Abstract
The heterogeneous group of oral bacteria within the sanguinis (sanguis) streptococci comprise members of the indigenous biota of the human oral cavity. While the association of Streptococcus sanguinis with bacterial endocarditis is well described in the literature, S. sanguinis is thought to play a benign, if not a beneficial, role in the oral cavity. Little is known, however, about the natural history of S. sanguinis and its specific relationship with other oral bacteria. As part of a longitudinal study concerning the transmission and acquisition of oral bacteria within mother-infant pairs, we examined the initial acquisition of S. sanguinis and described its colonization relative to tooth emergence and its proportions in plaque and saliva as a function of other biological events, including subsequent colonization with mutans streptococci. A second cohort of infants was recruited to define the taxonomic affiliation of S. sanguinis. We found that the colonization of the S. sanguinis occurs during a discrete “window of infectivity” at a median age of 9 months in the infants. Its colonization is tooth dependent and correlated to the time of tooth emergence; its proportions in saliva increase as new teeth emerge. In addition, early colonization of S. sanguinis and its elevated levels in the oral cavity were correlated to a significant delay in the colonization of mutans streptococci. Underpinning this apparent antagonism between S. sanguinis and mutans streptococci is the observation that after mutans streptococci colonize the infant, the levels of S. sanguinis decrease. Children who do not harbor detectable levels of mutans streptococci have significantly higher levels of S. sanguinis in their saliva than do children colonized with mutans streptococci. Collectively, these findings suggest that the colonization of S. sanguinis may influence the subsequent colonization of mutans streptococci, and this in turn may suggest several ecological approaches toward controlling dental caries.
The heterogeneous group of oral streptococci comprising the sanguinis streptococci are members of the human indigenous biota. (The previously recognized species of the genus Streptococcus named “sanguis” has recently been changed to “sanguinis” so as to conform to the rules of Latin grammar [32]). S. sanguinis is recognized not only for its historical association with life-threatening bacterial endocarditis, but also because of its putative antagonistic role in dental caries (20) and periodontal diseases (27). In terms of the former, S. sanguinis may compete with the mutans streptococci for colonization sites on tooth surfaces, since both groups of bacteria require the presence of teeth for colonization (6, 7) and may exhibit direct biochemical antagonism in situ (33). Because the cariogenic potential of S. sanguinis is deemed low compared to that of the mutans streptococci, several investigators have suggested that the S. mutans/S. sanguinis ratio may serve as an indicator for caries risk, i.e., the smaller the ratio, the lesser the risk of caries (12, 19, 20). In another study, however, a caries-predictive role for the S. mutans/S. sanguinis ratio could not be demonstrated (3).
Carlsson and coworkers were among the first to describe both the taxonomic and ecological features of S. sanguinis in the oral cavity (4–7). In fact, it was the Carlsson group that made the key observation that S. sanguinis did not colonize infants until after the emergence of teeth (7) and that colonization by S. sanguinis precedes that by mutans streptococci.
As part of a longitudinal study involving the acquisition of indigenous oral microbes in an infant population, the aim of the present study was to determine when S. sanguinis is acquired by infants and whether this acquisition period is discrete, as in the case of the mutans streptococci (8). Since oral colonization with S. sanguinis precedes that with mutans streptococci, it seems reasonable to speculate that the former event may influence the latter, especially considering that the colonization site of both organisms is tooth surfaces. Accordingly, we wanted to explore the relationship between the temporal and quantitative aspects of colonization by S. sanguinis in relation to subsequent colonization by mutans streptococci. Here, we show that infants acquire S. sanguinis during a discrete “window of infectivity” and that early colonization by S. sanguinis in infants results in delayed colonization by mutans streptococci.
MATERIALS AND METHODS
Study populations.
The study populations are composed of two distinct and temporally separate cohorts. The first cohort (natural history cohort), in which acquisition of S. sanguinis in infants was monitored longitudinally from 1984 to 1989, was described previously (8, 11, 38). Briefly, mothers-to-be were recruited from the Maternal and Infant Care program of the Jefferson County Health Department in Birmingham, Ala. Following birth and for the next 3 years, oral bacteriological samples were obtained at 3-month intervals from 48 mother-infant pairs (29 black and 19 white; 25 female and 23 male infants). The mothers' age averaged 23.3 ± 3.7 years (standard deviation [SD]) with a mean DMFS (decayed, missing, filled surfaces) score of 34.4 (SD, ±17.8). Complete data for S. sanguinis colonization were available for 45 of 48 mother-infant pairs. This cohort also included eight infants who acquired S. sanguinis but did not acquire mutans streptococci, as reported in our previous study (8). The second cohort (taxonomy cohort), derived from the same health center population of mother-infant pairs, was recruited in 1994 for the purpose of ascertaining the taxonomic characterization of isolates presumed to be S. sanguinis from the natural history cohort. This confirmation became necessary as the taxonomy of S. sanguinis used to characterize the natural history cohort was emended based on advances in genetic and biochemical definition of the viridans streptococci published subsequent to the natural history study (1, 10, 13, 15, 35, 37). The taxonomy cohort consisted of 38 infants, aged 6 to 36 months old, from whom 291 isolates presumed to be S. sanguinis (median of seven isolates per infant) were selected based on the same criteria as the natural history cohort and then further characterized based on the criteria given below. Written informed consent was obtained from all subjects in compliance with the Institutional Review Boards of the University of Alabama at Birmingham and the Jefferson County Health Department.
