Abstract
Although there is substantial evidence that type IV pili purified from diarrhea-associated Aeromonas species (designated Bfp for bundle-forming pilus) are intestinal colonization factors (S. M. Kirov, L. A. O'Donovan, and K. Sanderson, Infect. Immun. 67:5447–5454, 1999), nothing is known regarding the function of a second family of Aeromonas type IV pili (designated Tap for type IV Aeromonas pilus), identified following the cloning of a pilus biogenesis gene cluster tapABCD. Related pilus gene clusters are widely conserved among gram-negative bacteria, but their significance for virulence has been controversial. To investigate the role of Tap pili in Aeromonas pathogenesis, mutants of Aeromonas strains (a fish isolate of A. hydrophila and a human dysenteric isolate of A. veronii bv. sobria) were prepared by insertional inactivation of the tapA gene which encodes the type IV pilus subunit protein, TapA. Exotoxic activities were unaffected by the mutation in tapA. Inactivation of tapA had no effect on the bacterial adherence of these two isolates to HEp-2 cells. For the A. veronii bv. sobria isolate, adhesion to Henle 407 intestinal cells and to human intestinal tissue was also unaffected. There was no significant effect on the duration of colonization or incidence of diarrhea when the A. veronii bv. sobria strain was tested in the removable intestinal tie adult rabbit diarrhea model or on its ability to colonize infant mice. Evidence was obtained that demonstrated that TapA was expressed by both Aeromonas species and was present on the cell surface, although if assembled into pili this pilus type appears to be an uncommon one under standard bacterial growth conditions. Further studies into factors which may influence Tap expression are required, but the present study suggests that Tap pili may not be as significant as Bfp pili for Aeromonas intestinal colonization.
Aeromonas bacteria (aeromonads) are ubiquitous water-borne organisms that are also found in many foods. Strains of some Aeromonas species (primarily A. hydrophila HG1, A. veronii bv. sobria HG8/10, and A. caviae HG4) cause human gastroenteritis (“summer diarrhea”), particularly in children. They also cause more serious infections, such as septicemia and meningitis, in immunocompromised individuals (21, 25). Recently, aeromonads have been linked to cases of hemolytic-uremic syndrome (7, 9). Disease-associated strains possess a number of significant virulence determinants, including the ability to produce type IV pilus adhesins (14–16, 20, 30, 31) and the pore-forming toxin “aerolysin” (18, 50). Many Aeromonas strains grow at refrigeration temperatures, increasing concern about food-borne transmission (25, 26). Yet relatively little is known of the pathogenic mechanisms of Aeromonas species, and it is not possible to identify virulent strains definitively.
Colonization of the intestinal tract is likely to be a critical step in the disease process. Type IV pilus adhesins are essential for the colonization of the intestine by enteropathogens, such as Vibrio cholerae, enterotoxigenic Escherichia coli, and enteropathogenic E. coli (47). Our past studies have shown that gastroenteritis-associated Aeromonas species have the potential to express at least two distinct families of type IV pili (5). The predominant pilus type expressed on fecal isolates of A. veronii bv. sobria and A. caviae grown under standard in vitro conditions is the bundle-forming pilus (Bfp) (30, 31). Bfp pili have also been isolated from a strain of A. hydrophila, but fecal isolates of this species are often heavily piliated and express numerous type I pili (17). Bfp pili exhibit N-terminal sequence homology with the mannose-sensitive hemagglutinin pilus of V. cholerae. They have been purified from all Aeromonas species associated with diarrhea, but as yet this pilus type has not been genetically characterized (29). A second type IV Aeromonas pilus (Tap) was identified following the cloning of a biogenesis gene cluster (tapABCD) from a strain of A. hydrophila (strain Ah65) (42). Subsequent cloning of the tap cluster from a Bfp-positive strain of A. veronii bv. sobria (strain BC88) proved definitively that this cluster encoded a pilus type distinct from the purified Bfp pilus family. Tap pili differ from Bfp pili in their N-terminal sequences and molecular weights. They exhibit highest homology with the type IV pili of Pseudomonas aeruginosa and pathogenic Neisseria species (5). The tapA gene encodes the subunit protein. The tapB and tapC genes are probably involved in pilus biogenesis (49). The protein encoded by tapD is a type IV prepilin peptidase/N-methyltransferase which is responsible for the processing of several components of the general secretion pathway which mediate secretion of extracellular proteins, including aerolysin. TapD cleaves the 6-amino-acid leader peptide from prepilin and catalyzes the methylation of the N-terminal residue (19, 42, 47). Similar pilus gene clusters have been identified in V. cholerae (pil cluster), P. aeruginosa, and several other gram-negative bacteria. For many of these organisms, the encoded type IV pili have been shown to be important virulence factors, but for V. cholerae epithelial cell adherence or intestinal colonization functions could not be attributed to the pilus structure encoded by the pil cluster (10).
While there is increasing evidence that the Aeromonas Bfp pili are intestinal colonization factors (29), the significance of Tap pili for Aeromonas virulence is unknown. The aim of this study was to investigate the role of Tap pili in the pathogenesis of Aeromonas infection. The adhesive abilities of isogenic tapA mutant strains of A. hydrophila Ah65 and the dysenteric isolate of A. veronii bv. sobria, strain BC88, to HEp-2 cells were compared with those of the respective wild-type strains. The latter strain and its tapA mutant were also compared for their ability to adhere to intestinal cells and tissue and to produce diarrhea and/or colonize animals (rabbits and infant mice). Wild-type and selected tapA and tapD mutant strains were used to examine TapA expression using polyclonal antisera raised against His-Tag–TapA fusion proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Additional details of the Aeromonas strains examined are given in the Results section. Aeromonas strains were grown (37°C, 18 to 24 h) from storage on tryptone soy agar (TSA) supplemented with 6.0 g of yeast extract L21 (TSAY; Oxoid, Basingstoke, United Kingdom) per liter. For genetic manipulations, Aeromonas veronii bv. sobria strains were grown in brain heart infusion broth (BHIB; Oxoid), tryptone soy broth (TSB; Oxoid), or tryptone soy broth containing yeast extract (TSBY; Oxoid), as described above. A. hydrophila Ah65N was routinely grown at 22°C in TSB or BHIB. Transconjugants were grown on nutrient broth agar (NBA; Oxoid) or brain heart infusion agar (BHIA; Oxoid). E. coli was grown in Luria-Bertani (LB) medium (45). For Escherichia coli, the following antibiotic concentrations (in micrograms/milliliter) were used: spectinomycin, 50; carbenicillin, 100; ampicillin, 150; and chloramphenicol, 30. For Aeromonas, the antibiotic concentrations (in micrograms per milliliter) were: spectinomycin, 50; ampicillin, 150; chloramphenicol, 2.5; and nalidixic acid, 5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a final concentration of 1 mM. For exotoxin assays, Aeromonas spp. were grown in TSBY, at 37°C, for 24 h with shaking. For rabbit pathogenicity and mouse colonization experiments, Aeromonas wild-type and tapA mutant strains, and the E. coli K-12 surgical control strain EC101, were grown from stored cultures on BHIA at 35°C for 24 h. Log-phase cultures were then prepared in BHIB at 35°C for 18 h (static).
TABLE 1.
