Abstract
Viruses and their hosts can undergo coevolutionary arms races where hosts evolve increased resistance and viruses evolve counter-resistance. Given these arms race dynamics (ARD), both players are predicted to evolve along a single trajectory as more recently evolved genotypes replace their predecessors. By coupling phenotypic and genomic analyses of coevolving populations of bacteriophage λ and Escherichia coli, we find conflicting evidence for ARD. Virus-host infection phenotypes fit the ARD model, yet genomic analyses revealed fluctuating selection dynamics. Rather than coevolution unfolding along a single trajectory, cryptic genetic variation emerges and is maintained at low frequency for generations until it eventually supplants dominant lineages. These observations suggest a hybrid ‘leapfrog’ dynamic, revealing weaknesses in the predictive power of standard co-evolutionary models. The findings shed light on the mechanisms that structure coevolving ecological networks and reveal the limits of using phenotypic or genomic data alone to differentiate coevolutionary dynamics.
INTRODUCTION
Bacteria and their viruses (phage) are the two most abundant and genetically diverse groups of organisms on Earth (Clokie et al., 2011; Thomas et al., 2011; Torsvik et al., 2002). Together, they form complex networks of interactions whose structures have important implications that extend beyond the microbial world. For example when viruses lyse bacteria, they can also redirect organic matter towards the microbial loop and away from higher tropic levels, potentially reducing the productivity of macroscopic organisms (Brum et al., 2016; Fuhrman, 1999; Suttle, 2007; Weitz & Wilhelm, 2012; Wilhelm & Suttle, 1999). Altogether, changes in phage-bacteria infection networks that arise due to coevolution may have ripple effects throughout ecological communities and associated ecosystems.
Viral infection selects for the evolution of phage resistance amongst bacteria (Luria & Delbrück, 1943) that then triggers corresponding evolution of the phage to counter the resistance (Labrie et al., 2010; Luria, 1945). This dynamic triggers cycles of coevolution between phage and bacteria that can continually remodel their interaction network (Hampton et al., 2020), impacting ecosystem processes, the stability of ecological communities and maintenance of microbial diversity (Bohannan & Lenski, 2000; Buckling & Rainey, 2002; Rodriguez-Valera et al., 2009; Stern & Sorek, 2011). There is thus a growing interest in studying the dynamics of phage-bacteria coevolution and understanding the molecular and ecological mechanisms that shape their coevolving networks (Koskella & Brockhurst, 2014).
A starting point to study phage-bacteria coevolution is to characterise how their interactions change over time (Childs et al., 2012; Valverde et al., 2017; Weinberger et al., 2012; Weitz et al., 2005). Models of coevolution typically predict two types of dynamics (Agrawal & Lively, 2002; Weitz et al., 2013). First, arms race dynamics (ARD) where bacteria evolve resistance to an increasing number of phage and phage counter by expanding their host range. For the bacteria, this leads to an escalation where increasingly resistant bacteria replace their less-resistant predecessors through selective sweeps. The increase in resistance is expected to cause rapid bacterial genomic divergence and an imbalanced phylogenetic pattern with a single pronounced branch (Ebert & Fields, 2020). Similarly, as the phage broadens its host range, the most recently evolved type is expected to supplant its predecessors, resulting in the formation of a similarly imbalanced phylogeny, as previously observed (Scanlan et al., 2011). A second model of coevolution is based on the evolution of specialised interactions, often described as lock and key interactions (Weitz et al., 2013). In this model, as co-evolution progresses, bacteria gain resistance to contemporary phage, but lose resistance to phage encountered in the past. Likewise, as phage evolve counter-defences, they lose the ability to infect other host genotypes. Under this model, host genotypes rise and fall according to how abundant their corresponding parasite genotypes are, while parasite genotypes track the abundance of their hosts creating a feedback loop and fluctuating selection dynamics (FSD; Gandon et al., 2008; Sasaki, 2000). FSD produces negative frequency-dependent selection, a type of balancing selection, that is expected to promote diversification and the formation of a balanced phylogeny with multiple branches (Ebert & Fields, 2020). ARD and FSD represent distinct archetypes of possible coevolutionary dynamics and models have been proposed that span the space between the two end points (Agrawal & Lively, 2002; Leung & Weitz, 2016).
One way to gain insight on whether phage and bacteria coevolve according to ARD or FSD is to quantify their interaction networks (phage-bacteria interaction networks; PBINs) and test for non-random nested and modular patterns (Flores et al., 2011). Nestedness measures the extent to which interaction patterns form strict hierarchical subsets, analogous to nesting Russian dolls (Almeida-Neto et al., 2008). This pattern can be produced by ARD since at each step the phage adds on to its existing host range in a way that encapsulates its ancestors’ ranges; likewise, bacteria evolve resistance to previously evolved phage encapsulating its ancestors’ range of phage to which it is resistant. Modularity arises in bipartite networks when groups of phage and bacteria tend to interact significantly more often within clusters than between clusters (Barber, 2007). This pattern is consistent with FSD where interactions are expected to be highly specialised. The majority of PBINs are significantly nested supporting the prominence of ARD; however, some PBINs are modular (Flores et al., 2011), and while rare, specialised interactions have been documented to evolve during phage-bacteria coevolution (Gómez & Buckling, 2011).
The connections between interaction network structure and the dynamics are influenced by network topology; however, other aspects of phage and bacterial biology will influence their dynamics. For example if increases in resistance and/or host range incur a fitness cost, then even nested networks can result in negative frequency-dependent selection and FSD (Agrawal & Lively, 2002; Sasaki, 2000). Given this complication, it is not surprising that the lines delineating ARD and FSD can become blurred. For example nested patterns at short spatial scales can give way to modular patterns at large spatial scales (Flores et al., 2013), and ARD has been shown to give way to FSD during advanced stages of coevolution (Flores et al., 2013; Hall et al., 2011). More generally, the specificity of interaction alone need not necessarily dictate the outcome of coevolutionary dynamics when ecological dynamics are incorporated. Instead, arms races can give way to specialised interactions (Weitz et al., 2005), FSD can arise even in the absence of pre-defined explicit ecological interactions (Best et al., 2017), and finally, the incorporation of explicit ecological feedback can often dampen or transform coevolutionary dynamics (Ashby et al., 2019). Together, this variation and scale-dependence provide a glimpse of the challenges in connecting coevolutionary process to phenotypic outcome, especially in cases where population abundances jointly change along with genotype dynamics.
