseases.
With the advancement in sequencing technology, entire genome DNA sequencing and exome sequencing are being performed with patient samples. However, most of these studies are focused on mutation analysis that alters the amino acid sequence in exons. Although these mutations may create new splice sites or affect splicing regulatory elements, which may affect splicing, these aspects are overlooked. The mutation (c.2652 C > T; pV884V) located in JAK3 exon 19 does not change the amino acid sequence but activates a new 5’ss in exon 19, resulting in premature termination and no detectable protein expression73. Additionally, despite the functional importance of splicing isoforms, transcriptome analysis is largely focused on changes in total mRNA expression levels, and analysis based on splicing variants is rarely performed. However, in-depth studies on alternative splicing have reported that aberrant splicing events influence the onset and progression of various diseases. Short-read RNA sequencing, currently the most commonly used method of studying alternative splicing, is difficult to use to fully assemble or characterize complex isoforms due to the limited read length that can be accommodated161–163. Long-read RNA sequencing or third-generation sequencing, such as that enabled via an Iso-Seq platform developed by Pacific Bioscience and a direct RNA-seq platform developed by Oxford Nanopore Technology, shows great potential to overcome the issues of short-read RNA sequencing164. Compared to short-read RNA sequencing, long-read RNA sequencing increases mapping certainty, de novo assembly, isoform identification, and differential alternative splicing event detection. However, long-read transcriptomics is associated with high per-read costs, high error rates, and potential sequencing coverage bias; therefore, work is still needed to improve the accuracy of isoform expression quantification161,165. In addition to sequencing analysis, to distinguish between driver splicing, which is directly related to disease, and passenger splicing, which is not directly related to disease, the basic mechanism of alternative splicing and its functional role of resulting splicing isoforms should be further evaluated. These types of systematic studies will expand our understanding of pathogenesis associated with alternative splicing event and will help in developing targeted therapeutics.
Author contributions
K.K.K. conceptualized the manuscript; S.C., N.C., and K.K.K. drafted the manuscript; S.C. and N.C. prepared the figures; K.K.K., S.C., and N.C. helped prepare the discussion and edited the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF-2022R1A2C1003870 and NRF-2021R1I1A1A01051949).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sunkyung Choi, Namjoon Cho.
References
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Antebi YE, Nandagopal N, Elowitz MB. An operational view of intercellular signaling pathways. Curr. Opin. Syst. Biol. 2017;1:16–24. doi: 10.1016/j.coisb.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi S, et al. RBFOX2-regulated TEAD1 alternative splicing plays a pivotal role in Hippo-YAP signaling. Nucleic Acids Res. 2022;50:8658–8673. doi: 10.1093/nar/gkac509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan Y, et al. Characterization of Sin1 Isoforms Reveals an mTOR-Dependent and Independent Function of Sin1gamma. PLoS One. 2015;10:e0135017. doi: 10.1371/journal.pone.0135017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aitken A. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin. Cell Dev. Biol. 2011;22:673–680. doi: 10.1016/j.semcdb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Chan IH, Privalsky ML. Isoform-specific transcriptional activity of overlapping target genes that respond to thyroid hormone receptors alpha1 and beta1. Mol. Endocrinol. 2009;23:1758–1775. doi: 10.1210/me.2009-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon H, et al. Effects of PTCs on nonsense-mediated mRNA decay are dependent on PTC location. Oncol. Lett. 2017;13:1944–1948. doi: 10.3892/ol.2017.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 11.Bonnal SC, Lopez-Oreja I, Valcarcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat. Rev. Clin. Oncol. 2020;17:457–474. doi: 10.1038/s41571-020-0350-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: A systematic review. Signal Transduct. Target. Ther. 2021;6:78. doi: 10.1038/s41392-021-00486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 14.Shenasa H, Movassat M, Forouzmand E, Hertel KJ. Allosteric regulation of U1 snRNP by splicing regulatory proteins controls spliceosomal assembly. RNA. 2020;26:1389–1399. doi: 10.1261/rna.075135.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 16.Long JC, Caceres JF. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, Rio DC. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caceres JF, Kornblihtt AR. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/S0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, et al. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res. 2014;42:4019–4030. doi: 10.1093/nar/gkt1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-T. [DOI] [PubMed] [Google Scholar]
