Abstract
Gene regulation in eukaryotes requires the controlled access of sequence-specific transcription factors (TFs) to their sites in a chromatin landscape dominated by nucleosomes. Nucleosomes are refractory to TF binding, and often must be removed from regulatory regions. Recent genomic studies together with in vitro measurements suggest that the nucleosome barrier to TF binding is modulated by dynamic nucleosome unwrapping governed by ATP-dependent chromatin remodelers. Genome-wide occupancy and regulation of subnucleosomal intermediates have gained recent attention with the application of high-resolution approaches for precision mapping of protein-DNA interactions. Here we summarize recent findings on nucleosome substructures and TF-binding dynamics, and highlight how unwrapped nucleosomal intermediates provide a novel signature of active chromatin.
Keywords: Structural epigenomics, transcription factors, ATP-dependent remodeling, nucleosome dynamics, fragile nucleosome
Nucleosome dynamics determine chromatin accessibility
Nucleosomes are the fundamental repeating structural units that package eukaryotic genomes into chromatin. A nucleosome core particle (NCP) consists of ~147 base pairs (bp) of DNA making 1.67 left-handed helical turns around a histone protein octamer, consisting of a central tetramer of histones H3 and H4 ([H3-H4]2) flanked by dimers of histones H2A and H2B on either side [1]. Nucleosomes are positioned in the genome such that protein-binding sites are either exposed or occluded. Nucleosomes blocking DNA-binding of transcription factors (TFs) and components of the RNA- and DNA-polymerase machineries present formidable barriers to transcription, DNA replication, recombination, and repair [2]. Nucleosomes therefore must be dynamically remodeled in response to developmental and environmental cues for DNA-templated processes to occur. At dynamic regions of genomes, which include promoters, enhancers, DNA replication origins, and DNA-damage sites, nucleosomes undergo rapid turnover [3–7], incorporation of variant histones [8, 9], and other structural disruptions [10–13]. Precise positions of nucleosomes within the genome, their composition, stability, and structural conformations, are key features of cell-type specific gene-expression programs [14–17]. Moreover, the propagation and maintenance of DNA accessibility or inaccessibility patterns across mitotic cell divisions form, in part, the basis of the concept of cellular epigenetic inheritance [18].
Whereas transcriptionally inactive or repressed regions of the genome are compacted by chromatin-associated protein complexes and linker histones that regularly space nucleosomes, functional TF-binding sites are typically located within nucleosome-free or depleted regions (NFR or NDR, see Glossary). However, recent studies profiling chromatin components genome-wide now suggest that TF-binding sites within transcriptionally active promoters and enhancers from diverse eukaryotes, including yeast [10], flies [19, 20], worms [21], and mammals [22–24], are not always nucleosome-free. Two alternative scenarios have been proposed to explain how TFs may overcome the nucleosome barrier. A special class of pioneer TFs may target sites on the nucleosome surface directly by virtue of their ability to bind to nucleosomes, as in cases of activating developmentally repressed genes [24–26]. Alternatively, TFs may bind their sites by virtue of high-affinity binding and/or high concentration [27] using the window of opportunity created by nucleosome turnover, or dynamic conformational transitions that transiently expose DNA, as proposed by Widom and colleagues [28]. TF-binding would then drive the conformational equilibrium toward the unwrapped, accessible state [29]. Factors influencing the biophysical properties of a nucleosome, for example, post-translational modifications (PTMs) such as acetylation and phosphorylation of histone H3 near the nucleosomal DNA entry/exit site, enhance DNA accessibility for TF-binding, whereas the linker histone H1 may antagonize site-exposure [30, 31]. Here, we summarize recent evidence that suggests a model whereby site exposure catalyzed by ATP-dependent chromatin remodelers cause nucleosomes over regulatory regions to be partially unwrapped, thereby facilitating TF-binding and relieving the nucleosomal barrier to RNA polymerase. Much of our mechanistic understanding of the basic enzymatic machineries that modulate nucleosome dynamics come from studies with the budding yeast Saccharomyces cerevisiae. These studies support an emerging concept in which unwrapped nucleosomes and alternative nucleosome structural intermediates represent novel structural epigenomic “signatures” of active chromatin.
Promoter nucleosome organization and dynamic intermediates
Global mapping of nucleosome positions and chromatin accessibility has revealed a somewhat stereotypical arrangement of nucleosomes at distinct functional regions of the genome, such as relative to the transcription-start site (TSS) of genes (Figure 1A). The major approach for determining genome-wide positions of nucleosomes involves digesting chromatin with micrococcal nuclease (MNase) followed by deep-sequencing of DNA fragments that are protected from MNase digestion (MNase-seq) [32, 33] (Table 1). MNase is an endo-/exonuclease that processively digests DNA that is not protected by a histone octamer or a DNA-binding protein, including linker-DNA regions between nucleosomes. Nucleosome positions are inferred based on sequence alignments of DNA fragments that are ~150 bp in size. The best characterized of all genomes for nucleosome organization is that of budding yeast (reviewed in [33, 34]). TSSs of active genes in yeast are located about one helical turn inside of a strongly positioned nucleosome (designated as the +1 nucleosome). Another strongly positioned nucleosome (the −1 nucleosome) is located at a variable but locus-specific distance upstream. The regions in between −1 and +1 nucleosomes are largely devoid of histones [35, 36], accordingly called NDRs. NDRs constituting the core of active gene promoters are sites of TF-occupancy, transcription pre-initiation complex (PIC) assembly, and RNA polymerase II (RNAPII) loading. Beside promoters, NDRs are also prominent at origins of DNA replication and enhancer regions in multicellular organisms.
Figure 1: Promoter nucleosome organization is regulated by many factors.
