Summary
Agrobacterium tumefaciens causes crown gall disease in plants by the horizontal transfer of oncogenic DNA. The conjugation is mediated by the VirB/D4 type 4 secretion system (T4SS) that assembles an extracellular filament, the T-pilus, and is involved in mating pair formation between A. tumefaciens and the recipient plant cell. Here, we present a 3 Å cryo-EM structure of the T-pilus, solved by helical reconstruction. Our structure reveals that the T-pilus is a stoichiometric assembly of the VirB2 major pilin and phosphatidylglycerol (PG) phospholipid with 5-start helical symmetry. We show that PG-headgroups and the positively charged Arg 91 residues of VirB2 protomers form extensive electrostatic interactions in the lumen of the T-pilus. Mutagenesis of Arg 91 abolished pilus formation. While our T-pilus structure is architecturally similar to previously published conjugative pili structures, T-pilus lumen is narrower and positively charged, raising questions whether the T-pilus is a conduit for ssDNA transfer.
Keywords: Agrobacterium tumefaciens, VirB/D4 T4SS, Bacterial conjugation, T-pilus, helical filament, cryo-EM
Graphical Abstract
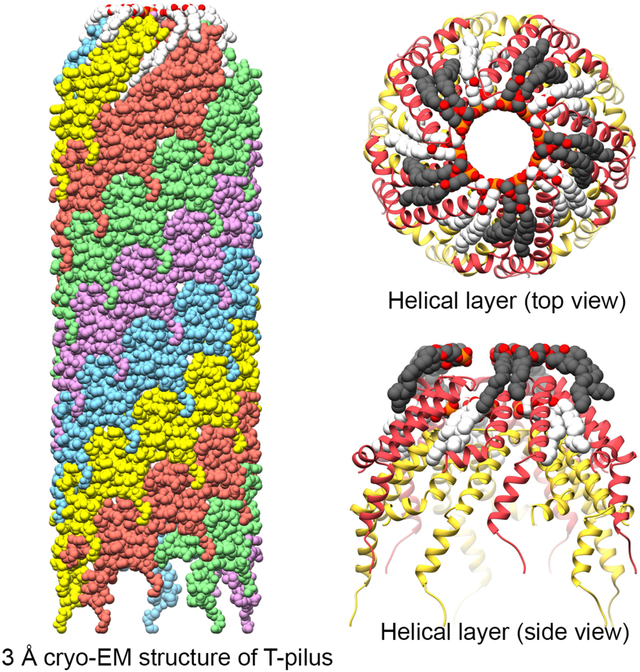
eTOC
The Agrobacterium tumefaciens T-pilus is involved in interkingdom conjugation. Kreida & Akihiro et al. report the cryo-EM structure of the assembled T-pilus, composed of VirB2 proteins and PG. The structure suggests that the T-pilus is an unlikely conduit for ssDNA transfer and will help in elucidating its role in conjugation.
Introduction
Agrobacterium tumefaciens is a plant pathogen that, upon sensing plant wounds, horizontally transfers a segment of tumor-inducing (Ti-) plasmid DNA that is integrated into the host’s genome by nonhomologous recombination. Induction of these genes causes tumorous swelling of the plant tissue, known as crown gall, and the production of opines that can be taken up and catabolized by the bacterium.1–3 A. tumefaciens is routinely used in biotechnology to generate transgenic plants,4 and in the genetic manipulation of fungi.5–7 This plasmid transfer is an example of bacterial conjugation. Extensive study since its discovery in Escherichia coli8 has revealed that conjugation is widespread in prokaryotes and may be of narrow- (between the same bacterial species) or broad- (across phyla or kingdoms) host range.9 Broadly speaking, conjugation allows prokaryotes to adapt to or, as in the case of the Ti-plasmid of A. tumefaciens, create a specific environmental niche, and thus conjugation plays an important role in bacterial evolution and pathogenicity.10
Bacterial conjugation is mediated by the type IV secretion system (T4SS).11 The conjugative T4SS in A. tumefaciens is encoded by the virulence (vir) region of the Ti plasmid and is comprised of 12 proteins (VirB1-VirB11 and VirD4)12,13 that form an envelope-spanning complex and an extracellular filament (T-pilus). The T4SS is directly involved in mating-pair formation and transfer of Ti-plasmid DNA and effector proteins.2,11 The T-pilus is primarily made up of VirB2 (major pilin) and decorated by a tipprotein, VirB5 (minor pilin).14–16 The exact role of the T-pilus in conjugation is unknown. However, a number of mutant alleles in VirB9–11 (reviewed in17) have identified uncoupling phenotypes that are virulent (albeit reduced compared to wild type) but with no detectable pili (Vir+, Pil−). Functional VirB2 was required for virulence in the case of the uncoupling alleles in VirB9 and VirB11; and certain singleamino acid substitutions in VirB2 alone have resulted in the Vir+, Pil− phenotype.18–21 Together this suggests that VirB2 may have multiple roles besides serving as the major pilin subunit of the T-pilus.
Previous structural and biochemical studies of three conjugative pili (pED208 of Salmonella Typhi, F-pilus of E. coli and pKpQIL of Klebsiella pneumoniae) have revealed that they are a 1:1 stoichiometric assemblies of protein and phosphatidylglycerol (PG) phospholipid. All three are so-called sex-pili that are involved in horizontal transfer of conjugative plasmids between bacterial species. They form hollow tube-like structures with outer diameters of 83–87 Å and luminal diameters of 25–28 Å, and with a net-negative luminal charge. The pED208 and F-pilus are highly homologous. The pKpQIL pilus differs mainly in rotational symmetry, i.e. the number of parallel helices in the filamentous structure. The pED208 and F-pilus both have 5-fold rotational symmetry while the pKpQIL is composed of one continuous pilus (1-fold symmetry).22,23
F-pilin subunits are acetylated at the N-terminus following proteolytic cleavage of the propilin.24,25 T-pilins are reported to undergo a covalent cyclization between the N- and C-terminal residues of the mature pilin.26 It is reasonable to assume that this difference may impact pilin packing in the assembled pilus. T-pili have been observed to be particularly resistant to harsh environmental conditions,27 but the structural basis remains unclear.
Here, using cryo-electron microscopy (cryo-EM), we reveal the structure of the T-pilus at 3.0 Å resolution. Our structure, consistent with other conjugative pili, reveals that the T-pilus is a 1:1 stoichiometric assembly of VirB2 pilin proteins and PG phospholipids. The structure is similar to the three reported structures in the near-luminal region that forms the hydrophobic core of the filament as well as the 5-fold symmetry that was shown for the pED208 and F-pilus.22 However, our structure shows a neutral luminal charge with positively charged sidechains protruding towards the luminal core. Moreover, the lumen is narrower compared to other reported structures. Surprisingly, we do not observe cyclization of VirB2 subunits as expected.
Results
The T-pilus is a five-start helix
T-pilus filaments were obtained by growing the nopaline-type pTiC58-derived strain A. tumefaciens NT1REB(pJK270)28 in the presence of acetosyringone as described previously29 (see also STAR Methods). Purified filaments were analyzed by SDS-PAGE (Figure 1A). The gel band, corresponding to the size of processed pilin, 7.2 kDa30 was excised and identified as major pilin VirB2 by in-gel digestion with chymotrypsin followed by mass-spectrometry with sequence coverage of the mature pilin.
Figure 1. Structure determination of the T-pilus.
