Abstract
The COVID-19 pandemic has significantly impacted human health for three years. To mitigate the spread of SARS-CoV-2, the development of neutralizing antibodies has been accelerated, including the exploration of alternative antibody formats such as single-domain antibodies. In this study, we identified variable new antigen receptors (VNARs) specific for the receptor binding domain (RBD) of SARS-CoV-2 by immunizing a banded houndshark (Triakis scyllium) with recombinant wild-type RBD. Notably, the CoV2NAR-1 clone showed high binding affinities in the nanomolar range to various RBDs and demonstrated neutralizing activity against SARS-CoV-2 pseudoviruses. These results highlight the potential of the banded houndshark as an animal model for the development of VNAR-based therapeutics or diagnostics against future pandemics.
Keywords: Variable new antigen receptor (VNAR), Phage display immune library, Banded houndshark (Triakis scyllium), SARS-CoV-2, COVID-19, Single domain antibody
1. Introduction
The COVID-19 pandemic caused by SARS-CoV-2 has resulted in a significant global health crisis, with over 760 million cases and 6.9 million deaths reported worldwide [1]. To combat the pandemic, researchers have sought to develop antibodies against the SARS-CoV-2 receptor binding domain (RBD) for use in viral therapeutics and diagnostics [2,3], including single-domain antibodies (sdAbs). SdAbs are antibody fragments of heavy chain-only antibodies derived from camelid IgG2 and IgG3 [[4], [5], [6], [7], [8], [9], [10]], or cartilaginous fish immunoglobulin new antigen receptor (IgNAR) [[11], [12], [13], [14], [15], [16]]. SdAbs obtained from camelids or cartilaginous fishes are known as nanobodies or VNARs, respectively. SdAbs possess several advantageous properties that make them attractive for use in therapeutics and diagnostics, including small size, high physicochemical stability, solubility, specificity for cryptic epitopes, low immunogenicity, ease of protein engineering, more efficient cell penetration, and low production cost [17,18]. Because of these advantages, some researchers have attempted to develop multivalent [6,14,16] or nasal drugs [7] using SARS-CoV-2 RBD-specific sdAbs.
A primary reason medical and biotechnological researchers prefer nanobodies is the relatively limited availability of animal models for discovering VNARs [19]. Although various IgNARs have been discovered in numerous cartilaginous fish species [20], the six reported SARS-CoV-2 specific VNARs were derived exclusively from the nurse shark (Ginglymostoma cirratum) [11,12,16], white-spotted bamboo shark (Chiloscyllium plagiosum) [13,14], and a synthetic library [15]. Nurse and white-spotted bamboo sharks are suitable animal models for immunization; however, these species have not been found in the coastal areas of Korea [21]. Nurse shark is restricted to the western and eastern Atlantic areas, while white-spotted bamboo shark has been found naturally in the Indian Ocean and central Indo-Pacific [21]. Therefore, we selected banded houndshark (Triakis scyllium) as another animal model for discovering SARS-CoV-2 RBD-specific VNARs. Banded houndsharks frequently inhabit the coastal waters of Korea [21], pose no threat to humans, and adapt well to captivity [22,23]. Interestingly, immunization of banded houndsharks has not been previously reported [19], although their IgNARs have been identified [[24], [25], [26]]. In this study, we isolated several SARS-CoV-2 RBD-specific VNARs from the banded houndshark, including CoV2NAR-1, which exhibited high binding affinity and neutralizing activity against SARS-CoV-2 pseudoviruses. These findings suggest that the banded houndshark could serve as a valuable animal model for the development of VNAR-based therapeutics or diagnostics to combat against future pandemics.