While the phenotypic predictions for ARD and FSD are often tested, assessments of the phylogenomic predictions are not as common (ARD: imbalanced, FSD: balanced) and we are unaware of an example where PBINs have been coupled with phylogenomic analyses (note that the link between evolutionary interactions and phylogenomic structure is relatively well developed in studies of virus infection of human and animal hosts, sensu Koelle et al., 2006; Volz et al., 2013). Here, we utilised a model phage-bacteria coevolutionary system; bacteriophage λ and its host, Escherichia coli. When these species are cultured under certain laboratory conditions, they rapidly coevolve (Meyer et al., 2012; Meyer & Lenski, 2020). E. coli is known to evolve resistance through mutations in the regulatory gene malT that suppress expression of the host receptor, the outer-membrane protein LamB. λ counters this by evolving mutations in the binding domain of its host recognition protein J that allows it to use a new receptor, OmpF. E. coli then evolves additional mutations in OmpF or in an inner-membrane protein complex, ManXYZ, that transports λ DNA into the cytoplasm (Erni et al., 1987; Esquinas-Rychen & Erni, 2001). While much is already known about the molecular details of their coevolution, little is known on the joint changes in interaction networks and genetic relatedness.
For this study, we revived cryopreserved samples from a previously reported coevolution experiment (Meyer et al., 2012). We focussed our analyses on a single replicate; the first experimental community in which λ evolved to use OmpF. We isolated a total of 50 bacteria and 44 phage spread across multiple time points. Next, we constructed a PBIN of all combinations of pairwise phage and bacteria interactions and used multiple analyses to characterise their coevolution based on phenotypes. All three phenotype-based analyses suggested that viruses and microbes engage in ARD. In parallel, we sequenced the full genomes of each isolate and reconstructed the isolates’ phylogenetic relationships. The genome sequences revealed a phylogenetic pattern that was inconsistent with the ARD model. Our study demonstrates that phenotypic or genomic analyses alone are not sufficient to test hypothesis on coevolutionary dynamics and reveals a hybrid type of coevolutionary dynamics that we refer to as leapfrog dynamics (LFD).
MATERIALS AND METHODS
Details on the initial coevolution experiment previously published
Meyer et al. (2012) performed the original coevolution experiment with E. coli B strain REL606 and a lytic bacteriophage λ strain, cI26. This λ strain was chosen because it cannot initiate lysogeny, a life cycle phase where λ confers immunity to additional λ infections. By choosing a lytic strain, we forced the bacteria to evolve genetic resistance. E. coli and λ were cocultured in a carbon-limited minimal glucose media at 37°C for 37 days (Meyer et al., 2012). At the end of each day, 1% of the community was transferred to new flasks with fresh media, and, weekly, 2 ml of the community was preserved by adding ~15% of glycerol and freezing the mixture at −80°C.
Isolation of host and phage clones
We randomly isolated ten host and eleven phage individuals from different timepoints from the cryopreserved samples. In total, 50 strains of E. coli and 44 strains of λ were isolated from days 8, 15, 22, 28 and 37 of the experiment (no phage were detected on day 37). Bacteria were isolated by streaking onto Luria–Bertani (LB) agar plates (Sambrook & Russell, 2001) and randomly picking 10 colonies. These colonies were re-streaked three times and grown overnight in liquid LB to create stocks. Phage were isolated by plating an appropriate dilution of the population onto overlay plates (Adams, 1959) with the sensitive ancestral bacteria, REL606 and randomly picking 11 plaques. We confirmed the suitability of REL606 for sampling phage diversity and present those findings later in the Results subsection Phage-bacteria infection network. These plaques were grown overnight with REL606 in LBM9 medium, and stocks were created using chloroform isolation technique (Meyer et al., 2012). All phage and bacteria stocks were stored at −80°C with the addition of 15% v/v glycerol.
Pairwise infection assays and efficiency of plating
We performed quantitative, pairwise infection assays for all combinations of host strains and phage strains that were isolated. Specifically, seven serial 1/10th dilutions were made of each phage isolate. 2 μl of each dilution plus undiluted phage stock was spotted on top of different host strain lawns including ancestor REL606. Thus, a total of 8 × 44 spots of phage were plated on 51 different types of bacterial lawns, leading to a total of 17,952 pairwise infections. This design allowed us to quantify phage infectivity of all possible pairs by calculating efficiency of plating (EOP), defined as the ratio of density of phage isolate calculated on a coevolved isolate to the density of phage calculated on the REL606 ancestor.
Analysis of PBIN nestedness and modularity
The BiMat software was used to assess the nestedness and modularity of the PBIN (Flores et al., 2016). The raw EOP value matrix was binarised into 0 for EOP = 0 and 1 for EOP > 0, and then BiMat was run with default settings. Here we report the statistics for a conservative version of the analysis where the rows and columns that contained all zeros were removed from the matrix to reduce any bias these entries cause in establishing significant nested patterns.
Resistance and infectivity calculations
For a total number of n host samples and m phage samples, we denote the EOP value for the ith host sample against jth phage sample as eij where i ∈ [1, n] and j ∈ [1, m]; n = 50 and m = 44. We denote the five checkpoint days of day 8, 15, 22, 28 and 37 for host by k, where k = 1, 2, 3, 4, 5, and the four checkpoint days of day 8, 15, 22 and 28 for phage by l where l = 1, 2, 3, 4. Host resistance for a host sample i is calculated as
where 1X is the indicator function and measures the number of phage strains that the host is resistant to. The host range of a phage sample j is calculated as
which measures the number of host strains that the phage can successfully infect. The resistance percentage of host for each day is calculated as
where denotes the range of the host sample that belongs to the kth sample checkpoint and denotes the cardinality of the set , that is the number of host samples at the kth checkpoint. Likewise, the host range percentage of phage for each day is calculated as
where denotes the range of the phage sample that belongs to the lth checkpoint and denotes the cardinality of the set , that is the number of phage samples at the l th checkpoint.
Genomic DNA preparation for sequencing
The λ genome extraction technique used was previously reported in (Meyer et al., 2016). To summarise, λ particles were concentrated using PEG precipitation, the phage were treated with DNase I to remove free-floating DNA not protected by phage capsids, the DNase is denatured with heat, which also releases capsid-enclosed phage DNA. The DNA was extracted using Invitrogen's PureLink kit. E. coli genomic DNA was extracted and purified from a 1 ml sample of culture by using PureLink kit. Genomic DNA was further processed by fragmenting the DNA and attaching adapters and barcodes using a method outlined in (Baym et al., 2015). Sequencing was done at UC San Diego IGM Genomics using paired-end Illumina HiSeq 4000 platform.
Construction of mutation profile tables
After collecting the raw sequencing reads, we removed the adapters using cutadapt (Martin, 2011) and performed quality control (QC) for each isolated strain using FastQC (Andrews, 2010). The QC filtered sequencing reads were then analysed using breseq (v0.32.1; Deatherage & Barrick, 2014). We ran breseq in the consensus mode with default parameters except for the consensus-frequency-cutoff, which was set to 0.5.
