Abstract
Retinal diseases are a leading cause of blindness in developed countries, accounting for the largest share of visually impaired children, working-age adults (inherited retinal disease), and elderly individuals (age-related macular degeneration). These conditions need specialised clinicians to interpret multimodal retinal imaging, with diagnosis and intervention potentially delayed. With an increasing and ageing population, this is becoming a global health priority. One solution is the development of artificial intelligence (AI) software to facilitate rapid data processing. Herein, we review research offering decision support for the diagnosis, classification, monitoring, and treatment of retinal disease using AI. We have prioritised diabetic retinopathy, age-related macular degeneration, inherited retinal disease, and retinopathy of prematurity. There is cautious optimism that these algorithms will be integrated into routine clinical practice to facilitate access to vision-saving treatments, improve efficiency of healthcare systems, and assist clinicians in processing the ever-increasing volume of multimodal data, thereby also liberating time for doctor-patient interaction and co-development of personalised management plans.
Keywords: Retina, Artificial intelligence, Age-related macular dystrophy, Inherited retinal disease, Diabetic retinopathy
Introduction
Retinal diseases are a significant cause of visual impairment and blindness, both in adults (secondary to age-related macular degeneration (AMD) and diabetic retinopathy (DR)) [1] and in children (due to inherited retinal disorders (IRD) and retinopathy of prematurity (ROP)) [2]. Diagnosing these conditions usually involves multimodal testing and multiple consultations with retina specialists, often not available in a timely manner, which can result in delays in sight-saving treatments. For rare diseases, it can take several years for a final diagnosis (‘diagnostic odyssey’), resulting in uncertainty about the prognosis and delay in appropriate care.
Healthcare data increases by approximately 50% every year, making it one of the fastest-growing digital areas [3]. Genomic data alone is as demanding in terms of data acquisition, storage, distribution, and analysis as astronomy or social media content [4]. Ophthalmology is one of the leading data generators, with 30 million optical coherence tomography (OCT) scans performed yearly in the USA [5]. This ever-increasing vast amount of data, alongside the development of cutting-edge digital technology, has made ophthalmology a pioneer in digital innovation and healthcare artificial intelligence (AI).
AI has been rapidly developing in multiple areas of medicine, including, dermatologist-level performance at detecting skin cancer [6], highly accurate classification of pulmonary tuberculosis [7], and genetic variant calling and classification [8]. AI-based ophthalmology telemedicine has been beneficial during the COVID-19 pandemic [9], and remote evaluation and analysis of retinal imaging may be useful in decreasing diagnostic time and facilitating triaging and classification [10, 11].
Development of highly sensitive and sensible AI-based tools requires transdisciplinary collaboration between clinicians and software engineers. Herein, we will provide an overview of current methodologies used in AI system development and validation and focus on clinical application in prioritising retinal diseases.
AI methodology overview
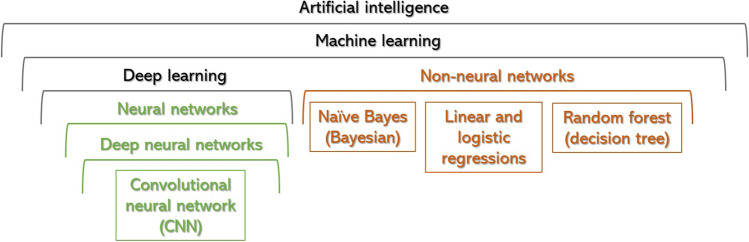
The most common techniques to develop AI-based healthcare tools will be summarised below and in Figs. 1 and 2:
AI is a phenomenon in which non-living entities mimic human intelligence [12]. It is an umbrella term encompassing a spectrum of computing programs. ‘Rule-based’, ‘hard-coded’ or ‘symbolic AI’ has existed for many decades and is the basis of any software system, from a traffic light management system to the autopilot flying every plane. In healthcare, symbolic AI has multiple applications, e.g. calculating cardiovascular risk index or eGFR.
Machine learning (ML) is an AI subfield in which a program achieves a task by being exposed to vast volumes of data and gradually learning to recognise patterns within the data, allocating data to distinct classes [13]. It involves ‘soft coding’, which means that the model learns from examples instead of being programmed with rules [12]. ML models can be supervised (based on data labelled by humans), unsupervised (i.e., grouping features within categories), or reinforcement learning (the system accumulates its own feedback to improve through a reward function) [14]. In medicine, supervision is the most common.
Nonneural network-supervised ML algorithms are useful in healthcare for prediction modelling and evaluating associations and best-fitted lines between two (linear regression, parametric) or multiple variables (random forest, non-parametric). The latter combines different inputs using a network of flowcharts (known as decision trees); each tree creates an outcome, and a collective one will be made by combining all the singular outputs [15]. Non-neural networks are often combined with deep neural network (DNN) architectures and achieve improved performance (Fig. 1) [16].
Deep learning (DL) is a subdivision of ML, defined by the presence of multiple layers of artificial neural networks (ANN) [17]. An ANN is composed of an input layer of multiple nodes—‘artificial neurons’—that represent characteristics to be analysed, e.g. pixels on an image, diagnoses (International Classification of Disease (ICD) coded), age, nucleotide changes, etc.; connected to one or more hidden layers that sum and analyse all inputs, and transmit a final value to an output layer (Fig. 2A).
DNN corresponds to multi-layered DL algorithms (with often over 100 hidden layers), which are currently the gold standard for image classification [15]. As more layers are added, an iterative training phenomenon starts occurring, by which deep layers combine stimuli sent from other layers and design new stimuli, improving the output layer and ultimately leading to better diagnoses [8].
Convolutional neural network (CNN) is a type of DNN particularly useful for image and video analysis [15]. These algorithms divide the files into pixels, convert them into numbers or symbols, analyse them by multiple convolutional layers that filter, merge, mask, and/or multiply features, and feed the results to a dense neural network that will create an output layer [18]. Fully convolutional networks (FCN) feed the output layers themselves, without the final step of dense layers (Fig. 2A) [17].
Fig. 1.
Diagram of artificial intelligence algorithms, subfields, and mechanisms
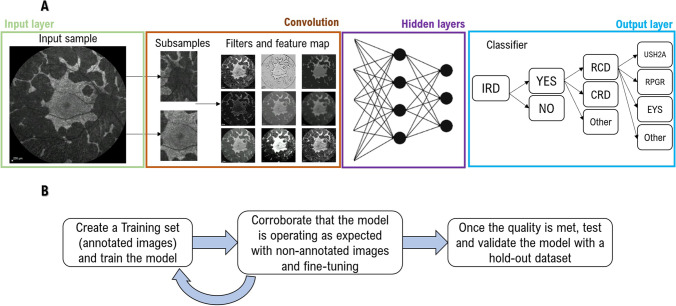
Fig. 2.
A Overview of a convolutional neural network (CNN). The process starts with an input layer, typically an image or video, that gets divided into subsamples and/or pixels and is analysed by multiple convolutional layers that filter, mask, or multiply features and feed the results to a dense neural network of multiple nodes ‘artificial neurons’. Each one represents a characteristic to be analysed (e.g., pixels, diagnoses, age, contrast, etc.) and is connected to hidden layers that sum and analyse all inputs, combining the received stimuli and designing a new one, leading to an improved output layer and final diagnosis. B The process of developing a supervised AI model. First, a training set needs to be created, and these images are used to train the model to interpret the different features; after this, a separate, non-annotated dataset (validation set) is presented to the model to try it, whilst still fine-tuning its configuration; and lastly, the algorithm is tested on new data, evaluating its overall performance
Useful concepts to better understand AI literature
There are different types of models, depending on the outcome prediction. (i) Classification models apply to categorical outputs, such as classifying retinal images into with or without DR; (ii) segmentation models are specialised for image processing and analysis, detecting presence or absence of features (e.g., intraretinal fluid), segmenting images into known anatomical correlates, or classifying them into diagnostic categories; (iii) regression models for when a quantitative output is needed, such as predict central macular thickness from an OCT file [19, 20].
Different performance metrics are used to present results of each model type. Dice similarity coefficient (dice score) and intraclass correlation coefficient (ICC) are metrics of segmentation accuracy suitable for evaluating performance of image segmentation DL algorithms, ranging from 0 to 1 [21]. There are multiple performance metrics for classification and regression model algorithms, such as (i) receiver operating characteristic (ROC) curves, that plot true positives (sensitivity) against false positives (1 = specificity) [22]; (ii) the area under the curve (AUC, also known as AUROC) ranging from 0 to 1, with 1 indicating a perfect algorithm [23]; (iii) precision-recall curves (PRC), which associate positive predictive value with sensitivity (also known as recall or true positive rate) [24]; (iv) the accuracy statistical score; (v) absolute difference; (vi) Pearson’s correlation between said parameters (the latter also goes from 0 and 1) [23].
The process of developing a supervised AI model generally involves three stages: (1) training, when the network is provided labelled images; (2) fine-tuning, where the model starts aiding the manual annotation and human graders to correct it and improve it; (3) validation or testing, where the algorithm is tested on a hold-out dataset annotated by human graders and kept separate from the training dataset (internal validation). External validation on datasets of completely independent origin than the training dataset is the gold standard for validation/performance evaluation, indicating generalizability (Fig. 2B). [23]
Selected retinal diseases for which AI-based tools have been developed
Diabetic retinopathy (DR)
Recent studies have shown that AI-based DR screening systems can achieve adequate levels of safety [25–29]. These algorithms include classical expert-designed image analysis, mathematical morphology, and transformations [30–33]. One of the approaches tested was to classify colour fundus images from training datasets into referable DR (moderate or advanced stage) or non-referable DR (no or mild DR, Table 1). These studies either built their own CNNs or used pretrained ones like AlexNet [34], Inception V3 [35], Inception-Resnet-V2 [36], and Resnet152 [37]. Other studies tried to detect DR based on fixed features such as red lesions [38,39], microaneurysms [40], exudates, and blood-vessel segmentation [41, 42]. Lastly, other groups introduced a method to detect DR and diabetic macular oedema (DMO) using a CNN model, being able to detect the exact stage of DR; these studies are summarised in Table 1 [49–60, 95].