Bacterial strains and plasmids used or constructed in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| A. hydrophila | ||
| Ah65N | Wild type, nalidixic acid resistant | 42 |
| Ah65N-D5 | Ah65N tapD mutant, chloramphenicol resistant | 42 |
| Ah65N-AΩ18 | Ah65N tapA mutant, streptomycin and spectinomycin resistant | This study |
| Ah65N-DΩ33.2 | Ah65N tapD mutant, streptomycin and spectinomycin resistant | This study |
| A. veronii bv. sobria | ||
| BC88 | Wild type, dysenteric isolate | 30 |
| BC88tapAΩ | BC88 tapA mutant, streptomycin and spectinomycin resistant | This study |
| E. coli | ||
| DH5α | F−supE44 lacU169 (φlacZΔM15)hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | BRL |
| S17-1λpir | supE44 hsdR endA1 recA thi pro RP4-2-Tc::Mu-kan::Tn7 (λpir) | 46 |
| BL21(DE3) | F−ompT hsdSBdcm gal (DE3) | Novagen |
| EC101 | Wild-type K-12 strain, avirulent laboratory strain used as a negative control in rabbit pathogenicity experiments | 1, 2 |
| Plasmids | ||
| Cloning vectors | ||
| pBluescript II SK(−) | Ampicillin-resistant cloning vector | Stratagene |
| pET15b | Ampicillin-resistant His-Tag cloning vector | Novagen |
| pJQ200KS/SK | Gentamicin-resistant broad-host-range sacB suicide vector | 43 |
| pMMB67EH/HE.cam | Chloramphenicol- and ampicillin-resistant broad-host-range vectors, lacIq/tac promoter | 22 |
| pEP185.2 | Chloramphenicol-resistant suicide vector, λpir dependent | 24 |
| Recombinant plasmids | ||
| pUC19Ω | 2.0-kb SmaI fragment (Ω) from pHP45 cloned into pUC19 (Specr) | 33 |
| pCP1065 | 1.0-kb BamHI fragment from Ah65N in pBluescript II SK(−) (tapA) | 42 |
| pCP1140 | 1.2-kb NruI fragment in pMMB67EH.cam (tapD) | 42 |
| pCP1147 | 3.3-kb HindIII-EcoRI fragment in pBluescript II SK(−) (tapDΩ) | 42 |
| pCP1178 | BamHI 10-mer in NruI site of tapA in pCP1065 | This study |
| pCP1179 | 1.0-kb BamHI fragment from pCP1065 cloned into BamHI site of pBluescript II SK(−) containing a blunted PstI site (tapA) | This study |
| pCP1180 | Ω interposon from pUC19Ω in PstI site of tapA in pCP1179 | This study |
| pCP1182 | 3.1-kb SalI-XbaI fragment from pCP1180 in pJQ200KS (tapAΩ) | This study |
| pCP1183 | 0.7-kb BamHI fragment from pCP1178 in pET15b (truncated tapA) | This study |
| pCP1190 | 3.3-kb HindIII-XbaI fragment from pCP1147 in pJQ200SK (tapDΩ) | This study |
| pCP1194 | 0.65-kb KpnI fragment from pCP1065 in pMMB67HE.cam (tapA) | This study |
| pTB012 | 1.0-kb BamHI fragment (tapA) from BC88 cloned into pGEM-3Zf(+) | 5 |
| pTB028 | 0.45-kb PCR product of BC88 cloned into pET15b (TapA minus the leader sequence) | This study |
| pMS012 | 1.0-kb BamHI fragment (tapA) from pTB012 cloned in BglII site of pEP185.2 | This study |
| pMS012Ω | 2.1-kb SmaI fragment (Ω) from pUC19Ω cloned into the blunted PstI site of pMS012 | This study |
DNA preparation and manipulations.
For small-scale plasmid preparations, E. coli DH5α served as the host strain, and the alkaline lysis procedure was followed (6). Restriction endonuclease digestion, ligation, and transformation and DNA electrophoresis were performed as described by Sambrook et al. (45). Plasmids were introduced from E. coli S17-1λpir into A. hydrophila and A. veronii bv. sobria by conjugation.
Construction of tap mutant strains.
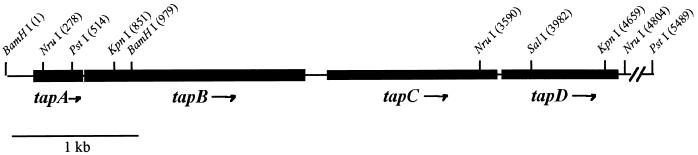
A map of the tap gene cluster of A. hydrophila Ah65 is shown in Fig. 1. Ah65 strains with mutations in tapA and tapD (Ah65N-AΩ18 and Ah65N-DΩ33.2, respectively) were constructed. These were prepared by allelic exchange of the wild-type copies of these genes with Ω interposon-disrupted copies encoding spectinomycin-streptomycin resistance. To create the tapA mutation, the Ω interposon from pUC19Ω was cloned as a SmaI fragment into a blunted PstI site within tapA resulting in pCP1180. A 3.1-kb SalI-XbaI fragment [both sites originating from the pBluescript II SK(−) polylinker] from pCP1180 carrying the tapAΩ gene was then inserted into pJQ200KS digested with the same enzymes, generating pCP1182.
FIG. 1.
The tap gene cluster of A. hydrophila Ah65 is encoded on a 5.5-kb DNA fragment as shown. Arrows indicate the direction of transcription of the open reading frames. Restriction sites of interest are indicated.
To create the tapD mutation, the Ω interposon was cloned as a SmaI fragment into an end-filled ClaI site within the tapD gene resulting in pCP1147 (42). A 3.3-kb end-filled HindIII-XbaI fragment [both sites originating from the pBluescript II SK(−) polylinker] from pCP1147 carrying the tapDΩ gene was inserted into pJQ200KS digested with SmaI and XbaI, generating pCP1190. To construct the tapA and tapD mutant strains, pCP1182 and pCP1190 were transformed into E. coli S17-1λpir, then introduced into A. hydrophila Ah65N by conjugation. Transconjugants were plated on NBA containing nalidixic acid, spectinomycin, and 5% (wt/vol) sucrose to induce expression of the lethal sacB gene product (11). Growth in the presence of sucrose and spectinomycin requires recombination of the Ω-carrying gene into the chromosome with subsequent loss of plasmid sequences. Gentamicin-sensitive candidates were examined by Southern blot analysis (Genius System; Boehringer Mannheim, Ind.) using appropriate digoxigenin-labeled probes: a 1.0-kb BamHI tapA fragment or a 0.7-kb internal SalI-KpnI fragment of tapD (Fig. 1). Construction of mutant Ah65N-D5 has been described previously (42). Ah65N-D5 and Ah65N-DΩ33.2 were indistinguishable phenotypically (no type IV peptidase activity, nonhemolytic on blood agar, and aerolysin in the periplasmic space).
The allelic exchange method was also used to prepare the tapA mutant strain of A. veronii bv. sobria strain BC88. In brief, it was constructed by inserting a 1.0-kb BamHI fragment (tapA) from pTB012 into the BglII site of the suicide vector, pEP185.2, resulting in pMS012. The Ω interposon (from plasmid pUC19Ω) was then inserted as a 2.1-kb SmaI fragment into the blunted PstI site of tapA, producing pMS012Ω. Plasmid pMS012Ω was transformed into E. coli S17-1λpir and mobilized into A. veronii bv. sobria strain BC88 by conjugation. (Plasmids derived from the suicide vector, pEP185.2, require the λpir gene product for replication, so selection in the absence of λpir requires recombination of the selectable marker into the chromosome.) Transconjugants were selected on BHIA containing ampicillin and spectinomycin. Identification of double recombinants, where the wild-type copy of tapA was completely replaced with tapAΩ, was determined by Southern hybridization of chromosomal DNA from potential mutants using a digoxigenin-labeled 1.0-kb BamHI fragment containing tapA as a probe.
In vitro characterization of the tapA mutant strain of A. veronii bv. sobria BC88.