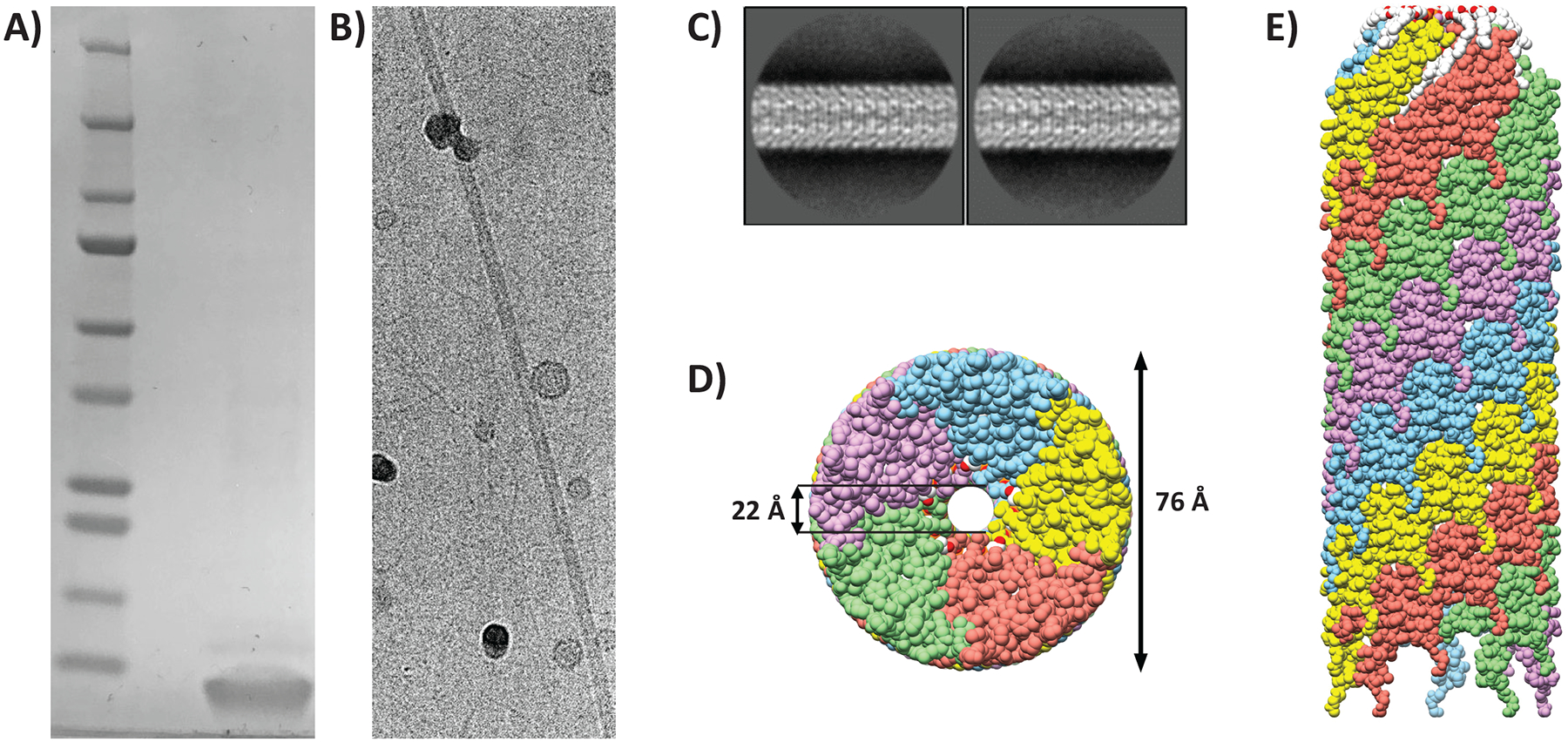
(A) SDS-PAGE of purified VirB2 protein stained with Coomassie (lane 2). Mature VirB2 has a predicted MW of 7.2 kDa. Precision plus molecular weight marker was used (lane 1). (B) Cryo-electron micrograph of intact purified T-pilus (C) Representative 2D-class averages from Relion with a 300 pixel box size. (D) Bottom view of the filament model; coloured by individual VirB2 molecules. (E) Side-view of the filament. The filament is built up by five parallel helices, each of these have a unique colour. The top of the represented filament is towards the tip (as determined before52).
Cryo-EM data on purified pili were collected using a 300 keV Titan Krios microscope with a pixel size of 0.416 Å (super-resolution), later binned to 0.832 Å during processing. Electron micrographs revealed intact pili with lengths of >100 nm (Figure 1B). The data was processed within the Relion framework.31,32 Particles were extracted as overlapping boxes of 300 pixels with a shift of 14 Å and the structure was solved by helical reconstruction. The shift was an estimation of the helical rise by inspection of the 2D-class averages. Representative 2D class-averages are shown in Figure 1C. A total of 3543 boxes were used for the final reconstruction at 3.0 Å (map:map FSC at 0.143) (Figure S1 A–B, Table S1), with applied C5 symmetry (5 parallel helices), a rise of 13.4 Å and helical twist of 32.6°.
The T-pilus is a tightly packed stoichiometric assembly of VirB2 and phospholipid.
The filament appears as a hollow tube with an outer diameter of 76 Å and luminal diameter of 22 Å (Figure 1D and E). The luminal diameter does not include Arg 91 sidechains, which protrude into the lumen but appear flexible. Segmentation of the map density showed that the filament is composed of two distinct densities that appear in the same stoichiometry (Figure 2A). Protein density dominated by alpha-helices fills most of the space and spans the filament at an angle (Figure 2B). This density was assigned to VirB2 (major pilin). The second density resembled a phospholipid (Figure 2A and C) with a small head-group appearing near the lumen and two hydrophobic tails extending along the VirB2 subunits. The lipid was identified as PG 16:1_18:1 (see Lipid determination section below).
Figure 2. The T-pilus is composed of VirB2 and PG-phospholipid.
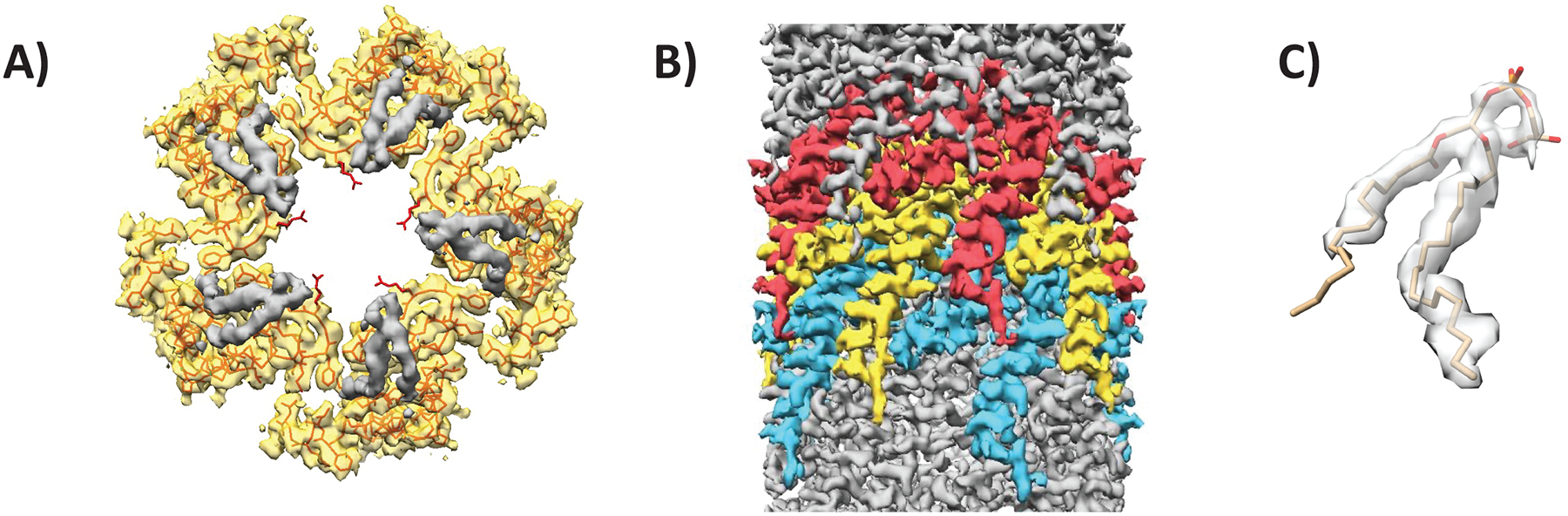
(A) Map density of a helical layer as seen from the top. VirB2-assigned density in yellow with the protein chain represented as a string. Bulky sidechains are shown. PG-assigned density is coloured in grey. Sidechain of Arg 91 is seen extending into the lumen. (B) The filament seen from the side with three layers of VirB2. (C) Map/model coverage of a single PG molecule.
The T-pilus ultrastructure can be described as layered along the helical axis (Figure 3A and B). Each layer contains five copies of VirB2 and five phospholipids. As implied by the helical parameters, the layer is related to the layer above or below by a rotational angle of 32.6° and a rise of 13.4 Å (Figure 3C–E). The rotation positions the phospholipids of layer n+1 (towards the pilus tip) directly above VirB2 protomers of layer n. The result of these helical operations is that each VirB2 subunit has a large interaction surface with acyl chains of four surrounding phospholipid molecules (Figure 4A–C). Similarly, each phospholipid molecule interacts with four neighboring VirB2 monomers forming an intricate interaction network. Moreover, each VirB2 protomer forms extensive hydrophobic interactions, as well as hydrogen bonds via Asn 62 and Gln 73 to neighboring VirB2 protomers (Figure 4D).
Figure 3. Helical symmetry of the T-pilus.