Table 1.
Artificial intelligence in retinal disease—methods, cohorts, and overall results
| Condition | Imaging analysed | Database (n) | AI tool | Task | Performance (metrics provided by each paper) | Publication |
|---|---|---|---|---|---|---|
| DR | Colour | DiaretDB0 (130), DiaretDB1 (89), and DrimDB (125) | CNN | Referable/non-referable DR |
Accuracy 99.17% (DiaretDB0), 98.53% (DiaretDB1), 99.18% (DrimDB) Sensitivity 100% (DiaretDB0), 99.2% (DiaretDB1), 100% (DrimDB) Specificity 98.4% (DiaretDB0), 97.97% (DiaretDB1), 98.44% (DrimDB) |
Adem et al. [41] |
| Colour | Kaggle (88,702), DiaretDB1 (89), and E-ophtha (107,799) | CNN | Referable/non-referable DR | AUC 0.954 (Kaggle), 0.949 (E-ophtha) | Quellec et al. [43] | |
| Colour | Kaggle (35,000) | CNN-ResNet34 | Referable/non-referable DR |
Sensitivity 85% Specificity 86% |
Esfahani et al. [44] | |
| Colour | Messidor-2 (1748), Kaggle (88,702), and DR2 (520) | CNN | Referable/non-referable DR | Accuracy 98.2% (Messidor-2), 98% (DR2) | Pires et al. [45] | |
| Colour | Own dataset (30,244) | CNN (Inception V3, Inception-Resnet-V2, and Resnet152) | Referable/non-referable DR |
AUC 0.946 Accuracy 88.21% Sensitivity 85.57% Specificity 90.85% |
Jiang et al. [46] | |
| Colour | Own dataset (60,000) and STARE (131) | CNN (WP-CNN, ResNet, SeNet, and DenseNet) | Referable/non-referable DR |
AUC 0.9823 (Own dataset), 0.951 (STARE) Accuracy 94.23% (Own dataset), 90.84% (STARE) Sensitivity 90.94% (Own dataset) Specificity 90.85% (Own dataset) |
Liu YP et al. [47] | |
| Colour | DIARETDB1 (89), DIARETDB0 (130), Kaggle (15,919), Messidor (1200), Messidor-2 (874), IDRiD (103), and DDR (4105) | CNN (VGG16, custom CNN) | Referable/non-referable DR |
AUC 0.786 (Kaggle, Messidor), 0.764 (Messidor-2), 0.912 (IDRiD, DDR) Accuracy 82.1% (Kaggle, Messidor), 91.1% (Messidor-2), 94% (IDRiD, DDR) |
Zago et al. [48] | |
| Colour | Messidor-2 (1748) | CNN | Different clinical stages of DR |
AUC 0.98 Sensitivity 96.8% Specificity 87% |
Abramoff et al. [49] | |
| Colour | Kaggle (80,000) | CNN | Different clinical stages of DR |
Accuracy 75% Sensitivity 30% Specificity 95% |
Pratt et al. [50] | |
| Colour | Kaggle (2000) | DNN, CNN (VGGNET architecture), BNN | Different clinical stages of DR | Accuracy: BNN = 42%, DNN = 86.3%, CNN = 78.3% | Dutta S et al. [51] | |
| Colour | Kaggle (166) | CNN (InceptionNet V3, AlexNet, and VGG16) | Different clinical stages of DR | Accuracy: AlexNet = 37.43%, VGG16 = 50.03%, InceptionNet V3 = 63.23% | Wang X et al. [52] | |
| Colour | Kaggle (35,126) | CNN (AlexNet, VggNet, GoogleNet, and ResNet) | Different clinical stages of DR |
AUC 0.9786 (VggNet) Accuracy 95.68% (VggNet) Sensitivity 90.78% (VggNet) Specificity 97.43% (VggNet) |
Wan S et al. [53] | |
| Colour | MESSIDOR (1200) | CNN (AlexNet, VggNet16, custom CNN) | Different clinical stages of DR |
Accuracy 98.15% Sensitivity 98.94% Specificity 97.87% |
Mobben-ur-Rehman et al. [54] | |
| Colour | Own dataset (13,767) | CNN (ResNet50, InceptionV3, InceptionResNetV2, Xception, and DenseNets) | Different clinical stages of DR | 96.5%, 98.1%, 98.9% | Zhang et al. [55] | |
| Colour | Kaggle (22,700) and IDRiD (516) | CNN (AlexNet) | Different clinical stages of DR | Accuracy 90.07% | Harangi et al. [56] | |
| Colour | DDR (13,673) | CNN (GoogLeNet, ResNet-18, DenseNet-121, VGG-16, and SE-BN-Inception) | Different clinical stages of DR | Accuracy 82.84% | Li T et al. [57] | |
| Colour | Messidor (1190) | CNN (modified Alexnet) | Different clinical stages of DR |
Accuracy 96.35% Sensitivity 92.35% Specificity 97.45% |
Shanthi T et al. [58] | |
| Colour | Own dataset (9194) and Messidor (1200) | CNN | Different clinical stages of DR |
Accuracy 92.95% (own dataset) Sensitivity 99.39% (own dataset), 99.93% (Messidor) Specificity 92.59% (own dataset), 96.2% (Messidor) |
Wang J et al. [59] | |
| Colour | Messidor (1200) and IDRiD (516) | CNN (ResNet50) | Different clinical stages of DR |
AUC 0.963 (Messidor) Accuracy 92.6% (Messidor), 65.1% (IDRiD) Sensitivity 92% (Messidor) |
Li X et al. [60] | |
| AMD | Colour | 407 eyes with nonadvanced AMD | DL | Distinguishes between low and high-risk AMD by quantifying drusen location, area, and size | For drusen area: ICC > 0.85; for diameter: ICC = 0.69; for AMD risk assessment: ROC = 0.948 and 0.954 | Van Grinsven et al. [61] |
| Colour, OCT, and IR | 278 eyes with/without reticular pseudodrusen (RPD) | DL | Automatic quantification of RPD | ROC = 0.94 and 0.958; κ agreement = 0.911; ICC = 0.704 | Van Grinsven et al. [62] | |
| Colour | 2951 subjects from AREDS (834 progressors) | DL | Association between genetic variants and transition to advanced AMD | AUC: 5 years = 0.885; 10 years = 0.915 | Seddon et al. [63] | |
| Colour and OCT | 280 eyes from 140 participants | DL | Prediction of progression to late AMD | AUC = 0.85 | Wu et al. [64] | |
| Colour and microperimetry | 280 eyes from 140 participants | DL | Predictive value of pointwise sensitivity and low luminance deficits for AMD progression | AUC = 0.8 | Wu et al. [65] | |
| Colour | > 4600 participants from AREDS | DL | Predict progression to advanced dry or wet AMD | Accuracy = 0.86 (1 year) and 0.86 (2 years); specificity = 0.85 (1 year) and 0.84 (2 years); sensitivity = 0.91 (1 year) and 0.92 (2 years) | Bhuiyan et al. [66] | |
| Colour | 1351 subjects from AREDS (> 31,000 images) | DL | Predict progression to advanced dry or wet AMD | AUC = 0.85 | Yan et al. [67] | |
| Colour | 67,401 colour fundus images from 4613 study participants | DL | Estimate 5-year risk of progression to wet AMD and geographic atrophy based on 9-step AREDS severity scale | Weighted κ scores = 0.77 for the 4-step and 0.74 for the 9-step AMD severity scales | Burlina et al. [68] | |
| Colour | 4507 AREDS participants and 2169 BMES participants | DL | Validation of a risk scoring system for prediction of progression | Sensitivity = 0.87; specificity = 0.73 | Chiu et al. [69] | |
| OCT | 2795 patients | DL | Prediction to nAMD within a 6-month window | AUC = 0.74 (conversion scan ground truth) and 0.886 (1st injection ground truth) | Yim et al. [70] | |
| OCT | 671 AMD fellow eyes with 13,954 observations | DL | Predict progression to wet AMD | AUC = 0.96 ± 0.02 (3 months); 0.97 ± 0.02 (21 months) | Banerjee et al. [71] | |
| OCT | 686 fellow eyes with non-neovascular AMD at baseline | DL | Predict conversion from non-neovascular to neovascular AMD | Drusen are within 3 mm of fovea (HR = 1.45); mean drusen reflectivity (HR = 3.97) | Hallak et al. [72] | |
| OCT | 2146 OCT scans of 330 AMD eyes (244 patients) | DL | Predict neovascular AMD progression within 5 years | AUC = 0.74 (5 years), 0.92 (11 months), 0.86 (16 months), 0.7 (18 months), and 0.79 (48 months) | de Sisternes et al. [73] | |
| OCT | 71 eyes of patients with early AMD and contralateral neovascular AMD (9088 OCT B-scans) | CNN | Prediction of conversion from early/intermediate to advanced neovascular AMD | AUC = 0.87 (VGG16) and 0.91 (AMDnet) | Russakoff et al. [74] | |
| OCT | 495 eyes | DL | Predictive model to assess risk of conversion to advanced AMD | AUC = 0.68 for CNV and 0.8 for geographic atrophy | Schmidt-Erfurth et al. [75] | |
| OCT | 2712 OCT B-scans | DL | Segmentation of features associated with AMD | Dice = 0.63 ± 0.15; ICC = 0.66 ± 0.22 | Liefers et al. [76] | |
| OCT | 930 OCT B-scans from 93 eyes of patients with neovascular AMD | CNN | Segmentation of features associated with neovascular AMD | Dice = 0.78 (IRF), 0.82 (SRF), 0.75 (SHRM), and 0.8 (PED); ICCs = 0.98 (IRF), 0.98 (SRF), 0.97 (SHRM), and 0.98 (PED) | Lee et al. [77] | |
| RP | Colour | 1128 RP and 517 healthy | CNN | Diagnose RP | AUROC 96.74% | Chen et al. [78] |
| Colour | 99 RP and 21 healthy | FCN | Diagnose RP | Accuracy 99.52% | Arsalan et al. [79] | |
| RP, best disease (BD), and Stargardt | FAF | 73 healthy, 125 Stargardt, 160 RP, 125 BD | CNN | Classify images into each group | Accuracy 0.95 | Miere et al. [80] |
