Abstract
Alzheimer’s disease is the most common neurodegenerative disease, characterized by dementia and premature death. Early-onset familial Alzheimer’s disease is caused in part by pathogenic variants in presenilin 1 (PSEN1) and presenilin 2 (PSEN2), and alternative splicing of these two genes has been implicated in both familial and sporadic Alzheimer’s disease. Here, we leveraged targeted isoform-sequencing to characterize thousands of complete PSEN1 and PSEN2 transcripts in the prefrontal cortex of individuals with sporadic Alzheimer’s disease, familial Alzheimer’s disease (carrying PSEN1 and PSEN2 variants), and controls. Our results reveal alternative splicing patterns of PSEN2 specific to sporadic Alzheimer’s disease, including a human-specific cryptic exon present in intron 9 of PSEN2 as well as a 77 bp intron retention product before exon 6 that are both significantly elevated in sporadic Alzheimer’s disease samples, alongside a significantly lower percentage of canonical full-length PSEN2 transcripts versus familial Alzheimer’s disease samples and controls. Both alternatively spliced products are predicted to generate a prematurely truncated PSEN2 protein and were corroborated in an independent cerebellum RNA-sequencing dataset. In addition, our data in PSEN variant carriers is consistent with the hypothesis that PSEN1 and PSEN2 variants need to produce full-length but variant proteins to contribute to the onset of Alzheimer’s disease, although intriguingly there were far fewer full-length transcripts carrying pathogenic alleles versus wild-type alleles in PSEN2 variant carriers. Finally, we identify frequent RNA editing at Alu elements present in an extended 3′ untranslated region in PSEN2. Overall, this work expands the understanding of PSEN1 and PSEN2 variants in Alzheimer’s disease, shows that transcript differences in PSEN2 may play a role in sporadic Alzheimer’s disease, and suggests novel mechanisms of Alzheimer’s disease pathogenesis.
Keywords: Alzheimer’s disease, long-read Iso-Seq, alternative splicing, PSEN1, PSEN2
Course et al. perform targeted long-read isoform-sequencing of PSEN1 and PSEN2 in prefrontal cortex from individuals with sporadic Alzheimer’s disease, familial Alzheimer’s disease with PSEN1 and PSEN2 pathogenic variants, and controls. The results reveal aberrant splicing of PSEN2 that is unique to sporadic Alzheimer’s disease.
Introduction
Ranking as the most common form of both dementia and neurodegeneration, Alzheimer’s disease is progressively debilitating and uniformly fatal. Almost six million Americans have the disease—a number that will continue to escalate as our population ages—and still there remains no effective treatment. To better develop targeted interventions, we sought to uncover the complex transcriptional changes that may take place in Alzheimer’s disease. Here, we study transcripts produced by presenilin 1 (PSEN1) and presenilin 2 (PSEN2).
Originally identified through linkage analyses, autosomal dominant variants in PSEN1, PSEN2, and amyloid precursor protein (APP) lead to early-onset Alzheimer’s disease (onset <65 years of age).1–3 These three risk genes are mechanistically united in the following way: either of the two presenilins form part of the γ-secretase complex, which cleaves APP to liberate a product called amyloid beta (Aβ) peptide. Though the mechanisms by which variants in PSEN1 and PSEN2 lead to Alzheimer’s disease are still debated, it is commonly thought that changes in PSEN1 or PSEN2 bias APP cleavage to form versions of Aβ peptide that are more prone to aggregation. Aggregation of Aβ, in turn, is a hallmark pathology of Alzheimer’s disease.4–7
In addition to playing a role in early-onset Alzheimer’s disease, variants in PSEN1 and PSEN2 can also increase risk for, or cause, late-onset Alzheimer’s disease.8–12 It is generally thought that all disease-causing variants in these genes impair normal function of the γ-secretase complex, resulting in abnormal cleavage of APP and aggregation of Aβ. Tellingly, of all identified Alzheimer’s disease-related PSEN1 variants (there are now over 200),13 not one leads solely to a truncation or absence of protein product.14PSEN2 variants are not as common as those in PSEN1 (19 identified so far),13 but they similarly do not appear to lead to haploinsufficiency. These observations have led to a controversial hypothesis that PSEN1 and PSEN2 produce full-length but variant proteins in the context of Alzheimer’s disease. Determining whether this hypothesis is true would notably increase our understanding of Alzheimer’s disease pathogenesis.
Recently, we characterized the impact of an Alzheimer’s disease-related PSEN2 variant, PSEN2 K115Efs.15 This variant is a 2 bp deletion, which should lead to a frameshift and premature stop codon, and therefore be the first known case of a loss-of-function variant in PSEN2. Instead, however, we detected a novel intronic splice acceptor site that added 77 bp to the transcript, which would restore the reading frame and generate a full-length PSEN2 product with 25 extra amino acids.16 This transcript was specific to patient brain tissue and was not observed in fibroblasts. We also observed the same intron retention splicing pattern in brain tissues from patients with sporadic Alzheimer’s disease (i.e. lacking the 2 bp deletion) and observed use of this alternative splice site in RNA-sequencing data of anterior frontal cortex in the Mount Sinai Brain Bank, leading us to hypothesize that alternative splicing of familial Alzheimer’s disease risk genes may also play a role in sporadic Alzheimer’s disease.
The reading frame of PSEN2 K115Efs could also be restored by the omission of exon 6. Alternative splicing leading to skipping of PSEN2 exon 6 occurs under hypoxic conditions—a condition increasingly implicated in Alzheimer’s disease17—and the truncated splicing product is elevated in the brains of patients with sporadic Alzheimer’s disease.18 This splicing event is thought to decrease the unfolded protein response and increase γ-secretase activity.19 Splicing is altered by high mobility group AT-hook 1 (HMGA1), which binds exon 6 and causes it to be skipped, resulting in a premature stop codon and thus a truncated protein product.20,21 PSEN2 K115Efs acts in a manner similar to PSEN2 transcripts with exon 6 spliced out—that is, both form similar truncated products—but together, they form an almost full-length PSEN2 product, lacking exon 6.
In this study, we set out to characterize full-length transcripts of PSEN1 and PSEN2 in brain tissue from individuals with familial and sporadic Alzheimer’s disease as well as controls using PacBio Isoform-Sequencing (Iso-Seq). PacBio Iso-Seq is a state-of-the-art technique that uses single molecule, real-time (SMRT) sequencing to completely sequence all targeted mRNAs up to 20 kilobases long.22 The long-read capability of PacBio Iso-Seq is the only way to fully capture all exons in combination with pathogenic variants in PSEN1 (2776 bp) and PSEN2 (2249 bp) transcripts. Unlike short-read sequencing, it allows us to identify complete novel transcripts and alternative splicing events, as well as quantify differences in transcript abundance between samples.
Materials and methods
Samples used
The University of Washington Alzheimer’s Disease Research Center (UW ADRC) Neuropathology Core provided post-mortem prefrontal cortex samples from the 20 cases listed in Table 1. All samples were collected following informed consent approved by the UW Institutional Review Board. Control individuals were recruited through the Adult Changes in Thought study from Kaiser Permanente Washington,23 a population-based cohort study in which participants 65 years or older undergo cognitive screening every 2 years. Tissues used for this study were collected during rapid brain autopsy and flash frozen in liquid nitrogen or supercooled isopentane. Here, two controls, two sporadic Alzheimer’s disease and two PSEN1 samples were frozen in isopentane.
Table 1.
