Abstract
Phenotypic analysis of Escherichia coli strains causing bacteremia in cancer patients suggests that they possess specific virulence properties. To investigate this hypothesis, we compared the frequency of the virulence-related genes cnf1, cnf2, papC, hlyC, and iut in 155 E. coli strains isolated from hospitalized cancer patients with epidemiologically unrelated cases of bacteremia to their frequency in 70 E. coli strains isolated from the feces of healthy unrelated volunteers. Of the blood isolates, 24, 37, and 26% were positive for cnf1, papC, and hlyC, respectively, versus only 6, 17, and 6% of the fecal isolates (P < 0.05 in all instances). By contrast, 47% of both isolates carried the iut gene. The patients' clinical characteristics did not significantly influence these frequencies. The presence on various pathogenicity islands (PAIs) of a combination of the cnf1, papC, and hlyC genes on the chromosome was strongly suggested by Southern blotting of pulsed-field gel electrophoresis (PFGE) patterns with specific DNA probes. The phylogenetic relatedness among 60 strains carrying three, two, one, or no virulence genes and 6 ECOR strains included as references was determined by neighbor joining, the unweighted pair-group method with arithmetic mean, and Wagner analysis of the randomly amplified polymorphic DNA (RAPD) patterns generated by 11 primers. Identification of a major cluster including 96.4% of the strains carrying the cnf1, papC, and hlyC genes and ECOR subgroup B2 strains suggested that the virulent E. coli strains causing bacteremia in cancer patients are closely related to ECOR B2 strains. The presence in the E. coli population surveyed of a strong linkage disequilibrium, and especially of a highly significant correlation between PFGE and RAPD genetic distances, confirms that clonal propagation has a major impact on the E. coli population structure. Nevertheless, low bootstrap values in the phylogenetic tree suggested that frequent genetic exchange inhibits the individualization of discrete genetic lineages, which are stable on an evolutionary scale.
Patients with cancer are highly susceptible to bacterial infections, particularly during the periods of severe neutropenia that follow anticancer chemotherapy. Bloodstream infections caused by gram-negative bacteria are among the most frequent of these infections and are associated with a high mortality rate (1). Most of the gram-negative bacteria that cause these bloodstream infections originate in the intestinal flora (55) and reach the blood by a mechanism called bacterial translocation (for a review, see reference 4). In experimental models, bacterial translocation is enhanced by intestinal colonization and overgrowth of the translocating bacteria, by immunosuppression of the host, and by alterations of the intestinal mucosa (for a review, see reference 4), all of which may occur in cancer patients.
Escherichia coli is the most common gram-negative species isolated from cancer patients with bacteremia (1, 55). Phenotypic studies performed on a series of strains isolated from the blood of patients with various underlying conditions suggest that the E. coli strains isolated from such patients express traits associated with virulence more frequently than do the strains isolated from the intestinal flora of healthy volunteers. These traits include O serogroup (13), alpha-hemolysin (2, 7, 13, 43), type B carboxylesterase (13, 21), cytotoxic necrotizing factor (CNF) (13), aerobactin (2, 13, 43), and colicin (13). This was confirmed by genotypic studies. For instance, hly, the operon encoding alpha-hemolysin, aero, the operon which encodes aerobactin, cnf1, which encodes CNF1, and pap, the operon which encodes pyelonephritis adhesin pili, were found preferentially in E. coli strains isolated from patients with urinary tract infections (8, 43, 64); pap, hly, aero, and afa, the operon encoding afimbrial adhesins, were found in patients with neonatal meningitis (5); and pap, hly, aero, and sfa, the operon which encode S fimbria adhesins, were found in patients with bacteremia (35, 43). cnf2, the gene encoding CNF2, has been found only in strains isolated from animals (45).
Virulence genes are often grouped in a single segment of the bacterial chromosome, constituting a distinct functional genetic unit called a pathogenicity island (PAI) (10, 24). PAIs have now been isolated from many bacterial species (for a recent review, see reference 25). They are often larger than 30 kb, have a G+C content different from that of the host organism, are often inserted into tRNA genes, may be flanked by insertion sequence elements, may be lost by deletion at high frequency, and, typically, are present more often in pathogenic than in less pathogenic strains within the same species (25). E. coli PAIs have been studied chiefly in uropathogenic and enteropathogenic strains. Several PAIs, all carrying hly genes and numbered I, II, IV, and V, have been described in uropathogenic strains (10, 54). In addition to hly, PAI II contains prf, encoding the P-related fimbriae (10), PAI IV contains pap (54), and PAI V contains both prs encoding a P-related sequence and cnf1 (10). PAI III, a newly renamed locus of enterocyte effacement, has been identified in enteropathogenic E. coli strains. It carries a specific set of virulence genes comprising sepA through sepI, encoding a type III secretion system protein, espA and espB, encoding secreted proteins involved in signal transduction, and eaeA, encoding intimin (37, 38). So far, the presence of PAIs has not been reported in E. coli strains causing bloodstream infections.