| Stargardt | OCT | 102 healthy (33 participants) and 647 Stargardt (60 patients) | CNN | Differentiate between Stargardt and healthy | Accuracy 99.6% | Shah et al. [81] |
| BVMD and AVMD | FAF and OCT | 118 BVMD eyes and 96 AVMD eyes | CNN | Differentiate between BVMD and AVMD | AUROC 0.880 | Crincoli et al. [23] |
| Stargardt and PRPH2-related pattern dystrophy | FAF |
304 Stargardt (40 patients) and 66 PRPH2 (9 patients) |
CNN | Differentiate between Stargardt and PRPH2-related pattern dystrophy | AUROC 0.890 | Miere et al. [82] |
| ABCA4-, RP1L1-, and EYS-related retinopathy | OCT | 58 IRD and 17 healthy | DL | Predict causative gene | ABCA4 100% accuracy; RP1L1 66.7 to 87.5%; EYS 82.4 to 100%; healthy 73.7 to 100% | Fujinami-Yokokawa et al. [83] |
| Stargardt disease | FAF | 47 images (24 patients) | CNN | Segment flecks | Dice score: 0.54 ± 0.14 for diffuse speckled patterns; 0.71 ± 0.08 for discrete flecks | Charng et al. [84] |
| Stargardt disease and AMD | FAF |
320 healthy 320 AMD and 100 Stargardt |
CNN & FCN | Detect and segment atrophy |
Atrophy screening: AMD 0.98 accuracy; Stargardt 0.95 Segmentation: AMD overlapping ratio of 0.89 ± 0.06; Stargardt: 0.78 ± 0.17 |
Wang et al. [85] |
| Stargardt and pattern dystrophy | FAF | 110 AMD, 204 Stargardt, and pattern dystrophy | CNN | Differentiate between AMD and IRD-associated macular atrophy | AUROC 0.981 | Miere et al. [86] |
| Stargardt disease | OCT | 87 scan sets (22 patients) | FCN | Detect outer and inner limits of the retina | Mean difference: 2.10 µm and 0.059 mm3 in central macular thickness and volume between model and annotators | Kugelman et al. [87] |
| AOSLO | 142 controls and 148 Stargardt | FCN | Identify cone photoreceptors | Dice score: 0.9431 ± 0.0482 | Davidson et al. [88] | |
| RP and CHM | OCT | 300 B-scans with RP and 300 with CHM | FCN | EZ segmentation | Similarity of 0.894 ± 0.102 automatic vs manual grading for RP; 0.912 ± 0.055 for CHM | Camino et al. [89] |
| CHM | OCT | 16 eyes CHM and 5 healthy | Nonneural (RF) | EZ segmentation | 0.876 ± 0.066 Jaccard similarity index | Wang et al. [90] |
| USH2A-related RP | OCT | 126 volume scans (126 patients) | CNN | EZ segmentation | Dice score 0.79 ± 0.27 | Loo et al. [91] |
| OCT | 86 volume scans (86 patients) | CNN | EZ segmentation | Dice score 0.867 ± 0.105 | Wang et al. [92] | |
| RP | OCT and IR | 2918 (314 patients) | CNN and FCN | Predict VA below or above 20/40 | AUROC 0.85 | Liu et al. [93] |
| Blue cone monochromacy (BCM) | OCT | 26 IRD, 16 BCM, 3 normal (patients) | Nonneural | Predict foveal sensitivity and VA | 0.174 RMSE for VA and 2.91 for sensitivity | Sumaroka et al. [94] |
RP, retinitis pigmentosa; CNN, convolutional neural network; AUROC, area under the receiver operating characteristic; FROC, free-response receiver operating characteristics; ICC, intraclass correlation coefficients; FCN, fully convolutional network; FAF, fundus autofluorescence; OCT, optical coherence tomography; BVMD, best vitelliform macular dystrophy; HR, hazard ratio; AVMD, adult-onset vitelliform macular dystrophy; DL, deep learning; AMD, age-related macular degeneration; IRD, inherited retinal dystrophy; AREDS, age-related eye disease study; BMES, Blue Mountains Eye Study; AOSLO, adaptive optics scanning laser ophthalmoscopy; CHM, choroideremia; EZ, ellipsoid zone; VA, visual acuity; RMSE, root-mean-square error; IRF, intraretinal fluid; SHRM, subretinal hyperreflective material; PED, pigment epithelial detachment; SRF, subretinal fluid
Wong et al. [96] developed a model to classify DR stages based on microaneurysms and haemorrhages, while others used exudates, blood vessel mapping, and the optic disc. [97, 98] The sensitivity of automatic DR screening has been reported as ranging from 75 to 94.7%, with comparable specificity and accuracy [99]. Several publicly available retinal datasets have been used to train, validate, and test these AI systems, and also to compare performance against other systems; namely, DIARETDB1, Kaggle, E-ophtha, DDR, DRIVE, HRF, Messidor, Messidor-2, STARE, CHASE DB1, Indian Diabetic Retinopathy Image Dataset (IDRiD), ROC, and DR2 [57, 100–108]. Several studies have used these datasets to detect red lesions, microaneurysms, DR lesions, exudates, individual DR stages, and blood vessel segmentation [38, 40, 41, 43, 52, 109, 110].
Another area of focus is the detection of DMO, currently assessed by OCT as the gold standard. AI-based groups have tried detecting DMO from colour fundus photography based on exudates and accurate identification of the macula. Automated detection via OCT imaging is ongoing, focusing on retinal layer segmentation [111, 112] and specific lesion (e.g. cysts) identification [113–118]. Recently, DL has also been used to detect macular thickening based on colour photographs, and it has been found to be comparable to OCT-measured thickness [119].
Multiple programs have tried to use AI-based methods in population-based screening for DR. The United States Food and Drug Administration (US FDA) has recently approved IDx-DR, a CNN for screening DR stages in adults aged 22 years or older [49, 120]. Initial versions of IDx-DR have been evaluated as part of the Iowa Detection Programme and have shown good results in White, North African, and Sub-Saharan populations [25]. Similar software, like the RetmarkerDR in Portugal and EyeArt in Canada, have been tested in local screening programs [121, 122]. Multiple South-Asian eye institutes are also involved in development and validation of AI-based algorithms in DR [95, 123, 124]. Recently, a Singapore-based DL tool has shown comparable diagnostic accuracy to manual grading, and a semi-automated DL model involving a secondary human assessment may prove to be the most cost-effective model [125, 126]. Their real-world performance remains to be tested [127].
Age-related macular degeneration (AMD)
The use of AI with DL tools has great potential in AMD, both for diagnostic purposes—while allowing for a more efficient and accurate approach—to prognostication of affected individuals and perhaps to directly determine (predict) efficacy of treatments. The most common imaging modalities being explored in the field of AI for AMD are OCT, colour fundus image, and fundus autofluorescence (FAF). OCT-angiography (OCTA) has also been used in DL approaches to diagnose and classify AMD, achieving high accuracy and sensitivity [128, 129]. Due to the huge number of studies, selected key ones will be discussed, with a summary of a broad range of studies in Table 1.
One of the first attempts to evaluate ML algorithms in risk assessment of AMD was a European study by van Grinsven et al. that aimed to detect and quantify drusen on colour fundus photographs in eyes without and with early to moderate AMD [61]. This study demonstrated that the proposed system was in keeping with experienced human observers in detecting the presence of drusen as well as estimating the area, with an ICC greater than 0.85. For AMD risk assessment, it achieved a ROC of 0.948 and 0.954—similar performance to human graders. Subsequently, the same group explored another algorithm for automatic detection of reticular pseudodrusen (RPD) [62]. This followed a multimodal imaging approach using colour fundus, FAF, and near-infrared images, with automated quantification having similar performance to the observers.
In 2018, Schmidt-Erfuth et al. evaluated the predictive potential of ML in terms of best-corrected visual acuity (BCVA) by analysing OCT volume scan features—intraretinal fluid (IRF), subretinal fluid (SRF), and pigment epithelial detachment (PED) [130]. A modest correlation was found between BCVA and OCT at baseline (R2 = 0.21), while functional outcome prediction accuracy increased in linear fashion. The same group then explored automated quantification of fluid volumes using a DL method and a CNN, using OCT data from the HARBOR study (NCT00891735) for neovascular AMD (nAMD) [131]. Retinal fluid volumes (IRF, SRF, and PED) were then validated by the authors as important biomarkers in nAMD [132].
