Abstract
Porphyromonas gingivalis is a gram-negative, black-pigmented anaerobe that has been associated with advanced periodontal disease. The genome of P. gingivalis has the potential to produce a number of virulence determinants including proteases, hemagglutinins, hemolysin, invasion-associated proteins, and products of the pathogenicity island ragAB; however, little is known about how their expression is controlled. Periodontal pockets experience a higher temperature during inflammation, and this elevated temperature may influence the pathogenicity of P. gingivalis by changing its patterns of gene expression. In this study, RNA has been isolated from cells of P. gingivalis grown to steady state at temperatures of 37, 39, and 41°C under hemin excess conditions (pH 7.0) in a chemostat. The RNA was subjected to PCR amplification following reverse transcription, using various combinations of randomly selected oligonucleotide primers. Reproducible RNA fingerprints have been obtained; however, differences were demonstrated in the RNA profiles of cells grown at the three temperatures, indicating differences in gene expression. Several PCR fragments were isolated that appeared to represent temperature-regulated genes. The nucleotide sequence of one of these has been identified as part of the ragAB locus, which codes for both a 55-kDa immunodominant antigen (RagB) and a homologue of the family of TonB-linked outer membrane receptors (RagA). These data indicate that expression of ragAB may be modulated in response to changes in temperature and that this may suggest a mechanism of evading the host response in the inflamed periodontal pocket.
Porphyromonas gingivalis is a gram-negative anaerobic oral bacterium that is strongly implicated in the etiology of advanced periodontal diseases in humans (27). These diseases are chronic inflammatory conditions of the supporting tissues of the teeth, which can lead to the destruction of the periodontium, including alveolar bone, and tooth loss. Although the microflora from deep periodontal pockets is diverse, P. gingivalis is frequently isolated in large numbers (9) and is detected only occasionally, and at low levels, at clinically sound sites (8, 37).
The relationship between the subgingival microflora and the host in health and disease is complex. In disease, there is a shift in the balance of the microflora and the proportions of obligately anaerobic and proteolytic bacteria increase (25). Tissue destruction is a consequence of both the direct action of individual bacteria and the indirect effects of the host inflammatory response to this microbial challenge (4). The expression of bacterial virulence is frequently modulated by the prevailing environmental conditions. To ensure survival, the cell requires a means of environmental sensing and response and an efficient mechanism of coordinating the response at the level of transcription. For example, in Shigella flexneri, the causative agent of bacillary dysentry, the invasive phenotype depends on the expression of genes carried on a high-molecular-mass virulence plasmid. Transcription of these genes is regulated in response to changes in temperature and osmolality such that expression occurs under the conditions found in the lower gut of the host. This is achieved through the deployment of transcriptional activators which are specific to the virulence gene locus as well as through more global regulatory circuitry involving DNA supercoiling and the distribution of abundant nuclear binding proteins (5). Environmental parameters liable to modulate gene expression in periodontal tissues will vary according to the inflammatory status of the site. For example, both pH and temperature can rise during inflammation (6, 7), while the increased flow of gingival crevicular fluid will not only introduce components of the host defenses but also provide an array of potentially novel nutrients, including heme-containing macromolecules, for subgingival bacteria. P. gingivalis produces a range of putative virulence factors, e.g., proteases, lipopolysaccharide, hemagglutinins, and adhesins (12), whose expression is environmentally regulated (21, 22, 24). It is likely that there are other genes, which remain to be identified, whose expression is also influenced by changes in environment.
Several approaches to the global identification of environmentally regulated genes have been developed. Many of these rely on the analysis of RNA populations isolated from cells grown under different conditions. Moreover, a genetic system, termed in vivo expression technology, has been developed to enable the identification of genes that are specifically expressed by pathogenic bacteria when infecting host tissues (20). Using this approach, genes that are specifically expressed during infection of mice by Salmonella enterica serovar Typhimurium (11) or Staphylococcus aureus (18) have been identified. Methods based on subtractive hybridization of RNA molecules have also been used with both prokaryotes and eukaryotes to detect differentially expressed genes. This approach was used to identify a gene induced in Mycobacterium avium cells phagocytosed by macrophages (29). However, this latter approach has two principal drawbacks: it usually cannot be used to identify genes expressed at low levels, and it often will not identify changes in the level of expression, i.e., up- or down-regulation. More recently, attempts have been made to adapt for use with prokaryotes the differential-display approach developed by Liang and Pardee (17) for eukaryotic RNA. Essentially, this is a two-stage procedure involving reverse transcription of RNA from two or more cell populations followed by PCR amplification using randomly selected oligonucleotides. The resulting DNA profiles are analyzed by polyacrylamide gel electrophoresis to obtain a fingerprint of the genes expressed. Using a modification of this approach, Wong and McClelland (36) were able to identify genes regulated by oxygen stress in S. enterica serovar Typhimurium, and Kwaik and Pederson (14) used a similar approach for the identification of macrophage-induced genes of Legionella pneumophila. More recently, a technique for identifying differentially expressed mRNA in bacteria, using customized amplification libraries, has been reported (2). We have further developed the differential-display procedure to include reverse transcription with randomly selected primers. This modification allows cDNA synthesis from RNA targets lacking a poly(A) tail.