Samples used in study
| Cohort | Age at death (years) | Sex | PMI (hours) | Cognitive status | PSEN variant, if applicable |
APOE status | Braak score | CERAD score |
|---|---|---|---|---|---|---|---|---|
| Control | 88 | F | 3.50 | No dementia | ε3/ε3 | III | 0 | |
| Control | 90 | M | 5.00 | No dementia | ε3/ε3 | III | 0 | |
| Control | 88 | F | 3.18 | No dementia | ε2/ε3 | II | 0 | |
| Control | 90 | F | 4.50 | No dementia | ε2/ε3 | III | 0 | |
| Control | 78 | M | 10.02 | No dementia | ε3/ε3 | 0 | 0 | |
| Control | 83 | F | 2.67 | No dementia | ε3/ε3 | III | 0 | |
| PSEN | 43 | M | 2.50 | Dementia | PSEN1, I143T | ε3/ε4 | VI | 3 |
| PSEN | 55 | M | 3.12 | Dementia | PSEN1, S212Y | ε2/ε4 | VI | 3 |
| PSEN | 58 | M | 6.33 | Dementia | PSEN2, N141I | ε3/ε3 | VI | 3 |
| PSEN | 73 | M | 5.50 | Dementia | PSEN2, N141I | VI | 3 | |
| PSEN | 67 | F | 10.18 | Dementia | PSEN2, K115Efs | ε3/ε3 | VI | 3 |
| PSEN | 40 | F | 4.65 | Dementia | PSEN1, V272A | VI | 3 | |
| PSEN | 38 | M | 9.85 | Dementia | PSEN1, M146L | VI | 3 | |
| Sporadic | 90+ | M | 6.00 | Dementia | ε3/ε4 | VI | 3 | |
| Sporadic | 90+ | F | 4.50 | Dementia | ε3/ε3 | V | 3 | |
| Sporadic | 88 | F | 3.33 | Dementia | ε3/ε3 | VI | 3 | |
| Sporadic | 84 | F | 4.47 | Dementia | ε2/ε4 | V | 3 | |
| Sporadic | 90+ | F | 5.50 | Dementia | ε3/ε4 | VI | 3 | |
| Sporadic | 81 | F | 4.50 | Dementia | ε3/ε4 | VI | 3 | |
| Sporadic | 60 | M | 8.68 | Dementia | VI | 3 |
APOE = Apolipoprotein E; CERAD = Consortium to Establish a Registry for Alzheimer’s Disease; F = female; M = male; PMI = post-mortem interval; PSEN = presenilin.
RNA extraction and Iso-Seq
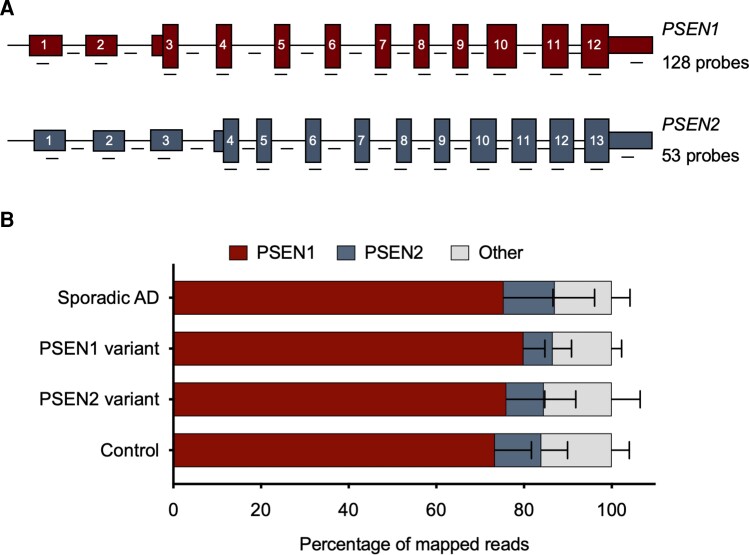
RNA was extracted from brain tissue samples using the RNeasy Lipid Tissue Mini Kit (Qiagen). Then 300 ng of RNA was then converted to cDNA, hybridized and prepared for Iso-Seq following a protocol from PacBio called ‘Iso-Seq Express Capture Using IDT xGen Lockdown Probes’. Briefly, RNA was converted into cDNA and amplified using a combination of the NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module (New England Biolabs), the Iso-Seq Express Oligo Kit (PacBio), and barcoded primers (found in Appendix 3 of the PacBio protocol). RNA integrity number scores for each sample varied on the basis of the quality of the original tissue. Samples were then pooled into two groups based on higher or lower RNA integrity number scores (researchers were blinded to sample identity), and these two pools were hybridized to a custom panel of xGen Lockdown Probes (Integrated DNA Technologies) using the xGen Lockdown Hybridization and Wash Kit (Integrated DNA Technologies). In total, the probe panel contained 181 probes tiling both exonic and intronic regions of PSEN1 (128 probes) and PSEN2 (53 probes) at approximately equal intervals (Fig. 1Aand Supplementary Table 1). After 15 cycles of amplification of the captured cDNA, pools were combined (after confirming that both DNA pools had similar quality control scores) and submitted to UW PacBio Sequencing Services for library preparation and running on PacBio Sequel II for 1 SMRT Cell 8M with a 30-h video time, using SQII v.2.0 polymerase and v.2.0 sequencing chemistry.
Figure 1.
Iso-Seq reads mapping to PSEN1 and PSEN2. (A) Schematic of probe location and number of probes used per gene. (B) Iso-Seq reads mapping to PSEN1 and PSEN2. Reads that mapped to PSEN1 and PSEN2 were not significantly different across cohorts (P = 0.62 for PSEN1 and P = 0.53 for PSEN2, one-way ANOVA). Bars are mean and standard deviation. Sporadic Alzheimer’s disease (AD) n = 7, PSEN variant carrier n = 7, control n = 6.
Iso-Seq analysis
We received full-length transcript information in FASTQ files, which were initially assessed using the standard Iso-Seq Analysis pipeline in SMRT Link (PacBio, v.10.1.0.119588). Briefly, barcoded reads were sorted, and barcodes and poly-A sequences were clipped using the Lima SMRT Analysis tool (v.2.2.0). Reads with a quality score of 20 or greater were mapped to the human GRCh38 genome using pbmm2 (v.1.3.0) and minimap2 (v.2.23) to generate a set of BAM files.
We counted the number of reads per sample with a transcript origin in PSEN1 (GRCh38 coordinates chr14:73,136,507–73,223,691) and PSEN2 (chr1:226,870,616–226,896,098). Of these, we quantified the number of reads that contained both canonical and alternative splice isoforms. To obtain total reads, we counted the number of reads that mapped to transcript coordinates. To ascertain full-length PSEN1 and PSEN2 transcripts, we extracted reads that contained the expected start and stop codon sequences. For uniquely mapped reads, all identical isoforms were counted as one.
RNA editing analysis
Iso-Seq reads that mapped to the AluJb (chr1:226896582–226896815) element in the PSEN2 3′ untranslated region (UTR) were assessed for the number of guanine residues at each adenine position. The percentage of total A-to-G changes was then quantified.
RNA-seq analysis
Mayo Clinic sample RNA-seq BAM files from the cerebellum and temporal cortex were downloaded from Accelerating Medicines Partnership Program for Alzheimer’s Disease, Synapse ID syn5550404. Percentage splice inclusion of alternative splice products was measured by calculating junction spanning reads across canonical and alternative splice junctions, confirmed by quantification from Sashimi plots generated by Integrated Genomics Viewer. Due to increased read depth at these positions, RNA editing was calculated across both AluJb (chr1:226896582-226896815) and AluY (chr1:226898626-226898913) elements in the PSEN2 3′ UTR.
Statistical analysis
Statistical analyses were performed using Prism v.9.2.0 (GraphPad Software). The Shapiro–Wilk test (n < 8) or D’Agostino–Pearson omnibus K2 test (n > 8) was first used to determine whether data were normally distributed. A one-way ANOVA was used to compare groups of more than two with parametric distribution. If the ANOVA gave P < 0.05, this analysis was then followed by Tukey’s multiple comparisons. To analyse correlations, a Spearman correlation coefficient was used for non-parametric data and a Pearson correlation coefficient was used for parametric data.
Data availability
RNA sequence reads from the Mayo Clinic are available from synapse.org with accession number syn5550404.
Results
Obtaining long-read transcripts of PSEN1 and PSEN2
To observe PSEN1 and PSEN2 transcripts in individuals with Alzheimer’s disease and controls, we first prepared cDNA from 20 samples. These samples were composed of prefrontal cortex tissues, from six non-demented donors with low or absent Alzheimer’s disease neuropathological change (referred to as ‘controls’), seven individuals with both familial Alzheimer’s disease and a PSEN1 or PSEN2 variant, and seven individuals with neuropathologically confirmed sporadic Alzheimer’s disease (Table 1). Of the seven samples from individuals with PSEN1 or PSEN2 variants, four had PSEN1 variants (all different), and three had PSEN2 variants, one of which was the K115Efs variant and two of which were N141I variants. Together, we refer to these samples as ‘PSEN variant carriers’. As noted later in this study, we did observe that one of our sporadic Alzheimer’s disease samples carried a PSEN1 variant of unknown significance, and we continued to consider this sample sporadic, based on the known literature of this variant of unknown significance (the individual’s late age of death at >90 years old also supports this decision). These samples were all obtained within a post-mortem interval of less than 12 h. Controls and sporadic Alzheimer’s disease samples were reasonably age- and sex-matched. Due to the early-onset nature of familial Alzheimer’s disease, we could not obtain age-matched controls for the PSEN variant carriers (Table 1).