A more recent study attempted to introduce an AI system that combines 3D OCT images and automatic tissue maps in individuals with unilateral nAMD to predict progression in the contralateral eye [70]. It achieved a sensitivity of 80% at 55% specificity and 34% specificity at 90% sensitivity while being able to identify high-risk groups and changes in anatomy before conversion to nAMD, outperforming 5 out of 6 experts. Also, the age-related eye disease studies (AREDS and AREDS2) used DL algorithms and survival analysis to predict risk of late AMD, which achieved high prognostic accuracy [133].
Several segmentation models have been described in AMD. In 2018, De Fauw et al. created a landmark OCT image segmentation model that utilised a DL framework to perform segmentation and automated diagnosis of retinal diseases [134]. Subsequently, Liefers et al. validated a DL model for segmentation of retinal features specifically in individuals with atrophic AMD and nAMD, with results comparable to independent observers [135]. A further automated segmentation algorithm with a CNN has been explored to quantify IRF, SRF, PED, and subretinal hyperreflective material (SHRM) in nAMD [136]. There was good agreement for both the segmentation and detection of lesions between clinicians and the network (dice scores ≥ 0.75 for all features). Two applications with validated automated DL segmentation algorithms are currently commercially available: RetinAI (Medical AG, Switzerland) and RetInSight (Vienna, Austria) [137].
Dry AMD with geographic atrophy (GA) has also been actively investigated. Zhang et al. developed a DL model that segments and classifies GA on OCT images, achieving similar performance to manual specialist assessment [138]. Another group segmented GA in both OCT and FAF images and had reasonable agreement, with better performance (highest dice) in FAF [139]. GA algorithms have also been used to predict VA, with certain features such as photoreceptor degeneration having high predictive significance [140].
Inherited retinal disorders (IRD)
AI algorithms using multimodal imaging techniques have been developed to facilitate the diagnosis [78], classification [80], decipher the genetic aetiology [83], and measure the progression rate of IRD [89, 84].
Chen et al. have developed a CNN that detects if a patient has retinitis pigmentosa (RP) by analysing colour fundus images, with an overall accuracy of 96% (versus 81.5% from four ophthalmology experts) [78]. Another group proposed an FCN that detects pigment in colour images and diagnoses RP with an accuracy of 99.5% [79].
To predict aetiologies, Miere et al. have created a CNN model that can distinguish between FAF images from patients with Stargardt disease (STGD), RP, and best disease (BD), with an overall accuracy of 0.95 [80]. Furthermore, Fujinami-Yokokawa et al. used OCT images to predict causative genes (ABCA4, RP1L1, and EYS) through a DL platform [83]. They achieved an accuracy of 100% for ABCA4, 66.7 to 87.5% for RP1L1, 82.4 to 100% for EYS, and 73.7 to 100% for healthy control images. Miere et al. also created a CNN that is able to outperform specialists in distinguishing between FAF images of STGD and PRPH2-related macular dystrophy (AUROC 0.890 versus experts 0.816) [82]. Shah et al. also achieved an accuracy of 99.6% with a model distinguishing between OCT images from patients with STGD and controls [81]. Crincoli et al. combined image processing with a CNN to differentiate between BD and adult-onset vitelliform macular dystrophy using FAF and OCT images, with an AUROC of 0.880 [23]. Moreover, this endeavour has been recently markedly upscaled by Pontikos et al. to differentiate between 36 gene classes by exploiting multimodal imaging [141]. However, further development is needed, given more than 300 genes are known to cause IRD to date.
STGD is the most prevalent inherited macular dystrophy, and it can affect both children and adults, with multiple ongoing clinical trials [142]. Charng et al. developed a CNN algorithm that segments flecks and is able to monitor their progression over time [84]. They obtained an overall agreement between manual and automatic segmentation of 0.54 ± 0.14 dice score for diffuse speckled patterns and 0.71 ± 0.08 for discrete flecks. Wang et al. also used FAF images, detecting and quantifying areas of atrophy in STGD and AMD [85]. They obtained an accuracy of 0.98 for differentiating normal eyes from those with AMD-related atrophy and 0.95 for eyes with STGD. Atrophic areas were also segmented manually and automatically, with an overlap ratio of 0.89 ± 0.06 in AMD and 0.78 ± 0.17 in STGD [85]. Miere et al. also assessed atrophy and developed a CNN that differentiates between FAF images with GA secondary to AMD and IRD-associated, with an AUROC of 0.981 [86].
Automatic macular OCT segmentation by the device manufacturers is often inaccurate in IRD, requiring manual correction in over one-third of scans [143]. OCT images of STGD were used to create an improved DL-based algorithm that is able to segment the inner and outer retinal limits, providing faster and better macular thickness and volume quantification [87]. Lastly, adaptive optics scanning light ophthalmoscopy images of STGD have also been used to develop an FCN that is able to accurately count macular cones (dice score: 0.9431 ± 0.0482) [88].
Other tools are being designed to assess disease severity and potentially have applications in determining eligibility for interventional trials. A CNN has been developed by Camino et al. that segments preserved EZ area on OCT images from patients with RP and choroideremia (CHM) [89]. This tool reached 0.894 ± 0.102 similarity between automatic and manual grading for RP and 0.912 ± 0.055 for CHM. Loo et al. also targeted EZ segmentation and validated their algorithm for macular telangiectasia in patients with USH2A-related RP, with excellent applicability (dice score 0.79 ± 0.27) [91]. Similarly, Wang et al. also tested an EZ segmentation CNN in USH2A-RP and obtained a Dice score of 0.867 ± 0.105 [92]. CHM EZ segmentation was then attempted by Wang et al. through a nonneural random forest approach and reached a Jaccard similarity index between manual and automated segmentation of 0.876 ± 0.066 [90].
Predicting VA based on OCT and infrared images in RP has been assessed by Liu et al. They were able to determine if a patient with RP had VA below or above 20/40, with an AUC of 0.85 [93]. Sumaroka et al. also developed a nonneural network to predict foveal sensitivity (Humphrey visual field testing), VA, and possible outcome of therapy in patients with blue cone monochromacy based on OCT scans, with good results [94].
Retinopathy of prematurity (ROP)
ROP is an important cause of preventable childhood blindness worldwide [144]. ROP causes abnormal blood vessel growth and can be detected by trained ophthalmologists using indirect ophthalmoscopy, with access to adequate, timely screening potentially limited due to the requirement of highly trained personnel and equipment. DL-based detection and staging of ROP[145] by evaluation of posterior pole fundus images has been attempted with high sensitivity and specificity [146]. Authors have developed a ROP vascular severity score with good correlation with the labels set by the International Classification of Retinopathy of Prematurity committee [147]. The DeepROP score [148] and i-ROP DL system are DL algorithms developed to evaluate clinically significant severe ROP at the posterior pole [149]. ROP plus disease, a more aggressive form of ROP, is often difficult to diagnose given the lack of consensus among ophthalmologists; several authors have evaluated automated algorithms that may be able to objectively diagnose plus disease [150–153].
These study limitations are the review of the literature in a non-systematic approach, possibly leading to some papers being omitted or not adequately prioritised; and editorial restrictions, which prevented us from doing a comprehensive review of AI applications in all retinal disorders. Substantial research has been undertaken in other fields of medical retina (e.g., uveitis and oncology), which will be reviewed in a subsequent project. [154, 155]
Concluding remarks and future directions
Retinal disease has been at the forefront of AI in ophthalmology, with the first AI-related publication being on DR. Since then, research groups focusing their efforts on AI have multiplied around the world, targeting all aspects of the patient journey, including diagnosis, triage, and prognostication, by leveraging multiple imaging (and functional) modalities, as well as a range of AI tools. A shortage of medical professionals is anticipated in the short term, likely further increasing healthcare inequalities and challenging our ability to improve care for preventable diseases [156]. AI represents one important approach to help meet these challenges and moreover facilitate improvements in patient care—both at the individual level with more timely, accurate, and bespoke management, as well as population-level, large-scale healthcare. Ever-improving DNN and CNN algorithms can become a helping hand for healthcare to lean on towards meeting current capability endpoints.
Despite the huge promise, many challenges remain for AI in ophthalmology, including, (i) the need for larger, more diverse, and representative datasets that fully represent real life, (ii) the closer collaboration by experts (both national and international) to develop disease-specific consensus and subsequently provide a comprehensive large volume of image grading, and (iii) greater synergy between healthcare professionals, patients, and data scientists, communicating and improving the software interface as it is being iteratively created, and ensuring it complements the human interaction that underpins the practice of medicine, rather than seeking to replace it [157]. Further uses of AI are yet to be explored, such as multimodal inputs to determine the best candidates for interventional clinical trials, the selection of the ideal anti-VEGF and therapeutic scheme in nAMD, and the estimation of functional impairment based on structural parameters for IRD, among others.
The future of healthcare will increasingly incorporate the advantages that AI can provide to improve the lives of our patients and no doubt perform assessments quicker and more accurately than retina specialists can currently sustainably provide, allowing us to spend more time being better clinicians and scientists. Nevertheless, as always with new technology, there will be new learnings and surprises along the way.
Funding
This study was funded by grants from the Wellcome Trust [099173/Z/12/Z], the National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, Moorfields Eye Charity, Retina UK, and the Foundation Fighting Blindness (no specific grant/award number for the latter). NP is funded by an NIHR AI Award (AI_AWARD02488). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Declarations
Ethical approval
This article does not contain any studies with animals or human participants performed by any of the authors.
Conflict of interest
The authors alone are responsible for the content and writing of this article. MM consults for MeiraGTx Ltd. All other authors certify that they have no affiliations with or involvement in any organisation or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; expert testimony or patent-licensing arrangements) or non-financial interest (such as personal or professional relationships, affiliations, knowledge, or beliefs) in the subject matter or materials discussed in this manuscript.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Konstantinos Balaskas, Email: k.balaskas@nhs.net.