The aim of this study was to demonstrate environmentally regulated gene expression in P. gingivalis by comparing RNA fingerprints of cells grown at different temperatures. Using temperatures corresponding to those found in the gingival crevice in health and disease, it was hypothesized that genes involved in the process of disease progression could be identified.
MATERIALS AND METHODS
Bacterial culture conditions.
P. gingivalis W50 was grown in a 2-liter-capacity chemostat (FT Applikon, Scheidom, The Netherlands) operated at a working volume of 700 ml as described previously (28). The pH of the culture was maintained at 7.0 ± 0.1 by the automatic addition of 1 M NaOH and 0.5 M HCl, and the temperature was controlled at 37 ± 0.1°C. The culture vessel was sparged with a gas mixture of oxygen-free nitrogen (95%, vol/vol) and carbon dioxide (5%, vol/vol) to maintain anaerobic conditions; once bacterial growth was initiated, the Eh of the culture fell to −350 mV and this value was maintained throughout the cultivation process. The medium was brain heart infusion broth (Oxoid, Basingstoke, United Kingdom) supplemented with 5 mg of hemin (Sigma, Gillingham, United Kingdom) per liter to achieve hemin excess. The medium flow rate was adjusted to give a dilution rate, D, of 0.1 h−1, corresponding to a mean generation time of 6.9 h. P. gingivalis W50 was grown to late logarithmic phase in anaerobic batch culture at 37°C, and 100 ml of the culture was used to inoculate the chemostat. The medium was introduced initially very slowly and left overnight to reach the required working volume of 700 ml; once this value was attained, the medium flow rate was increased to give the required dilution rate (D = 0.1 h−1). In subsequent experiments, chemostat cultures were started at 37 ± 0.1°C and then increased to either 39 ± 0.1 or 41 ± 0.1°C. At each temperature, the chemostat was allowed to achieve a steady state (10 culture volume changes, i.e., 3 to 4 days) after inoculation, and samples were taken from steady-state cultures for analysis over 6 days.
Estimation of biomass.
The biomass of the culture was determined by daily measurements of the optical density at 540 nm, dry weight, and viable counts of the culture, as described previously (21).
Total RNA extraction.
Fresh culture (1.5 ml) was removed directly from the chemostat at each steady state and centrifuged at 11,600 × g in a microcentrifuge at 4°C for 5 min. The pelleted cells were mixed with total RNA isolation reagent (Advanced Biotechnologies, Leatherhead, United Kingdom), and the RNA was extracted as specified by the manufacturer. RNA samples (5 μg) were resolved in denaturing formaldehyde-agarose gels by electrophoresis (33).
Differential-display PCR.
RNA was treated with RQ1 DNAse (Promega, Southampton, United Kingdom) as recommended by the manufacturer. RQ1-treated RNA samples (0.2 μg) were used as templates for the synthesis of cDNA with 100 U of Superscript II reverse transcriptase (Gibco-BRL, Paisley, United Kingdom) with one or more arbitrarily chosen primers (0.2 μM) in a reaction volume of 10 μl as specified by the manufacturer. A 2-μl aliquot of the cDNA was then subjected to 40 cycles of PCR amplification in the presence of arbitrarily chosen random primers and [α-32P]dCTP using 1 U of AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.). The primers used in this study were chosen randomly from a commercially available RNA fingerprinting kit (Clontech, Basingstoke, United Kingdom) and were originally designed for differential display of eukaryotic cDNA: P4, 5′-ATTAACCCTCACTAAATGCTGGTAG-3′; P5, 5′-ATTAACCCTCACTAAAGATCTGACTG-3′; P7, 5′-ATTAACCCTCACTAAATGCTGTATG-3′; and P8, 5′-ATTAACCCTCACTAAATGGAGCTGG-3′. Thermal cycling was carried out at 94°C for 30 s, 40°C for 30 s, and 72°C for 2 min. In each reaction, the same primers were used for PCR that had been used for cDNA synthesis. Following the cycling reactions, the labeled PCR products were separated by electrophoresis on 6% (wt/vol) denaturing polyacrylamide gels. The gels were dried down on filter paper and subjected to autoradiography with Fuji-RX medical X-ray film. To orientate the gel and the autoradiograph, radioactive ink was spotted onto the borders of the dried-down gel.
Characterization of differentially expressed products.