We obtained PSEN1- and PSEN2-specific transcripts from the cDNA by hybridization capture, and then submitted these samples for SMRT sequencing. We favoured the use of hybridization probes over PCR amplification of transcripts to ensure that we would be able to observe all alternative splice isoforms, including truncated isoforms. Our hybridization probes corresponded to both coding sequence and genomic region (Supplementary Table 1), so that we could identify potential intron retention products. The number of probes used per gene and a schematic of probe location is provided in Fig. 1A. A step in cDNA conversion involving selection for poly-A tails suggests that our findings here reflect transcripts that have undergone mRNA processing.
Alignment to PSEN1 and PSEN2
On average, we obtained 81 034 ± 36 374 mapped reads per sample (range 28 444–167 125). Of these, 75.8 ± 8.68% mapped to PSEN1 (range 53.8–86.1%) and 9.86 ± 6.91% mapped to PSEN2 (range 2.69–31.5%). Values were not significantly different across cohorts (Fig. 1B; P = 0.62 for PSEN1 and P = 0.53 for PSEN2, one-way ANOVA). Analysis of uniquely mapped reads instead of total reads led to similar results (Supplementary Fig. 1). The higher mapping rate for PSEN1 may reflect an increased abundance of PSEN1 in the frontal cortex or could be due to the higher number of probes used for PSEN1 mRNA versus PSEN2 mRNA (Fig. 1A). The reads that did not map to PSEN1 or PSEN2 were evenly distributed throughout the genome, with no evidence for biased enrichment of a particular gene when analysing unique reads (Supplementary Fig. 2A), and some preferential amplification of proteolipid protein 1 (PLP1) and myelin basic protein in just two samples when analysing total reads (Supplementary Fig. 2B). The distribution of reads was comparable between cohorts.
PSEN1 isoforms
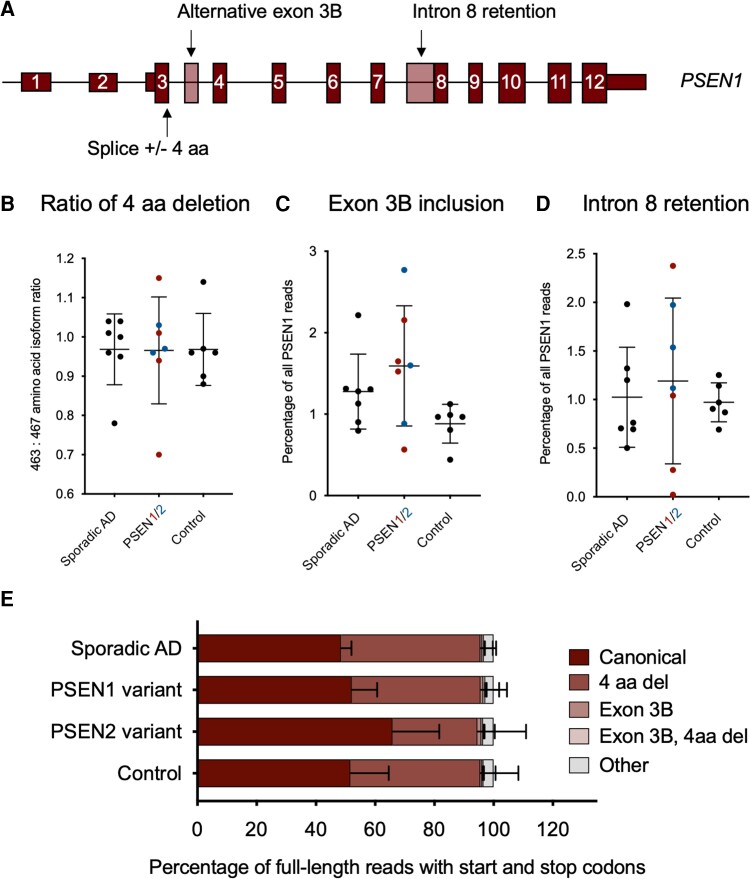
The generation of appropriately spliced and full-length PSEN1 was largely efficient, with ∼75% of reads encoding for an in-frame protein product. A minimum of 16 000 canonical full-length transcripts of PSEN1 that encoded for an in-frame protein product were found in each sample. Importantly, in the PSEN variant carriers, roughly half of full-length transcripts of PSEN1 were observed to contain the pathogenic variant.
Two predominant splice isoforms were detected, corresponding to the omission or inclusion of 12 bp (the amino acids VRSQ) at the end of exon 3 of 12 (first coding exon) of the PSEN1 transcript (Fig. 2A). The abundance of the transcript without these 12 bp (463 aa; ENST00000357710.8) versus with these nucleotides (467 aa; ENST00000324501.10) was comparable across sporadic Alzheimer's disease, PSEN variant carriers and controls, with short and long isoforms each occurring at the same rate (Fig. 2B; P = 1.00, one-way ANOVA). Transcripts missing the 12 bp are thought to be the alternative transcripts, as these nucleotides are conserved in PSEN2 transcripts.
Figure 2.
Isoforms of PSEN1. (A) Schematic showing the three most predominant isoforms identified for PSEN1. (B) Ratio of 4 amino acid (aa) deletion to inclusion isoform of PSEN1. Abundance (percentage of total PSEN1 transcripts) of PSEN1 (C) exon 3b inclusion isoform, and (D) intron 8 retention isoform, across cohorts. No isoform exhibited significantly different ratio or abundance across cohorts (P > 0.05, one-way ANOVA, for each). (E) Distribution of various PSEN1 isoforms as a percentage of all full-length transcripts with canonical start and stop codons. All bars are mean and standard deviation. Sporadic Alzheimer’s disease (AD) n = 7, PSEN variant carrier n = 7, control n = 6.
The third most common isoform from our analysis, however, was not annotated. This isoform of PSEN1 included a 130 bp alternatively spliced exon in intron 3 (GRCh38 chr14:73,160,009–73,160,138) (Fig. 2A). The extra exon (exon 3B) is composed of portions of AluJb and L1MB2 repeat elements and introduces a frameshift and premature termination codon at amino acid 61. If an internal methionine start codon is used, the N-terminally truncated protein product would be 384 amino acids long. The inclusion of exon 3B remained the same between controls, PSEN variant carriers, and sporadic Alzheimer’s disease samples (Fig. 2C; P = 0.084, one-way ANOVA). One of the next most abundant isoforms contains an alternative splice acceptor site in intron 8 that leads to an intron retention product and is present in ∼1% of reads for all samples. Again, this number remained the same across all three cohorts (Fig. 2D; P = 0.79, one-way ANOVA). All full-length transcripts carrying each of these alternative splice events in an otherwise canonical sequence are quantified as a percentage of total full-length PSEN1 isoforms observed across cohorts in Fig. 2E. Analysis of uniquely mapped reads gave similar results for all PSEN1 isoforms (Supplementary Fig. 2).