Michel Michaelides, Email: michel.michaelides@ucl.ac.uk.
References
- 1.Causes of blindness and vision impairment in and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the right to sight: an analysis for the Global Burden of Disease Study. Lancet Glob Health. 2020;9:e144–e160. doi: 10.1016/S2214-109X(20)30489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solebo AL, Teoh L, Rahi J. Epidemiology of blindness in children. Arch Dis Child. 2017;102:853–857. doi: 10.1136/archdischild-2016-310532. [DOI] [PubMed] [Google Scholar]
- 3.(https://www.cycloneinteractive.com/cyclone/assets/File/digital-universe-healthcare-vertical-report-ar.pdf)
- 4.Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ et al (2015) Big data: astronomical or genomical? PLoS Biol 13(7):e1002195. 10.1371/journal.pbio.1002195 [DOI] [PMC free article] [PubMed]
- 5.Ting DSW, Lin H, Ruamviboonsuk P, et al. Artificial intelligence, the internet of things, and virtual clinics: ophthalmology at the digital translation forefront. Lancet Digit Health. 2020;2:e8–e9. doi: 10.1016/S2589-7500(19)30217-1. [DOI] [PubMed] [Google Scholar]
- 6.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 8.Dias R, Torkamani A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019;11:1–12. doi: 10.1186/s13073-019-0689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah P, Mishra D, Shanmugam M, et al. Acceptability of artificial intelligence-based retina screening in general population. Indian J Ophthalmol. 2022;70:1140–1144. doi: 10.4103/ijo.IJO_1840_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong L, He W, Zhang R et al (2022) Artificial intelligence for screening of multiple retinal and optic nerve diseases. JAMA Netw Open 5(5):e229960. 10.1001/jamanetworkopen.2022.9960 [DOI] [PMC free article] [PubMed]
- 11.Tan TE, Chan HW, Singh M, Wong TY, Pulido JS, Michaelides M, Sohn EH, Ting D (2021) Artificial intelligence for diagnosis of inherited retinal disease: an exciting opportunity and one step forward. Br J Ophthalmol 105(9):1187–1189. 10.1136/bjophthalmol-2021-319365 [DOI] [PubMed]
- 12.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 13.El Naqa I, Murphy MJ (2015) What is machine learning?. Springer International Publishing, Berlin, pp 3–11
- 14.Tizhoosh HR (2005, December) Reinforcement learning based on actions and opposite actions. In International conference on artificial intelligence and machine learning, vol 414
- 15.Rashidi HH, Tran NK, Betts EV, et al. Artificial intelligence and machine learning in pathology: the present landscape of supervised methods. Acad Pathol. 2019;6:2374289519873088. doi: 10.1177/2374289519873088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghods A, Cook DJ. A survey of deep network techniques all classifiers can adopt. Data Min Knowl Discov. 2021;35:46–87. doi: 10.1007/s10618-020-00722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci. 2020;111:1452–1460. doi: 10.1111/cas.14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhillon A, Verma GK. Convolutional neural network: a review of models, methodologies and applications to object detection. Progress in Artificial Intelligence. 2020;9:85–112. doi: 10.1007/s13748-019-00203-0. [DOI] [Google Scholar]
- 19.Ferizi U, Honig S, Chang G. Artificial intelligence, osteoporosis and fragility fractures. Curr Opin Rheumatol. 2019;31:368–375. doi: 10.1097/BOR.0000000000000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols JA, Herbert Chan HW, Baker MAB. Machine learning: applications of artificial intelligence to imaging and diagnosis. Biophys Rev. 2019;11:111–118. doi: 10.1007/s12551-018-0449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose AM, Shah AZ, Waseem NH, et al. Expression of PRPF31 and TFPT: regulation in health and retinal disease. Hum Mol Genet. 2012;21:4126–4137. doi: 10.1093/hmg/dds242. [DOI] [PubMed] [Google Scholar]
- 22.Goldhagen BE, Al-khersan H. Diving deep into deep learning: an update on artificial intelligence in retina. Curr Ophthalmol Rep. 2020;8:121–128. doi: 10.1007/s40135-020-00240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crincoli E, Zhao Z, Querques G, et al. Deep learning to distinguish best vitelliform macular dystrophy (BVMD) from adult-onset vitelliform macular degeneration (AVMD) Sci Rep. 2022;12:12745. doi: 10.1038/s41598-022-16980-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chicco D. Ten quick tips for machine learning in computational biology. BioData Min. 2017;10:1–17. doi: 10.1186/s13040-017-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abràmoff MD, Folk JC, Han DP, et al. Automated analysis of retinal images for detection of referable diabetic retinopathy. JAMA Ophthalmol. 2013;131:351–357. doi: 10.1001/jamaophthalmol.2013.1743. [DOI] [PubMed] [Google Scholar]
- 26.Fleming AD, Goatman KA, Philip S, et al. Automated grading for diabetic retinopathy: a large-scale audit using arbitration by clinical experts. Br J Ophthalmol. 2010;94:1606–1610. doi: 10.1136/bjo.2009.176784. [DOI] [PubMed] [Google Scholar]
- 27.Hansen MB, Abràmoff MD, Folk JC, Mathenge W, Bastawrous A, Peto T (2015) Results of automated retinal image analysis for detection of diabetic retinopathy from the nakuru study, Kenya. PLoS ONE 10(10):e0139148. 10.1371/journal.pone.0139148 [DOI] [PMC free article] [PubMed]
- 28.Roychowdhury S, Koozekanani DD, Parhi KK. DREAM: diabetic retinopathy analysis using machine learning. IEEE J Biomed Health Inform. 2014;18:1717–1728. doi: 10.1109/JBHI.2013.2294635. [DOI] [PubMed] [Google Scholar]
- 29.Trucco E, Ruggeri A, Karnowski T, et al. Validating retinal fundus image analysis algorithms: issues and a proposal. Invest Ophthalmol Vis Sci. 2013;54:3546–3559. doi: 10.1167/iovs.12-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quellec G, Lamard M, Cazuguel G, et al. Automated assessment of diabetic retinopathy severity using content-based image retrieval in multimodal fundus photographs. Invest Ophthalmol Vis Sci. 2011;52:8342–8348. doi: 10.1167/iovs.11-7418. [DOI] [PubMed] [Google Scholar]
- 31.Abràmoff MD, Reinhardt JM, Russell SR, et al. Automated early detection of diabetic retinopathy. Ophthalmology. 2010;117:1147–1154. doi: 10.1016/j.ophtha.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaum E, Karnowski TP, Govindasamy VP, Abdelrahman M, Tobin KW (2008) Automated diagnosis of retinopathy by content-based image retrieval. Retina 28(10):1463–1477. 10.1097/IAE.0b013e31818356dd [DOI] [PubMed]
- 33.Fleming AD, Goatman KA, Philip S, et al. The role of haemorrhage and exudate detection in automated grading of diabetic retinopathy. Br J Ophthalmol. 2010;94:706–711. doi: 10.1136/bjo.2008.149807. [DOI] [PubMed] [Google Scholar]
- 34.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 35.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, pp 2818–2826
- 36.Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2017) Inception-v4, Inception-ResNet and the impact of residual connections on learning. Proceedings of the AAAI Conference on Artificial Intelligence. AAAI 31(1). 10.1609/aaai.v31i1.11231
- 37.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, pp 770–778
- 38.Orlando JI, Prokofyeva E, del Fresno M, Blaschko MB. An ensemble deep learning based approach for red lesion detection in fundus images. Comput Methods Programs Biomed. 2018;153:115–127. doi: 10.1016/j.cmpb.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 39.Yan Y, Gong J, Liu Y (2019) A novel deep learning method for red lesions detection using hybrid feature. In: 2019 Chinese Control And Decision Conference (CCDC), Nanchang, China, pp 2287–2292. 10.1109/CCDC.2019.8833190
- 40.Chudzik P, Majumdar S, Calivá F, et al. Microaneurysm detection using fully convolutional neural networks. Comput Methods Programs Biomed. 2018;158:185–192. doi: 10.1016/j.cmpb.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 41.Adem K. Exudate detection for diabetic retinopathy with circular Hough transformation and convolutional neural networks. Expert Syst Appl. 2018;114:289–295. doi: 10.1016/j.eswa.2018.07.053. [DOI] [Google Scholar]