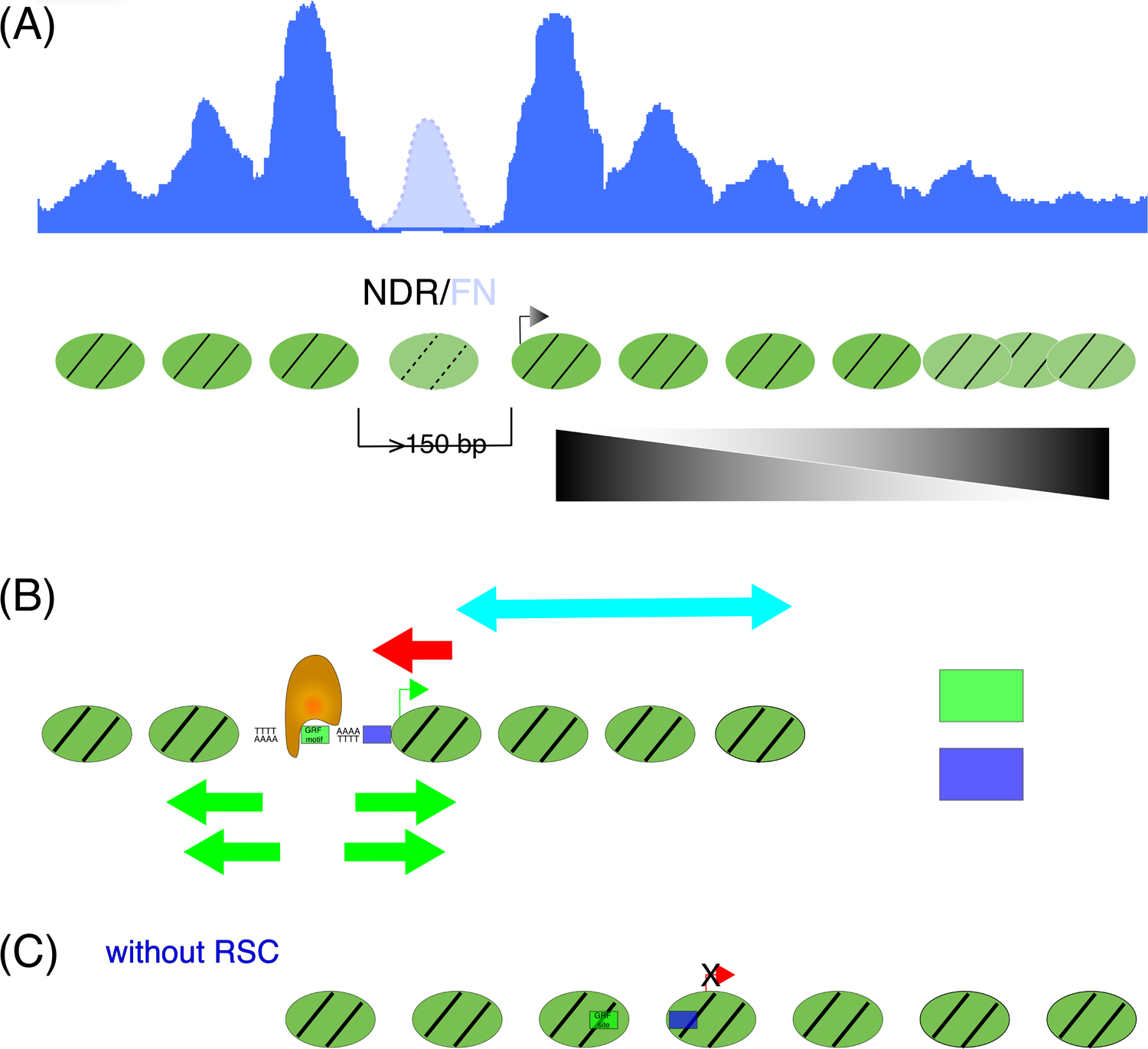
(A) Nucleosome organization relative to the TSS (shown by an arrow) of a representative transcriptionally active gene in budding yeast. Strongly positioned nucleosomes flank a NDR, and positioning decreases towards the gene-body (“fuzziness”). A dynamic MNase-sensitive nucleosome or a fragile nucleosome (FN, shown with broken margins) is often occupies the NDR when the gap between the strongly positioned −1 and +1 nucleosomes is larger than 150 bp. The top panel shows the readout from a representative genomic nucleosome profiling experiment (for example MNase-seq). Positioned nucleosomes are seen as discrete peaks (blue). The FN peak (cyan) is seen with partial MNase-digestion of chromatin, but is lost upon extensive digestion. (B) Transcriptionally active or “open” promoter architecture is the result of a tightly regulated interplay of DNA sequence, GRFs, and ATP-dependent nucleosome remodelers. In this configuration, TF-binding sites and the TATA-box are accessible, and the TSS (green arrow) is in a permissive position. (C) In the absence of RSC function, nucleosomes intrude into the NDR-space resulting in occlusion of TF-binding motifs, the TATA box, and the TSS (red arrow). Increased spacing (longer linker DNA) between nucleosomes is typical of these repressed or “closed” promoters.
Table 1:
Genomic approaches discussed in this review for assaying nucleosome or TF dynamics
| Method(s) | Brief description | Notes | Ref(s). |
|---|---|---|---|
| Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) | Antibody-based enrichment of target-protein associated DNA from formaldehyde-crosslinked and sonicated chromatin. |
|
[105–107] |
| MNase digestion followed by deep sequencing (MNase-seq) | Enzymatic digestion of chromatin and paired-end (commonly) sequencing of DNA-ends protected from digestion by DNA-bound proteins. Can be combined with ChIP (MNase-ChIP-seq or ORGANIC) of histones/TF/remodelers for high-resolution mapping of nucleosomal and non-nucleosomal footprints. |
|
[19, 33, 34, 38, 57, 61, 62, 84, 108] |
| ChIP followed by exonuclease digestion and deep sequencing (ChIP-exo) | Immunoprecipitated DNA fragments are trimmed up to the protein-DNA crosslink using an exonuclease. |
|
[13, 109] |
| Cleavage under targets and release using nuclease (CUT&RUN) | Targeted chromatin cleavage and release under native conditions by tethering a Protein A-MNase fusion to antibody bound to epitopes in intact permeabilized cells or nuclei. Can be followed by ChIP for histones and histone PTMs under native conditions to directly profile co-occupancies (CUT&RUN.ChIP) |
|
[10, 68, 110, 111] |
| Histone (H3 or H4)-anchored chemical cleavage mapping | DNA proximal residue Ser 47 in H4 or Gln 85 in H3 is mutated to a Cys (H4S47C or H3Q85C) and derivatized with a phenanthroline ligand. The phenanthroline chelates a copper ion, which in the presence of peroxide cleaves DNA backbone in the local vicinity of the modified histone residue. |
|
[12, 23, 36] |
| Methidiumpropyl-EDTA (MPE)-seq | MPE-Fe(II) complex intercalates with DNA and generates single- and doublestranded DNA breaks in the presence of oxygen. MPE-Fe(II) preferentially cleaves linker DNA and NDRs. |
|
[22] |
| Mapping in vivo nascent chromatin with EdU and sequencing (MINCE-seq) or nascent chromatin avidin pull-down (NChAP), and replication-coupled assay for transposase-accessible chromatin (repli-ATAC-seq) | Replicating DNA incorporates 5-ethynyl-2’-deoxyuridine (EdU) during a pulse labeling step and EdU-labelled DNA is enriched by covalently attaching biotin azide to EdU followed by streptavidin pull-down for deep sequencing. Nucleosome occupancy and accessibility of EdU labeled chromatin is assessed based on protection from MNase digestion (MINCE-seq and NChAP) or accessibility for transposition (repli-ATAC-seq). |
|
[66, 86, 102, 103] |
The best understood model for promoter nucleosome organization is that NFRs are formed largely due to poly(dA:dT) sequence tracts that intrinsically disfavor stable nucleosome formation [37], and to binding of sequence-specific TFs such as the general regulatory transcription factors (GRFs) in yeast [38–40] that may directly compete with histones [27]. Proteins occupying the NDRs act as static barriers, and nucleosomes are statistically positioned between barriers [41]. A histone H3-directed chemical cleavage method for mapping single nucleosome dyads (imaginary axis of symmetry between the two halves of a nucleosome) with unprecedented accuracy has revealed that each nucleosome around the NDRs can occupy preferred positions that differ by integral multiples of the DNA helical repeat (~10 bp), and specific positions are influenced by ATP-dependent nucleosome remodelers (Box 1) [36]. The essential Remodeling the Structure of Chromatin (RSC ) remodeling complex utilizes poly(dA:dT) motifs to directionally remodel nucleosomes and remove them from TF-binding sites to create NDRs, and also sets the positions of the +1 and −1 nucleosomes [38–40] (Figure 1B). This function is likely partially redundant with that of INO80, and also ISWI family remodelers (ISW2 and ISW1a) (Box 1), which utilize their DNA-length sensing properties [42, 43] to reposition nucleosomes to fixed distances from a GRF barrier. Chd1 and ISWI remodelers further adjust nucleosome spacing (distance between two consecutive nucleosomes) within the phased arrays by using the tightly positioned +1 nucleosome as a point of reference [40, 44, 45]. NDR width and nucleosome spacing within an array are sensitive to the level of transcription of a gene and even long-range inter-nucleosome interactions [36, 46, 47]. Strong phasing of nucleosomes is also observed around the binding sites for the insulator protein CCCTC-binding factor (CTCF) in metazoans [15, 48].
Box1: ATP-dependent chromatin remodelers.