Phylogenomics
Due to the prevalence of large insertions and deletions in the host genomes, conventional nucleotide substitution models were not suitable for estimating the host phylogenetic tree. However, such models were suitable for estimating the maximum-likelihood phylogenetic tree for phage genomes. As a result, two different approaches were taken to reconstruct the evolutionary trajectories of the host and virus. To construct the phage phylogeny, multiple sequence alignments were performed for all recovered genomes and the ancestral genome using mafft (v7.305b; Katoh et al., 2002) with default settings except that ‘retree’ was set to 2 and ‘maxiterate’ was set to 1000. A maximum-likelihood tree was constructed using raxml-ng (Stamatakis, 2014). Finally, the TreeTime (Sagulenko et al., 2018) program was used to generate the phylogenetic tree.
To reconstruct the host's phylogeny, we constructed a Hamming distance matrix to calculate genetic distances between different host isolates. Neighbour-joining (NJ) trees were then built based on the hamming distance matrix using T-REX (Makarenkov, 2001). Finally, the TreeTime program was used to build the host phylogenetic tree.
Whole genome whole population sequencing
We sequenced the full population of phage and bacteria from day 8 of the experiment to 3726-fold and 142-fold coverage respectively. To do this, λ and E. coli populations were revived by growing 120 μl of frozen stock of the whole community in the laboratory conditions from the original experiment (Meyer et al., 2012). Phage and bacteria were then separated, and their genomic DNA was extracted similarly as described before for clonal stocks. UC San Diego IGM Genomics prepared genomic libraries using NexteraXT kit and sequenced the samples using 75 base single reads on the Illumina HiSeq 4000 platform. breseq v0.32.1 was used in polymorphism mode with default settings to analyse whole population sequencing data of day 8 and construct the mutation profile tables.
RESULTS
Phage-bacteria infection network
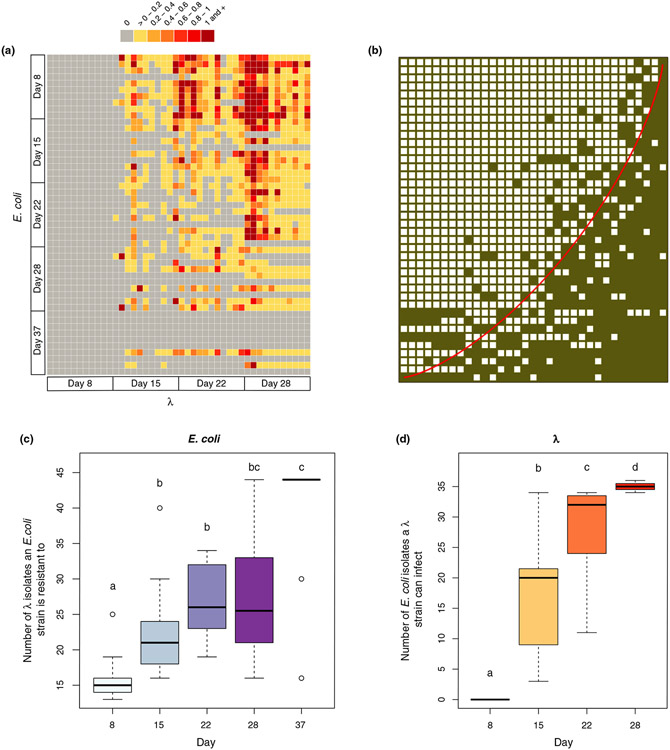
The pairwise interaction study revealed multiple λ genotypes with phenotypically distinct host ranges, and E. coli genotypes that vary in resistance (Figure 1a). We found that the interactions were highly nested (Figure 1b) and had a low level of modularity (Figure S1). In line with ARD, the levels within the nested structure are correlated with time (E. coli evolved increasing resistance (Figure 1c, , F1,48 = 51.01, p = 4.453e-09 for linear model: response ~time), and λ gained increasing host range and infectivity (Figure 1d, , F1,42 = 188.1, p = 2.2e-16) through the course of the study). Note, all mean EOP values were zero for day 8 phage because all isolated hosts were resistant to all day 8 phage (Figures 1a and 2a).
FIGURE 1.
Host resistance and phage infectivity measured by pairwise plaque assays. (A) Phage-bacteria infection network where the colour of each cell is determined by the efficiency of plating (EOP) values obtained for that host-phage interaction pair; grey cells represent no infection by λ on the given Escherichia coli strain, yellow represents low infectivity and red represents high infectivity. (B) The original network in (a) reassembled using the software BiMat to visualise maximal nestedness (Flores et al., 2016). Filled squares indicate a combination of host and phage that result in successful interactions (EOP >0), and the red line highlights the isocline using the nestedness temperature calculator algorithm. The nestedness value of the network utilises the nestedness based on overlapping and decreasing fill metric, which was significantly greater than the null expectation when constraining the fill of the bipartite network (measured value of nestedness 0.839 vs. null value of nestedness 0.638 ± 0.011 based on 200 trials). (C) Boxplots showing the total number of λ isolates from all days that E. coli genotypes are resistant to across different sampling days. (D) Boxplots showing the total number of E. coli isolates from all days that λ genotypes can infect across different sampling days. Lowercase letters in (c) and (d) denote significant difference between different days via Tukey's honest significance test: (c) ANOVA: F4,45 = 13.3, p = 3.11e-07, (d) ANOVA: F3,40 = 67.05, p = 1.17e-15. A simple linear regression model with time as the predictor variable was also used to test if E. coli evolved increasing resistance in (c) and λ evolved increasing host range in (d) (statistics in the main text)
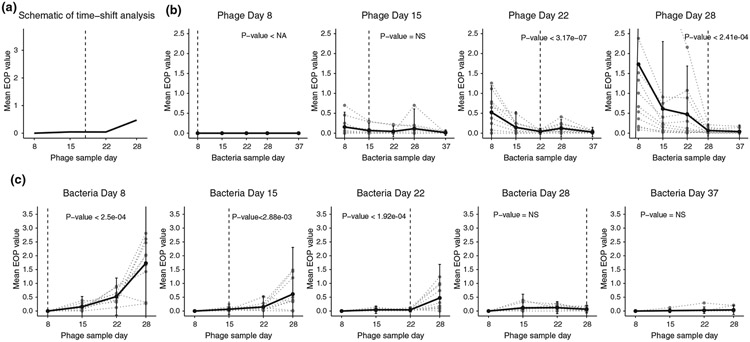
FIGURE 2.