- 42.Wang H, Yuan G, Zhao X, Peng L, Wang Z, He Y, Qu C, Peng Z (2020) Hard exudate detection based on deep model learned information and multi-feature joint representation for diabetic retinopathy screening. Comput Methods Prog Biomed 191:105398. 10.1016/j.cmpb.2020.105398 [DOI] [PubMed]
- 43.Quellec G, Charrière K, Boudi Y, et al. Deep image mining for diabetic retinopathy screening. Med Image Anal. 2017;39:178–193. doi: 10.1016/j.media.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Esfahani M T, Ghaderi M, Kafiyeh R (2018) Classification of diabetic and normal fundus images using new deep learning method. Leonardo Electron J Pract Technol 17(32):233–248
- 45.Pires R, Avila S, Wainer J, Valle E, Abramoff MD, Rocha A (2019) A data-driven approach to referable diabetic retinopathy detection. Artif Intell Med 96:93–106. 10.1016/j.artmed.2019.03.009 [DOI] [PubMed]
- 46.Jiang H, Yang K, Gao M, Zhang D, Ma H, Qian W (2019) An interpretable ensemble deep learning model for diabetic retinopathy disease classification. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, pp 2045–2048. 10.1109/EMBC.2019.8857160 [DOI] [PubMed]
- 47.Liu YP, Li Z, Xu C, Li J, Liang R (2019) Referable diabetic retinopathy identification from eye fundus images with weighted path for convolutional neural network. Artif Intell Med 99:101694. 10.1016/j.artmed.2019.07.002 [DOI] [PubMed]
- 48.Zago GT, Andreão RV, Dorizzi B, Teatini Salles EO (2020) Diabetic retinopathy detection using red lesion localization and convolutional neural networks. Comput Biol Med 116:103537. 10.1016/j.compbiomed.2019.103537 [DOI] [PubMed]
- 49.Abràmoff MD, Lou Y, Erginay A, et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci. 2016;57:5200–5206. doi: 10.1167/iovs.16-19964. [DOI] [PubMed] [Google Scholar]
- 50.Pratt H, Coenen F, Broadbent DM, et al. Convolutional neural networks for diabetic retinopathy. Procedia Comput Sci. 2016;90:200–205. doi: 10.1016/j.procs.2016.07.014. [DOI] [Google Scholar]
- 51.Dutta S, Manideep BC, Basha SM, Caytiles RD, Iyengar NCSN (2018) Classification of diabetic retinopathy images by using deep learning models. Int J Grid Distrib Comput 11(1):89–106. 10.14257/ijgdc.2018.11.1.09
- 52.Wang X, Lu Y, Wang Y, Chen W-B (2018) Diabetic retinopathy stage classification using convolutional neural networks. In 2018 IEEE International Conference on Information Reuse and Integration (IRI), Salt Lake City, pp 465–471. 10.1109/IRI.2018.00074
- 53.Wan S, Liang Y, Zhang Y. Deep convolutional neural networks for diabetic retinopathy detection by image classification. Comput Electr Eng. 2018;72:274–282. doi: 10.1016/j.compeleceng.2018.07.042. [DOI] [Google Scholar]
- 54.ur Rehman M, Abbas Z, Khan SH, Ghani SH, Najam (2018) Diabetic retinopathy fundus image classification using discrete wavelet transform. In: 2018 2nd International Conference on Engineering Innovation (ICEI), Bangkok, pp 75-80. 10.1109/ICEI18.2018.8448628
- 55.Zhang W, Zhong J, Yang S, et al. Automated identification and grading system of diabetic retinopathy using deep neural networks. Knowl Based Syst. 2019;175:12–25. doi: 10.1016/j.knosys.2019.03.016. [DOI] [Google Scholar]
- 56.Harangi B, Toth J, Baran A, Hajdu A (2019) Automatic screening of fundus images using a combination of convolutional neural network and hand-crafted features. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, pp 2699–2702. 10.1109/EMBC.2019.8857073 [DOI] [PubMed]
- 57.Li T, Gao Y, Wang K, et al. Diagnostic assessment of deep learning algorithms for diabetic retinopathy screening. Inf Sci (N Y) 2019;501:511–522. doi: 10.1016/j.ins.2019.06.011. [DOI] [Google Scholar]
- 58.Shanthi T, Sabeenian RS. Modified Alexnet architecture for classification of diabetic retinopathy images. Comput Electr Eng. 2019;76:56–64. doi: 10.1016/j.compeleceng.2019.03.004. [DOI] [Google Scholar]
- 59.Wang J, Luo J, Liu B, et al. Automated diabetic retinopathy grading and lesion detection based on the modified R-FCN object-detection algorithm. IET Comput Vision. 2020;14:1–8. doi: 10.1049/iet-cvi.2018.5508. [DOI] [Google Scholar]
- 60.Li X, Hu X, Yu L, et al. CANet: cross-disease attention network for joint diabetic retinopathy and diabetic macular edema grading. IEEE Trans Med Imaging. 2020;39:1483–1493. doi: 10.1109/TMI.2019.2951844. [DOI] [PubMed] [Google Scholar]
- 61.van Grinsven MJJP, Lechanteur YTE, van de Ven JPH, et al. Automatic drusen quantification and risk assessment of age-related macular degeneration on color fundus images. Invest Ophthalmol Vis Sci. 2013;54:3019–3027. doi: 10.1167/iovs.12-11449. [DOI] [PubMed] [Google Scholar]
- 62.Van Grinsven MJJP, Buitendijk GHS, Brussee C, et al. Automatic identification of reticular pseudodrusen using multimodal retinal image analysis. Invest Ophthalmol Vis Sci. 2015;56:633–639. doi: 10.1167/iovs.14-15019. [DOI] [PubMed] [Google Scholar]
- 63.Seddon JM, Silver RE, Kwong M, Rosner B. Risk prediction for progression of macular degeneration: 10 common and rare genetic variants, demographic, environmental, and macular covariates. Investig Opthalmol Vis Sci. 2015;56(4):2192. doi: 10.1167/iovs.14-15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Z, Bogunović H, Asgari R, Schmidt-Erfurth U, Guymer RH. Predicting progression of age-related macular degeneration using OCT and fundus photography. Ophthalmol Retina. 2021;5(2):118–125. doi: 10.1016/j.oret.2020.06.026. [DOI] [PubMed] [Google Scholar]
- 65.Wu Z, Luu CD, Hodgson LA, et al. Examining the added value of microperimetry and low luminance deficit for predicting progression in age-related macular degeneration. Br J Ophthalmol. 2021;105(5):711–715. doi: 10.1136/bjophthalmol-2020-315935. [DOI] [PubMed] [Google Scholar]
- 66.Bhuiyan A, Wong TY, Ting DSW, Govindaiah A, Souied EH, Smith RT. Artificial intelligence to stratify severity of age-related macular degeneration (AMD) and predict risk of progression to late AMD. Transl Vis Sci Technol. 2020;9(2):25. doi: 10.1167/tvst.9.2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan Q, Weeks DE, Xin H et al (2020) Deep-learning based prediction of late age-related macular degeneration progression. Nat Mach Intell 2(2):141–150. 10.1038/s42256-020-0154-9 [DOI] [PMC free article] [PubMed]
- 68.Burlina PM, Joshi N, Pacheco KD, Freund DE, Kong J, Bressler NM. Use of deep learning for detailed severity characterization and estimation of 5-year risk among patients with age-related macular degeneration. JAMA Ophthalmol. 2018;136(12):1359. doi: 10.1001/jamaophthalmol.2018.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiu CJ, Mitchell P, Klein R, et al. A risk score for the prediction of advanced age-related macular degeneration. Ophthalmology. 2014;121(7):1421–1427. doi: 10.1016/j.ophtha.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yim J, Chopra R, Spitz T, et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat Med. 2020;26:892–899. doi: 10.1038/s41591-020-0867-7. [DOI] [PubMed] [Google Scholar]
- 71.Banerjee I, de Sisternes L, Hallak JA, et al. Prediction of age-related macular degeneration disease using a sequential deep learning approach on longitudinal SD-OCT imaging biomarkers. Sci Rep. 2020;10(1):15434. doi: 10.1038/s41598-020-72359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hallak JA, de Sisternes L, Osborne A, Yaspan B, Rubin DL, Leng T. Imaging, genetic, and demographic factors associated with conversion to neovascular age-related macular degeneration. JAMAOphthalmol. 2019;137(7):738. doi: 10.1001/jamaophthalmol.2019.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Sisternes L, Simon N, Tibshirani R, Leng T, Rubin DL. Quantitative SD-OCT imaging biomarkers as indicators of age-related macular degeneration progression. Investig Opthalmol Vis Sci. 2014;55(11):7093. doi: 10.1167/iovs.14-14918. [DOI] [PubMed] [Google Scholar]
- 74.Russakoff DB, Lamin A, Oakley JD, Dubis AM, Sivaprasad S. Deep learning for prediction of AMD progression: a pilot study. Investig Opthalmol Vis Sci. 2019;60(2):712. doi: 10.1167/iovs.18-25325. [DOI] [PubMed] [Google Scholar]