ATP-dependent remodelers are evolutionarily conserved multi-subunit protein complexes belonging to four major subfamilies within the Snf2 family: mating type switching/sucrose non-fermenting (SWI/SNF), imitation switch (ISWI), chromodomain helicase (CHD), and inositol requiring (INO80) (reviewed in [92]). Each remodeling complex catalytic subunit has a common bipartite DExx and HELICc domain structure (also called the motor) harboring the ATP hydrolysis and DNA translocation activities. The motor is capable of translocating on nucleosomal DNA while hydrolyzing ATP, resulting in DNA moving through nucleosomes, which is tuned to cause specific changes in nucleosome organization such as nucleosome spacing (ISWI, CHD and INO80), octamer eviction (SWI/SNF), or histone dimer exchange (INO80). Additional domains within the catalytic subunit with specific chromatin binding properties regulate the motor and remodeling activities, and often serve as docking sites for additional subunits. Members in each subfamily generally form lineage-specific complexes with both unique and shared additional subunits that can interact with DNA, TFs, histones, and histone PTMs, determining their specificity and serving as regulatory modules [92, 93]. The SWI/SNF and INO80 families are the most complex in terms of subunit composition across species.
The INO80, ISWI, and CHD remodelers have partially redundant roles in spacing nucleosome at promoters or coding regions of genes [40, 78]. Activities of these remodelers are tightly regulated by the length of linker DNA connecting nucleosomes. ISWI activity is also stimulated by the histone H4 tails, and is sensitive to its acetylation status. SWI/SNF remodelers recognize acetylated H3 tails via their bromodomains, and other than RSC, SWI/SNF remodelers are considered to have context-specific roles in activating or repressing gene expression [92]. In addition to evicting nucleosomes, SWI/SNF is able to slide nucleosomes past a bound transcription factor, likely evicting it [94], and metazoan SWI/SNF also can evict Polycomb Repressive Complex 1 [95]. Beside their roles in transcription, INO80 and SWI/SNF remodelers help maintain genome integrity by removing nucleosomes for DNA-replication fork progression, DNA-damage repair, and recombination. The versatile functions of ATP-dependent remodelers are manifested in their roles in development and their mutation or misregulation in diseases such as cancer [96, 97].
Promoter proximal +1 and −1 nucleosomes are dynamic and show high rates of histone turnover at active genes [3]. The most prominent example of turnover is the replacement of histone H2A with the replication-independent variant H2A.Z (reviewed in [49]) catalyzed by another nucleosome remodeler SWR1, which recognizes an NDR [50], such that +1 and −1 positions with respect to the NDRs are selectively enriched for H2A.Z [10]. Antagonistic activities of RSC and ISWI remodelers [51] appear to cause a +1 nucleosome to toggle between two or three rotational positions that are either permissive (RSC) or refractory (ISW2) to transcription [40]. RSC action is also critical for positioning +1 nucleosomes such that TATA or TATA-like elements are accessible [39] (Figure 1C).
Recent high-resolution mapping approaches have characterized intermediates of +1 and −1 nucleosomes, showing that these positions have structural variants (Figure 2). In one study, chemical modification of a specific amino acid residue within histone H4 (H4S47) converting it into a site-specific DNA cleavage reagent was used to map the precise contact points of H4S47 in both halves of each nucleosome genome-wide [12]. It was found that a subset of +1 and −1 nucleosomes in yeast does not show H4-DNA interaction on one side of the dyad axis, while both halves were protected from MNase digestion, suggesting that these nucleosomes are asymmetrically distorted. Remarkably, these nucleosomes are sites of RSC occupancy. RSC binding engulfs and unwraps a nucleosome up to the dyad axis by electrostatic attraction of nucleosomal DNA to its inner surface, which upon ATP hydrolysis and RSC translocation become susceptible to nuclease cleavage [52]. Promoter proximal asymmetric nucleosomes in yeast therefore resemble a RSC-engulfed nucleosome [53] (Figure 2B). Further analysis revealed that the distorted side of the dyad axis with fewer histone H4 contacts show increased protection from MNase digestion compared to the other side, likely due to steric occlusion from binding to RSC [12]. A more recent study showed that a fraction of RSC-bound +1 and −1 nucleosomes display subnucleosomal (≤ 120 bp) protections, suggesting that they are partially unwrapped from the edges (Figures 2C, D), consistent with RSC peeling off DNA from the nucleosome surface or generating a hexasomal intermediate (a nucleosome lacking one H2A/H2B dimer) during remodeling [10]. Partially unwrapped +1 and −1 nucleosomes are enriched for histone H2A.Z, for histone H3 with trimethylation of lysine 4 (H3K4me3), and for histone H4 with acetylated N-terminal tails, all features associated with active transcription [10]. Asymmetrically distorted and partially-unwrapped nucleosomes may represent distinct intermediates of the RSC-nucleosome remodeling cycle.
Figure 2: Promoter nucleosome substructures.
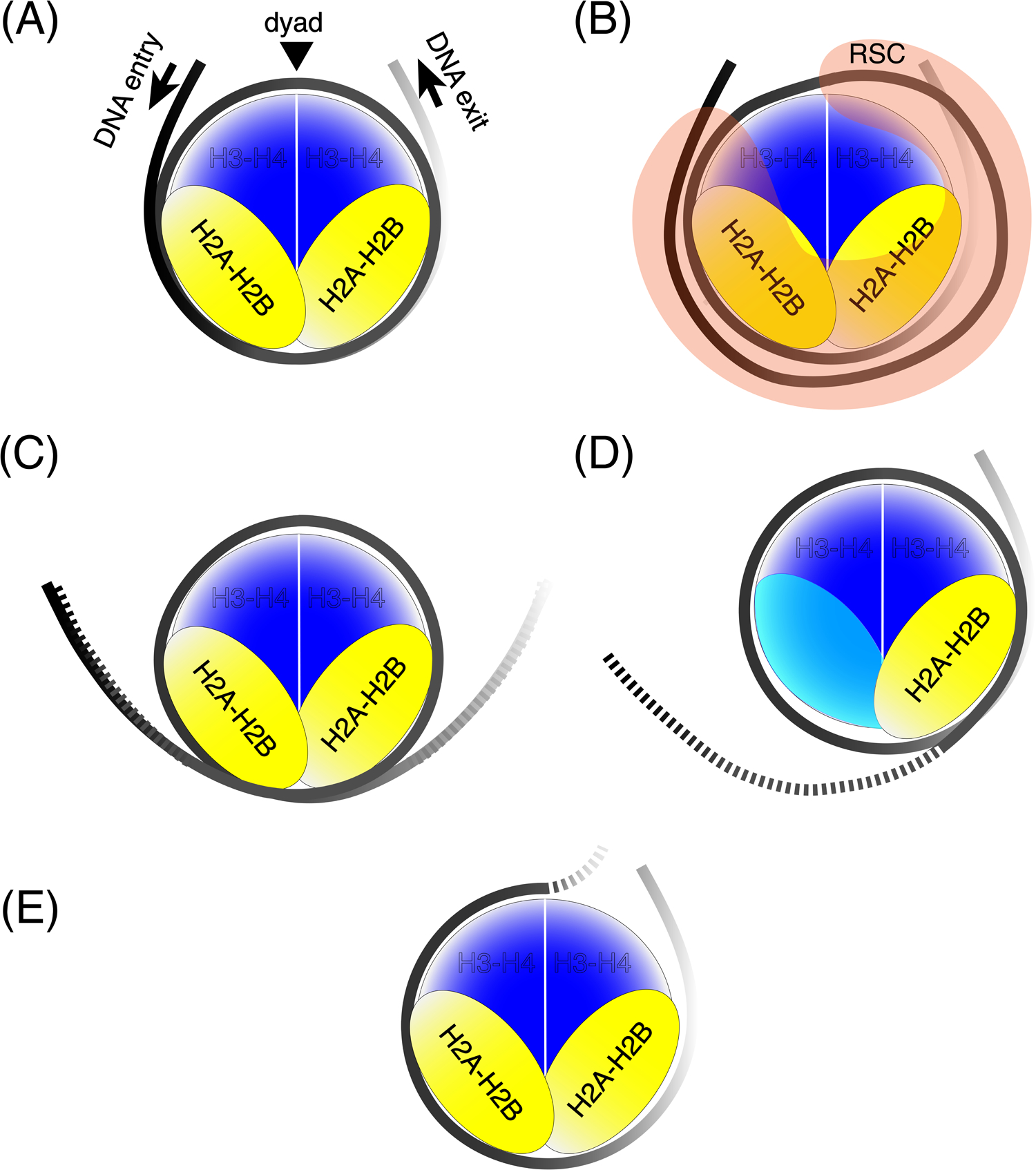
(A) Canonical nucleosomes have ~147 bp of DNA wrapped around the histone octamer with close to two helical turns of DNA, forming two distinct gyres. The dyad axis (an imaginary axis of symmetry) runs through the center of the nucleosomal DNA. (B) RSC engulfing a nucleosome asymmetrically distorts histone-DNA interactions along one DNA gyre, but the distorted DNA is protected from nuclease cleavage by RSC. (C) Partial unwrapping of DNA from either side (DNA entry and exit) results in increased sensitivity to nucleases (depicted as broken lines). (D) Loss of one of the two H2A-H2B dimers results in a hexasome with increased nuclease sensitivity of DNA close to the edge from where the dimer is lost. (E) Histone-DNA interactions can be lost from an entire gyre (one half of the nucleosome) asymmetrically.
