Abstract
Pseudomonas aeruginosa is the nosocomial bacterial pathogen most commonly isolated from the respiratory tract. Animal models of this infection are extremely valuable for studies of virulence and immunity. We thus evaluated the utility of a simple model of acute pneumonia for analyzing P. aeruginosa virulence by characterizing the course of bacterial infection in BALB/c mice following application of bacteria to the nares of anesthetized animals. Bacterial aspiration into the lungs was rapid, and 67 to 100% of the inoculum could be recovered within minutes from the lungs, with 0.1 to 1% of the inoculum found intracellularly shortly after infection. At later time points up to 10% of the bacteria were intracellular, as revealed by gentamicin exclusion assays on single-cell suspensions of infected lungs. Expression of exoenzyme U (ExoU) by P. aeruginosa is associated with a cytotoxic effect on epithelial cells in vitro and virulence in animal models. Insertional mutations in the exoU gene confer a noncytotoxic phenotype on mutant strains and decrease virulence for animals. We used the model of acute pneumonia to determine whether introduction of the exoU gene into noncytotoxic strains of P. aeruginosa lacking this gene affected virulence. Seven phenotypically noncytotoxic P. aeruginosa strains were transformed with pUCP19exoUspcU which carries the exoU gene and its associated chaperone. Three of these strains became cytotoxic to cultured epithelial cells in vitro. These strains all secreted ExoU, as confirmed by detection of the ExoU protein with specific antisera. The 50% lethal dose of exoU-expressing strains was significantly lower for all three P. aeruginosa isolates carrying plasmid pUCP19exoUspcU than for the isogenic exoU-negative strains. mRNA specific for ExoU was readily detected in the lungs of animals infected with the transformed P. aeruginosa strains. Introduction of the exoU gene confers a cytotoxic phenotype on some, but not all, otherwise-noncytotoxic P. aeruginosa strains and, for recombinant strains that could express ExoU, there was markedly increased virulence in a murine model of acute pneumonia and systemic spread.
Pseudomonas aeruginosa infection occurs when normal defense mechanisms are impaired or in cases of extensive tissue damage. Extracellular virulence factors including proteases, cytotoxins, phospholipases, pili, flagella, and smooth lipopolysaccharides have been shown to contribute to virulence in various animal models (18, 25, 26). Proteins exported by the type III secretion system, notably, exoenzyme S (ExoS), ExoT, and ExoU, have toxic effects on cells in culture (3, 7, 14, 24, 27, 28) and are thought to be important virulence factors of P. aeruginosa. Disruption of the pscC gene (a member of the secretin family of proteins needed for secretion of the exoenzyme proteins) by insertion of Tn1 (29) reduced the virulence of cytotoxic strain PA 388 in burn wound infections in mice (18). This disruption did not affect levels of the mutant strain in a rat model of chronic lung infection, although there was a reduction in the amount of lung damage (19). In contrast, disruption of exsA in strain PAO1 had no effect in a neonatal mouse model of acute pneumonia (26). With another cytotoxic and highly virulent P. aeruginosa strain, PA103, disruption of the exoU gene resulted in a loss of cytotoxicity and reduced virulence in a murine acute lung infection model (3), a finding also reported by Hauser et al. (10), who designated the gene as pepA in their study. In a related study, Kurahashi et al. (15) used a PA103 strain with an interrupted exoT gene and a deleted exoU gene and showed a loss of the ability of the strain to induce systemic inflammation and septic shock following instillation into the lungs of rabbits. These authors concluded that in P. aeruginosa strains expressing ExoU the cytotoxin may cause epithelial cell damage in the lung contributing to the subsequent release of inflammatory mediators into the systemic circulation that give rise to inflammation and septic shock.
These results clearly indicate that ExoU is an important virulence factor for P. aeruginosa strains that contain the gene and secrete the protein. However, not all clinical isolates of P. aeruginosa make ExoU (5, 11); thus, serious infection can develop without relying on this factor. An additional way to evaluate the role of a virulence factor such as ExoU in pathogenesis is to introduce the DNA for this protein into strains that lack it and determine whether there is a gain of virulence by the transformed strain. Evaluations of transformed strains for increased virulence can be hampered, however, if an appropriate animal model is not available with sufficient sensitivity to measure the increase in pathogenic capacity of the strains. To address these issues in the context of P. aeruginosa virulence and pathogenesis, we evaluated the phenotypic properties and virulence of noncytotoxic, exoU-negative strains of P. aeruginosa and isogenic strains transformed with DNA, allowing for expression of the ExoU cytotoxin, in a simple model of acute pneumonia in mice. Application of P. aeruginosa to the nares of anesthetized mice resulted in rapid aspiration of most of the inoculum to the lungs, rapid internalization of a portion of the inoculum into lung cells, and death from acute pneumonia and sepsis within 24 to 48 h. Critically important, the model was highly sensitive to changes in virulence following transformation of three noncytotoxic P. aeruginosa strains with the exoU gene and its associated chaperon, with the ExoU-secreting transformants having dramatic reductions in 50% lethal dose (LD50) values.
MATERIALS AND METHODS
Bacterial strains.