2. Results and discussion
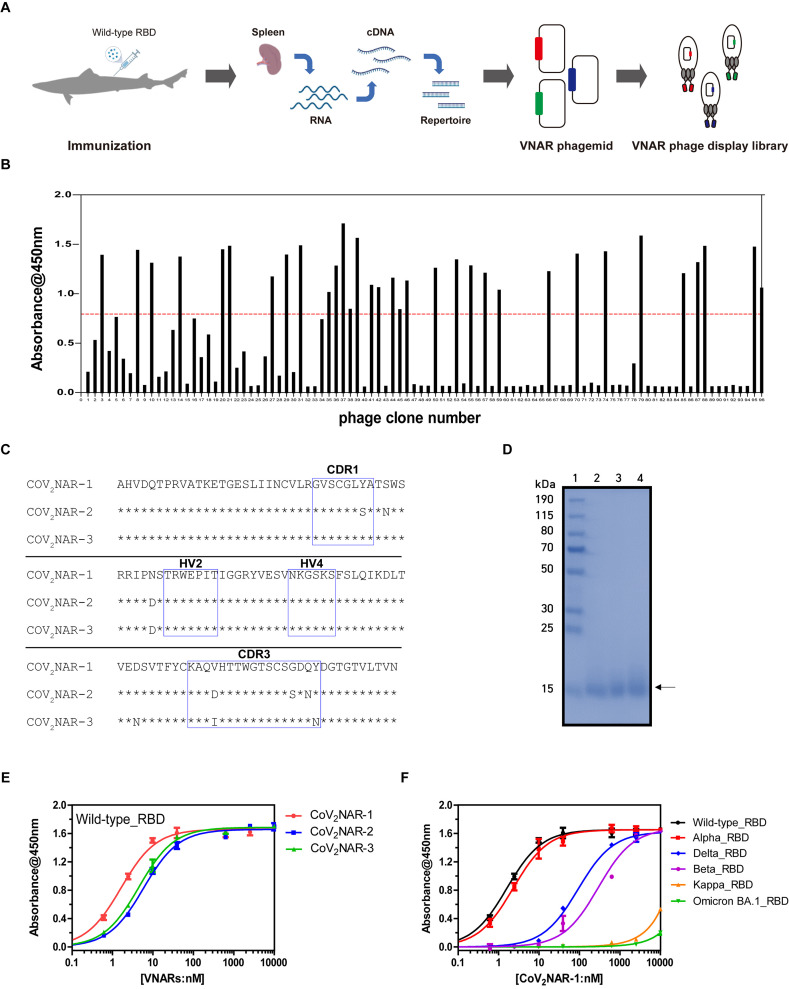
To generate immunized VNAR libraries against SARS-CoV-2 RBD, a banded houndshark was immunized with recombinant SARS-CoV-2 wild-type RBD protein. After the three consecutive immunizations, the shark exhibited an appropriate anti-serum titer (Supplementary Fig. 1). Total RNA was then extracted from the shark spleen and converted to cDNA to construct a phage display immune library (Fig. 1 A). After analyzing 45 randomly picked colonies from the 107 diluted immune libraries, it was found that each of the 45 colony sequences was unique, indicating a library size of 4.5 × 108 as determined by the number of transformants (Supplementary Fig. 2).
Fig. 1.
Identification of banded houndshark derived VNAR against SARS-CoV-2 RBD. (A) Schematic figure of phage library construction from immunized sharks. (B) Identification of wild-type RBD specific VNAR by the periplasmic extract ELISA. The red dashed line indicates a threshold for the selection of RBD-specific VNARs. (C) Amino acid sequence alignment of CoV2NARs. Asterisks indicate amino acid sequences consistent with CoV2NAR-1. Blue boxes represent CDRs and HVs determined according to the previous reference [24]. (D) SDS-PAGE analysis of purified CoV2NAR-1,2,3. The lanes of SDS-PAGE are as follows: lane 1, protein size marker (Thermo Fisher); lane 2, CoV2NAR-1; lane 3, CoV2NAR-2; and lane 4, CoV2NAR-3. Binding affinity analysis of (E) purified CoV2NARs against the wild-type RBD and (F) CoV2NAR-1 against the RBDs from wild-type and its variants (Alpha, Beta, Delta, Kappa, and Omicron BA.1) by ELISA. Error bars are ±standard deviation for triplicate experiments.