Following realignment of the developed autoradiograph with the dried gel, DNA corresponding to the bands that appeared to be temperature regulated was excised from the acrylamide gel. The excised DNA-gel slice was boiled for 10 min in 100 μl of PCR grade water and then briefly centrifuged in a bench microcentrifuge. The supernatant was transferred to a fresh tube, and the DNA was precipitated by the addition of 10 μl of 3 M sodium acetate, 5 μl of glycogen (10 mg ml−1), and 450 μl of ethanol. After incubation at −80°C for 30 min, the samples were centrifuged at 10,000 × g in a bench centrifuge for 10 min. The pellets were briefly dried and then resuspended in 10 μl of PCR grade water. A 2-μl aliquot of the boiled sample was then reamplified by PCR using the appropriate primers. Reamplified DNA fragments were then cloned into the plasmid vector pGEM-T (Promega, Madison, Wis.) as recommended by the manufacturer.
Northern blot analysis.
To confirm that the clones obtained represented temperature-regulated genes, the cloned cDNA fragments were labeled with [32P]dCTP using a random-prime labeling system (Gibco-BRL). Labeled DNA fragments were hybridized to Northern blots of P. gingivalis RNA (5 μg per lane) from each of the different cultures. Hybridization was carried out at 65°C for 18 h in 5× SSPE (0.9 M NaCl, 0.05 M sodium phosphate, 0.005 M EDTA [pH 7.7]) plus 5× Denhardt's solution (0.1% [wt/vol], bovine serum albumin, 0.1% [wt/vol] Ficoll, 0.1% [wt/vol] polyvinylpyrrolidone), containing sonicated salmon sperm DNA at 100 μg ml−1. The blots were washed in 2× SSPE–0.1% (wt/vol) sodium dodecyl sulfate (SDS) at room temperature for 30 min, and then given a more stringent wash with 0.2× SSPE–0.1% (wt/vol) SDS at 65°C for 30 min. The blots were then sealed in a plastic bag and subjected to autoradiography.
DNA sequence analysis.
Clones containing cDNA from temperature-regulated genes were sequenced using a T7-polymerase sequencing kit (Pharmacia, St. Albans, United Kingdom). The nucleotide sequences obtained were used to screen the GenBank and EMBL databases in an attempt to identify the transcripts by using the BLASTN and BLASTX programs available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/).
Western blot analysis.
Western blot analyses were performed to confirm the differential expression of selected proteins by P. gingivalis during growth at different temperatures. Bacterial pellets from 1.5 ml of culture were harvested by centrifugation and solubilized in sample-loading buffer (0.5 M Tris [pH 6.8], 10% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.05% bromophenol blue) after the addition of N α-p-tosyl-l-lysine chloromethyl ketone to a final concentration of 1 mM. Following protein estimation using a Lowry Micro Method protein assay kit (Sigma), 20 μg of protein was analyzed by SDS-polyacrylamide gel electrophoresis (15). Proteins were transferred to nitrocellulose by the method of Towbin et al. (35). The blotting buffer contained 25 mM Tris, 192 mM glycine, and 10% (vol/vol) methanol, and the transfer was performed at 70 V for 1 h. Membranes were incubated with the primary antibody DRU 55.5, which reacts with a 55-kDa antigen, RagB (23), at a 1:100 dilution. Horseradish peroxidase-conjugated rabbit anti-mouse secondary antibody (1:200 dilution) was used, and antibody binding sites were visualized using 0.04% (wt/vol) 3-amino-9-ethyl carbazole in 5.0% dimethyl formamide–95% aqueous sodium acetate solution (10 mM; pH 5.0).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported in this paper are AJ242672, AJ242673, and AJ242674.
RESULTS
Growth of P. gingivalis at different temperatures.
P. gingivalis W50 grew well and achieved a steady state at each of the temperatures imposed, although optimal growth occurred at 37°C as judged by dry weight and viable counts (28). No significant difference in viable counts was seen between cultures grown at 37, 39, and 41°C.
Random-primed cDNA synthesis using P. gingivalis RNA.