Sample procurement and bacteriology.
Bacterial samples were obtained from saliva and from the teeth, when present, in both cohorts. Unstimulated saliva samples were collected with a sterile cotton swab from the sublingual area of the mouth until saturated. Samples from the teeth (plaque) were taken with a sterile toothpick; the toothpick was placed in each approximal site and then passed along the gingival margin into the next approximal site of both upper and lower teeth. Swab and toothpick samples were placed into separate 1.0-ml reduced transport fluid vials (29) and then processed as previously described (8). Appropriate dilutions of saliva and plaque samples were plated onto MM10-sucrose agar (28). After 3 days of anaerobic incubation (85% N2, 10% CO2, and 5% H2), colonies presumed to be S. sanguinis were selected from MM10-sucrose agar based on their firm, adherent, star-shaped colony morphology (20, 28). Discrete colonies were then isolated from subcultures and placed in the appropriate medium for the detection of hydrolysis of arginine and lack of fermentation of mannitol. (Mannitol fermentation differentiates mutans streptococci from S. sanguinis.) This characterization is consistent with the original classification of Carlsson and coinvestigators as group I:B of S. sanguis (4, 7). As these isolates were not saved after the biochemical tests were done, and as new criteria became available for species determination of S. sanguinis (13, 15, 24, 35), the same initial screening procedures were performed on isolates from the taxonomy cohort (characteristic morphology on MM10-sucrose medium, hydrolysis of arginine, and failure to ferment mannitol), but unlike isolates from the natural history cohort, these were frozen for more detailed biochemical and genetic characterization. These additional tests included an extended panel of biochemical assays described by Whiley and Beighton (35) and Kilian and coworkers (15), i.e., fermentation of amydalin, inulin, melibiose, raffinose, and sorbitol; hydrolysis of arginine and esculin; and production of H2O2. Enzymatic reactions, including β-d-fucosidase, α-l-fucosidase, β-d-glucosidase, α-d-glucosidase, α-d-galactosidase, and β-d-N-acetylgalactosminidase, as described by Whiley et al. (36), supplemented the battery of tests. All 291 isolates were subjected to these tests. As controls, the prototype strain of S. sanguinis (ATCC 10556), along with prototypic strains of the Streptococcus species S. gordonii, S. parasanguis, S. oralis, and S. mitis, were subjected to the same battery of tests.
In addition, the DNA coding for 16S rRNA (rDNA) loci of 20 randomly chosen strains of the 291 isolates placed within one of the four S. sanguinis biovars (15) were obtained via PCR using custom-designed primers. The primers encompassed one of the variable regions of the 16S rDNA locus from various streptococci (nucleotides 9 to 369, Escherichia coli numbering) aligned and analyzed by Bentley and coworkers (2) and available from GenBank. These primers (5′-GGCTCAGGACGAACGCTGGCG-3′ and 5′-ACGCGGCGTTGCTCGGTCAGG-3′), produced a single amplicon of approximately 360 bp. This number of nucleotides proved sufficient to determine each strain's phylogenetic affiliation (described below). At least two amplicons from different reactions were then sequenced in both directions directly from the PCR after purification (Qiagene Quick PCR Purification kit; Qiagene, Chatsworth, Calif.). The phylogenetic affiliation of each of the 20 sequences was determined from the 16S rRNA database (Ribosomal Project Database [21]).
Definitions and statistical methods.
The time to initial colonization of S. sanguinis was defined as the first positive detection of S. sanguinis in either plaque or saliva samples. The Wilcoxon sign rank test was used to test the difference between the time that the first S. sanguinis appeared in plaque versus its first appearance in saliva. The time of initial colonization by mutans streptococci was also available for each infant; these data were published in a previous report (8).