ChIP-exo mapping (Table 1) of the limits of specific histone-DNA interactions genome-wide in yeast suggests that ~10% of +1 nucleosomes are asymmetric in terms of DNA associated with either half of the nucleosome (Figures 2D, E). Partial loss of histone-DNA contacts along one gyre of DNA suggest that these altered nucleosomes may exist as hexasomes or half-nucleosomes (where the DNA is in contact with one H2A/H2B dimer and one H3/H4 dimer) [13]. Likewise, it was observed in Drosophila melanogaster S2 cells that torsional strain generated by elongating RNAPII on +1 nucleosomes induce asymmetrically unwrapped hexasomal intermediates that require the histone chaperone FACT (facilitates chromatin transcription) for stability [11]. Hexasomes and unwrapped nucleosomes are more permissive to transcription by RNAPII than intact nucleosomes [54].
TSSs of active genes in multicellular organisms may be up to ~150 bp upstream of the +1 nucleosome edge, and in mammals, are also found to be populated by subnucleosome-sized particles as determined by MNase digestion of chromatin [14]. Furthermore, human H2A.Z nucleosomes show a higher susceptibility to asymmetric internal cleavage by MNase and thus protect shorter DNA compared to H2A nucleosomes [55]. Subnucleosomal particles represent informative signatures of dynamic and transcriptionally active promoters, analogous to well-studied histone PTMs, and are likely to be the subject of future investigations.
NDRs are dynamic: partially unwrapped nucleosomes over TF-binding sites
Dynamically unwrapped nucleosomes are also observed over sequence-specific TF-binding sites; a classic example being the galactose-inducible GAL1-GAL10 upstream activating sequence (UASg) containing Gal4 binding sites in yeast [56, 57]. A subnucleosome-size particle associated with ~120 bp DNA was first identified over the Gal4 binding sites based on its susceptibility to MNase digestion – observed with limited MNase digestion, but lost with longer digestion. These Gal4 binding sites are adjacent to CCGG motifs, which are binding sites for the Rsc3 subunit of the RSC nucleosome remodeling complex [58], and the MNase-sensitive/accessible particle coincided with RSC occupancy. It was proposed that prior to galactose induction, RSC engulfment and partial unwinding of a H2A.Z-containing nucleosome at UASg renders the Gal4 activator binding sites more accessible [56]. Analysis of DNA fragment sizes obtained from differential MNase digestion revealed an inverse relationship between nucleosome and RSC occupancy at this locus, suggesting that RSC action evicts nucleosomes. These particles are referred to as fragile nucleosomes (FNs) owing to their nucleosome-like footprint and MNase sensitivity [57, 59]. Systematic analysis of differential sensitivity of nucleosome-like particles to MNase digestion showed the existence of FNs throughout the yeast genome, remarkably over sites previously annotated to be nucleosome-free, including promoter NDRs immediately upstream of TSSs and +1 nucleosomes [38, 47, 59]. FN occupancy within a promoter closely corresponds with the distance between the edges of the MNase-resistant −1 and +1 nucleosomes, and requires at least 150 base pairs – just enough for a nucleosome to form [38]. This set of sites accounts for ~1/3 of all protein-coding gene promoters in yeast, mostly associated with highly transcribed house-keeping genes [47]. FNs occupy one or more sites for sequence-specific binding of GRFs ARS-binding factor 1 (Abf1), RNAPI enhancer binding protein (Reb1), or Repressor/Activator site binding protein (Rap1) [38], and strongly correlate with Rsc3-binding CGCG motifs and poly-(dA:dT) motifs implicated in directional RSC remodeling [39, 40, 60]. However, chromatin immunoprecipitation (ChIP)-based approaches [13, 61] and histone H3 or H4-directed chemical cleavage [36, 61] (Table 1) did not detect histones over MNase-sensitive sites in promoters, raising the question of whether MNase-sensitive footprints over promoters represent nucleosomes or instead non-nucleosomal proteins such as the GRFs or RSC [61, 62].
This issue was resolved by CUT&RUN profiling (Table 1), which showed that FN occupancy strongly correlates with RSC-bound ~100 bp particles, and using CUT&RUN supernatants for ChIP revealed that FN particles contain histones [10]. The subnucleosome-size of RSC-associated particles suggest that these are partially unwrapped nucleosomal intermediates (Figure 2), and possibly hexasomes, which protect ~110 bp of DNA from MNase in vitro [63]. FNs occlude GRF-binding sites, and remarkably, a fraction of the GRFs Abf1 and Reb1 bound to their sites are also associated with partially-unwrapped nucleosomes, suggesting that RSC-remodeling unravels nucleosomes to facilitate binding of the GRFs [10] (Figure 3). In multicellular eukaryotes, TF-binding sites that are dispersed across promoters, enhancers, and super-enhancers are also often found to co-map with nucleosomes [14–17, 20, 24]. For example, tissue-specific enhancers that bind the FoxA2 pioneer TF in mouse liver cells were found to be selectively enriched for MNase-sensitive nucleosomes [24]. Likewise, genome-wide approaches leveraging the precision of chemical cleavage mapping of nucleosomes in mouse embryonic stem cells (ESCs) indicate subnucleosome occupancy ubiquitously at promoters and over binding sites for pluripotency TFs and CTCF [22, 23]. Similar to FNs in yeast, subnucleosome-like particles over TF-binding sites in mammalian cells, Drosophila (fly) embryo, and Caenorhabditis elegans (worms) are sensitive to the extent of MNase digestion and have smaller footprints than canonical nucleosomes [19, 22–24], thereby suggesting a conserved role of nucleosome unwrapping for TF-binding. Interestingly, rapid transcription induction of some stress-response genes in Drosophila is associated with increase in MNase-sensitivity of nucleosomes at promoters and enhancers, but not nucleosome removal [20].
Figure 3(key figure): A model for catalyzed nucleosome-unwrapping facilitating TF-binding.
