Abstract
Most organisms adjust their development according to the environmental conditions. For the majority, this implies the sensing of alterations to cell walls caused by different cues. Despite the relevance of this process, few molecular players involved in cell wall sensing are known and characterized. Here, we show that the wall-associated kinase-like protein RESISTANCE TO FUSARIUM OXYSPORUM 1 (RFO1) is required for plant growth and early defense against Fusarium oxysporum and functions by sensing changes in the pectin methylation levels in the cell wall. The RFO1 dwell time at the plasma membrane is affected by the pectin methylation status at the cell wall, regulating MITOGEN-ACTIVATED PROTEIN KINASE and gene expression. We show that the extracellular domain of RFO1 binds de-methylated pectin in vitro, whose distribution in the cell wall is altered during F. oxysporum infection. Further analyses also indicate that RFO1 is required for the BR-dependent plant growth alteration in response to inhibition of pectin de-methyl-esterase activity at the cell wall. Collectively, our work demonstrates that RFO1 is a sensor of the pectin methylation status that plays a unique dual role in plant growth and defense against vascular pathogens.
This study identifies RFO1, a wall-associated kinase-like protein, as a sensor of the pectin methylation status of the cell wall. RFO1 was shown to mediate plant responses to altered pectin methylation levels in the cell wall caused by Fusarium oxysporum infection. It also functions as a novel positive regulator of BR signaling required for normal growth under conditions of altered PME activity.
Introduction
Most cells have a wall as their outermost layer surroundsing, and connected with their plasma membrane (PM). The cell wall allows expansion of the cell while preventing its rupture. Thus, the wall is indispensable for the survival of the cell and, in multicellular organisms, permits cell adhesion. The main properties of cell walls (i.e., strength and extensibility) rely on chemical interactions among the polysaccharides that build the core structure of the wall, as well as the capacity to rearrange these constituents rapidly at a subcellular level (Bidhendi and Geitmann, 2016). This is particularly relevant for plants, which need to adjust their development in response to a constantly changing environment (Vaahtera et al., 2019).
The plant cell wall core is composed of the polysaccharides cellulose, hemicelluloses, and pectins. As the most hydrophilic and flexible cell wall polysaccharide, pectins have a central role in plant growth and acclimation to the environment. Pectins have a homogalacturonan (HG) backbone that makes them structurally significantly different from the β-1,4-Glc-based cellulose and hemicellulose polysaccharides (Albersheim et al., 1996; Mohnen, 2008). The pectin polygalacturonic acid core is secreted heavily decorated with methyl and/or acetyl groups (Mohnen, 2008; Anderson, 2016). Once at the cell wall, extracellular enzymes called PECTIN METHYLESTERASES (PMEs) and PECTIN ACETYLESTERASES gradually remove those decorations, while PME inhibitors (PMEIs) counter the action of PMEs by directly inhibiting their catalytic domains (Coculo and Lionetti, 2022). These changes in pectin decorations modify the physicochemical properties and, consequently, the structure and function of the cell wall. Moreover, PME activity is involved in plant immunity, as it leads to cell wall reinforcement release of MeOH, and it indirectly promotes the generation of oligogalacturonides (OGs), both of which are pectin-derived damage-associated molecular patterns (DAMPs) (Bethke et al., 2014; Lionetti, 2015; Del Corpo et al., 2020). Therefore, perturbing the delicate balance between methylated and demethylated pectin (hereafter mPectin and dmPectin, respectively) has severe consequences for plant growth and defense (Wolf et al., 2012; Fan et al., 2017; Engelsdorf et al., 2018; Feng et al., 2018; Liu et al., 2018). These developmental responses to pectin alterations start with cellular monitoring of the state of its cell wall through PM-localized receptors, among other sensors (Ringli, 2010). In the context of plant growth, the Arabidopsis PM-localized receptor kinase FERONIA has recently been shown to regulate pavement cell morphogenesis upon binding to highly dmPectin (Lin et al., 2022). In Arabidopsis, the PM receptors BRASSINOSTEROID INSENSITIVE 1 (BRI1) and RECEPTOR-LIKE PROTEIN 44 (RLP44), together with the BRI1 co-receptor BRI1-Associated Kinase 1 (BAK1), have been reported to coordinately mediate plant responses to abnormally high levels of mPectin present at the cell wall (Wolf et al., 2012, 2014; Glöckner et al., 2022). Similarly, the WALL-ASSOCIATED KINASEs (WAKs) and WAK-likes (WAKLs) participate in controlling cell expansion (Lally et al., 2001; Wagner and Kohorn, 2001; Wu et al., 2020; Li et al., 2021), presumably by sensing pectin perturbations. Indeed, WAK1 and WAK2 were reported to bind and respond to dmPectin but not highly mPectin or branched pectin (He et al., 1996; Anderson et al., 2001; Decreux et al., 2006; Kohorn et al., 2009).
WAKs have also been shown to function as sensors of OGs released upon wounding and pathogen infection (Brutus et al., 2010). Additionally, transcriptomic data indicate that most WAKs and WAKLs are important for plant defense (Ferrari et al., 2013; Menna et al., 2021). This function is demonstrated by a high and increasing number of reports in different crops, including wheat (Saintenac et al., 2018; Dmochowska-Boguta et al., 2020), cotton (Wang et al., 2020; Yang et al., 2021), and tomato (Rosli et al., 2013). Although WAKLs contain similar extracellular domains to those present in WAKs, their association with the plant cell wall has not been previously demonstrated (Verica and He, 2002; Verica et al., 2003). In addition, recent studies in cotton and tomato suggest that the role of WAKs and WAKLs in signal transduction expand beyond that of responding to pectin changes (Wang et al., 2020; Zhang et al., 2020; Qi et al., 2021). The cotton GhWAK7A seems to be needed for chitin but not OG perception during infection with vascular fungi (Wang et al., 2020). The tomato TaWAK7D, which is required for defense against a necrotrophic fungus, appears to participate in plant responses to both chitin and pectin (Qi et al., 2021). Moreover, WAK1 and other members of the family are suggested to participate in flagellin-triggered immunity (Zhang et al., 2020). Among the WAKs and WAKLs identified to play a role in Arabidopsis responses toward pathogens, WAKL22 was found to be required for resistance to Fusarium oxysporum (Fo) and Verticillium spp. wilts and was therefore named RESISTANCE TO FUSARIUM OXYSPORUM 1 (RFO1) (Verica and He, 2002; Diener and Ausubel, 2005).
Root vascular fungi such as F. oxysporum and Verticillium dahliae are among the most detrimental pathogens worldwide, affecting high-value crops and plants in natural ecosystems (Klosterman et al., 2009; Gordon, 2017). F. oxysporum and V. dahliae infections begin at the epidermis and progress to the xylem mainly intercellularly; i.e., through the apoplast. Once in the root vasculature, the fungi proliferate, blocking the xylem and causing wilting and, ultimately, plant death (Bishop and Cooper, 1983). In their path toward the root vasculature, F. oxysporum and V. dahliae modify and degrade the plant cell walls through the secretion of a wide range of cell-wall-modifying enzymes (Jonkers and Rep, 2009; Glass et al., 2013; Bravo-Ruiz et al., 2017), including PMEs, pectin and pectate lyases, and polygalacturonases (Cooper and Wood, 1975; Durrands and Cooper, 1988; Di Pietro and Roncero, 1998; Huertas-González et al., 1999; García-Maceira et al., 2000; García-Maceira et al., 2001; Bravo Ruiz et al., 2016; Fan et al., 2017; Yang et al., 2018; Safran et al., 2021; Gámez-Arjona et al., 2022). It is vital for a host plant’s survival to sense these pectin modifications, either directly or through the sensing of pectin degradation products, as part of their response to these root vascular pathogens and to mediate growth-defense processes. However, no PM-localized protein has been identified to play this vital function. To fill this gap in knowledge, we characterized the role of RFO1 as a putative sensor of the pectin methylation status at the cell wall. We link RFO’s function to the detection of altered pectin methylation at the cell wall produced during F. oxysporum infection and generated by PME inhibition. The results presented here indicate that RFO1 has a dual role in pectin sensing during both plant development and defense.
Results
RFO1 is a PM-localized protein necessary for root resistance to F. oxysporum
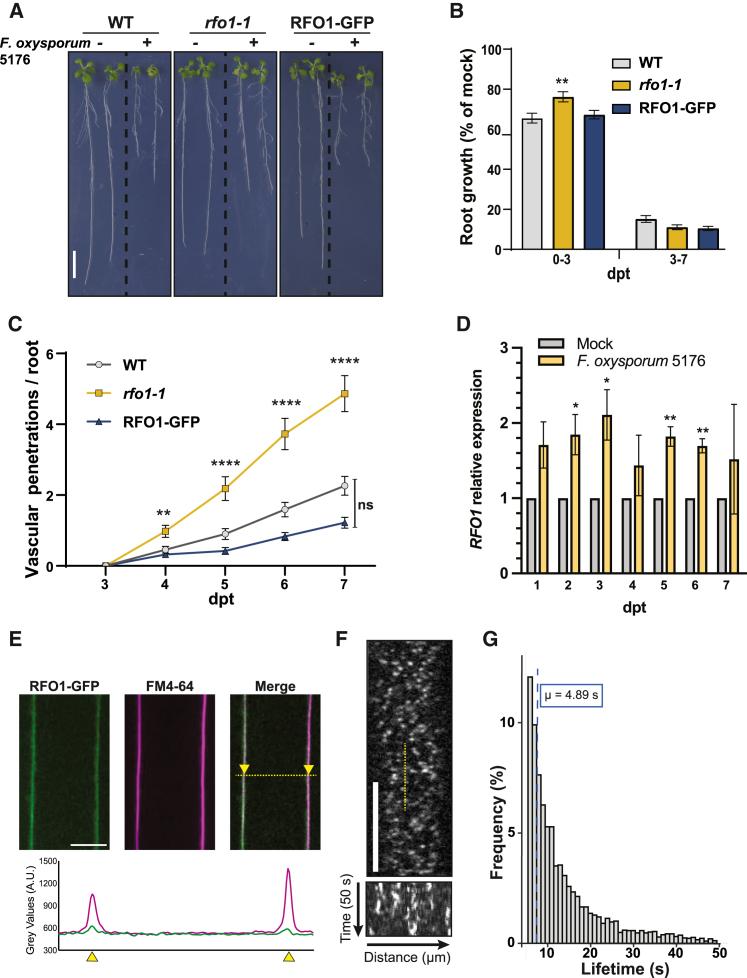
To evaluate the role of RFO1 in plant defense against F. oxysporum in the root, we used the model pathosystem Arabidopsis–F. oxysporum5176 to characterize the infection phenotype of the mutant rfo1-1 following a previously described plate infection assay employing F. oxysporum5176 pSIX1::GFP (Kesten et al., 2019; Huerta et al., 2020). While there were no growth differences between the Col-0 wild-type (WT) control and rfo1-1 under mock treatment conditions, the mutant showed less root growth inhibition during the first days of the interaction with the fungus (0–3 days post transfer [dpt] to plates containing F. oxysporum microconidia) than WT. (Figure 1A and 1B, Supplemental Figure 1A and 1B). Additionally, rfo1-1 plants showed increased vascular colonization compared with WT, starting at 4 dpt (Figure 1C). To confirm RFO1 function in plant response to root colonization by F. oxysporum5176, we complemented rfo1-1 plants with a C-terminal-tagged GFP fusion driven by the RFO1 promoter (pRFO1::RFO1-GFP). The resulting RFO1-GFP-complemented line showed no altered phenotypes compared with rfo1-1 and WT plants (Supplemental Figure 1A and 1B), but the rfo1-1 susceptibility phenotype was restored to the WT phenotype (Figure 1D), confirming the functionality of the complemented line. In addition, the expression of RFO1 increased by two-fold in Fo-infected WT roots at 2, 3, 5, and 6 dpt (Figure 1D). Our results suggest that rfo1-1 susceptibility to F. oxysporum might be a consequence of RFO1’s role in root resistance to F. oxysporum before the fungus reaches the vasculature. Increased RFO1 levels seem to be required by the plant to slow down the entrance of F. oxysporum into the xylem, inducing growth-defense tradeoffs.
Figure 1.
The plasma membrane-localized RFO1 is required for plant resistance to F. oxysporum5176 pSIX1::GFP during root colonization.
(A) Representative images of 8-day-old Mock (−) or F. oxysporum5176 pSIX1::GFP infected (+) wild-type (WT; Col-0), rfo1-1, and RFO1-GFP (rfo1-1 pRFO1::RFO1-GFP) seedlings after 3 days post transfer (dpt) to the corresponding treatment. Scale bar, 10 mm.
(B) Root growth of F. oxysporum5176-infected seedlings relative to Mock-treated ones, as shown in (A), between 0 and 3 days post treatment with F. oxysporum5176 (dpt) and 3–7 dpt. Data represent the mean ± SE of N > 40 seedlings per genotype from N = 3 independent replicates. Repeated measures ANOVA with Dunnett’s multiple-comparison test, ∗∗p < 0.01. Significance shown compared with WT.