In one sample annotated as sporadic, we identified a 3 bp deletion in exon 4 of PSEN1 (p.Asp40del; GRCh38 coordinates chr14:73,170,825–73,170,827). This 3 bp deletion has been previously observed in Alzheimer’s disease cases24,25 and is present in gnomAD (v.3.1.2) in 18 individuals (allele frequency of 0.0001183), indicating that it is currently a variant of unknown significance. In a recent systematic study, this variant was deemed a risk factor for Alzheimer's disease.26
PSEN2 isoforms
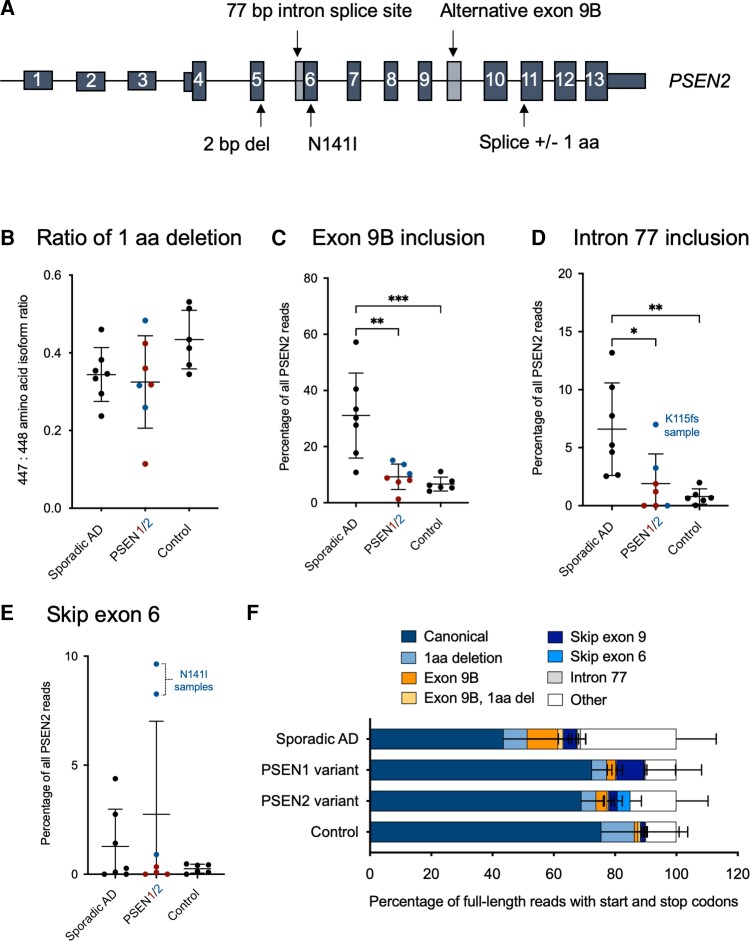
Alignment of Iso-Seq reads to PSEN2 revealed a more heterogeneous population of transcripts than those observed for PSEN1 (Fig. 3A). The generation of an appropriately spliced and full-length PSEN2 was slightly less efficient than for PSEN1, with 62% of reads encoding for an in-frame protein product. At least 882 canonical full-length transcripts of PSEN2 were found in each sample. Like PSEN1, full-length transcripts contained both wild-type and pathogenic variant alleles for each of the PSEN variant carriers.
Figure 3.
Isoforms of PSEN2. (A) Schematic showing the three most predominant isoforms identified for PSEN2, as well as the 77 bp intron splice site and the location of the N141I variant. (B) Ratio of 1 amino acid (aa) deletion to inclusion isoform of PSEN2. Abundance (percentage of total PSEN2 transcripts) of PSEN2 (C) exon 9b inclusion isoform, (D) intron 77 inclusion isoform, and (E) exon 6 skipping events, across cohorts. (F) Distribution of various PSEN2 isoforms as a percentage of all full-length transcripts with canonical start and stop codons. P-values were determined by a one-way ANOVA, followed by Tukey’s multiple comparisons. *P < 0.05, **P < 0.01, ***P < 0.001. All bars are mean and standard deviation. Sporadic Alzheimer’s disease (AD) n = 7, PSEN variant carrier n = 7, control n = 6.
Again, after the canonical PSEN2 transcript, the second most abundant transcript that spanned from start to stop codons was one that contained an alternative splice site—an alternative acceptor site in this instance—at the beginning of exon 11 of 13, which resulted in the removal of 3 bp (1 amino acid) from the total protein product. Sporadic Alzheimer’s disease samples, controls and PSEN variant carriers all had similar levels of this isoform, with the short isoform occurring less often than the long isoform (Fig. 3B; P = 0.11, one-way ANOVA).
Also, like PSEN1, the third most abundant transcript was non-productive (i.e. a premature stop codon is introduced). A 1657 bp transcript was detected in 4.8% of full-length reads across all samples. This alternative exon appears between exons 9 and 10, herein referred to as exon 9B (Fig. 3A). The exon is 310 bp, flanked by canonical splice sites, contains stop codons in all three reading frames and is predicted to produce a protein truncated at 321 amino acids. Reads mapping to the exon 9B region accounted for over 50% of total PSEN2 reads for one sporadic sample and were significantly more abundant in sporadic Alzheimer’s disease than in controls (P = 0.0007) or PSEN variant carriers (P = 0.0013), while PSEN variant carriers exhibited similar levels as controls (Fig. 3C; P = 0.88, one-way ANOVA followed by Tukey’s multiple comparisons). A known isoform of PSEN2 (ENST00000487450.1) lists the presence of exon 9B, though mRNA abundance stemming from this isoform is low based on GTEx data.27 We observed several other transcript isoforms that contained exon 9B in combination with intron retention and alternative splice products present in introns 8 and 9, many of which were enriched in sporadic Alzheimer’s disease (Supplementary Fig. 3). Exon 9B inclusion was not correlated with age at death in sporadic or any other cases [Supplementary Fig. 4; r = 0.24, P (two-tailed) = 0.30, Spearman correlation].
A fourth PSEN2 isoform was also detected corresponding to the inclusion of 77 bp of intronic sequence prior to exon 6 (Fig. 3A). We previously identified the presence of this 77 bp intron product in the context of a 2 bp deletion.16 This transcript is a full-length mRNA including start and stop codons and all exons in between, but due to a frame shift, is not predicted to form a full-length protein product. Again, sporadic Alzheimer’s disease samples had a significantly higher abundance of this 77 bp intron product as compared to controls (P = 0.0050) or PSEN variant carriers (P = 0.017), while PSEN variant carriers exhibited similar levels as controls (Fig. 3D; P = 0.76, one-way ANOVA followed by Tukey’s multiple comparisons). Average percentages were 6.59% for sporadic Alzheimer’s disease, 1.91% in PSEN (although three samples had zero reads) and 0.79% in controls. The highest level of the 77 bp intron product in the PSEN variant carriers was observed in the PSEN2 2 bp deletion (PSEN2 K115Efs; highlighted in Fig. 3D), which corroborates our previous work.16
Exon 6 skipped products are not a common feature of PSEN2 Iso-Seq data
An exon 6 skipped product, termed PS2V, is the subject of several previous reports.18,21,28,29 This exon skipping is induced by hypoxia and mediated by binding of HMGA1.20,21 Here, we find few examples of reads that skip exon 6, including in the 2 bp deletion sample, where deletion of exon 6 would restore full-length protein production (minus 48 aa).16 Abundance of transcripts exhibiting exon 6 skipping remained the same across cohorts (Fig. 3E; P = 0.28, one-way ANOVA). A notable exception to this lack of PS2V was in the two carriers of the PSEN2 N141I pathogenic variant. In each, 8.26–9.64% of all reads that mapped to PSEN2 contained a splice product skipping exon 6 (highlighted in Fig. 3E). Interestingly, amino acid 141 is in the middle of exon 6, suggesting that the nucleotide variant may influence alternative splicing of exon 6 (Fig. 3A). The binding site consensus sequence of HMGA1 is CUGCUACAAG at the end of exon 6 (amino acids 163–166), distal to the N141 codon (Supplementary Fig. 5).
In one of the PSEN2 N141I carriers, we identified a heterozygous position (rs1046240) in exon 5. Using this information, we could deduce that all 479 reads that skipped exon 6 also contained the alternate allele at rs1046240 and therefore were in cis with the 141I allele. For the second N141I individual, however, we did not identify heterozygous positions in the mRNA that could be leveraged for a similar analysis.
Finally, full-length PSEN2 transcripts carrying each of these mentioned alternative splice events on their own were quantified as a percentage of all full-length PSEN2 isoforms (containing canonical start and stop codons and an otherwise canonical transcript) observed across cohorts in Fig. 3F. These data include a splice isoform that skips exon 9, though this isoform maintains the PSEN2 reading frame and is not significantly different between cohorts (Supplementary Fig. 3). In contrast to PSEN1, the proportion of full-length reads for PSEN2 is significantly lower in sporadic Alzheimer’s disease cases relative to controls (P = 0.0055) or PSEN variant carriers (P = 0.013, one-way ANOVA followed by Tukey’s multiple comparisons). Of note, the ‘other’ group in Fig. 3F includes various combinations of alternative exons and intron retention products whose commonality involves the sharing of exon 9B sequence (Supplementary Fig. 3). Analysis of uniquely mapped reads led to similar results for all PSEN2 isoforms (Supplementary Fig. 2).