Clinical isolates of P. aeruginosa from bacteremic patients were used to determine the presence of the exoU gene. Laboratory strain PAO1 was originally obtained from Michael Vasil, Denver, Colo. Strain PAO6ad (Lanyi serogroup 06ad) was supplied by B. Lanyi, Budapest, Hungary (16), and the noncytotoxic corneal isolate, strain 6294, and the cytotoxic corneal isolate, strain 6077, were clinical isolates from patients with ulcerative keratitis.
Vectors, determination of exoU in clinical isolates, and transformation of bacterial strains.
The exoU gene, its chaperone spcU, and flanking DNA were cloned by Frank and colleagues into plasmid pUCP19 to create plasmid pUCP19exoUspcU (4), which they kindly supplied for this study. The cloning vector pUCP19 was also introduced into P. aeruginosa strains, and these transformed strains were used as controls. The clinical isolates of P. aeruginosa were tested for the presence of exoU by PCR. Chromosomal DNA was extracted from bacterial cells with the use of a commercial kit (QIAamp Tissue Kit; Qiagen, Valencia, Calif.). Then, 30 ng of DNA was used in a PCR reaction to detect a 428-bp internal sequence of exoU using primers 5′-GGGAATACTTTCCGGGAAGTT-3′ and 5′-CGATCTCGCTGCTAATGTGTT-3′. The PCR reaction was performed using 32 cycles each of 94°C for 30 s, 59°C for 60 s, and 72°C for 90 s. Results were visualized by electrophoresis in a 1% agarose gel followed by ethidium bromide staining. P. aeruginosa strains negative for the exoU gene were then transformed with pUCP19exoUspcU or the control plasmid pUCP19 by electroporation. Approximately 1010 CFU of bacteria were made electrocompetent by repeated washing steps in 1 ml of ice-cold deionized H2O. After the last washing, distilled H2O was replaced by 10% ice-cold glycerol, and a final centrifugation of the cells was performed. Bacteria were then suspended in 100 μl of 10% glycerol, and 1 μl of either plasmid pUCP19exoUspcU or plasmid pUCP19 was carefully pipetted into 40 μl of bacterial suspension and transferred into an electroporation cuvette with a 2-mm gap. Electroporation was carried out at 1.8 kV, 25 mF, and 200 Ω; 900 μl of SOC medium (23) was added, and transformed bacteria were incubated with rotation at 37°C for 1 h. Transformed bacteria were then plated on L-agar plates containing 400 μg of carbenicillin/ml. After 18 h of incubation at 37°C, single colonies were picked and screened for the presence of the correct plasmids, which, if present, were extracted from 3-ml bacterial cultures grown overnight in Luria-Bertani (LB) broth containing 400 μg of carbenicillin/ml using the Qiagen Plasmid Miniprep Kit (Qiagen Plasmid Mini Kit). The amount of recovered DNA was measured by UV spectrophotometry. A total of 0.5 to 1 μg of DNA was digested with BamHI, resulting in either linearization of pUCP19 or the liberation of a 6.5-kb exoU spcU fragment from pUCP19exoUspcU. DNA fragments were visualized after electrophoresis in a 1% agarose gel followed by staining with ethidium bromide.
Antiserum to ExoU.
The exoU gene was amplified from plasmid pUCP19exoUspcU by PCR with primers 5′-GGATCCATGCATATCCAATCGTTGGG-3′ and 5′-GCGGCCGCTGTGAACTCCTTATTCCGCC-3′, and the resultant product was ligated into the TA cloning vector, pCRII (Invitrogen, San Diego, Calif.), which was transformed into competent Escherichia coli INVαf′ cells for cloning. The recombinant plasmid was recovered, verified to contain full-length exoU by digestion with BamHI and NotI, and cloned into the histidine (His)-tagged expression vector pET24a. After transformation into E. coli BL21(DR3)/pLYSS, the recombinant His-tagged ExoU protein was found to be present in the insoluble fraction, which was obtained from the E. coli cells by freeze-thawing. This fraction was added to a nickel affinity column (His Bind Purification Kit; Novagen, Inc., Madison, Wis.) and washed extensively with binding buffer, and the recombinant protein was released with an elution buffer containing 6 M urea, 1 M imidazole, 0.5 M NaCl, and 20 mM Tris-HCl at pH 7.9. Recovered material was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining of the gel, and a single band containing the 70-kDa ExoU protein was found to be present. The purified, recombinant ExoU protein was used to immunize a rabbit (10 μg in Freund complete adjuvant given subcutaneously, followed by two subsequent doses of 10 μg/week in saline given intravenously). The resulting antiserum was analyzed by enzyme-linked immunosorbent assay (ELISA) and Western blot for activity. A high ELISA titer (>2,500) was detected, and the serum reacted specifically by Western blot with a single band in crude extracts of P. aeruginosa cells expressing ExoU as well as with the purified recombinant protein.
Detection of ExoU protein in recombinant strains.
P. aeruginosa strains carrying the cloning vector or pUCP19exoUspcU were grown in LB broth supplemented with 10 mM nitrilotriacetic acid (Sigma) and 400 μg of carbenicillin/ml. Supernatants were recovered, proteins were precipitated by the addition of ammonium sulfate to a 55% (vol/vol) saturation final concentration, and the precipitate was recovered and dissolved in one-tenth the original volume, using PBS, and then dialyzed against PBS and used in an immuno-dot blot assay as described earlier (13).
In vitro cytotoxicity assay.