(C) Cumulative number of F. oxysporum5176 vascular penetrations per root in plants infected as depicted in (A) from 3 to 7 dpt. Data represent the mean ± SE of N > 40 seedlings per genotype from N = 3 independent replicates. RM ANOVA with Dunnett’s multiple-comparison test, ∗∗p < 0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001. ns, no significance. Significance shown compared with WT.
(D)RFO1 expression relative to GADPH in Mock and F. oxysporum5176-infected WT roots at 2 and 3 dpt, as shown in (A). Bars represent the mean ± SE of N = 3 independent replicates, normalized to mock values. t-test, ∗p < 0.05, ∗∗p < 0.001.
(E) Representative longitudinal optical cross section (top) of an elongating epidermal cell from a 5-day-old RFO1-GFP root (green) after staining with FM4-64 (magenta), with the corresponding plot of GFP and RFP gray values (bottom) taken along the labeled dashed yellow line. The arrows represent the margins of the PM. Scale bar, 10 μm.
(F) Representative spinning-disc confocal image of a 5-day-old RFO1-GFP root elongating epidermal cell and a kymograph along the length of the dashed yellow line from the corresponding video (n = 50 frames, t = 50 s) showing RFO1 localization at discrete PM foci. Scale bar, 5 μm.
(G) Dwell time frequencies of RFO1-GFP foci at the PM as depicted in (F). Mean (μ) is shown by a dashed blue line centered at 4.89 s. Data are derived from N > 4000 particles from 10 cells across three roots.
To characterize the in vivo transcriptional activation of RFO1, plants were generated expressing RFP fused to a C-terminal N7-nuclear localization signal under the control of the RFO1 promoter (pRFO1::RFP-N7). Expression of RFP-N7 was observed in, but was not limited to, root epidermal cells in mock-treated roots and roots exposed to F. oxysporum5176 pSIX1::GFP microconidia and imaged after 1 day (Kesten et al., 2019; Huerta et al., 2020) (Supplemental Figure 1C and 1D). The nuclear-localized RFP signal was significantly enhanced in root epidermal cells of the root tip exposed to F. oxysporum5176 pSIX1::GFP pre-germinated microconidia, compared with the mock-treated root (95.4 ± 8.2 a.u. for mock vs. 110.0 ± 23.3 a.u. for F. oxysporum5176). These results suggest that there is increased RFO1 expression upon F. oxysporum infection, corroborating the results obtained by qPCR (Figure 1D).
The localization of RFO1 at the PM was confirmed through observation of optical longitudinal cross sections of RFO1-GFP root cells stained with the fluorescent lipophilic dye FM4-64 (Miyake and McNeil, 1995). Composite overlays of these sections demonstrated that RFO1-GFP and FM4-64 signals co-localize at the PM (Figure 1E). Spinning-disk confocal microscopy followed by single-particle tracking showed that RFO1-GFP is organized in discrete foci at the PM with an average dwell time of 4.89 s (Figure 1F and 1G, Supplemental Figure 1C; Supplemental Video 1). These dynamics of the RFO1 foci is characteristic of free diffusion, as demonstrated by the linear fit of particle mean squared displacement (MSD) plots (Supplemental Figure 1D). Our results confirm the predicted localization of RFO1 at the PM and describe its particle dynamics.
The video depicts the dynamic behavior of RFO1-GFP expressed in rfo1-1 in Arabidopsis root epidermal cells acquired using spinning-disc confocal microscopy. Foci identified by Trackmate using the LAP tracker to be analyzed are indicated in the right panel. This video corresponds to Figure 1F and 1G.
RFO1 is required for plant growth responses to perturbation of pectin methylation at the cell wall
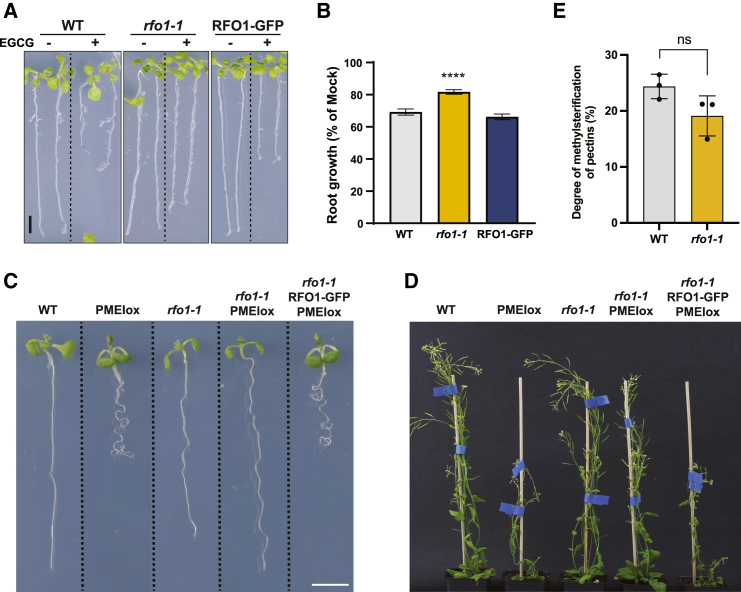
Extracellular galacturonan-binding (GUB) and epidermal growth factor (EGF)-like domains found in WAKs and WAKLs, including RFO1 (Supplemental Figure 2A), have been implicated in WAK perception of dmPectin (He et al., 1996; Wagner and Kohorn, 2001). To determine if RFO1 can mediate plant response to perturbations in pectin methylation, plants were treated with epigallocatechin gallate (EGCG), a pharmacological inhibitor of PMEs at the cell wall (Lewis et al., 2008). EGCG treatment induces root growth inhibition in Arabidopsis seedlings, presumably through its role in decreasing the levels of dmPectin in the cell wall (Lewis et al., 2008). rfo1-1 seedlings showed significantly decreased root growth inhibition upon EGCG treatment compared with WT and RFO1-GFP plants (Figure 2A and 2B), suggesting that RFO1 is required for plant responses to EGCG. To ensure that the lower sensitivity of rfo1-1 to EGCG was due to PME inhibition and not to any off-target effects of the drug, rfo1-1 was introduced into the PMEIox mutant background (Wolf et al., 2012), which over-expresses PMEI5. The overexpression of PMEI5 activity at the cell wall and increases the levels of mPectin. The resulting rfo1-1 PMEIox plants were phenotypically very similar to WT, indicating that rfo1-1 almost completely suppressed the PMEI5 overexpression phenotypes (Figure 2C and 2D). In addition, when RFO1-GFP was introduced into the rfo1-1 PMEIox background, the PMEIox-induced pectin perturbation phenotypes were restored (Figure 2C and 2D). Quantification of the degree of pectin methylesterification in WT and rfo1-1 roots revealed no significant influence of RFO1 on pectin decoration (Figure 2E). Altogether, these data indicate that RFO1 is required for mediating plant responses to chemical or genetic perturbation of pectin methylation at the cell wall.
Figure 2.
rfo1-1 mutant shows altered plant response to perturbation of pectin methylation at the cell wall.
(A) Representative image of 8-day-old WT (Col-0), rfo1-1, and RFO1-GFP (rfo1-1 pRFO1::RFO1-GFP) seedlings after a 48-h Mock (−) or 6.25 μM EGCG (+) treatment. Scale bar, 5 mm.
(B) Root growth of seedlings exposed to EGCG relative to Mock-treated ones as depicted in (A). Data represent the mean ± SE of N ≥ 38 seedlings per genotype from four independent replicates. One-way ANOVA with Dunnett’s multiple-comparison test, ∗∗∗∗p < 0.0001. Significance shown compared with WT.
(C and D) Representative images of WT, PMEIox, rfo1-1, rfo1-1 PMEIox, and rfo1-1 RFO1-GFP PMEIox 8-day-old seedlings (C) or 8-week-old plants (D). In (C), scale bar, 5 mm.
(E) Determination of the degree of methylesterification of pectins using FTIR in 8-day-old WT and rfo1-1 roots. Values correspond to means ± SE of three biological replicates, each one containing two technical replicates.
The extracellular domain of RFO1 in Col-0 plants is necessary for the defense against F. oxysporum and for detecting alterations in the pectin methylation status
Previous studies demonstrated that the Arabidopsis Ty-0 ecotype variant of RFO1 (RFO1Ty−0) is impaired in its ability to confer resistance against F. oxysporum (Diener and Ausubel, 2005). Using our plant infection assay, we confirmed this observation at the root level using RFO1Ty−0-GFP (rfo1-1 RFO1Ty−0-GFP) plants (Supplemental Figure 2B). In addition, RFOTy−0-GFP plants were less sensitive than WT to the root inhibition induced by EGCG (Supplemental Figure 2C and 2D), and plants expressing RFOTy−0-GFP in the rfo1-1 PMEIox backgrounddid not suffer from PMEIox growth defects (Supplemental Figure 2E), resembling in both cases the rfo1-1 mutant. These results suggest that RFOTy−0 is not involved in plant resistance against F. oxysporum in the root and is also not required for sensing altered pectin methylation status at the cell wall (Supplemental Figure 2C and 2D).
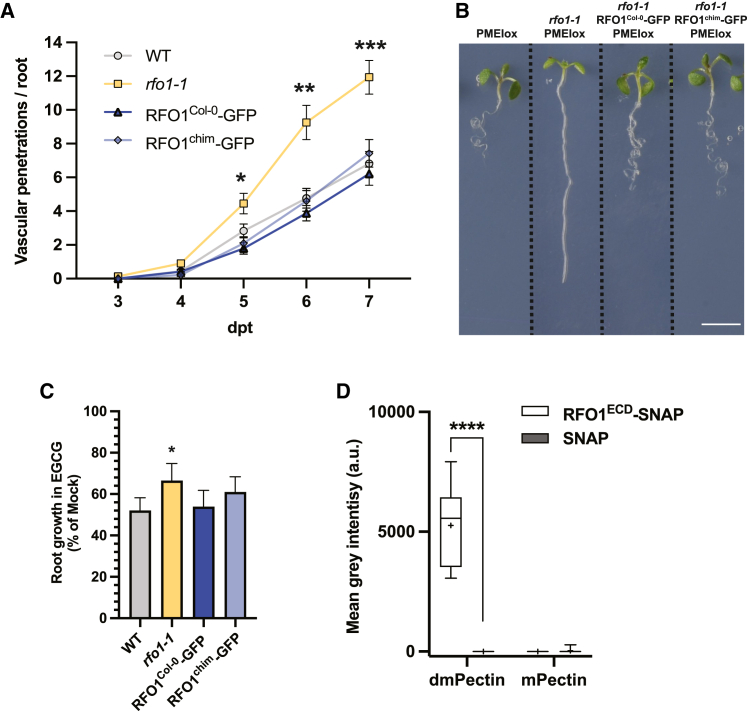
To understand which domain of RFO1Col−0 is essential for pectin-integrity sensing and defense against Fo, we generated a chimeric RFO1 protein through the fusion of the extracellular and the transmembrane domains of RFO1Col−0 and the cytosolic domain of RFO1Ty−0. The resulting chimeric protein (chim) was placed under the control of the RFO1Col−0 promoter (pRFO1::RFO1chim-GFP) and was expressed in the rfo1-1 background (RFO1chim-GFP). RFO1chim-GFP was found to be localized at the PM of root epidermal cells, where it was organized as foci similar to RFO1Col−0-GFP and RFO1Ty−0-GFP particles (Supplemental Figure 2F). Furthermore, RFO1chim-GFP roots did not show growth alterations (Supplemental Figure 2G). RFO1chim-GFP could restore the rfo1-1 susceptibility to F. oxysporum5176 to WT levels, as observed in RFO1Col−0-GFP plants (Figure 3A). In addition, the F1 rfo1-1 (−/−) RFO1chim-GFP (+/−) PMEIox (+/−) plants were phenotypically indistinguishable from PMEIox and rfo1-1 RFO1Col−0-GFP PMEIox lines (Figure 3B), indicating that the extracellular domain of RFO1Col−0 is important for perceiving the pectin modifications caused by PMEI5 overexpression (Figure 3B). Contrary to roots expressing RFO1Ty−0-GFP, those expressing the chimera variant showed sensitivity to EGCG similar to that of roots expressing RFO1Col−0-GFP (Figure 3C and Supplemental Figure 2B and 2C). These data suggest that the RFO1Col−0 ectodomain is sufficient for maintaining RFO1 functionality in defense against F. oxysporum5176 pSIX1::GFP and for plant response to modifications in the pectin methylation status at the cell wall. Moreover, the protein sequence differences observed between Col-0 and Ty-0 in the non-kinase region of RFO1 reveal potential amino acids involved in sensing the pectin methylation status of the cell wall in Arabidopsis (Supplemental Figure 2H).
Figure 3.
The extracellular domain of RFO1Col−0 binds commercial and Arabidopsis cell wall-derived dmPectin.