Bacterium-free broth supernatants from the wild-type and tapA mutant strains of A. veronii bv. sobria strain BC88 were examined for hemolytic activity against rabbit red blood cells, cytotoxic activity for Vero cells, and enterotoxic activity in suckling mice as described elsewhere (26, 32). The hemolysin titer was recorded as the last broth dilution showing 50% hemolysis, while the cytotoxin titer was recorded as the last broth dilution causing ≥50% of the cells to round up or die.
Construction of expression plasmids.
To construct a tapA-overexpressing plasmid of A. hydrophila strain Ah65, a KpnI site was created upstream of the coding region (GGAACC changed to GGTACC at positions 202 to 207 of the tapABCD sequence; EMBL-GenBank-DDBJ Data Libraries accession number U20255) by PCR using Kpn-A (5′-CAC TTC CCA GGT ACC AAG GAC AAA A-3′) and T3 (5′-ATT AAC CCT CAC TAA AG-3′) as primers and pCP1065 as a template. The 0.79-kb product was digested with KpnI, producing a 0.65-kb fragment that was inserted into pMMB67HE.cam to generate pCP1194. To construct a His-Tag–tapA fusion plasmid, a BamHI site was introduced into pCP1065 by inserting a BamHI linker (10-mer) into the NruI site of tapA, generating pCP1178. Next, the 0.7-kb BamHI fragment from pCP1178, containing a truncated tapA gene (corresponding to amino acids 11 to 136 of the mature pilin), was inserted in frame into the His-Tag cloning vector, pET15b (Novagen), resulting in pCP1183.
For A. veronii bv. sobria strain BC88 the tapA open reading frame was amplified from pTB012 using primers PO13 (5′-TGA AGA AAC AAC ATA TGT TTT TAC CCT TAT TG-3′) and PO14 (5′-CTA TTA GAT CTA GAG GTC ATT ATT TGG-3′). PO13 was designed to incorporate a NdeI site after the TapA leader sequence, so that this sequence could be removed following digestion with NdeI. The 0.45-kb PCR product was digested with NdeI and BglII and cloned into the NdeI and BamHI sites of pET15b, resulting in pTB028.
Construction of a tapD-overexpressing plasmid has been described elsewhere (42). In brief, the tapD gene was cloned as a 1.2-kb NruI fragment into SmaI-digested pMMB67EH.cam (22), resulting in pCP1140, in which the inducible transcription of tapD is under the control of the tac promoter.
Purification of His-Tag–TapA fusion proteins and production of anti-TapA antisera.
To prepare antisera against TapA from A. hydrophila Ah65 and A. veronii bv. sobria strain BC88, overnight cultures of E. coli BL21(DE3) harboring pCP1183 or pTB028 were inoculated (1:100) into 50 ml of LB broth containing carbenicillin and grown at 37°C. When the cultures reached an optical density at 600 nm (OD600) of ∼0.6, IPTG was added, and the cells were grown for 5 h at 37°C with vigorous shaking. Preparation of lysates and purification of the ∼15-kDa His-Tag–TapA fusion proteins were carried out according to the manufacturer's protocol (Xpress System; Invitrogen). Due to the insolubility of the A. veronii bv. sobria His-Tag–TapA fusion, it was necessary to purify this protein by successive solubilization in urea (44). The TapA protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gel slice containing it was emulsified in 1 ml of complete Freund's adjuvant. Antisera were prepared either in New Zealand White rabbits by R & R Rabbitry Research Development (for Ah65 TapA) or in New Zealand White rabbits held at the University of Tasmania Animal Facility (for BC88 TapA) by comparable methods. Briefly, rabbits (n = 3) were administered ∼100 μg of His-Tag–TapA protein subcutaneously into five sites. Booster injections of the SDS-PAGE His-Tag–TapA protein band in incomplete Freund's adjuvant were given at days 21, 35, and 49. Trial bleeds were collected on days 28, 42, and 56, and a volume bleed was done at day 63. Serum from the volume bleed of the rabbit showing the highest reactivity was used in this study. In both cases, antisera were rendered specific for TapA by absorption against an acetone powder of Ah65-AΩ18 or BC88tapAΩ.
Cell fractionation and Western blotting.
Whole-cell lysate samples were prepared by mixing 10 μl of an overnight TSB culture with 2.5 μl of 5× sample buffer (0.3125 M Tris-HCl, pH 6.8; 50% [vol/vol] glycerol; 10% [wt/vol] SDS; 0.25% [vol/vol] 2-mercaptoethanol; 0.5% [wt/vol] bromophenol blue). For cell fractionation experiments, periplasmic contents were extracted by osmotic shock (52). Briefly, overnight cultures grown under the appropriate conditions were diluted 1:20 in TSB and incubated until the cultures reached an OD600 of ∼2. Bacteria from a 2.5-ml sample were recovered by centrifugation (15,000 × g, 5 min). They were gently resuspended in 1 ml of ice-cold 33 mM Tris-HCl (pH 8)–1 mM EDTA–0.5 M sucrose and held on ice for 10 min. The cells were pelleted at low speed (6,000 × g, 2 min) and then gently resuspended in 1 ml of ice-cold 0.5 mM MgCl2 and held on ice for 10 min. After centrifugation (4,000 × g, 2 min), the periplasmic contents were recovered in the supernatant. The cytoplasmic contents and membrane fractions were then extracted from the pellet (35). In brief, the pellet was resuspended in 150 μl of 100 mM Tris-HCl (pH 8)–0.5 mM EDTA–0.5 M sucrose containing 15 μl of a 2-mg/ml mixture of lysozyme and 150 μl of cold distilled H2O and held on ice for 5 min before centrifugation (15,000 × g, 3 min, 4°C). The pellet was subsequently resuspended in 1 ml of 10 mM Tris-HCl (pH 8) and subjected to three freeze-thaw cycles in liquid nitrogen, after which 33 μl of 1 M MgCl2 containing 10 μl of a 1-mg/ml mixture of DNase I was added. After centrifugation (15,000 × g, 25 min, 4°C), the cytoplasmic contents were recovered in the supernatant, and the pellet contained the membrane fractions. For each of the fractionated samples, 20-μl aliquots were mixed with 5 μl of 5× sample buffer. SDS-PAGE was performed as described by Laemmli using discontinuous 15 or 18.5% acrylamide gels (34). The proteins were transferred to nitrocellulose (48), incubated with anti-TapA polyclonal antiserum (see above), and visualized with goat anti-rabbit alkaline phosphatase conjugate (Promega).
Electron microscopy.
Bacterial cells were negatively stained with either 2% phosphotungstic acid (pH 7.2) on parlodion-coated grids or 1% uranyl acetate on Formvar-coated copper grids (28, 30). They were examined with a JEOL 100-B transmission electron microscope operated at 60 kV or a Philips 410 electron microscope at 80 kV. For immune electron microscopy (IEM), bacteria on Formvar-coated grids were washed briefly in a drop of TTB buffer (20 mM Tris-HCl, 25 mM NaCl, 0.1% [wt/vol] bovine serum albumin, 0.05% [vol/vol] Tween 20; pH 8.2) and floated on a drop of 5% bovine serum albumin in TTB for 15 min. The grids were then washed three times in TTB and reacted with 10-fold dilutions of TapA antiserum (1:10 to 1:1,000) for 60 min. Grids were again washed (three times in TTB). They were then exposed (60 min) to goat anti-rabbit immunoglobulin G conjugated with 10-nm gold particles (BioCell, Cardiff, United Kingdom) diluted 1:50 in TTB. The grids were subsequently washed and negatively stained. A 1:100 dilution of Bfp antiserum served as a positive control for the IEM (30).
Purification of pili.