To identify RBD-specific VNARs, biopanning was conducted using the recombinant wild-type RBD as the target antigen (Supplementary Methods). After completing three rounds of biopanning, 33 clones of M13 phages displaying RBD-specific VNARs were isolated via enzyme-linked immunosorbent assay (ELISA) using periplasmic extract (Fig. 1B). Among the 33 clones, 31 were turned out to be derived from a single enriched clone, CoV2NAR-1 with identical nucleotide sequences of complementarity determining regions (CDRs) and hypervariable region (HV), while the remaining two clones, CoV2NAR-2 and CoV2NAR-3 displayed slightly different CDRs and framework sequences (Fig. 1C). To determine their binding affinity to the recombinant wild-type RBD, CoV2NAR-1, -2, and -3 were expressed in Escherichia coli and purified using Ni-NTA chromatography (Fig. 1D). The purified CoV2NAR-1, -2, and -3 demonstrated strong binding affinities to the wild-type RBD, with 50% maximal effective concentration (EC50) values of 1.6 nM, 5.8 nM, and 4.5 nM, respectively (Fig. 1E). CoV2NAR-1 exhibited the highest binding affinity to the recombinant wild-type RBD (Fig. 1E) and also demonstrated broad binding affinities to RBDs of SARS-CoV-2 Alpha, Beta, and Delta variants (Fig. 1F and Table 1 ). These findings indicate that the RBD specific VNAR clones were successfully enriched by biopanning using the banded houndshark immunized phage display library.
Table 1.
Binding affinities of CoV2NAR-1 against the recombinant RBD from SARS-CoV-2 and its variants.
ND: Not determined.
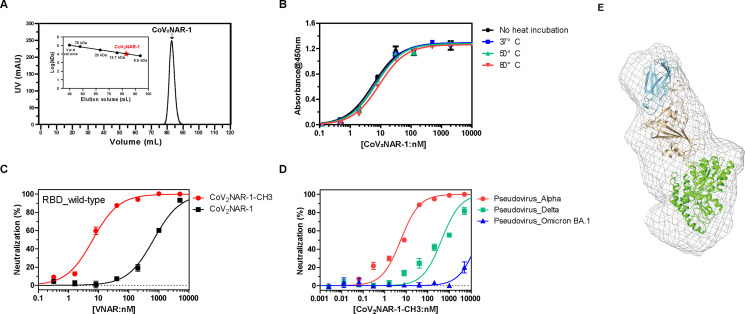
To investigate the physicochemical property and in vitro efficacy of CoV2NAR-1, we conducted various biophysical and biochemical studies shown in Fig. 2 . Protein homogeneity analysis using size-exclusion chromatography showed that the Ni-NTA purified CoV2NAR-1 was monomeric, with no detectable oligomers or higher-order aggregates (Fig. 2A). Heat-treated CoV2NAR-1 retained its binding affinity even after incubation at 80 °C for 1 h (Fig. 2B), indicating great thermal stability. CoV2NAR-1 was also found to neutralize wild-type SARS-CoV-2 pseudovirus with a half-maximal inhibitory concentration (IC50) of 660 nM in a pseudotyped virus neutralization assay (Fig. 2C). To improve the IC50, we produced a bivalent CoV2NAR-1 by utilizing the human IgG1 Fc CH3 domain (Supplementary Fig. 3), which is a small-sized dimeric fused protein capable of inducing avidity [27] while retaining the advantageous features of VNAR such as small size and cell permeability. The IC50 of bivalent CoV2NAR-1 was remarkably improved by two orders of magnitude compared to that of monovalent CoV2NAR-1 (Fig. 2C), indicating that the bivalent or multivalent form offers an excellent platform for developing SARS-CoV-2 therapeutics using VNAR. Additionally, bivalent CoV2NAR-1 exhibited broad neutralizing activities against SARS-CoV-2 Alpha and Delta variants with the half-maximal inhibitory concentration (IC50) in the nanomolar range (Fig. 2D), demonstrating its high potential as a therapeutic agent against diverse SARS-CoV-2 variants. Small-angle X-ray scattering measurement of the CoV2NAR-1/MBP fused RBD complex (Fig. 2E and Supplementary Table 2) revealed that CoV2NAR-1 binds to two previously reported epitope groups of RBD (group A and B) [28], which significantly overlap with the ACE2 binding site and include RBD mutation sites, such as K417 (group A) and E484 (group B) [28]. These findings suggest that RBD mutations at K417 and E484 could potentially affect the binding affinities of CoV2NAR-1 to Beta (3 mutations in RBD including K417N and E484K), Kappa (2 mutations in RBD including E484Q), and Omicron BA.1 (15 mutations in RBD including K417N, E484A) variants.
Fig. 2.