(A) Cumulative number of vascular penetrations observed in WT (Col-0), rfo1-1, RFO1Col−0-GFP (rfo1-1 RFO1Col−0-GFP), and RFO1chim-GFP (rfo1-1 RFO1chim-GFP) roots between 3 and 7 dpt to plates containing F. oxysporum5176 pSIX1GFP microconidia. Data represent the mean ± SE of N ≥ 10 seedlings per genotype from three independent replicates. RM ANOVA with Tukey’s multi-comparison post hoc test, ∗∗p < 0.01, ∗∗∗p < 0.001. Significance shown compared with WT.
(B) Representative images of 8-day-old WT, PMEIox, rfo1-1, rfo1-1 PMEIox, rfo1-1 RFO1Col−0-GFP PMEIox, and rfo1-1 RFO1chim-GFP PMEIox seedlings. Scale bar, 5 mm.
(C) Relative root growth of EGCG-treated WT, rfo1-1, RFO1Col−0-GFP, and RFO1chim-GFP roots as a percentage of Mock. Bars represent the mean ± SE of >15 seedlings per genotype from four biological replicates. One-way ANOVA with Dunnett’s multiple-comparison post hoc test, ∗∗∗∗p < 0.0001. Significance indicated is compared with WT.
(D) Quantification of dot immunobinding assay intensities in mean gray values from membranes of immobilized commercial demethylesterified pectin (dmPectin) and methylated pectin (mPectin) probed with RFO1ECD-SNAP or SNAP recombinant proteins as shown in Supplemental Figure 5A. Boxplots: centerlines show the medians; means are marked by +; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 12 from ≥3 independent replicates, two-way ANOVA with Tukey’s multiple-comparison test, ∗∗∗∗p < 0.0001.
The RFO1 ectodomain binds demethylated pectin in vitro
To test the role of the extracellular domain of RFO1 in plant response to different pectin methylation levels, we performed dot immunobinding assays. We produced dmPectin from apple-derived mPectin following a previously described method (Liners et al., 1992; Kohorn et al., 2009) and confirmed the complete removal of methyl groups using antibodies against cell wall-derived mPectin (JIM7; Knox et al., 1990; Supplemental Figure 3). Both compounds were immobilized on nitrocellulose membranes that were exposed to either the recombinant C-terminal SNAP-tagged extracellular domain of RFO1 (RFO1ECD) or recombinant SNAP-tag, as a negative binding control (Supplemental Figures 4 and 5A). The immunoassays demonstrated in vitro binding of RFO1ECD to dmPectin but not to mPectin, while little to no binding was observed when using the SNAP alone (Figure 3D and Supplemental Figure 5A). Both RFO1ECD-SNAP and SNAP bound plant cell wall glycoprotein (GP) extracts (Supplemental Figure 5B), demonstrating that the SNAP control protein allows us to differentiate specific (dmPectin; Figure 3D and Supplemental Figure 5A) from non-specific (GP; Supplemental Figure 5B) in vitro binding to cell wall components. We also tested RFO1ECD-dmPectin interaction using thermal shift assays (TSAs),which cover a narrower window of affinities compared with dot blot assays (∼100 nM, ∼100 μM; Layton and Hellinga, 2011). TSAs revealed no increase of RFO1ECD melting temperature in the presence of de-esterified pectin (Supplemental Figure 6), indicating that RFO1ECD cannot bind dmPectin with high affinity. Our data point toward the low-affinity recognition of dmPectin by RFO1ECD.
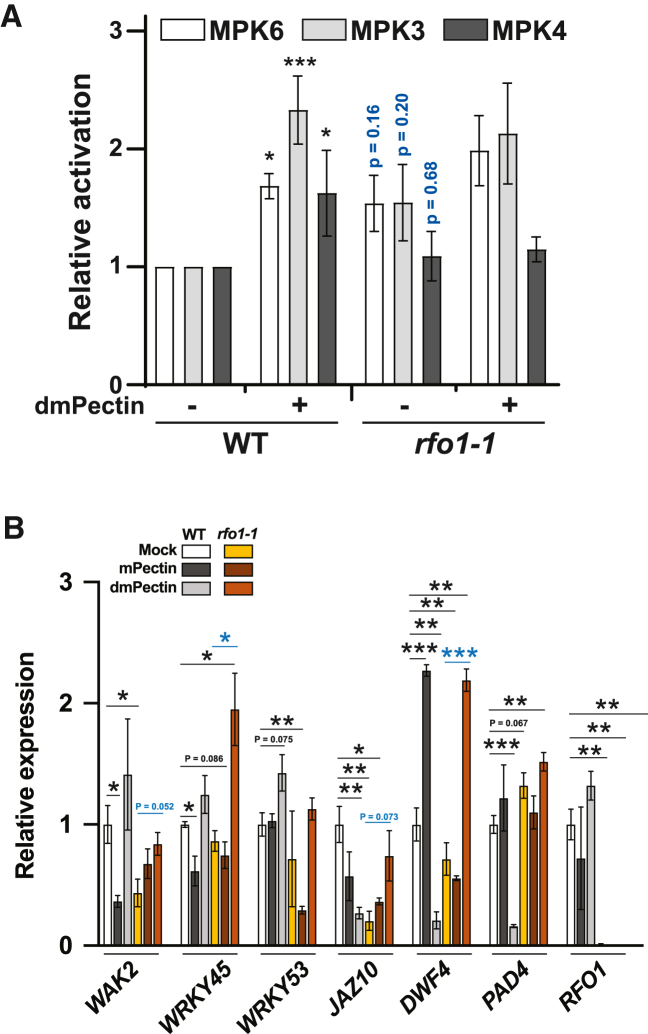
Figure 5.
rfo1-1 has altered basal and dmPectin-induced MAPK phosphorylation levels and gene expression activation.
(A) Quantification of relative MAPK activation in blots as shown in Supplemental Figure 8A. Within each blot, relative MAPK activation was determined by calculating active to total MAPK intensities per sample, and these active/total data were normalized to the untreated WT. Data represent the mean ± SE of N = 5 independent replicates. Multiple-comparison t-tests were performed for WT or rfo1-1 samples with respect to their mock-treated control (black) or between WT and rfo1-1 samples (blue), ∗p < 0.05, ∗∗∗p < 0.001.
(B)WAK2, WRKY45, WRKY53, JAZ10, DWF4, PAD4, and RFO1 gene expression relative to GAPDH in 8-day-old roots of Mock-, mPectin-, or dmPectin-treated WT and rfo1-1 plants. The expression values were then normalized to Mock WT. Data represent the mean ± SE of N = 3 independent replicates. Data were analyzed by t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Asterisks in black indicate the difference compared with WT Mock. Asterisks in blue indicate the differences between rfo1-1 Mock versus rfo1-1 treatment (mPectin or dmPectin).
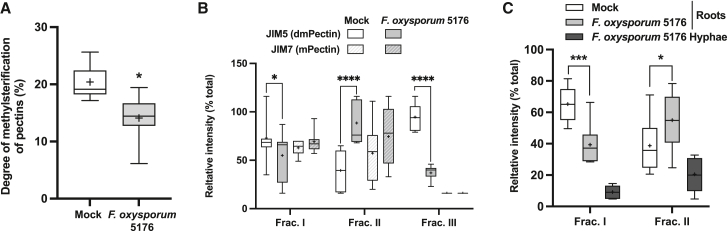
To clarify the role of RFO1 during plant defense against F. oxysporum5176, we quantified the changes in the degree of methylesterification of pectins (DM) by Fourier-transform infrared spectroscopy (FTIR) in mock- and Fo-infected roots at 4 dpt. We observed a significant reduction of the DM after F. oxysporum5176 infection (Figure 4A). In addition, we inspected the pectin methylation status of mock- and F. oxysporum5176-infected roots in three cell wall soluble fractions. This fractionation method allows for the sequential extraction of cell wall polysaccharides based on their chemical linkages. The chelating agent imidazol extracts mainly homogalacturonans (fraction I), boron linkages are broken using carbonate generating a solution enriched in rhamnogalacturonans (fraction II), and KOH is used to extract mainly hemicelluloses and left-over pectins (fraction III) (Hotchkiss and Hicks, 1990; Mort et al., 1991). The obtained fractions were consequently immobilized onto nitrocellulose membranes and probed with antibodies against cell wall-derived dmPectin (JIM5; Casero and Knox, 1995) and mPectin (JIM7; Knox et al., 1990). JIM5 binding was lower in fractions I and III, while it was higher in fraction II of infected roots compared with mock samples (Figure 4B and Supplemental Figure 7A). No differences were observed in the capacity of JIM7 to bind the different fractions in mock- or fungal-infected roots. As fraction III is extracted using a dilute base that removes all pectin methyl groups, the reduction in JIM5 binding and the absence of JIM7 binding to this fraction might be a result of a reduction in total pectin (Figure 4B and Supplemental Figure 7A). Our data indicate that F. oxysporum5176 infection specifically modifies the levels of dmPectins found in fractions I and II while leaving the levels of mPectin mostly unaffected.
Figure 4.
F. oxysporum5176 infection reduces the cell wall pectin methylation level in roots, which seems to be detected by RFO1.
(A) Determination of the degree of methylesterification of pectins using FTIR in mock- and F. oxysporum5176-infected roots at 4 dpt. Values correspond to means ± SE of three biological replicates, each one containing at least four technical replicates. Data were analyzed by t-test, ∗p < 0.05.
(B) Quantification of dot immunobinding assays from membranes of immobilized plant cell wall fractions from Mock- and F. oxysporum5176-infected WT roots at 4 dpt and probed with JIM5 (anti-dmPectin) or JIM7 (anti-mPectin) antibodies as shown in Supplemental Figure 7A.
(C) Quantification of dot immunobinding assay using root cell wall fractions I and II as described in (B) and F. oxysporum5176 hyphae cell wall fractions probed with RFO1ECD-SNAP recombinant protein as shown in Supplemental Figure 7B.
(B and C) Boxplots: centerlines show the medians; means are marked by +; box limits indicate the 25th and 75th percentiles; whiskers extend to the minimum and maximum. N ≥ 12 from ≥3 independent replicates. For each replicate, data are reported as the percentage that each fraction represents from the total gray value of the blot. Two-way ANOVA with Tukey’s multiple-comparison test, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Further dot immunobinding assays were performed to determine if RFO1ECD binds to cell wall fractions in a dmPectin-dependent manner, using cell wall fractions obtained from mock- and F. oxysporum5176-infected roots. The cell walls of in vitro-grown F. oxysporum5176 hyphae were fractionated in a similar way to root cell walls and included as a negative control. RFO1ECD binding to the immobilized cell wall fractions correlated with the changes observed in dmPectin levels within fractions I and II after F. oxysporum5176 infection (Figure 4C and Supplemental Figure 7B), determined by the aforementioned JIM5/JIM7 immunobinding assays (Figure 4B and Supplemental Figure 7A). Fraction III was not included in the assay, as it does not help to differentiate the pectin methylation state at the cell wall (Supplemental Figure 7A). The SNAP negative control showed no differential pattern of binding to dmPectin within cell wall fractions (Supplemental Figure 7C and 7D). Together, these data indicate that levels of dmPectin in root cell wall fractions are altered during F. oxysporum infection and that RFO1 acts in sensing these modifications through differential binding to dmPectin.
RFO1 triggers MAPK signaling and gene expression activation in response to altered dmPectin levels
Phospho-activation of MITOGEN-ACTIVATED PROTEIN KINASEs (MAPKs) is required for WAK2-mediated signaling in plants upon dmPectin sensing (Kohorn et al., 2009). We tested whether RFO1 is similarly responsible for MAPK activation upon dmPectin sensing. The phosphorylation levels of MAPK6 and MAPK3 in WT roots increased after 30 min of exogenous dmPectin treatment, suggesting that dmPectin signals regulate MAPK cascades (Figure 5A and 5B), as previously suggested for MAPK3 (Kohorn et al., 2009). In contrast, rfo1-1 roots showed increased basal MAPK phosphorylation but not the same degree of activation after treatment with dmPectin (Figure 5A and 5B). These results suggest that loss of RFO1 triggers increased basal MAPK phosphorylation and prevents upregulation of phosphorylation in response to dmPectin.