T84 colon carcinoma cells were maintained and passed at 37°C in 5% CO2 in a 1:1 mixture of Dulbecco modified Eagle medium supplemented with 4.5 g of glucose and Ham's F-12 medium per liter, 10% non-heat-inactivated fetal bovine serum, and 1% l-glutamine. Freshly passed cells were cultured in 96-well plates (Falcon; Becton Dickinson, Franklin Lakes, N.J.) and used in experiments after a confluent monolayer had formed. After cells were washed once with phosphate-buffered saline (PBS), 200 μl of transformed P. aeruginosa strains at a concentration of approximately 107 CFU/ml, suspended in a culture medium containing 400 μg of carbenicillin/ml, was added. Three wells of cells were used for each strain in each experiment. Control samples were incubated with culture medium and carbenicillin alone. After incubation for 3 h, the medium was removed and the cell layer was washed once with PBS to remove most of the nonassociated bacteria. Then, 50 μl of trypan blue was added for 90 s and removed, and the cells were washed once with PBS. The extent of cell damage was scored on a scale of 1 to 4, with 4 representing the amount of cytotoxicity exhibited by the exoU-positive cytotoxic P. aeruginosa strain 6077. The amount of cell damage caused by the noncytotoxic strain 6294 was represented by a score of 1. This method had been found to correlate well with the results of quantitative assessment by chromium release assays (6).
Experimental pneumonia in mice.
Two murine models of acute P. aeruginosa pneumonia were used to evaluate pathogenesis. For bacterial inocula, transformed P. aeruginosa strains were grown on L agar containing 400 μg of carbenicillin/ml (i.e., with antibiotic). Wild-type clinical and laboratory strains were grown without additional antibiotic (i.e., without antibiotic). Bacteria from this plate were inoculated into LB broth ± 400 μg of carbenicillin/ml at an optical density at 650 nm (OD650) of 0.1 and grown to an OD650 of 0.5 with rotation at 37°C. Bacteria were recovered by centrifugation and resuspended to an OD650 of 0.4 in 1% proteose peptone with 400 μg of carbenicillin/ml.
Infection of neonatal mice with P. aeruginosa by nasal application was performed as described previously (20, 21, 25). For adult mice, 6- to 8-week-old female BALB/c mice were anesthetized by intraperitoneal administration of a freshly prepared mixture of ketamine hydrochloride (65 mg/kg) and xylazine (13 mg/kg). With mice held in an upright position, 10 μl of a bacterial suspension was placed on each nostril (20 μl total). Animals were either observed for survival for up to 72 h or sacrificed at various time periods up to 24 h after infection for determination of CFU in tissues. Lungs, spleen, and a 200- to 300-mg portion of the liver were surgically removed, weighed, and homogenized in 1 ml of proteose peptone on ice. Serial 10-fold dilutions were performed in 1% proteose peptone, and 100 μl of diluted bacterial suspensions was plated on MacConkey agar plates at 37°C for 18 to 24 h. The resultant CFU were calculated as the level of bacterial infection per gram of homogenized tissue. For determination of intracellular P. aeruginosa, lungs were aseptically removed and single cell suspensions were made by forcing the tissue through first a 100- and then an 80-mesh sterile screen into tissue culture medium containing 300 μg of gentamicin/ml. Large tissue fragments were allowed to settle, and the suspended cells were pipetted into another tube and incubated in the antibiotic for 1 h at 37°C. The cells were washed to remove the gentamicin and lysed in 0.5% Triton X-100 to release intracellular bacteria, which were quantified by serial dilution and plating as described above. Student t tests were used for two-way comparisons of tissue levels of P. aeruginosa, and logistic regression for parallel bioassays was used to test for differences in the LD50s.
Expression of ExoU in vivo.
BALB/c mice were challenged with 2 × 106 of transformed bacteria as described above, and 24 h later animals were sacrificed, lungs were surgically removed and homogenized, and total RNA was extracted with a commercial kit (RNeasy; Qiagen). cDNA was transcribed with reverse transcriptase from 2 μg of total RNA (SuperScriptII; Gibco-BRL/Life Technologies, Rockville, Md.). A total of 30 ng of cDNA was added to a PCR reaction that included primers specific to exoU and identical to the ones mentioned above. cDNA was amplified at 94°C for 30 s, 59°C for 30 s, and 72°C for 60 s for a total of 35 cycles. Primers amplifying a 314-bp rpoB fragment (5′-CCGATAAGGAGTTCTTCGGGT-3′ and 5′-GAACACGATCTCGTCGGTTAC-3′) served as a quality control. DNA was separated on a 2% agarose gel and stained with ethidium bromide.
RESULTS
Characterization of the adult murine model of acute pneumonia.
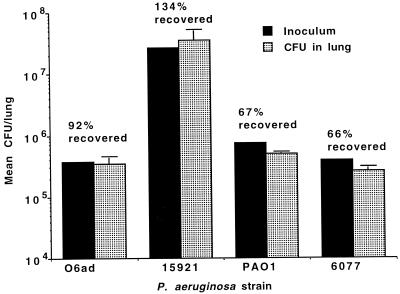
George et al. (8, 9) have previously used nasal application of P. aeruginosa in mice to evaluate bacterial virulence. Using a similar approach we initially characterized the model to assess its utility and sensitivity for the evaluation of P. aeruginosa virulence. Placement of 20 μl of bacterial suspension onto the nose of anesthetized BALB/c mice was found to be a reliable and reproducible way to initiate acute pneumonia. Mice infected intranasally showed rapid aspiration of 67 to >100% of the inoculum to the lungs (Fig. 1). Recovery of >100% of the inoculum is attributable to small dilution and plating errors inherent in enumeration of large numbers of organisms. This level of infection was detected within the time it took to inoculate the mice, sacrifice them, and remove the lungs for analysis (ca. 10 to 15 min). It is interesting that, when P. aeruginosa was applied to the nares of unanesthetized adult mice, we found no aspiration of bacteria to the lungs and no subsequent infection.