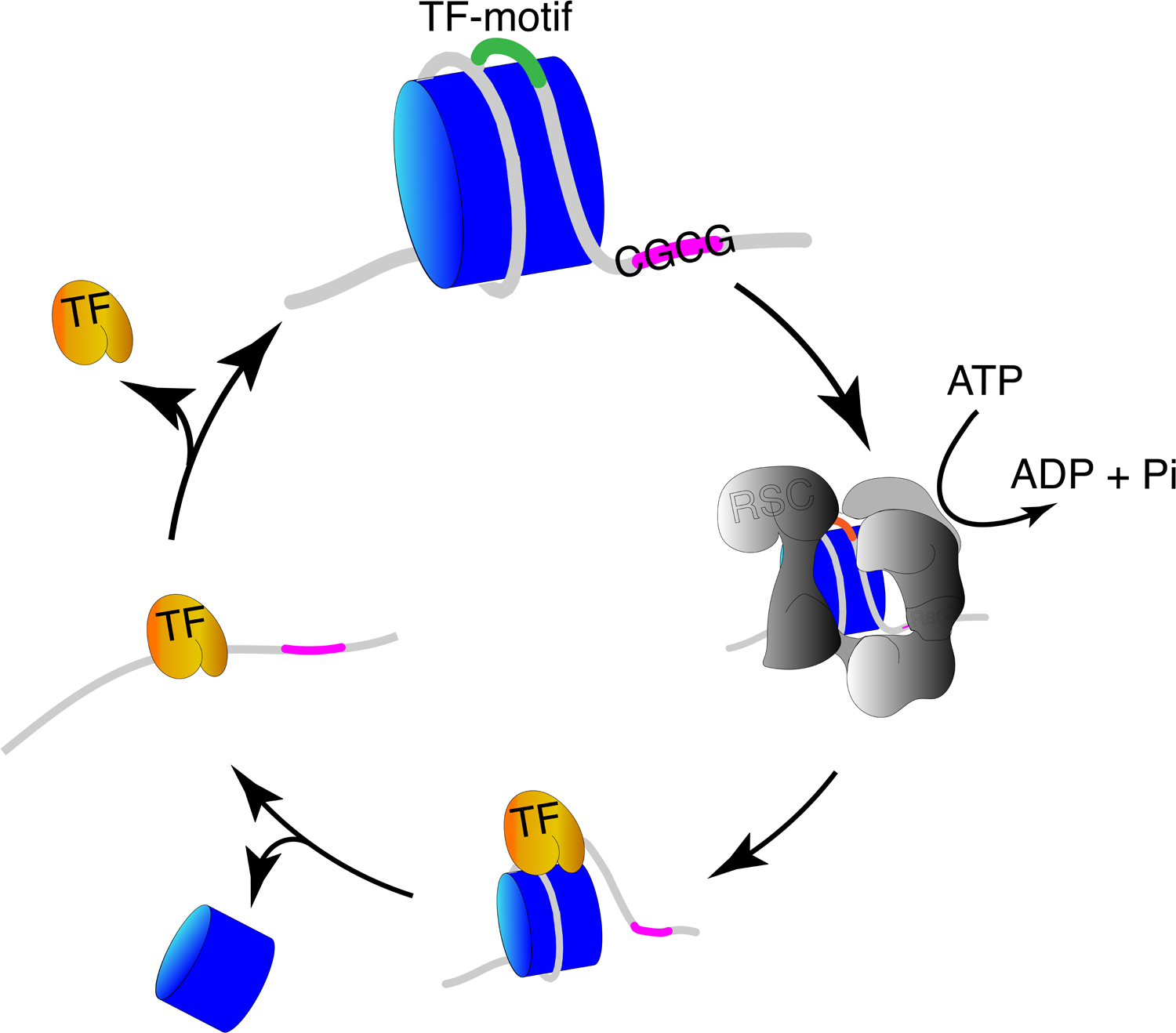
A SWI/SNF remodeler first engulfs a promoter nucleosome behind the replication fork. The remodeler uses energy from ATP hydrolysis to partially unwrap the nucleosome so as to expose the TF-binding motif. TFs (such as the GRFs in yeast) binding to exposed sequence motifs within unwrapped nucleosomes trap the nucleosome in a partially unwrapped state, but the unstable nucleosome is eventually displaced. TFs binding to non-nucleosomal DNA have short dwell-time, and a new nucleosome is deposited, occluding the site for a displaced TF, thus constituting a dynamic cycle.
The nucleosome remodeler RSC can destabilize or evict a nucleosome core from the DNA, causing selective removal of promoter nucleosomes, particularly at the −1 position and at NDRs from in vitro reconstituted or purified chromatin in a sequence-dependent manner [40, 60, 64]. Perhaps the transition from a nucleosome-occupied to nucleosome-depleted state at promoters occurs via a metastable partially unwrapped intermediate, of which RSC is a part [10]. RSC-mediated unwrapping and distortion of nucleosome dyads [52], as well as GRF-binding, might explain the absence of H3/H4-DNA contacts within FN particles [12, 61]. Consistent with RSC-mediated dynamics at NDRs, yeast promoters display a high rate of replication-independent histone turnover [65]. Depletion of RSC activity either by genetic deletion of non-essential subunits [51] or by nuclear depletion of the catalytic subunit Sth1 [38] cause shifts in nucleosome positioning into wide NDRs (200–300 bp) that are enriched for poly(dA:dT) and CGCG motifs (Figure 1C). Shrinking of NDRs after acute depletion of Sth1 using an auxin-induced degron occurs within 10 minutes, and is fully reversible upon reintroduction of Sth1 [66]. RSC-mediated NDR clearing occurs throughout the cell cycle, underlining the dynamic nature of promoter nucleosome organization [51, 66] (Figure 3).
Similarly to RSC in yeast, SWI/SNF family orthologs such as the Drosophila Brahma complex and mammalian BRG1/BRM-associated factor (BAF ) and Polybromo-associated BAF (PBAF) are enriched over promoters and enhancers [67]. Mouse ESCs and developing blastocysts show dependence on BAF for pluripotency factors Oct4 and Nanog binding [68, 69], and BAF activity is implicated in general for factor binding during reprograming [70], ESC pluripotency maintenance, and differentiation [71]. It is tempting to speculate that BAF remodeling may therefore facilitate TF-binding by generating unwrapped nucleosomal intermediates akin to what is observed in yeast. Whereas yeast RSC rapidly clears nucleosomes to generate NDRs, likely through sequence-specific recruitment to CGCG motifs in promoters by Rsc3 and its putative DNA-binding zinc-cluster, direct recruitment of other SWI/SNF remodelers is not evident. DNA binding mediated by a modified helix-turn-helix subdomain of the mammalian SWI/SNF subunit AT-rich interaction domain 1A (ARID1A) protein is important for chromatin interaction of the complex, but DNA-binding is not sequence-specific [72]. In this regard, Rsc3 was reported to be a nucleosome depleting factor like the GRFs using a reporter assay comparing a library of known TF-binding motifs and their sequence variants for the ability to restrict nucleosome occupancy [27]. Perhaps RSC components are recruited at Rsc3/Rsc30 bound sites during holocomplex assembly, consistent with the reorganization of a partial RSC complex containing Rsc3 to NDRs upon acute heat shock [73]. In contrast, SWI/SNF family complexes in multicellular eukaryotes likely cooperate with nucleosome-binding TFs and histone PTMs for site-specific recruitment. Examples include BAF recruitment by the forkhead box D3 factor (FOXD3) to promoters for ESC pluripotency, GATA binding factors (GATA1–3) recruitment to tissue-specific enhancers, and glucocorticoid receptor (GR) recruitment during steroid receptor signaling (reviewed in [74]). A model for coordinated action of pioneer factors, remodelers and other non-pioneering TFs proposes that pioneer factors recruit remodelers to displace nucleosomes and facilitate binding of non-pioneering TFs [24, 25, 74].
A fundamental difference between budding yeast and multicellular organisms is the absence of the canonical linker histone H1 and less condensed chromatin in yeast. The unusual linker histone Hho1 in yeast is structurally different from H1, and is much less abundant. However, an unusual high mobility group box (HMGB) family protein HMO1 considered to be functionally similar to the mammalian linker histone H1 in its ability to compact chromatin, is thought to stabilize non-canonical promoter nucleosomes (reviewed in [75]). Consistent with their function in chromatin opening, mammalian FoxA TFs have been suggested to antagonize H1-binding [24]. The action of mammalian nucleosome remodelers in the context of linker histone H1 and in cooperation with nucleosome-binding TFs is only just beginning to be understood [74, 76].