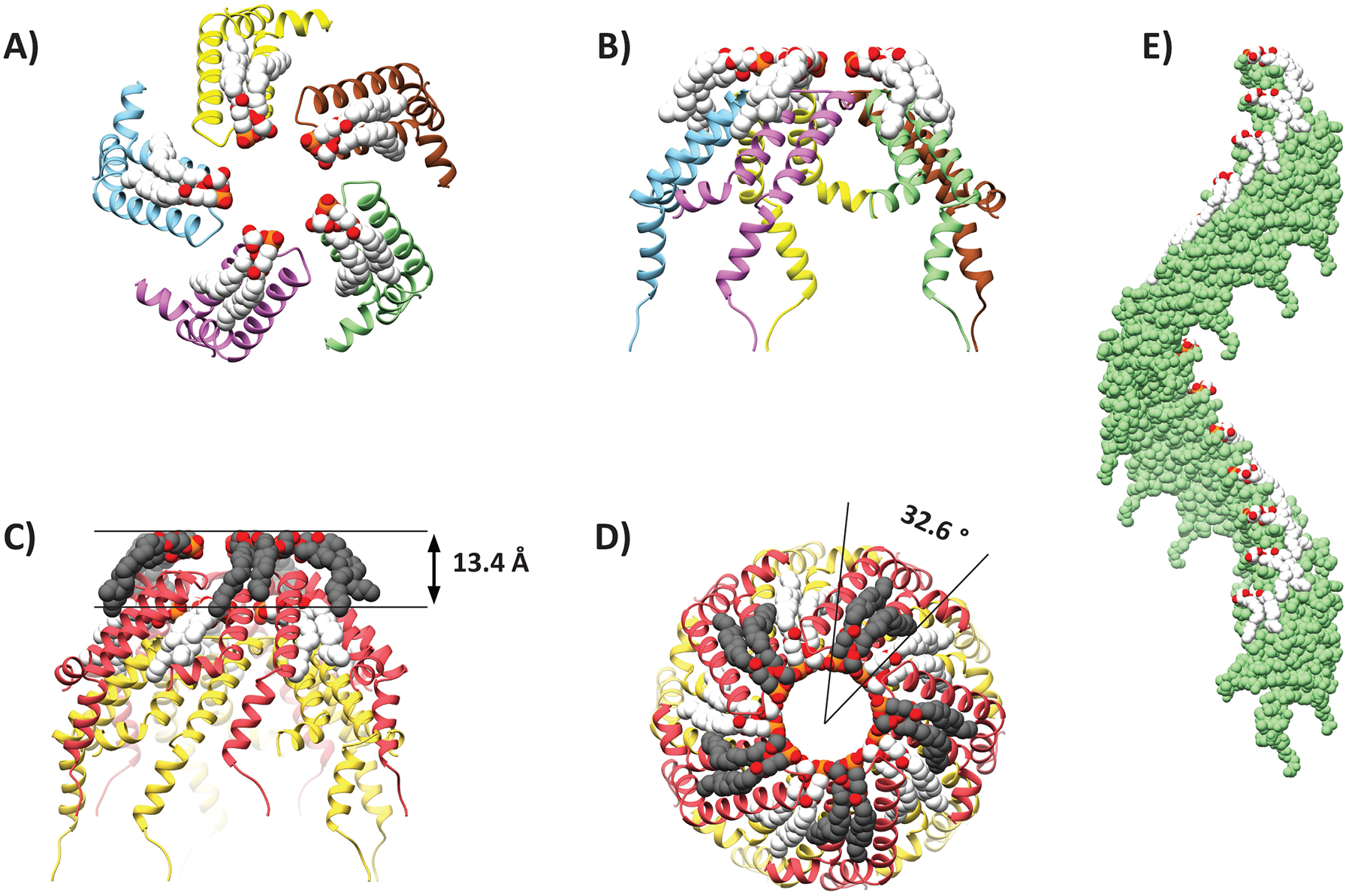
(A-B) Ribbon representation of the top and side view of a helical layer. (C-D) Representation of the helical rise and twist. (E) Helical axis of a single helix in the symmetry.
Figure 4. Interaction network between virB2 monomers and lipid molecules.
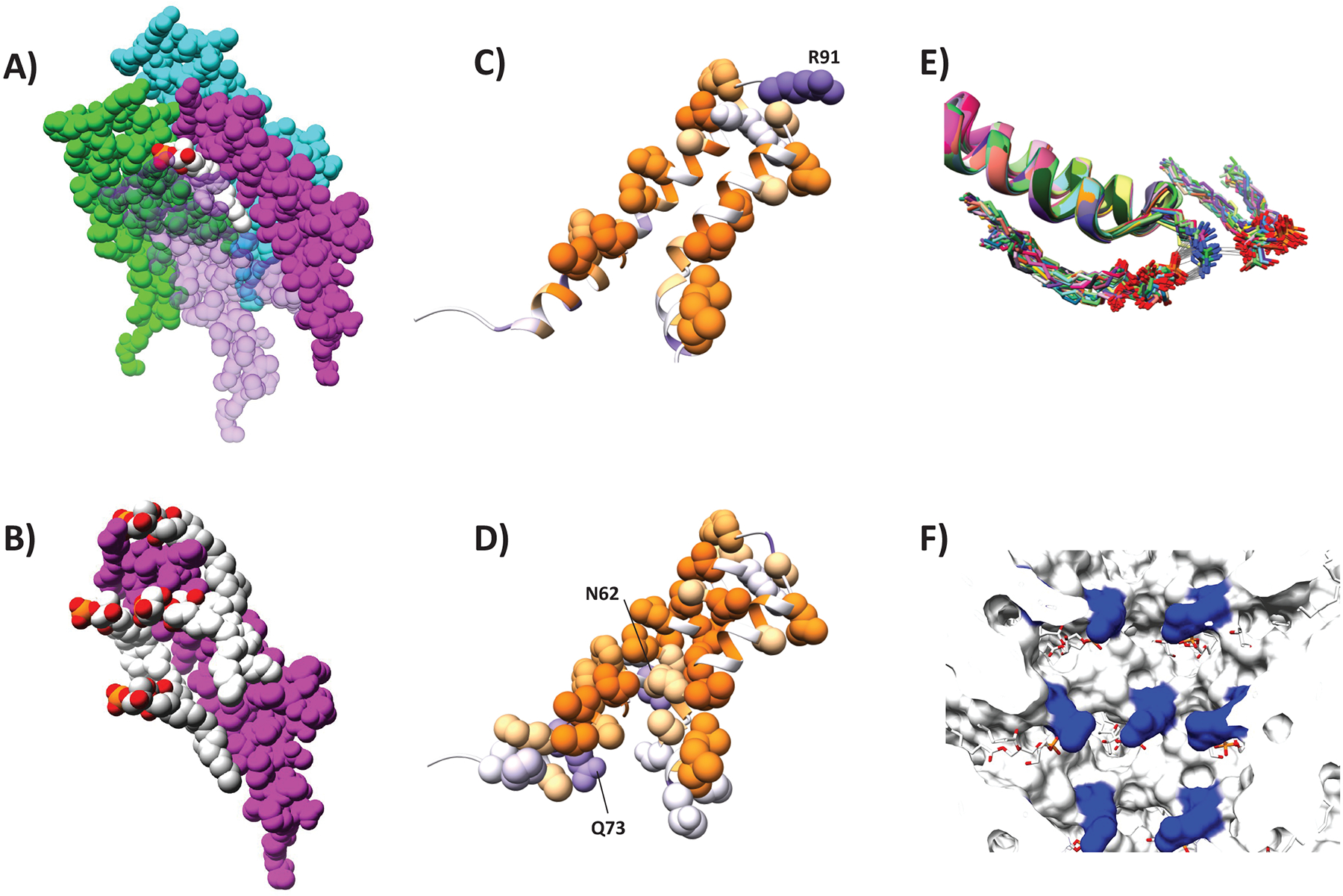
Space filling models showing that (A) each PG molecule (white) interacts with four VirB2 protomers (purple, cyan, green and light pink) and (B) each VirB2 protomer (purple interacts with four PG molecules (white and light gray). VirB2 Interface involved in (C) protein-lipid or (D) protein-protein interaction, colour-scaled by hydrophobicity (purple: hydrophilic, orange: hydrophobic). (E) Flexibility of Arg 91 as determined by MD simulations showing that it is possible for Arg 91 sidechains to interact with two lipidic head groups by electrostatic interaction and hydrogen bonds. 30 unfixed molecules from two independent MD simulations are overlayed. Gray lines indicate hydrogen bonds between lipids and VirB2 protomers. (F) View of the pilus lumen showing Arg 91 (blue) and phospholipids (white/red).
The VirB2 chains are somewhat U-shaped. The termini are located on the outward side of the filament and are not covalently joined – i.e. the VirB2 subunits are not cyclic in our structure as previously reported from mass spectrometry.26 Starting from the N-terminus, the chain runs towards the lumen and forms two helices – α1 and α2 (Figure S2A). The chain turns in the lumen and returns outwards via two helices α3a and α3b. As shown in Figure 4, residues along the length of each protein chain are involved in inter-protein or inter-lipid interactions with multiple proteins and lipids. α2 and α3a are anti-parallel to each other, largely hydrophobic and buried in the structure. The residues separating the two helices form a luminal loop (residues 89–93) that holds Arg 91, the only positively charged residue in the lumen (Figure 4C,E–F). The position of the Arg 91 sidechain is not well resolved the map density (Figure 2A). Molecular dynamics simulations (see STAR Methods) showed that the position of the arginine sidechain fluctuates so that the guanidino groups can interact with two separate phosphate groups (the only negatively charged functional groups in the lumen) in the helical assembly (Figure 4E). This results in a network of electrostatic interactions in the lumen with no net charge (Figure 4F, Figure S2 B–C).
The luminal loop is important for T-pilus formation and virulence
To investigate the importance of the VirB2 luminal loop, we generated mutant VirB2 proteins containing individual R91E, R91A and S93A mutations. We tested these mutants for their ability to form an active pilus using western blot and functional assays (Figure 5A). Mutant versions of VirB2 were expressed in A. tumefaciens PC1002 that contains a nonpolar deletion in virB2. Expression was monitored by Western blot using anti-VirB2 specific antibodies. The western blot showed no detectable band for R91E and R91A, indicating that these mutations destabilized VirB2 (Figure 5A). VirB2 S93A mutant was stable but accumulated at a lower level. Analysis with anti-VirB10 antibody showed that vir genes were induced in all mutants. Negative staining of cells grown under vir-inducing conditions showed that wild type cells and S93A displayed T-pili, whereas no pili were observed in R91A (n=80). Representative examples are shown in Figure S3.
Figure 5. Importance of luminal loop in structure and function of the T-pilus.