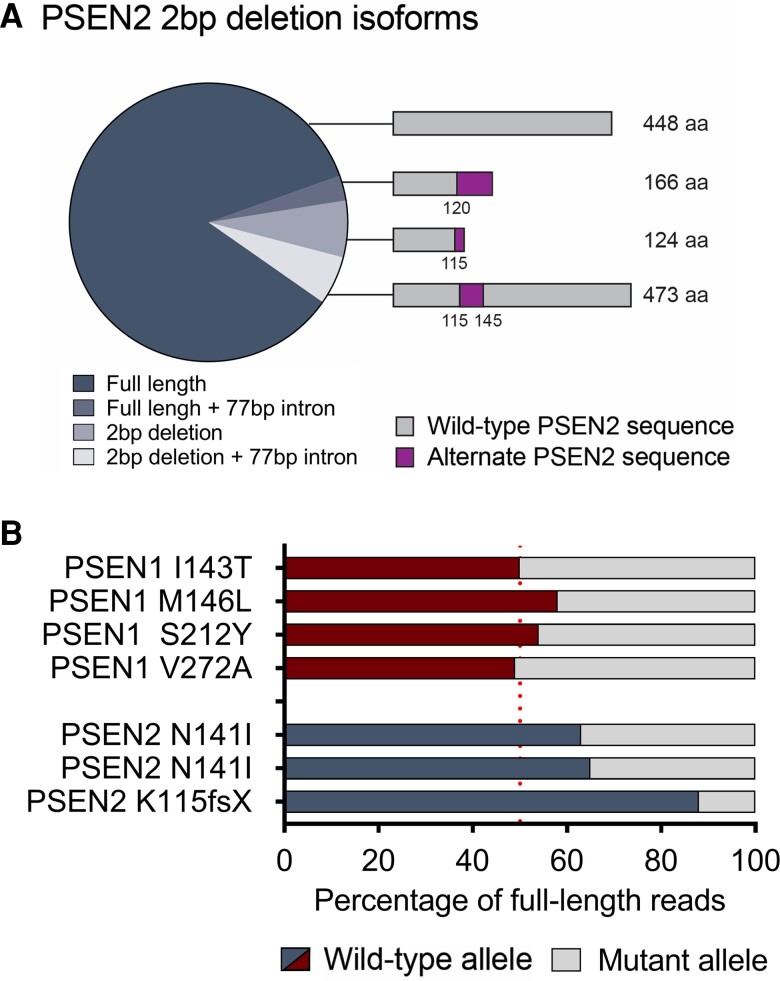
Allelic bias at PSEN1 and PSEN2 pathogenic variant positions
Long-read Iso-Seq also afforded us the ability to examine allelic bias in PSEN variant carriers. An unresolved question from our previous work was whether the 2 bp deletion could produce a full-length in-frame product in combination with the 77 bp insertion in intron 6. Aligning reads from this individual revealed that the wild-type allele accounted for 88% of the reads (2858 reads for full-length PSEN2 and 98 reads including the 77 bp intronic sequence). Of the 412 reads with a 2 bp deletion, about half spliced to the canonical exon 6 junction and continued to the last exon of the transcript, which is predicted to yield a truncated 124 amino acid protein product (Fig. 4A). However, 190 additional reads included the 77 bp product, generating a full-length transcript with 25 additional amino acids inserted after exon 5.
Figure 4.
Allelic bias at PSEN1 and PSEN2 pathogenic variant positions. (A) Percentage of PSEN2 isoforms with or without the 2 bp deletion and 77 bp intron retention. (B) Percentage of transcripts with the wild-type allele or the PSEN variant (mutant allele) present. Sporadic Alzheimer’s disease n = 7, PSEN variant carrier n = 7, control n = 6. aa = amino acids.
The PSEN2 2 bp deletion allele (i.e. the K115Efs variant) only contributes to 12% of total PSEN2 transcripts, therefore some nonsense-mediated decay is probably active. The other two PSEN2 samples (both with the N141I variant) displayed only 35–37% of transcripts with pathogenic variants, further indicating that decreased stability of the variant allele (probably due to nonsense-mediated decay) may take place. For PSEN1, the pathogenic variant was present in transcripts in roughly equal amounts at 42–51% of the time (Fig. 4B).
Confirmation of PSEN1 and PSEN2 alternative splicing events in independent RNA-seq datasets
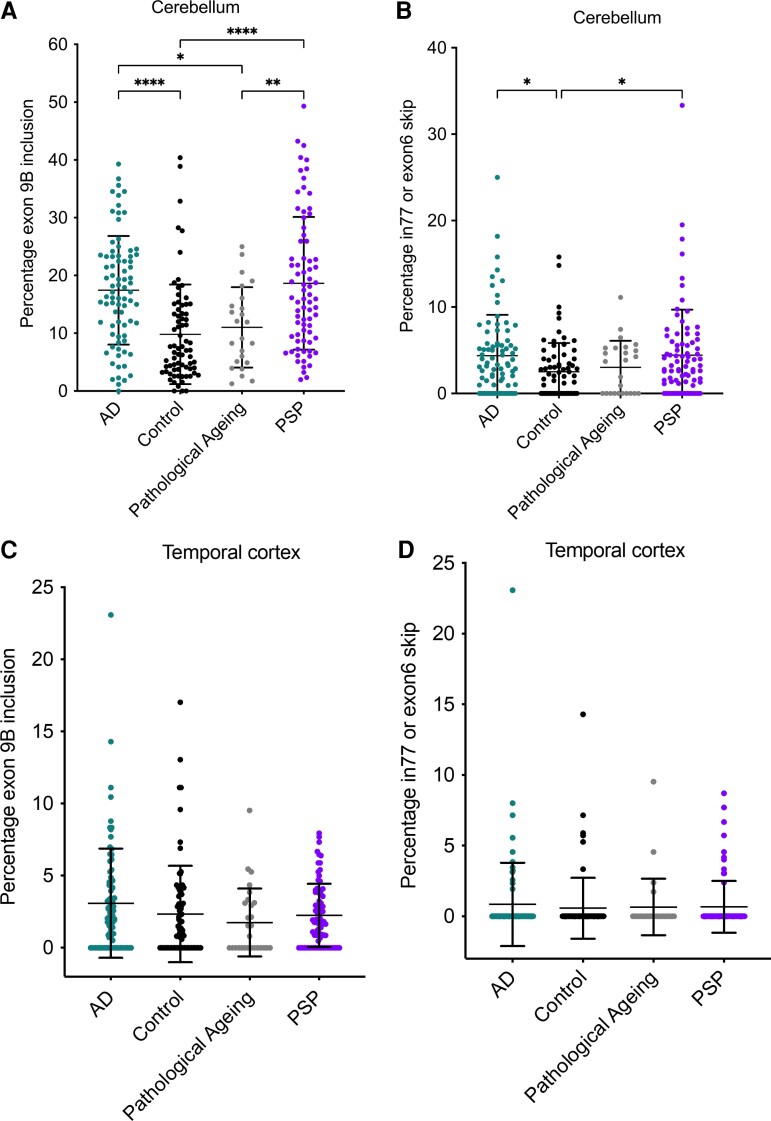
To determine whether the PSEN2 exon 9B splice product is a prominent feature of Alzheimer's disease, we queried RNA-seq datasets made available through the Accelerating Medicines Partnership Program for Alzheimer’s Disease. We determined that the exon 9B splice products were highest in the cerebellum from the Mayo Clinic,30 which corresponds to increased expression of PSEN2 in the cerebellum and cerebellar hemisphere relative to other brain regions based on GTEx data.27 The Mayo Clinic dataset contains RNA-seq data from the cerebellum for 82 individuals with sporadic Alzheimer's disease, 76 controls, 25 individuals with pathological ageing (Aβ deposits but no cognitive impairment) and 81 individuals with progressive supranuclear palsy. Exon 9B was included 17.4% of the time on average in Alzheimer’s disease samples, significantly higher than in controls (9.81%, P < 0.0001) and in the pathological ageing cohort (11.0%, P = 0.024) but not different than progressive supranuclear palsy samples (Fig. 5A; 18.6%, P = 0.87, one-way ANOVA followed by Tukey’s multiple comparisons).
Figure 5.