Time-shift analysis results from different checkpoints. (a) Schematic for the time-shift analysis that compares the mean efficiency of plating (EOP) from hosts or phage interacting with their counterparts from the past, contemporary and the future. (b) Time-shift results from phage checkpoints day 8, 15, 22 and 28 respectively. The grey dotted line shows the time-shift curve for each individual phage and the black line shows the average. The vertical dashed line represents the phage sample day. The p-values shown here are the maximum p-value from one-sided paired t-tests comparing the initial checkpoints with each of the later checkpoints. (c) Time-shift results from host checkpoints day 8, 15, 22, 28 and 37 respectively. The grey dotted line shows the time-shift curve for each individual host and the black line shows the average. The vertical dashed line represents the host sample day. The p-values shown here are the maximum p-value from one-sided paired t-tests comparing the final checkpoints with each of the previous checkpoints
We further performed a time-shift analysis using EOP values to determine how phage infectivity varies when presented with past, contemporary, or future bacteria (Figure 2a; Gaba & Ebert, 2009). ARD predicts that phage will be able to infect past and contemporary, but not future hosts, while FSD predicts that phage will be best at infecting contemporary hosts. The time-shift analysis was conducted for each λ isolate by calculating its mean EOP value for all 10 bacterial isolates on each day (Figure 2b). This analysis was repeated for the bacteria using the same EOP data but by calculating levels of resistance to λ isolates from different time points (Figure 2c). In line with ARD, λ isolates from days 22 and 28 had higher infectivity on past hosts than contemporary or future hosts (Figure 2b). The analysis for day 15 phage was inconclusive because the variation in EOP between isolates was too large to detect statistically significant differences in average EOP across time. The pattern for bacteria was also in line with ARD: isolates from days 8, 15 and 22, had lower resistance (higher EOP) for phage samples from the future versus the phage isolated from the same time or in the past (Figure 2c). A full time-shift analysis could not be conducted for isolates from days 28 and 37 since the phage went extinct between days 28 and 37; however, E. coli isolates from these time points were the most resistant.
At all timepoints, phages were isolated using the ancestral host (REL606) in an effort to minimise sampling bias. In previous studies we found that all evolved phages were able to infect the ancestor (Flores et al., 2011). However, if phages evolve to specialise on coevolved bacterial genotypes in line with FSD, the phages may lose the ability to infect the bacterial ancestor and would not be sampled, thereby artificially reducing support for FSD. To test whether phages evolved that lost the ability to infect REL606, we isolated phages using lawns of all unique E. coli genotypes (16 unique genotypes, however no plaques formed on 3). Eight phages were sampled from each host yielding a sample size of 104 phage isolates. Each phage isolate was able to form plaques on REL606 (Table S3). An additional efficiency of plaquing analyses was performed for one phage isolate from each host. This analysis revealed that most phages were more likely to produce plaques on REL606 than on the coevolved hosts from which they were isolated (Table S3). Together, these results suggest that REL606 was suitable for sampling the coevolved phage diversity in this system.
Genome sequencing
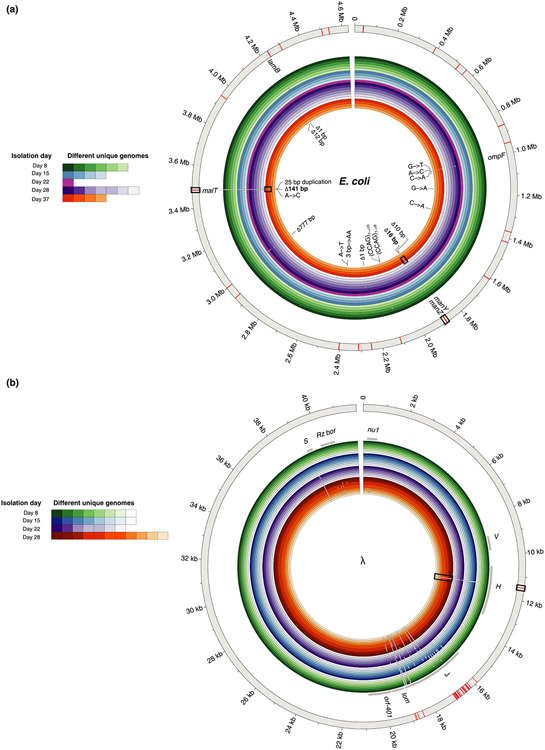
The genome sequencing revealed 22 unique E. coli genomes and 34 unique phage genomes. Among the E. coli strains, we found a total of 18 unique mutations: six missense mutations, one nonsense mutation, one intergenic point mutation, seven deletions and three duplications (Figure 3a). The most abundant mutation that occurred in 38 out of 50 host genomes was a frameshift mutation caused by a 25-base duplication in the malT gene, in line with the original study (Meyer et al., 2012). Disruptions in malT interferes with the expression of LamB protein which ancestral λ needs to bind to E. coli cells. We also observed one isolate with a lamB mutation (1-base deletion) in lieu of the typical malT mutation. The most resistant E. coli strains on day 37 have multiple mutations that are expected to confer resistance; a malT deletion, a non-synonymous change in ompF, and a deletion in manZ (Burmeister et al., 2021; Meyer et al., 2012).
FIGURE 3.
Genomic diversity in clones isolated from different days and whole population sequencing for (a) Escherichia coli and (b) λ. The outermost grey ring represents the reference genome. The inner coloured rings represent the isolates sequenced from different time points (outer rings are genomes isolated from earlier time points). Shades within each colour depict unique genomes sequenced from each time point. White gaps in the genomic rings indicate the location of mutations. All 18 unique mutations found in clonal isolates have been labelled for E. coli in (a); however, due to the large number of mutations in λ, only the gene names that harbour mutations from clonal isolates have been identified (grey bars). Note that white gaps corresponding to mutations are larger than a single base, so occasionally a single gap represents multiple adjacent mutations. The red bars in the outermost grey ring indicate the placement of mutations uncovered by the whole population sequencing at day 8. The mutations that become dominant at later stages of coevolution and were also found in day 8 population sequencing have been highlighted with rectangular boxes
A common bacterial mutation observed in our data set was a 777 bp deletion caused by the excision of an IS element. Despite occurring in 25 genomes, this deletion was not observed in our previous studies and does not affect any genes known to protect against λ infection; insB-22 which encodes for IS1 protein InsB, insA-22 which encodes for IS1 protein InsA and ECB_02825 which encodes for a pyrophosphorylase (Maynard et al., 2010). Similar IS mutations are known to occur at high rates (Cooper et al., 2001) and the deletion was only observed in genomes with a malT mutation, suggesting it was a neutral genomic hitchhiker. However, when we tested this hypothesis in a follow-up experiment, we discovered that this conclusion was wrong. The deletion enhances resistance when it co-occurs with the malT mutation, signifying the deletion was adaptive and epistatic with malT mutations (Figure S2). We are unsure of how the deletion confers resistance; however, this result suggests that experimental evolution could be used to reveal novel interactions in the λ-E. coli interactome.