Both parametric and nonparametric tests were employed based upon the distribution of the variable being analyzed. A univariate procedure was used to test the assumption of normalcy and to evaluate the distribution of the variables. All plaque and salivary levels of the oral bacteria studied were transformed to log10 prior to statistical analyses to normalize the variance (9). The dependent variable for all analyses, unless stated otherwise, was the time of initial colonization by S. sanguinis. The independent variables were gender, race, time of mutans streptococci colonization, first tooth emergence, dental caries, and dental and microbiological status of the mother. The Spearman and Pearson correlation analyses, chi square, Fisher exact tests, and multiple regression analysis with stepwise selection procedures were used in data analyses (the test used for each analysis is given in the Results). The Cox proportionate hazard model was used to predict the time of oral colonization of mutans streptococci using the time until first colonization with S. sanguinis, the levels of S. sanguinis in plaque and saliva samples of infants, and the proportion of salivary and plaque S. sanguinis to the total cultivable bacteria in the respective sample as independent variables. The time to colonization for S. sanguinis was based on either a plaque or saliva sample, whichever detected S. sanguinis first in chronological order. Different models were constructed using the stepwise technique. The fit of the data to the final model was tested using the Hosmer and Limeshaw method (14). The estimates presented in relation to the levels and proportions of S. sanguinis are based on salivary levels.
For all statistical tests, type I error probability less than or equal to 0.05 was considered significant.
RESULTS
Taxonomic characterization of S. sanguinis.
In the original natural history study, we selected and biochemically defined S. sanguinis at the time using a simple identification scheme based on the existing literature (7). Subsequent to that study, emended taxonomic definitions became available, making it necessary to reconfirm our original characterization of S. sanguinis. In addition, and independent from taxonomic realignments, the species name was changed to “sanguinis.” Because only a few isolates of strains presumed to be S. sanguinis were saved from the natural history cohort, we enrolled a second group of infants (taxonomy cohort) for the purpose of confirming that the isolates that we described in the natural history cohort were, indeed, S. sanguinis. Isolates selected by the same approach as for the original natural history cohort (i.e., selection based upon colony morphology, cleavage of arginine, and production of H2O2) were characterized using the biochemical tests described by Kilian and coworkers and Whiley and Beighton (15, 35). We found that 98.6% (291 of 295) of the isolates we had selected as S. sanguinis from MM10-sucrose medium were indeed S. sanguinis based upon their ability to cleave arginine and produce H2O2. These finding were further supported by subsequent tests that separated these 291 isolates into one of four biovars of S. sanguinis (35) or, for three isolates, S. gordonii or S. parasanguinis. Interestingly, the prototype strain ATCC 10556 exhibited a biochemical profile that better fit biovar 4 than the biovar 1 group originally proposed by Kilian et al. (15). Twenty isolates of S. sanguinis from 20 individuals were chosen at random from each of the four biovar groups (4 to 7 isolates per biovar). PCR amplicons from a variable region of the 16S rDNA locus were purified and then sequenced. Comparison of the sequences revealed that each of the 20 selected strains bore the highest phylogenetic affiliation with the 16S rDNA sequence of the S. sanguinis prototype strain ATCC 10566 archived in the ribosomal DNA database (21). The similarity (Sab) index (21) ranged from 0.73 to 1.00, with a mean value of 0.96. Interesting, the strain with the lowest Sab (0.73) still showed its highest similarity with the 16S rDNA locus of ATCC 10556. Together with the biochemical phenotypes, these results demonstrated that the isolates selected by distinct colony morphology on MM10-sucrose medium and the hydrolysis of arginine bore the closest affiliation with the prototype strain S. sanguinis ATCC 10566 compared to any other members of the viridans streptococci.
Initial colonization of S. sanguinis in infants from the natural history cohort.
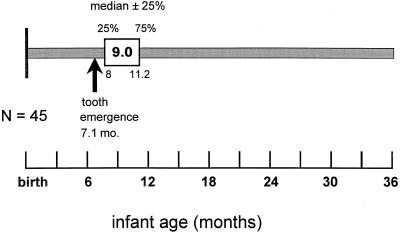
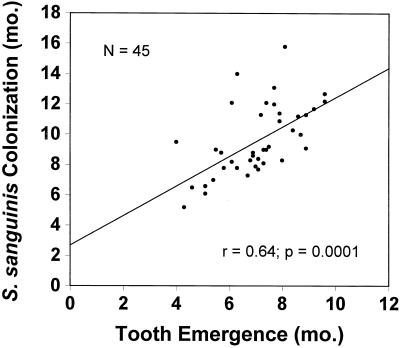
The median age of colonization by S. sanguinis in this infant population was 9.0 months (Fig. 1, Table 1). All 45 infants acquired S. sanguinis sometime after the emergence of their primary teeth. Twenty-five percent of the infants had acquired S. sanguinis by 8.0 months of age, and 75% had S. sanguinis by 11.4 months. The means, SDs, and ranges for initial detection of S. sanguinis in saliva or plaque are given in the table. The median age of the emergence of the first tooth was 7.1 months (range, 3.9 to 9.5 months). As shown in Fig. 2, the time of initial colonization by S. sanguinis was significantly correlated to the infant's age at first tooth emergence (r = 0.64; P = 0.0001; Spearman correlation analysis).