A non-canonical nucleosome-like particle called the “prenucleosome” has been described in vitro as a nucleosome-assembly intermediate containing the full complement of a histone octamer, but requires remodeling by ISWI or Chd remodelers for maturation [77]. Strikingly, prenucleosomes protect ~80 bp of DNA from MNase digestion, and structurally resemble nucleosome-like particles at NDRs of active promoters in yeast. It can be envisioned that prenucleosomes, like fragile-nucleosomes, are dynamic intermediates of nucleosome disassembly and reassembly at active promoters, that can be more readily disrupted by TFs or chromatin remodelers.
Unwrapped nucleosomes and TF-binding dynamics
Is catalyzed nucleosome unwrapping a prerequisite for TF-binding? The first reports of unwrapped nucleosomes associated with the Gal4 activator binding sites [56] and yeast stress-response gene promoters [59] suggested that nucleosome unwrapping exposes sequences so that TFs can readily bind. However, TFs occupy their cognate sites in the absence of RSC, albeit with reduced affinity in some cases [39, 56]. At the large majority of yeast promoters, a GRF and RSC can bind independently of each other as shown by depletion experiments, suggesting that independent sets of sequence motifs drive promoter DNA access by these factors [39]. Interestingly, depletion of Abf1 or Reb1 have mixed and locus-specific effects on FN occupancy; at some promoters they seem to destabilize FNs, while at others, GRF-binding is required for the stability of the partially unwrapped state [38]. It is likely that coordinated and opposing functions of multiple ATP-dependent remodelers and GRFs [78], all seemingly occupying and acting on shared genomic loci based on bulk mapping experiments [40, 67, 79], make results from depletion experiments ambiguous.
Recent sensitive biophysical analyses including single-molecule measurements of the binding and dissociation kinetics, as well as dwell times of TFs in vitro, perhaps provide a guideline based on which TF-nucleosome dynamics can be broadly classified into at least two categories [80–82]. Occlusion of binding sites by nucleosomes generally lower TF association rates compared to free DNA [81, 82]. TFs take advantage of nucleosome unwrapping that transiently expose binding sites for chromatin access [29]. TFs such as Gal4 rapidly dissociate when bound within a nucleosome due to steric interference, resulting in orders-of-magnitude shorter dwell-times than when bound to free DNA [82]. In contrast, GRFs bound to nucleosomes have lower dissociation rates that compensate for their lower association rates compared to binding to free DNA [81]. Binding of GRFs Reb1 and Cbf1 to nucleosomal DNA traps the nucleosome in a partially-unwrapped state to remain bound for much longer durations than when bound to free DNA in vitro [81]. However, in vivo, GRF motifs within the FN promoters tend to be close to the center of the annotated FN sites [38]. GRFs may take advantage of thermal fluctuations such as nucleosome breathing [83] or sequence-dependent site exposure [84] to access sites close to the edges of nucleosomes, likely attributable to Reb1 binding to the edge of −1 nucleosomes [85], but it is apparently the action of RSC that renders DNA within the FN core accessible to the GRFs. The apparently equal affinity of GRFs for nucleosomal and non-nucleosomal DNA is a characteristic of metazoan pioneering TFs such as FoxA1/2, which is shown to bind nucleosomes stably in vitro (reviewed in [25]).
The organization of nucleosomes, TFs, and other chromatin proteins is disrupted during the catastrophic process of DNA replication. Nucleosomes and TF-binding reestablish de novo in the wake of replication forks as a result of a dynamic of competition between histones and TFs (Box 2) that is likely dependent on TF-binding affinity and concentration, as well as SWI/SNF remodeling [86].
Box2: Replication of promoter and enhancer landscapes.
Maintenance of lineage-specific transcriptional programs across mitotic divisions requires both replication of DNA and restoration of the parental chromatin landscape. Passage of the DNA-replication fork is a disruptive process that separates DNA strands, displacing histones, TFs, remodelers, and other DNA-bound protein factors. Chromatin maturation entails reestablishment of the steady-state chromatin landscape, including re-positioning of nucleosomes relative to DNA sequences, re-binding of TFs to their cognate sites, and restoration of histone-PTM patterns. During chromatin assembly in the wake of replication fork passage, parental (H3-H4)2 tetramers from displaced histone octamers are equally redistributed to both replicating strands (leading and lagging), orchestrated by replication-associated histone chaperones [98, 99]. H2A-H2B dimers are then added to complete the nucleosomes, and chromatin gaps are filled by new histone deposition. Orchestrated reassembly of the daughter chromatids ensures that that both daughter cells maintain the gene expression program of their lineage. This is achieved by equal redistribution of parental histone PTMs such that cognate modifiers can be recruited to restore modification patterns [18], and perhaps also by ensuring that displaced TFs have equal chances of rebinding to either replicating daughter chromatid. Exceptions are seen, as in the case of asymmetric stem cell division where the daughter stem cell – but not the differentiating daughter cell – predominantly retains parental histones [100].
Although histones redeposit immediately in the wake of replication fork, recent evidence reveals a surprising dynamic of competition between histones and TFs at newly-replicated regulatory elements. In mouse ESCs [101] and Drosophila S2 cells [86], nucleosomes pervasively fill up promoter and enhancer regions post-replication at the expense of TF-binding and steady-state nucleosome positions, and NDRs are not restored for at least 1–2 hours after replication fork passage. Consequently, newly-replicated metazoan chromatin is transcriptionally silent [101]. Restoration requires TF-binding, action of chromatin remodelers, and transcriptional restart. Nucleosome positions at super-enhancers containing clusters of TF-binding sites are restored much faster [101], perhaps due to a mass-action effect of multiple TFs dynamically competing against histones [27]. In sharp contrast, NDRs and TF-binding in yeast reestablishes within minutes of replication fork passage [102–104], evidently facilitated by Snf2 subfamily remodelers [86]. It is likely that RSC in yeast is rapidly recruited back to newly-replicated promoter CGCG-motifs in one of the two daughter chromatids, and may be shuffled between two sites held in proximity by the replication fork. Animal orthologs of RSC in contrast may depend on re-associating TFs for recruitment, thus accounting for the delay in chromatin maturation.
Concluding remarks
It is now well-established that nucleosome positioning, distinct types of histone isoforms (variants), and PTMs of histones (the roles of histone PTMs in nucleosome dynamics is reviewed in [87]) profoundly modulate functional chromatin states. Analogous to histone modification signatures, structural variations of dynamic nucleosomes provide physical signatures of active chromatin. The application of high-resolution genomics and sensitive biophysical assays to the study of nucleosome unwrapping has begun to reveal how the nucleosome barrier to transcriptional activation and elongation is modulated, resulting in a deeper mechanistic understanding of gene regulation. Genome-wide mapping of nucleosome unwrapping promises to have translational application, for example, as a surrogate for profiling gene expression in cell-free DNA [11].
How pioneer TFs invade the chromatin landscape is a much-debated active area of research [27, 88], and the interplay between pioneer factors and ATP-dependent remodeling in displacing nucleosomes is just beginning to emerge [74, 76]. Both remodelers and TFs appear to utilize nucleosome unwrapping to pioneer chromatin opening, although not necessarily in a mutually exclusive manner. Unwrapped nucleosomes at sites for pioneering likely poise genes for future context-specific activation in response to environmental signals and developmental cues [21]. While we have discussed the role of SWI/SNF family remodelers in this context, the potential role of other ATP-dependent remodelers in nucleosome unwrapping has not been thoroughly examined (see Outstanding questions). INO80 is a likely candidate as this remodeler also unwraps part of the nucleosome [89], can by itself generate NDRs and position −1 and +1 nucleosomes in reconstituted yeast chromatin [40], and chromatin accessibility at pluripotency factor binding sites is significantly decreased after INO80 depletion [90]. As we learn more about the basic mechanisms of nucleosome unwrapping involving chromatin remodelers and pioneer factors, we expect that this knowledge will extend to the study of developmental processes, including the action of repressors [91] and the involvement of histone modifications in the propagation of cellular memory [18].