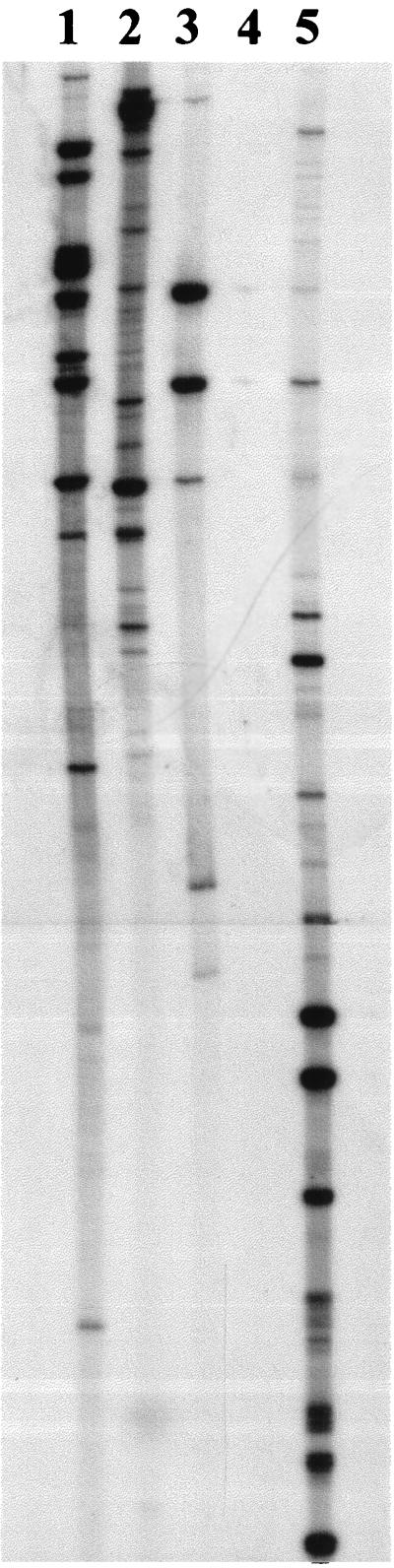
To determine that cDNA could be synthesized from P. gingivalis RNA, a series of reactions was performed with a range of different primers in the presence of [α-32P]dCTP. Negative control reactions with RNA but lacking in primer were also included to ensure that subsequent PCR products resulted only from newly synthesized cDNA. To confirm that synthesis was occurring during the reverse transcription, control reactions were set up for each RNA sample in which the incorporation of [α-32P]dCTP was measured. The percent incorporation was routinely on the order of 5% of the total available [32P]dCTP in these reactions. The cDNA samples were subjected to random-primed PCR. Figure 1 shows the results obtained with several arbitrarily chosen primers either individually or as a mixture. In each lane, with the exception of the negative control, there are several bands on the autoradiograph. The number of bands generated appeared to be variable. Primers P5 and P7 both gave approximately 20 bands each, whereas P8 gave only about 6 bands. A mixture containing four primers (P4, P5, P7, and P8) gave the largest number of bands, but it was noticeable that the majority of the fragments generated in this reaction were smaller than when the primers were used individually. The absence of bands in the negative control (lane 4) confirmed that the bands in the other lanes were derived from newly synthesized cDNA and were not due to contamination of the RNA samples with residual DNA.
FIG. 1.
Arbitrarily primed reverse transcription-PCR of P. gingivalis RNA from a culture grown at 37°C. Lanes: 1, P5; 2, P7; 3, P8; 4, negative control (RNA subjected to reverse transcription in the absence of primers); 5, mix of P4, P5, P7, and P8.
Reproducibility of differential-display PCR.
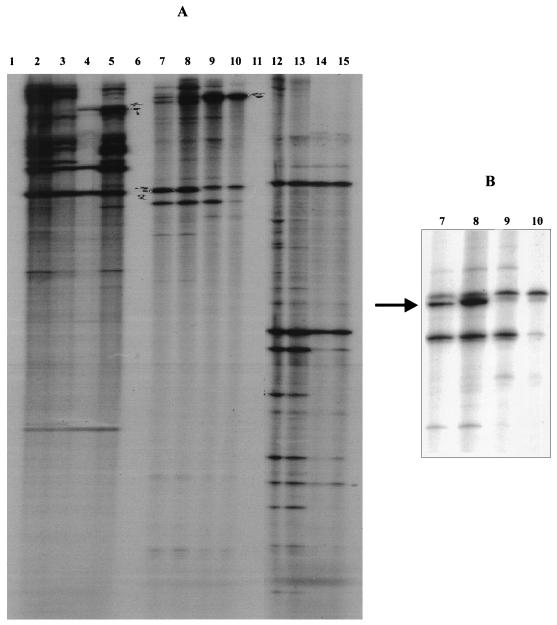
To determine whether the pattern of bands obtained was reproducible, duplicate cDNA samples obtained from RNA from cells grown at either 37 or 41°C were compared. cDNA was synthesized in the presence of [α-32P]dCTP using either primers P5 or P7 or a mix of primers P4, P5, P7, and P8. The cDNA synthesis was monitored by the level of incorporation of [α-32P] dCTP. The duplicate samples were then subjected to PCR analysis, using the same primers, to generate an RNA fingerprint of the samples. Figure 2 shows the pattern of bands obtained. The results show that the data are reproducible, since the duplicate samples gave similar profiles. For example, lanes 7 and 8 gave identical profiles, as did lanes 9 and 10. The samples shown in lanes 7 to 10 represent fingerprints obtained from four different culture samples. Examination of the fingerprints from each of these samples indicates that the majority of bands are common to all lanes when amplified using the same oligonucleotide primer.