In the λ genomes, we found a total of 176 unique mutations: 53 non-synonymous point mutations, 87 synonymous point mutations, 2 insertions, 3 deletions and 31 intergenic mutations (Figure 3b). While this level of molecular evolution may seem surprisingly high for such a short-term experiment, similar levels have been observed for other phage evolving in the laboratory (Wichman et al., 2005). 116 of these mutations were in the host-recognition gene J. The J protein lies at the end of the phage's tail, and initiates infection by binding to E. coli's LamB protein. Some of these J mutations have been shown to increase adsorption rates to LamB and allow λ to exploit a novel receptor, OmpF (Burmeister et al., 2016; Maddamsetti et al., 2018; Petrie et al., 2018). Interestingly, our extensive sequencing effort revealed a mutation in another tail fibre protein called H (C → T substitution at nucleotide position 11,451). This mutation rises late in the experiment and likely plays a role in expanding λ’s host range. The tape-measure gene H helps determine the length of λ’s tail, and mutations in this gene have been shown to increase λ’s host range (Scandella & Arber, 1976).
Phylogenomic reconstruction of coevolution
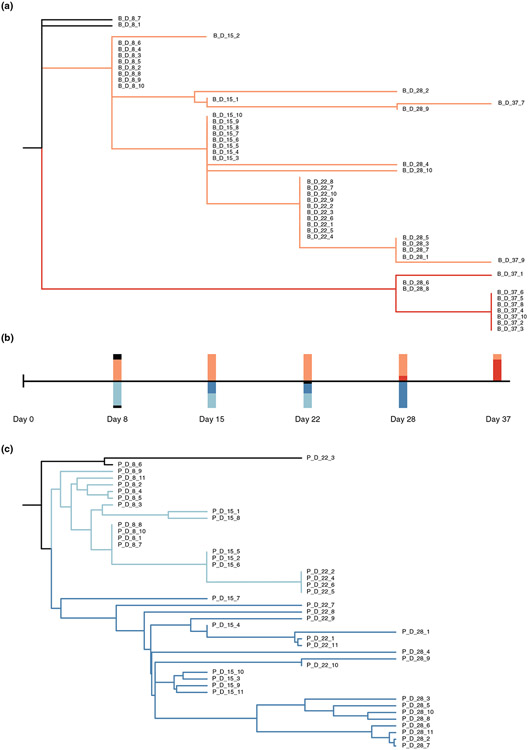
Even though multiple analysis of the phenotypic data supported the ARD model for coevolution, the macro pattern produced by the phylogenies are in line with predictions of FSD (Figure 4), suggesting a hybrid dynamic. The phylogenies of both E. coli and λ show that multiple lineages coexist for weeks, rather than a single dominant branch. Another unexpected observation was that the bacteria that had acquired the highest level of resistance at the end of the experiment on day 37 was most closely related to a common ancestor of isolates identified at the early stages of the experiment (on day 8) than isolates from time points closer to day 37 (e.g. days 28, 22 or 15; compare Figures 1a with 4a). This finding suggests that a rare lineage leapt ahead of the dominant lineage, a process we term LFD. Similarly, for λ, we found that that the clade dominant at the final timepoint with the broadest host range was more closely related to wildtype λ than the clade dominant at preceding timepoints (comparison of Figures 1a with 4c). For both species, the clades that win out later in the arms race appeared to persist as cryptic subpopulations early in the coevolutionary experiment. Within the larger LFD pattern, there appears to be progressive evolution of phage and bacteria within each lineage in line with ARD. This suggests that selection is directional during coevolution to favour increased resistance and host-range. Sometimes selection may favour the rise of variants pulled from the dominant lineage, and other times selection favours variants from cryptic lineages (consistent with earlier findings of non-monotonic dynamics, Betts et al., 2014).
FIGURE 4.
Reconstructed phylogenetic trees of the host and phage. (a) The host phylogenetic tree based on host mutation profiles. All completely resistant host strains are located on the red branch. Bars above the time scale in (b) represents the proportion of host strains from each coloured branch across different checkpoints. (c) The phage phylogenetic tree based on the phage mutation profiles. All day 28 phage strains are located on the dark blue branch. Bars below the time scale in (b) represents the proportion of phage strains from each coloured branch across different checkpoints
Whole population sequencing at an early stage of coevolution
To test the key prediction of LFD that cryptic lineages coexist with dominant lineages and can supply the genetic reservoir used for later stages of coevolution, we sequenced full populations of E. coli and λ from day 8 and searched for mutations that rose to prominence at the end of the study. For E. coli, we searched for two mutations: a Δ16 bp deletion at position 1,882,915 in manZ and a non-synonymous mutation at 1,003,271 in ompF. These mutations are present in most of the day 37 isolates and disruptions to both genes separately have been shown to block λ infection (Borin et al., 2021; Burmeister et al., 2021; Meyer et al., 2012), suggesting their combination should cause high levels of resistance. The Δ16 bp deletion in manZ was detected, but not the ompF mutation (Table S1). We also found a 141 bp deletion in malT that co-occurs in the day 37 genomes with the manZ mutation. The malT deletion was at the same frequency and the manZ mutation, suggesting that these mutations were indeed linked and that they evolved sometime before day 8. For λ, we focused on the mutation in H that rises to dominance between days 22 and 28. Indeed, this specific H mutation was present at day 8 (Table S2). Unlike E. coli, we did not find any other mutations present in the day 28 isolates, suggesting that the H mutation was the first adaptation to occur in this lineage.
Besides revealing the eventual mutations associated with the prevailing bacterial lineages of the arms race, we also discovered greater genetic diversity via whole population sequencing than through isolate sequencing (as anticipated). Whole genome sequencing revealed 52 unique mutations in E. coli and 38 mutations in λ from full population sequencing compared to 7 and 30 through isolate sampling respectively. The combined sequencing strategy suggests that there is significant genetic diversity generated at the earliest phases of the arms race that can fuel subsequent adaptation as the host (for phage) or phage (for hosts) change as a result of coevolution.