FIG. 1.
Rapid aspiration of four different P. aeruginosa strains from the nares to the lungs after application of the indicated inoculum in a 20-μl volume to the nares of anesthetized mice. Bars represent the mean, and the error bars show the standard deviations. The percentage of the inoculum that was recovered from the lungs is indicated above each pair of bars.
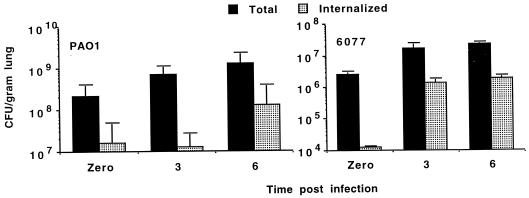
Comparisons were then made between noncytotoxic P. aeruginosa strain PAO1 and cytotoxic strain 6077 inoculated into murine nares at a dose determined in preliminary experiments to be just above that needed to kill all infected mice (Fig. 2). Comparisons of the total and internalized CFU/gram of lung tissue in mice sacrificed shortly after infection (time zero) or 3 or 6 h after infection showed progressive increases in bacterial levels in the tissue. There was evidence of rapid internalization of a portion of the P. aeruginosa inoculum, with up to 1% of the inoculum apparently intracellular, as evidenced by resistance to gentamicin killing in single-cell suspensions of lungs (Fig. 2) and up to 10% of the inoculum resistant to killing by gentamicin, and presumably intracellular, by 3 to 6 h (Fig. 2). Specific cell types ingesting the P. aeruginosa bacteria were not investigated. Spleens and livers were generally sterile in mice sacrificed prior to 6 h postinfection, but at this and subsequent time points P. aeruginosa was recovered in increasing numbers from these tissues (data not shown). We noted also that, in mice given a lethal inocula of P. aeruginosa, the levels of bacteria in the lungs and extrapulmonary tissues 6 h after infection were predictive of a lethal or nonlethal outcome: levels of P. aeruginosa in lungs of mice given lethal inocula and sacrificed at 6 h after infection were found to exceed the inocula (Fig. 2), and there was always evidence of extrapulmonary infection. In contrast, mice given sublethal inocula showed a decrease in the level of bacteria in the lung, compared with the initial inoculum, by 6 h after infection, and there was rarely evidence of extrapulmonary infection at this time (data not shown).
FIG. 2.
Comparison of total and internalized (resistant to gentamicin in single cell suspensions of lung) noncytotoxic P. aeruginosa PAO1 (left) and cytotoxic P. aeruginosa 6077 (right) cells at the indicated times after application to the nares of anesthetized mice. Bars represent the mean CFU, and the error bars show the standard deviations. The inoculum for P. aeruginosa PAO1 was 3 × 108 CFU/nose; the inoculum for P. aeruginosa 6077 was 3 × 106 CFU/nose.
Comparison of P. aeruginosa pathogenesis in neonatal and adult mice.
Tang et al. (25, 26) described the utility of application of P. aeruginosa into the nares of unanesthetized neonatal mice for evaluation of pathogenesis. However, in this model mortality was reported to range from 0 to 60% depending on the strain of P. aeruginosa used, while unanesthetized adult mice tolerated doses of up to 1010 CFU/mouse without effect (25). Thus, virulence in the neonatal mice is usually measured by the histologic appearance of lung tissue or by bacterial loads in tissues. Since it appeared that anesthetized adult mice manifested a greater degree of mortality following nasal application of P. aeruginosa than awake neonatal mice, we determined the CFU/gram of tissue and LD50 in 7-day-old neonatal BALB/c mice. A smaller but nonetheless substantial proportion of the inoculum applied to the neonatal nares reached the lungs quickly (mean, 22.7 ± 0.9% for strain PAO1) than was seen with anesthetized adult mice (Fig. 1 and 2). However, the neonatal mice rapidly cleared inocula of P. aeruginosa strain PAO1 of <2 × 108 CFU/mouse, and there was no mortality. Thus, anesthetized adult mice succumb more readily to P. aeruginosa infection than awake neonatal mice, a result likely due to the greater ability of P. aeruginosa to enter adult lungs following nasal application.
Detection of exoU in clinical isolates of P. aeruginosa.