RNA splicing in Mayo Clinic RNA-seq samples. Proportion of spliced reads in cerebellum samples for PSEN2 (A) exon 9B, and (B) 77 bp intron retention product or exon 6 skipping. Proportion of spliced reads in temporal cortex samples for PSEN2 (C) exon 9B, and (D) 77 bp intron retention or exon 6 skipping products. PSP = progressive supranuclear palsy. P-values were determined by a one-way ANOVA, followed by Tukey’s multiple comparisons. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All bars are mean and standard deviation. Sporadic Alzheimer’s disease (AD) n = 82, control n = 76, pathological ageing n = 25, progressive supranuclear palsy n = 81 for cerebellum samples. Sporadic AD n = 82, control n = 74, pathological ageing n = 28, progressive supranuclear palsy n = 80 for temporal cortex samples.
In addition, we observed both exon 6 skipping and 77 bp intron inclusion alternative splice events at exon 6. Alternative splicing at exon 6 was higher in Alzheimer’s disease samples versus controls (4.37% versus 2.52%; P = 0.045), as well as in progressive supranuclear palsy versus controls (4.42% versus 2.52%; P = 0.037), but not for Alzheimer’s disease versus pathological ageing (P = 0.54) or progressive supranuclear palsy (Fig. 6B; P = 1.00, one-way ANOVA followed by Tukey’s multiple comparisons). As in our Iso-Seq data, the 1 amino acid deletion splice product was not significantly different in PSEN2 in Alzheimer’s disease versus other samples (Supplementary Fig. 6A; P = 0.059) nor was splicing different for the PSEN1 4 aa deletion product (Supplementary Fig. 6B; P = 0.75), or inclusion of alternative exon 3B (Supplementary Fig. 6C, P = 0.70, one-way ANOVA). We found that exon 9B inclusion did not correlate with age at death [Supplementary Fig. 7; P (two-tailed) > 0.10 for all cohorts].
Figure 6.
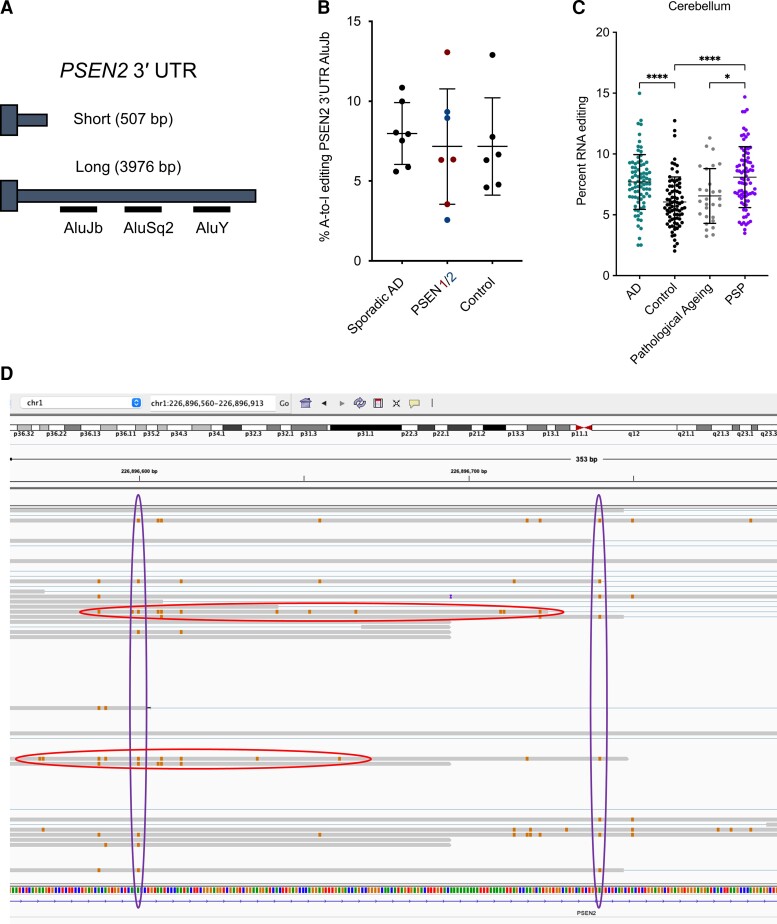
RNA editing in PSEN2 3′ UTR. (A) Schematic showing two 3′ UTRs identified for PSEN2, short (without Alu elements) and long (with Alu elements). (B) Percentage of A-to-I editing of the AluJb and AluY sites, across cohorts. Sporadic Alzheimer’s disease (AD) n = 7, PSEN variant carrier n = 7, control n = 6. (C) Level of RNA editing in Alu elements in the PSEN2 3′ UTR. P-values were determined by a one-way ANOVA, followed by Tukey’s multiple comparisons. *P < 0.05, ****P < 0.0001. All bars are mean and standard deviation. Sporadic Alzheimer’s disease n = 82, control n = 76, pathological ageing n = 25, progressive supranuclear palsy n = 81. (D) Examples of individual reads with several edits (circled horizontally) and individual residues with multiple edits (circled vertically).
RNA from the temporal cortex was also sequenced for many of the same samples in the Mayo Clinic dataset (82 Alzheimer’s disease, 74 controls, 28 pathological ageing and 80 progressive supranuclear palsy). Here, we found fewer reads that mapped to the exon 9B splice junction, with several samples having no reads at all. Alzheimer’s disease samples averaged 3.09% splicing to exon 9B relative to 2.34% in controls, 1.75% in pathological ageing and 2.25% in progressive supranuclear palsy (Fig. 5C). Levels of this transcript were not significantly different between cohorts (P = 0.15, one-way ANOVA). The number of PSEN2 exon 6 skipping or 77 bp intron inclusion events likewise were minimal in the temporal cortex, and not significantly different between cohorts (Fig. 5D; P = 0.90, one-way ANOVA).
An extended PSEN2 3′ UTR is subject to RNA editing
During our analysis of PSEN2 N141I transcripts, we observed a series of A-to-G changes present in the 3′ UTR sequence. Upon further inspection of these variants, we found they were scattered across all reads in a non-allelic manner (i.e. they were not specific to one haplotype). The A-to-G nature suggested instead that these are RNA editing events, reinforced by their enriched abundance at AluJb and AluY short interspersed nuclear elements in the 3′ UTR sequence. Adenosine deaminases acting on RNA-mediated editing of adenosine to inosine nucleosides can be detected by the presence of A-to-G mismatches in the cDNA as reverse transcription enzymes incorporate guanidine residues at inosine positions.
We identified two 3′ UTRs for PSEN2 (a short 3′ UTR of 507 bp and long 3′ UTR of 2976 bp; Fig. 6A). The long 3′ UTR included the Alu elements and is observed about one-third of the time across samples. The level of RNA editing at Alu elements in the 3′ UTR of PSEN2 was comparable in sporadic Alzheimer’s disease cases, PSEN variant carriers and controls (Fig. 6B; P = 0.69, one-way ANOVA). However, the larger dataset from the Mayo Clinic also permitted evaluation of RNA editing at Alu sites in the 3′ UTR of PSEN2. Here, we found that RNA editing was significantly enriched in both Alzheimer’s disease and progressive supranuclear palsy samples relative to controls (P < 0.0001 for both), and in progressive supranuclear palsy relative to pathological ageing samples (Fig. 6C; P = 0.011, one-way ANOVA followed by Tukey’s multiple comparisons). We detected the presence of both individual reads with several edits, and individual residues with several edits (examples shown in Fig. 6D).
Motivated by our identification of RNA editing in the 3′ UTR of PSEN2, we revisited alternative exons 3B in PSEN1 and 9B in PSEN2, both of which include sections of repetitive elements. The PSEN1 exon 3B sequence includes portions of AluJB and L1MB2 long interspersed nuclear element (LINE) repeats. There are several RNA edits in this region, although the level of editing is low across sporadic AD, PSEN variant carriers and controls. The alternative PSEN2 acceptor site in exon 9B is in an L2 LINE. While we did not detect RNA editing events, it is possible that some of the adjacent intronic sequence is edited, thus strengthening the splice acceptor sequence.