A. hydrophila strains containing plasmids of interest were streaked from −80°C glycerol stocks onto TSA containing appropriate antibiotics and grown at 22°C overnight. For each strain, a single colony was inoculated into 10 ml of TSB containing antibiotics and incubated overnight at 22°C without shaking. Eight 1-liter flasks each containing 500 ml of TSB plus antibiotics were inoculated with 1 ml from the 10-ml overnight culture and then incubated at 22°C for 72 h (static). After the bacteria were harvested, the pili were sheared and purified according to a procedure described elsewhere (30). For Western blotting of pilus preparations, samples containing 1 μg of total protein in 10 μl of 0.5 M Tris-HCl (pH 7.5) were mixed with 2.5 μl of 5× sample buffer.
Adhesion assays.
Adhesion of bacteria to HEp-2 epithelial cells, Henle 407 intestinal cells, and fresh human intestinal tissue was assessed by bright-field microscopy (8, 27, 29). In brief, 1-ml aliquots of 5 × 106 CFU were inoculated onto the semiconfluent coverslip cultures of the cell lines grown in Eagle minimal essential medium containing 5 to 10% fetal calf serum (MEM-FCS) or onto fresh samples of intestinal tissue in MEM-FCS in 24-well tissue culture plates. After incubation (60 min, 37°C, 5% CO2), nonadherent bacteria were removed by washing (four times in phosphate-buffered saline [PBS]). The cell monolayers were fixed with 3:1 methanolacetic acid (1 ml, 5 min), stained with May-Grünwald and Giemsa stains (BDH, Poole, United Kingdom), and mounted for counting. At least three coverslip cultures were assayed for each strain in each experiment. Intestinal specimens were fixed in formalin after washing and then embedded in paraffin and sectioned. Sections (∼10 μm) on glass slides were deparaffinized, hydrated, and stained with hematoxylin-eosin for light microscopic examination (29).
For A. hydrophila Ah65, the adherence ability was also assessed by quantitative bacterial plate counts (40). In brief, the monolayers were incubated with bacteria as described above and then washed four times with PBS. Cell-associated bacteria were released by treatment with 0.1% Triton X-100, and plate counts were performed on TSA. The percentage of bacteria recovered relative to the initial inoculum was then determined.
Removable intestinal tie adult rabbit diarrhea (RITARD) model.
Wild-type and tapA mutant Aeromonas strains were tested in New Zealand White rabbits (1,250 to 1,650 g; 7 to 9 weeks of age; 3 to 4 weeks postweaning) according to the protocol of Pazzaglia et al. (41). Each strain was tested in nine rabbits. Five control rabbits received E. coli EC101. Each rabbit received 1010 CFU in 10 ml of BHIB injected into the jejunum close to the ligament of Treitz. Animals were monitored for 7 days for diarrheal symptoms and shedding of Aeromonas organisms in feces. The animals were sacrificed on day 8 postchallenge.
Infant mouse colonization.
BALB/c mice, obtained from a breeding colony held at the University of Tasmania, were inoculated orally with bacteria under test conditions according to the protocol of Attridge et al. (3). Log-phase cultures of the wild-type and mutant Aeromonas strains were diluted to obtain a culture containing 2 × 108 CFU per ml of each strain, and 5 μl of blue food coloring was added to facilitate the monitoring of the inoculation procedure. Three- to five-day-old infant mice were taken from their mothers 4 h prior to oral infection. They were inoculated by gastric lavage with 50 μl of bacterial suspension (∼107 CFU per mouse) and held at 25°C for 24 h, after which time they were sacrificed and their intestines were removed. The intestines were homogenized in 5 ml of PBS, and Aeromonas bacteria were quantitated by plate counts of serial dilutions of these homogenates on TSAY.
For competition assays, log-phase wild-type and mutant cultures were diluted to ∼2 × 107 CFU per ml, and a suspension containing equal volumes of the wild-type and mutant strains was prepared (107 CFU of each strain). Each mouse received 50 μl (∼106 organisms in total) of this suspension by intragastric lavage, as described above. The precise input ratio was determined retrospectively by plating dilutions of the suspension on TSAY (total bacteria) and on TSAY containing 50 μg of spectinomycin (mutant bacteria) per ml. Mice were sacrificed after 24 h, intestinal homogenates prepared, and Aeromonas numbers were quantitated on selective media as described above. The colonization index was calculated as the ratio of wild-type to mutant colonies following 24 h of incubation.
Statistical analysis.
The differences in adherence to cell lines by wild-type and tapA mutant strains and between groups of mice inoculated with these strains were analyzed by the Student's t test using Microsoft Excel software.
RESULTS
Construction of mutant strains.
Initially, tapA and tapD mutants (Ah65N-AΩ18 and Ah65N-DΩ33.2, respectively) of A. hydrophila Ah65N were constructed as described in Materials and Methods. Strain Ah65 was originally isolated from rainbow trout (Salmo gairdneri) and called “A. hydrophila” (36). Its 16S ribosomal DNA sequences, however, were identical to those of the HG2 definition strain (A. M. Carnahan, personal communication). Ribotyping was unable to identify it definitively but putatively classified it as belonging to HG3 (M. Altwegg, personal communication). The strain was relatively poorly adherent to epithelial and intestinal cell lines and thus proved a poor choice for in vivo experiments designed to investigate the role of Tap in intestinal colonization and virulence. Hence, a tapA mutant strain of a dysenteric isolate of A. veronii bv. sobria (strain BC88) was subsequently constructed for such functional investigations. Strain BC88 was originally isolated (in 1983) at the Princess Margaret Hospital, Perth, Western Australia, from the stool of a child with bloody diarrhea. The virulence-associated factors of this strain included the ability to produce enterotoxin (positive suckling mouse assay), cytotoxin (Vero cell assay), and hemolysin (titer of >512 versus rabbit erythrocytes). It was also able to invade HEp-2 cells and was highly adhesive to epithelial and intestinal cell lines and intestinal tissue. In addition, it was the strain from which we had purified and characterized the type IV bundle-forming pilus colonization factor (29, 30).
Exotoxic activities of A. veronii bv. sobria strain BC88 and A. hydrophila tapA mutant strains.
Mutation in tapA did not affect the ability of strain BC88 to produce exotoxic activities. The hemolytic titer of the mutant was 1,024, as was the titer of the wild-type strain (200 μl of broth supernatant in the first well, doubling dilutions in PBS; 37°C, 1 h; 4°C, 1 h). Cytotoxic titers of both strains for Vero cells were identical at 128 (50 μl of broth supernatant in 150 μl of MEM in the first well; doubling dilutions in MEM; 40 min, 37°C, 5% CO2). Supernatants (100 μl) from both strains also gave positive results (intestinal-weight/remaining-body-weight ratios of 0.087 and 0.085 for the wild-type and mutant strains, respectively) in the suckling mouse enterotoxin assay (eight mice per group). The hemolytic titers in the tapA mutant of A. hydrophila Ah65 were not significantly different from those of the wild-type strain. However, mutation in tapD decreased the hemolytic activity titer in broth supernatants from 128 in the wild-type to 0 (42).
Effect of tapA mutation on epithelial and intestinal cell adhesion.
A. hydrophila Ah65 was relatively poorly adherent to HEp-2 cells (<8 bacteria per cell). Studies of quantitative counts of bacterial adherence to HEp-2 cells, however, showed no difference between numbers of wild-type (Ah65N) or tapA mutant (Ah65N-AΩ18) bacteria recovered from cells. The percentages (means ± standard deviations) of cell-associated bacteria were 17.6 ± 2.4 and 19.4 ± 1.8 for the wild-type and tapA mutant strains, respectively.