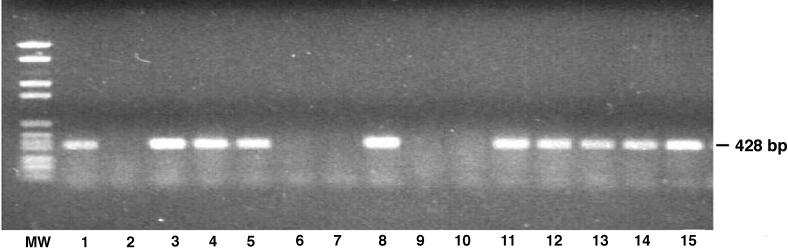
Among 14 clinical isolates of P. aeruginosa, 9 strains had the exoU gene, whereas in the other 5 there was no detectable exoU even after repeated PCR evaluations (Fig. 3). All five of these strains, as well as noncytotoxic strains PAO1 and PAO6ad, were transformed with plasmid pUCP19exoUspcU or pUCP19, and all recombinant strains contained the correct plasmid after transformation, as confirmed by plasmid extraction and restriction enzyme analysis.
FIG. 3.
Agarose gel stained with ethidium bromide showing the presence or absence of the 428-bp amplified exoU gene fragment in 14 clinical isolates of P. aeruginosa. Lanes: MW, molecular weight marker, 1, positive control pUCP19exoUspcU; 2, strain 45203; 3, strain 9156; 4, strain 56184; 5, strain Weaver; 6, strain 15921; 7, strain 1597; 8, strain Becker; 9, strain 29185; 10, strain 9882; 11, strain Rhodes; 12, strain 1947; 13, strain 9326; 14, strain 05074; 15, strain 3006.
Detection of cytotoxicity and expression of exoU in transformed P. aeruginosa strains.
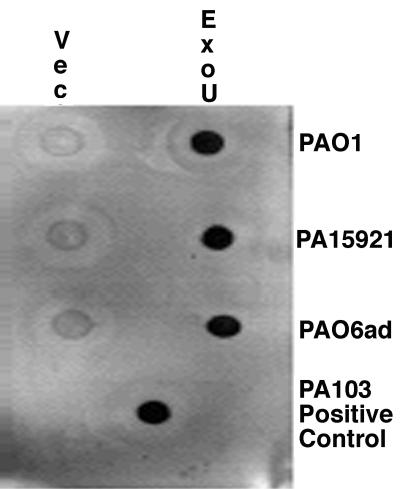
All transformed strains were tested for in vitro cytotoxic activity on T84 human colon carcinoma cells. Only three of the transformants, strains PAO1(pUCP19exoUspcU), PA06ad(pUCP19exoUspcU), and 15921(pUCP19exoUspcU) were cytotoxic. The other transformants containing pUCP19exoUspcU failed to show cytotoxic activity. The three recombinant, cytotoxic strains all expressed a protein in extracellular culture supernatants strongly reactive with the ExoU-specific antiserum (Fig. 4), whereas there was no reactive protein in any of the other strains carrying pUCP19exoUspcU but lacking a cytotoxic phenotype (not shown). The three recombinant strains positive for ExoU expression by immuno-dot blot also had in their culture supernatants the appropriately sized 70-kDa band reactive with the ExoU-specific antiserum in a Western blot (not shown).
FIG. 4.
Immuno-dot blot of expression of recombinant ExoU protein in culture supernates of P. aeruginosa PAO1, 15921, and PAO6ad carrying either the control, vector plasmid pUCP19 (Vec), or the plasmid containing the exoU gene, pUCP19exoUspcU (ExoU).
Evaluation of the role of ExoU in P. aeruginosa virulence.
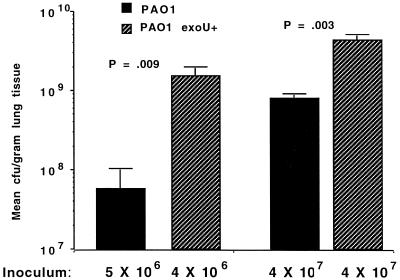
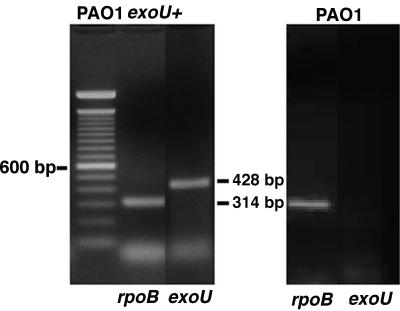
Pilot experiments comparing cytotoxic and noncytotoxic, nonisogenic strains of P. aeruginosa suggested that expression of ExoU enhanced bacterial virulence in the acute pneumonia model, as all cytotoxic strains tested (i.e., 6077 and 103) had LD50 values of <5 × 106 CFU/mouse, whereas noncytotoxic strains generally had LD50 values at least 1 log higher. To formally evaluate the role of ExoU in virulence, the three pairs of cytotoxic and noncytotoxic P. aeruginosa strains isogenic for the plasmid containing the exoU gene and the cloning vector plasmid were inoculated at various doses onto the nares of anesthetized, adult BALB/c mice. Comparisons were made between the bacterial loads in the lung and extrapulmonary bacteremic spread after infection was established, and the LD50 values were determined. Groups of five animals each were sacrificed 18 to 24 h after infection. In all cases, the CFU/gram of lung tissue was significantly higher in the lungs of animals infected with P. aeruginosa carrying the exoU gene; data from animals infected with two doses of isogenic PAO1 are shown in Fig. 5. mRNA for the ExoU protein was detected by reverse transcription-PCR (RT-PCR) in the lungs of mice infected with P. aeruginosa carrying the exoU gene but not in the lungs of animals infected with P. aeruginosa lacking the exoU gene (Fig. 6). Extrapulmonary infection in the spleens and livers was routinely observed in animals given lethal doses of P. aeruginosa intranasally, e.g., infected livers and spleens were found in those mice inoculated with >106 CFU of P. aeruginosa strains carrying the exoU gene and >5 × 107 CFU of strains lacking the exoU gene (not shown).