FIG. 2.
Differential display of RNA from P. gingivalis grown at 37 and 41°C. (A) Lanes 1, 6, and 11 contain negative controls (RNA subjected to reverse transcription in the absence of primers); lanes 1 to 5, P5; lanes 6 to 10, P7; lanes 11 to 15, a mix of primers P4, P5, P7, and P8. Lanes 2, 3, 7, 8, 12, 13 contain RNA from a 37°C culture; lanes 4, 5, 9, 10, 14, and 15 contain RNA from a 41°C culture. (B) Close-up view of a region of the gel showing a transcript in lanes 7 and 8 (indicated by an arrow) obtained with primer P7, which is apparently down-regulated in the 41°C samples in lanes 9 and 10.
Characterization of differentially expressed transcripts.
Differences in profile between the samples grown under different conditions were found. Closer examination of the fingerprints in Fig. 2 indicates that there are some bands that appear to be present (or present in markedly different quantities) in pairs of samples from cultures grown at 37°C compared with the samples grown at 41°C. The bands indicated by the arrow in the inset figure indicate the presence of DNA fragments that appear to be less abundant in the cultures grown at 41°C. Bands that represented potential temperature-regulated transcripts were excised from the gel, and the DNA was extracted and reamplified. The products of the reamplification were analyzed by agarose gel electrophoresis to determine the sizes of the fragments. The concentration of DNA in the PCR mixtures was then estimated by visual comparison with known standards, and ligations containing a molar ratio of approximately 1:1 (PCR product to pGEM-T) were set up and used to transform competent Escherichia coli (strain JM109) cells. Over 400 ampicillin-resistant colonies were obtained with each transformation, of which between 30 and 70% appeared to be recombinants, as judged by blue/white selection on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside plus isopropyl-β-d-thiogalactopyranoside (X-Gal/IPTG) (33). Plasmid DNA from six putative recombinants from each of the ligations was obtained and digested with the restriction endonuclease HaeIII to confirm that each isolate from the same ligation contained a fragment with an identical restriction fingerprint. HaeIII was chosen because it recognizes a 4-base sequence in DNA and therefore digests most DNA templates into multiple fragments. Different DNA fragments are likely to give completely different digestion profiles. Northern blots of RNA from P. gingivalis grown at 37, 39, and 41°C were probed with three radiolabeled, amplified clone inserts to confirm that they were differentially expressed at the three temperatures. RNA was first quantified by UV spectrophotometry followed by visualization on ethidium bromide-stained gels to ensure equal loadings on the gels. Additionally, blots were probed with a nondifferentially expressed clone to demonstrate standardized loadings on the gels. A total of three apparently differentially expressed cDNA fragments were identified in experiments using the primers shown. The cloned inserts derived from the three differentially expressed transcripts were sequenced using the universal M13 primer. This step was carried out to obtain sufficient sequence data to facilitate screening for homologous sequences in the nucleic acid sequence databases. The sequences were used to perform BLASTN searches of the GenBank and EMBL databases BLASTX searches were also carried out on the sequences. Two of the sequences failed to demonstrate homology to any sequences in the databases. However, the sequence of clone pBB240 showed 99% homology to a portion of the recently identified ragA locus of P. gingivalis. The sequence obtained is shown in Fig. 3, together with the deduced amino acid sequence.
FIG. 3.
Partial nucleotide sequence and deduced amino acid translation of the insert cloned into pBB240.
RagAB expression at 37, 39, and 41°C.
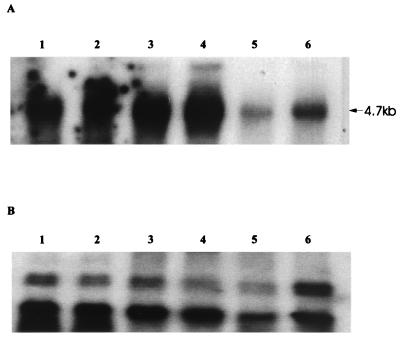
Figure 4 shows the hybridization of the 1.6-kb PCR-amplified product of ragA (10) to RNA samples isolated from P. gingivalis grown at different temperatures. This probe gives a single fragment of the expected size, 4.7 kb. The samples grown at 39°C show a slight increase in expression compared to those grown at 37°C, whereas at 41°C there is a clear reduction in the levels of transcript. The panel showing the control hybridization, using a probe generated from one of the non-temperature-regulated fragments, confirms that approximately equal amounts of mRNA were loaded in each lane. This probe (pBB241), which is derived from a cDNA for a protein with no known function, was used because it has repeatedly been shown to give a uniform hybridization signal with RNA from cultures grown at different temperatures (data not shown).
FIG. 4.