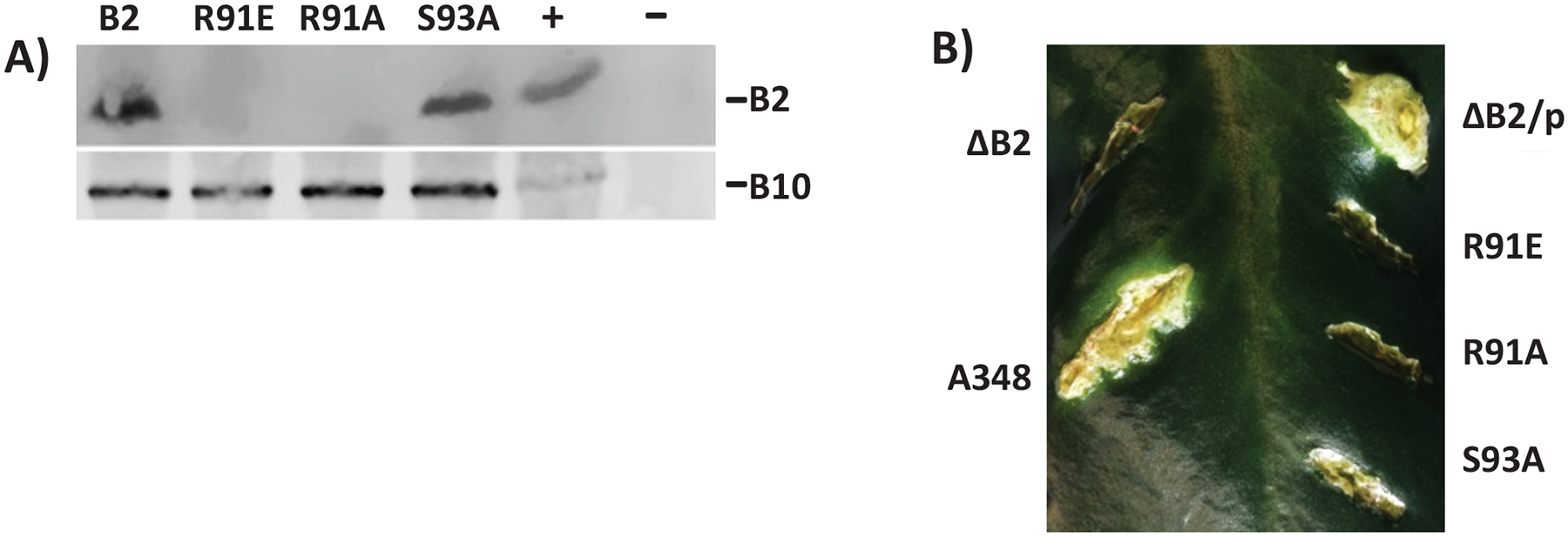
(A) Analysis of VirB2 mutant protein accumulation by western blot assays. A. tumefaciens expressing virB2 or its mutants were analyzed by SDS-PAGE followed by western blot assays using anti-VirB2 antibodies. The blot was reprobed with anti-VirB10 antibodies as a control for induction of the vir genes. -, -Ti plasmid; +, pTi. (B) Effect on mutations in VirB2 Arginine 91 and Serine 93 on A. tumefaciens virulence. The effect of a mutation in virB2 was monitored by complementation assays. Agrobacterium PC1002 (ΔB2) and its derivatives harbouring a plasmid that expressed virB2 (p) or its mutant derivatives B2R91E (R91E), B2R91A (R91A) or B2S93A (S93A) were used to infect K. daigremontiana leaves.
The Kalanchoe daigremontiana leaf infection assay is a standard functional assay used for monitoring Vir protein function.33 The results of the functional assays correlated well with the pilus protein expression levels. Strains with VirB2 mutants R91E and R91A failed to form tumors indicating both mutants were non-functional in DNA transfer. The VirB2 S93A mutant is functional since bacteria producing this protein showed reduced virulence (Figure 5B). The reduction in virulence can be due to its lower abundance in the cell and/or a negative effect on its function.
The T-pilus incorporates PG
The lipid was identified by shotgun lipidomics spectrometry, in which the lipid content of purified pili was compared to that of a negative control. The negative control was produced by growing A. tumefaciens under non-inducing conditions and subjecting the cells to the same purification procedure as for the pili-containing sample. Samples were analyzed in triplicates from three independent preparations and the presence or absence of pili was verified by negative staining.
Lipids were extracted from the sample using a modified Bligh and Dyer extraction procedure and were analyzed for lipid content.34,35 Lipid standards of 1450 lipid species were added to annotate lipids in biological samples. An independent experiment in which different concentrations of the same preparation (pili and negative control, respectively) were analyzed for lipid content (data not shown). Lipid species that did not increase in abundance with increasing sample concentration were considered contaminants (e.g. from tubes, lipid extraction solutions) and disregarded from analysis. A number of PE species (see STAR Methods) were not annotated in the selection experiment but appeared in the analytical experiment that was done in triplicates. This group included PLs containing heavily unsaturated acyl chains 16:3 and 20:5 that are unlikely and unreported to be present in A. tumefaciens. As there was no unbiased way of excluding these, they were included in the analysis. They were unchanged between pili sample and negative control. The selection yielded 24 non-contaminant lipid species in the samples, all belonging to PE, PG and PC classes. (Table S2).
If the T-pilus itself incorporates a lipid, it would be expected that a specific lipid species would increase as a fraction of the total composition in the pili-containing sample compared to the negative control. PG 16:1_18:1 stands out from the 24 selected lipid species as it shows a 9.2-fold increase in the pili-containing sample (MANOVA, p<0.001) (Figure S4A). It was necessary to mathematically quantify how important this increase was compared to all changes in the composition to a draw a conclusion. Unaveraged data was therefore transformed to centered log-ratios (CLRs) and the variance was calculated for each lipid species from all 6 observations (3 from pili and 3 from negative control). A large difference between induced and non-induced cells would thus result in a high variance. The variance of each lipid species was then divided by the sum of that of all species and normalized to 100% (contribution to total variance) (Figure S4B). This metric showed that the observed increase in PG 16:1_18:1 accounted for 37% of the total variance in the dataset, which is large considering that 24 lipids species were included. The analysis further revealed that PG 18:1_18:1 increases 3-fold (p<0.001) and that this increase accounted for 13% of the total variance in the dataset. Since PG 16:1_18:1 and PG 18:1_18:1 are chemically similar, a model in which the T-pilus incorporates both species is likely and would explain 50% of the total variance in the dataset. We therefore propose that the T-pilus is promiscuous and incorporates both these lipids, but preferentially PG 16:1_18:1, which was modelled into the structure.
Discussion
Conjugative pili have been classified by morphology and general functional characteristics. So-called F-type pili (e.g., pED208, F-pilus) and pKpQIL-pili are characterized by thin, long and flexible filaments that promote efficient conjugation in both solid and liquid environments and have a narrow host-range. In contrast, the P-type pili (which includes the T-pilus) are generally thicker, shorter and more rigid, are more efficient in solid environments and have a broader host-range.36,37 These traits should be seen as general trends that reveal evolutionary origins and exceptions exist, e.g., the T-pilus is thinner than the F-pilus and forms long filaments (Figure S3).
Our near-atomic resolution structure of the T-pilus revealed that it is a stoichiometric assembly of protein and PG phospholipid, similar to the F-type pili (pED208, F-pilus and pKpQIL conjugative plasmid-derived pili), described earlier.22,23 Since this is the fourth conjugative pilus structure solved to date, it seems likely that all conjugative T4SS pili are stoichiometric protein-lipid assemblies. The phospholipids have a clear structural role in the T-pilus and can be likened to putty that fills the gaps between VirB2 subunits. Moreover, positively charged residues in the luminal loop (a conserved feature in VirB2/TraA pilins38) and the negatively charged phospholipid headgroups create a network of alternating positive and negative charge that may have a function in stabilizing the structure of the lumen. Beside this structural role, we speculate that the lipids may play functional roles in establishing contact (or fusing) with the target membrane or facilitate movement of pilin subunits back into the membrane during pilus retraction. This should be the subject of future investigations.
VirB2 adopts a distinct fold with structural homology to that of the other known conjugative pili structures. Structural comparison between the T- and F-pili (PDB ID: 5LEG) revealed that they are architecturally similar in the near-luminal and buried regions composed of the parallel helices α2 and α3 (Figure 6). However, VirB2 has an elongated α1 and a 72 αkink in α3 at Gly 109. G109A substitution was previously shown to abolish pilus formation and virulence,21 suggesting that division of α3 into two helices (here called α3a and α3b) is important for T-pilus stability. The structural difference between T- and F-pili pilins likely causes the difference in packing of the assembled filament. The result is that the T-pilus is denser (with smaller filament and luminal diameters) than the F-pilus despite the subunits being of similar size and molecular weight. The luminal diameters of the F-family pili are 28 Å, whereas that of the T-pilus was 22 Å, not including the Arg 91 sidechains. The net charge of the lumen is also different. Whereas known conjugative pili have a net-negative luminal charge, the T-pilus has a net neutral charge, but with positively charged sidechains extending towards the center of the lumen (Figure S2). As the positive charge would attract the negatively charged ssDNA, it is unclear whether this would facilitate ssDNA transfer.
Figure 6. Structural Comparison of the T- and F-pilus.