FIG. 5.
Comparison of CFU/gram of lung tissue 14 to 18 h after intranasal infection of anesthetized mice with isogenic P. aeruginosa PAO1(pUCP19) or PAO1(pUCP19exoUspcU). Bars indicate the means, and the error bars show the standard deviations. P values were determined by unpaired Student t tests.
FIG. 6.
Demonstration of elaboration of mRNA for ExoU in infected lung tissue of mice. RT-PCR of lung tissue from mice infected with either the exoU+ PAO1(pUCP19exoUspcU) strain or the parental strain carrying the cloning vector, PAO1(pUCP19). mRNA from tissue was reverse transcribed and amplified with primers specific to the rpoB gene to yield a product of 314 bp or with primers specific to the exoU gene to yield a product of 428 bp. Molecular weight markers on the left are oligonucleotides differing by 100 bp.
After application of various doses of cytotoxic and noncytotoxic transformed strains to groups of four or five animals and a follow-up period of 72 h to observe for death, the LD50s were calculated and compared by logistic regression for parallel bioassays (Table 1). In all cases a significant increase in virulence was associated with expression of ExoU, in the range of 20- to >50-fold decreases in the LD50. The LD50 for P. aeruginosa strain 15921 lacking the exoU gene could not be calculated since there were insufficient deaths for an accurate LD50 determination in mice given intranasal doses as high as 109 CFU. When one considers that the LD50 was lowered by 2 × 107 to 9 × 107 CFU of P. aeruginosa for strains expressing ExoU (Table 1), the marked contribution of ExoU to P. aeruginosa virulence in this animal model can readily be appreciated.
TABLE 1.
LD50 values after 72 h of infection comparing three strains of P. aeruginosa carrying either pUCP19 or pUCP19exoUspcU after intranasal application to anesthetized mice
| P. aeruginosa strain | Plasmid | LD50 CFU | Pa |
|---|---|---|---|
| PAO1 | pUCP19 | 2.8 × 107 | |
| pUCP19exoUspcU | 7.1 × 105 | 0.047 | |
| PAO6ad | pUCP19 | 2.2 × 107 | |
| pUCP19exoUspcU | 1.4 × 106 | 0.0001 | |
| 15921 | pUCP19 | >108b | |
| pUCP19exoUspcU | 3.6 × 106 | 0.0001 |
P was determined by logistic regression for parallel bioassays.
The LD50 for 15921(pUCP19) could not be calculated since the maximal dose applied of 109 CFU/mouse caused insufficient mortality.
DISCUSSION
We used and further characterized a murine model of acute P. aeruginosa pneumonia following application of bacteria to the nares of anesthetized animals and found that simply placing two 10-μl volumes of bacterial suspensions in each nostril resulted in a reliable and reproducible induction of pneumonia and systemic spread. Between 67 and 100% of the inoculated P. aeruginosa CFU were recovered from the lungs minutes after infection, and up to 1% of the inoculum was immediately taken up by respiratory cells, as evidenced by bacterial resistance to killing by gentamicin in single cell suspensions of infected lungs. The specific cells ingesting the P. aeruginosa were not determined, although phagocytes usually rapidly kill P. aeruginosa following ingestion (17). Lethal doses of P. aeruginosa resulted in increasing levels of bacteria in the lungs over a 24-h period and extrapulmonary spread to the spleen and liver by 6 h after infection. For ExoU-expressing strains, LD50 values in the range of 105 to 106 CFU per mouse were determined, indicating that a fairly low P. aeruginosa inoculum can be applied to the noses of intact mice to achieve a lethal infection. The utility of this simple model, its sensitivity for measuring virulence properties for many P. aeruginosa strains, and its clear relevance to P. aeruginosa respiratory infections should make it a highly useful tool for determinations of P. aeruginosa virulence as well as host immune effectors relevant to P. aeruginosa respiratory tract colonization and initial infection.
Comolli et al. (2) recently reported on the use of this model to measure the virulence of P. aeruginosa strains deficient in the pilT or pilU genes whose products contribute to the pilus-mediated twitching motility of this organism. Previously, Tang et al. (25) reported a reduction in virulence in the neonatal mouse model of pneumonia of mutant P. aeruginosa strains unable to produce pili. However, Comolli et al. (2) found no effect on either lung levels of P. aeruginosa or mortality from loss of the pilT or pilU genes but did find decreased levels of the mutant organism in the liver. As shown here, other extrapulmonary tissues such as the spleen are also infected, so the lower levels of the mutant strains in the liver found in the study of Comolli et al. (2) may merely have been due to a shift of the mutant strains toward infection of other tissues. As we found that extrapulmonary infection correlated with mortality, the lack of a difference in mortality between wild-type and pilT or pilU mutant strains suggests little role for pilus-mediated twitching motility in the dissemination of P. aeruginosa from the lung to extrapulmonary tissues in this mouse model.