FIG. 1.
Window of infectivity for S. sanguinis. Infants became colonized by S. sanguinis at a median age of 9.0 months. Colonization followed the emergence of primary teeth; the first tooth emerged at a median age of 7.1 months.
TABLE 1.
Age of infants at initial detection of S. sanguinis in either plaque or saliva samples
| Sample | Age (mo)
|
||
|---|---|---|---|
| Mean ± SD | Median | Range | |
| Saliva | 13.9 ± 6.0 | 12.2 | 4.0–29.5 |
| Plaque | 10.6 ± 5.2 | 9.1 | 5.2–36.5 |
| Either | 9.7 ± 3.1 | 9.0 | 3.9–20.9 |
FIG. 2.
Relationship between the age at first tooth emergence and colonization of the infant by S. sanguinis. MS, mutans streptococci. The correlation was statistically significant (r = 0.64, P = 0.0001).
The time of the initial detection of S. sanguinis in plaque preceded its detection in unstimulated saliva by an average of 4.4 months. More specifically, S. sanguinis was first detected in plaque of infants at a median age of 9.0 months (mean, 9.6; SD, ±2.3) and in saliva at a median age of 12.7 months (mean, 14.0; SD, ±5.7). This difference was statistically significant (P = 0.0001; Wilcoxon sign-rank test). Over all the samples from infants colonized with S. sanguinis, the average level of S. sanguinis was 2.0 × 105 CFU/ml of unstimulated saliva. This comprised 0.1% of the total cultivable bacteria in saliva. In plaque, S. sanguinis comprised 10% (±18.3%) of the total cultivable bacteria from dentate infants. As new teeth emerged in the infants, the levels of S. sanguinis in saliva increased (r = 0.89; P = 0.0001; Pearson correlation analysis). There were no racial or gender differences in any of the measured aspects of initial acquisition of S. sanguinis.
Relationship between colonization by S. sanguinis and mutans streptococci.
Because colonization by S. sanguinis not only precedes that by mutans streptococci but, like that by mutans streptococci, is dependent on the presence of teeth, we wondered whether colonization with S. sanguinis influenced subsequent colonization with mutans streptococci. In addition, S. sanguinis is thought to be an antagonist of mutans streptococci. To address this query, we employed Cox regression analysis (stepwise selection) using the time of oral colonization of mutans streptococci as the dependent variable and time of infection and proportions of S. sanguinis to total cultivable bacteria, race, gender, first tooth emergence, and mother's caries experience as the independent variables. Infants with higher pre-mutans streptococci proportions of S. sanguinis to total cultivable bacteria in saliva exhibited a significant delay in the time to colonization by mutans streptococci compared with infants who had lower proportions (29 versus 23 months; risk ratio = 0.91; P = 0.02; Cox regression analysis). Moreover, the time until S. sanguinis colonization significantly influences the time until mutans streptococci colonization, i.e., early S. sanguinis colonization correlates to late mutans streptococci colonization (risk ratio = 0.82; P = 0.03; Cox regression analysis).
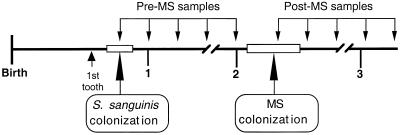
To further examine the possible antagonism between S. sanguinis and mutans streptococci, we averaged the levels of S. sanguinis in saliva for periods before and after colonization with mutans streptococci (Fig. 3). On average, the pre- and post-mutans streptococci saliva samples were comprised of 6.8 and 4.8 samples, respectively, from 37 infants. (Eight infants did not harbor detectable levels of mutans streptococci.) We then compared pre-mutans streptococci levels (4.5 × 105 CFU/ml) to post-mutans streptococci levels (1.9 × 105 CFU/ml) and found that the S. sanguinis levels were significantly greater (P = 0.05, paired t test) in saliva before the initial colonization by mutans streptococci than after. As expected, pre- and post-mutans streptococci levels of S. sanguinis within each individual were positively correlated with each other (r = 0.43; P = 0.01). Interestingly, the eight infants who appeared to be free of mutans streptococci exhibited higher average levels of S. sanguinis in their saliva (5.7 × 105 CFU/ml) than did the 37 mutans streptococci-infected infants (4.6 × 105 CFU/ml); this difference was also statistically significant (P = 0.03, Wilcoxon rank sum test).