- 75.Schmidt-Erfurth U, Waldstein SM, Klimscha S, et al. Prediction of individual disease conversion in early AMD using artificial intelligence. Investig Opthalmol Vis Sci. 2018;59(8):3199. doi: 10.1167/iovs.18-24106. [DOI] [PubMed] [Google Scholar]
- 76.Liefers B, Taylor P, Alsaedi A, et al. Quantification of key retinal features in early and late age-related macular degeneration using deep learning. Am J Ophthalmol. 2021;226:1–12. doi: 10.1016/j.ajo.2020.12.034. [DOI] [PubMed] [Google Scholar]
- 77.Lee H, Kang KE, Chung H, Kim HC. Automated segmentation of lesions including subretinal hyperreflective material in neovascular age-related macular degeneration. Am J Ophthalmol. 2018;191:64–75. doi: 10.1016/j.ajo.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 78.Chen T-C, Lim WS, Wang VY, et al. Artificial intelligence–assisted early detection of retinitis pigmentosa — the most common inherited retinal degeneration. J Digit Imaging. 2021;34:948–958. doi: 10.1007/s10278-021-00479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arsalan M, Baek NR, Owais M, Mahmood T, Park KR (2020) Deep learning-based detection of pigment signs for analysis and diagnosis of retinitis pigmentosa. Sensors 20(12):3454. 10.3390/s20123454 [DOI] [PMC free article] [PubMed]
- 80.Miere A, Le Meur T, Bitton K, Pallone C, Semoun O, Capuano V, Colantuono D, Taibouni K, Chenoune Y, Astroz P, Berlemont S, Petit E, Souied E (2020) Deep learning-based classification of inherited retinal diseases using fundus autofluorescence. J Clin Med 9(10):3303. 10.3390/jcm9103303 [DOI] [PMC free article] [PubMed]
- 81.Shah M, Roomans Ledo A, Rittscher J. Automated classification of normal and Stargardt disease optical coherence tomography images using deep learning. Acta Ophthalmol. 2020;98:e715–e721. doi: 10.1111/aos.14353. [DOI] [PubMed] [Google Scholar]
- 82.Miere A, Zambrowski O, Kessler A, Mehanna C-J, Pallone C, Seknazi D, Denys P, Amoroso F, Petit E, Souied EH (2021) Deep learning to distinguish abca4-related stargardt disease from prph2-related pseudo-stargardt pattern dystrophy. J Clin Med 10(24):5742. 10.3390/s20123454 [DOI] [PMC free article] [PubMed]
- 83.Fujinami-Yokokawa Y, Pontikos N, Yang L, et al. Prediction of causative genes in inherited retinal disorders from spectral-domain optical coherence tomography utilizing deep learning techniques. J Ophthalmol. 2019;2019:1691064. doi: 10.1155/2019/1691064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Charng J, Xiao D, Mehdizadeh M, et al. Deep learning segmentation of hyperautofluorescent fleck lesions in Stargardt disease. Sci Rep. 2020;10:16491. doi: 10.1038/s41598-020-73339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z, Sadda SR, Hu Z (2019) Deep learning for automated screening and semantic segmentation of age-related and juvenile atrophic macular degeneration. In: Proc. SPIE 10950, Medical Imaging 2019: Computer-Aided Diagnosis, 109501Q. 10.1117/12.2511538
- 86.Miere A, Capuano V, Kessler A, Zambrowski O, Jung C, Colantuono D, Pallone C, Semoun O, Petit E, Souied E (2021) Deep learning-based classification of retinal atrophy using fundus autofluorescence imaging. Comput Biol Med 130:104198. 10.1016/j.compbiomed.2020.104198 [DOI] [PubMed]
- 87.Kugelman J, Alonso-Caneiro D, Chen Y, et al. Retinal boundary segmentation in Stargardt disease optical coherence tomography images using automated deep learning. Transl Vis Sci Technol. 2020;9:12. doi: 10.1167/tvst.9.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davidson B, Kalitzeos A, Carroll J, et al. Automatic cone photoreceptor localisation in healthy and Stargardt afflicted retinas using deep learning. Sci Rep. 2018;8:7911. doi: 10.1038/s41598-018-26350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Camino A, Wang Z, Wang J, et al. Deep learning for the segmentation of preserved photoreceptors on en face optical coherence tomography in two inherited retinal diseases. Biomed Opt Express. 2018;9:3092–3105. doi: 10.1364/BOE.9.003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Z, Camino A, Hagag AM et al (2018) Automated detection of preserved photoreceptor on optical coherence tomography in choroideremia based on machine learning. J Biophotonics 11:e201700313. 10.1002/jbio.201700313 [DOI] [PMC free article] [PubMed]
- 91.Loo J, Jaffe GJ, Duncan JL, et al. Validation of a deep learning-based algorithm for segmentation of the ellipsoid zone on optical coherence tomography images of an USH2A-related retinal degeneration clinical trial. Retina. 2022;42:1347–1355. doi: 10.1097/IAE.0000000000003448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang YZ, Birch DG (2022) Performance of deep learning models in automatic measurement of ellipsoid zone area on baseline optical coherence tomography (OCT) images from the rate of progression of USH2A-Related retinal degeneration (RUSH2A) study. Front Med (Lausanne) 9:932498. 10.3389/fmed.2022.932498 [DOI] [PMC free article] [PubMed]
- 93.Liu TYA, Ling C, Hahn L et al (2022) Prediction of visual impairment in retinitis pigmentosa using deep learning and multimodal fundus images. Br J Ophthalmol. 10.1136/bjo-2021-320897 [DOI] [PMC free article] [PubMed]
- 94.Sumaroka A, Cideciyan AV, Sheplock R, et al. Foveal therapy in blue cone monochromacy: predictions of visual potential from artificial intelligence. Front Neurosci. 2020;14:800. doi: 10.3389/fnins.2020.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 96.Wong LY, Acharya R, Venkatesh YV, Chee C, Min LC. Identification of different stages of diabetic retinopathy using retinal optical images. Inf Sci. 2008;178(106):21. [Google Scholar]
- 97.Imani E, Pourreza H-R, Banaee T. Fully automated diabetic retinopathy screening using morphological component analysis. Comput Med Imaging Graph. 2015;43:78–88. doi: 10.1016/j.compmedimag.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 98.Yazid H, Arof H, Isa HM. Automated identification of exudates and optic disc based on inverse surface thresholding. J Med Syst. 2012;36:1997–2004. doi: 10.1007/s10916-011-9659-4. [DOI] [PubMed] [Google Scholar]
- 99.Niemeijer M, Abràmoff MD, van Ginneken B. Fast detection of the optic disc and fovea in color fundus photographs. Med Image Anal. 2009;13:859–870. doi: 10.1016/j.media.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kauppi T, Pietilä J, Kalesnykiene V, Kamarainen JK, Lensu L, Sorri I, Raninen A, Voutilainen R, Uusitalo H, Kälviäinen H (2007, September) The diaretdb1 diabetic retinopathy database and evaluation protocol. In BMVC 1(1):10
- 101.Kaggle dataset [Online]. Available, https://kaggle.com/c/diabetic-retinopat hy-detection
- 102.Decencière E, Cazuguel G, Zhang X, et al. TeleOphta: Machine learning and image processing methods for teleophthalmology. IRBM. 2013;34:196–203. doi: 10.1016/j.irbm.2013.01.010. [DOI] [Google Scholar]
- 103.Staal J, Abràmoff MD, Niemeijer M, et al. Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging. 2004;23:501–509. doi: 10.1109/TMI.2004.825627. [DOI] [PubMed] [Google Scholar]
- 104.Decencière E, Zhang X, Cazuguel G, et al. Feedback on a publicly distributed image database: the Messidor database. Image Analysis & Stereology. 2014;33:231. doi: 10.5566/ias.1155. [DOI] [Google Scholar]
- 105.Hoover AD, Kouznetsova V, Goldbaum M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans Med Imaging. 2000;19:203–210. doi: 10.1109/42.845178. [DOI] [PubMed] [Google Scholar]
- 106.Owen CG, Rudnicka AR, Mullen R, et al. Measuring retinal vessel tortuosity in 10-year-old children: validation of the Computer-Assisted Image Analysis of the Retina (CAIAR) program. Invest Ophthalmol Vis Sci. 2009;50:2004–2010. doi: 10.1167/iovs.08-3018. [DOI] [PubMed] [Google Scholar]
- 107.Porwal P, Pachade S, Kamble R, et al. Indian Diabetic Retinopathy Image Dataset (IDRiD): a database for diabetic retinopathy screening research. Data (Basel) 2018;3:25. doi: 10.3390/data3030025. [DOI] [Google Scholar]
- 108.ROC dataset [Online]. Available, http://roc.healthcare.uiowa.edu. [36] DR2 [Online]. Available, https://figshare.com/articles/Advancing_Bag_of_Visual_Words_Representations_for_Lesion_Classification_in_Retinal_Images/953671 .