Mutation of tapA also had no effect on the ability of A. veronii bv. sobria strain BC88 to adhere to epithelial and intestinal cell lines or to fresh human intestinal tissue. Values from the cell line adhesion assays for this strain are summarized as follows. Levels of adhesion to HEp-2 cells and Henle 407 cells were 10.1 ± 0.7 and 13.3 ± 2.1 bacteria/cell, respectively, for the wild-type strain and 11.1 ± 1.6 (P = 0.759, not significant, Student's t test) and 16.2 ± 1.3 (P = 0.129, not significant, Student's t test) bacteria/cell, respectively, for the tapAΩ strain. Each value represents the mean number of bacteria per cell of three coverslip cultures ± the standard deviation. The adhesion to fresh intestinal tissue was not quantitated, but light microscopic examination showed both strains adhered well.
Virulence of the tapA mutant of A. veronii biovar sobria BC88 in the rabbit (RITARD) model.
Wild-type and tapA mutant strains of A. veronii bv. sobria BC88 were compared for virulence in the RITARD model. The results are summarized in Table 2.
TABLE 2.
Comparison of A. veronii bv. sobria strain BC88 wild-type and tapA mutant strains in the RITARD model
| Strain | Total no. of rabbits | No. of animals shedding Aeromonas organisms | No. of animals with diarrhea |
|---|---|---|---|
| A. veronii bv. sobria wild type | 9 | 7a | 6 |
| A. veronii bv. sobria tapAΩ mutant | 9 | 7b | 4 |
| E. coli EC101 | 5 (12)c | 0 (0) |
Three rabbits shed for 1 day, one shed for 2 days, and three shed for 3 days; one rabbit (no diarrhea) died on day 3.
One rabbit shed for 1 day, three shed for 2 days, two shed for 3 days, and one shed for 5 days; one rabbit (mild diarrhea) died on day 1.
Five rabbits were included as controls in this experiment; 12 control animals were from other experiments.
Both wild-type and mutant strains of Aeromonas were shed for periods ranging from 1 to 5 days. In only one rabbit (inoculated with the wild-type strain) were aeromonads recovered at sacrifice by day 8. None of five surgical control rabbits that received E. coli EC101 developed diarrheal symptoms. Moreover, review of the incidence of transient diarrhea in such control animals (n = 12) from previous RITARD experiments showed that none of these animals had developed diarrhea. Ten animals that received the Aeromonas strains, however, developed mild diarrhea 24 to 48 h after bacterial inoculation. Six had received the wild-type strain, and four had received the tapA mutant strain. The diarrhea was transient and resolved after 1 day. One animal in each of the test groups died. Mutation in tapA, therefore, had no major effect on the virulence of strain BC88 for rabbits.
Effect of tapA mutation on the intestinal colonization of infant mice by A. veronii bv. sobria BC88.
Wild-type and tapA mutant strains were also compared for their ability to colonize the intestines of infant mice. Strains were administered alone or in competition experiments in which both strains were administered to the same animal (Fig. 2). In the former case, the colonization of infant mice by A. veronii bv. sobria strain BC88 wild-type and tapA mutant strains yielded the following recovery results: wild-type strain (inoculum of 1.4 × 107), 3.1 × 107 ± 4.4 × 107 CFU; and tapAΩ mutant strain (inoculum of 2.1 × 107), 2.6 × 107 ± 2.0 × 107 CFU. The inoculum in each case was determined retrospectively by use of plate counts. The recovery was calculated as the total number of bacteria (CFU ± the standard deviation) recovered from intestinal tissue after 24 h (six mice per group).
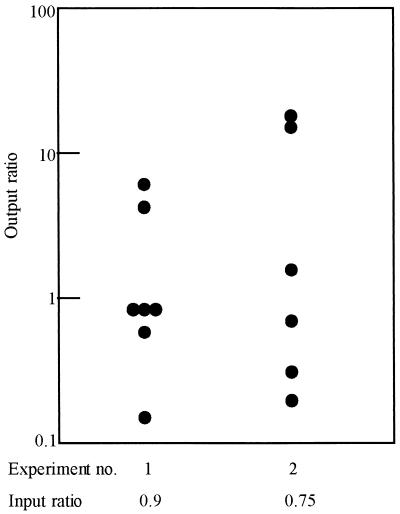
FIG. 2.
Comparative ability of A. veronii bv. sobria strain BC88 wild-type and tapA mutant strains to colonize infant mice. Mice were fed mixed suspensions of wild-type and mutant bacteria. Input ratios are indicated below each test group (two experiments). Each point represents the wild-type/mutant output ratio of bacteria recovered from the intestines of individual mice (seven and six mice for experiments 1 and 2, respectively).
When administered alone, the tapA mutant strain (BC88tapAΩ) was able to colonize infant mice at a level comparable to the level of colonization by the wild-type strain. Both strains exhibited some variability in their levels of colonization in different mice. However, for all mice, each strain was recovered at a level ranging from 4 × 106 to 9.6 × 107 CFU.
In competitive colonization experiments, the wild-type and mutant strains were administered together in a 1:1 ratio to individual mice. As shown in Fig. 2, in two experiments the output ratio of wild-type and mutant bacteria recovered from each mouse was comparable to the input ratio, indicating that the loss of TapA did not affect colonizing ability.
Examination of TapA expression.
To evaluate the significance of these functional investigations, it was important to determine whether TapA was expressed and able to be assembled on the bacterial cell surface. To this end, antiserum to TapA of A. hydrophila Ah65N and A. veronii bv. sobria strain BC88 were prepared using His-Tag–TapA fusion proteins as described in Materials and Methods. The TapA proteins of these two strains are antigenically distinct. Hence, it was necessary to prepare antiserum to TapA of each strain. For A. hydrophila strain Ah65N, anti-TapA serum was used to examine the wild type and the tapA and tapD mutants and their respective complemented mutant strains for Tap pilin expression by using Western blotting. More limited experiments were done with A. veronii bv. sobria strain BC88. A tapA complemented strain of this organism was not examined, nor was a TapD mutant. However, bacterial shearing experiments and IEM were performed with both strains to investigate whether Tap pili were assembled on the cell surface.
Figure 3 shows the results for A. hydrophila Ah65N. Whole-cell lysates of overnight TSB cultures were analyzed by Western blot analysis using the anti-TapA polyclonal antiserum raised against purified His-Tag–TapA fusion protein of this strain. The vector (pMMB67HE.cam) alone was introduced into the wild type (Ah65N) and the tapA mutant strain (Ah65N-AΩ18) (shown in Fig. 3) as controls. The complemented mutant strain was prepared by introducing the plasmid expressing TapA (pCP1194) into Ah65N-AΩ18.
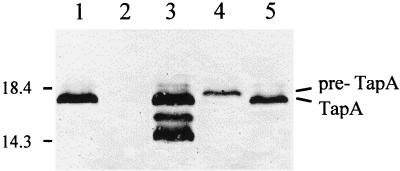
FIG. 3.
Western immunoblot of whole-cell extracts from A. hydrophila Ah65N probed with Ah65 TapA polyclonal antiserum. Lane 1, Ah65N (wild-type) plus pMMB67HE.cam (vector); lane 2, Ah65N-AΩ18 (tapA mutant) plus pMMB67HE.cam; lane 3, Ah65N-AΩ18 (pCP1194) (tapA complemented strain); lane 4, Ah65N-DΩ33.2 (tapD mutant) plus pMMB67HE.cam; lane 5, Ah65N-DΩ33.2 (pCP1140) (tapD complemented strain). Numbers at the left represent protein molecular masses in kilodaltons.