OUTSTANDING QUESTIONS BOX.
Do unwrapped nucleosomes function as molecular rheostats for gene expression?
Is the absence of sequence-specific targeting of SWI/SNF in multicellular organisms a benefit for developmental regulation of gene expression?
Are unwrapped nucleosomal intermediates found at other dynamic regions of the genome such as around DNA replication origins?
How do TFs trap nucleosomes in a partially unwrapped state? Does that require interactions with nucleosomal histones as suggested for the metazoan FoxA factors?
Are chromatin remodelers other than SWI/SNF complexes involved in nucleosome unwrapping?
Are nucleosomes with particular histone variants or PTMs preferentially unwrapped?
HIGHLIGHTS.
High-resolution mapping uncovers dynamically unwrapped nucleosomes at regulatory regions.
Dynamic nucleosome unwrapping is a general feature of transcriptional regulation.
ATP-dependent chromatin remodelers catalyze nucleosome unwrapping that may facilitate transcription factor binding.
Unwrapped nucleosomal intermediates provide a novel physical signature of active chromatin.
Glossary
- DNA entry/exit
Sites at the periphery of a nucleosome ~73 bp from the dyad in either direction
- Dyad
The midpoint of DNA wrapped around a histone octamer. An imaginary pseudo-two-fold axis of symmetry (dyad axis) runs through the dyad, dividing the nucleosome into mirrored halves
- Fragile nucleosomes
Nucleosomes that are especially susceptible to MNase digestion
- GRFs
General Regulatory Factors are DNA-binding proteins known to regulate nucleosome positioning at yeast gene promoters, and transcription
- Insulator proteins
Sequence-specific DNA-binding proteins in multicellular eukaryotes, that block long-range intra-chromosomal interactions, such as enhancer-promoter interactions and the “spreading” of heterochromatin, thereby demarcating functionally and topologically distinct chromatin domains
- Linker histone
A basic protein that binds to linker DNA separating nucleosomes. Linker histones bind to the nucleosome core particle around the DNA entry and exit sites, reducing the conformational flexibility of nucleosomes and inducing chromatin compaction
- NFR and NDR
A region of DNA that is devoid of nucleosomes, such as promoter regions. Nucleosome-free regions (NFRs) are constitutive, often resulting from DNA sequences that are relatively refractory to nucleosome formation. Nucleosome-depleted regions (NDRs) are formed and regulated by the action of chromatin remodelers and TFs, and are often linked to the transcriptional activity of a gene
- Pioneer TFs
Transcription factors that can directly bind condensed chromatin to initiate chromatin opening
- Super-enhancers
Regulatory regions of the genome where multiple enhancers are clustered
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Davey CA et al. (2002) Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol 319 (5), 1097–113. [DOI] [PubMed] [Google Scholar]
- 2.Talbert PB et al. (2019) Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat Rev Genet 20 (5), 283–297. [DOI] [PubMed] [Google Scholar]
- 3.Deal RB et al. (2010) Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328 (5982), 1161–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teves SS and Henikoff S (2011) Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev 25 (22), 2387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deaton AM et al. (2016) Enhancer regions show high histone H3.3 turnover that changes during differentiation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraushaar DC et al. (2013) Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol 14 (10), R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svensson JP et al. (2015) A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin. Genome Res 25 (6), 872–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talbert PB and Henikoff S (2014) Environmental responses mediated by histone variants. Trends Cell Biol 24 (11), 642–50. [DOI] [PubMed] [Google Scholar]
- 9.Talbert PB and Henikoff S (2017) Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Biol 18 (2), 115–126. [DOI] [PubMed] [Google Scholar]
- 10.Brahma S and Henikoff S (2019) RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast. Mol Cell 73 (2), 238–249 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramachandran S et al. (2017) Transcription and Remodeling Produce Asymmetrically Unwrapped Nucleosomal Intermediates. Mol Cell 68 (6), 1038–1053 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran S et al. (2015) Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res 25 (3), 381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HS et al. (2014) Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 159 (6), 1377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai B et al. (2018) Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562 (7726), 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teif VB et al. (2012) Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol 19 (11), 1185–92. [DOI] [PubMed] [Google Scholar]
- 16.West JA et al. (2014) Nucleosomal occupancy changes locally over key regulatory regions during cell differentiation and reprogramming. Nat Commun 5, 4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W et al. (2016) Nucleosome positioning changes during human embryonic stem cell differentiation. Epigenetics 11 (6), 426–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinberg D and Vales LD (2018) Chromatin domains rich in inheritance. Science 361 (6397), 33–34. [DOI] [PubMed] [Google Scholar]
- 19.Chereji RV et al. (2016) Genome-wide profiling of nucleosome sensitivity and chromatin accessibility in Drosophila melanogaster. Nucleic Acids Res 44 (3), 1036–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller B et al. (2017) Widespread changes in nucleosome accessibility without changes in nucleosome occupancy during a rapid transcriptional induction. Genes Dev 31 (5), 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffers TE and Lieb JD (2017) Nucleosome fragility is associated with future transcriptional response to developmental cues and stress in C. elegans. Genome Res 27 (1), 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii H et al. (2015) MPE-seq, a new method for the genome-wide analysis of chromatin structure. Proc Natl Acad Sci U S A 112 (27), E3457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voong LN et al. (2016) Insights into Nucleosome Organization in Mouse Embryonic Stem Cells through Chemical Mapping. Cell 167 (6), 1555–1570 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwafuchi-Doi M et al. (2016) The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol Cell 62 (1), 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaret KS and Mango SE (2016) Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 37, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soufi A et al. (2015) Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161 (3), 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan C et al. (2018) Systematic Study of Nucleosome-Displacing Factors in Budding Yeast. Mol Cell 71 (2), 294–305 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polach KJ and Widom J (1995) Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol 254 (2), 130–49. [DOI] [PubMed] [Google Scholar]
- 29.Li G and Widom J (2004) Nucleosomes facilitate their own invasion. Nat Struct Mol Biol 11 (8), 763–9. [DOI] [PubMed] [Google Scholar]
- 30.Bernier M et al. (2015) Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nature Communications 6 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brehove M et al. (2015) Histone Core Phosphorylation Regulates DNA Accessibility. Journal of Biological Chemistry 290 (37), 22612–22621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teif VB (2016) Nucleosome positioning: resources and tools online. Brief Bioinform 17 (5), 745–57. [DOI] [PubMed] [Google Scholar]
- 33.Lieleg C et al. (2015) Nucleosome positioning in yeasts: methods, maps, and mechanisms. Chromosoma 124 (2), 131–51. [DOI] [PubMed] [Google Scholar]
- 34.Chereji RV and Clark DJ (2018) Major Determinants of Nucleosome Positioning. Biophys J 114 (10), 2279–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CK et al. (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36 (8), 900–5. [DOI] [PubMed] [Google Scholar]
- 36.Chereji RV et al. (2018) Precise genome-wide mapping of single nucleosomes and linkers in vivo. Genome Biol 19 (1), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal E and Widom J (2009) Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol 19 (1), 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubik S et al. (2015) Nucleosome Stability Distinguishes Two Different Promoter Types at All Protein-Coding Genes in Yeast. Mol Cell 60 (3), 422–34. [DOI] [PubMed] [Google Scholar]
- 39.Kubik S et al. (2018) Sequence-Directed Action of RSC Remodeler and General Regulatory Factors Modulates +1 Nucleosome Position to Facilitate Transcription. Mol Cell 71 (1), 89–102 e5. [DOI] [PubMed] [Google Scholar]
- 40.Krietenstein N et al. (2016) Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell 167 (3), 709–721 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chereji RV et al. (2011) Statistical mechanics of nucleosome ordering by chromatin-structure-induced two-body interactions. Phys Rev E Stat Nonlin Soft Matter Phys 83 (5 Pt 1), 050903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brahma S et al. (2018) The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling. Nat Commun 9 (1), 3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada K et al. (2011) Structure and mechanism of the chromatin remodelling factor ISW1a. Nature 472 (7344), 448–53. [DOI] [PubMed] [Google Scholar]