The population structure of E. coli, as represented by the ECOR collection (41), is thought to be clonal, since it comprises five major clonal groups called A, B1, B2, D, and E (26). Most E. coli blood isolates belong to the B2 ECOR group (48). This is also true for E. coli strains isolated from patients with neonatal meningitis (5). Recently, it was shown that the B2 and D ECOR strains carried virulence genes, including hly, pap, sfa, and kps, a cluster of genes encoding proteins involved in export of type II capsular polysaccharides, more often than did strains from the other ECOR groups (11). It was also reported that a set of virulence genes comprising pap, hly, and sfa was preferentially distributed in a major cluster of strains causing bloodstream infections in patients with various underlying conditions (35). All these studies suggest that a cluster of E. coli strains acquired virulence genes by horizontal transfer, thus defining a highly virulent group. The link between the B2 phylogenetic group and the virulence in E. coli strains causing extraintestinal infection was recently confirmed in studies of experimentally infected mice (47).
In the present study, we attempted to determine whether a clustered distribution of virulence genes can be found in E. coli strains causing bacteremia in cancer patients, and we used new approaches to explore further the general problem of the population structure of E. coli and its mode of evolution.
MATERIALS AND METHODS
Bacterial strains and patient characteristics.
We studied 155 strains of E. coli isolated between April 1992 and December 1993 from consecutive patients with bacteremia who were hospitalized at Institut Gustave-Roussy, a 420-bed French cancer referral center. The major underlying diseases were leukemia in 8 patients (5.2%), lymphoma in 17 (11%), solid tumors in 114 (73.5%) and unknown in 16 (10.3%). Neutropenia was defined as a leukocyte count lower than 500/mm3 at the onset of bacteremia, and a urinary source of bacteremia was suspected when more than 105 CFU of E. coli per ml was isolated from urine samples. We also studied 70 E. coli strains isolated from the feces of healthy volunteers living in the community. For each fecal sample, one colony was chosen at random and it was assumed that the isolates of E. coli so obtained were representative of the bacterial population present in the feces of the volunteers at that time.
E. coli reference strains isolated from humans or animals, with or without known virulence genes, were used as positive and negative controls for the PCR studies. Six E. coli strains from the ECOR collection (41) were used as reference strains for the phylogenetic study, and three unrelated strains of enterobacteria were taken as the outgroup (Table 1).
TABLE 1.
Characteristics of the E. coli reference strains isolated from humans and animals
| Strain | Phenotype | ECOR subgroup | Host | Reference |
|---|---|---|---|---|
| J96 | Cnf1, Pap, Hly | NDa | Human | 29 |
| 1520 | Cnf1 | ND | Dog | 45 |
| BM4.1 | Cnf1 | ND | Calf | 45 |
| BM2.1 | Cnf1 | ND | ND | 45 |
| JL21+ | Cnf2 | ND | Calf | 45 |
| S5 | Cnf2, Aero | ND | Lamb | 45 |
| BM2.10 | Cnf1, Aero | ND | Calf | 45 |
| HB101 | No known virulence factor | ND | Human | 45 |
| JL21− | JL21 without Cnf2 | ND | Calf | 45 |
| ECOR4 | ND | A | Human | 42 |
| ECOR25 | ND | A | Dog | 42 |
| ECOR45 | ND | B1 | Pig | 42 |
| ECOR48 | Pap, Hly | D | Human | 42 |
| ECOR52 | Cnf1, Pap, Hly | B2 | Orangutan | 42 |
| ECOR59 | ND | B2 | Human | 42 |
ND, not determined.
Bacterial cultures and DNA extraction.
E. coli strains were grown on Luria-Bertani agar or in Luria-Bertani broth at 37°C overnight. Genomic DNA was extracted by rapid lysis, as described previously (50), and used as the target for PCR assays.
PCR materials and methods.
Primers were used to amplify intragenic fragments of the genes cnf1 (15), cnf2 (46), papC (32), hlyC (17), iut (representative of the aerobactin operon) (13b), and the tRNA pheR, which encodes phenylalanine tRNA and which is often the site of insertion from PAI V. Details of primer sequences and predicted sizes of the amplified products are given in Table 2. PCR amplification of bacterial DNA extracts was performed in 25-μl volumes containing 5 μl of cell lysate for detecting cnf1 and cnf2 (51) and papC (32). For detection of hlyC and iut, PCRs were performed using 15 mM Tris-HCl (pH 8.3), 40 mM KCl, 200 μM each deoxynucleoside triphosphate (Boehringer Mannheim, Meylan, France), 1 μM each primer, and 1 U of Ampli-Taq DNA polymerase (Perkin-Elmer) per reaction. The concentration of MgCl2 optimized for each PCR was 4 mM for hlyC and 1 mM for iut. PCR amplification consisted of one cycle at 94°C for 5 min, 28 cycles of denaturation at 94°C for 45 s, annealing at 69°C for 40 s, and extension at 72°C for 1 min, and one cycle of 72°C for 5 min in a Thermocycler 9600 (Perkin-Elmer/Cetus). For the detection of tRNA pheR, the PCR mixtures differed from those of hlyC and iut in that Ampli-Taq DNA was replaced by DNA Taq polymerase (Boehringer Mannheim). PCRs were performed in a final volume of 50 μl containing 5 μl of cell lysate. The concentration of MgCl2 was 1 mM. PCR amplification consisted of one cycle at 94°C for 5 min, 25 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 45 s, and extension 72°C for 1 min, and one cycle of 72°C for 10 min. Aliquots (10 μl) of the final reaction mixtures underwent standard gel electrophoresis in 2% agarose. Amplified DNA fragments of specific sizes were detected by UV fluorescence after being stained with ethidium bromide.
TABLE 2.