FIG. 3.
Study design of the natural history study: sampling periods relative to the infant time line. Bacteriological samples were taken at 3-month intervals. Pre- and post-mutans streptococci levels of S. sanguinis/total cultivable bacteria in saliva were averaged and compared relative to the time to colonization by the mutans streptococci.
Relationship between sanguinis streptococcus colonization and caries.
We compared the time to initial colonization with S. sanguinis and its levels in both saliva and plaque to caries experience at 2 and 3 years of age, but failed to show a significant correlation, perhaps due to the low prevalence of caries in this population of children at 2 and 3 years of age (10 and 23%, respectively.)
DISCUSSION
Within this population of urban infants from Birmingham, Ala., initial colonization by S. sanguinis occurs during a discrete window of infectivity, around 9 months of age. Acquisition during a discrete window period is analogous to what was observed for the acquisition of mutans streptococci, which occurred around a median age of 26 months (8). Also similar to mutans streptococci, colonization by S. sanguinis follows and is significantly correlated to the emergence of primary teeth. Unlike colonization by mutans streptococci (8), however, all of the infants eventually acquired S. sanguinis, albeit some 12% of the infants did so later than the median window period shown in Fig. 1. The earlier colonization with S. sanguinis than with mutans streptococci may reflect the greater affinity for attachment of S. sanguinis to tooth surfaces than mutans streptococci (34).
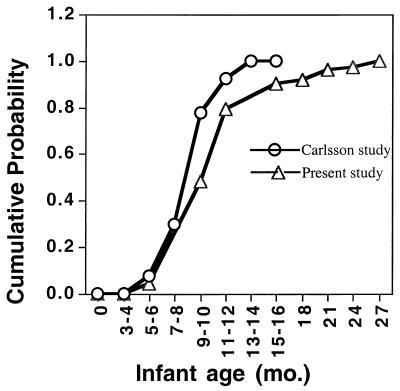
The present study confirms and extends the pioneering work of Carlsson and coworkers (7), who first showed that the colonization of both S. sanguinis and mutans streptococci is dependent upon the presence of teeth. Extrapolation from the original data of Carlsson (7) shows close agreement with data from the present study as evident when comparing the cumulative probability of infection curves (Fig. 4). Also in agreement with Carlsson's group is the strong correlation between time to colonization of S. sanguinis and the time of emergence of the primary dentition (Fig. 1). That colonization with S. sanguinis is dependent upon the presence of teeth is further supported in the present study, which shows that detection of S. sanguinis in plaque precedes its detection in saliva by more than 4 months. In another study in Boston area infants, Smith and coworkers (26) reported that 7 of 14 (50%) infants were colonized by S. sanguinis by 12 months of age. This observation agrees well with our observation that S. sanguinis was detected at a median age of 12 months in unstimulated saliva.
FIG. 4.
Cumulative probability of infection by S. sanguinis in 45 infants. Data from the present study (▵) are compared to data extrapolated from the study of Carlsson and coworkers (○) (7).
The notion of a time-dependent window for acquisition of members of the oral biota requires an understanding of the limitations in defining such a window. Populations may differ in time to acquisition based upon environmental and developmental exposures, such as sucrose consumption and enamel hypoplasia, to name only two possible predeterminants of early colonization. The method of detection (e.g., cultural, PCR, or DNA probes) dictates time to infection, but methods can vary widely. Moreover, a possibly more correct designation for a window period would be window of colonization rather than infectivity, because our methods indicate colonization only after sufficient levels are present for detection by cultural methods. More sensitive methods (e.g., PCR and DNA probes) would likely detect S. sanguinis, S. mutans, or other oral indigenous biota at a much earlier times than detection after colonization, perhaps as early as just after birth. Our own work detected mutans streptococci in predentate infants using PCR to spaB (18), suggesting that reservoirs of mutans streptococci other than tooth surfaces exist. It may be that initial infection occurs at or around birth, while colonization occurs as a result of the presence of the right environmental conditions, e.g., teeth.
The time of initial detection of oral bacteria is also dependent upon the design of the study, that is, longitudinal versus cross-sectional. Study results may differ based on this factor alone. We prefer longitudinal surveys, as sustained colonization can be distinguished from transient infections, but longitudinal studies are expensive and loss to follow-up is often a problem, especially with the mobility inherent in urban populations.
