Abstract
Background
Biomarkers that provide insight into drivers of aging are needed for people with human immunodeficiency virus (PWH). The study objective was to determine if epigenetic age acceleration (EAA) markers are associated with physiologic frailty measured by the Veterans Aging Cohort Study (VACS) Index and predict all-cause mortality for PWH.
Methods
Epigenome-wide DNA methylation was profiled in VACS total white blood cell samples collected during 2005–2007 from 531 PWH to generate 6 established markers of EAA. The association of each EAA marker was tested with VACS Index 2.0. All-cause mortality was assessed over 10 years. For each EAA marker, the hazard ratio per increased year was determined using Cox regression. To evaluate mortality discrimination, C-statistics were derived.
Results
Participants were mostly men (98.5%) and non-Hispanic Black (84.4%), with a mean age of 52.4 years (standard deviation [SD], 7.8 years). Mean VACS Index score was 59.3 (SD, 16.4) and 136 deaths occurred over a median follow-up of 8.7 years. Grim age acceleration (AA), PhenoAA, HannumAA, and extrinsic epigenetic AA were associated with the VACS Index and mortality. HorvathAA and intrinsic epigenetic AA were not associated with either outcome. GrimAA had the greatest mortality discrimination among EAA markers and predicted mortality independently of the VACS Index. One-year increase in GrimAA was associated with a 1-point increase in VACS Index and a 10% increased hazard for mortality.
Conclusions
The observed associations between EAA markers with physiologic frailty and mortality support future research to provide mechanistic insight into the accelerated aging process and inform interventions tailored to PWH for promoting increased healthspan.
Keywords: DNA methylation, epigenetics, aging, frailty, mortality
The observed associations between epigenetic age acceleration markers with physiologic frailty and mortality provide mechanistic insight into accelerated aging in people with HIV (PWH) and support future interventions tailored to PWH for prevention and treatment of age-related disease.
Despite improved survival from viral suppression, people with human immunodeficiency virus (PWH) remain disproportionately burdened with age-related conditions. Numerous intrinsic mechanisms of biological aging as well as extrinsic structural determinants of health influence this process, underlying the challenge of developing a holistic prognostic indicator. The Veterans Aging Cohort Study (VACS) Index provides robust evidence for predicting age-related outcomes, including hospitalization [1], bone fractures [2], and all-cause mortality [3], and thus is viewed as a measure of physiologic frailty [4]. The VACS Index is based on readily available clinical laboratory results, facilitating its application in clinical care and research [5]. While the VACS Index is useful for distinguishing multiorgan injury, biomarkers that can also provide insight into biological aging mechanisms would help identify at-risk PWH and inform the development of targeted strategies for healthy aging.
Epigenetics is the study of DNA modifications that affect gene transcription without altering the genetic sequence [6]. DNA methylation (DNAm) is intrinsic to the biological aging process, changing over the life course across an array of disease conditions, and is sensitive to both positive (eg, physical activity) and negative (eg, smoking) environmental stressors [7–10]. An epigenome-wide association study (EWAS) provided the first evidence that DNAm could be used to evaluate accelerated aging in PWH, showing age-associated methylation patterns in PWH [11]. Epigenetic clocks are defined as multivariate age predictors that utilize methylation levels at specific cytosine residues of cytosine-phosphate-guanine dinucleotides (CpGs) to estimate chronological age and mortality risk [12]. Among the present epigenetic clocks [13], Horvath’s epigenetic clock, based on 353 CpGs, has been widely studied using the predicted epigenetic age, Horvath DNAm age [14]. In people without human immunodeficiency virus (HIV), Horvath DNAm age predicted mortality [15, 16] and correlated with a frailty index but not telomere length [17]. In PWH, we and others found that Horvath DNAm age is increased compared to sex-, race/ethnicity-, and chronological age–matched adults without HIV [18, 19].