Oligonucleotide primers used to amplify virulence-associated genes
| Target gene | Oligonucleotide sequence (5′→3′) | Predicted size of amplified product (bp) | Reference |
|---|---|---|---|
| cnf1 | CATGCTTCTTCCTCAGTAGC | 255 | 51 |
| GTCTAAAAGGGGGGCAGCCA | |||
| cnf2 | CTTCTTCAGTTGTTCCTCCG | 583 | 51 |
| TAAGGCGGGCACAAACCAGA | |||
| hlyC | GCCAGAACAGTGCTTATCCT | 1,576 | This work |
| CCAGTTCCCCATTACACAGA | |||
| iut | AGGTGACGGCTTTCTCTTCG | 189 | This work |
| TCTTGTTGTTCAGCCCTCCG | |||
| papC | GACGGCTGTACTGCAGGGTGTGGCG | 328 | 32 |
| ATATCCTTTCTGCAGGGATGCAATA | |||
| tRNA pheR | AGCCGCGCTTTGGTACAGTAGC | 1,168 | This work |
| CAGATGGAACTGGTGCTGGAAGG |
PFGE typing.
Epidemiological relationships between strains were assessed by studying the pulsed-field gel electrophoresis (PFGE) patterns of genomic DNA after restriction by XbaI, SfiI, and NotI (Boehringer, Mannheim, Germany), as described previously (22, 23). These enzymes do not have restriction sites within the sequences of the five virulence genes under study. An XbaI digest of Salmonella enterica serovar Indiana (T461) DNA was used as a reference pattern for strain analysis and band size determination. PFGE patterns were compared using Gel-Compar software (Applied Maths, Kortrijk, Belgium). Strains with more than seven distinct restriction fragments were considered different, as described previously (56).
Southern blot hybridization.
PFGE digests were transferred to nylon membranes (Amersham) and hybridized with digoxigenin-labeled probes which were generated by the specific primers of the cnf1, papC, and hlyC genes as recommended by the manufacturer (Boehringer Mannheim).
Relationships among strains from our collection.
A subset of 60 strains from our collection were selected so as to include representative combinations of the virulence genes under study. We chose 19, 13, 8, and 20 bacteremic strains among those that carried three, two, one, and no virulence genes, respectively. DNA, extracted as described previously (33), was subjected to 11 different randomly amplified polymorphic DNA (RAPD) assays using the following commercially available 10-mer primers: A1, A2, A3, A7, A9, A10, A13, A16, A17, A19, and A20 (kit A from Operons Technologies, Alameda, Calif.). Preliminary results (not shown) showed that these primers generated distinct reproducible RAPD patterns. RAPD assays were performed in duplicate in 61-μl reaction volumes containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 100 μM each deoxynucleoside triphosphate (Boehringer), 200 nM primer, 0.9 U of DNA Taq polymerase (Boehringer), and 10 ng of bacterial DNA. A PTC 100 DNA Engine machine (MJ Research, Watertown, Mass.) was used for amplification, which comprised 45 cycles, each consisting of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C. Amplified fragments underwent standard gel electrophoresis in 1.6% agarose and were detected by UV fluorescence after staining with ethidium bromide. Only the major bands of the resulting patterns were taken into account in the analysis (63).
We used two previously described genetic population tests, namely, the f and g tests (62), both based on the assessment of linkage disequilibrium (i.e., the nonrandom association of genotypes occurring at different loci) by Monte Carlo simulations with 104 iterations. As a null hypothesis, both tests take panmixia, a situation in which genetic exchange occurs randomly. Means to avoid biases due to geographical and/or temporal separation have been proposed (57, 61). The f test only evaluates the probability of observing, by chance, a linkage disequilibrium as high as or higher than the one actually observed in the sample, whereas the g test also evaluates the correlation between the genetic distances calculated with two independent sets of genetic markers (here PFGE and RAPD patterns). In a panmixia, these two sets of markers recombine independently and therefore do not correlate. The correlation is evaluated by the nonparametric Mantel test (34), which does not require an assumed degree of freedom in sample.
The unweighted pair group method with arithmetic mean (53), neighbor-joining, and Wagner phylogenetic methods derived from the PHYLIP software package (19) were used to assess the phylogenetic relationships between the strains. S. enterica serovar Indiana and Citrobacter freundii were chosen as outgroups to root trees because they are closely related to the genus Escherichia. Only branch topologies found similar by all the above three methods were considered significant, because agreement between the results of several phylogenetic methods is a strong indication that phylogeny has been correctly determined (28, 31). The robustness of the branches was estimated by the bootstrap method (18).
RESULTS
Prevalence of virulence genes.
The cnf1, papC, and hlyC genes, but not the iut gene, were detected in a significantly larger number of E. coli blood isolates from cancer patients than in fecal isolates from healthy volunteers (Table 3). The cnf2 gene was not detected in any human strains, as reported previously (45). As shown in Table 4, four different combinations of the genes under study were identified. Only the association of cnf1, papC, and hlyC occurred in significantly more blood than fecal isolates (19 and 4.2%, respectively; P < 0.05). Conversely, the absence of all virulence genes as detected here was noted in more fecal than blood isolates (81 and 57%, respectively; P < 0.05) (data not shown).
TABLE 3.
Prevalence of virulence genes in E. coli strains isolated from the blood of cancer patients and the feces of healthy volunteers
| Target gene | No. (%) of positive strains in:
|
P valuea | |
|---|---|---|---|
| Blood (n = 155) | Feces (n = 70) | ||
| cnfI | 37 (24) | 4 (6) | <0.05 |
| papC | 58 (37) | 12 (17) | <0.05 |
| hlyC | 40 (26) | 4 (6) | <0.05 |
| iut | 73 (47) | 33 (47) | NSb |
Chi-square test.