DISCUSSION
Through large scale phenotypic assays and whole genome sequencing, we were able to test existing paradigmatic models of coevolution and learn that λ and E. coli's coevolutionary dynamics possessed characteristics of both ARD and FSD. Three complimentary phenotypic analyses in Figures 1 and 2 suggested that coevolution between λ and E. coli followed ARD. However, the phylogenetic pattern revealed by whole genome sequencing did not reveal a series of selective sweeps in line with ARD, rather multiple lineages coexisted consistent with balancing selection that arises from FSD (Figure 3). These observations lead us to develop a hybrid conceptual model to characterise λ and E. coli's coevolution: which we term LFD. In this model, parasite genotypes with ever-expanding host ranges are selected and hosts with ever-increasing resistance are favoured. However, there is a genetically diverse pool of hosts and parasites that evolve early, and on occasion, rare individuals with advantageous phenotypes drawn from this pool replace the dominant strains. This is a hybrid model because the phylogenomics pattern points to FSD, yet selection at the phenotypic level operates similarly to the ARD model.
Arms race dynamics models fall short in making accurate predictions for the phylogenies likely because of simplifying assumptions about the genetics of host range expansion and resistance. Evolutionary models tend to assume that mutations have additive effects on phenotypes (as highlighted in Weitz et al., 2013). Applied to ARD, this would mean that the phage with the broadest host range is likely to expand its host range faster than lagging genotypes, and for bacteria, the strain with the greatest resistance is most likely to acquire the next level of resistance and outcompete other strains. This genetic architecture favours the evolution of directed phylogenies with one dominant branch. Instead, λ J mutations are known to possess high-order epistasis and are non-additive (Maddamsetti et al., 2018; Meyer et al., 2016). This may allow rare lineages with certain combinations of mutations to suddenly leap ahead if new modifications have synergistic interactions with pre-existing mutations. Non-additivity was also discovered in the bacteria with respect to the interactions between malT− and the Δ777 mutations (Figure S2). A second problem with evolutionary models is the assumption of small effect-size mutations since mutations like the malT mutation can cause nearly complete resistance to some λ phage (Figure 1a). Acquiring such large effect mutations could also help lineages leap ahead. Lastly, these models typically do not incorporate recombination, which was observed within this phage population to cause sudden increases in fitness (Borin et al., 2021) and could contribute to the phage's ability to leap ahead. In contrast, evidence of recombination was not observed in the bacterial genome sequences, consistent with earlier findings in which recombination is not known to occur in this strain of E. coli (Souza et al., 1997).
A key question left unanswered by this study is how multiple lineages persisted in this population for long durations. An explanation is clonal interference where multiple lineages of nearly equal fitness contend with one another for prolonged periods of time until one eventually wins out (Good et al., 2017; Maddamsetti et al., 2015). A second is that trade-offs between host range and other viral traits for phage, and resistance and competitive fitness for bacteria (Bohannan et al., 2002; Lenski & Levin, 1985; Meyer & Lenski, 2020), could explain the evolution of genomic diversity. Indeed, trade-offs between host range and λ stability were previously observed (Petrie et al., 2018); however, we were unable to detect trade-offs in E. coli for phage resistance by pairwise competition experiments. Future studies with higher resolution sampling (e.g. daily) at greater depths (e.g. full communities as performed for day 8) will improve quantification of the timing of the selection of novel mutations, frequency changes in mutations over time and genomic linkages (e.g. Lang et al., 2013). This information would help reveal the ecological context in which mutations emerge, genotypes change in frequency and potentially uncover hidden trade-offs.
It remains unclear how prevalent LFD is in coevolving host–parasite systems. For this work we chose to intensively sample a single coevolving community because in previous studies we characterised dynamics in many populations (Meyer et al., 2012). Increased sampling depth allowed us to reveal LFD; however, we do not know how common it is for cryptic diversity to arise early during coevolution and then to fuel the late-stage dynamics. A related phage evolution experiment revealed a relatively large number of cooccurring genetic variants from a single time point, but it is not known what contribution this variation played in the coevolutionary dynamics (Paterson et al., 2010). Recent genomic barcoding technologies have allowed more in-depth sampling of genetic diversity in microbial populations. This increased resolution has revealed the rapid evolution of an enormous number of adaptive mutation, much more than previously anticipated (Levy et al., 2015). This result and our findings suggest that cryptic genetic variation may play a larger role during evolution than is currently appreciated.
One of the challenges in establishing a link between pattern and process is that the nested pattern typically associated with FSD is ubiquitous in PBINs (Flores et al., 2011) and many other ecological networks (Bascompte et al., 2003; Guimarães et al., 2007; James et al., 2012). One hypothesis is that the structure is determined by the genetics of the interactions as in the gene-for-gene mechanism of coevolution (Weitz et al., 2013). An alternative hypothesis is ecological: nestedness emerges because of how the selection steadily shifts during an arms race to promote incremental increases in host range and resistance. The conventional wisdom is that genetics constrains coevolution and controls the pattern formation since mutations and evolution are thought to occur more slowly than changes in ecology. The coevolution experiment studied here was initiated with small populations of isogenic stocks of λ and E. coli that should have favoured genetic control because the genetic variation had to evolve de novo. However, significant genetic variation was generated at the earliest phase of the arms race that appeared to remain cryptic in an ecological sense for many generations, suggesting that the ecology controlled the dynamics, not the availability of genetic variation.
These results provide a cautionary tale for over-interpreting data derived from a single source—either genotypic or phenotypic. Our initial prediction before performing genomic analysis was that λ-E. coli coevolution under these laboratory conditions would fit the ARD model. However, this proved to be erroneous due to the large amount of cryptic genetic variation and its role in driving late stages of coevolution. This finding corroborates earlier suggestions that linking process and pattern may require examination of time-shift experiments at varying time scales to detect coevolutionary signals (Betts et al., 2014). A second observation also revealed a potential limitation of using PBIN data for making dynamical predictions. Based on the matrix in Figure 1a one would predict that the phage should go extinct on day 8. Still, λ maintained a population due to a phenomenon known as ‘leaky-resistance’ (Chaudhry et al., 2018). Leaky resistance occurs when a small number of resistant hosts revert to sensitive, thereby sustaining phage in the population until phage evolve to gain access to a new receptor, for example OmpF in the case of phage λ (Meyer et al., 2012). Together, the mechanisms of LFD and leaky-resistance show that host-phage dynamics revealed from PBINs alone miss the rich dynamics occurring at lower frequencies in the population.
Analogous to our warning about relying solely on phenotypic information, the same is true for genomic data. J’s large genetic diversity and the phylogenomic pattern of both E. coli and λ suggest FSD; however, selection is directional during coevolution in line with ARD. As genome sequencing becomes more affordable, researchers may forgo laborious phenotypic assays, assuming that the genomic data provide greater resolution. However, as shown here and argued in a recent review (Ebert & Fields, 2020), integration of phenotypic and genomic are required to test hypothesis about host–parasite coevolution.