Characterization of CoV2NAR-1. (A) Size exclusion profile of CoV2NAR-1 using Hiload Superdex 75 16/600 (Cytiva). To obtain the calibration curve for Hiload Superdex 75, conalbumin (75 kDa), carbonic anhydrase (29 kDa), ribonuclease A (13.7 kDa), and aprotinin (6.5 kDa) in Gel Filtration LMW Calibration Kit (Cytiva) were used as size markers. The estimated molecular mass of the CoV2NAR-1 was 10.4 kDa. (B) Binding affinity analysis of purified CoV2NAR-1 against the wild-type RBD after acute heat challenge by ELISA. CoV2NAR-1 was incubated at different temperatures for 1 h. The binding affinities of the heat-treated CoV2NAR-1 against the wild-type RBD were then measured by ELISA. Neutralization efficiency of CoV2NAR-1 against (C) a pseudo-SARS-CoV-2 wild-type and (D) its variants. IC50 for SARS-CoV-2 pseudoviruses were mediated by CoV2NAR-1 in HEK293T-ACE2-TMPRSS2 cell. Error bars are ±standard deviation for triplicate experiments. (E) SAXS-based rigid body model of the CoV2NAR-1 in complex with MBP fused wild-type RBD. Sky blue, brown, and green cartoons indicate CoV2NAR-1, wild-type RBD, and MBP, respectively.
VNARs offer several advantages over conventional IgG antibodies, including a small size that allows for more efficient tissue penetration, high stability, and specificity for cryptic epitopes [18]. Since antigen-specific VNARs from immunized libraries generally have higher binding affinities than naïve and synthetic libraries [19], there is a growing demand for validated animal models that can produce antigen-specific VNARs to broaden the scope of VNAR-based research. In this study, we constructed an immunized library from banded houndshark for the first time to identify VNARs against SARS-CoV-2 RBD. Using recombinant RBD affinity-based screening, we discovered CoV2NAR-1, which displayed excellent physicochemical properties, potent neutralizing activities, and broad binding affinities to RBDs from both the wild-type and its variants. Furthermore, in silico immunogenicity assessments for 27 human leukocyte antigen (HLA)-types indicated that peptides containing the CoV2NAR-1 sequence were even less immunogenic than peptides derived from trastuzumab (anti-HER2: Herceptin®), which is a therapeutic IgG antibody currently used in clinical practice (Supplementary Fig. 4). To date, only semi-synthetic libraries have been constructed from banded houndshark [25,26]. Therefore, our study demonstrates the potential of the banded houndshark as an alternative animal model for the development of VNAR-based therapeutics or diagnostics against various pathogens, including but not limited to emerging viruses, and provides a new avenue for the discovery of novel VNAR-based biologics.
Credit author statement
Woo Sung Kim, performed the experiments and performed the data analysis. drafted the manuscript. Hee Do Chae, performed the experiments and performed the data analysis. Inji Jung, performed the experiments and performed the data analysis. Won-Kyu Lee, performed the experiments and performed the data analysis. Woo Jun Lee, performed the experiments and performed the data analysis. Jisun Lee, performed the experiment and performed the data analysis. Yegin Gong, performed the experiments and performed the data analysis. Dohyun Lee, performed the experiments and performed the data analysis. Byeong-Won Kim, performed the experiments and performed the data analysis. Jaehyeon Hwang, provided reagents and analytical tools. Dae-Hyuk Kweon, provided reagents and analytical tools. Jin-Koo Kim. provided reagents and analytical tools. Sang Taek Jung, drafted the manuscript. revised the manuscript. Jung-Hyun Na, drafted the manuscript. revised the manuscript. conceived the idea of this study. All authors approved the final version of the manuscript.
Declaration of competing interest
A patent related to this work has been filed by the authors except Yejin Gong, Byeong-Won Kim, Jaehyeon Hwang, and Dae-Hyuk Kweon.
Acknowledgments
This work was supported by the National Research Foundation (NRF) of Korea grant (RS-2023-00214327) funded by the Korea government (Ministry of Science and ICT). This work was also supported by a grant (21172MFDS724) from Ministry of Food and Drug Safety in 2021.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fsi.2023.108807.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.World Health Organization COVID-19 Dashboard. Geneva: https://covid19.who.int/(Accessed on 26 April 2023).