NS, not significant.
TABLE 4.
Combination of virulence genes identified in E. coli strains isolated from the blood of cancer patients and the feces of healthy volunteers
| Gene combination | % (no.) of positive strains in:
|
P valuea | |
|---|---|---|---|
| Blood (n = 155) | Feces (n = 70) | ||
| No known virulence factors | 57 (89) | 81.4 (57) | <0.05 |
| cnf1 + papC + hlyC | 19 (30) | 4.2 (3) | <0.05 |
| cnf1 + papC | 0.6 (1) | 0 | |
| cnf1 + hlyC | 4 (6) | 0 | |
| papC + hlyC | 1.2 (2) | 1.4 (2) | NSb |
| cnf1 | 0 | 0 | |
| papC | 16 (25) | 11.4 (8) | NS |
| hlyC | 1.2 (2) | 0 | |
Chi-square test.
NS, not significant.
Clinical data analysis.
We did not find any significant differences between the number of virulence factors in strains recovered from patients with or without the clinical characteristics studied, which included the number of circulating polymorphonuclear leukocytes at the onset of bacteremia, the underlying disease, and the association of bacteremia with a positive urine culture (data not shown).
Epidemiological relationships between E. coli strains.
Digestion of E. coli DNA with XbaI, SfiI, and NotI produced 16 to 20 well-resolved fragments ranging from 20 to 700 kb (data not shown). PFGE patterns were highly heterogeneous among the different strains. PFGE resolved at least seven distinct fragments when the patterns were compared pairwise. Thus, on the basis of widely accepted interpretative criteria (56), we concluded that the strains were epidemiologically unrelated.
Physical location of virulence genes.
The results of Southern blotting of PFGE patterns with the hlyC, cnf1, and papC probes suggested that these three genes were physically linked in 22 of the 32 strains (69%) carrying the three virulence genes (Table 4). The 32 strains were found in either blood or fecal isolates. In 18 of 22 strains (82%), the three probes hybridized on the same restriction fragment with all three restriction enzymes. For the 10 strains carrying two virulence genes (Table 4), the probes hybridized with all the restriction enzymes in 7 of the 10 strains (70%). The sizes of the hybridization bands in the strains carrying three and two virulence genes were smaller than 190 kb in 10 of 22 strains (45%) and 4 of 10 strains (40%), respectively. This cutoff limit was chosen because the largest previously described PAI in E. coli, PAI II, is 190 kb long (9). Among the 22 strains carrying three virulences genes, the tRNA pheR was conserved in size in only 2 strains (10%). In comparison, 5 of 12 strains (42%) carrying no studied virulence genes had tRNA pheR.
Population genetic and phylogenetic analysis.
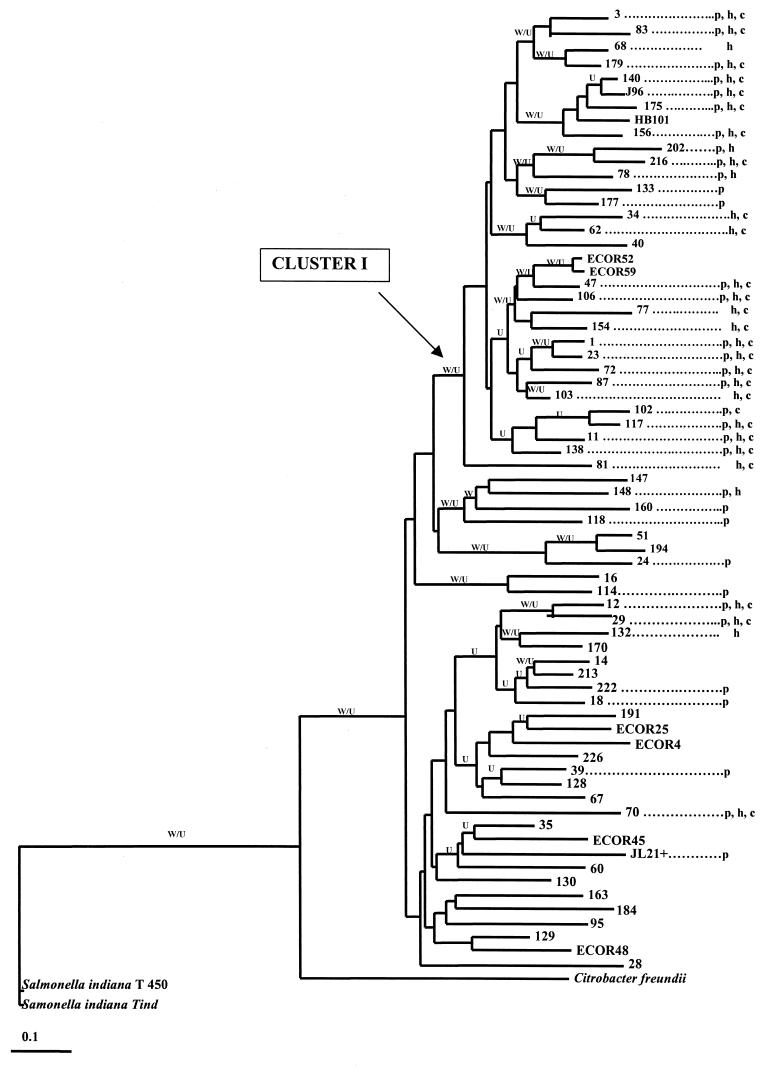
The RAPD patterns generated by each primer were polymorphic (results not shown), and there was a highly significant correlation between RAPD and PFGE genetic distances (r = 0.33, P < 10−4; Mantel nonparametric test). The linkage disequilibrium f and g tests were significant for both the overall sample, except for the reference stocks and outgroups (f < 10−4, g < 10−4) and for a main cluster referred to below as cluster I (f = 1.3 × 10−3, g < 10−4) and shown in Fig. 1. However, f and g test P values were not significant when the strains not included in cluster I were analyzed separately.