As shown in the present study, the levels of S. sanguinis increase with the age of the infant. After the mutans streptococci colonize the oral cavity, however, the levels of S. sanguinis decrease. We attribute this initial increase in S. sanguinis prior to the colonization of mutans streptococci to the availability of new colonization sites as primary teeth emerge. Other investigators have reported an increase in total streptococci correlated with an increase in the number of teeth present (31), but the same investigators did not show that this increase was correlated to increases in S. sanguinis. These conflicting data may be due to the fact that Tappuni and Challacombe (31) reported a low isolation frequency of only 5.4% for S. sanguinis from dentate infants aged 1 to 3.4 years. In contrast, we show that 100% of dentate infants were colonized by S. sanguinis prior to 18 months of age. These authors also isolated S. sanguinis from predentate infants (1.7%), as did we (2%), but our subsequent samples failed to document persistent colonization prior to tooth emergence.
To some extent, differences in findings among the various studies may arise from the definition of the species comprising the sanguinis complex. As a result of recent reexamination of the oral streptococci, the group of oral bacteria formerly classified as S. sanguis have now been divided into at least two species, S. sanguinis and S. gordonii, each species having three to four biotypes or biovars (15, 35). This reclassification is not without controversy, however. The three authoritative reviews involved with reexamination of the classification of viridans streptococci lack agreement, particularly with regard to the grouping of S. sanguinis (10, 15, 35). This ambiguity is further heightened by the fact that the genetic determinants of the four biovars of the newly designated S. sanguinis vary to the extent that they may constitute separate species (10, 22). Our data confirm this lack of genetic homogeneity in that the 16S rDNA sequences varied as much within biovars as between biovars and there was no apparent overlap or clustering within biovars and their 16S rDNA loci (Y. Pan, Y. Li, and P. W. Caufield, unpublished data). In fact, Willcox (37) argued for at least nine species within the “S. sanguinis group” based on biochemical and genetic data. Nonetheless, the isolates we selected based primarily on colony morphology bore the highest phylogenetic affiliation with the S. sanguinis prototype strain, ATCC 10556. This affiliation may be somewhat misleading, however, because only a few sequences from the 16S rDNA locus of S. sanguinis are present in the GenBank database. It can be argued that the genetic data support splitting the sanguinis streptococci into at least two genetic groups based on 16S rDNA locus diversity alone. Clearly, the sanguinis group of streptococci that we describe here is well defined in terms of their discrete ecological characteristics (i.e., colonization site and time of acquisition) as well as their phylogenetic affiliation. We and others (23) suggest that similarity indices based only on genetic data (e.g., DNA-DNA hybridization and rRNA) using more or less arbitrary cutoff points for defining a species may be inappropriate when applied to an ecologically defined population of bacteria.
Perhaps the most interesting finding of this study, and of possible clinical relevance, is the relationship between levels and time of colonization of S. sanguinis and subsequent colonization of mutans streptococci. Serial samples of saliva, which contained higher levels of S. sanguinis, were associated with a 6-month delay in the colonization of mutans streptococci. Consistent with this finding is our observation that early colonization of S. sanguinis in an infant results in the later colonization of mutans streptococci. The notion that delayed colonization of mutans streptococci may lead to less caries has been demonstrated by Köhler and coworkers (16). Whether the early introduction of S. sanguinis into the oral cavity of infants or increasing its relative proportions by artificial means could affect subsequent mutans streptococci colonization or caries has yet to be demonstrated. The concept of effector or probiotic therapy to displace another organism has precedence, however (25). Efforts to artificially implant oral streptococci in the oral cavity to serve as an effector strain have been proposed by others (30).
Another interesting observation was that the colonization of mutans streptococci might have adversely influenced the S. sanguinis levels, since they were shown to drop significantly following colonization of the mutans streptococci. This finding is consistent with the notion that S. sanguinis and the mutans streptococci antagonize or compete with each other in the oral cavity. Further supporting this possible antagonism is the observation that the eight infants who did not acquire mutans streptococci during the window period had significantly higher mean levels of S. sanguinis than mutans streptococcus-infected infants (5.7 × 105 CFU/ml versus 4.6 × 105 CFU/ml; P = 0.03). Taken as a whole, the data here indicate that S. sanguinis and the mutans streptococci compete for colonization of the infant and that one affects the colonization of the other.