In a more recent approach, DNAm age is regressed on chronologic age, allowing the result to be centered around zero with positive values reflecting accelerated aging and quantitated in years of epigenetic age acceleration (EAA) [13]. EAA markers include those based on Horvath DNAm age and Hannum DNAm age in addition to newer versions of predicted epigenetic age, Grim age and Pheno age. These EAA markers are greater in PWH compared with adults without HIV [20–22] and are reduced by antiretroviral therapy (ART) [23]. The utility of EAA markers as a clinical prognostic indicator remains unclear. Small studies have shown associations of EAA markers with cognitive function and HIV-associated neurocognitive disorder [20, 24, 25] but not the Fried frailty phenotype [20]. In a longitudinal study over 4 years, changes in EAA markers were variable, and baseline values did not predict incident HIV-related and unrelated clinical events [26]. Collectively, the capacity of current EAA markers to predict mortality in PWH has yet to be established. The objective of this study was to determine if markers of EAA are associated with physiologic frailty as measured by the VACS Index and predict all-cause mortality in PWH.
METHODS
The VACS is a well-defined cohort of prospectively enrolled Veterans with and without HIV infection from 8 Veterans Affairs Medical Centers [27]. Blood samples for this analysis were collected between 2005 and 2007 from a subset of participants. Total white blood cells were isolated at the time of sample collection for DNA extraction. This study included 533 samples with available DNA methylation data from participants with HIV infection. Two participants who did not have all laboratory components for calculation of the VACS Index within 180 days of the sample collection were excluded, resulting in an analytical set of 531 participants. All-cause mortality was determined from the Veterans Health Administration (VHA) vital status file, which uses inputs from the Social Security Administration death master file, the Beneficiary Identification and Records Locator Subsystem, and the VHA Medical Statistical Analysis Systems inpatient datasets.
VACS Index
The VACS Index is based on age and routine clinical laboratory values that predict adverse clinical outcomes including hospitalization and death in diverse patient populations with and without HIV infection. The most recent iteration, VACS Index 2.0, including body mass index, albumin, and total white blood cell count [28], was used based on superior mortality discrimination over version 1.0 [3]. The VACS Index 2.0 score was calculated from laboratory and body mass index data within 180 days of the blood sampling.
DNA Methylation Assay and Data Processing
The Illumina Infinium HumanMethylation 450 (450K) BeadChip was used to measure blood-based DNA methylation profile. BeadStudio software was used to generate the raw β values, which are the proportion of methylation intensity of a site (ranging from 0 to 1). As previously described [29], 927 CpG sites were removed from the analysis due to high missing rate (>5%). Another 35 605 CpG sites were removed because their probes contained single-nucleotide polymorphisms within 10 base pairs of the CpG site. An additional 24 729 CpG sites were removed because their probes mapped to multiple locations in the genome. We also excluded samples with >10% missing rate of methylation data and any results with a detection P > .01. We further performed quantile normalization and batch effect corrections using the minfi package available in R [30].
DNA Methylation Age and Age Acceleration Estimation
We first calculated 4 measurements of DNAm age based on Horvath DNAm age, Hannum DNAm age, DNAm Pheno age, and DNAm Grim age. The Horvath epigenetic clock and Hannum epigenetic clock were the first developed biomarkers of DNAm age and have a demonstrated robust relationship with chronological age and mortality [15, 16]. The more recently developed DNAm Pheno age has shown strong associations with all-cause mortality and aging-related morbidity [31], whereas DNAm Grim age performed especially well in predicting coronary heart disease and all-type cancer [32], as well as all-cause mortality [33]. All measures of DNAm ages were calculated through the online calculator (https://dnamage.genetics.ucla.edu) developed by Horvath’s group [14].
We calculated 6 markers of EAA from the residuals resulting from regressing DNAm age calculations (Horvath DNAm age, Hannum DNAm age, Pheno age, and Grim age) on chronological age in a linear model: Horvath DNAm age acceleration (HorvathAA), Hannum DNAm age acceleration (HannumAA), intrinsic epigenetic age acceleration (IEAA), extrinsic epigenetic age acceleration (EEAA), Pheno age acceleration (PhenoAA), and Grim age acceleration (GrimAA). IEAA is defined as the residual from regressing Horvath DNAm age on chronological age after adjusting for cell types including CD4 and naive CD8 T cells, CD8+CD28–CD45RA– (memory and effector T cells), plasmablasts, monocytes, granulocytes, and natural killer cells. Similarly, EEAA is calculated using Hannum DNAm age, adjusting for cell types including naive CD8, plasmablasts, and CD8+CD28–CD45RA–. The remaining 4 age acceleration markers were age-adjusted residuals without additional adjustment for the proportion of cell types.