FIG. 1.
Rooted neighbor-joining tree generated by the phylogenetic relationships among 61 E. coli strains isolated from the blood of cancer patients or the feces of healthy volunteers. The letters W and U indicate branches that were also found by the Wagner and UPGMA methods, respectively. Strains numbered up to 155 were blood isolates from cancer patients, and those with higher numbers were fecal isolates from healthy volunteers. p, h, and c indicates carriage of the papC, hlyC, and cnf1 genes, respectively.
Cluster I contained 29 strains, 28 (96.5%) of which carried at least one virulence gene. This cluster also included the two B2 subgroup strains of the ECOR collection and reference strains HB101 and J96. Note that 16 of 19 strains (84%) from our collection carrying the association of papC, hlyC, and cnf1 genes were included in this cluster whereas 19 of 20 of our isolates (95%) that carried no virulence genes were found outside the cluster, like the ECOR strains from groups A, B1, and D. Cluster I was identified by the three phylogenetic methods used. However, the robustness of bootstrap analysis of this cluster was low (0.24).
DISCUSSION
In this study, we found that the E. coli strains isolated from cancer patients with bacteremia carried the virulence-associated papC, hlyC, and cnf1 genes more frequently than did strains isolated from the feces of healthy volunteers. This result might be considered somewhat unexpected since cancer patients are often severely immunocompromised and thus could have been infected by much less virulent strains. The frequencies of the papC, hlyC, cnf1, and iut genes in both fecal and blood isolates were similar to those reported previously (35, 40). The high frequency of carriage of the cnf1 gene in our collection of E. coli blood isolates was also in agreement with a previous report on the phenotypic expression of CNF in these strains (6), suggesting that cnf1 plays a role in the pathogenicity of these isolates (16, 20, 27). In the present study, the iut gene, which can be either plasmid or chromosome borne (30, 39) and has never been found within a PAI (25), was detected with a non-significantly different frequency in blood and fecal isolates, in contrast with our previous observation concerning aerobactin production in a similar collection of blood isolates from cancer patients (2). However, the prevalence of aerobactin expression we observed in the later collection was dependent on the technique used, since it ranged from 52% using a chemical assay to 89% using a bioassay (2). Other workers reported that the prevalence of the aero operon in E. coli blood isolates varied from 53% of African strains to 81% of American strains (35). The cause of these variations remains unclear, but their existence makes the role of aerobactin production uncertain in the pathogenesis of E. coli bloodstream infections in humans.
Our finding that one or more of the other three virulence genes detected were present in significantly more blood isolates from cancer patients than in fecal isolates from healthy volunteers suggested that they might be important in the pathogenesis of bacteremia. Note that the prevalence of these genes was not significantly different among the strains from bacteremia patients, whatever their underlying disease and immune status, as judged by the number of circulating polymorphonuclear leukocytes, or from the possible urinary source of some cases of bacteremia. This suggests that bacterial factors were of major importance in the pathogenesis of this kind of infection. However, 57% of the bacteremic strains had no virulence factors. It is possible that the corresponding patients had specific immune defects not recorded in this retrospective study. These strains without virulence factors did not group in any specific clone, suggesting that the belonged to the general E. coli intestinal populations of the patients.
To establish whether the virulence genes papC, hlyC, and cnf1 are linked in the same segment of the chromosome, suggesting their inclusion in a PAI, we hybridized the PFGE patterns generated by three restriction enzymes with specific probes, as did others (9, 54). In 22 of the 32 strains carrying the combination of papC, hlyC, and cnf1 genes (69%), these three genes were indeed located on a single restriction fragment, regardless of the restriction enzyme used. The 98% DNA sequence homology between papC and prs suggested that we had detected prs in E. coli J96 and that the association of papC, hlyC, and cnf1 found in our strains was probably part of a PAI which is very similar, and perhaps identical, to PAI V. No tRNA pheR was detected in 20 of 22 strains (90%), carrying the combination of papC, hlyC, and cnf1, suggesting that this combination of genes was probably located in a PAI, such as PAI V. However, in some of our isolates carrying the three virulence genes, the probes hybridized on a band as small as 87 kb, whereas the reported size of PAI V is 110 kb (9, 54), suggesting that one of the three restriction enzymes used had a restriction site(s) in PAI V. This could not be formally confirmed, because the full sequence of PAI V is not available. In some strains carrying two virulence genes, the probes for papC and hlyC also hybridized on a DNA fragment smaller than 190 kb. This is compatible with the 170-kb long PAI IV, as described in E. coli J96 (9). Finally, to our knowledge, the combination of cnf1 and hlyC without papC, which we found in six strains, has not been previously reported in a PAI.