In summary, by studying coevolving phage and bacterial populations with both phenotypic and genomic approaches we revealed coevolutionary patterns that are not wholly explained by archetypical models of coevolution. We found that phenotypic or genomic assays alone fall short in characterising the underlying nature of coevolution—given the potential for cryptic genetic variation to fundamentally alter coevolutionary trajectories. We also showed that highly organised ecological pattern like nestedness can emerge despite the apparent absence of fundamental genetic constraints, demonstrating the power of selection in driving emergent ecological patterns. Moving forward, these results suggest the critical need to incorporate both phenotypic and phylogenomic approaches in evaluating phage-bacteria coevolution.
Supplementary Material
ACKNOWLEDGEMENT
We thank Morgan Mouchka for her help with sequencing genomes of the whole community.
Funding information
We thank the NSF-DEB 1934515 (to JRM) and ARO W911NF1910384 (to JSW) for financial support.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing interests.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
DATA AVAILABILITY STATEMENT
All sequencing data have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA791964. Data that support the findings of this study are deposited at Dryad at https://doi.org/10.6076/D1S596 and all code is available at GitHub at https://github.com/anigupta12/leapfrog-dynamics-paper. Biological material is available from J.R.M. under a material transfer agreement with UC San Diego (http://blink.ucsd.edu/research/conducting-research/mta/index.html).
REFERENCES
- Adams MH (1959) Bacteriophages. New York: Interscience Publishers Inc. [Google Scholar]
- Agrawal A & Lively CM (2002) Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evolutionary Ecology Research, 4, 79–90. [Google Scholar]
- Almeida-Neto M, Guimarães P, Guimarães PR Jr, Loyola RD & Ulrich W (2008) A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos, 117, 1227–1239. [Google Scholar]
- Andrews S (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Accessed May 2018].
- Ashby B, Iritani R, Best A, White A & Boots M (2019) Understanding the role of eco-evolutionary feedbacks in host-parasite coevolution. Journal of Theoretical Biology, 464, 115–125. [DOI] [PubMed] [Google Scholar]
- Barber MJ (2007) Modularity and community detection in bipartite networks. Physical Review E, 76, 066102. [DOI] [PubMed] [Google Scholar]
- Bascompte J, Jordano P, Melián CJ & Olesen JM (2003) The nested assembly of plant-animal mutualistic networks. Proceedings of the National Academy of Sciences of the United States of America, 100, 9383–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM & Kishony R (2015) Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One, 10(5), e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best A, Ashby B, White A, Bowers R, Buckling A, Koskella B et al. (2017) Host-parasite fluctuating selection in the absence of specificity. Proceedings of the Royal Society B: Biological Sciences, 284, 20171615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts A, Kaltz O & Hochberg ME (2014) Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proceedings of the National Academy of Sciences of the United States of America, 111, 11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannan BJM, Kerr B, Jessup CM, Hughes JB & Sandvik G (2002) Trade-offs and coexistence in microbial microcosms. Antonie van Leeuwenhoek, 81, 107–115. [DOI] [PubMed] [Google Scholar]
- Bohannan BJM & Lenski RE (2000) Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecology Letters, 3, 362–377. [Google Scholar]
- Borin JM, Avrani S, Barrick JE, Petrie KL & Meyer JR (2021) Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proceedings of the National Academy of Sciences of the United States of America, 118, e2104592118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brum JR, Hurwitz BL, Schofield O, Ducklow HW & Sullivan MB (2016) Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. The ISME Journal, 10, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A & Rainey PB (2002) The role of parasites in sympatric and allopatric host diversification. Nature, 420, 496–499. [DOI] [PubMed] [Google Scholar]
- Burmeister AR, Lenski RE & Meyer JR (2016) Host coevolution alters the adaptive landscape of a virus. Proceedings of the Royal Society B: Biological Sciences, 283(1839), 20161528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister AR, Sullivan RM, Gallie J & Lenski RE (2021) Sustained coevolution of phage Lambda and Escherichia coli involves inner- as well as outer-membrane defences and counter-defences. Microbiology, 167, 001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry WN, Pleška M, Shah NN, Weiss H, McCall IC, Meyer JR et al. (2018) Leaky resistance and the conditions for the existence of lytic bacteriophage. PLOS Biology, 16, e2005971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs LM, Held NL, Young MJ, Whitaker RJ & Weitz JS (2012) Multiscale model of CRISPR-induced coevolutionary dynamics: diversification at the interface of Lamarck and Darwin. Evolution, 66, 2015–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie MRJ, Millard AD, Letarov AV & Heaphy S (2011) Phages in nature. Bacteriophage, 1, 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M & Lenski RE (2001) Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. Journal of Bacteriology, 183, 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE & Barrick JE (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods in Molecular Biology, 1151, 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D & Fields PD (2020) Host–parasite co-evolution and its genomic signature. Nature Reviews Genetics, 21, 754–768. [DOI] [PubMed] [Google Scholar]
- Erni B, Zanolari B & Kocher HP (1987) The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. Journal of Biological Chemistry, 262, 5238–5247. [PubMed] [Google Scholar]
- Esquinas-Rychen M & Erni B (2001) Facilitation of bacteriophage lambda DNA injection by inner membrane proteins of the bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS). Journal of Molecular Microbiology and Biotechnology, 3, 361–370. [PubMed] [Google Scholar]
- Flores CO, Meyer JR, Valverde S, Farr L & Weitz JS (2011) Statistical structure of host-phage interactions. Proceedings of the National Academy of Sciences of the United States of America, 108, E288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores CO, Poisot T, Valverde S & Weitz JS (2016) BiMat: a MATLAB package to facilitate the analysis of bipartite networks. Methods in Ecology and Evolution, 7, 127–132. [Google Scholar]
- Flores CO, Valverde S & Weitz JS (2013) Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phages. The ISME Journal, 7, 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA (1999) Marine viruses and their biogeochemical and ecological effects. Nature, 399, 541–548. [DOI] [PubMed] [Google Scholar]
- Gaba S & Ebert D (2009) Time-shift experiments as a tool to study antagonistic coevolution. Trends in Ecology & Evolution, 24, 226–232. [DOI] [PubMed] [Google Scholar]
- Gandon S, Buckling A, Decaestecker E & Day T (2008) Host-parasite coevolution and patterns of adaptation across time and space. Journal of Evolutionary Biology, 21, 1861–1866. [DOI] [PubMed] [Google Scholar]
- Gómez P & Buckling A (2011) Bacteria-phage antagonistic coevolution in soil. Science, 332, 106–109. [DOI] [PubMed] [Google Scholar]
- Good BH, McDonald MJ, Barrick JE, Lenski RE & Desai MM (2017) The dynamics of molecular evolution over 60,000 generations. Nature, 551, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães PR Jr, Sazima C, dos Reis SF & Sazima I (2007) The nested structure of marine cleaning symbiosis: is it like flowers and bees? Biology Letters, 3, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AR, Scanlan PD, Morgan AD & Buckling A (2011) Host-parasite coevolutionary arms races give way to fluctuating selection. Ecology Letters, 14, 635–642. [DOI] [PubMed] [Google Scholar]