Activation of MAPK signaling pathways is involved in the regulation of gene expression upon abiotic and biotic stress and in response to pectin oligomers (Aziz et al., 2004; Nakagami et al., 2005; Ren et al., 2008; Fan et al., 2017; Devendrakumar et al., 2018). To further characterize the role of the RFO1-mediated signaling cascade in Arabidopsis response to dmPectin, we quantified the expression of genes involved in pectin sensing and defense against Fo. These include the dmPectin and OG receptor WAK2, the defense-related transcriptional regulators WRKY45 and WRKY53, and the phytohormone regulatory/biosynthesis genes JASMONATE-ZIM-DOMAIN 10 (JAZ10), DWARF 4 (DWF4), and PHYTOALEXIN DEFICIENT 4 (PAD4) (Choe et al., 1998; Zhou et al., 1998; Kohorn et al., 2009; Wolf et al., 2014; Lyons et al., 2015; Menna et al., 2021). Our data show an opposite response of WT roots to dmPectin and mPectin regarding differential expression of certain genes. The receptor WAK2 and the Fo-induced defense regulator WRKY45 were downregulated in response to mPectin, while thy showed an upregulation tendency in response to dmPectin. On the contrary, the expression of the phytohormone-related genes DWF4 (brassinosteroids) and PAD4 (salicylic acid) increased under mPectin treatment and decreased in dmPectin-treated roots (Figure 5B). The jasmonic acid (JA) response gene JAZ10 was downregulated in WT plants under both treatments, while WRKY53 expression was not altered upon either of the Pectin treatments (Figure 5B and Supplemental Figure 8B). Regarding the effects of rfo1-1 mutation in this process, we detected a constitutive lower expression level of WAK2 and JAZ10 and a higher expression level of PAD4 (p = 0.067) in the mutant compared with WT mock samples (Figure 5B). A similar relative expression pattern was observed for WAK2 and PAD4 in WT mPectin-treated roots (Figure 5B). In addition, no significant transcriptional changes were detected in rfo1-1 roots exposed to mPectin. These results suggest that loss of RFO1 function triggers a constitutive transcriptional changes similar to those driven by exogenous mPectin, which are not further modified when rfo1-1 is exposed to exogenous mPectin. On the other hand, significant upregulation in the expression of most tested genes was observed in rfo1-1 roots exposed to dmPectin (Figure 5B and Supplemental Figure 8B), indicating that RFO1 participates in the dmPectin-driven response in Arabidopsis roots. The expression of was not altered by mPectin or dmPectin treatments (Figure 5B).
Taken together, our data indicate that RFO1 might regulate the expression of specific genes in response to changes of pectin methylation through the regulation of MAPK3 and MAPK6 phosphorylation status.
RFO1 lifetime at the PM is altered by dmPectin fluxes
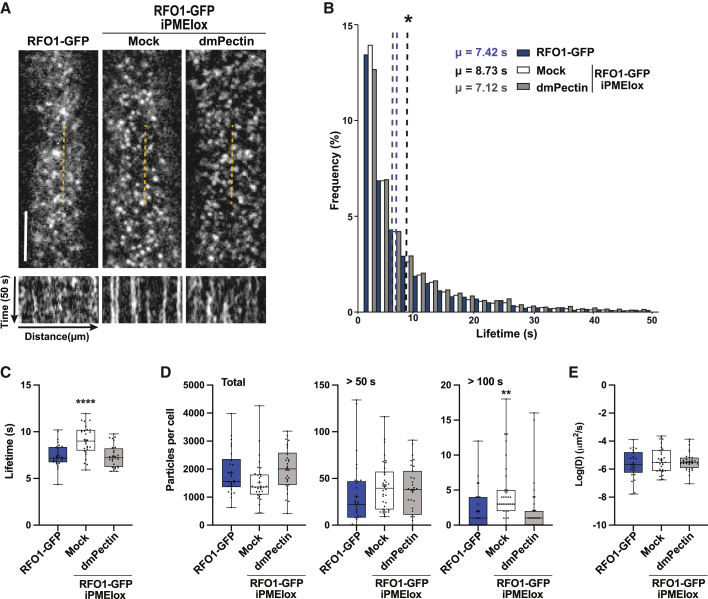
Signal perception can modify the nanoscale organization of receptors at the PM, influencing the corresponding downstream signaling pathways (Jaillais and Ott, 2020; Pan et al., 2020; Gronnier et al., 2022). Thus, we tested whether the RFO1 dwell time and dynamics at the PM are altered by sensing dmPectin (Supplemental Video 1). To do this, a previously characterized estradiol-inducible construct overexpressing PMEI5 (iPMEIox; Wolf et al., 2012) was introduced into the RFO1-GFP plant background (rfo1-1 RFO1-GFP iPMEIox). The presence of RFO1 in the PM was studied using time-lapse videos of RFO1-GFP over 3 min (Supplemental Video 2), from which kymographs were obtained to quantify the dwell time of the corresponding foci. These kymograph analyses revealed an increase in the dwell time of the RFO1 particles at the PM when PMEI5 was overexpressed (7.42 ± 0.24 s and 8.73 ± 0.49 s in control and iPMEIox background, respectively). This effect was reversed by treatment with dmPectin (7.12 ± 0.34 s) (Figure 6A and 6B). Single-particle tracking confirmed the kymograph observations (Figure 6C) and showed that, although the number of RFO1-GFP particles at the PM did not differ between conditions, PMEI5 overexpression increased the number of middle- (>50 s) and long-lived (>100 s) RFO1 particles (Figure 6D). Co-treatment with dmPectin also reversed the effect of iPMEIox (Figure 6B–6D), suggesting that an increase in the mPectin/dmPectin ratio increases the dwell time of RFO1-GFP at the PM. On the other hand, the instantaneous diffusion coefficients of RFO1-GFP particles were not significantly different between genotypes or treatments (Figure 6E). These data indicate that in vivo RFO1-GFP particle dwell time but not lateral diffusion at the PM is dependent on the pectin methylation status.
Figure 6.
RFO1-GFP particle dwell time at the PM is altered by the perturbation of cell wall pectin methylation.
(A) Representative spinning-disc confocal images (top) of a 5-day-old root elongating epidermal cell and kymographs (bottom) along the length of the dashed yellow lines from the corresponding video (n = 50 frames, 1 frame/s) of RFO1-GFP (rfo1-1 pRFO1RFO1-GFP) and RFO1-GFP iPMEIox (rfo1-1 pRFO1RFO1-GFP iPMEIox) seedlings treated with 12 μM estradiol for 2 h. RFO1-GFP iPMEIox seedlings were co-treated with Mock (−) or 10 μg/ml of dmPectin (+). Unless specified, all data derived from 180-s videos taken at 1 frame/s. Scale bar, 5 μm.
(B) Dwell time frequencies of RFO1-GFP particles at the PM as depicted in (A). Means (μ) are shown by dashed lines. For each condition, data represent N > 12 000 particles from a minimum of three cells per root and three roots.
(C) Average RFO1-GFP particle dwell times at the PM.
(D) Quantification of the total number of RFO1-GFP particles at the PM (left), and particles with dwell times >50 s (middle) and >100 s (right).
(E) Average instantaneous diffusion coefficients of RFO1-GFP particles at the PM. (C–E) Data represent the mean ± SE of N > 25 cells per condition from three independent replicates using three cells per root, three roots per condition, and tracking ≥1072 particles/cell, obtained from videos as described in (A). One-way ANOVA with Dunnett’s multiple-comparison test, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
The video depicts the dynamic behavior of RFO1-GFP expressed in Arabidopsis rfo1-1 root epidermal cells acquired using spinning-disc confocal microscopy. The pectin methylation level in the cell wall was modulated by introducing the transgene iPMEIox, which was activated using estradiol (mock; middle panel) to reduce the dmPectin levels. This effect was compensated for by adding dmPectin (right panel). The control line is shown in the left panel. Scale bar, 5 μm. This video corresponds to Figure 6.
RFO1 is necessary to regulate the PMEIox-induced BR signaling response
Our data indicate that RFO1 is required for plant response to PME-induced pectin perturbation (Figure 2), similar to the previously described PM-localized receptors RLP44 and BRI1 (Wolf et al., 2014). Thus, to test the potential role of RFO1 in BR signaling to maintain cell wall integrity, we investigated the expression of BR-signaling-related genes such as BR6ox2, CPD, CYP90D1, DWF4, and PME3 (Wolf et al., 2012) in WT, PMEIox, and rfo1-1 PMEIox seedlings. The overexpression of PMEI5 significantly reduced the expression of BR6ox2 and DWF4 and partially attenuated the expression of CPD, CYP90D1, and PMEI3 (Supplemental Figure 9A), confirming previous results (Wolf et al., 2014). The transcriptional levels of these genes were partially restored to WT levels in the rfo1-1 PMEIox background (Supplemental Figure 9A). These data suggest that RFO1, like RLP44 and BRI1, participates in regulating BR signaling in response to inhibition of pectin de-methylesterification in the cell wall. Additionally, we explored the role of some main BR signaling players involved in plant defense against F. oxysporum5176 using the hypomorphic BRI1 mutant bri1-301 (Xu et al., 2008) and the bak1-5 mutant (Schwessinger et al., 2011). Plate infection assays demonstrated that bri1-301 had increased resistance and bak1-5 was more susceptible to F. oxysporum5176 vascular colonization (Supplemental Figure 9B), suggesting a positive role of BAK1 in defense against this fungus opposite to that of the activated BRI1 receptor.
Discussion
The capacity of organisms to integrate external and internal cues for proper development in a constantly changing environment is essential for their survival, yet is far from being completely understood. A critical element for this phenotypic plasticity is the ability to respond to the changes in cell wall properties caused by those cues. Effective sensing of plant cell wall integrity becomes even more important during root colonization by vascular pathogens such as Fo. These microbes extensively remodel plant cell walls, and the host needs to balance defense and growth (Lionetti and Métraux, 2014; Vaahtera et al., 2019; Dora et al., 2022). Previous studies demonstrated that RLK, WAK, and WAKL receptors are involved in detecting changes in pectin in the cell wall during plant growth and defense responses, but the molecular mechanisms behind these functions have remained unresolved. Our data demonstrate that the PM-localized WAKL, RFO1, binds dmPectin in vitro and reacts in vivo to increase dmPectin/mPectin ratios in the cell wall. This capacity to sense the pectin methylation status is required for both plant development and resistance to root vascular fungi (Figure 6).
Since its identification as an essential element of Arabidopsis resistance to F. oxysporum and V. dahliae in 2005 (Verica and He, 2002; Diener and Ausubel, 2005), RFO1 has not been further characterized. Here we expand the knowledge of its molecular function at a high spatial and temporal resolution. Our work sheds light on RFO1’s role in root response to vascular fungi before the intruder reaches the xylem (Figure 1A–1C). F. oxysporum infection causes root growth inhibition similar to that observed after other microbe infections and cellular damage: microbe- and damage-associated molecular patterns (MAMPs and DAMPs), respectively (Gómez-Gómez et al., 1999; Poncini et al., 2017; Kesten et al., 2019; Huerta et al., 2020). The absence of RFO1 caused a delay in the root growth inhibition induced by Fo, as reported for MAMP-receptor mutants exposed to their corresponding MAMPs. The PM localization of RFO1 (Figure 1E–1G and Supplemental Figure 1C–1F) hints that its function is important to detect the pathogen after the first interaction and slow down its growth toward the vasculature.
Inhibition of PME activity, by PMEI overexpression or exposure to EGCG and the consequent alteration of the mPectin/dmPectin ratio, also impairs root development (Wolf et al., 2012). These phenotypes were almost completely restored to WT ones by the rfo1-1 mutation (Figure 2), indicating that RFO1 is crucial to respond to changes in pectin methylation in the cell wall. Root vascular fungi also alter the cell wall pectin methylation state during plant colonization (Fan et al., 2017). We have confirmed that F. oxysporum5176 infection reduces the pectin methylesterification of Arabidopsis roots (Figure 4A), generating putative signals perceived by RFO1. The Arabidopsis ecotype analysis presented here and based on previous studies of natural variation in plant defense to F. oxysporum (Diener and Ausubel, 2005) indicates that RFO1 might be involved in plant defense to F. oxysporum by recognizing Fo-induced alterations to the pectin methylation status through its ECD (Figure 3A–3C and Supplemental Figure 2). The same RFO1ECD is involved in plant response to genetic and chemical PME inhibition (Figure 3A–3C and Supplemental Figure 2). In vitro experiments suggest that RFO1ECD might interact with dmPectin with low affinity. Our data showed that F. oxysporum infection altered the pectin methylation status in the soluble fractions of root cell walls (Figure 3, Supplemental Figure 6, and Supplemental Figure 4). Importantly, since the concentration of pectins in the cell wall is high and the dmPectin/mPectin ratio changes dynamically, a high affinity sensor would not be able to perceive significant changes because of constant saturation, making a lower affinity sensor more suitable to perceive these changes. In planta experiments where we altered the pectin methylation state in the plant cell wall (Figures 2, 3, 5, and 6) support the notion that RFO1 has the capacity to react to changes in pectin demethylation status and, therefore, has a role as dmPectin/mPectin sensor at the cell wall.
Further research will be necessary to better understand the molecular basis of the role of RFO1ECD in recognizing areas enriched in dmPectin in the cell wall, for example, by determining the complex structure, investigating its connection with local dmPectin/mPectin changes at the cell wall at the highest spatial resolution (Francoz et al., 2019; Haas et al., 2020), and evaluating the function of its allelic diversity within Arabidopsis ecotypes. Furthermore, the ability of RFO1 to provide broad resistance against F. oxysporum and Verticillium spp. in Arabidopsis, reinforces the idea of RFO1 as a sensor of pectin integrity rather than a recognizer of specific fungal factors (Li et al., 2020).