To ascertain whether the virulence factors were randomly distributed over the entire spectrum of E. coli genotypes, we analyzed the population structure and phylogenetic relationships in a subset of representative E. coli isolates. For this purpose, we used the multiprimer RAPD method, which has been successfully applied to the study of the population genetics of several microbial species such as Candida albicans (49, 58), Trypanosoma cruzi (12, 13a), and Leishmania spp. (3). This method generates results equivalent to those of the reference multilocus enzyme electrophoresis method (63) but is less time-consuming and easier to perform. We found that 85% of the E. coli strains that carried the combination of papC, hlyC, and cnf1 were located in the same genetic cluster, together with the two ECOR subgroup B2 strains. However, the robustness of this cluster is not supported by bootstrap analysis. Thus, although the strains inside the cluster tend to exhibit significantly greater genetic similarity than those outside it, it would hazardous to consider this cluster a real discrete phylogenetic entity.
The cluster included most of the strains that carried at least one virulence gene, whether they were isolated from blood or feces. This observation, combined with the very sporadic occurrence of virulence genes among the strains outside the cluster, suggested that the distribution of virulence genes was not random but was preferentially linked to the cluster.
However, since the cluster also contained the two strains of the ECOR subgroup B2, it probably corresponds to the clonal subgroup B2 of the ECOR collection. In two recent investigations, the pap, hly, kps, and sfa genes were detected mainly in the strains from this subgroup (11, 47). Also, a nonrandom distribution of virulence genes was reported among clinical strains isolated from patients with bloodstream infections (35) and neonatal meningitis (5). In a collection of American and African E. coli strains isolated from patients with bloodstream infections, pap, hly, and sfa genes were found in a major cluster (35), which was probably identical to the cluster described here. It was recently shown that E. coli strains which were isolated from patients with neonatal meningitis and carried the ibe10 gene encoding brain endothelial cell invasion, the pap, afa, sfa/foc, and hly genes were distributed mainly in a genetic cluster corresponding to the B2 subgroup of the ECOR collection (5). Thus, whatever their clinical or geographic origin, most strains from this subgroup are likely to carry virulence genes. Since we found that the virulence genes carried by strains from cluster I were likely to be part of PAI IV or V, we suggest that the distribution of PAI IV and V is preferentially linked to strains from the B2 subgroup. Also, since PAIs are suspected to originate from species other than E. coli by horizontal transfer and since PAIs are integrated into specific regions of the bacterial chromosome such as the tRNA locus, we suggest that B2 subgroup strains are more prone to integrate PAIs into their bacterial chromosome than are strains outside this subgroup.
As regards the general problem of the E. coli population structure, the strong linkage disequilibrium found in the 60 isolates tested and in cluster I confirms the hypothesis that clonal propagation has a major impact on E. coli genotype distribution (44, 52). This linkage disequilibrium persisted even when the genotypes rather than the strains were taken as units of analysis. This indicates that our population of E. coli does not correspond to the model of epidemic clonality, as proposed for Neisseria meningitidis (36). Our results also do not support the hypothesis that this sample of E. coli is divided into discrete genetic entities called discrete typing units (59, 60). The only strong bootstrap value that we found was in fact related to the entire E. coli group. All the other branches were weakly supported by bootstrap analysis, which suggests that genetic exchange inhibits strictly separate evolution of the subgroups. This picture is comparable to the picture observed for T. cruzi, a parasitic protozoan which is responsible for Chagas' disease and is a typical case of a structured species whose pattern is probably attributable to long-term clonal evolution (57). In this parasite, the discreteness of the genetic lines that subdivide the species is much more apparent than in E. coli. Pseudomonas aeruginosa, in contrast, exhibits a picture of panmixia, with no apparent linkage disequilibrium (14). The frequency of genetic recombination therefore seems much stronger in this species than in E. coli.
In conclusion, the present results suggest that even in immunosuppressed patients with cancer, specific strain characteristics are of major importance in the pathogenesis of E. coli bacteremia.
ACKNOWLEDGMENTS
We thank P. Boquet (Nice, France) for helpful discussions and J. Craig and M. Dreyfus for English revision.
This work was supported in part by Contrat de Recherche Clinique Institut Gustave Roussy 96.1.