Discussion
In this study, we used long-read Iso-Seq to identify transcript variation in PSEN1 and PSEN2 in cases of sporadic and familial Alzheimer’s disease. Our Iso-Seq findings for PSEN1 match its established gain-of-function role in Alzheimer’s disease. We identified equivalent expression of wild-type and variant alleles for four different PSEN1 pathogenic variants that were analysed. We observed a high degree of splicing fidelity—even in aged individuals—with little to no splicing differences between sporadic Alzheimer’s disease versus controls (Fig. 2). In contrast, our findings illuminate a previously under-appreciated role for PSEN2 alternative splicing in Alzheimer’s disease. Patients with the N141I Volga German pathogenic variant were biased towards skipping of exon 6 and loss of the variant allele, skewing the ratio of mapped reads towards wild-type PSEN2. The skipping of exon 6—which would lead to a premature termination codon—indicates that a partial loss-of-function property of PSEN2 may be implicated. How the variant might promote exon 6 skipping is currently unknown, though the variant does not overlap an established binding site for HMGA1, which can promote exon 6 skipping. In addition, we found increased expression of the wild-type PSEN2 allele in a carrier of the K115Efs variant, while the PSEN2 isoforms associated with the 2 bp deletion were often linked to a 77 bp intron retention product, leading to an in-frame protein product and confirming our previous work.16
We also identified significant transcript differences in sporadic Alzheimer’s disease cases versus controls and pathogenic PSEN variant carriers, including more non-productive transcripts (Fig. 3). While we would have predicted that individuals with pathogenic variants in PSEN2 had the most disrupted splicing of PSEN2 mRNA, instead we found that it was in the sporadic Alzheimer’s disease cases that demonstrated the greatest abundance of non-productive intron 77 retention and exon 9B products (Fig. 3). In general, many alternative splice events were observed for PSEN2 that were enriched in sporadic Alzheimer’s disease. Our results are consistent with the hypothesis that PSEN1 and PSEN2 produce full-length but variant proteins to contribute to Alzheimer’s disease. In addition, these data indicate that alternative splicing of PSEN2 is not unique to Alzheimer’s disease variants, but rather that transcript differences in PSEN2 may play an important role in the onset of sporadic Alzheimer’s disease.
PSEN1 and PSEN2 are highly similar (65% identical at the amino acid level). Despite their similarities, many more pathogenic variants have been documented in PSEN1 than in PSEN2.8–11 This phenomenon may be explained by differences in cellular localization (for example, PSEN2 plays a selective role in microglia),31–33 subcellular localization34 or function (PSEN1 has a greater effect on calcium signalling).35 It is also possible that pathogenic variants in PSEN2 are more prevalent than currently thought, but that cases with these variants are considered sporadic due to their later age of onset.36 A modest shift to PSEN2 transcripts with premature termination codons emerges both in individuals with PSEN2 pathogenic variants and in sporadic Alzheimer’s disease. Whether these non-productive isoforms have a pathogenic role is currently unclear. However, the commonality is distinct from PSEN1, where ample evidence demonstrates that loss-of-function variants appear unrelated to Alzheimer’s disease aetiology and instead predispose individuals to familial acne inversa.37 The evidence for or against PSEN2 loss-of-function variants in Alzheimer’s disease pathology is less clear—they are not implicated in acne inversa for instance—and our data suggest that further investigation is warranted.
Our confirmation that the PSEN2 exon 9B splice product is elevated in sporadic Alzheimer’s disease cases in an external RNA-seq dataset (Fig. 5) is consistent with the idea that the exon 9B product plays a role in Alzheimer’s disease aetiology, and further experimental evidence of functionality could point to elimination of the exon 9B product as a potential therapeutic target. However, the fact that exon 9B inclusion is highest in the cerebellum is surprising, considering that the cerebellum is relatively spared from Alzheimer’s disease pathology until late stages of disease. Even more surprising is the similar inclusion of this splice product in progressive supranuclear palsy cerebellum samples (Fig. 6A). Progressive supranuclear palsy exhibits tau pathology but not Aβ aggregation,38 which differentiates it from Alzheimer’s disease pathology. The significant upregulation of exon 9B isoform in the progressive supranuclear palsy samples thus presents a potential connection between PSEN2 and tau. Perhaps tau aggregation and mis-localization may promote exon 9B inclusion by sequestering splice factors.39 However, given that PSEN pathogenic variant carriers also do not harbour increased exon 9B inclusion products, additional genetic and/or environmental risk factors are probably involved in sporadic Alzheimer’s disease.
These findings indicate that Iso-Seq of other brain regions that have selective vulnerability in Alzheimer’s disease or ageing is warranted to identify the proportion of reads that incorporate PSEN2 exon 9B. In addition, because our starting tissue was bulk brain tissue, we cannot yet make conclusions about the cell-type specificity of these transcripts, which would be similarly illuminating. These findings also demonstrate the value of examining full-length transcripts, as other databases created using short-read sequencing, such as the Mount Sinai Brain Bank or from the Religious Orders Study and Rush Memory and Aging Project,40,41 do not exhibit widespread inclusion of the intron 77 bp or exon 9B product. Of note, exon 9B splice junctions are not conserved in other species, including mice and non-human primates. The human-specific nature of exon 9B may help explain why mouse models of sporadic Alzheimer’s disease have been difficult to establish.42
The discovery of Alu short interspersed nuclear elements and LINEs in a long 3′ UTR of PSEN2 raises intriguing questions about the cell-specific usage of these isoforms. RNA editing of Alu repeat-containing 3′ UTRs invokes a proposed mechanism to restrict the transcripts to the nucleus: if they are exported to the cytoplasm, they could precipitate a potent immune response.43 If the edited mRNA is exported to the cytoplasm, reduced levels of polysomes are found on the edited RNA,44 indicating that this isoform of PSEN2 may have reduced translation capacity.
Finally, the presence of alternative splice products with non-productive exons deep within traditional introns of both PSEN1 and PSEN2 suggests that intronic variants surrounding these sites may promote inclusion of these non-productive exons and act as risk factors for Alzheimer’s disease. The increasing number of whole genome sequences available for Alzheimer’s disease allows us now to screen for these rare variants and test for cryptic exon inclusion in functional assays, such as minigene reporters.
Using long-read sequencing technology, we have considerably expanded our understanding of PSEN1 and PSEN2 alternative splicing—which is consistent with the hypothesis that pathogenic variants in PSEN1 and PSEN2 lead to disease not through haploinsufficiency but rather an alteration of function—and identified novel splice isoforms associated with disease. Specifically, we found several transcript differences in PSEN2. Future work will help reveal whether they play an important role in the onset of sporadic Alzheimer’s disease–the form of which accounts for >95% of Alzheimer’s disease cases.45 Our findings also reveal a direct connection between sporadic and familial Alzheimer’s disease, strengthening the amyloid cascade hypothesis that Alzheimer’s disease is caused by disrupted production or clearance of APP cleavage products.
Supplementary Material
Acknowledgements
Support for using patient samples was provided by Aimee Shantz at UW Neuropathology. Advice for preparing samples for Iso-Seq was provided by Katy Munson, Jason Underwood, Jenny Ekholm, Ting Hon and Elizabeth Tseng at PacBio. We thank the critical contributions of brain donors and their families for their generosity and willingness to contribute to scientific research.
Contributor Information
Meredith M Course, Division of Medical Genetics, University of Washington School of Medicine, Seattle, WA 98195, USA; Department of Molecular Biology, Colorado College, Colorado Springs, CO 80903, USA.
Kathryn Gudsnuk, Division of Medical Genetics, University of Washington School of Medicine, Seattle, WA 98195, USA.
C Dirk Keene, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA 98195, USA.
Thomas D Bird, Division of Medical Genetics, University of Washington School of Medicine, Seattle, WA 98195, USA; Northwest Mental Illness Research, Education and Clinical Centers, VA Puget Sound Health Care System, Seattle, WA 98108, USA; Geriatrics Research Education and Clinical Center, Puget Sound VA Medical Center, Seattle, WA 98108, USA; Department of Neurology, University of Washington School of Medicine, Seattle, WA 98195, USA.
Suman Jayadev, Division of Medical Genetics, University of Washington School of Medicine, Seattle, WA 98195, USA; Department of Neurology, University of Washington School of Medicine, Seattle, WA 98195, USA.
Paul N Valdmanis, Division of Medical Genetics, University of Washington School of Medicine, Seattle, WA 98195, USA.
Funding
This work was supported by a development project grant awarded to M.M.C. through the UW ADRC, and by the UW ADRC Neuropathology Core, which is funded by the National Institute on Aging (NIA) (P30 AG066509). This work was also supported by the ACT (Adult Changes in Thought) study (funded by U19 AG066567) and the Nancy and Buster Alvord Endowment to CDK and R01NS122766, funded by the National Institute of Neurological Disorders and Stroke (NINDS) and the NIA, to P.N.V.