From these data, we speculate that early colonization of S. sanguinis could delay the colonization of the mutans streptococci. This, in turn, could result in a reduction in dental caries, as delayed colonization of mutans streptococci has been associated with lower caries scores (16). Repeating these studies in a more caries-active population may bear out this relationship. If such a relationship exists, one might also predict caries risk based on S. sanguinis levels or time to initial infection. Further manipulation of this relationship may involve the early introduction of S. sanguinis into the mouths of infants, serving as an effector strain, possibility reducing the risk for future caries. What remains, and is currently under investigation, is determining the source of S. sanguinis to the infant. If the mother is the primary source of S. sanguinis, as has been shown in the case of mutans streptococci (17), then measures that foster the transmission of S. sanguinis to her infants may be a viable means of protecting the infant from mutans streptococci infection and caries. Caution is warranted here, however, because the sanguinis streptococci have been implicated with life-threatening diseases, including bacterial endocarditis.
ACKNOWLEDGMENTS
Support for this study came from USPHS research grants RFP 5-83-3R, RO3 DE10224, and P50 DE11147 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 20892.
We thank Zhenmei Lu and Winnie Lee for their technical assistance in the area of clinical microbiology.
REFERENCES
- 1.Beighton D, Hardie J M, Whiley R A. A scheme for the identification of viridans streptococci. J Med Microbiol. 1991;35:367–372. doi: 10.1099/00222615-35-6-367. [DOI] [PubMed] [Google Scholar]
- 2.Bentley R W, Leigh J A, Collins M D. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991;41:487–494. doi: 10.1099/00207713-41-4-487. [DOI] [PubMed] [Google Scholar]
- 3.Burt B A, Loesche W J, Eklund S A, Earnest R W. Stability of Streptococcus mutans and its relationship to caries in a child population over 2 years. Caries Res. 1983;17:532–542. doi: 10.1159/000260714. [DOI] [PubMed] [Google Scholar]
- 4.Carlsson J. A numerical taxonomic study of human oral streptococci. Odontol Revy. 1968;19:137–160. [PubMed] [Google Scholar]
- 5.Carlsson J. Zooglea-forming streptococci, resembling Streptococcus sanguis, isolated from dental plaque in man. Odontol Revy. 1965;16:348–358. [PubMed] [Google Scholar]
- 6.Carlsson J, Soderholm G, Almfeldt I. Prevalence of Streptococcus sanguis and Streptococcus mutans in the mouth of persons wearing full-dentures. Arch Oral Biol. 1969;14:243–249. doi: 10.1016/0003-9969(69)90226-x. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson J, Grahnen H, Jonsson G, Wikner S. Establishment of Streptococcus sanguis in the mouths of infants. Arch Oral Biol. 1970;15:1143–1148. doi: 10.1016/0003-9969(70)90005-1. [DOI] [PubMed] [Google Scholar]
- 8.Caufield P W, Cutter G R, Dasanayake A P. Initial acquisition of mutans streptococci infections in infants: evidence for a discrete window of infectivity. J Dent Res. 1993;72:37–45. doi: 10.1177/00220345930720010501. [DOI] [PubMed] [Google Scholar]
- 9.Caufield P W, Gibbons R J. Suppression of Streptococcus mutans in the mouths of humans by a dental prophylaxis and topically-applied iodine. J Dent Res. 1979;58:1317–1326. doi: 10.1177/00220345790580040301. [DOI] [PubMed] [Google Scholar]
- 10.Coykendall A L. Classification and identification of the viridans streptococci. Clin Microbiol Rev. 1989;2:315–328. doi: 10.1128/cmr.2.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasanayake A P, Caufield P W, Cutter G R, Stiles H M. Transmission of mutans streptococci to infants following short term application of iodine-NaF solution to mothers' dentition. Comm Dent Oral Epidemiol. 1993;21:136–142. doi: 10.1111/j.1600-0528.1993.tb00738.x. [DOI] [PubMed] [Google Scholar]
- 12.de Stoppelaar J D, van Houte J, Backer-Dirks O. The relationship between extracellular polysaccharide-producing streptococci and smooth surface caries in 13-year-old children. Caries Res. 1969;3:190–199. doi: 10.1159/000259582. [DOI] [PubMed] [Google Scholar]