An ∼17-kDa protein was recognized by the anti-TapA antibody in the wild-type strain Ah65N(pMMB67HE.cam) (Fig. 3, lane 1). However, no such band was seen with whole-cell lysates prepared from the tapA mutant strain Ah65N-AΩ18(pMMB67HE.cam) (Fig. 3, lane 2). When Ah65N-AΩ18 was complemented with the TapA-expressing plasmid (pCP1194), the protein band was again detected (Fig. 3, lane 3). Some lower-molecular-weight species (probably breakdown products associated with the overexpression of TapA) were also seen in Ah65N-AΩ18 (Fig. 3, lane 3) and Ah65N wild type (data not shown) complemented with pCP1194.
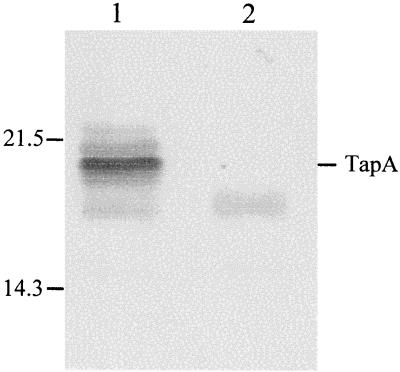
For A. veronii bv. sobria strain BC88, it was similarly demonstrated that a protein band (∼20 kDa) reacted with the BC88 TapA antiserum in cell lysates of the wild-type strain (BC88). This band was, however, absent in the tapA mutant strain (BC88tapAΩ) (Fig. 4). The apparent molecular mass of TapA of strain BC88 was higher than that of TapA from strain Ah65N, as expected from previous studies (5).
FIG. 4.
Western immunoblot of A. veronii bv. sobria strain BC88 whole-cell extracts probed with BC88 TapA polyclonal antiserum. Lane 1, BC88 (wild-type); lane 2, BC88tapAΩ (tapA mutant). Numbers at the left represent protein molecular masses in kilodaltons.
Effect of mutation of tapD on the production of TapA by A. hydrophila Ah65N.
Whole-cell lysates from Ah65N-DΩ33.2 (tapD mutant), containing either the vector, pMMB67HE.cam alone, or a TapD-expressing plasmid (pCP1140) were also examined by Western blotting (Fig. 3, lanes 4 and 5, respectively) to determine if, as for other type IV pilus gene homologs, the type IV peptidase encoded by tapD processes the tapA prepilin into a form that can be assembled into a pilus structure.
In the tapD mutant strain, Ah65N-DΩ33.2, (Fig. 3, lane 4), the protein detected by the anti-TapA antibody had a slightly higher molecular mass than the protein detected in the wild-type strain (Fig. 3, lane 1). Complementation of the tapD mutant strain (TapD-expressing plasmid, pCP1140 introduced in Ah65N-DΩ33.2) again resulted in an ∼17-kDa band (Fig. 3, lane 5). These observations are consistent with the larger band (lane 4) being the precursor form of the pilin protein, pre-TapA, which TapD processes into the mature ∼17-kDa pilin (42, 47).
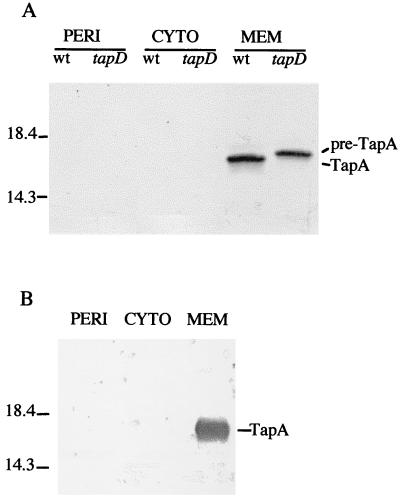
To determine where the pilin localizes within the cell, A. hydrophila Ah65N (wild-type) and Ah65N-D5 (tapD mutant) and A. veronii bv. sobria BC88 (wild-type) bacteria were fractionated into periplasmic, cytoplasmic, and membrane components. These fractions were analyzed by Western blotting using the appropriate TapA antiserum. The mature form of TapA is localized in the membranes in both Ah65N and BC88 wild-type strains (Fig. 5). In the tapD mutant of Ah65N, the precursor form of TapA is also found only in the membrane (Fig. 5A). Overall, these results demonstrate that TapA is expressed in Ah65N and BC88 and that the precursor form of the protein, pre-TapA, is processed in Ah65N by TapD to the mature pilin species.
FIG. 5.
Western immunoblot of the cellular fractions of A. hydrophila Ah65N and A. veronii bv. sobria strain BC88. (A) wt, Ah65N (wild type); tapD, Ah65N-D5 (tapD mutant). (B) wt, BC88 (wild type); PERI, periplasm; CYTO, cytoplasm; MEM, membranes. Unprocessed pre-TapA and mature TapA pilin species are indicated on the right, with molecular mass standards on the left.
Detection of TapA on the bacterial cell surface.
To determine if TapA is assembled into pili on the cell surface, electron microscopic examination of wild-type A. hydrophila Ah65N and isogenic tapA and tapD mutant strains (Ah65N-AΩ18 and Ah65N-D5) was undertaken. Bacteria were grown on TSAY at 22°C and negatively stained. All three strains displayed numerous pili. IEM with anti-TapA serum could not establish whether any of the filamentous structures were Tap pili and, hence, whether they were missing in mutant strains (data not shown). Previous studies with A. hydrophila isolates have shown that the vast majority of pili on the surface of this species are “short-rigid,” type I pili (13, 17, 28).
Fecal isolates of A. veronii bv. sobria are generally poorly piliated and express few (<20), long, wavy pili on the cell surface. Our past studies have shown that for A. veronii bv. sobria strain BC88 grown in TSBY at 22°C the vast majority of these are Bfp type IV pili. However, on TSAY at 22°C the proportion of unlabeled pili seen on IEM with anti-Bfp serum was substantially higher (30). IEM with anti-TapA serum failed to demonstrate the labeling of pili on the cell surface of bacteria grown under either of these conditions, however.
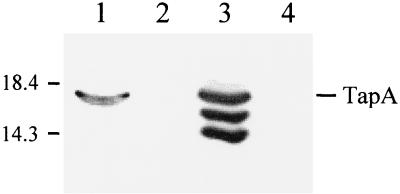
An alternative approach to detect whether Tap pili were present was to mechanically shear surface-associated structures from the bacterial cell surface and then compare mutant and wild-type strains for TapA by Western blotting. When A. hydrophila Ah65N (wild type) and Ah65N-AΩ18 (tapA mutant) were compared in this way, TapA was demonstrated in the sample from Ah65N but not the sample from Ah65N-AΩ18 (Fig. 6, lanes 1 and 2). When the tapA mutation in Ah65N-AΩ18 was complemented in trans with the TapA-overproducing plasmid (pCP1194), TapA was recovered after shearing as with the wild-type strain Ah65N (Fig. 6, lane 3). Hence, TapA was present in the filamentous preparation. To confirm that it was from the cell surface and not a result of contamination from other cellular fractions during the shearing procedure, surface-associated structures were sheared from Ah65N-DΩ33.2 (tapD mutant), where TapA is expressed but remains membrane-associated (see above). No TapA was detected in the sample from Ah65N-DΩ33.2 (Fig. 6, lane 4). These results suggest that Tap pili are assembled on the cell surface.
FIG. 6.
Western immunoblot of pili sheared from A. hydrophila strain Ah65N wild-type and mutant strains probed with TapA polyclonal antiserum. Lane 1, Ah65N (wild-type); lane 2, Ah65N-AΩ18 (tapA mutant); lane 3, Ah65N-AΩ18 + pCP1194 (tapA complemented strain); lane 4, Ah65N-DΩ33.2 (tapD mutant). The TapA pilin species is indicated on the right, molecular mass standards on the left.