Statistical Analysis
Linear regression was performed for each EAA marker as the independent variable and VACS Index as the dependent variable. Age acceleration was deemed to be present (>0) or absent (≤0) based on the value of the EAA marker. Survival analyses included Kaplan-Meier plots for the presence and absence of age acceleration. In the case of the VACS Index score, the median value was used as the cutoff point to define comparison groups. Cox regression was used to determine the hazard ratio (HR) for all-cause death per increased unit of each EAA marker (1 year). Based on prior work [3], the VACS Index score was coded in 5-point increments in the Cox models. The proportional hazards assumption was confirmed by test of Schoenfeld residuals. Additional analyses to assess discrimination for mortality were conducted; Harrell C-statistic was calculated for each Cox model and compared. Cox regression with adjustment for the VACS Index was performed for each EAA marker to determine whether prognostic accuracy improved. The association of EAA markers with participant baseline characteristics was tested by Student t test or χ2 test. Post hoc analysis included Cox regression with adjustment for smoking exposure.
RESULTS
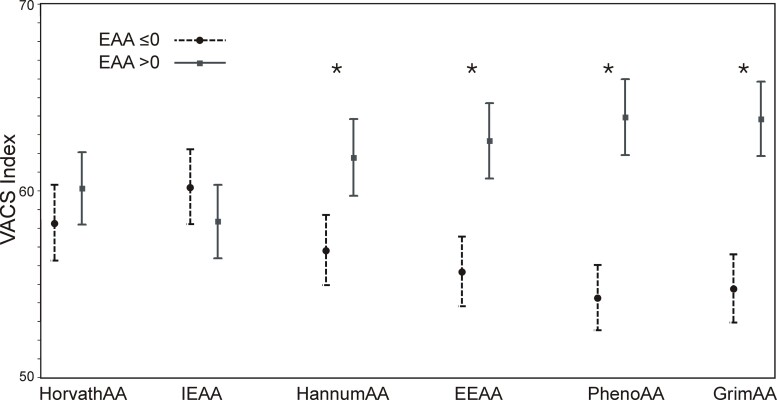
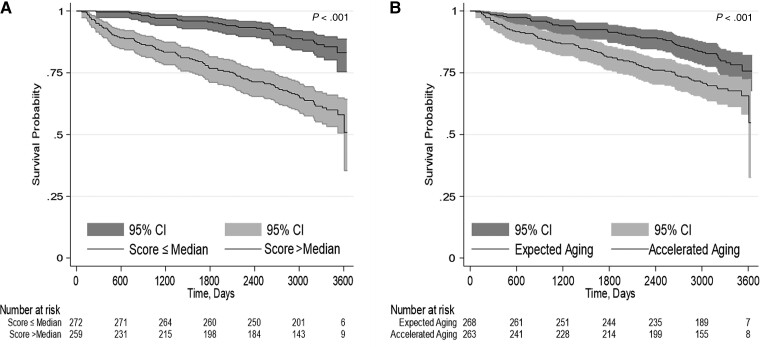
Table 1 describes the baseline characteristics of the study population. Participants were predominantly men (98.5%) and of non-Hispanic Black race/ethnicity (84.4%) with a mean age of 52.4 years (standard deviation [SD], 7.8 years). The VACS Index mean score was 59.3 (SD, 16.4). There were 136 deaths during a median follow-up of 8.7 years (interquartile range, 7.7–9.4 years). Four of the 6 EAA markers were significantly associated with the VACS Index: HannumAA, EEAA, GrimAA, and PhenoAA (Table 2). GrimAA demonstrated the strongest association; each year increase in GrimAA was associated with approximately 1-point increase in VACS Index (β = 1.18 [95% confidence interval {CI}, .91–1.45]; P < .0001). HorvathAA and IEAA, both based on the Horvath epigenic clock, were not significantly associated with the VACS Index. Similarly, the mean VACS Index was significantly greater if age acceleration was present (EAA >0) compared with absence (EAA ≤0) based on the markers HannumAA, EEAA, GrimAA, and PhenoAA (Figure 1). HannumAA, EEAA, GrimAA, and PhenoAA were significantly associated with all-cause mortality, but HorvathAA and IEAA were not (Table 3). GrimAA had the greatest mortality discrimination among the EAA markers, but less than the VACS Index (Table 