- Hampton HG, Watson BNJ & Fineran PC (2020) The arms race between bacteria and their phage foes. Nature, 577, 327–336. [DOI] [PubMed] [Google Scholar]
- James A, Pitchford JW & Plank MJ (2012) Disentangling nestedness from models of ecological complexity. Nature, 487, 227–230. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K & Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle K, Cobey S, Grenfell B & Pascual M (2006) Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science, 314, 1898. [DOI] [PubMed] [Google Scholar]
- Koskella B & Brockhurst MA (2014) Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiology Reviews, 38, 916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE & Moineau S (2010) Bacteriophage resistance mechanisms. Nature Reviews Microbiology, 8, 317–327. [DOI] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D et al. (2013) Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature, 500, 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE & Levin BR (1985) Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. The American Naturalist, 125, 585–602. [Google Scholar]
- Leung CY & Weitz JS (2016) Conflicting attachment and the growth of bipartite networks. Physical Review E, 93, 032303. [DOI] [PubMed] [Google Scholar]
- Levy SF, Blundell JR, Venkataram S, Petrov DA, Fisher DS & Sherlock G (2015) Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature, 519, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE (1945) Mutations of bacterial viruses affecting their host range. Genetics, 30, 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE & Delbrück M (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics, 28, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddamsetti R, Johnson DT, Spielman SJ, Petrie KL, Marks DS & Meyer JR (2018) Gain-of-function experiments with bacteriophage lambda uncover residues under diversifying selection in nature. Evolution, 72, 2234–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddamsetti R, Lenski RE & Barrick JE (2015) Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics, 200, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenkov V (2001) T-REX: reconstructing and visualizing phylogenetic trees and reticulation networks. Bioinformatics, 17, 664–668. [DOI] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet Journal, 17, 10–12. [Google Scholar]
- Maynard ND, Birch EW, Sanghvi JC, Chen L, Gutschow MV & Covert MW (2010) A forward-genetic screen and dynamic analysis of lambda phage host-dependencies reveals an extensive interaction network and a new anti-viral strategy. Plos Genetics, 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Dobias DT, Medina SJ, Servilio L, Gupta A & Lenski RE (2016) Ecological speciation of bacteriophage lambda in allopatry and sympatry. Science, 354, 1301–1304. [DOI] [PubMed] [Google Scholar]
- Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT & Lenski RE (2012) Repeatability and contingency in the evolution of a key innovation in phage lambda. Science, 335, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR & Lenski RE (2020) Subtle environmental differences have cascading effects on the ecology and evolution of a model microbial community. In: Banzhaf W, Cheng BHC, Deb K, Holekamp KE, Lenski RE, Ofria C, Pennock RT, Punch WF & Whittaker DJ (Eds.) Evolution in action: past, present and future: a Festschrift in Honor of Erik D. Goodman. Cham: Springer International Publishing, pp. 273–288. [Google Scholar]
- Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR et al. (2010) Antagonistic coevolution accelerates molecular evolution. Nature, 464, 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie KL, Palmer ND, Johnson DT, Medina SJ, Yan SJ, Li V et al. (2018) Destabilizing mutations encode nongenetic variation that drives evolutionary innovation. Science, 359, 1542–1545. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F et al. (2009) Explaining microbial population genomics through phage predation. Nature Reviews Microbiology, 7, 828–836. [DOI] [PubMed] [Google Scholar]
- Sagulenko P, Puller V & Neher RA (2018) TreeTime: maximum-likelihood phylodynamic analysis. Virus Evolution, 4, vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J & Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sasaki A (2000) Host-parasite coevolution in a multilocus gene-for-gene system. Proceedings of the Royal Society of London. Series B: Biological Sciences, 267, 2183–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandella D & Arber W (1976) Phage λ DNA injection into Escherichia coli pel—mutants is restored by mutations in phage genes V or H. Virology, 69, 206–215. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Hall AR, Lopez-Pascua LDC & Buckling A (2011) Genetic basis of infectivity evolution in a bacteriophage. Molecular Ecology, 20, 981–989. [DOI] [PubMed] [Google Scholar]
- Souza V, Turner PE & Lenski RE (1997) Long-term experimental evolution in Escherichia coli. V. Effects of recombination with immigrant genotypes on the rate of bacterial evolution. Journal of Evolutionary Biology, 10, 743–769. [Google Scholar]
- Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A & Sorek R (2011) The phage-host arms race: shaping the evolution of microbes. BioEssays, 33, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA (2007) Marine viruses—major players in the global ecosystem. Nature Reviews Microbiology, 5, 801–812. [DOI] [PubMed] [Google Scholar]
- Thomas R, Berdjeb L, Sime-Ngando T & Jacquet S (2011) Viral abundance, production, decay rates and life strategies (lysogeny versus lysis) in Lake Bourget (France). Environmental Microbiology, 13, 616–630. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Øvreås L & Thingstad TF (2002) Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science, 296, 1064–1066. [DOI] [PubMed] [Google Scholar]
- Valverde S, Elena SF & Solé R (2017) Spatially induced nestedness in a neutral model of phage-bacteria networks. Virus Evolution, 3, vex021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz EM, Koelle K & Bedford T (2013) Viral phylodynamics. PLoS Computational Biology, 9, e1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger AD, Sun CL, Pluciński MM, Denef VJ, Thomas BC, Horvath P et al. (2012) Persisting viral sequences shape microbial CRISPR-based immunity. PLOS Computational Biology, 8, e1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz JS, Hartman H & Levin SA (2005) Coevolutionary arms races between bacteria and bacteriophage. Proceedings of the National Academy of Sciences of the United States of America, 102, 9535–9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB et al. (2013) Phage-bacteria infection networks. Trends in Microbiology, 21, 82–91. [DOI] [PubMed] [Google Scholar]
- Weitz JS & Wilhelm SW (2012) Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biology Reports, 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman HA, Millstein J & Bull JJ (2005) Adaptive molecular evolution for 13,000 phage generations. Genetics, 170, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm SW & Suttle CA (1999) Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. BioScience, 49, 781–788. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA791964. Data that support the findings of this study are deposited at Dryad at https://doi.org/10.6076/D1S596 and all code is available at GitHub at https://github.com/anigupta12/leapfrog-dynamics-paper. Biological material is available from J.R.M. under a material transfer agreement with UC San Diego (http://blink.ucsd.edu/research/conducting-research/mta/index.html).