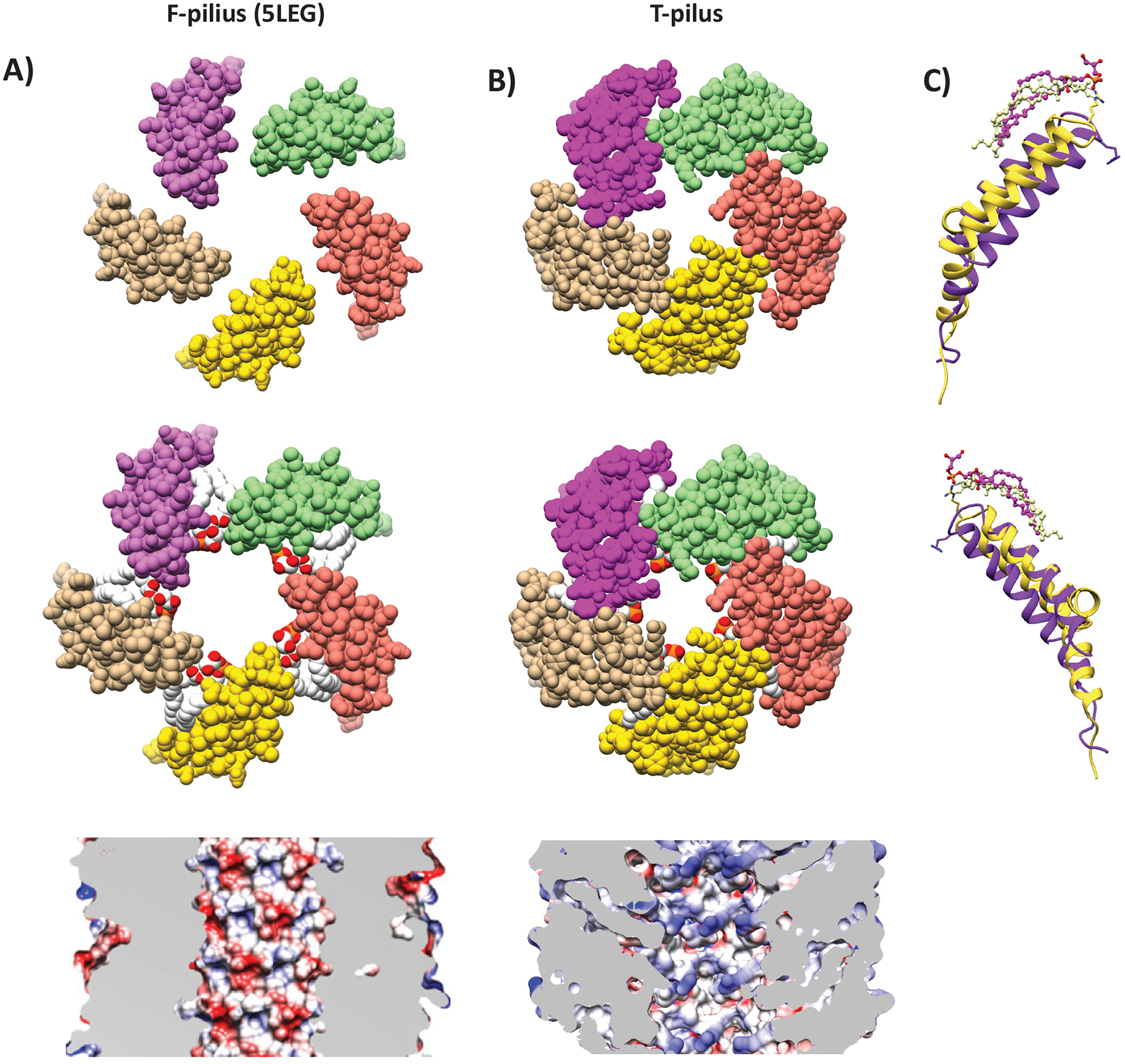
Helical cross-sections showing organization of pilins and phospholipids in (A) F-pilus (PDB ID: 5LEG) and (B) T-pilus. The T-pilus adopts a denser structure with narrower lumen. The F-pilus lumen is moderately negatively charged (reproduced from Costa et al., 2016), whereas T-pilus lumen is neutral. (C) Structural overlay and secondary structure annotation of T-pilus VirB2 (yellow) and F-pilus TraA (purple). Phospholipids from equivalent positions in the helical packing are displayed.
Surprisingly, VirB2 did not form cyclic subunits as seen previously by mass-spectrometry of purified T-pili.26 The first and last three residues of mature VirB2 were not modeled due to poor map density at the termini, but the modelled termini were ~30 Å apart in the structure - too far to be connected by a cyclic linkage. Cyclic VirB2 may thus represent an intermediate form that is later linearized or, as VirB2 appears to have multiple roles during mating pair formation, a different functional form.
The T-pilus has 5-fold rotational symmetry, just like pED208 and F-pilus, but different from the pKpQIL conjugative pilus. The recent cryo-EM structure of the R388 conjugative T4SS exhibited a previously unseen stalk complex in the periplasm between the inner- and outer membrane core complexes (OMCC).39 The stalk was made up of pentamers of VirB6 and thus matched the 5-fold rotational symmetry of the F-like pili. A model was therefore proposed that the pilus is formed on top of the VirB6 platform, with the pentameric VirB5 tip being pushed up through the OMCC by polymerization of VirB2. This dictates the filament must be narrower than the periplasmic cavity of the OMCC pore to be able to fit and be 5-fold symmetric. The OMCC of VirB/D4 was previously determined to have an inner diameter of 82 Å.40 The T-pilus has the required 5-fold symmetry and diameter of only 76 Å. The structure is thus in agreement with the model of pilus assembly proposed by Macé et al.39
The role of the luminal Arg 91 is interesting. The model presented here suggests that it has a structural role in creating a patchwork of electrostatic interactions with the lipid headgroups. It has previously been shown that R91A substitution results in loss of virulence and deficiency of VirB2 pre-pilins, suggesting that it has a role in folding in the inner membrane.21 These findings are supported by our K. daigremontiana leaf infection assay. Substitution of Arg 91 to either alanine or glutamic acid resulted in no detectable pili or virulence. Furthermore, S93A substitution, a residue of the luminal loop that is not involved in lipid interaction, resulted in reduced virulence and reduced extracellular pilus formation. These effects could be attributed to structural changes of the luminal loop, which effect helical packing (as was observed in23 and T-pilus stability.
PG was also found in F-type pili, suggesting it to be a conserved feature in conjugative T4SS pili. In support of this identification, A. tumefaciens contains two virulence genes, lpiA (a Lysyl-PG synthase) and acvB/VirJ (Lysyl-PG hydrolases), that are involved in PG maturation in the inner membrane.41–43 In C58-derived A. tumefaciens strains these genes are located chromosomally in the same operon and are, like the T-pilus machine, both induced at low pH.44–46 Moreover, the octopoine pTiA6 plasmid contains VirJ (an acvB homologue) directly under the control of VirA/VirG, the two-component system that is induced by certain plant phenols (e.g acetosyringotone) and controls the transcription of Vir genes.47 These PG-maturation genes are thus co-induced with the T-pilus machine. Interestingly, acvB inactivation was shown not to affect cellular levels of VirB8 and VirB9 (used as markers for T4SS) but reduced levels of the minor pilin VirB5 (VirB2 levels were not monitored). These findings together suggest that PG maturation has an impact on pilus formation, which is explained by our findings. We note that VirA/VirG-induced PG maturation and VirB2 folding both occur in the inner membrane and VirB2 extraction was proposed to be mediated by VirB4.41,48 It is thus possible that VirB2 and PG are removed from the inner membrane together for assembly into the pilus in a 1:1 ratio. Phosphatidylcholine (PC) synthase has also been identified as a critical virulence factor.49 However, PC-deficient mutants did not produce T4SS machines and showed no sensitivity to acetosyringone,50 suggesting that PC-deficiency has an upstream effect on VirA/VirG regulation of the vir operon.
In a contemporary study, Amro et al also resolved the structure of the A. tumefaciens T-pilus at comparable 3.2 Å resolution.51 While their structure and ours are very similar, they identified the lipid as PC. PC species were present in the pili-containing fraction of our experiment, but their part of the composition did not increase substantially compared to the negative control (Table S2), causing us to reject PC as a candidate for the lipid. Notably, PG species were not identified at all in A. tumefaciens membranes in this study; however, A. tumefaciens has been shown to contain PG species, (49 and this study) and as stated above, holds genes for PG maturation. The absence of PG in their study may therefore be due to strain-related differences, or perhaps their experimental setup did not allow detection of PG.
Pili make part of prokaryotes’ toolbox to interact with their environment. While the similarities of the T-pilus structure presented here with that of other conjugative pili is striking and suggest a conserved function, the differences may indicate interesting specializations.