The immunobinding and TSA assays suggest the low-affinity recognition of dmPectin by RFO1 (Figure 2 and Supplemental Figure 6). Similar results have been reported using analogous immunoassays when studying another member of the WAK family, WAK1 (Feng et al., 2018). This and our data suggest that members of the WAK and WAK-like family have the ability to interact with pectin in vitro and share the preference for dmPectin over mPectin as a ligand (Figure 3E, 3F, and 6A; Decreux and Messiaen, 2005; Kohorn et al., 2009). RFO1-mediated perception of dmPectin promotes increased MAPK3 and MAPK6 phosphorylation and transcriptional changes (Figure 4). Unlike RFO1, other members of the WAK family, such as WAK2, mediate the perception of dmPectin through activation of MAPK3 but not MAPK6 (Kohorn et al., 2009), suggesting an expanded role of RFO1 in pectin sensing. In addition, RFO1 modulates the expression of WAK2 in response to altered pectin methylation status, suggesting a role for RFO1 upstream of this receptor in pectin-integrity sensing. Moreover, the presence of RFO1 in the PM is dependent on the amount of dmPectin in the cell wall. The short-term and local reduction of dmPectin in PMEIox-induced seedlings increased the dwell time of RFO1 at the PM (Figure 6), potentially altering RFO1’s ability to interact with signaling partners to regulate downstream signaling cascades (Figure 6B and 6C). A similar mechanism has been reported for other PM receptors such as BRI1 and PEPR1 (Gadeyne et al., 2014; Ortiz-Morea et al., 2016; Zhou et al., 2018; Ma et al., 2020). Considering the role of WAK2 in plant responses to alterations in pectin methylation, it could work together with RFO1 in coordinating plant responses to alterations in pectin methylation at the cell wall (Kohorn et al., 2009; Wolf et al., 2014), a scenario that needs to be addressed with further investigations.
With regard to plant defense, the absence of RFO1 led to the misregulation of the JA transcriptional suppressor, JAZ10, and the salicylic acid (SA) activation gene, PAD4, probably contributing to the susceptibility phenotypes observed in rfo1-1. (Figure 5B). Both pathways have been shown to be regulated through MAPK activation (Brodersen et al., 2006; Hettenhausen et al., 2015). Furthermore, the JA signaling pathway has recently been shown to participate in Arabidopsis root defense against F. oxysporum vascular penetration (Menna et al., 2021). Therefore, the activation of the JA and SA signaling pathways during RFO1-mediated dmPectin sensing could serve to boost defense responses against root vascular fungi, as shown for other WAKs in cotton (Yang et al., 2021). Importantly, although rfo1-1 responses to PME inhibition mimics those reported for bri1-301 (Figure 2 and Supplemental Figure 9A; Wolf et al., 2012; 2014), this is not the case in the defense against Fo, where bri1-301 is more resistant (Supplemental Figure 9B). Considering the increased susceptibility to the fungus observed in bak1-5 (Supplemental Figure 9B), we hypothesize that RFO1 participates in a BAK1-dependent Fo-defense signaling cascade at least partially different from that activated via BAK1-BRI1 in response to PME inhibition (Figure 6C). Importantly, RFO1 expression in WT plants was not affected by external dmPectin or mPectin (Figure 5B), indicating that its upregulation during F. oxysporum5176 infection (Figure 1D) is a response to modifications of pectin methylation more complex than those mimicked with commercial products. Indeed, in the Arabidopsis–Botrytis cinerea and banana–F. oxysporum pathosystems, PMEIs were described as modulators of PME activity and pectin methylesterification levels at later stages of infection to prevent microbial pectinase degradation (Fan et al., 2017; Lionetti et al., 2017). Therefore, RFO1 may also perceive a putative PMEI-dependent pectin methylation ratio maintenance when F. oxysporum has highly colonized the plant. Another, not exclusive, possibility is that, as suggested for other WAKs, RFO1 might contribute to plant response to an additional signal together with dmPectin, such as OGs or MAMPs, to distinguish biotic stress from cell expansion cues (Kohorn et al., 2014; Wang et al., 2020; Qi et al., 2021). These hypotheses need to be further explored.
Taken together, our work shows a reduction in pectin methylation in the cell wall of roots upon F. oxysporum infection. Our data indicate that RFO1, a previously uncharacterized WAKL, participates in reading those pectin alterations, partially explaining its role in plant defense. We also show that pectin methylation changes altered RFO1’s dwell time at the PM, showing for the first time that a cell wall sensor dynamically responds to cell wall-derived signals. RFO1-dmPectin interaction initiates the activation of a MAPK-dependent signaling cascade and the upregulation of defense players and receptors involved in pectin perception (Figure 7). Additionally, RFO1 is a novel positive regulator of BR signaling required for normal growth upon alterations of PME activity, and it also seems to be required for a BAK1-dependent defense against F. oxysporum infection. In conclusion, RFO1’s dual functionality in development and defense seems to contribute to ensuring cell wall homeostasis during plant growth and response to biotic stress.
Figure 7.
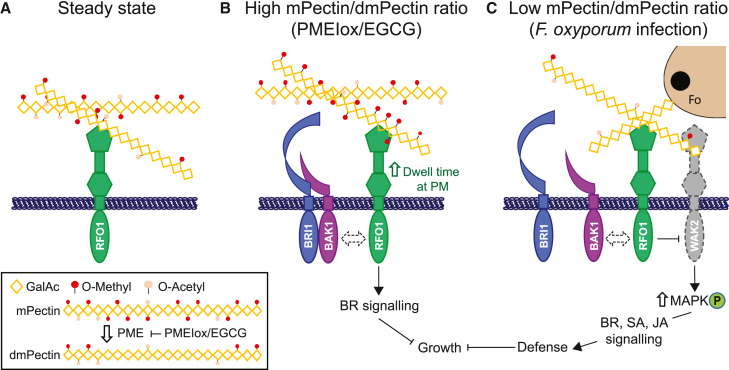
model illustrating how RFO1 functions at the plasma membrane.
(A) Pectin secreted to the cell wall is heavily methyl- and acetyl-esterified. RFO1 acts as a sensor of the methyl-esterification level of pectins.
(B) When the ratio of mPectin/dmPectin increases at the cell wall, as observed in plants overexpressing PMEI (PMEIox) or during EGCG treatment, RFO1 increases its dwell time at the PM and triggers BR signaling to cause reduced plant growth. RFO1 may be part of the BRI1-BAK1 complex in this signaling process (dashed double arrow).
(C) When the ratio of mPectin/dmPectin is low, as observed during F. oxysporum (Fo) infection or exogenous dmPectin treatment, RFO1 might release signaling partner(s), such as WAK2 (dashed), to increase MAPK activation and downstream expression of defense-related genes. In the absence of RFO1, those potential signaling partners (i.e., WAK2) are constitutively free of RFO1-mediated repression, leading to increased basal levels of MAPK phosphorylation. RFO1 sensing of mPectin/dmPectin ratio is required to upregulate the MAPK signaling cascade and thereby activate defense responses. BAK1 liberation from BRI1 after F. oxysporum infection is required for plant defense against this pathogen and might enable direct or indirect association of BAK1 with RFO1 (dashed double arrow).
Methods
Plant material and growth
Arabidopsis thaliana plants used in these experiments, which were all the Col-0 ecotype unless otherwise indicated (Ty-0), were grown in a 16-h light (24°C)/8-h dark (21°C) cycle. In vitro experiments were carried out in 1/2 MS medium plates (pH 5.7, not buffered; Duchefa #M0222.0025) supplemented with 1% (w/v) sucrose (when indicated) and 0.9% bacteriological grade agar (Difco #214530). The rfo1-1 (SALK_077975; Diener and Ausubel, 2005), PMEIox (Wolf et al., 2012), bri1-301 (Xu et al., 2008), and bak1-5 (Schwessinger et al., 2011) lines were previously described. rfo1-1 pRFO1::RFO1-GFP, rfo1-1 pRFO1::RFO1Ty−0-GFP, rfo1-1 pRFO1::RFO1chim-GFP, and pRFO1:RFP-N7 plants (see section “generation of constructs”') were obtained by transformation according to standard procedures (Hellens et al., 2000). Transgenic lines were selected on soil based on resistance to 0.2% BASTA. Resulting heterozygous seedlings were screened for GFP or RFP fluorescence before continuing to select homozygous lines through BASTA selection and genotyping.
Fungal strains, culture conditions, and plate infection assays
F. oxysporum f.sp. conglutinans5176 pSIX1::GFP (Kesten et al., 2019) was used in this study. Fungal strains were cultured, microconidia were obtained, and plate infection assays were performed as previously described (Huerta et al., 2020).
Generation of constructs
The pRFO1::RFO1-GFP construct was generated and assembled using Gateway cloning. To obtain the pRFO1-GFP destination vector, an 833-bp promoter region upstream of the RFO1 (AT1G79670) open reading frame (ORF) was amplified from Col-0 gDNA using primers 1 and 2 (Supplemental Table 2) and ligated (NEBuilder #E5520S) into the Gateway pUBC-GFP plasmid (Grefen et al., 2010) after removal of the original UBQ promoter by digestion (SacI + XhoI). The RFO1 ORF was amplified from Col-0 gDNA using primers 3 and 4 (Supplemental Table 2) and cloned into the pENTR/TOPO-D/SD Gateway vector according to the standard TOPO PCR cloning protocol. A Gateway LR reaction was performed using the resulting pRFO1-GFP destination and pENTR-RFO1 plasmids and transformed into DH5ɑ Escherichia coli cells. pRFO1::RFO1-GFP E. coli transformants were selected through spectinomycin selection.
pRFO1::RFO1Ty−0-GFP and pRFO1::RFO1chim-GFP constructs were generated using the same method described above. The RFO1Ty−0 ORF was amplified from the Arabidopsis Ty-0 ecotype cDNA using primers 5 and 4 (Supplemental Table 2). The RFO1chim chimera was generated by amplifying the Col-0 RFO1 ectodomain and transmembrane domains using primers 3 and 7, and the Ty-0 RFO1 cytosolic domain using primers 6 and 4. The resulting two fragments were then ligated through an elongation PCR employing primers 3 and 4. A Gateway LB reaction was performed using the pRFO1-GFP destination vector, and the resulting pENTR-RFO1Ty-0 and pENTR-RFO1chim plasmids were transformed into DH5ɑ E. coli cells. Transformants were selected through spectinomycin selection.
A new vector, which we named pNew, was generated by digesting pGreenII with NotI and EcoRI. A new insertion was designed and ordered as a gene fragment containing the A. thaliana ubiquitin-10 promoter (Grefen et al., 2010) followed by a multiple cloning site and the A. thaliana HSP terminator (Nagaya et al., 2010) for convenient insertion of genes under the control of the ubiquitin-10 or native promoter by cutting the vector with either BamHI and XbaI or EcoRI and XbaI. The pRFO1::RFP-N7 construct was generated using primers 8–13 (Supplemental Table 2) by amplifying the RFO1 promoter as described above, the RFP from pUBN-RFP-Dest (Grefen et al., 2010), and the N7 nuclear targeting sequence from AT4G19150. The fragments were cloned into predigested pNew (EcoR1 and Xba1) and ligated in a one-step ligation (NEBuilder #E5520S). The resulting construct was then transformed into DH5ɑ E. coli cells and selected for resistance to kanamycin.
All the above constructs were transformed into electrocompetent GV31 Agrobacterium tumefaciens (together with pSOUP for pRFO1::RFP-N7) and selected using the appropriate antibiotic plus 25 μg/ml gentamicin and rifampicin.
FTIR spectroscopy
Alcohol insoluble residues (AIRs) of the various samples were analyzed with attenuated total reflection (ATR) mode on a Shimadzu IRAffinity-1S. The IR spectrum was obtained using the software LabSolutions IR (MIRacle10 (Ge)) in transmission mode after performing 40 scans with 4 cm−1 resolution (700 and 4000 cm−1 range). The degree of esterification (DM) is defined as the ratio (%) of the absorbance at 1740 cm−1 (esterified carboxyl groups of the pectin molecules) over the sum of the absorbance of the bands at 1740 and 1630 cm−1 (vibration of carboxyl groups of (1,4)-α-d-GalUA) using the equation DM=(A1740/(A1740 + A1630)) × 100 as previously described (Leroux et al., 2015). Commercial pectins with determined DM were used as controls. For each biological replicate, four spectra were acquired.