- 25.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 28.Gupta S, Takebe N, Lorusso P. Targeting the Hedgehog pathway in cancer. Ther. Adv. Med. Oncol. 2010;2:237–250. doi: 10.1177/1758834010366430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carballo GB, Honorato JR, de Lopes GPF, Spohr T. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018;16:11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skoda AM, et al. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci. 2018;18:8–20. doi: 10.17305/bjbms.2018.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeng KS, et al. Sonic Hedgehog signaling pathway as a potential target to inhibit the progression of hepatocellular carcinoma. Oncol. Lett. 2019;18:4377–4384. doi: 10.3892/ol.2019.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotulak-Chrzaszcz A, Kmiec Z, Wierzbicki PM. Sonic Hedgehog signaling pathway in gynecological and genitourinary cancer (Review) Int. J. Mol. Med. 2021;47:106. doi: 10.3892/ijmm.2021.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C, Qi Y, Sun Z. The role of sonic hedgehog pathway in the development of the central nervous system and aging-related neurodegenerative diseases. Front. Mol. Biosci. 2021;8:711710. doi: 10.3389/fmolb.2021.711710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu F, Zhang Y, Sun B, McMahon AP, Wang Y. Hedgehog signaling: From basic biology to cancer therapy. Cell Chem. Biol. 2017;24:252–280. doi: 10.1016/j.chembiol.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palaniswamy R, Teglund S, Lauth M, Zaphiropoulos PG, Shimokawa T. Genetic variations regulate alternative splicing in the 5’ untranslated regions of the mouse glioma-associated oncogene 1, Gli1. BMC Mol. Biol. 2010;11:32. doi: 10.1186/1471-2199-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimokawa T, Rahnama F, Zaphiropoulos PG. A novel first exon of the Patched1 gene is upregulated by Hedgehog signaling resulting in a protein with pathway inhibitory functions. FEBS Lett. 2004;578:157–162. doi: 10.1016/j.febslet.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Agren M, Kogerman P, Kleman MI, Wessling M, Toftgard R. Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene. 2004;330:101–114. doi: 10.1016/j.gene.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Kogerman P, et al. Alternative first exons of PTCH1 are differentially regulated in vivo and may confer different functions to the PTCH1 protein. Oncogene. 2002;21:6007–6016. doi: 10.1038/sj.onc.1205865. [DOI] [PubMed] [Google Scholar]
- 39.Shimokawa T, et al. Distinct roles of first exon variants of the tumor-suppressor Patched1 in Hedgehog signaling. Oncogene. 2007;26:4889–4896. doi: 10.1038/sj.onc.1210301. [DOI] [PubMed] [Google Scholar]
- 40.Nagao K, et al. Detecting tissue-specific alternative splicing and disease-associated aberrant splicing of the PTCH gene with exon junction microarrays. Hum. Mol. Genet. 2005;14:3379–3388. doi: 10.1093/hmg/ddi369. [DOI] [PubMed] [Google Scholar]
- 41.Rahnama F, Toftgard R, Zaphiropoulos PG. Distinct roles of PTCH2 splice variants in Hedgehog signalling. Biochem. J. 2004;378:325–334. doi: 10.1042/bj20031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang XQ, Rothnagel JA. Post-transcriptional regulation of the gli1 oncogene by the expression of alternative 5’ untranslated regions. J. Biol. Chem. 2001;276:1311–1316. doi: 10.1074/jbc.M005191200. [DOI] [PubMed] [Google Scholar]