STAR ★METHODS
RESOURCE AVAILABILITY
Lead contact
Further inquiries and request for data, strains and resources should be directed to the lead contact Debnath Ghosal, debnath.ghosal@unimelb.edu.au
Materials availability
A. tumefaciens stains and plasmids generated in this study (see Key Resource Table) are available upon request from the lead contact.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-VirB2 | Mossey et al., 2010.33 | N/A |
| Bacterial and virus strains | ||
| A. tumefaciens NT1REB(pJK270) | Chesnokova et al., 1997.28 | N/A |
| A. tumefaciens PC1002 | Berger and Christie, 1994.53 | N/A |
| Biological samples | ||
| Kalanchoe daigremontiana | Prof. Eugene W. Nester, Dept. of Microbiology & Immunology, University of Seattle, WA. | N/A |
| Critical commercial assays | ||
| QuickChangee II site-directed mutagenesis kit | Agilent Technologies | Cat#200523 |
| Deposited data | ||
| Cryo-EM map of the T-pilus | This study | EMD-28957 |
| Structure model of the T-pilus | This study | PDB ID: 8FAI |
| Oligonucleotides | ||
| Primers: see VirB2 mutagenesis section | Integrated DNA Technologies inc., Coralville, IA. | N/A |
| Recombinant DNA | ||
| pAD1412 | Mossey et al., 2010.33 | N/A |
| pAD1891 | Mossey et al., 2010.33 | N/A |
| pAD2161 (B2wt) | This study | N/A |
| pAD2162 (B2R91E) | This study | N/A |
| pAD2163 (B2R91A) | This study | N/A |
| pAD2164 (B2S93A) | This study | N/A |
| Software and algorithms | ||
| R version 4.1.2 | R Core Team, 2013.66 | http://www.r-project.org |
| Serial-EM | Mastronarde 2005.54 | http://bio3d.colorado.edu/SerialEM |
| Relion 4.0beta2 | Scheres, 2012.32 | https://relion.readthedocs.io |
| Motioncorr2 | Zheng et al., 2017.55 | https://relion.readthedocs.io |
| CTFFIND-4.1 | Rohou and Grigorieff, 2015.56 | https://grigoriefflab.umassmed.edu/ |
| SWISS-MODEL | Guex et al., 2009.57 | https://swissmodel.expasy.org |
| Rosetta3 | Leaver-Fay et al., 2011.58 | https://www.rosettacommons.org |
| Coot | Emsley et al., 2010.59 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| ChimeraX | Goddard et al., 2018.60 | https://www.cgl.ucsf.edu/chimerax/ |
| Phenix | Adams et al., 2011.62 | https://phenix-online.org |
| NAMD | Philips et al., 2020.63 | http://www.ks.uiuc.edu/Research/namd/ |
Data and code availability
The Cryo-EM map and model have been deposited to EMBD (ID: EMD-28957) and RCSB (PDB ID: 8FAI), respectively. The raw data is available upon request.
This paper does not report original code.
Any additional information required to reanalyze the data, including lipidomics raw data, presented in this study is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
A. tumefaciens strains
The nopaline-type pTiC58-derived strain A. tumefaciens NT1REB(pJK270)28 was used for pili purification for cryo-EM and shotgun lipidomics. This strain contains mutations that impair flagellum formation (known as a “bald” strain), which was verified by negative staining of cells. For functional assays, WB and negative staining of cells the PC1002 stain was used. This strain is derivative of the wild-type Agrobacterium strain A348 that carries the octopine type Ti-plasmid pTiA6NC and has a nonpolar deletion in virB2.53 Cells were grown in AB medium (1 gL−1 NH4Cl, 0.3 Gl−1 MgSO4 7 × H2O, 0.15 gL−1 KCl, 10 mgL−1 CaCl2, 2.5 mgL−1 FeSO4 7xH2O, 3.9 gL−1 MES, 10 gL−1 Glucose, 10 mM Na/KPO4,) at pH of 5.5–5.8. For vir induction, cells were grown on 1.5% bacto agar AB-plates with 300 μM acetosyringone.
Kalanchoe daigremontiana
K. daigremontiana plants were originally obtained from the Nester lab (Prof. Eugene W. Nester, Dept. of Microbiology & Immunology, University of Washington, Seattle, WA). Kalanchoe plants are easy to grow and infect. They are very sensitive to Agrobacterium.
METHOD DETAILS
A. tumefaciens growth
A. tumefaciens cells were grown and purified under conditions similar to those described previously.29 An overnight culture in LB was used to inoculate AB/pH 5.5 buffer to an OD600 of 0.1. Cells were grown at 28°C in a shake-incubator at 220 rpm. For (non-induced) negative control, 2.4 L of culture was grown for 72 h. For induced cells, a 100 mL of culture was grown for 5 h, then centrifuged at 4,000 × g (20 min) at room temperature and the pellet resuspended in ¼ of the original volume in fresh AB/pH 5.5 buffer. 0.5 mL of this suspension was spread per each 15 mm petri dishes containing AB/pH 5.5 buffer, 1.5% Bacto Agar (Difco), 300 μM acetosyringone (Sigma). 40 such plates were made for each preparation. The plates were incubated at 22°C (72 hours).
Pili purification for cryo-EM and lipid analysis
The A. tumefaciens NT1REB(pJK270) strain28 was used for pili purification. Cells were grown as described above. Induced and non-induced cells were harvested by centrifugation at 6,000 × g at 4°C. Induced cells were scraped from plates and resuspended in 50 mM MES, pH 5.5 prior to centrifugation.
Cells were resuspended in 12 mL 50 mM MES and passed through a 26-gauge syringe needle ten times in and out. This procedure causes the cells to shed its pili. The suspension was centrifuged at 10,000 × g at 4°C (1 h). The supernatant was then ultracentrifuged at 100,000 × g (1.5 h). The resulting pellet was resuspended in 50 mM MES pH 5.5. The pilin was identified as VirB2 by mass-spec (Q-Exactive HF, LCMS) at the Caltech Proteome Exploration Laboratory. Presence or absence of pilus filaments was confirmed by negative staining on formvar grids with 3% uranyl acetate (30 s).
Sample preparation and data collection for Cryo-EM
Pili sample was cryogenically frozen on a FEI Vitrobot MK4, set to 22 °C and 100% humidity. 3 uL of sample was blotted on glow-discharged UltraAuFoil EM-grids (R1.2/1.3, 300 mesh, Electron Microscopy Sciences), with 3 s of wait time and 7 sec of blot time. Cryo-electron microscopy was done in the Beckman Institute Resource Center for Transmission Electron Microscopy at Caltech on a 300 kV FEI Titan Krios equipped with a Gatan K3 direct detector and Gatan energy filter. Acquisition software was Serial-EM.54 Movies of 40 frames were collected at a calibrated pixel size of 0.416 Å (super res), with a total dose of 60 e−/Å2. The defocus range was set to −0.6 to −2.0.
Helical reconstruction and data processing
The data was processed in Relion by helical reconstruction,31,32 using Motioncorr2 for motion correction55 and CTFFIND-4.156 for CTF-estimation. The data was downsized to 0.832 Å and overlapping 300×300 pixel boxes were extracted with a 14 Å shift (~equivalent to a helical rise). A 7 nm cylinder was use as a starting model for 3D classification. The initial helical rise and pitch were estimated using the 2D class averages. 3D classification was then used to screen the different combination of twist and n-fold symmetry that would be possible given the estimated helical rise and pitch. The highest resolving 3D class averages were 3D-refined and visually inspected in Chimera for features expected at the given resolution. 3543 boxes were used in the final reconstruction with a rise of 13.4 Å, helical twist of 32.6° and C5 symmetry.
Model building and molecular dynamics simulations
The Initial model was generated with SWISS-MODEL57 from the mature VirB2 sequence and fitted to the density map using Rosetta3.58 The resulting model was iteratively refined with COOT59; molecular dynamics flexible fitting (MDFF), using the ChimeraX extension ISOLDE60,61; and Phenix.62
Molecular Dynamics simulations were performed in NAMD63 to estimate the dynamics of the Arg 91 sidechain. The simulations included 5 layers of proteins and lipids, explicit waters and 100 mM NaCl. The top and bottom layers were fixed. All Cα atoms in the protein, except for the five residues surrounding Arg 91; and all non-H atoms in the lipids were under weak harmonic constraints with k = 0.3 kcal mol−1 Å−2. The simulations were performed at 300 K for 1 nsec. Two independent simulations were performed.
virB2 mutagenesis
Mutations in virB2 were introduced using QuikChange II site-directed mutagenesis kit (Agilent Technologies). Plasmid pAD1891 DNA, a pUC118 derivative with a virDp-virB2 chimeric gene was used as template DNA. Mutations were introduced by PCR amplification using primer pairs (mutations in bold):
GTTCGGGGAGGCTTCGCTTGGGCTG and GCGAAGCCTCCCCGAACATCCAGGAG (R91E); GTTCGGGGCGGCTTCGCTTGGGC and GCGAAGCCGCCCCGAACATCCAGGAG (R91A) or GGGCGGGCAGCGCTTGGGCTGGTTG and CCCAAGCGCTGCCCGCCCGAACATCC (S93A).
The mutations were confirmed by DNA sequence analysis of the entire genes. The plasmids were fused to the plasmid pAD141233 to construct the wide-host-range derivatives. Plasmids were introduced into Agrobacterium PC1002 by electroporation. PC1002 has a nonpolar deletion in virB2.53 The Vir proteins of the Ti-plasmids are highly similar and are functionally equivalent in functional studies. The PTiA6NC VirB2 mature peptide is nearly identical to the pTiC58 VirB2 and only differ at the C-terminal end (K115Q, G121S of the mature protein). Both A. tumefaciens PC1002 an NT1REB(pJK270) used in structural studies also have a near-identical chromosome. The NT1REB has an additional mutation that abolishes flagellum formation.28
Protein analysis and functional assays:
Complementation assays were used to monitor the effect of a mutation on virB2 function.33 Agrobacterium PC1002 derivatives harboring a plasmid expressing virB2 or its mutant were used to infect K. daigremontiana leaves. A fully expanded young leaf was wounded by scratching the surface 2–3 times with a sterile toothpick. 5 uL of A. tumefaciens cells grown overnight were then gently spread over the wound site. Tumor formation was monitored 3 weeks after infection.