Northern blot analysis of RNA from P. gingivalis grown at different temperatures. (A) Hybridization with the 1.6-kb ragA probe (10); (B) hybridization with a control probe (pBB241). Lanes: 1 and 2, 37°C; 3 and 4, 39°C; 5 and 6, 41°C. The arrow at the side of panel A indicates the position of the 4.7-kb ragAB transcript.
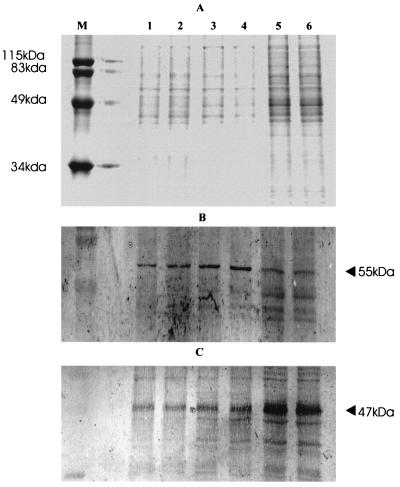
The ragA gene is part of a small operon and is cotranscribed with the gene for the 55-kDa antigen RagB, for which a monoclonal antibody, DRU 55.5, is available. To determine if the protein levels of RagB are affected by temperature, Western blot analysis was performed on protein samples prepared from P. gingivalis cells grown under different culture conditions (Fig. 5). These results confirmed that the protein levels of RagB are reduced when the temperature of the culture is increased to 41°C. As a control, the levels of the 47-kDa antigen of P. gingivalis, a surface-bound glutamate dehydrogenase which is not regulated by changes in temperature (28), were analyzed in the same samples. The stained gel indicates that the protein loadings for the 41°C samples were higher than those for the other two temperatures. This was confirmed by the blot that was reacted with the antibody to the 47-kDa antigen, which also shows slightly higher levels in the 41°C samples. The anti-RagB antibody, however, clearly shows a reduced reaction with the 41°C samples. These results confirm that while the levels of the 47-kDa antigen are not affected by temperature, the levels of RagB are reduced at 41°C.
FIG. 5.
SDS-polyacrylamide gel electrophoresis of P. gingivalis cell extracts, from cultures grown at different temperatures, and Western blot analysis with monoclonal antibodies to RagB and the 47-kDa surface antigen (28). Lanes: M, molecular size markers; 1 and 2, 37°C; 3 and 4, 39°C; 5 and 6, 41°C. (A) Coomassie blue-stained gel; (B) anti-RagB monoclonal antibody; (C) anti-47-kDa monoclonal antibody. The arrowhead in panel B indicates the 55-kDa RagB and the arrowhead in panel C indicates the 47-kDa antigen.
DISCUSSION
The ability to respond rapidly to changes in the local environment can be critical to the growth of opportunistic pathogens in the host. To date, the influence of the environment has been restricted to a few targeted known genes, and as such the global effects of environment on the regulation of genes have not been studied in great detail. As a consequence, new methods are being developed to achieve a broader understanding of the effects of specific environmental conditions on changes in total gene expression (2, 14, 36).
Previous studies have shown that a number of known genes are environmentally regulated in P. gingivalis. For example, increases in hemin concentration, pH, and temperature regulate protease production (21, 22, 24, 28); temperature also regulates fimbrial and superoxide dismutase expression (3). The way in which an organism adapts to its environment will be reflected at the level of gene expression, and these changes may also be reflected in an altered pathogenic potential. It is important, therefore, to identify the changes in gene expression that result from environmental change and to determine whether these changes are responsible for or contribute to such changes in pathogenicity. This study has demonstrated that arbitrarily selected oligonucleotides can prime the synthesis of cDNA from P. gingivalis total RNA and can be also used for subsequent PCR amplification. The procedure described here is reproducible and enables the isolation of differentially expressed genes. It is likely that some of the bands seen on the autoradiographs may be due to priming synthesis from rRNA. This is unlikely to lead to the increased isolation of false positives, since the levels of rRNA do not usually vary in cells under the growth conditions used here. It is possible, however, that amplification of rRNA may lead to an overestimation of genome coverage when using this method for differential display. The success of this approach has, however, been confirmed by the identification of a temperature-regulated antigen in this study. The ability to identify environmentally regulated genes is an important tool in our understanding of the ways in which organisms respond to the changing environment. However, studies on bacterial mRNA have generally been hampered by the difficulty in identifying polyadenylated RNA in prokaryotic species. Although several reports have suggested that prokaryote mRNA is sometimes polyadenylated (13, 32), this has not yet been shown to be the general case. Identifying differentially regulated genes in eukaryotes by using differential display is technically more simple, since the poly(A) tail both serves as a means of mRNA isolation and acts as a priming site for first-strand cDNA synthesis. This approach has been widely used in eukaryotes to identify tissue-specific gene expression (1, 16, 26, 34, 38). The arbitrarily primed cDNA synthesis and PCR approach used in this study and the similar approaches used by others should facilitate the wider use of this approach to the study of bacterial mRNA. Using the differential-display approach to RNA analysis, we have identified a transcript fragment representing ragA, which has been characterized as a component of a polycistronic mRNA coded for by the ragAB locus.