- 109.Xu K, Feng D, Mi H (2017) Deep convolutional neural network-based early automated detection of diabetic retinopathy using fundus image. Molecules 22(12):2054. 10.3390/molecules22122054 [DOI] [PMC free article] [PubMed]
- 110.Oliveira A, Pereira S, Silva CA. Retinal vessel segmentation based on fully convolutional neural networks. Expert Syst Appl. 2018;112:229–242. doi: 10.1016/j.eswa.2018.06.034. [DOI] [Google Scholar]
- 111.Mayer MA, Hornegger J, Mardin CY, Tornow RP. Retinal nerve fiber layer segmentation on FD-OCT scans of normal subjects and glaucoma patients. Biomed Opt Express. 2010;1:1358–1383. doi: 10.1364/BOE.1.001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garvin MK, Abramoff MD, Kardon R, et al. Intraretinal layer segmentation of macular optical coherence tomography images using optimal 3-D graph search. IEEE Trans Med Imaging. 2008;27:1495–1505. doi: 10.1109/TMI.2008.923966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Quellec G, Lee K, Dolejsi M, et al. Three-dimensional analysis of retinal layer texture: identification of fluid-filled regions in SD-OCT of the macula. IEEE Trans Med Imaging. 2010;29:1321–1330. doi: 10.1109/TMI.2010.2047023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schlegl T, Glodan A-M, Podkowinski D, Waldstein SM, Gerendas BS, Schmidt-Erfurth U, Langs G. Automatic segmentation and classifcation of intraretinal cystoid fuid and subretinal fuid in 3d-oct using convolutional neural networks. Investig Ophthalmol Vis Sci. 2015;56(7):5920. [Google Scholar]
- 115.Fu D, Tong H, Zheng S, et al. Retinal status analysis method based on feature extraction and quantitative grading in OCT images. Biomed Eng Online. 2016;15:87. doi: 10.1186/s12938-016-0206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Srinivasan PP, Kim LA, Mettu PS, et al. Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images. Biomed Opt Express. 2014;5:3568. doi: 10.1364/BOE.5.003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Venhuizen FG, van Ginneken B, Bloemen B, van Grinsven MJJP, Philipsen R, Hoyng C, Theelen T, Sánchez CI (2015) Automated age-related macular degeneration classification in OCT using unsupervised feature learning. In: Proc. SPIE 9414, Medical Imaging 2015: Computer-Aided Diagnosis, 94141I. 10.1117/12.2081521
- 118.Alsaih K, Lemaitre G, Rastgoo M, et al. Machine learning techniques for diabetic macular edema (DME) classification on SD-OCT images. Biomed Eng Online. 2017;16:68. doi: 10.1186/s12938-017-0352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arcadu F, Benmansour F, Maunz A, et al. Deep learning predicts OCT measures of diabetic macular thickening from color fundus photographs. Investigative Opthalmology & Visual Science. 2019;60:852. doi: 10.1167/iovs.18-25634. [DOI] [PubMed] [Google Scholar]
- 120.Abràmoff MD, Lavin PT, Birch M, et al. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit Med. 2018;1:39. doi: 10.1038/s41746-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ribeiro L, Oliveira C, M, Neves C, Ramos J, D, Ferreira H, Cunha-Vaz J (2015) Screening for diabetic retinopathy in the central region of portugal. Added Value of Automated ‘Disease/No Disease' Grading. Ophthalmologica 233:96–103. 10.1159/000368426 [DOI] [PubMed]
- 122.Rajalakshmi R, Subashini R, Anjana RM, Mohan V. Automated diabetic retinopathy detection in smartphone-based fundus photography using artificial intelligence. Eye (Lond) 2018;32:1138–1144. doi: 10.1038/s41433-018-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gulshan V, Rajan RP, Widner K, et al. Performance of a deep-learning algorithm vs manual grading for detecting diabetic retinopathy in India. JAMA Ophthalmol. 2019;137:987–993. doi: 10.1001/jamaophthalmol.2019.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dismuke C. Progress in examining cost-effectiveness of AI in diabetic retinopathy screening. Lancet Digit Health. 2020;2:e212–e213. doi: 10.1016/S2589-7500(20)30077-7. [DOI] [PubMed] [Google Scholar]
- 125.Ting DSW, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xie Y, Nguyen QD, Hamzah H, et al. Artificial intelligence for teleophthalmology-based diabetic retinopathy screening in a national programme: an economic analysis modelling study. Lancet Digit Health. 2020;2:e240–e249. doi: 10.1016/S2589-7500(20)30060-1. [DOI] [PubMed] [Google Scholar]
- 127.Lee AY, Yanagihara RT, Lee CS, et al. Multicenter, head-to-head, real-world validation study of seven automated artificial intelligence diabetic retinopathy screening systems. Diabetes Care. 2021;44:1168–1175. doi: 10.2337/dc20-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang J, Hormel TT, Gao L, et al. Automated diagnosis and segmentation of choroidal neovascularization in OCT angiography using deep learning. Biomed Opt Express. 2020;11:927–944. doi: 10.1364/BOE.379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vaghefi E, Hill S, Kersten HM, Squirrell D. Multimodal retinal image analysis via deep learning for the diagnosis of intermediate dry age-related macular degeneration: a feasibility study. J Ophthalmol. 2020;2020:7493419. doi: 10.1155/2020/7493419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmidt-Erfurth U, Bogunovic H, Sadeghipour A, et al. Machine learning to analyze the prognostic value of current imaging biomarkers in neovascular age-related macular degeneration. Ophthalmol Retina. 2018;2:24–30. doi: 10.1016/j.oret.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 131.Schmidt-Erfurth U, Vogl W-D, Jampol LM, Bogunović H. Application of automated quantification of fluid volumes to anti–VEGF therapy of neovascular age-related macular degeneration. Ophthalmology. 2020;127:1211–1219. doi: 10.1016/j.ophtha.2020.03.010. [DOI] [PubMed] [Google Scholar]
- 132.Keenan TDL, Chakravarthy U, Loewenstein A, et al. Automated quantitative assessment of retinal fluid volumes as important biomarkers in neovascular age-related macular degeneration. Am J Ophthalmol. 2021;224:267–281. doi: 10.1016/j.ajo.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peng Y, Keenan TD, Chen Q, et al. Predicting risk of late age-related macular degeneration using deep learning. NPJ Digit Med. 2020;3:1–10. doi: 10.1038/s41746-020-00317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 135.Liefers B, Taylor P, Alsaedi A, et al. Quantification of key retinal features in early and late age-related macular degeneration using deep learning. Am J Ophthalmol. 2021;226:1–12. doi: 10.1016/j.ajo.2020.12.034. [DOI] [PubMed] [Google Scholar]
- 136.Lee H, Kang KE, Chung H, Kim HC. Automated segmentation of lesions including subretinal hyperreflective material in neovascular age-related macular degeneration. Am J Ophthalmol. 2018;191:64–75. doi: 10.1016/j.ajo.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 137.Gerendas BS, Sadeghipour A, Michl M, et al. Validation of an automated fluid algorithm on real-world data of neovascular age-related macular degeneration over five years. Retina. 2022;42:1673–1682. doi: 10.1097/IAE.0000000000003557. [DOI] [PubMed] [Google Scholar]
- 138.Zhang G, Fu DJ, Liefers B, et al. Clinically relevant deep learning for detection and quantification of geographic atrophy from optical coherence tomography: a model development and external validation study. Lancet Digit Health. 2021;3:e665–e675. doi: 10.1016/S2589-7500(21)00134-5. [DOI] [PubMed] [Google Scholar]
- 139.Hu Z, Medioni GG, Hernandez M, et al. Segmentation of the geographic atrophy in spectral-domain optical coherence tomography and fundus autofluorescence images. Invest Ophthalmol Vis Sci. 2013;54:8375–8383. doi: 10.1167/iovs.13-12552. [DOI] [PubMed] [Google Scholar]
- 140.Balaskas K, Glinton S, Keenan TDL, et al. Prediction of visual function from automatically quantified optical coherence tomography biomarkers in patients with geographic atrophy using machine learning. Sci Rep. 2022;12:15565. doi: 10.1038/s41598-022-19413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pontikos N, Woof W, Veturi A et al (2022) Eye2Gene: prediction of causal inherited retinal disease gene from multimodal imaging using deep-learning. PREPRINT (Version 1) available at Research Square. 10.21203/rs.3.rs-2110140/v1
- 142.Tanna P, Strauss RW, Fujinami K, Michaelides M. Stargardt disease: clinical features, molecular genetics, animal models and therapeutic options. Br J Ophthalmol. 2017;101:25–30. doi: 10.1136/bjophthalmol-2016-308823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Strauss RW, Muñoz B, Wolfson Y, et al. Assessment of estimated retinal atrophy progression in Stargardt macular dystrophy using spectral-domain optical coherence tomography. Br J Ophthalmol. 2016;100:956–962. doi: 10.1136/bjophthalmol-2015-307035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wheatley CM, Dickinson JL, Mackey DA, et al. Retinopathy of prematurity: recent advances in our understanding. Br J Ophthalmol. 2002;86:696–700. doi: 10.1136/bjo.86.6.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Campbell JP, Kim SJ, Brown JM, et al. Evaluation of a deep learning-derived quantitative retinopathy of prematurity severity scale. Ophthalmology. 2021;128:1070–1076. doi: 10.1016/j.ophtha.2020.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang J, Liu Y, Mitsuhashi T, Matsuo T. Accuracy of deep learning algorithms for the diagnosis of retinopathy of prematurity by fundus images: a systematic review and meta-analysis. J Ophthalmol. 2021;2021:8883946. doi: 10.1155/2021/8883946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Campbell JP, Chiang MF, Chen JS, et al. Artificial intelligence for retinopathy of prematurity: validation of a vascular severity scale against international expert diagnosis. Ophthalmology. 2022;129:e69–e76. doi: 10.1016/j.ophtha.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li J, Huang K, Ju R, et al. Evaluation of artificial intelligence-based quantitative analysis to identify clinically significant severe retinopathy of prematurity. Retina. 2022;42:195–203. doi: 10.1097/IAE.0000000000003284. [DOI] [PubMed] [Google Scholar]
- 149.Redd TK, Campbell JP, Brown JM, et al. Evaluation of a deep learning image assessment system for detecting severe retinopathy of prematurity. Br J Ophthalmol. 2018 doi: 10.1136/bjophthalmol-2018-313156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tan Z, Simkin S, Lai C, Dai S. Deep learning algorithm for automated diagnosis of retinopathy of prematurity plus disease. Transl Vis Sci Technol. 2019;8:23. doi: 10.1167/tvst.8.6.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brown JM, Campbell JP, Beers A, et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018;136:803–810. doi: 10.1001/jamaophthalmol.2018.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang J, Ju R, Chen Y, et al. Automated retinopathy of prematurity screening using deep neural networks. EBioMedicine. 2018;35:361–368. doi: 10.1016/j.ebiom.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ting DSW, Wu W-C, Toth C. Deep learning for retinopathy of prematurity screening. Br J Ophthalmol. 2018 doi: 10.1136/bjophthalmol-2018-313290. [DOI] [PubMed] [Google Scholar]
- 154.Abellanas M, Elena MJ, Keane PA, et al. Artificial intelligence and imaging processing in optical coherence tomography and digital images in uveitis. Ocul Immunol Inflamm. 2022;30:675–681. doi: 10.1080/09273948.2022.2054433. [DOI] [PubMed] [Google Scholar]
- 155.Kaliki S, Vempuluru VS, Ghose N, et al. Artificial intelligence and machine learning in ocular oncology: retinoblastoma. Indian J Ophthalmol. 2023;71:424–430. doi: 10.4103/ijo.IJO_1393_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ting DSW, Al-Aswad LA. Augmented intelligence in ophthalmology: the six rights. Asia Pac J Ophthalmol (Phila) 2021;10:231–233. doi: 10.1097/APO.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ting DSW, Pasquale LR, Peng L, et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103:167–175. doi: 10.1136/bjophthalmol-2018-313173. [DOI] [PMC free article] [PubMed] [Google Scholar]