Recombinant protein production and purification
Codon-optimized synthetic genes for expression in Spodoptera frugiperda (Invitrogen GeneArt), coding for the ectodomain of RFO1 (residues 28–347; AT1G79670) with a SNAP-tag C-terminal fusion, were cloned into a modified pFastBac (Geneva Biotech) vector providing a tobacco etch virus (TEV) protease cleavable C-terminal StrepII-9xHis tag. For protein expression, Trichoplusia ni Tnao38 cells were infected with the RFO1-SNAP virus and incubated for 1 day at 28°C and an extra 2 days at 22°C at 110 rpm. The secreted RFO1-SNAP (under the 30K signal peptide) protein was purified from the supernatant by sequential Ni2+ (HisTrap excel; GE Healthcare; equilibrated in 25 mM KPi pH 7.8, 500 mM NaCl) and StrepII (Strep-Tactin Superflow high capacity [IBA Lifesciences] equilibrated in 25 mM Tris pH 8.0, 250 mM NaCl, 1 mM EDTA) affinity chromatography. The SNAP-tag was cloned into a modified pETM11 vector, providing a C-terminal TEV site and StrepII-9xHis tag. The SNAP protein was purified from E. coli cells following the same procedure described above. Both proteins were incubated with TEV protease to remove the tags. Proteins were further purified by size exclusion chromatography on a Superdex 200 increase 10/300 GL column (GE Healthcare), equilibrated in 20 mM HEPES pH 7.5, 150 mM NaCl (Supplemental Figure 4A and 4B). For biochemical experiments and quality control, proteins were concentrated using Amicon Ultra concentrators (Millipore, molecular weight cutoff 10 000 and 30 000 Da). Also, proteins were analyzed for purity and structural integrity by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (Supplemental Figure 4A and 4B).
Generation of dmPectin and mPectin, plant cell wall pectin fractions, and plant cell wall GP extracts
To produce dmPectin and mPectin samples, 1 mg/ml apple-derived pectin (Sigma #93854; 50%–75% esterification) was treated as previously reported (Liners et al., 1992; Kohorn et al., 2009): the pectin was dissolved in 50 mM NaOH and distilled water for production of dmPectin and mPectin, respectively, and placed on ice for 60 min. Then, samples were dialyzed overnight at 4°C against distilled water in Spectra/Por 3 pre-wet dialysis tubing (Spectrum #132720). Afterward, they were transferred to a 50-ml conical tube, flash-frozen, and lyophilized overnight. The lyophilized dmPectin and mPectin samples were then resuspended in 1 ml of 1.5 mM EDTA solution in water and stored at −20°C until use.
To produce cell wall fractions, cell wall AIR was first obtained as previously described (Menna et al., 2020) from mock- and F. oxysporum5176-infected Arabidopsis roots at 4 dpt. Fractionation of 10 mg of AIR proceeded as previously described (Hotchkiss and Hicks, 1990; Mort et al., 1991) with the following modifications. Fraction I was obtained by treating the AIR with 1 ml of a 50 mM imidazole and 20 mM NaBH4 solution at pH 7 at 4°C with rotation for 8 h. After centrifugation, the supernatant was transferred to a fresh tube, the pellet was treated once more as described above, and the supernatants were combined and used as fraction I. The remaining pellet was used to obtain fraction II as done for fraction I, but using 50 mM Na2CO3 and 20 mM NaBH4 solution instead of imidazole. The resulting pellet after two rounds of fraction II extraction was incubated with 1 M KOH and 20 mM NaBH4 to obtain fraction III, following the same steps described for the previous fractions. The three fractions were neutralized using glacial acetic acid before transferring them to Spectra/Por 3 pre-wet dialysis tubing for overnight dialysis at 4°C against distilled water. After dialysis, samples were transferred to a pre-weighed 50-ml conical tube, flash-frozen, and lyophilized overnight. The lyophilized cell wall fractions were then weighed and resuspended with distilled water to a concentration of 1 mg/ml and stored at −20°C until further use.
For cell wall GP extracts, leaves of 6-week-old WT plants were harvested and processed as previously described (Menna et al., 2021). The final lyophilized cell wall GPs were weighed, flash-frozen in liquid nitrogen, and stored at −80°C until use.
Dot immunobinding assays
For analysis of RFO1-SNAP and SNAP recombinant protein interaction with pectins and plant cell wall-derived samples, 15 μg of plant cell wall material (fractions or GP extracts) per sample were immobilized in triplicate on nitrocellulose membranes using a Hybri-dot manifold (BRL #1050MM) under light vacuum and air-dried at room temperature for 30 min. The dot blot membranes were blocked with 5% bovine serum albumin (Sigma #A9647) for 1 h, quickly washed in a sodium acetate buffer (NaAc 20 mM; pH 5.4), and incubated overnight at 4°C with fresh 300 μM solutions of RFO1-SNAP or SNAP in NaAc buffer. The blots were washed three times with NaAc buffer and incubated with ɑSNAP antibody (NEB P9310S) at a 1:5000 dilution in TBST (25 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.05% Tween 20) for 1 h. After washing, the membranes were probed with rabbit–horseradish peroxidase (HRP) antibody (Azure #AC2114) at a 1:10 000 dilution in TBST for 1 h, washed in TBST for 4 h, and developed with 1 ml of the HRP substrate. Chemiluminescence was detected for 500 s using a Bio-Rad ChemiDoc Touch imaging system. Dot blot images were analyzed using Fiji according to the protocol Dot Blot Analysis (nih.gov) with the following modifications. Background subtraction was performed using a 20-pixel-radius rolling ball. Data were derived from Arabidopsis cell wall components. For the data quantification, we obtained the percentage (%) that each fraction represents of the total gray value of the blot. For data derived from pure pectin samples, the average gray values were shown unnormalized.
For blots using JIM5 and JIM7 antibodies, 15 μg of carbohydrate material (mPectin, dmPectin, and the different root or F. oxysporum5176 cell wall fractions) per sample were immobilized in triplicate on nitrocellulose membranes, blocked, and washed as indicated above. The dot blot membranes were incubated for 1 h with JIM5 (Megazyme AB-JIM5) or JIM7 (Megazyme AB-JIM7) antibodies at a 1:100 dilution in TBST, washed three times in TBST, and probed with rat-HRP antibody (Azure AC2118) at a 1:10 000 dilution in TBST for 1 h. The blots were washed in TBST for 1 h before being exposed to the HRP substrate and analyzed following the aforementioned protocol.
TSA
TSAs were performed as described in Huynh and Partch (2015). Samples were prepared in a final volume of 30 μl with the following composition. Protein was diluted to a concentration of 5 μM in 20 mM MES, 50 mM NaCl, pH 6. The concentration of de-esterified apple-derived pectin (Sigma 93854) is indicated in the figure. Pectin was diluted first in 50 mM NaOH and then dialyzed in water. SYPRO Orange (Molecular Probes, CA) fluorescent probe was used at a 5× concentration. Samples were loaded in MicroAmp Fast 96-Well Reaction Plates (0.1ml, Applied Biosystems; Thermo Scientific) and sealed with MicroAmp Optical Adhesive Film (Applied Biosystems; Thermo Scientific). Plates were then inserted in a QuantStudio 3 real-time PCR machine (Applied Biosystems; Thermo Scientific) to run the assay. The temperature was increased at a rate of 0.5°C per minute between 30°C and 75°C. The fluorescence at 530 nm was recorded every minute. The negative of the derivative of the fluorescence (F) as a function of the temperature (T) (−dF/dT) was plotted, and the minimum was used to determine the Tm.
EGCG root growth inhibition assays
Two 5-day-old seedlings grown on 1/2 MS agar +1% sucrose plates were scanned and transferred to 12-well plates containing 2 ml of ½ MS + 1% sucrose with or without 6.25 μM EGCG (Sigma #E4143) and grown for 48 h under continuous light at 21°C with shaking at 70 rpm. Root length at the start of the experiment and after 48 h of treatment was measured using the SNT plugin of the Fiji application. Root growth inhibition of EGCG-treated seedlings was calculated by dividing the average root length increase of every pair of roots treated with EGCG by the average root length increase of every pair of mock-treated roots of the corresponding genotype.
Assessment of MAPK activation in response to dmPectin
Three or four 8-day-old seedlings were transferred from agar plates to six-well plates containing 2 ml of 1/2 MS, left to equilibrate at room temperature for 1 h, and treated for 30 min with 20 μl of 1.5 mM EDTA with or without 1 mg/ml dmPectin, to get a final concentration of 10 μg/ml dmPectin. Three or four roots per genotype and condition were ground with Laemmli sample buffer plus silica sand using a mortar and pestle. The resulting extract was transferred to a centrifuge tube and boiled at 80°C for 5 min, and centrifuged; 20 μl of each supernatant was subjected to SDS-PAGE on a 10% polyacrylamide gel. Proteins were transferred to nitrocellulose membranes using the Bio-Rad Trans-Blot Turbo transfer system and protocol with a10-min transfer at 2.5 V, blocked for 1 h using 5% milk protein in TBST, quickly washed with TBST, and incubated with P-p44/42 MAPK antibody (Cell Signal #9101) at a 1:500 dilution in TBST for 1 h to determine activated MAPK levels. Then, the blots were washed with TBST, incubated with rabbit-HRP antibody (Azure #AC2114) at a 1:10 000 dilution in TBST for 1 h, washed in TBST for 1 h, and treated with 1 ml of HRP substrate. Chemiluminescence was detected for 60 s using a Bio-Rad ChemiDoc Touch imaging system. Dot blot images were analyzed in Fiji using the Gel Tools plugin after background subtraction using a 20-pixel-radius rolling ball. To quantify the total MAPK protein levels in these samples, blots were then stripped for 30 min using 0.2 M glycine pH 2.2, blocked with 5% milk protein, incubated with p44/42 MAPK antibody (Cell Signal #9102), and subjected to the same washing, treatment, and imaging as above. The relative MAPK activation of each sample was normalized to the total MAPK intensity values per condition, genotype, and blot, and then compared with WT.
Gene expression analysis by qRT-PCR
For gene expression analysis in response to dmPectin and mPectin, 100 mg of root material per condition and genotype grown and treated as described above (see section “assessment of MAPK activation in response to dmPectin”) was collected. For RFO1 expression quantification during F. oxysporum5176 infection, roots of mock- and F. oxysporum5176-infected WT seedlings were harvested at 1–7 dpt. For expression analysis of PME3- and BR-related genes, 100 mg of root tissue was harvested. In all cases, total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen #74904) and treated with RNase-Free DNase Set (Qiagen #79254).
The first-strand cDNAs synthesized using the Maxima H Master Mix (Thermo #M1681) were amplified by RT-qPCR using SYBR green master mix (AB #4309155) in a 10-μl reaction using the primers indicated in Supplemental Table 2: RFO1 (primers 37 and 38), WAK2 (primers 39 and 40; Wolf et al., 2014), WRKY45 (primers 41 and 42; Souza et al., 2017), WRKY53 (primers 43 and –44; Masachis et al., 2016), JAZ10 (primers 31 and 32; Liu et al., 2016), DWF4 (primers 27 and 28; Wolf et al., 2014), PAD4 (primers 35 and 36; Chen et al., 2015), BR6ox2 (primers 21 and 22; Shimada et al., 2003), CPD (primers 23 and 24; Shimada et al., 2003), CYP90D1 (primers 25 and 26; Shimada et al., 2003), PME3 (primers 35 and 36; Bethke et al., 2014), and the reference gene GAPDH (primers 29 and 30; Czechowski et al., 2005), using a LightCycler 480 II (Roche). No-template controls and melting curves were examined to ensure no contamination and primer–dimer formation were present. The 2ΔCT method was used to quantify the relative expression of each gene (Schmittgen and Livak, 2008).
Spinning disk live-cell imaging and single-particle tracking analysis
Roots of 5-day-old seedlings were covered with a 1% agarose cushion as described previously (Gutierrez et al., 2009). rfo1-1 RFO1-GFP and rfo1-1 RFO1-GFP iPMEIox seedlings were transferred to six-well plates containing 2 ml of 1/2 MS with 12 μM β-estradiol (Sigma #E8875; in DMSO) and 20 μl of 1.5 mM EDTA with or without 1 mg/ml dmPectin dissolved in it, and incubated for 2 h before imaging. RFO1-GFP particles were imaged with a CSU-W1 Yokogawa spinning-disk head fitted to a Nikon Eclipse Ti-E-inverted microscope with a CFI PlanApo × 100 NA 1.40 oil immersion objective and two iXon Ultra EM-CCD cameras (Andor, GB); a ×1.2 lens between the spinning disk and camera was used. For this system, GFP was imaged using a 488-nm solid-state diode laser and a 525/50-nm emission filter; RFP was detected with a 561-nm solid-state diode laser and a 630/75-nm emission filter. Time-lapse images were processed and analyzed with Fiji (Schindelin et al., 2012). Before particle tracking, drifts were corrected by using the plugin StackReg (Thevenaz et al., 1998). Backgrounds were subtracted by the Subtract Background tool (rolling ball radius, 40 pixels). The tool Walking Average was additionally applied, averaging three frames. Single-particle tracking was performed using the Fiji application, Trackmate (Tinevez et al., 2017), using the LAP tracker with a particle radius of 0.4 μm and an initial threshold of 15. Auto-filtering based on signal quality was performed for each image. Cells were imaged for 180 s at 1 frame/s, 800-ms exposure, and a gain of 250. The generated spot and track data from Trackmate were used for dwell time, MSD, and log diffusion coefficient (Log(D)) calculations. All data analyses were performed using R (version 4.0.3) with R Studio (version 1.1.463), and images were made with ggplot2. For dwell time analysis, the particles already present at the PM when the video started and those that were still visible when the video finished were not considered for the analysis. Only tracks starting 1 s after the start of imaging and ending 1 s before the end of the imaging were included in the dwell time analysis. Middle- (>50 s) and long-lived (>100 s) tracks were counted separately and compared with all filtered tracks from the dwell time analysis above. MSD values were derived from the spot data of all spots in a given video. MSD and Log(D) were calculated as previously described (Wagner et al., 2017), using the following formulas:
| (Equation 1) |
| (Equation 2) |
Confocal microscopy
Root epidermal cells expressing pRFO1:RFP-N7 were imaged with a Zeiss 780 confocal laser scanning microscope equipped with a 20× 0.5 NA objective. RFP was visualized using 561-nm laser excitation and 592–754-nm spectral detection. z stack images were taken, and image tiling and stitching was performed with the ZEN software (Zeiss). The images were further processed and analyzed with Fiji.