REFERENCES
- 1.Andremont A, Lancar R, Lê N A, Hattchouel J M, Baron S, Tavakoli T, Daniel M F, Tancrède C, Lê M G. Secular trends in mortality associated with bloodstream infections in 4268 patients hospitalized in a cancer referral center between 1975 and 1989. Clin Microbiol Infect. 1996;1:160–167. doi: 10.1111/j.1469-0691.1996.tb00547.x. [DOI] [PubMed] [Google Scholar]
- 2.Aumont P, Enard C, Expert D, Pieddeloup C, Tancrède C, Andremont A. Production of hemolysin, aerobactin and enterobactin by strains of Escherichia coli causing bacteremia in cancer patients, and their resistance to human serum. Res Microbiol. 1989;140:21–26. doi: 10.1016/0923-2508(89)90055-7. [DOI] [PubMed] [Google Scholar]
- 3.Banuls A, Guerrini F, Pont F L, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol. 1997;44:408–411. doi: 10.1111/j.1550-7408.1997.tb05716.x. [DOI] [PubMed] [Google Scholar]
- 4.Berg R. Bacterial translocation from the gastrointestinal tract. Trends Microbiol. 1995;3:149–154. doi: 10.1016/s0966-842x(00)88906-4. [DOI] [PubMed] [Google Scholar]
- 5.Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177:642–650. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- 6.Blanco J, Alonso M, Gonzalez E, Blanco M, Garabal J. Virulence factors of bacteraemic Escherichia coli with particular reference to production of cytotoxic necrotising factor (CNF) by P-fimbriate strains. J Med Microbiol. 1990;31:175–183. doi: 10.1099/00222615-31-3-175. [DOI] [PubMed] [Google Scholar]
- 7.Blanco J, Blanco M, Alonso M, Blanco J, Gonzalez E, Garabal J. Characteristics of haemolytic Escherichia coli with particular reference to production of cytotoxic necrotizing factor type 1 (CNF1) Res Microbiol. 1992;143:869–878. doi: 10.1016/0923-2508(92)90074-x. [DOI] [PubMed] [Google Scholar]
- 8.Blanco M, Blanco J, Alonso M, Blanco J. Virulence factors and O groups of Escherichia coli isolates from patients with acute pyelonephritis, cystitis and asymptomatic bacteriuria. Eur J Epidemiol. 1996;12:191–198. doi: 10.1007/BF00145506. [DOI] [PubMed] [Google Scholar]
- 9.Blum G, Falbo V, Caprioli A, Hacker J. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett. 1995;126:189–195. doi: 10.1111/j.1574-6968.1995.tb07415.x. [DOI] [PubMed] [Google Scholar]
- 10.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd E, Hartl D. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brisse S, Barnabé C, Banuls A, Sidibe I, Noel S, Tibayrenc M. A phylogenetic analysis of the Trypanosoma cruzi genome project CL Brener reference strain by multilocus enzyme electrophoresis and multiprimer random amplified polymorphic DNA fingerprinting. Mol Biochem Parasitol. 1998;92:253–263. doi: 10.1016/s0166-6851(98)00005-x. [DOI] [PubMed] [Google Scholar]
- 13.Cherifi A, Contrepois M, Picard B, Goullet P, de Rycke J, Fairbrother J, Barnouin J. Factors and markers of virulence in Escherichia coli from human septicemia. FEMS Microbiol Lett. 1990;58:279–283. doi: 10.1111/j.1574-6968.1990.tb13989.x. [DOI] [PubMed] [Google Scholar]
- 13a.de Lana M, Pinto A, Barnabé C, Quesney V, Noel S, Tibayrenc M. Trypanosoma cruzi: compared vectorial transmissibility of three major clonal genotypes by triatoma infestans. Exp Parasitol. 1998;90:20–25. doi: 10.1006/expr.1998.4304. [DOI] [PubMed] [Google Scholar]
- 13b.de Lorenzo V, Bindereif A, Paw B, Neilands J. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol. 1986;165:570–578. doi: 10.1128/jb.165.2.570-578.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denamur E, Picard B, Decoux G, Denis J, Elion J. The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Microbiol Lett. 1993;110:275–280. doi: 10.1111/j.1574-6968.1993.tb06334.x. [DOI] [PubMed] [Google Scholar]
- 15.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun. 1993;61:4909–4914. doi: 10.1128/iai.61.11.4909-4914.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falzano L, Fiorentini C, Donelli G, Michel E, Kocks C, Cossart P, Cabanie L, Oswald E, Boquet P. Induction of phagocytic behaviour in human epithelial cells by Escherichia coli cytotoxic necrotizing factor type 1. Mol Microbiol. 1993;9:1247–1254. doi: 10.1111/j.1365-2958.1993.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 17.Felmlee T, Pellett S, Welch R A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163:94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein J. Confidence limits of phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. PHYLIP: Phylogenetic Inference Package, version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 20.Fiorentini C, Matarrese P, Straface E, Falzano L, Fabbri A, Donelli G, Cossarizza A, Boquet P, Malorni W. Toxin-induced activation of Rho GTP-binding protein increases Bcl-2 expression and influences mitochondrial homeostasis. Exp Cell Res. 1998;242:341–350. doi: 10.1006/excr.1998.4057. [DOI] [PubMed] [Google Scholar]
- 21.Goullet P, Picard B. Electrophoretic typing of Escherichia coli esterases in septicemia. Presse Med. 1984;13:1079–1081. [PubMed] [Google Scholar]
- 22.Grothues D, Koopmann U, van der Hardt H, Tümmler B. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol. 1988;26:1973–1977. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grothues D, Tümmler B. New approaches in genome analysis by pulsed-field gel electrophoresis: application to the analysis of Pseudomonas species. Mol Microbiol. 1991;5:2763–2776. doi: 10.1111/j.1365-2958.1991.tb01985.x. [DOI] [PubMed] [Google Scholar]
- 24.Hacker J, Bender L, Ott M, Wingender J, Lund B, Marre R, Goebel W. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- 25.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 26.Herzer P, Inouye S, Inouye M, Whittam T. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofman P, Flatau G, Selva E, Gauthier M, Negrate G L, Fiorentini C, Rossi B, Boquet P. Escherichia coli cytotoxic necrotizing factor 1 effaces microvilli and decreases transmigration of polymorphonuclear leukocytes in intestinal T84 epithelial cell monolayers. Infect Immun. 1998;66:2494–2500. doi: 10.1128/iai.66.6.2494-2500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huelsenbeck J P, Hillis D M. Success of phylogenetic methods in the four-taxon case. Syst Biol. 1993;42:247–264. [Google Scholar]