DISCUSSION
This study has examined the expression and functional significance of a second Aeromonas type IV pilus, Tap, identified following the cloning of its biogenesis gene cluster, tapABCD. This pilus gene cluster is widespread in Aeromonas species and has homologs in a number of other gram-negative bacteria (4, 10). Tap pili are distinct from the Bfp type IV pili which are expressed on diarrhea-associated Aeromonas species and are known to be important intestinal cell adhesins and colonization factors (14, 15, 20, 29, 37). In contrast to Bfp pili, Tap pili have never been isolated from Aeromonas species, and there have been no previous studies to investigate their significance for Aeromonas virulence. For P. aeruginosa, related pili are a major virulence-associated adhesin (12) and also play an important role in microcolony formation in biofilms (39). For V. cholerae, however, for which the organization of the homologous gene cluster, pilABCD, shows a striking similarity to that of the Aeromonas tap gene cluster (genes grouped together and transcribed in the same direction), this pilus type is reportedly not important for adhesion to HEp-2 cells or for the colonization of infant mice (10). Moreover, it is not required for V. cholerae adherence to some solid substrates, questioning its role in adherence in the environment, despite its 100% conservation between the classical and El Tor biotypes (51).
We prepared and used specific mutants of two Aeromonas strains (A. hydrophila Ah65, the strain from which the tap gene cluster was originally cloned, and a dysenteric, fecal isolate of A. veronii bv. sobria, strain BC88) to investigate the expression and function of Tap pili. The A. veronii bv. sobria strain was chosen for the in vivo functional studies because the poor cell adhesion and low virulence of strain Ah65 made it unsuitable for in vivo studies. It was established that mutations in tapA of this strain did not affect the production of exotoxins considered important for diarrhea induction.
Inactivation of the pilus subunit gene, tapA, had no effect on the adherence ability of either of the above Aeromonas strains to HEp-2 cells. For A. veronii bv. sobria strain BC88, adherence to the intestinal cell line (Henle 407) and intestinal tissue was also not significantly different for the wild type and for the tapA mutant strain. Since type I and Bfp pili are the predominant pilus types seen on these bacterial strains (17, 30), this result may not be entirely unexpected. Experiments aimed at visualizing Tap pili on the bacterial cell surface by IEM were not successful, despite the growth of A. veronii bv. sobria strain BC88 under conditions previously shown to increase non-Bfp pilus expression (30). It is possible that the TapA antisera did not recognize the proteins in their native conformation. (TapA antisera were prepared against denatured recombinant proteins prepared by SDS-PAGE.) Furthermore, it is possible that the His-Tag sequence or the absence of disulfide bonds in the recombinant proteins may have altered their folding and, hence, their antigenicity compared to the native proteins. However, expression studies did establish that TapA was produced and present at the cell surface. Western blots of wild-type and tapD mutant and complemented strains established that TapA is processed by the type IV leader peptidase/N-methyltransferase, TapD and localizes in the cell membrane. Shearing experiments using Western blot comparisons of TapA from wild-type and mutant strains suggested that TapA was most likely present in the form of pili. There are other possible explanations for the detection of TapA in sheared preparations from the wild type but not the tapD mutant. TapA pilin may not be assembled into pili in Aeromonas species but may be more surface accessible in the wild type compared to a mutant carrying a lesion in the type IV peptidase. In this case, failure to cleave off the prepilin leader peptide would prevent it from crossing into the outer membrane. However, this is unlikely since other investigators have shown that processed and unprocessed pilin is distributed equally in the cytoplasmic and outer membranes in P. aeruginosa (38). Another possible explanation is that the assembly of Tap pili is regulated by, or requires the presence of, another protein with a role similar to that postulated for PilC in the assembly of Neisseria gonorrhoeae type IV pili (23). Under the conditions tested here, this protein may not be expressed in sufficient quantity to promote assembly of large numbers of Tap pili on the cell surface. In any event it is clear that if Tap pili are present on the surface of Aeromonas spp. they exist in only small numbers under standard bacterial growth conditions.
For Tap pili to play a role in vivo, conditions in the intestine should favor expression. However, in in vivo experiments, mutation of TapA did not affect intestinal colonization of rabbits or infant mice or significantly alter the ability of Aeromonas spp. to cause diarrheal symptoms in the RITARD model. Symptoms caused by A. veronii bv. sobria strain BC88 in rabbits were not severe in comparison to effects observed with other enteropathogenic bacteria such as V. cholerae (2) and Providencia alcalifaciens (1). Nevertheless, both the wild-type and mutant strains caused significant, short-lived diarrhea (55%, 10 of 18 rabbits overall inoculated with either strain) compared with the E. coli surgical controls which showed no symptoms. In two mouse models, there was also no evidence that the mutation decreased colonization ability. These latter results are in agreement with those of the V. cholerae pilA mutant studies in mice (10). Discernible, short-lived differences in colonization could have been missed in these in vivo studies, however, given the presence of the Bfp pilus intestinal colonization factor. Mutagenesis of the latter awaits the cloning of the Bfp pilin gene.
Further studies are required to identify factors that may influence the expression of Tap pili and to determine why the genes encoding them (and related pili in V. cholerae) are so widely conserved. The widespread distribution of the tap gene cluster in all Aeromonas species (including nonclinical species) and the results obtained in this study, however, suggest that Tap pili are not as significant as Bfp for intestinal colonization by diarrheagenic Aeromonas species.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Health and Medical Research Council of Australia (project no. 981530). C. M. Pepe was supported by a National Research Council-NOAA Research Associateship and an NOAA Cooperative Education and Research Program grant (NA67FE0396) to Faye M. Dong of the University of Washington, School of Fisheries.
We thank Adrian Kelleher for his contributions to the exotoxin assays and Korshed Alam for his help with the RITARD studies.