The most striking results were obtained by comparing the virulence and lethality of P. aeruginosa strains isogenic for expression of the ExoU cytotoxin. In a small sample of 14 blood isolates, 5 did not have the exoU gene, but this small sample is not likely to be representative of clinical isolates of P. aeruginosa. When these five noncytotoxic strains were transformed with plasmid pUCP19exoUspcU, only one strain became cytotoxic and expressed ExoU. Two other noncytotoxic laboratory strains, PAO1 and PAO6ad, became cytotoxic when transformed with pUCP19exoUspcU. Thus, we had three isogenic strains for comparisons. The inability of some strains transformed with pUCP19exoUspcU to express ExoU is not understood at this time but may be due to the complexity of the type III secretion apparatus needed to export ExoU. When the three noncytotoxic strains of P. aeruginosa that could be complemented to a cytotoxic phenotype with the exoU gene were compared for virulence and LD50 values, strains carrying the exoU gene exhibited a statistically significant enhanced virulence. This finding confirms the previous work of Finck-Barbancon et al. (3), Hauser et al. (10), and Wiener-Kronish and colleagues (15), who used P. aeruginosa strains with an interrupted exoU gene to document a role in virulence for this factor. Our work extends these findings by showing that transformation of P. aeruginosa with pUCP19exoUspcU can confer cytotoxicity due to ExoU expression on some strains and can also result in a significant gain of virulence when evaluated in an acute lung infection model of mice. Taken together, these findings all suggest that, when expressed, ExoU plays an important role in virulence of P. aeruginosa. However, it must also be appreciated that numerous clinical isolates of P. aeruginosa lacking the exoU gene are recovered from patients. For example, Hirakata et al. (11) recently reported that only 4 of 32 P. aeruginosa blood isolates and 4 of 45 respiratory isolates were cytotoxic and possessed exoU. Therefore, in the absence of the exoU gene, other virulence factors of P. aeruginosa, such as exoS, which is present in the chromosome when exoU is not (5), can contribute to P. aeruginosa infection. Nonetheless, based on evaluations in animal models, the subset of strains of P. aeruginosa producing ExoU seem to be more virulent.
We also showed here that anesthetized adult mice are more susceptible to P. aeruginosa lung infection following nasal application than were awake neonatal mice. Comparable levels of anesthesia with adequate recovery are difficult to induce in neonatal mice with available veterinary anesthetics (unpublished observation), and unanesthetized adult mice did not aspirate the intranasal inoculum of P. aeruginosa to the lungs. Therefore, we made virulence comparisons between anesthetized adult mice and awake neonatal mice that we and others have previously used to study P. aeruginosa virulence (20, 21, 25). While younger animals are generally considered to be more susceptible to infection, we found the opposite to be the case here. In addition to BALB/c mice, we found that most other common laboratory strains of mice (C3H, C57BL/6 and Swiss-Webster) are susceptible to intranasal P. aeruginosa infection at levels comparable to those of the BALB/c mice reported here (unpublished observation).
Overall, application of P. aeruginosa to the nares of anesthetized adult mice was found to be a reliable means to produce P. aeruginosa pneumonia and systemic spread in these animals. Furthermore, adult animals were more sensitive to P. aeruginosa infection than neonatal animals due to their better ability to aspirate the initial inoculum into the lungs in a short time period. Relatively modest inocula (<5 × 106 CFU/animal) of cytotoxic strains were required to achieve a potent pathologic effect, and these inocula may reasonably reflect levels of P. aeruginosa aspirated into the lungs of humans who get P. aeruginosa infections. The model confirmed a potent role for the ExoU cytotoxin in P. aeruginosa pathogenesis, reducing LD50 levels significantly, particularly when viewed in the context of the absolute reduction in CFU of P. aeruginosa needed for a lethal infection when isogenic ExoU-positive and -negative strains were compared. We also found that P. aeruginosa strain PAO1 could be transformed to a cytotoxic phenotype with plasmid pUCP19exoUspcU, resulting in a 39-fold reduction in the LD50. As PAO1 is often used in virulence studies in animals, the availability of a relevant animal model to study pathogenesis of this strain as both a cytotoxic and a noncytotoxic variant should be of value in defining the role of other P. aeruginosa factors in disease. However, since variability in the virulence of different P. aeruginosa strains designated as PAO1 has been found (22), it is not certain they are all the same strain. Consequently, not all strains designated PAO1 may express ExoU from pUCP19exoUspcU as did the one reported here. Since P. aeruginosa is the most common bacterial pathogen isolated from respiratory specimens of patients in intensive care units (1, 12), the mouse model described here should be of value in evaluations of bacterial and host factors relevant to pathogenesis and immunity in acute P. aeruginosa pneumonia.
ACKNOWLEDGMENTS
We thank Dara Frank for providing plasmids pUCP19 and pUCP19exoUspcU and for many helpful suggestions with the manuscript.
This work was supported by NIH grants AI22535 and AI22806.