RagB has been identified as the 55-kDa immunodominant antigen of P. gingivalis strains W50 and W83 and has been implicated in the destructive disease process of pathogenic strains of P. gingivalis (10). The mobility of RagB on Western blots derived from two-dimensional gel electrophoresis of P. gingivalis (30) may be comparable to that of the 50-kDa protein identified by Lu and McBride (19) that was down-regulated in cultures of P. gingivalis shifted from 37 to 42°C. These authors showed that a shift in the culture temperature resulted in both up- and down-regulation of specific polypeptides. The present study has shown that the differential-display technique can be useful in identifying genes that are both up- and down-regulated. RagA shows extensive sequence homology to susC of Bacteroides thetaiotaomicron (31), which plays an important role in the metabolism of polysaccharides. Both RagA and RagB are believed to be membrane proteins and may act as targets for the host cellular immune response. The finding that these genes may be down-regulated by an increase in temperature, comparable to that found during tissue inflammation, may be indicative of one of the mechanisms whereby P. gingivalis avoids the host response. It has been suggested (28) that P. gingivalis may decrease the expression of certain virulence factors, in particular extracellular proteases, at elevated temperatures in order to reduce the intensity of the host response. As a result, adopting a less inflammatory phenotype may enable P. gingivalis to maintain population levels under hostile conditions. RagB is a significant target of the serum immunoglobulin G antibody response of patients with periodontal disease (10). It is possible that a down-regulation of the ragAB operon will help the organism to evade host responses. Further analysis of how this regulation occurs at the molecular level will further our understanding of the pathogenicity of this organism.
In summary, techniques have been developed that permit the study of changing patterns of gene expression by analyzing bacterial RNA by using arbitrarily primed reverse transcription-PCR. The data have shown that the ragAB operon of P. gingivalis is regulated by changes in temperature and confirm that this approach can be used to identify genes that are environmentally controlled. It is likely that studies of this type can be used to identify genes important for adaptation to a range of environments.
REFERENCES
- 1.Aiello L P, Robinson G S, Lin Y W, Nishio Y, King G L. Identification of multiple genes in bovine retinal pericytes altered by exposure to elevated levels of glucose by using messenger RNA differential display. Proc Natl Acad Sci USA. 1997;91:6231–6235. doi: 10.1073/pnas.91.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland D, Kramnik I, Weisbrod T R, Otsubo L, Cerny R, Miller L P, Jacobs W R, Jr, Bloom B R. Identification of differentially-expressed mRNA in prokaryotic organisms by customised amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1998;95:13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano A, Sharma A, Sojar H T, Kuramitsu H K, Genco R J. Effects of temperature stress on expression of fimbriae and superoxide-dismutase by Porphyromonas gingivalis. Infect Immun. 1994;62:4682–4685. doi: 10.1128/iai.62.10.4682-4685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darveau R P, Tanner A, Page R C. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 5.Dorman C J. DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology. 1995;141:1271–1280. doi: 10.1099/13500872-141-6-1271. [DOI] [PubMed] [Google Scholar]
- 6.Eggert F M, Drewell L, Bigelow J A, Speck J E, Goldner M. The pH of gingival crevices and periodontal pockets in children, teenagers and adults. Arch Oral Biol. 1991;36:233–238. doi: 10.1016/0003-9969(91)90091-8. [DOI] [PubMed] [Google Scholar]
- 7.Fedi P F, Jr, Killoy W J. Temperature differences at periodontal sites in health and disease. J Periodontol. 1992;63:24–27. doi: 10.1902/jop.1992.63.1.24. [DOI] [PubMed] [Google Scholar]
- 8.Gmur R, Guggenheim B. Interdental supragingival plaque—a natural habitat of Actinobacillus acinomycetemcomitans, Bacteroides forsythus, Campylobacter rectus and Prevotella nigrescens. J Dent Res. 1994;73:1421–1428. doi: 10.1177/00220345940730080501. [DOI] [PubMed] [Google Scholar]
- 9.Haffajee A D, Socransky S S. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 10.Hanley S A, Aduse-Opoku J, Curtis M A. A 55kDa immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect Immun. 1999;67:1157–1171. doi: 10.1128/iai.67.3.1157-1171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt S C, Bramanti T E. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit Rev Oral Biol Med. 1991;2:177–281. doi: 10.1177/10454411910020020301. [DOI] [PubMed] [Google Scholar]