- 29.Hull R, Gill R, Hsu P, Minshew B, Falkow S. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun. 1981;33:933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson S, Tullus K, Wretlind B, Brauner A. Aerobactin-mediated uptake of iron by strains of Escherichia coli causing acute pyelonephritis and bacteremia. J Infect. 1988;16:147–152. doi: 10.1016/s0163-4453(88)93947-3. [DOI] [PubMed] [Google Scholar]
- 31.Kim J. Improving of phylogenetic estimation by combining different methods. Syst Biol. 1993;42:331–340. [Google Scholar]
- 32.LeBouguenec C, Archambaud M, Labigne A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol. 1992;30:1189–1193. doi: 10.1128/jcm.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 34.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 35.Maslow J, Whittam T, Gilks C, Wilson R, Mulligan M, Adams K, Arbeit R. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maynard Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Nat Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDaniel T, Jarvis K, Donnenberg M, Kaper J. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDaniel T, Kaper J. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 39.McDougall S, Neilands J. Plasmid-and chromosome-coded aerobactin synthesis in enteric bacteria: insertion sequences flank operon in plasmid-mediated systems. J Bacteriol. 1984;159:300–305. doi: 10.1128/jb.159.1.300-305.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhldorfer I, Blum G, Donohue-Rolfe A, Heier H, Olschlager T, Tschape H, Wallner U, Hacker J. Characterization of Escherichia coli strains isolated from environmental water habitats and from stool samples of healthy volunteers. Res Microbiol. 1996;147:625–635. doi: 10.1016/0923-2508(96)84019-8. [DOI] [PubMed] [Google Scholar]
- 41.Ochman H, Selander R. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochman H, Whittam T, Caugant D, Selander R. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 43.Opal S, Cross A, Gemski P, Lyhte L. Aerobactin and alpha-hemolysin as virulence determinants in Escherichia coli isolated from human blood, urine, and stool. J Infect Dis. 1990;161:794–796. doi: 10.1093/infdis/161.4.794. [DOI] [PubMed] [Google Scholar]
- 44.Orskov F, Orskov I. From the National Institutes of Health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the enterobacteriaceae and other bacteria. J Infect Dis. 1983;48:346–357. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- 45.Oswald E, de Rycke J, Lintermans P, van Muylem K, Mainil J, Daube G, Pohl P. Virulence factors associated with cytotoxic necrotizing factor type two in bovine diarrheic and septicemic strains of Escherichia coli. J Clin Microbiol. 1991;29:2522–2527. doi: 10.1128/jcm.29.11.2522-2527.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oswald E, Sugai M, Labigne A, Wu H C, Fiorentini C, Boquet P, O'Brien A D. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc Natl Acad Sci USA. 1994;91:3814–3818. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picard B, Garcia J S, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picard B, Goullet P. Correlation between electrophoretic types B1 and B2 of carboxylesterase B and host-dependent factors in Escherichia coli septicaemia. Epidemiol Infect. 1988;100:51–61. doi: 10.1017/s0950268800065559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pujol C, Joly S, Lockhart S, Noel S, Tibayrenc M, Soll D. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saulnier P, Bourneix C, Prevost G, Andremont A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saulnier P, Chachaty E, Hilali F, Andremont A. Single-step polymerase chain reaction for combined gene detection and epidemiological typing in three bacterial models. FEMS Microbiol Lett. 1997;150:311–316. doi: 10.1111/j.1574-6968.1997.tb10386.x. [DOI] [PubMed] [Google Scholar]
- 52.Selander R, Levin B. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210:545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- 53.Sneath P H A, Sokal R R. Numerical taxonomy. The principle and practice of numerical classification. San Francisco, Calif: W. H. Freeman & Co.; 1973. [Google Scholar]
- 54.Swenson D, Bukanov N, Berg D, Welch R. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tancrède C, Andremont A. Bacterial translocation and gram-negative bacteremia in patients with hematological malignancies. J Infect Dis. 1985;152:99–103. doi: 10.1093/infdis/152.1.99. [DOI] [PubMed] [Google Scholar]
- 56.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995;36:47–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 58.Tibayrenc M. Are Candida albicans natural populations subdivided? Trends Microbiol. 1997;5:253–254. doi: 10.1016/S0966-842X(97)01068-8. [DOI] [PubMed] [Google Scholar]
- 59.Tibayrenc M. Beyond strain typing and molecular epidemiology: integrated genetic epidemiology of infectious diseases. Parasitol Today. 1998;14:323–329. doi: 10.1016/s0169-4758(98)01286-1. [DOI] [PubMed] [Google Scholar]
- 60.Tibayrenc M. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int J Parasitol. 1998;28:85–104. doi: 10.1016/s0020-7519(97)00180-x. [DOI] [PubMed] [Google Scholar]
- 61.Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Breniere S, Darde M, Ayala F. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci USA. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tibayrenc M, Kjellberg F, Ayala F. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tibayrenc M, Neubauer K, Barnabe C, Guerrini F, Skarecky D, Ayala F J. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zingler G, Ott M, Blum G, Falkenhagen U, Naumann G, Sokolowska-Kohler W, Hacker J. Clonal analysis of Escherichia coli serotype O6 strains from urinary tract infections. Microb Pathog. 1992;12:299–310. doi: 10.1016/0882-4010(92)90048-s. [DOI] [PubMed] [Google Scholar]