REFERENCES
- 1.Albert M J, Alam K, Ansaruzzamzn M, Islam M M, Rahman A S M H, Haider K, Nahar S, Ryan N, Montaro J, Mathan M M. Pathogenesis of Providencia alcalifaciens-induced diarrhea. Infect Immun. 1992;60:5017–5024. doi: 10.1128/iai.60.12.5017-5024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert M J, Alam K, Rahman A S M H, Huda S, Sack R B. Lack of cross-protection against diarrhea due to Vibrio cholerae O139 Bengal. J Infect Dis. 1994;169:709–710. doi: 10.1093/infdis/169.3.709. [DOI] [PubMed] [Google Scholar]
- 3.Attridge S R, Voss E, Manning P A. The role of toxin-coregulated pili in the pathogenesis of Vibrio cholerae O1 El Tor. Microb Pathog. 1993;15:421–431. doi: 10.1006/mpat.1993.1091. [DOI] [PubMed] [Google Scholar]
- 4.Barnett T C, Kirov S M. The type IV Aeromonas pilus (Tap) gene cluster is widely conserved in Aeromonas species. Microb Pathog. 1999;26:77–84. doi: 10.1006/mpat.1998.0252. [DOI] [PubMed] [Google Scholar]
- 5.Barnett T C, Kirov S M, Strom M S, Sanderson K. Aeromonas spp. possess at least two distinct type IV pilus families. Microb Pathog. 1997;23:241–247. doi: 10.1006/mpat.1997.0152. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogdanovic R, Cobeljic M, Markovic M, Nikolic V, Ognjanovic M, Makic D. Haemolytic-uraemic syndrome associated with Aeromonas hydrophila enterocolitis. Pediatr Nephrol. 1991;5:293–295. doi: 10.1007/BF00867480. [DOI] [PubMed] [Google Scholar]
- 8.Carrello A, Silburn K A, Budden J R, Chang B J. Adhesion of clinical and environmental Aeromonas isolates to HEp-2 cells. J Med Microbiol. 1988;26:19–27. doi: 10.1099/00222615-26-1-19. [DOI] [PubMed] [Google Scholar]
- 9.Fang J S, Chen J B, Chen W J, Hsu K T. Haemolytic-uraemic syndrome in an adult male with Aeromonas hydrophila enterocolitis. Nephrol Dial Transplant. 1999;14:439–440. doi: 10.1093/ndt/14.2.439. [DOI] [PubMed] [Google Scholar]
- 10.Fullner K J, Mekalanos J J. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect Immun. 1999;67:1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gay P, LeCoq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 13.Ho A S Y, Mietzner T A, Smith A J, Schoolnik G K. The pili of Aeromonas hydrophila: identification of an environmentally regulated “mini pilin.”. J Exp Med. 1990;172:795–806. doi: 10.1084/jem.172.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hokama A, Honma Y, Nakasone N. Pili of an Aeromonas hydrophila strain as a possible colonization factor. Microbiol Immunol. 1990;34:901–915. doi: 10.1111/j.1348-0421.1990.tb01069.x. [DOI] [PubMed] [Google Scholar]
- 15.Hokama A, Iwanaga M. Purification and characterization of Aeromonas sobria pili, a possible colonization factor. Infect Immun. 1991;59:3478–3483. doi: 10.1128/iai.59.10.3478-3483.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hokama A, Iwanaga M. Purification and characterization of Aeromonas sobria Ae24 pili: a possible new colonization factor. Microb Pathog. 1992;13:325–334. doi: 10.1016/0882-4010(92)90042-m. [DOI] [PubMed] [Google Scholar]
- 17.Honma Y, Nakasone N. Pili of Aeromonas hydrophila: purification, characterization, and biological role. Microbiol Immunol. 1990;34:83–98. doi: 10.1111/j.1348-0421.1990.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 18.Howard S P, Buckley J T. Molecular cloning and expression in Escherichia coli of the structural gene for the hemolytic toxin aerolysin from Aeromonas hydrophila. Mol Gen Genet. 1986;204:289–295. doi: 10.1007/BF00425512. [DOI] [PubMed] [Google Scholar]
- 19.Howard S P, Critch J, Bedi A. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol. 1993;175:6695–6703. doi: 10.1128/jb.175.20.6695-6703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwanaga M, Hokama A. Characterization of Aeromonas sobria TAP13 pili: a possible new colonization factor. J Gen Microbiol. 1992;138:1913–1919. doi: 10.1099/00221287-138-9-1913. [DOI] [PubMed] [Google Scholar]
- 21.Janda J M, Abbott S L. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations and unanswered questions. Clin Infect Dis. 1998;27:332–344. doi: 10.1086/514652. [DOI] [PubMed] [Google Scholar]
- 22.Jiang B, Howard S P. The Aeromonas hydrophila exeE gene, required for secretion and normal biogenesis, is a member of the general secretion pathway. Mol Microbiol. 1992;6:1351–1361. doi: 10.1111/j.1365-2958.1992.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 23.Jonsson A B, Nyberg G, Normark S. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 1991;10:477–488. doi: 10.1002/j.1460-2075.1991.tb07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 25.Kirov S M. Aeromonas and Plesiomonas. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: American Society for Microbiology; 1997. pp. 265–287. [Google Scholar]
- 26.Kirov S M, Ardestani E K, Hayward L J. The growth and expression of virulence factors at refrigeration temperature by Aeromonas strains isolated from foods. Int J Food Microbiol. 1993;20:159–168. doi: 10.1016/0168-1605(93)90108-s. [DOI] [PubMed] [Google Scholar]
- 27.Kirov S M, Hayward L J, Nerrie M A. Adhesion of Aeromonas sp. to cell lines used as models for intestinal adhesion. Epidemiol Infect. 1995;115:465–473. doi: 10.1017/s0950268800058623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirov S M, Jacobs I, Hayward L J, Hapin R. Electron microscopic examination of factors influencing the expression of filamentous surface structures on clinical and environmental isolates of Aeromonas veronii biovar sobria. Microbiol Immunol. 1995;39:329–338. doi: 10.1111/j.1348-0421.1995.tb02209.x. [DOI] [PubMed] [Google Scholar]
- 29.Kirov S M, O'Donovan L A, Sanderson K. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect Immun. 1999;67:5447–5454. doi: 10.1128/iai.67.10.5447-5454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirov S M, Sanderson K. Characterization of a type IV bundle-forming pilus (SFP) from a gastroenteritis-associated strain of Aeromonas veronii biovar sobria. Microb Pathog. 1996;21:23–34. doi: 10.1006/mpat.1996.0039. [DOI] [PubMed] [Google Scholar]
- 31.Kirov S M, Sanderson K, Dickson T C. Characterisation of a type IV pilus produced by Aeromonas caviae. J Med Microbiol. 1998;47:527–531. doi: 10.1099/00222615-47-6-527. [DOI] [PubMed] [Google Scholar]
- 32.Kirov S M, Rees B, Wellock R C, Goldsmid J M, Van Galen A D. Virulence characteristics of Aeromonas spp. in relation to source and biotype. J Clin Microbiol. 1986;24:827–834. doi: 10.1128/jcm.24.5.827-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koga T, Ishimoto K, Lory S. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosa pili. Infect Immun. 1993;61:1371–1377. doi: 10.1128/iai.61.4.1371-1377.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 36.McIntyre S, Trust T J, Buckley J T. Identification and characterization of outer membrane fragments released by Aeromonas sp. Can J Biochem. 1980;58:1018–1025. doi: 10.1139/o80-138. [DOI] [PubMed] [Google Scholar]
- 37.Nakasone N, Iwanaga M, Yamashiro T, Nakashima K, Albert M J. Aeromonas trota strains, which agglutinate with Vibrio cholerae O139 Bengal antiserum, possess a serologically distinct fimbrial colonization factor. Microbiology. 1996;142:309–313. doi: 10.1099/13500872-142-2-309. [DOI] [PubMed] [Google Scholar]
- 38.Nunn D N, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 40.Paranjpye R N, Lara J C, Pepe J C, Pepe C M, Strom M S. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect Immun. 1998;66:5659–5668. doi: 10.1128/iai.66.12.5659-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pazzaglia G, Sack R B, Bourgeois A L, Froehlich J, Eckstein J. Diarrhea and intestinal invasiveness of Aeromonas strains in the removable intestinal tie rabbit model. Infect Immun. 1990;58:1924–1931. doi: 10.1128/iai.58.6.1924-1931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pepe C M, Eklund M W, Strom M S. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol Microbiol. 1996;194:857–869. doi: 10.1046/j.1365-2958.1996.431958.x. [DOI] [PubMed] [Google Scholar]
- 43.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 44.Reischl U. Purification and immunological characterization of recombinant antigens expressed in the form of insoluble aggregates (inclusion bodies) In: Reischl U, editor. Methods in molecular medicine. 13. Molecular diagnosis of infectious diseases. Totowa, N.J: Humana Press, Inc.; 1998. pp. 331–343. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 47.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 48.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner L R, Lara J C, Nunn D N, Lory S. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:4962–4969. doi: 10.1128/jb.175.16.4962-4969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Goot F G, Pattus F, Parker M, Buckley J T. The cytolytic toxin aerolysin: from soluble form to the transmembrane channel. Toxicology. 1994;87:19–28. doi: 10.1016/0300-483x(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 51.Watnick P I, Fullner K J, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yim H H, Villarejo M. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol. 1992;174:3637–3644. doi: 10.1128/jb.174.11.3637-3644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]