All graphs were plotted using Prism 9 (GraphPad Software).
Funding
The work described in this paper was supported by the Swiss National Foundation and the Heinz Imhof Foundation to C.S.R. (2-72160-16 to A.I.H., SNF 31003A_163065/1 and SNF 310030_184769 to G.S.), the Peter und Traudl Engelhorn Stiftung to C.K., and the European Research Council (ERC) grant agreement no. 716358 and the Fondation Philanthropique Famille Sandoz to J.S.
Author contributions
A.I.H. and C.S.R. conceived the study;. A.I.H., C.S.R., J.C.M., J.P., and J.S. designed experiments;. K.C. and R.S. provided essential material;. A.I.H., J.C.M., G.S.A., J.S.N., S.B., C. P.-R., and T.A. performed the experiments;. A.I.H., J.C.M., G.S.A., J.P., and J.S.N. and analyzed the data;. A.I.H., J.C.M., G.S.A., and C.S.R. wrote the manuscript with comments from all the other authors.
Acknowledgments
We would like to thank Dr. Sebastian Wolf (University of Heidelberg, DE) for donating plant material used in this work, Dr. Alexandra Menna (ETH Zurich, CH) for the generation of plant glycoprotein GP extracts, Dr. Julien Gronier (UZH, CH) for his advice regarding single-particle tracking, Dr Caroline A. Ahad Hadad (UPJV, FR) for help with FTIR analyses, and the ScopeM (ETH Zurich, CH) for their assistance and service in the microscopy of plant cells. We also are very grateful to the Plant Cell Biology laboratory at ETHZ, especially to Dr. Francisco Gámez-Arjona and Dr. Alfonso González de la Rubia, and to Dr. Sebastian Wolf, Prof. Olivier Voinnet (ETH Zurich, CH), and Prof. Cyril Zipfel (UZH, CH) for fruitful scientific discussion. No conflict of interest is declared.
Published: March 31, 2023
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Molecular Plant Online.
Supplemental information
References
- Albersheim P., Darvill A.G., O’Neill M.A., Schols H.A., Voragen A.G.J. In: Progress in Biotechnology. Visser J., Voragen A.G.J., editors. 1996. An hypothesis: the same six polysaccharides are components of the primary cell walls of all higher plants; pp. 47–55. [Google Scholar]
- Anderson C.T. We be jammin’: an update on pectin biosynthesis, trafficking and dynamics. J. Exp. Bot. 2016;67:495–502. doi: 10.1093/jxb/erv501. [DOI] [PubMed] [Google Scholar]
- Anderson C.M., Wagner T.A., Perret M., He Z.H., He D., Kohorn B.D. WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 2001;47:197–206. [PubMed] [Google Scholar]
- Aziz A., Heyraud A., Lambert B. Oligogalacturonide signal transduction, induction of defense-related responses and protection of grapevine against Botrytis cinerea. Planta. 2004;218:767–774. doi: 10.1007/s00425-003-1153-x. [DOI] [PubMed] [Google Scholar]
- Bethke G., Grundman R.E., Sreekanta S., Truman W., Katagiri F., Glazebrook J. Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol. 2014;164:1093–1107. doi: 10.1104/pp.113.227637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidhendi A.J., Geitmann A. Relating the mechanics of the primary plant cell wall to morphogenesis. J. Exp. Bot. 2016;67:449–461. doi: 10.1093/jxb/erv535. [DOI] [PubMed] [Google Scholar]
- Bishop C.D., Cooper R.M. An ultrastructural study of root invasion in three vascular wilt diseases. Physiol. Plant Pathol. 1983;22:15–IN13. [Google Scholar]
- Bravo Ruiz G., Di Pietro A., Roncero M.I.G. Combined action of the major secreted exo- and endopolygalacturonases is required for full virulence of Fusarium oxysporum. Mol. Plant Pathol. 2016;17:339–353. doi: 10.1111/mpp.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Ruiz G., Sassi A.H., Marcet-Houben M., Di Pietro A., Gargouri A., Gabaldon T., Roncero M.I.G. Regulatory mechanisms of a highly pectinolytic mutant of penicillium occitanis and functional analysis of a candidate gene in the plant pathogen Fusarium oxysporum. Front. Microbiol. 2017;8:1627. doi: 10.3389/fmicb.2017.01627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Bjørn Nielsen H., Zhu S., Newman M.-A., Shokat K.M., Rietz S., Parker J., Mundy J. Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 2006;47:532–546. doi: 10.1111/j.1365-313X.2006.02806.x. [DOI] [PubMed] [Google Scholar]
- Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA. 2010;107:9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero P.J., Knox J.P. The monoclonal antibody JIM5 indicates patterns of pectin deposition in relation to pit fields at the plasma-membrane-face of tomato pericarp cell walls. Protoplasma. 1995;188:133–137. [Google Scholar]
- Chen Q.-F., Xu L., Tan W.-J., Chen L., Qi H., Xie L.-J., Chen M.-X., Liu B.-Y., Yu L.-J., Yao N., et al. Disruption of the Arabidopsis defense regulator genes SAG101, EDS1, and PAD4 confers enhanced freezing tolerance. Mol. Plant. 2015;8:1536–1549. doi: 10.1016/j.molp.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Dilkes B.P., Fujioka S., Takatsuto S. The Plant Advance. 1998. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coculo D., Lionetti V. The plant invertase/pectin methylesterase inhibitor superfamily. Front. Plant Sci. 2022;13:863892. doi: 10.3389/fpls.2022.863892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R.M., Wood R.K.S. Regulation of synthesis of cell wall degrading enzymes by Veticillium albo-atrum and Fusarium oxysporum f. sp. lycopersici. Physiol. Plant Pathol. 1975;5:135–156. [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decreux A., Messiaen J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005;46:268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- Decreux A., Thomas A., Spies B., Brasseur R., Van Cutsem P., Messiaen J. In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry. 2006;67:1068–1079. doi: 10.1016/j.phytochem.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Del Corpo D., Fullone M.R., Miele R., Lafond M., Pontiggia D., Grisel S., Kieffer-Jaquinod S., Giardina T., Bellincampi D., Lionetti V. AtPME17 is a functional Arabidopsis thaliana pectin methylesterase regulated by its PRO region that triggers PME activity in the resistance to Botrytis cinerea. Mol. Plant Pathol. 2020;21:1620–1633. doi: 10.1111/mpp.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasi Devendrakumar K., Li X., Zhang Y. MAP kinase signalling: interplays between plant PAMP- and effector-triggered immunity. Cell. Mol. Life Sci. 2018;75:2981–2989. doi: 10.1007/s00018-018-2839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener A.C., Ausubel F.M. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics. 2005;171:305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A., Roncero M.I. Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 1998;11:91–98. doi: 10.1094/MPMI.1998.11.2.91. [DOI] [PubMed] [Google Scholar]
- Dmochowska-Boguta M., Kloc Y., Zielezinski A., Werecki P., Nadolska-Orczyk A., Karlowski W.M., Orczyk W. TaWAK6 encoding wall-associated kinase is involved in wheat resistance to leaf rust similar to adult plant resistance. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dora S., Terrett O.M., Sánchez-Rodríguez C. Plant-microbe interactions in the apoplast: communication at the plant cell wall. Plant Cell. 2022;34:1532–1550. doi: 10.1093/plcell/koac040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrands P.K., Cooper R.M. The role of pectinases in vascular wilt disease as determined by defined mutants of Verticillium albo-atrum. Physiol. Mol. Plant Pathol. 1988;32:363–371. [Google Scholar]
- Engelsdorf T., Gigli-Bisceglia N., Veerabagu M., McKenna J.F., Vaahtera L., Augstein F., Van der Does D., Zipfel C., Hamann T. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aao3070. [DOI] [PubMed] [Google Scholar]
- Fan H., Dong H., Xu C., Liu J., Hu B., Ye J., Mai G., Li H. Pectin methylesterases contribute the pathogenic differences between races 1 and 4 of Fusarium oxysporum f. sp. cubense. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-13625-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Kita D., Peaucelle A., Cartwright H.N., Doan V., Duan Q., Liu M.-C., Maman J., Steinhorst L., Schmitz-Thom I., et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 2018;28:666–675.e5. doi: 10.1016/j.cub.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Savatin D.V., Sicilia F., Gramegna G., Cervone F., Lorenzo G.D. Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013;4:49. doi: 10.3389/fpls.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz E., Ranocha P., Le Ru A., Martinez Y., Fourquaux I., Jauneau A., Dunand C., Burlat V. Pectin demethylesterification generates platforms that anchor peroxidases to remodel plant cell wall domains. Dev. Cell. 2019;48:261–276.e8. doi: 10.1016/j.devcel.2018.11.016. [DOI] [PubMed] [Google Scholar]
- Gadeyne A., Sánchez-Rodríguez C., Vanneste S., Di Rubbo S., Zauber H., Vanneste K., Van Leene J., De Winne N., Eeckhout D., Persiau G., et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell. 2014;156:691–704. doi: 10.1016/j.cell.2014.01.039. [DOI] [PubMed] [Google Scholar]
- Gámez-Arjona F.M., Vitale S., Voxeur A., Dora S., Müller S., Sancho-Andrés G., Montesinos J.C., Di Pietro A., Sánchez-Rodríguez C. Impairment of the cellulose degradation machinery enhances Fusarium oxysporum virulence but limits its reproductive fitness. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abl9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Maceira F.I., Di Pietro A., Roncero M.I. Cloning and disruption of pgx4 encoding an in planta expressed exopolygalacturonase from Fusarium oxysporum. Mol. Plant Microbe Interact. 2000;13:359–365. doi: 10.1094/MPMI.2000.13.4.359. [DOI] [PubMed] [Google Scholar]
- García-Maceira F.I., Di Pietro A., Huertas-González M.D., Ruiz-Roldán M.C., Roncero M.I. Molecular characterization of an endopolygalacturonase from Fusarium oxysporum expressed during early stages of infection. Appl. Environ. Microbiol. 2001;67:2191–2196. doi: 10.1128/AEM.67.5.2191-2196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N.L., Schmoll M., Cate J.H.D., Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- Glöckner N., Zur Oven-Krockhaus S., Rohr L., Wackenhut F., Burmeister M., Wanke F., Holzwart E., Meixner A.J., Wolf S., Harter K. Three-fluorophore FRET enables the analysis of ternary protein association in living plant cells. Plants. 2022;11 doi: 10.3390/plants11192630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Felix G., Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- Gordon T.R. Fusarium oxysporum and the Fusarium wilt syndrome. Annu. Rev. Phytopathol. 2017;55:23–39. doi: 10.1146/annurev-phyto-080615-095919. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010;64:355–365. doi: 10.1111/j.1365-313X.2010.04322.x. [DOI] [PubMed] [Google Scholar]
- Gronnier J., Franck C.M., Stegmann M., DeFalco T.A., Abarca A., von Arx M., Dünser K., Lin W., Yang Z., Kleine-Vehn J., et al. Regulation of immune receptor kinase plasma membrane nanoscale organization by a plant peptide hormone and its receptors. Elife. 2022;11 doi: 10.7554/eLife.74162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M.C., Ehrhardt D.W. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- Haas K.T., Wightman R., Meyerowitz E.M., Peaucelle A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science. 2020;367:1003–1007. doi: 10.1126/science.aaz5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z.-H., Fujiki M., Kohorn B.D. A cell wall-associated, receptor-like protein kinase∗. J. Biol. Chem. 1996;271:19789–19793. doi: 10.1074/jbc.271.33.19789. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hettenhausen C., Schuman M.C., Wu J. MAPK signaling: a key element in plant defense response to insects. Insect Sci. 2015;22:157–164. doi: 10.1111/1744-7917.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss A.T., Jr., Hicks K.B. Analysis of oligogalacturonic acids with 50 or fewer residues by high-performance anion-exchange chromatography and pulsed amperometric detection. Anal. Biochem. 1990;184:200–206. doi: 10.1016/0003-2697(90)90669-z. [DOI] [PubMed] [Google Scholar]
- Huerta A.I., Kesten C., Menna A.L., Sancho-Andrés G., Sanchez-Rodriguez C. In-Plate quantitative characterization of Arabidopsis thaliana susceptibility to the fungal vascular pathogen Fusarium oxysporum. Curr. Protoc. Plant Biol. 2020;5 doi: 10.1002/cppb.20113. [DOI] [PubMed] [Google Scholar]
- Huertas-González M.D., Ruiz-Roldán M.C., García Maceira F.I., Roncero M.I., Di Pietro A. Cloning and characterization of pl1 encoding an in planta-secreted pectate lyase of Fusarium oxysporum. Curr. Genet. 1999;35:36–40. doi: 10.1007/s002940050430. [DOI] [PubMed] [Google Scholar]
- Huynh K., Partch C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015;79:28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Ott T. The nanoscale organization of the plasma membrane and its importance in signaling: a proteolipid perspective. Plant Physiol. 2020;182:1682–1696. doi: 10.1104/pp.19.01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers W., Rep M. Mutation of CRE1 in Fusarium oxysporum reverts the pathogenicity defects of the FRP1 deletion mutant. Mol. Microbiol. 2009;74:1100–1113. doi: 10.1111/j.1365-2958.2009.06922.x. [DOI] [PubMed] [Google Scholar]