- 43.Zaphiropoulos PG. Genetic variations and alternative splicing: The Glioma associated oncogene 1, GLI1. Front. Genet. 2012;3:119. doi: 10.3389/fgene.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimokawa T, et al. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J. Biol. Chem. 2008;283:14345–14354. doi: 10.1074/jbc.M800299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo HW, Zhu H, Cao X, Aldrich A, Ali-Osman F. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009;69:6790–6798. doi: 10.1158/0008-5472.CAN-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Speek M, Njunkova O, Pata I, Valdre E, Kogerman P. A potential role of alternative splicing in the regulation of the transcriptional activity of human GLI2 in gonadal tissues. BMC Mol. Biol. 2006;7:13. doi: 10.1186/1471-2199-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanimura A, Dan S, Yoshida M. Cloning of novel isoforms of the human Gli2 oncogene and their activities to enhance tax-dependent transcription of the human T-cell leukemia virus type 1 genome. J. Virol. 1998;72:3958–3964. doi: 10.1128/JVI.72.5.3958-3964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadam H, et al. GLI2 cell-specific activity is controlled at the level of transcription and RNA processing: Consequences to cancer metastasis. Biochim. Biophys. Acta. 2016;1862:46–55. doi: 10.1016/j.bbadis.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Siavriene E, et al. Novel GLI3 variant causes Greig cephalopolysyndactyly syndrome in three generations of a Lithuanian family. Mol. Genet. Genomic Med. 2019;7:e878. doi: 10.1002/mgg3.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bournazos AM, et al. Standardized practices for RNA diagnostics using clinically accessible specimens reclassifies 75% of putative splicing variants. Genet. Med. 2022;24:130–145. doi: 10.1016/j.gim.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 52.Ma S, Meng Z, Chen R, Guan KL. The hippo pathway: Biology and pathophysiology. Annu. Rev. Biochem. 2019;88:577–604. doi: 10.1146/annurev-biochem-013118-111829. [DOI] [PubMed] [Google Scholar]
- 53.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 55.Misra JR, Irvine KD. The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet. 2018;52:65–87. doi: 10.1146/annurev-genet-120417-031621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu CY, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J. Biol. Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaffney CJ, et al. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene. 2012;509:215–222. doi: 10.1016/j.gene.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finch-Edmondson ML, Strauss RP, Clayton JS, Yeoh GC, Callus BA. Splice variant insertions in the C-terminus impairs YAP’s transactivation domain. Biochem. Biophys. Rep. 2016;6:24–31. doi: 10.1016/j.bbrep.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das S, Anczukow O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi Y, et al. A splicing isoform of TEAD4 attenuates the Hippo-YAP signalling to inhibit tumour proliferation. Nat. Commun. 2016;7:ncomms11840. doi: 10.1038/ncomms11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bangru S, et al. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat. Struct. Mol. Biol. 2018;25:928–939. doi: 10.1038/s41594-018-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hyun J, et al. Epithelial splicing regulatory protein 2-mediated alternative splicing reprograms hepatocytes in severe alcoholic hepatitis. J. Clin. Invest. 2020;130:2129–2145. doi: 10.1172/JCI132691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hyun J, et al. Dysregulation of the ESRP2-NF2-YAP/TAZ axis promotes hepatobiliary carcinogenesis in non-alcoholic fatty liver disease. J. Hepatol. 2021;75:623–633. doi: 10.1016/j.jhep.2021.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qureshy Z, Johnson DE, Grandis JR. Targeting the JAK/STAT pathway in solid tumors. J. Cancer Metastasis Treat. 2020;6:27. [PMC free article] [PubMed] [Google Scholar]
- 67.O’Shea JJ, et al. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris R, Kershaw NJ, Babon JJ. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018;27:1984–2009. doi: 10.1002/pro.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct. Target. Ther. 2021;6:402. doi: 10.1038/s41392-021-00791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma W, et al. JAK2 exon 14 deletion in patients with chronic myeloproliferative neoplasms. PLoS One. 2010;5:e12165. doi: 10.1371/journal.pone.0012165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Catarsi P, et al. JAK2 exon 14 skipping in patients with primary myelofibrosis: A minor splice variant modulated by the JAK2-V617F allele burden. PLoS One. 2015;10:e0116636. doi: 10.1371/journal.pone.0116636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lai KS, et al. A kinase-deficient splice variant of the human JAK3 is expressed in hematopoietic and epithelial cancer cells. J. Biol. Chem. 1995;270:25028–25036. doi: 10.1074/jbc.270.42.25028. [DOI] [PubMed] [Google Scholar]