- 13.Gilmour M N, Whittam T, Kilian M, Selander R K. Genetic relationships among the oral streptococci. J Bacteriol. 1987;169:5247–5257. doi: 10.1128/jb.169.11.5247-5257.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosmer D W, Lemeshow S. The importance of assessing the fit of logistic regression models: a case study. Am J Public Health. 1991;81:1630–1635. doi: 10.2105/ajph.81.12.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilian M, Mikkelsen L, Henrichsen J. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis. Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- 16.Kohler B, Andreen I, Jonsson B. The earlier the colonization by mutans streptococci, the higher the caries prevalence at 4 years of age. Oral Microbiol Immunol. 1988;3:14. doi: 10.1111/j.1399-302x.1988.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Caufield P W. The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J Dent Res. 1995;74:681–685. doi: 10.1177/00220345950740020901. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Teague E, Zhuang Z, Caufield P W. Screening for the spaP gene of Streptococcus mutans in predentate infants. J Dent Res. 1997;76:102. [Google Scholar]
- 19.Loesche W J, Bhat M. Evaluation of diagnostic broths for Streptococcus mutans. In: Stiles H M, Loesche W J, O'Brien, editors. Microbial aspects of dental caries. Washington, D.C.: Informational Retrieval Inc.; 1976. pp. 291–301. [Google Scholar]
- 20.Loesche W J, Rowan J, Straffon L H, Loos P J. Association of Streptococcus mutans with human dental decay. Infect Immun. 1975;11:1252–1260. doi: 10.1128/iai.11.6.1252-1260.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyvad B, Kilian M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987;95:369–380. doi: 10.1111/j.1600-0722.1987.tb01627.x. [DOI] [PubMed] [Google Scholar]
- 23.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 24.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders E. Bacterial interference. I. Its occurrence among the respiratory tract flora and characterization of inhibition of group a streptococci by viridans streptococci. J Infect Dis. 1969;120:698–707. doi: 10.1093/infdis/120.6.698. [DOI] [PubMed] [Google Scholar]
- 26.Smith D J, Anderson J M, King W F, van Houte J, Taubman M A. Oral streptococcal colonization of infants. Oral Microbiol Immunol. 1993;8:1–4. doi: 10.1111/j.1399-302x.1993.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 27.Socransky S S, Haffajee A D, Dzink J L, Hillman J D. Associations between microbial species in subgingival plaque samples. Oral Microbiol Immunol. 1988;3:1–7. doi: 10.1111/j.1399-302x.1988.tb00596.x. [DOI] [PubMed] [Google Scholar]
- 28.Syed S A, Loesche W J. Efficiency of various growth media in recovering oral bacterial flora from human dental plaque. J Clin Microbiol. 1973;26:459–465. doi: 10.1128/am.26.4.459-465.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Syed S A, Loesche W J. Survival of human dental plaque flora in various transport media. Appl Microbiol. 1972;24:638–644. doi: 10.1128/am.24.4.638-644.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanzer J M, Kurasz A B, Clive J. Competitive displacement of mutans streptococci and inhibition of tooth decay by Streptococcus salivarius TOVE-R. Infect Immun. 1985;48:44–50. doi: 10.1128/iai.48.1.44-50.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tappuni A R, Challacombe S J. Distribution and isolation frequency of eight streptoccocal species in saliva from predentate and dentate children and adults. J Dent Res. 1993;72:31–36. doi: 10.1177/00220345930720010401. [DOI] [PubMed] [Google Scholar]
- 32.Truper H, Clari L D. Taxonomic note: necessary corrections of specific epithets formed as substantives (nouns) “in apposition.”. Int J Syst Bacteriol. 1997;47:908–909. [Google Scholar]
- 33.van der Hoeven J S, Camp P J. Mixed continuous cultures of Streptococcus mutans with Streptococcus sanguis or with Streptococcus oralis as a model to study the ecological effects of the lactoperoxidase system. Caries Res. 1993;27:26–30. doi: 10.1159/000261511. [DOI] [PubMed] [Google Scholar]
- 34.van Houte J, Gibbons R J, Banghart S B. Adherence as a determinant of the presence of Streptococcus salivarius and Streptococcus sanguis on the human tooth surface. Arch Oral Biol. 1970;15:1025–1034. doi: 10.1016/0003-9969(70)90115-9. [DOI] [PubMed] [Google Scholar]
- 35.Whiley R A, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol. 1998;13:195–216. doi: 10.1111/j.1399-302x.1998.tb00698.x. [DOI] [PubMed] [Google Scholar]
- 36.Whiley R A, Fraser H, Hardie J M, Beighton D. Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the “Streptococcus milleri group.”. J Clin Microbiol. 1990;28:1497–1501. doi: 10.1128/jcm.28.7.1497-1501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willcox M D P. Identification and classification of species within the Streptococcus sanguis group. Aust Dent J. 1996;41:107–112. doi: 10.1111/j.1834-7819.1996.tb05922.x. [DOI] [PubMed] [Google Scholar]
- 38.Wright J T, Cutter G R, Dasanayake A P, Stiles H M, Caufield P W. Effect of conventional dental restorative treatment on bacteria in saliva. Comm Dent Oral Epidemiol. 1992;20:138–143. doi: 10.1111/j.1600-0528.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]