- 13.Johnson M D, Popowski J, Cao G J, Shen P, Sarker N. Bacteriophage T7 mRNA is polyadenylated. Mol Microbiol. 1998;27:23–30. doi: 10.1046/j.1365-2958.1998.00649.x. [DOI] [PubMed] [Google Scholar]
- 14.Kwaik Y A, Pederson L L. The use of differential display-PCR to isolate and characterise a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Liang P, Bauer D, Averboukh L, Warthoe P, Rohrwild M, Muller H, Strauss M, Pardee A B. Analysis of altered gene-expression by differential display. Methods Enzymol. 1995;254:304–321. doi: 10.1016/0076-6879(95)54022-9. [DOI] [PubMed] [Google Scholar]
- 17.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 18.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in-vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 19.Lu B, McBride B C. Stress response of Porphyromonas gingivalis. Oral Microbiol Immunol. 1994;9:166–173. doi: 10.1111/j.1399-302x.1994.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 20.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh P D, McDermid A S, McKee A S, Baskerville A. The effect of growth rate and haemin on the virulence and proteolytic activity of Porphyromonas gingivalis. Microbiology. 1994;140:861–865. doi: 10.1099/00221287-140-4-861. [DOI] [PubMed] [Google Scholar]
- 22.McDermid A S, McKee A S, Marsh P D. Effect of environmental pH on enzyme activity and growth of Bacteroides gingivalis W50. Infect Immun. 1988;56:1096–1100. doi: 10.1128/iai.56.5.1096-1100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millar D J, Scott E E, Slaney J M, Benjamin P, U S, Curtis M A. Production and characterisation of monoclonal antibodies to the principle sonicate antigens of Porphyromonas gingivalis W50. FEMS Immunol Med Microbiol. 1993;7:211–222. doi: 10.1111/j.1574-695X.1993.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 24.Minhas T, Greenman J, Schaffer A G. Effects of mucin, haemoglobin and collagen on the maximum specific growth rate, biomass and hydrolytic enzyme production of Porphyromonas gingivalis in continuous culture. Microb Ecol Health Dis. 1991;4:311–318. [Google Scholar]
- 25.Moore W E, Moore L V. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 26.Nishio Y, Aiello L P, King G L. Glucose induced genes in bovine aortic smooth-muscle cells identified by messenger-RNA differential-display. FASEB J. 1994;8:103–106. doi: 10.1096/fasebj.8.1.8299882. [DOI] [PubMed] [Google Scholar]
- 27.Page R C. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodont Res. 1991;26:230–242. doi: 10.1111/j.1600-0765.1991.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 28.Percival R S, Marsh P D, Devine D A, Rangarajan M, Aduse-Opoku J, Shepherd P, Curtis M A. Effect of temperature on growth, haemagglutination and protease activity of Porphyromonas gingivalis. Infect Immun. 1999;67:1917–1921. doi: 10.1128/iai.67.4.1917-1921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plum G, Clark-Curtiss J E. Induction of Mycobacterium avium gene expression following phagocytosis by human macrophages. Infect Immun. 1994;62:476–483. doi: 10.1128/iai.62.2.476-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pridmore A M, Devine D A, Bonass W A, Silley P. Influence of sample preparation technique on two-dimensional gel electrophoresis of proteins from Porphyromonas gingivalis. Lett Appl Microbiol. 1999;28:245–249. doi: 10.1046/j.1365-2672.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- 31.Reeves A R, D'Elia J N, Frias J, Sayers A A. A Bacteroides thetaiotaomicron outer membrane protein that is essential for the utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rindi L, Lari N, Gil M G, Garzelli C. Oligo (dT)-primed synthesis of cDNA by reverse transcriptase in mycobacteria. Biochem Biophys Res Commun. 1998;248:216–218. doi: 10.1006/bbrc.1998.8948. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Sagar R, Anisowicz A, Neveu M, Liang P, Sotiropoulou G. Identification by differential-display of alpha-6 integrin as a candidate tumour-suppressor gene. FASEB J. 1993;7:964–970. doi: 10.1096/fasebj.7.10.8344495. [DOI] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong K K, McClelland M. Stress-inducible gene of Salmonella typhimurium identified by arbitrarily primed PCR of RNA. Proc Natl Acad Sci USA. 1994;91:639–643. doi: 10.1073/pnas.91.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmer W, Wilson M, Marsh P D, Newman H N, Bulman J. Porphyromonas gingivalis, Prevotella intermedia and Actinobacillus actinomycetemcomitans in the plaque of children without periodontitis. Microb Ecol Health Dis. 1991;4:329–336. [Google Scholar]
- 38.Zimmerman J W, Schultz R M. Analysis of gene expression in the preimplantation mouse embryo -use of messenger RNA differential display. Proc Natl Acad Sci USA. 1994;91:5456–5460. doi: 10.1073/pnas.91.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]