- 73.Platt CD, et al. Janus kinase 3 deficiency caused by a homozygous synonymous exonic mutation that creates a dominant splice site. J. Allergy Clin. Immunol. 2017;140:268–271 e6. doi: 10.1016/j.jaci.2016.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu K, et al. Isolation and expression analysis of STAT members from synechogobius hasta and their roles in leptin affecting lipid metabolism. Int. J. Mol. Sci. 2016;17:406. doi: 10.3390/ijms17030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song H, Yan YL, Titus T, He X, Postlethwait JH. The role of stat1b in zebrafish hematopoiesis. Mech. Dev. 2011;128:442–456. doi: 10.1016/j.mod.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang D, et al. A small amphipathic alpha-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 2000;19:392–399. doi: 10.1093/emboj/19.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caldenhoven E, et al. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 78.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 79.Parrini M, et al. The C-Terminal transactivation domain of STAT1 has a gene-specific role in transactivation and cofactor recruitment. Front. Immunol. 2018;9:2879. doi: 10.3389/fimmu.2018.02879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Semper C, et al. STAT1beta is not dominant negative and is capable of contributing to gamma interferon-dependent innate immunity. Mol. Cell. Biol. 2014;34:2235–2248. doi: 10.1128/MCB.00295-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meissl K, et al. STAT1 Isoforms Differentially Regulate NK Cell Maturation and Anti-tumor Activity. Front. Immunol. 2020;11:2189. doi: 10.3389/fimmu.2020.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vairo D, et al. Severe impairment of IFN-gamma and IFN-alpha responses in cells of a patient with a novel STAT1 splicing mutation. Blood. 2011;118:1806–1817. doi: 10.1182/blood-2011-01-330571. [DOI] [PubMed] [Google Scholar]
- 83.Maritano D, et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat. Immunol. 2004;5:401–409. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]
- 84.Ng IH, Bogoyevitch MA, Jans DA. Cytokine-induced slowing of STAT3 nuclear import; faster basal trafficking of the STAT3beta isoform. Traffic. 2014;15:946–960. doi: 10.1111/tra.12181. [DOI] [PubMed] [Google Scholar]
- 85.Turton KB, Annis DS, Rui L, Esnault S, Mosher DF. Ratios of Four STAT3 Splice Variants in Human Eosinophils and Diffuse Large B Cell Lymphoma Cells. PLoS One. 2015;10:e0127243. doi: 10.1371/journal.pone.0127243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng M, et al. A mix of S and DeltaS variants of STAT3 enable survival of activated B-cell-like diffuse large B-cell lymphoma cells in culture. Oncogenesis. 2016;4:e184. doi: 10.1038/oncsis.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoey T, et al. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. 2003;22:4237–4248. doi: 10.1093/emboj/cdg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barnabei L, Laplantine E, Mbongo W, Rieux-Laucat F, Weil R. NF-kappaB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 2021;12:716469. doi: 10.3389/fimmu.2021.716469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia L, et al. Role of the NFkappaB-signaling pathway in cancer. Onco. Targets Ther. 2018;11:2063–2073. doi: 10.2147/OTT.S161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 94.Albensi BC. What Is Nuclear Factor Kappa B (NF-kappaB) Doing in and to the Mitochondrion? Front. Cell Dev. Biol. 2019;7:154. doi: 10.3389/fcell.2019.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seitz C, Muller P, Krieg RC, Mannel DN, Hehlgans T. A novel p75TNF receptor isoform mediating NFkappa B activation. J. Biol. Chem. 2001;276:19390–19395. doi: 10.1074/jbc.M101336200. [DOI] [PubMed] [Google Scholar]
- 96.Haehnel V, Schwarzfischer L, Fenton MJ, Rehli M. Transcriptional regulation of the human toll-like receptor 2 gene in monocytes and macrophages. J. Immunol. 2002;168:5629–5637. doi: 10.4049/jimmunol.168.11.5629. [DOI] [PubMed] [Google Scholar]
- 97.Chang JS, et al. Myobacterium tuberculosis induces selective up-regulation of TLRs in the mononuclear leukocytes of patients with active pulmonary tuberculosis. J. Immunol. 2006;176:3010–3018. doi: 10.4049/jimmunol.176.5.3010. [DOI] [PubMed] [Google Scholar]