Accumulation of VirB2 mutant proteins was analyzed by western blot assays.33 A GST-VirB2 fusion protein containing the C-terminal 74 residues of VirB2 was used as antigen to raise polyclonal antibodies in rabbits. Antibodies were purified by affinity chromatography on an affigel – GST-VirB2 fusion column. Total proteins from induced Agrobacterium strains were separated by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane and probed with anti-VirB2 antibodies.
Negative staining and imaging of A. tumefaciens cells
A. tumefaciens cells were grown on plates in presence of acetosyringone as described above. Cells were collected after 72 hours using a sterile cotton swab and resuspended in 50 mM MES pH 5.5. Cells were not centrifuged to prevent pili breaking from the cells. 3 μL of this suspension was deposited on glow-discharged 300 mesh formvar/carbon copper grids (Electron Microscopy Sciences), washed once with water and stained with 0.5% w/v phosphotungstic acid, pH 5.5, for 30 s. Cells were imaged on a 120 kV FEI Tecnei Ti12 with LAB6 filament and Gatan Ultrascan 2k × 2k CCD.
Lipid data collection, selection and treatment
Lipid extraction and mass spectrometry lipidomics analysis.
Lipid Extraction and mass spectrometry was performed by UCLA Lipidomics. A modified Bligh and Dyer extraction35 was done to extract lipids from samples. Prior to extraction, 70 lipid standards across 17 subclasses were added to each sample (AB Sciex 5040156, Avanti 330827, Avanti 330830, Avanti, 791642). Extraction was performed twice, and the pooled organic layers were dried by Speedvac (Thermo, SPD300DDA) with ramp setting 4, 35°C for 45 min with a total run time of 90 min. Samples were resuspended in 1:1 methanol/dichloromethane with 10 mM ammonium acetate and transferred to robovials (Thermo, 10800107) for analysis.
Samples were analyzed by direct infusion on a Sciex 5500 with differential mobility device (DMS), tuned with EquiSPLASH LIPIDOMIX (Avanti 330731) with a targeted acquisition list of 1450 lipid species. Descriptions of instruments settings, MRM list and in-house data analysis workflow is available.34
Lipid selection and lipid data analysis
Three independently purified samples of purified pili and negative control were analyzed for lipid content. Lipids were selected for subsequent analysis by an independent experiment, in which dilution series (1x, 0.5x, 0.25x) of induced and non-induced pili preparation were analyzed for lipid content. Lipid species were selected if they increased in abundance (in nmoles) with increasing sample concentration. Some lipid species were not annotated in the selection experiment but appeared in different amounts in induced and non-induced cells and were included in the analysis. These were: PE 14:0_16:3, 14:0_18.2, 14:1_18:2, 15:0_18.2, 15:0_20.5, 16:0_16:2, 16:0_16:3, 16:0_17.1. The total number of selected lipid species were 24 (Table S2).
4 out of 144 measurements (2.78%) were missing values and needed to be generated to be able to log-transform the data for subsequent multivariate statistical analysis. The missing values were in species PE 14:0_16:2, PE 14:0_16:3 and PE 16:0_17:1. Visual inspection of the raw data showed that the main variation was between, rather than within triplicates of induced and non-induced cells. Moreover, existing data of affected lipid species did not suggest that the missing values were missing because of being below the detection limit. Replacing the missing values with low positive values would thus risk introducing false positives. Instead, they were estimated using existing measurements within the triplicates using the R-package zCompositions.64 SD was not calculated for these lipid species.
Data was treated as compositional and closed to 100% (table 1). For MANOVA and variance calculations, data was first transformed to centered logratios (CLRs).65 with weights proportional to species abundance (giving higher weight to more abundant species in the compositions). Variance was calculated for each lipid species as the variance of all observations (induced and non-induced cells, n = 6), divided by the sum of variance of all species and closed to 100%.
Quantification and statistical analysis
Shotgun lipidomics data was analyzed in R.66 SD calculation and MANOVA was performed in R. Missing values were estimated using the R-package zComposition.64 CLR transformation and variance calculations were done using the R package easyCODA.65 Plots were produced in ggplot2.67
Supplementary Material
Highlights.
Cryo-EM helical reconstruction of the A. tumefaciens T-pilus at 3 Å resolution
The T-pilus is a stoichiometric protein-phospholipid assembly
The T-pilus lumen is narrow and positively charged - an unlikely conduit of ssDNA
VirB2 monomers are not covalently joined at the termini in the assembled filament
Acknowledgements
We thank Dr. Songye Chen, Caltech cryo-EM facility for assistance during data collection; Dr. Tsui-Fen Chou, Dr. Brett Lomenick and Dr. Jeff Jones from Caltech Proteome Exploration Laboratory for conducting the protein mass-spec analysis. We also thank Dr. Kevin Williams and UCLA Lipidomics for performing lipid extraction, lipid data collection and giving valuable advice in lipidomic experimental design and data interpretation. This project was funded by a National Institutes of Health grant (R01 AI127401 to G.J.J), a National Health and Medical Research Council grant (APP1196924 to DG) and a Natural Sciences and Engineering Research Council of Canada Discovery grant (RGPIN 04345 to EIT). SK is supported by the Swedish Research Council (2019-06293).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Dessaux Y, Petit A, Farrand SK, and Murphy PJ (1998). Opines and Opine-Like Molecules Involved in Plant-Rhizobiaceae Interactions BT - The Rhizobiaceae: Molecular Biology of Model Plant-Associated Bacteria. In, Spaink HP, Kondorosi A, and Hooykaas PJJ, eds. (Springer; Netherlands: ), pp. 173–197. 10.1007/978-94-011-5060-6_9. [DOI] [Google Scholar]
- 2.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJJ, Farrand SK, and Winans SC (2000). The bases of crown gall tumorigenesis. J. Bacteriol 182, 3885–3895. 10.1128/JB.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Attikum H, Bundock P, and Hooykaas PJJ (2001). Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 20, 6550–6558. 10.1093/emboj/20.22.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang H-H, Yu M, and Lai E-M (2017). Agrobacterium-Mediated Plant Transformation: Biology and Applications. Arab. B 2017. 10.1199/tab.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Groot MJA, Bundock P, Hooykaas PJJ, and Beijersbergen AGM (1998). Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol 16, 839–842. 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 6.Bundock P, Den Dulk-Ras A, Beijersbergen A, and Hooykaas PJJ (1995). Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214. 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Idnurm A, Bailey AM, Cairns TC, Elliott CE, Foster GD, Ianiri G, and Jeon J (2017). A silver bullet in a golden age of functional genomics: The impact of agrobacterium-mediated transformation of fungi. Fungal Biol. Biotechnol 4, 1–28. 10.1186/s40694017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lederberg J, and Tatum EL (1946). Gene recombination in Escherichia coli. Nature 158, 558. [DOI] [PubMed] [Google Scholar]
- 9.Virolle C, Goldlust K, Djermoun S, Bigot S, and Lesterlin C (2020). Plasmid transfer by conjugation in gram-negative bacteria: From the cellular to the community level. Genes (Basel). 11, 1–33. 10.3390/genes11111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norman A, Hansen LH, and Sørensen SJ (2009). Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. B Biol. Sci 364, 2275–2289. 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa TRD, Harb L, Khara P, Zeng L, Hu B, and Christie PJ (2021). Type IV secretion systems: advances in structure, function, and activation. Mol. Microbiol 115, 436–452. 10.1111/mmi.14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirasu K, Morel P, and Kado CI (1990). Characterization of the virB operon of an Agrobacterium tumefaciens Ti plasmid: nucleotide sequence and protein analysis. Mol. Microbiol 4, 1153–1163. 10.1111/j.1365-2958.1990.tb00690.x. [DOI] [PubMed] [Google Scholar]
- 13.Kuldau GA, De Vos G, Owen J, McCaffrey G, and Zambryski P (1990). The virB operon of Agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. Mol. Gen. Genet. MGG 221, 256–266. 10.1007/BF00261729. [DOI] [PubMed] [Google Scholar]
- 14.Beijersbergen A, Smith SJ, and Hooykaas PJJ (1994). Localization and Topology of VirB Proteins of Agrobacterium tumefaciens. Plasmid 32, 212–218. 10.1006/plas.1994.1057. [DOI] [PubMed] [Google Scholar]