- 2.Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., Cathcart A.L., Hebner C.M., Sager J., Mogalian E., Tipple C., Peppercorn A., Alexander E., Pang P.S., Free A., Brinson C., Aldinger M., Shapiro A.E. Early treatment for covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 2021;385(21):1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 3.Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., Binshtein E., Shrihari S., Ostrowski M., Chu H.Y., Didier J.E., MacRenaris K.W., Jones T., Day S., Myers L., Eun-Hyung Lee F., Nguyen D.C., Sanz I., Martinez D.R., Rothlauf P.W., Bloyet L.-M., Whelan S.P.J., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26(9):1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pymm P., Adair A., Chan L.-J., Cooney J.P., Mordant F.L., Allison C.C., Lopez E., Haycroft E.R., O'Neill M.T., Tan L.L., Dietrich M.H., Drew D., Doerflinger M., Dengler M.A., Scott N.E., Wheatley A.K., Gherardin N.A., Venugopal H., Cromer D., Davenport M.P., Pickering R., Godfrey D.I., Purcell D.F.J., Kent S.J., Chung A.W., Subbarao K., Pellegrini M., Glukhova A., Tham W.-H. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. USA. 2021;118(19) doi: 10.1073/pnas.2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., Murrell B., McInerney G.M. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11(1):4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huo J., Mikolajek H., Le Bas A., Clark J.J., Sharma P., Kipar A., Dormon J., Norman C., Weckener M., Clare D.K., Harrison P.J., Tree J.A., Buttigieg K.R., Salguero F.J., Watson R., Knott D., Carnell O., Ngabo D., Elmore M.J., Fotheringham S., Harding A., Moynié L., Ward P.N., Dumoux M., Prince T., Hall Y., Hiscox J.A., Owen A., James W., Carroll M.W., Stewart J.P., Naismith J.H., Owens R.J. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021;12(1):5469. doi: 10.1038/s41467-021-25480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haga K., Takai-Todaka R., Matsumura Y., Song C., Takano T., Tojo T., Nagami A., Ishida Y., Masaki H., Tsuchiya M. Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 2021;17(10) doi: 10.1371/journal.ppat.1009542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 9.Ye G., Gallant J., Zheng J., Massey C., Shi K., Tai W., Odle A., Vickers M., Shang J., Wan Y., Du L., Aihara H., Perlman S., LeBeau A., Li F. The development of Nanosota-1 as anti-SARS-CoV-2 nanobody drug candidates. Elife. 2021;10 doi: 10.7554/eLife.64815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao H., Cai H., Li T., Zhou B., Qin W., Lavillette D., Li D. A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2. PLoS Pathog. 2021;17(3) doi: 10.1371/journal.ppat.1009328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauhar A., Privezentzev C.V., Demydchuk M., Gerlza T., Rieger J., Kungl A.J., Walsh F.S., Rutkowski J.L., Stocki P. Single domain shark VNAR antibodies neutralize SARS-CoV-2 infection in vitro. Faseb. J. 2021;35(11) doi: 10.1096/fj.202100986RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ubah O.C., Lake E.W., Gunaratne G.S., Gallant J.P., Fernie M., Robertson A.J., Marchant J.S., Bold T.D., Langlois R.A., Matchett W.E., Thiede J.M., Shi K., Yin L., Moeller N.H., Banerjee S., Ferguson L., Kovaleva M., Porter A.J., Aihara H., LeBeau A.M., Barelle C.J. Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography. Nat. Commun. 2021;12(1):7325. doi: 10.1038/s41467-021-27611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng B., Chen Z., Sun J., Xu T., Wang Q., Yi H., Niu X., Zhu J., Fan M., Hou R., Shao Y., Huang S., Li C., Hu P., Zheng P., He P., Luo J., Yan Q., Xiong X., Liu J., Zhao J., Chen L. A class of shark-derived single-domain antibodies can broadly neutralize SARS-related coronaviruses and the structural basis of neutralization and Omicron escape. Small Methods. 