- 44.Lieleg C et al. (2015) Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density. Mol Cell Biol 35 (9), 1588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ocampo J et al. (2016) The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res 44 (10), 4625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beshnova DA et al. (2014) Regulation of the nucleosome repeat length in vivo by the DNA sequence, protein concentrations and long-range interactions. PLoS Comput Biol 10 (7), e1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiner A et al. (2010) High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20 (1), 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valouev A et al. (2011) Determinants of nucleosome organization in primary human cells. Nature 474 (7352), 516–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giaimo BD et al. (2019) The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 12 (1), 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ranjan A et al. (2013) Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154 (6), 1232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parnell TJ et al. (2015) The chromatin remodelers RSC and ISW1 display functional and chromatin-based promoter antagonism. Elife 4, e06073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaban Y et al. (2008) Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol 15 (12), 1272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramachandran S and Henikoff S (2016) Nucleosome dynamics during chromatin remodeling in vivo. Nucleus 7 (1), 20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kireeva ML et al. (2002) Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell 9 (3), 541–52. [DOI] [PubMed] [Google Scholar]
- 55.Tolstorukov MY et al. (2009) Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res 19 (6), 967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Floer M et al. (2010) A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141 (3), 407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henikoff JG et al. (2011) Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci U S A 108 (45), 18318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badis G et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32 (6), 878–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xi Y et al. (2011) Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res 21 (5), 718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorch Y et al. (2014) Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev 28 (22), 2492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chereji RV et al. (2017) MNase-Sensitive Complexes in Yeast: Nucleosomes and Non-histone Barriers. Mol Cell 65 (3), 565–577 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kubik S et al. (2017) A Reply to “MNase-Sensitive Complexes in Yeast: Nucleosomes and Non-histone Barriers,” by Chereji et al. Mol Cell 65 (3), 578–580. [DOI] [PubMed] [Google Scholar]
- 63.Arimura Y et al. (2012) Structural analysis of the hexasome, lacking one histone H2A/H2B dimer from the conventional nucleosome. Biochemistry 51 (15), 3302–9. [DOI] [PubMed] [Google Scholar]
- 64.Cakiroglu A et al. (2019) Genome-wide reconstitution of chromatin transactions reveals that RSC preferentially disrupts H2AZ-containing nucleosomes. Genome Res 29 (6), 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dion MF et al. (2007) Dynamics of replication-independent histone turnover in budding yeast. Science 315 (5817), 1405–8. [DOI] [PubMed] [Google Scholar]
- 66.Klein-Brill A et al. (2019) Dynamics of Chromatin and Transcription during Transient Depletion of the RSC Chromatin Remodeling Complex. Cell Rep 26 (1), 279–292 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Dieuleveult M et al. (2016) Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530 (7588), 113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hainer SJ et al. (2019) Profiling of Pluripotency Factors in Single Cells and Early Embryos. Cell 177 (5), 1319–1329 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.King HW and Klose RJ (2017) The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singhal N et al. (2010) Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell 141 (6), 943–55. [DOI] [PubMed] [Google Scholar]
- 71.Ho L and Crabtree GR (2010) Chromatin remodelling during development. Nature 463 (7280), 474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandler RL et al. (2013) ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol 33 (2), 265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vinayachandran V et al. (2018) Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swinstead EE et al. (2016) Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: A new perspective: Multiple transcription factors can effect chromatin pioneer functions through dynamic interactions with ATP-dependent chromatin remodeling factors. Bioessays 38 (11), 1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panday A and Grove A (2017) Yeast HMO1: Linker Histone Reinvented. Microbiol Mol Biol Rev 81 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson TA et al. (2018) Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res 46 (1), 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fei J et al. (2015) The prenucleosome, a stable conformational isomer of the nucleosome. Genes Dev 29 (24), 2563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kubik S et al. (2019) Opposing chromatin remodelers control transcription initiation frequency and start site selection. Nat Struct Mol Biol 26 (8), 744–754. [DOI] [PubMed] [Google Scholar]
- 79.Yen K et al. (2012) Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149 (7), 1461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donovan BT et al. (2019) Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J 38 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Donovan BT et al. (2019) Dissociation rate compensation mechanism for budding yeast pioneer transcription factors. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo Y et al. (2014) Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res 42 (5), 3017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winogradoff D and Aksimentiev A (2019) Molecular Mechanism of Spontaneous Nucleosome Unraveling. J Mol Biol 431 (2), 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo D et al. (2018) MNase, as a probe to study the sequence-dependent site exposures in the +1 nucleosomes of yeast. Nucleic Acids Res 46 (14), 7124–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gutin J et al. (2018) Fine-Resolution Mapping of TF Binding and Chromatin Interactions. Cell Rep 22 (10), 2797–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramachandran S and Henikoff S (2016) Transcriptional Regulators Compete with Nucleosomes Post-replication. Cell 165 (3), 580–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bowman GD and Poirier MG (2015) Post-translational modifications of histones that influence nucleosome dynamics. Chem Rev 115 (6), 2274–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meers MP et al. (2019) Pioneer Factor-Nucleosome Binding Events during Differentiation Are Motif Encoded. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brahma S et al. (2017) INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun 8, 15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang L et al. (2014) INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14 (5), 575–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu N et al. (2018) Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell 173 (2), 430–442 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clapier CR et al. (2017) Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 18 (7), 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paul S and Bartholomew B (2018) Regulation of ATP-dependent chromatin remodelers: accelerators/brakes, anchors and sensors. Biochem Soc Trans 46 (6), 1423–1430. [DOI] [PubMed] [Google Scholar]
- 94.Li M et al. (2015) Dynamic regulation of transcription factors by nucleosome remodeling. Elife 4, e06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stanton BZ et al. (2017) Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nature Genetics 49, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hota SK and Bruneau BG (2016) ATP-dependent chromatin remodeling during mammalian development. Development 143 (16), 2882–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bracken AP et al. (2019) Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petryk N et al. (2018) MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361 (6409), 1389–1392. [DOI] [PubMed] [Google Scholar]
- 99.Yu C et al. (2018) A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361 (6409), 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie J et al. (2017) Breaking Symmetry - Asymmetric Histone Inheritance in Stem Cells. Trends Cell Biol 27 (7), 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stewart-Morgan KR et al. (2019) Transcription Restart Establishes Chromatin Accessibility after DNA Replication. Mol Cell. [DOI] [PubMed] [Google Scholar]
- 102.Fennessy RT and Owen-Hughes T (2016) Establishment of a promoter-based chromatin architecture on recently replicated DNA can accommodate variable inter-nucleosome spacing. Nucleic Acids Res 44 (15), 7189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vasseur P et al. (2016) Dynamics of Nucleosome Positioning Maturation following Genomic Replication. Cell Rep 16 (10), 2651–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yadav T and Whitehouse I (2016) Replication-Coupled Nucleosome Assembly and Positioning by ATP-Dependent Chromatin-Remodeling Enzymes. Cell Rep 15 (4), 715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jain D et al. (2015) Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res 43 (14), 6959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Policastro RA and Zentner GE (2018) Enzymatic methods for genome-wide profiling of protein binding sites. Brief Funct Genomics 17 (2), 138–145. [DOI] [PubMed] [Google Scholar]
- 107.Teytelman L et al. (2013) Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci U S A 110 (46), 18602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kasinathan S et al. (2014) High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods 11 (2), 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rossi MJ et al. (2018) Simplified ChIP-exo assays. Nat Commun 9 (1), 2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Janssens DH et al. (2018) Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin 11 (1), 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Skene PJ et al. (2018) Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13 (5), 1006–1019. [DOI] [PubMed] [Google Scholar]