REFERENCES
- 1.Banerjee S S, Emori T G, Culver D H, Gaynes R P, Jarvis W R, Horan T, Edwards J R, Tolson J, Henderson T, Martone W J. Secular trends in nosocomial primary bloodstream infections in the United States, 1980–1989. Am J Med. 1991;91(Suppl. 3B):86S–89S. doi: 10.1016/0002-9343(91)90349-3. [DOI] [PubMed] [Google Scholar]
- 2.Comolli J C, Hauser A R, Waite L, Whitchurch C B, Mattick J S, Engel J N. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect Immun. 1999;67:3625–3630. doi: 10.1128/iai.67.7.3625-3630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finck-Barbancon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 4.Finck-Barbancon V, Yahr T L, Frank D W. Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J Bacteriol. 1998;180:6224–6231. doi: 10.1128/jb.180.23.6224-6231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleiszig S M J, Wiener-Kronish J P, Miyazaki H, Vallas V, Mostov K E, Kanada D, Sawa T, Yen T S B, Frank D W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleiszig S M J, Zaidi T S, Preston M J, Grout M, Evans D J, Pier G B. Relationship between cytotoxicity and corneal epithelial cell invasion by clinical isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64:2288–2294. doi: 10.1128/iai.64.6.2288-2294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 8.George S E, Kohan M J, Gilmour M I, Taylor M S, Brooks H G, Creason J P, Claxton L D. Pulmonary clearance and inflammatory response in C3H/HeJ mice after intranasal exposure to Pseudomonas spp. Appl Environ Microbiol. 1993;59:3585–3591. doi: 10.1128/aem.59.11.3585-3591.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George S E, Kohan M J, Whitehouse D A, Creason J P, Kawanishi C Y, Sherwood R L, Claxton L D. Distribution, clearance, and mortality of environmental pseudomonads in mice upon intranasal exposure. Appl Environ Microbiol. 1991;57:2420–2425. doi: 10.1128/aem.57.8.2420-2425.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser A R, Kang P J, Engel J N. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 11.Hirakata Y, Finlay B B, Simpson D A, Kohno S, Kamihira S, Speert D P. Penetration of clinical isolates of Pseudomonas aeruginosa through MDCK epithelial cell monolayers. J Infect Dis. 2000;181:765–769. doi: 10.1086/315276. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis W R, Martone W J. Predominant pathogens in hospital infections. J Antimicrob Chemother. 1992;29(Suppl A):19–24. doi: 10.1093/jac/29.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 13.Kocharova N A, Knirel Y A, Shashkov A S, Kochetkova N K, Pier G B. Structure of an extracellular, cross reactive polysaccharide from Pseudomonas aeruginosa immunotype 4. J Biol Chem. 1988;263:11291–11295. [PubMed] [Google Scholar]
- 14.Kudoh I, Wienerkronish J P, Hashimoto S, Pittet J F, Frank D. Exoproduct secretions of Pseudomonas aeruginosa strains influence severity of alveolar epithelial injury. Am J Physiol-Lung Cell Mol Physiol. 1994;11:L551–L556. doi: 10.1152/ajplung.1994.267.5.L551. [DOI] [PubMed] [Google Scholar]
- 15.Kurahashi K, Kajikawa O, Sawa T, Ohara M, Gropper M A, Frank D W, Martin T R, Wiener-Kronish J P. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J Clin Investig. 1999;104:743–750. doi: 10.1172/JCI7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanyi B, Bergan T. Serological characterization of Pseudomonas aeruginosa. In: Bergan T, Norris J R, editors. Methods in microbiology. Vol. 10. London, England: Academic Press, Inc.; 1978. pp. 94–168. [Google Scholar]
- 17.Mizgerd J P, Brain J D. Reactive oxygen species in the killing of Pseudomonas aeruginosa by human leukocytes. Curr Microbiol. 1995;31:124–128. doi: 10.1007/BF00294288. [DOI] [PubMed] [Google Scholar]
- 18.Nicas T I, Bradley J, Lochner J E, Iglewski B H. The role of exoenzyme S in infections with Pseudomonas aeruginosa. J Infect Dis. 1985;152:716–721. doi: 10.1093/infdis/152.4.716. [DOI] [PubMed] [Google Scholar]
- 19.Nicas T I, Frank D W, Stnezel P, Lile J D, Iglewski B H. Role of exoenzyme S in chronic Pseudomonas aeruginosa lung infections. Eur J Clin Microbiol. 1985;4:174–179. doi: 10.1007/BF02013593. [DOI] [PubMed] [Google Scholar]
- 20.Pier G B, Grout M, Zaidi T S. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc Natl Acad Sci USA. 1997;94:12088–12093. doi: 10.1073/pnas.94.22.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pier G B, Grout M, Zaidi T S, Olsen J C, Johnson L G, Yankaskas J R, Goldberg J B. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preston M J, Fleiszig S M J, Zaidi T S, Goldberg J B, Shortridge V D, Vasil M L, Pier G B. Rapid and sensitive method for evaluating Pseudomonas aeruginosa virulence factors during corneal infections in mice. Infect Immun. 1995;63:3497–3501. doi: 10.1128/iai.63.9.3497-3501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sawa T, Ohara M, Kurahashi K, Twining S S, Frank D W, Doroques D B, Long T, Gropper M A, Wiener-Kronish J P. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect Immun. 1998;66:3242–3249. doi: 10.1128/iai.66.7.3242-3249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H, Kays M, Prince A. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect Immun. 1995;63:1278–1285. doi: 10.1128/iai.63.4.1278-1285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallis A J, Finck-Barbancon V, Yahr T L, Frank D W. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect Immun. 1999;67:2040–2044. doi: 10.1128/iai.67.4.2040-2044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallis A J, Yahr T L, Barbieri J T, Frank D W. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]