- 98.Hai T, et al. An alternative splice product of IkappaB kinase (IKKgamma), IKKgamma-delta, differentially mediates cytokine and human T-cell leukemia virus type 1 tax-induced NF-kappaB activation. J. Virol. 2006;80:4227–4241. doi: 10.1128/JVI.80.9.4227-4241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Orstavik KH, et al. Novel splicing mutation in the NEMO (IKK-gamma) gene with severe immunodeficiency and heterogeneity of X-chromosome inactivation. Am. J. Med. Genet. A. 2006;140:31–39. doi: 10.1002/ajmg.a.31026. [DOI] [PubMed] [Google Scholar]
- 100.Willems M, Dubois N, Musumeci L, Bours V, Robe PA. IkappaBzeta: An emerging player in cancer. Oncotarget. 2016;7:66310–66322. doi: 10.18632/oncotarget.11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamazaki S, Muta T, Matsuo S, Takeshige K. Stimulus-specific induction of a novel nuclear factor-kappaB regulator, IkappaB-zeta, via Toll/Interleukin-1 receptor is mediated by mRNA stabilization. J. Biol. Chem. 2005;280:1678–1687. doi: 10.1074/jbc.M409983200. [DOI] [PubMed] [Google Scholar]
- 102.Motoyama M, Yamazaki S, Eto-Kimura A, Takeshige K, Muta T. Positive and negative regulation of nuclear factor-kappaB-mediated transcription by IkappaB-zeta, an inducible nuclear protein. J. Biol. Chem. 2005;280:7444–7451. doi: 10.1074/jbc.M412738200. [DOI] [PubMed] [Google Scholar]
- 103.Phan HH, Cho K, Sainz-Lyon KS, Shin S, Greenhalgh DG. CD14-dependent modulation of NF-kappaB alternative splicing in the lung after burn injury. Gene. 2006;371:121–129. doi: 10.1016/j.gene.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 104.Song J, et al. Novel Form of Alternative Splicing of NFKB1. Its Role in Polycythemia and Adaptation to High Altitude in Andean Aymara. Blood. 2018;132:2316. doi: 10.1182/blood-2018-99-117463. [DOI] [Google Scholar]
- 105.Li J, et al. Biochemically deleterious human NFKB1 variants underlie an autosomal dominant form of common variable immunodeficiency. J. Exp. Med. 2021;218:e20210566. doi: 10.1084/jem.20210566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leeman JR, Weniger MA, Barth TF, Gilmore TD. Deletion analysis and alternative splicing define a transactivation inhibitory domain in human oncoprotein REL. Oncogene. 2008;27:6770–6781. doi: 10.1038/onc.2008.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou B, et al. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022;7:95. doi: 10.1038/s41392-022-00934-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen W, Huang J, Wang Y. Biological Significance of NOTCH Signaling Strength. Front. Cell Dev. Biol. 2021;9:652273. doi: 10.3389/fcell.2021.652273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Adamia S, et al. NOTCH2 and FLT3 gene mis-splicings are common events in patients with acute myeloid leukemia (AML): new potential targets in AML. Blood. 2014;123:2816–2825. doi: 10.1182/blood-2013-02-481507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hwang IS, Shin S, Min YH, Lee ST, Choi JR. NOTCH2 missplicing can occur in relation to apoptosis. Blood. 2015;126:1731–1732. doi: 10.1182/blood-2015-07-657825. [DOI] [PubMed] [Google Scholar]
- 111.Bianchi S, et al. First deep intronic mutation in the NOTCH3 gene in a family with late-onset CADASIL. Neurobiol. Aging. 2013;34:2234 e9–12. doi: 10.1016/j.neurobiolaging.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 112.Jespersen DS, et al. Expression of NOTCH3 exon 16 differentiates Diffuse Large B-cell Lymphoma into molecular subtypes and is associated with prognosis. Sci. Rep. 2019;9:335. doi: 10.1038/s41598-018-36680-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joutel A, et al. Splice site mutation causing a seven amino acid Notch3 in-frame deletion in CADASIL. Neurology. 2000;54:1874–1875. doi: 10.1212/WNL.54.9.1874. [DOI] [PubMed] [Google Scholar]
- 114.Aversa R, et al. Alternative splicing in adhesion- and motility-related genes in breast cancer. Int. J. Mol. Sci. 2016;17:121. doi: 10.3390/ijms17010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grim JE, et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J. Cell. Biol. 2008;181:913–920. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, et al. Multiple novel alternative splicing forms of FBXW7alpha have a translational modulatory function and show specific alteration in human cancer. PLoS One. 2012;7:e49453. doi: 10.1371/journal.pone.0049453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sionov RV, Netzer E, Shaulian E. Differential regulation of FBXW7 isoforms by various stress stimuli. Cell Cycle. 2013;12:3547–3554. doi: 10.4161/cc.26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim KK, Nam J, Mukouyama YS, Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J. Cell. Biol. 2013;200:443–458. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zong FY, et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang T, et al. NOVA1-Mediated SORBS2 Isoform Promotes Colorectal Cancer Migration by Activating the Notch Pathway. Front. Cell Dev. Biol. 2021;9:673873. doi: 10.3389/fcell.2021.673873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang F, Chen YG. Regulation of TGF-beta receptor activity. Cell Biosci. 2012;2:9. doi: 10.1186/2045-3701-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu S, Chen S, Zeng J. TGFbeta signaling: A complex role in tumorigenesis (Review) Mol. Med. Rep. 2018;17:699–704. doi: 10.3892/mmr.2017.7970. [DOI] [PubMed] [Google Scholar]
- 126.Fujiwara T, et al. Distinct variants affecting differential splicing of TGFBR1 exon 5 cause either Loeys-Dietz syndrome or multiple self-healing squamous epithelioma. Eur. J. Hum. Genet. 2018;26:1151–1158. doi: 10.1038/s41431-018-0127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen T, et al. An intronic variant of the TGFBR1 gene is associated with carcinomas of the kidney and bladder. Int. J. Cancer. 2004;112:420–425. doi: 10.1002/ijc.20419. [DOI] [PubMed] [Google Scholar]
- 128.Rotzer D, et al. Type III TGF-beta receptor-independent signalling of TGF-beta2 via TbetaRII-B, an alternatively spliced TGF-beta type II receptor. EMBO J. 2001;20:480–490. doi: 10.1093/emboj/20.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.del Re E, Babitt JL, Pirani A, Schneyer AL, Lin HY. In the absence of type III receptor, the transforming growth factor (TGF)-beta type II-B receptor requires the type I receptor to bind TGF-beta2. J. Biol. Chem. 2004;279:22765–22772. doi: 10.1074/jbc.M401350200. [DOI] [PubMed] [Google Scholar]
- 130.Yagi K, et al. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- 131.Tao S, Sampath K. Alternative splicing of SMADs in differentiation and tissue homeostasis. Dev. Growth Differ. 2010;52:335–342. doi: 10.1111/j.1440-169X.2009.01163.x. [DOI] [PubMed] [Google Scholar]
- 132.Ueberham U, et al. Smad2 isoforms are differentially expressed during mouse brain development and aging. Int. J. Dev. Neurosci. 2009;27:501–510. doi: 10.1016/j.ijdevneu.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 133.Kjellman C, et al. Identification and characterization of a human smad3 splicing variant lacking part of the linker region. Gene. 2004;327:141–152. doi: 10.1016/j.gene.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 134.Nishita M, Ueno N, Shibuya H. Smad8B, a Smad8 splice variant lacking the SSXS site that inhibits Smad8-mediated signalling. Genes Cells. 1999;4:583–591. doi: 10.1046/j.1365-2443.1999.00285.x. [DOI] [PubMed] [Google Scholar]
- 135.Liu J, et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022;7:3. doi: 10.1038/s41392-021-00762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Azbazdar Y, Karabicici M, Erdal E, Ozhan G. Regulation of Wnt Signaling Pathways at the Plasma Membrane and Their Misregulation in Cancer. Front. Cell Dev. Biol. 2021;9:631623. doi: 10.3389/fcell.2021.631623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patel S, Alam A, Pant R, Chattopadhyay S. Wnt Signaling and Its Significance Within the Tumor Microenvironment: Novel Therapeutic Insights. Front. Immunol. 2019;10:2872. doi: 10.3389/fimmu.2019.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Y, Wang X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J. Hematol. Oncol. 2020;13:165. doi: 10.1186/s13045-020-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rim EY, Clevers H, Nusse R. The Wnt Pathway: From Signaling Mechanisms to Synthetic Modulators. Annu. Rev. Biochem. 2022;91:571–598. doi: 10.1146/annurev-biochem-040320-103615. [DOI] [PubMed] [Google Scholar]
- 140.Charames GS, et al. A novel aberrant splice site mutation in the APC gene. J. Med. Genet. 2002;39:754–757. doi: 10.1136/jmg.39.10.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cheah PY, et al. A novel indel in exon 9 of APC upregulates a ‘skip exon 9’ isoform and causes very severe familial adenomatous polyposis. Eur. J. Hum. Genet. 2014;22:833–836. doi: 10.1038/ejhg.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wanitsuwan W, Vijasika S, Jirarattanasopa P, Horpaopan S. A distinct APC pathogenic germline variant identified in a southern Thai family with familial adenomatous polyposis. BMC Med. Genomics. 2021;14:87. doi: 10.1186/s12920-021-00933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Disciglio V, et al. APC Splicing Mutations Leading to In-Frame Exon 12 or Exon 13 Skipping Are Rare Events in FAP Pathogenesis and Define the Clinical Outcome. Genes (Basel) 2021;12:353. doi: 10.3390/genes12030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shafienia S, Mirzaei M, Kavosi A, Yavarian M. A novel splice-site mutation in the LRP5 gene causing Familial Exudative Vitreoretinopathy. Gene Reports. 2020;21:100801. doi: 10.1016/j.genrep.2020.100801. [DOI] [Google Scholar]