The results published here are also in part based on data obtained from the Alzheimer’s disease Knowledge Portal (https://adknowledgeportal.org). The Mayo RNAseq study data was led by Dr. Nilüfer Ertekin-Taner, Mayo Clinic, Jacksonville, FL, USA, as part of the multi-PI U01 AG046139 (MPIs Golde, Ertekin-Taner, Younkin, Price). Samples were provided from the following sources: The Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216 and R01 AG003949, NINDS grant R01 NS080820, CurePSP and support from the Mayo Foundation. Study data include samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. [DOI] [PubMed] [Google Scholar]
- 2. Levy-Lahad E, Wasco W, Poorkaj P, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. [DOI] [PubMed] [Google Scholar]
- 3. Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. [DOI] [PubMed] [Google Scholar]
- 4. Chávez-Gutiérrez L, Bammens L, Benilova I, et al. The mechanism of gamma-secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernandez MA, Klutkowski JA, Freret T, Wolfe MS. Alzheimer presenilin-1 mutations dramatically reduce trimming of long amyloid beta-peptides (abeta) by gamma-secretase to increase 42-to-40-residue abeta. J Biol Chem. 2014;289:31043–31052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Veugelen S, Saito T, Saido TC, Chavez-Gutierrez L, De Strooper B. Familial Alzheimer’s disease mutations in presenilin generate amyloidogenic abeta peptide seeds. Neuron. 2016;90:410–416. [DOI] [PubMed] [Google Scholar]
- 7. Szaruga M, Munteanu B, Lismont S, et al. Alzheimer’s-causing mutations shift abeta length by destabilizing gamma-secretase-abetan interactions. Cell. 2017;170:443–456.e14. [DOI] [PubMed] [Google Scholar]
- 8. Bird TD. Alzheimer disease overview. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews®. University of Washington; 2018. [Google Scholar]
- 9. Schellenberg GD, Montine TJ. The genetics and neuropathology of Alzheimer’s disease. Acta Neuropathol. 2012;124:305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016;12:733–748. [DOI] [PubMed] [Google Scholar]
- 11. Pottier C, Hannequin D, Coutant S, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17:875–879. [DOI] [PubMed] [Google Scholar]
- 12. Cruchaga C, Haller G, Chakraverty S, et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS ONE. 2012;7:e31039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lanoiselee H-M, Nicolas G, Wallon D, et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017;14:e1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Strooper B. Loss-of-function presenilin mutations in Alzheimer disease. Talking point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jayadev S, Leverenz JB, Steinbart E, et al. Alzheimer’s disease phenotypes and genotypes associated with mutations in presenilin 2. Brain. 2010;133:1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Braggin JE, Bucks SA, Course MM, et al. Alternative splicing in a presenilin 2 variant associated with Alzheimer disease. Ann Clin Transl Neurol. 2019;6:762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iadecola C. Neurovascular regulation in the normal brain and in Alzheimer’s disease. Nat Rev Neurosci. 2004;5:347–360. [DOI] [PubMed] [Google Scholar]
- 18. Sato N, Hori O, Yamaguchi A, et al. A novel presenilin-2 splice variant in human Alzheimer’s disease brain tissue. J Neurochem. 1999;72:2498–2505. [DOI] [PubMed] [Google Scholar]
- 19. Moussavi Nik SH, Newman M, Wilson L, et al. Alzheimer’s disease-related peptide PS2V plays ancient, conserved roles in suppression of the unfolded protein response under hypoxia and stimulation of gamma-secretase activity. Hum Mol Genet. 2015;24:3662–3678. [DOI] [PubMed] [Google Scholar]
- 20. Moussavi Nik SH, Newman M, Lardelli M. The response of HMGA1 to changes in oxygen availability is evolutionarily conserved. Exp Cell Res. 2011;317:1503–1512. [DOI] [PubMed] [Google Scholar]
- 21. Manabe T, Katayama T, Sato N, et al. Induced HMGA1a expression causes aberrant splicing of presenilin-2 pre-mRNA in sporadic Alzheimer’s disease. Cell Death Differ. 2003;10:698–708. [DOI] [PubMed] [Google Scholar]
- 22. Tseng E, Rowell WJ, Glenn OC, et al. The landscape of SNCA transcripts across synucleinopathies: New insights from long reads sequencing analysis. Front Genet. 2019;10:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kukull WA, Higdon R, Bowen JD, et al. Dementia and Alzheimer disease incidence: A prospective cohort study. Arch Neurol. 2002;59:1737–1746. [DOI] [PubMed] [Google Scholar]
- 24. Nicolas G, Wallon D, Charbonnier C, et al. Screening of dementia genes by whole-exome sequencing in early-onset Alzheimer disease: Input and lessons. Eur J Hum Genet. 2016;24:710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nygaard HB, Lippa CF, Mehdi D, Baehring JM. A novel presenilin 1 mutation in early-onset Alzheimer’s disease with prominent frontal features. Am J Alzheimers Dis Other Demen. 2014;29:433–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsu S, Pimenova AA, Hayes K, et al. Systematic validation of variants of unknown significance in APP, PSEN1 and PSEN2. Neurobiol Dis. 2020;139:104817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. GTEx Consortium . The GTEx Consortium Atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moussavi Nik SH, Porter T, Newman M, et al. Relevance of a truncated PRESENILIN 2 transcript to Alzheimer’s disease and neurodegeneration. J Alzheimers Dis. 2021;80:1479–1489. [DOI] [PubMed] [Google Scholar]
- 29. Smith MJ, Sharples RA, Evin G, et al. Expression of truncated presenilin 2 splice variant in Alzheimer’s disease, bipolar disorder, and schizophrenia brain cortex. Brain Res Mol Brain Res. 2004;127:128–135. [DOI] [PubMed] [Google Scholar]
- 30. Allen M, Carrasquillo MM, Funk C, et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data. 2016;3:160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fung S, Smith CL, Prater KE, et al. Early-onset familial Alzheimer disease variant PSEN2 N141I heterozygosity is associated with altered microglia phenotype. J Alzheimers Dis. 2020;77:675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jayadev S, Case A, Alajajian B, Eastman AJ, Moller T, Garden GA. Presenilin 2 influences miR146 level and activity in microglia. J Neurochem. 2013;127:592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jayadev S, Case A, Eastman AJ, et al. Presenilin 2 is the predominant gamma-secretase in microglia and modulates cytokine release. PLoS ONE. 2010;5:e15743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sannerud R, Esselens C, Ejsmont P, et al. Restricted location of PSEN2/gamma-secretase determines substrate specificity and generates an intracellular abeta pool. Cell. 2016;166:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Payne AJ, Kaja S, Koulen P. Regulation of ryanodine receptor-mediated calcium signaling by presenilins. Receptors Clin Investig. 2015;2:e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blauwendraat C, Wilke C, Jansen IE, et al. Pilot whole-exome sequencing of a German early-onset Alzheimer’s disease cohort reveals a substantial frequency of PSEN2 variants. Neurobiol Aging. 2016;37:208.e11–208.e17. [DOI] [PubMed] [Google Scholar]
- 37. Wang B, Yang W, Wen W, et al. Gamma-secretase gene mutations in familial acne inversa. Science. 2010;330:1065. [DOI] [PubMed] [Google Scholar]
- 38. Dickson DW, Ahmed Z, Algom AA, Tsuboi Y, Josephs KA. Neuropathology of variants of progressive supranuclear palsy. Curr Opin Neurol. 2010;23:394–400. [DOI] [PubMed] [Google Scholar]
- 39. Lester E, Ooi FK, Bakkar N, et al. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron. 2021;109:1675–1691.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious Orders Study and Rush Memory and Aging Project. J Alzheimers Dis. 2018;64:S161–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang M, Beckmann ND, Roussos P, et al. The mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci Data. 2018;5:180185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang L, Chen C, Mak MS, et al. Advance of sporadic Alzheimer’s disease animal models. Med Res Rev. 2020;40:431–458. [DOI] [PubMed] [Google Scholar]
- 43. Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35:377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hundley HA, Krauchuk AA, Bass BL. C. elegans and H. sapiens mRNAs with edited 3’ UTRs are present on polysomes. RNA. 2008;14:2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer’s disease. Nat Rev Dis Primers. 2015;1:15056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequence reads from the Mayo Clinic are available from synapse.org with accession number syn5550404.