2022;6(7) doi: 10.1002/smtd.202200387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y.L., Lin J.J., Ma H., Zhong N., Xie X.X., Yang Y., Zheng P., Zhang L.J., Jin T., Cao M.J. Screening and characterization of shark-derived VNARs against SARS-CoV-2 spike RBD protein. Int. J. Mol. Sci. 2022;23(18) doi: 10.3390/ijms231810904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdovino-Navarro B.J., Duenas S., Flores-Acosta G.I., Gasperin-Bulbarela J., Bernaldez-Sarabia J., Cabanillas-Bernal O., Cervantes-Luevano K.E., Licea-Navarro A.F. Neutralizing ability of a single domain VNAR antibody: in vitro neutralization of SARS-CoV-2 variants of concern. Int. J. Mol. Sci. 2022;23(20) doi: 10.3390/ijms232012267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W.-H., Hajduczki A., Martinez E.J., Bai H., Matz H., Hill T.M., Lewitus E., Chang W.C., Dawit L., Peterson C.E., Rees P.A., Ajayi A.B., Golub E.S., Swafford I., Dussupt V., David S., Mayer S.V., Soman S., Kuklis C., Corbitt C., King J., Choe M., Sankhala R.S., Thomas P.V., Zemil M., Wieczorek L., Hart T., Duso D., Kummer L., Yan L., Sterling S.L., Laing E.D., Broder C.C., Williams J.K., Davidson E., Doranz B.J., Krebs S.J., Polonis V.R., Paquin-Proulx D., Rolland M., Reiley W.W., Gromowski G.D., Modjarrad K., Dooley H., Joyce M.G. Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity. Nat. Commun. 2023;14(1):580. doi: 10.1038/s41467-023-36106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmsen M.M., De Haard H.J. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 2007;77(1):13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juma S.N., Gong X., Hu S., Lv Z., Shao J., Liu L., Chen G. Shark new antigen receptor (IgNAR): structure, characteristics and potential biomedical applications. Cells. 2021;10(5):1140. doi: 10.3390/cells10051140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei L., Wang M., Xiang H., Jiang Y., Gong J., Su D., Al Azad M.A.R., Dong H., Feng L., Wu J., Chan L.L., Yang N., Shi J. Bamboo shark as a small animal model for single domain antibody production. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalid Z., Chen Y., Yu D., Abbas M., Huan M., Naz Z., Mengist H.M., Cao M.J., Jin T. IgNAR antibody: structural features, diversity and applications. Fish Shellfish Immunol. 2022;121:467–477. doi: 10.1016/j.fsi.2022.01.027. [DOI] [PubMed] [Google Scholar]
- 21.Ebert D.A., Dando M., Fowler S., Jabado R. Princeton University Press; 2021. Sharks of the World: A Complete Guide. [Google Scholar]
- 22.Michael S.W. 1993. Reef Sharks and Rays of the World: A Guide to Their Identification, Behavior, and Ecology, Sea Challengers. [Google Scholar]
- 23.Rigby C.L., Walls R.H.L., Derrick D., Dyldin Y.V., Herman K., Ishihara H., Jeong C.-H., Semba Y., Tanaka S., Volvenko I.V., Yamaguchi A. 2021. Triakis Scyllium, the IUCN Red List of Threatened Species. [Google Scholar]
- 24.Honda Y., Kondo H., Caipang C.M., Hirono I., Aoki T. cDNA cloning of the immunoglobulin heavy chain genes in banded houndshark Triakis scyllium. Fish Shellfish Immunol. 2010;29(5):854–861. doi: 10.1016/j.fsi.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 25.Ohtani M., Hikima J., Jung T.S., Kondo H., Hirono I., Takeyama H., Aoki T. Variable domain antibodies specific for viral hemorrhagic septicemia virus (VHSV) selected from a randomized IgNAR phage display library. Fish Shellfish Immunol. 2013;34(2):724–728. doi: 10.1016/j.fsi.2012.11.041. [DOI] [PubMed] [Google Scholar]
- 26.Ohtani M., Hikima J., Jung T.S., Kondo H., Hirono I., Aoki T. Construction of an artificially randomized IgNAR phage display library: screening of variable regions that bind to hen egg white lysozyme. Mar. Biotechnol. 2013;15(1):56–62. doi: 10.1007/s10126-012-9456-1. [DOI] [PubMed] [Google Scholar]
- 27.Maute R.L., Gordon S.R., Mayer A.T., McCracken M.N., Natarajan A., Ring N.G., Kimura R., Tsai J.M., Manglik A., Kruse A.C., Gambhir S.S., Weissman I.L., Ring A.M. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc. Natl. Acad. Sci. USA. 2015;112(47):E6506–E6514. doi: 10.1073/pnas.1519623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.