- 15.Lai E-M, and Kado CI (1998). Processed VirB2 Is the Major Subunit of the Promiscuous Pilus of Agrobacterium tumefaciens. J. Bacteriol 180, 2711–2717. 10.1128/JB.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aly KA, and Baron C (2007). The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 153, 3766–3775. 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- 17.Li YG, and Christie PJ (2018). The Agrobacterium VirB/VirD4 T4SS: mechanism and architecture defined through in vivo mutagenesis and chimeric systems. Agrobacterium Biol., 233–260. 10.1007/82_2018_94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, and Christie PJ (2009). Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol 71, 779–794. 10.1111/j.13652958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakubowski SJ, Krishnamoorthy V, Cascales E, and Christie PJ (2004). Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol 341, 961–977. 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakubowski SJ, Cascales E, Krishnamoorthy V, and Christie PJ (2005). Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J. Bacteriol 187, 3486–3495. 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu HY, Chen CY, and Lai EM (2014). Expression and functional characterization of the Agrobacterium VirB2 amino acid substitution variants in T-pilus biogenesis, virulence, and transient transformation efficiency. PLoS One 9, 1–11. 10.1371/journal.pone.0101142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa TRD, Ilangovan A, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, and Waksman G (2016). Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex. Cell 166, 1436–1444.e10. 10.1016/j.cell.2016.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng W, Pena A, Low WW, Wong JLC, Frankel G, and Egelman EH (2020). Cryoelectron-Microscopic Structure of the pKpQIL Conjugative Pili from Carbapenem-Resistant Klebsiella pneumoniae. Structure 28, 1321–1328.e2. 10.1016/j.str.2020.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frost LS, Armstrong GD, Finlay BB, Edwards BF, and Paranchych W (1983). N-terminal amino acid sequencing of EDP208 conjugative pili. J. Bacteriol 153, 950–954. 10.1128/jb.153.2.950-954.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frost LS, Paranchych W, and Willetts NS (1984). DNA sequence of the F traALE region that includes the gene for F pilin. J. Bacteriol 160, 395–401. 10.1128/jb.160.1.395-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, and Lanka E (1999). Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem 274, 22548–22555. 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 27.Lai E-M, and Kado CI (2002). The Agrobacterium tumefaciens T pilus composed of cyclic T pilin is highly resilient to extreme environments. FEMS Microbiol. Lett 210, 111–114. 10.1016/S0378-1097(02)00601-8. [DOI] [PubMed] [Google Scholar]
- 28.Chesnokova O, Coutinho JB, Khan IH, Mikhail MS, and Kado CI (1997). Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol. Microbiol 23, 579–590. 10.1046/j.13652958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Zambryski PC, and Baron C (1999). Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol 181, 7485–7492. 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirasu K, and Kado CI (1993). Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol. Lett 111, 287–293. 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 31.He S, and Scheres SHW (2017). Helical reconstruction in RELION. J. Struct. Biol 198, 163–176. 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheres SHW (2012). RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mossey P, Hudacek A, and Das A (2010). Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J. Bacteriol 192, 2830–2838. 10.1128/JB.01331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su B, Bettcher LF, Hsieh W-Y, Hornburg D, Pearson MJ, Blomberg N, Giera M, Snyder MP, Raftery D, and Bensinger SJ (2021). A DMS shotgun lipidomics workflow application to facilitate high-throughput, comprehensive lipidomics. J. Am. Soc. Mass Spectrom 32, 2655–2663. 10.1021/jasms.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh W-Y, Williams KJ, Su B, and Bensinger SJ (2021). Profiling of mouse macrophage lipidome using direct infusion shotgun mass spectrometry. STAR Protoc. 2, 100235. 10.1016/j.xpro.2020.100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.María G, and de la Fernando C (2018). Natural and Artificial Strategies To Control the Conjugative Transmission of Plasmids. Microbiol. Spectr 6, 6.1.03. 10.1128/microbiolspec.MTBP-0015-2016. [DOI] [PubMed] [Google Scholar]
- 37.Christie PJ (2016). The Mosaic Type IV Secretion Systems. EcoSal Plus 7. 10.1128/ecosalplus.ESP-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai E-M, and Kado CI (2000). The T-pilus of Agrobacterium tumefaciens. 8, 361–369. 10.1016/S0966-842X(00)01802-3. [DOI] [PubMed] [Google Scholar]
- 39.Macé K, Vadakkepat AK, Redzej A, Lukoyanova N, Oomen C, Braun N, Ukleja M, Lu F, Costa TRD, Orlova EV, et al. (2022). Cryo-EM structure of a type IV secretion system. Nature 607. 10.1038/s41586-022-04859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon JE, Costa TRD, Patel RS, Gonzalez-Rivera C, Sarkar MK, Orlova EV, Waksman G, and Christie PJ (2017). Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation. Mol. Microbiol 105, 273–293. 10.1111/mmi.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groenewold MK, Hebecker S, Fritz C, Czolkoss S, Wiesselmann M, Heinz DW, Jahn D, Narberhaus F, Aktas M, and Moser J (2019). Virulence of Agrobacterium tumefaciens requires lipid homeostasis mediated by the lysyl‐phosphatidylglycerol hydrolase AcvB. Mol. Microbiol 111, 269–286. 10.1111/mmi.14154. [DOI] [PubMed] [Google Scholar]
- 42.Wirawan IGP, Hi Wan Kang, and Kojima M (1993). Isolation and characterization of a new chromosomal virulence gene of Agrobacterium tumefaciens. J. Bacteriol 175, 3208–3212. 10.1128/jb.175.10.3208-3212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan SQ, Jin S, Boulton MI, Hawes M, Gordon MP, and Nester EW (1995). An Agrobacterium virulence factor encoded by a Ti plasmid gene or a chromosomal gene is required for T-DNA transfer into plants. Mol. Microbiol 17, 259–269. 10.1111/j.1365-2958.1995.mmi_17020259.x. [DOI] [PubMed] [Google Scholar]
- 44.Vinuesa P, Neumann-Silkow F, Pacios-Bras C, Spaink HP, Martínez-Romero E, and Werner D (2003). Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol. Plant-Microbe Interact 16, 159–168. 10.1094/MPMI.2003.16.2.159. [DOI] [PubMed] [Google Scholar]
- 45.Winans SC (1990). Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J. Bacteriol 172, 2433–2438. 10.1128/jb.172.5.24332438.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCullen CA, and Binns AN (2006). Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol 22, 101–127. 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- 47.Kalogeraki VS, and Winans SC (1995). The octopine-type Ti plasmid pTiA6 of Agrobacterium tumefaciens contains a gene homologous to the chromosomal virulence gene acvB. J. Bacteriol 177, 892–897. 10.1128/jb.177.4.892-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerr JE, and Christie PJ (2010). Evidence for VirB4-mediated dislocation of membraneintegrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J. Bacteriol 192, 4923–4934. 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aktas M, Danne L, Möller P, and Narberhaus F (2014). Membrane lipids in Agrobacterium tumefaciens: Biosynthetic pathways and importance for pathogenesis. Front. Plant Sci 5, 1–13. 10.3389/fpls.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klüsener S, Hacker S, Tsai YL, Bandow JE, Gust R, Lai EM, and Narberhaus F (2010). Proteomic and transcriptomic characterization of a virulence-deficient phosphatidylcholinenegative Agrobacterium tumefaciens mutant. Mol. Genet. Genomics 283, 575–589. 10.1007/s00438-010-0542-7. [DOI] [PubMed] [Google Scholar]
- 51.Amro J, Black C, Jemouai Z, Rooney N, Daneault C, Zeytuni N, Ruiz M, Bui KH, and Baron C (2022). Cryo-EM structure of the Agrobacterium tumefaciens T-pilus reveals the importance of positive charges in the lumen. bioRxiv, 2022.04.28.489814. 10.1101/2022.04.28.489814. [DOI] [PubMed] [Google Scholar]
- 52.Meng R, Jiang M, Cui Z, Chang JY, Yang K, Jakana J, Yu X, Wang Z, Hu B, and Zhang J (2019). Structural basis for the adsorption of a single-stranded RNA bacteriophage. Nat. Commun 10. 10.1038/s41467-019-11126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berger BR, and Christie PJ (1994). Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol 176, 3646–3660. 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guex N, Peitsch MC, and Schwede T (2009). Automated comparative protein structure modeling with SWISS‐MODEL and Swiss‐PdbViewer: A historical perspective. Electrophoresis 30, S162–S173. 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 58.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman KW, Renfrew PD, Smith CA, and Sheffler W (2011). ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. In Methods in enzymology (Elsevier; ), pp. 545–574. 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Croll TI (2018). ISOLDE: a physically realistic environment for model building into lowresolution electron-density maps. Acta Crystallogr. Sect. D Struct. Biol 74, 519–530. 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Echols N, Headd JJ, Hung L-W, Jain S, Kapral GJ, and Kunstleve RWG (2011). The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106. 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phillips JC, Hardy DJ, Maia JDC, Stone JE, Ribeiro JV, Bernardi RC, Buch R, Fiorin G, Hénin J, and Jiang W (2020). Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys 153, 44130. 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palarea-Albaladejo J, and Martín-Fernández JA (2015). zCompositions — R package for multivariate imputation of left-censored data under a compositional approach. Chemom. Intell. Lab. Syst 143, 85–96. 10.1016/j.chemolab.2015.02.019. [DOI] [Google Scholar]
- 65.Greenacre M (2021). Compositional data analysis. Annu. Rev. Stat. its Appl 8, 271–299. [Google Scholar]
- 66.R Core Team RC (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. [Google Scholar]
- 67.Wickham H (2016). Data analysis. In ggplot2 (Springer; ), pp. 189–201. 10.1007/978-3-319-24277-4_9. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Cryo-EM map and model have been deposited to EMBD (ID: EMD-28957) and RCSB (PDB ID: 8FAI), respectively. The raw data is available upon request.
This paper does not report original code.
Any additional information required to reanalyze the data, including lipidomics raw data, presented in this study is available from the lead contact upon request.