- Kesten C., Gámez-Arjona F.M., Menna A., Scholl S., Dora S., Huerta A.I., Huang H.-Y., Tintor N., Kinoshita T., Rep M., et al. Pathogen-induced pH changes regulate the growth-defense balance in plants. EMBO J. 2019;38 doi: 10.15252/embj.2019101822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman S.J., Atallah Z.K., Vallad G.E., Subbarao K.V. Diversity, pathogenicity, and management of verticillium species. Annu. Rev. Phytopathol. 2009;47:39–62. doi: 10.1146/annurev-phyto-080508-081748. [DOI] [PubMed] [Google Scholar]
- Knox J.P., Linstead P.J., King J., Cooper C., Roberts K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta. 1990;181:512–521. doi: 10.1007/BF00193004. [DOI] [PubMed] [Google Scholar]
- Kohorn B.D., Johansen S., Shishido A., Todorova T., Martinez R., Defeo E., Obregon P. Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 2009;60:974–982. doi: 10.1111/j.1365-313X.2009.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B.D., Kohorn S.L., Saba N.J., Martinez V.M. Requirement for pectin methyl esterase and preference for fragmented over native pectins for wall-associated kinase-activated, EDS1/PAD4-dependent stress response in Arabidopsis. J. Biol. Chem. 2014;289:18978–18986. doi: 10.1074/jbc.M114.567545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lally D., Ingmire P., Tong H.Y., He Z.H. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 2001;13:1317–1331. doi: 10.1105/tpc.13.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton C.J., Hellinga H.W. Quantitation of protein–protein interactions by thermal stability shift analysis. Protein Sci. 2011;20:1439–1450. doi: 10.1002/pro.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux C., Bouton S., Kiefer-Meyer M.-C., Fabrice T.N., Mareck A., Guénin S., Fournet F., Ringli C., Pelloux J., Driouich A., et al. PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination. Plant Physiol. 2015;167:367–380. doi: 10.1104/pp.114.250928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K.C., Selzer T., Shahar C., Udi Y., Tworowski D., Sagi I. Inhibition of pectin methyl esterase activity by green tea catechins. Phytochemistry. 2008;69:2586–2592. doi: 10.1016/j.phytochem.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Li J., Cornelissen B., Rep M. Host-specificity factors in plant pathogenic fungi. Fungal Genet. Biol. 2020;144 doi: 10.1016/j.fgb.2020.103447. [DOI] [PubMed] [Google Scholar]
- Li L., Li K., Ali A., Guo Y. AtWAKL10, a cell wall associated receptor-like kinase, negatively regulates leaf senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22094885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Tang W., Pan X., Huang A., Gao X., Anderson C.T., Yang Z. Arabidopsis pavement cell morphogenesis requires FERONIA binding to pectin for activation of ROP GTPase signaling. Curr. Biol. 2022;32:497–507.e4. doi: 10.1016/j.cub.2021.11.030. [DOI] [PubMed] [Google Scholar]
- Liners F., Thibault J.F., Van Cutsem P. Influence of the degree of polymerization of oligogalacturonates and of esterification pattern of pectin on their recognition by monoclonal antibodies. Plant Physiol. 1992;99:1099–1104. doi: 10.1104/pp.99.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V. PECTOPLATE: the simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Front. Plant Sci. 2015;6:331. doi: 10.3389/fpls.2015.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V., Métraux J.P. Plant cell wall in pathogenesis, parasitism and symbiosis. Front. Plant Sci. 2014;5:612. doi: 10.3389/fpls.2014.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V., Fabri E., De Caroli M., Hansen A.R., Willats W.G.T., Piro G., Bellincampi D. Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to. Plant Physiol. 2017;173:1844–1863. doi: 10.1104/pp.16.01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Sonbol F.-M., Huot B., Gu Y., Withers J., Mwimba M., Yao J., He S.Y., Dong X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016;7 doi: 10.1038/ncomms13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Sun Y., Pei Y., Zhang X., Wang P., Li X., Li F., Hou Y. A pectin methylesterase inhibitor enhances resistance to verticillium wilt. Plant Physiol. 2018;176:2202–2220. doi: 10.1104/pp.17.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R., Stiller J., Powell J., Rusu A., Manners J.M., Kazan K. Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Claus L.A.N., Leslie M.E., Tao K., Wu Z., Liu J., Yu X., Li B., Zhou J., Savatin D.V., et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature. 2020;581:199–203. doi: 10.1038/s41586-020-2210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S., Segorbe D., Turrà D., Leon-Ruiz M., Fürst U., El Ghalid M., Leonard G., López-Berges M.S., Richards T.A., Felix G., Di Pietro A. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.43. [DOI] [PubMed] [Google Scholar]
- Menna A., Fischer-Stettler M., Pfister B., Andrés G.S., Holbrook-Smith D., Sánchez-Rodríguez C. Single-run HPLC quantification of plant cell wall monosaccharides. Bio. Protoc. 2020;10 doi: 10.21769/BioProtoc.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna A., Dora S., Sancho-Andrés G., Kashyap A., Meena M.K., Sklodowski K., Gasperini D., Coll N.S., Sánchez-Rodríguez C. A primary cell wall cellulose-dependent defense mechanism against vascular pathogens revealed by time-resolved dual transcriptomics. BMC Biol. 2021;19:1–20. doi: 10.1186/s12915-021-01100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K., McNeil P.L. Vesicle accumulation and exocytosis at sites of plasma membrane disruption. J. Cell Biol. 1995;131:1737–1745. doi: 10.1083/jcb.131.6.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Mort A.J., Moerschbacher B.M., Pierce M.L., Maness N.O. Problems encountered during the extraction, purification, and chromatography of pectic fragments, and some solutions to them. Carbohydr. Res. 1991;215:219–227. [Google Scholar]
- Nagaya S., Kawamura K., Shinmyo A., Kato K. The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 2010;51:328–332. doi: 10.1093/pcp/pcp188. [DOI] [PubMed] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Ortiz-Morea F.A., Savatin D.V., Dejonghe W., Kumar R., Luo Y., Adamowski M., Van den Begin J., Dressano K., Pereira de Oliveira G., Zhao X., et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc. Natl. Acad. Sci. USA. 2016;113:11028–11033. doi: 10.1073/pnas.1605588113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Fang L., Liu J., Senay-Aras B., Lin W., Zheng S., Zhang T., Guo J., Manor U., Van Norman J., et al. Auxin-induced signaling protein nanoclustering contributes to cell polarity formation. Nat. Commun. 2020;11:3914. doi: 10.1038/s41467-020-17602-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncini L., Wyrsch I., Dénervaud Tendon V., Vorley T., Boller T., Geldner N., Métraux J.P., Lehmann S. In roots of Arabidopsis thaliana, the damage-associated molecular pattern AtPep1 is a stronger elicitor of immune signalling than flg22 or the chitin heptamer. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Zhu X., Guo F., Lv L., Zhang Z. The wall-associated receptor-like kinase TaWAK7D is required for defense responses to rhizoctonia cerealis in wheat. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22115629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.-Y., Han L., Mao G., Glazebrook J., Zhang S. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2008;105:5638–5643. doi: 10.1073/pnas.0711301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C. Monitoring the outside: cell wall-sensing mechanisms. Plant Physiol. 2010;153:1445–1452. doi: 10.1104/pp.110.154518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosli H.G., Zheng Y., Pombo M.A., Zhong S., Bombarely A., Fei Z., Collmer A., Martin G.B. Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 2013;14:R139. doi: 10.1186/gb-2013-14-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran J., Habrylo O., Cherkaoui M., Lecomte S., Voxeur A., Pilard S., Bassard S., Pau-Roblot C., Mercadante D., Pelloux J., Sénéchal F. New insights into the specificity and processivity of two novel pectinases from Verticillium dahliae. Int. J. Biol. Macromol. 2021;176:165–176. doi: 10.1016/j.ijbiomac.2021.02.035. [DOI] [PubMed] [Google Scholar]
- Saintenac C., Lee W.-S., Cambon F., Rudd J.J., King R.C., Marande W., Powers S.J., Bergès H., Phillips A.L., Uauy C., et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 2018;50:368–374. doi: 10.1038/s41588-018-0051-x. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y., Goda H., Nakamura A., Takatsuto S., Fujioka S., Yoshida S. Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 2003;131:287–297. doi: 10.1104/pp.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza C.d.A., Li S., Lin A.Z., Boutrot F., Grossmann G., Zipfel C., Somerville S.C. Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol. 2017;173:2383–2398. doi: 10.1104/pp.16.01680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- Tinevez J.-Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Vaahtera L., Schulz J., Hamann T. Cell wall integrity maintenance during plant development and interaction with the environment. Native Plants. 2019;5:924–932. doi: 10.1038/s41477-019-0502-0. [DOI] [PubMed] [Google Scholar]
- Verica J.A., He Z.-H. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002;129:455–459. doi: 10.1104/pp.011028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica J.A., Chae L., Tong H., Ingmire P., He Z.-H. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 2003;133:1732–1746. doi: 10.1104/pp.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.A., Kohorn B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Kroll A., Haramagatti C.R., Lipinski H.-G., Wiemann M. Classification and segmentation of nanoparticle diffusion trajectories in cellular micro environments. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhou L., Jamieson P., Zhang L., Zhao Z., Babilonia K., Shao W., Wu L., Mustafa R., Amin I., et al. The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell. 2020;32:3978–4001. doi: 10.1105/tpc.19.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Mravec J., Greiner S., Mouille G., Höfte H. Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr. Biol. 2012;22:1732–1737. doi: 10.1016/j.cub.2012.07.036. [DOI] [PubMed] [Google Scholar]
- Wolf S., van der Does D., Ladwig F., Sticht C., Kolbeck A., Schürholz A.K., Augustin S., Keinath N., Rausch T., Greiner S., et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. USA. 2014;111:15261–15266. doi: 10.1073/pnas.1322979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Bacic A., Johnson K.L., Humphries J. 2020. The role of brachypodium distachyon wall-associated kinases (WAKs) in cell expansion and stress responses. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Huang J., Li B., Li J., Wang Y. Is kinase activity essential for biological functions of BRI1? Cell Res. 2008;18:472–478. doi: 10.1038/cr.2008.36. [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhang Y., Li B., Yang X., Dong Y., Qiu D. A Verticillium dahliae pectate lyase induces plant immune responses and contributes to virulence. Front. Plant Sci. 2018;9:1271. doi: 10.3389/fpls.2018.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Xie M., Wang X., Wang G., Zhang Y., Li Z., Ma Z. Identification of cell wall-associated kinases as important regulators involved in Gossypium hirsutum resistance to Verticillium dahliae. BMC Plant Biol. 2021;21:220. doi: 10.1186/s12870-021-02992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Pombo M.A., Rosli H.G., Martin G.B. Tomato wall-associated kinase SlWak1 depends on fls2/fls3 to promote apoplastic immune responses to Pseudomonas syringae. Plant Physiol. 2020;183:1869–1882. doi: 10.1104/pp.20.00144. –1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F., Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu D., Wang P., et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. USA. 2018;115:E1906–E1915. doi: 10.1073/pnas.1712251115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video depicts the dynamic behavior of RFO1-GFP expressed in rfo1-1 in Arabidopsis root epidermal cells acquired using spinning-disc confocal microscopy. Foci identified by Trackmate using the LAP tracker to be analyzed are indicated in the right panel. This video corresponds to Figure 1F and 1G.
The video depicts the dynamic behavior of RFO1-GFP expressed in Arabidopsis rfo1-1 root epidermal cells acquired using spinning-disc confocal microscopy. The pectin methylation level in the cell wall was modulated by introducing the transgene iPMEIox, which was activated using estradiol (mock; middle panel) to reduce the dmPectin levels. This effect was compensated for by adding dmPectin (right panel). The control line is shown in the left panel. Scale bar, 5 μm. This video corresponds to Figure 6.