- 145.Laine CM, et al. Novel mutations affecting LRP5 splicing in patients with osteoporosis-pseudoglioma syndrome (OPPG) Eur. J. Hum. Genet. 2011;19:875–881. doi: 10.1038/ejhg.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Korvala J, et al. Mutations in LRP5 cause primary osteoporosis without features of OI by reducing Wnt signaling activity. BMC Med. Genet. 2012;13:26. doi: 10.1186/1471-2350-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Little RD, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Boyden LM, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 149.Roman-Gomez J, et al. Transcriptional silencing of the Dickkopfs-3 (Dkk-3) gene by CpG hypermethylation in acute lymphoblastic leukaemia. Br. J. Cancer. 2004;91:707–713. doi: 10.1038/sj.bjc.6602008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kobayashi K, et al. Reduced expression of the REIC/Dkk-3 gene by promoter-hypermethylation in human tumor cells. Gene. 2002;282:151–158. doi: 10.1016/S0378-1119(01)00838-1. [DOI] [PubMed] [Google Scholar]
- 151.Kwok JB, et al. GSK3B polymorphisms alter transcription and splicing in Parkinson’s disease. Ann. Neurol. 2005;58:829–839. doi: 10.1002/ana.20691. [DOI] [PubMed] [Google Scholar]
- 152.Wang L, et al. Alternative splicing of the porcine glycogen synthase kinase 3beta (GSK-3beta) gene with differential expression patterns and regulatory functions. PLoS One. 2012;7:e40250. doi: 10.1371/journal.pone.0040250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Patturajan M, et al. DeltaNp63 induces beta-catenin nuclear accumulation and signaling. Cancer Cell. 2002;1:369–379. doi: 10.1016/S1535-6108(02)00057-0. [DOI] [PubMed] [Google Scholar]
- 154.Chakrabarti R, et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat. Cell Biol. 2014;16:1–13. doi: 10.1038/ncb3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brkic S, et al. Dual targeting of JAK2 and ERK interferes with the myeloproliferative neoplasm clone and enhances therapeutic efficacy. Leukemia. 2021;35:2875–2884. doi: 10.1038/s41375-021-01391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wolf A, et al. JAK2-V617F-induced MAPK activity is regulated by PI3K and acts synergistically with PI3K on the proliferation of JAK2-V617F-positive cells. JAKSTAT. 2013;2:e24574. doi: 10.4161/jkst.24574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Longhi MT, et al. PI3K-AKT, JAK2-STAT3 pathways and cell-cell contact regulate maspin subcellular localization. Cell Commun. Signal. 2021;19:86. doi: 10.1186/s12964-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang J, et al. Feedback activation of STAT3 limits the response to PI3K/AKT/mTOR inhibitors in PTEN-deficient cancer cells. Oncogenesis. 2021;10:8. doi: 10.1038/s41389-020-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vogt PK, Hart JR. PI3K and STAT3: A new alliance. Cancer Discov. 2011;1:481–486. doi: 10.1158/2159-8290.CD-11-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liang F, et al. The crosstalk between STAT3 and p53/RAS signaling controls cancer cell metastasis and cisplatin resistance via the Slug/MAPK/PI3K/AKT-mediated regulation of EMT and autophagy. Oncogenesis. 2019;8:59. doi: 10.1038/s41389-019-0165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Amarasinghe SL, et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tilgner H, Grubert F, Sharon D, Snyder MP. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. USA. 2014;111:9869–9874. doi: 10.1073/pnas.1400447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Steijger T, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods. 2013;10:1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pollard MO, Gurdasani D, Mentzer AJ, Porter T, Sandhu MS. Long reads: Their purpose and place. Hum. Mol. Genet. 2018;27:R234–R241. doi: 10.1093/hmg/ddy177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hu Y, et al. LIQA: Long-read isoform quantification and analysis. Genome Biol. 2021;22:182. doi: 10.1186/s13059-021-02399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]