3). Addition of the VACS Index increased the discrimination of the GrimAA Cox model (C-statistic, 0.62 [95% CI, .57–.67] vs 0.73 [95% CI, .69–.78]). Notably, only GrimAA remained significantly associated with mortality after adjustment for the VACS Index (Table 3). Ten-year prediction of all-cause mortality by the VACS Index and GrimAA is shown using Kaplan-Meier plots (Figure 2). Among clinical characteristics, lower CD4 cell count, detectable viral load, smoking exposure, alcohol abuse, and history of hepatitis C seropositivity were associated with age acceleration, defined as GrimAA >0 (Table 1). Similar results were found for CD4 cell count and viral load with other EAA measures, except that IEAA was not significant (Supplementary Table 1). Only GrimAA and PhenoAA were associated with smoking exposure.
Table 1.
Characteristics of Study Population With and Without Epigenetic Age Acceleration
| Characteristic | Total (N = 531) | Grim Age Accelerationa | ||
|---|---|---|---|---|
| Present (n = 263) | Absent (n = 268) | P Valueb | ||
| Age, y, mean (SD) | 52.4 (7.8) | 52.4 (6.9) | 52.4 (8.6) | .95 |
| Race/ethnicity | .01 | |||
| ȃBlack | 448 (83.4) | 227 (86.3) | 221 (82.5) | |
| ȃWhite | 56 (10.6) | 30 (11.4) | 26 (9.7) | |
| ȃHispanic or other | 27 (5.1) | 6 (2.3) | 21 (7.8) | |
| Sex, male | 523 (98.5) | 257 (97.7) | 266 (99.3) | .17 |
| Smoking, current or former | 419 (78.9) | 248 (94.3) | 171 (63.8) | <.001 |
| Alcohol current use | .01 | |||
| ȃNone | 189 (35.7) | 88 (33.5) | 101 (38.0) | |
| ȃNonhazardous | 109 (20.6) | 44 (16.7) | 65 (24.4) | |
| ȃHazardous use or alcohol abuse | 231 (43.7) | 131 (49.8) | 100 (37.6) | |
| HCV antibody positive | 301 (56.7) | 162 (61.6) | 139 (51.9) | .02 |
| Diabetes | 100 (18.8) | 51 (19.4) | 49 (18.3) | .74 |
| Prior AIDS-defining illness | 198 (37.3) | 118 (44.9) | 80 (29.9) | <.001 |
| On ART | 431 (81.2) | 215 (81.8) | 216 (80.6) | .73 |
| CD4 count, cells/µL, median (IQR) | 390 (326) | 368 (307) | 401 (343) | .01 |
| HIV viral load nondetectablec | 291 (54.8) | 123 (46.8) | 168 (62.7) | <.001 |
| VACS Index, median (IQR) | 58 (22) | 62 (22) | 54 (20) | <.001 |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ART, antiretroviral therapy; HCV, hepatitis C virus; HIV, human immunodeficiency virus; IQR, interquartile range; SD, standard deviation; VACS, Veterans Affairs Cohort Study.
Grim age acceleration present (>0) or absent (≤0).
Student t test or Wilcoxon rank-sum used for continuous variables; χ2 test or Fisher exact test used for categorical variables.
Nondetectable: HIV type 1 RNA <75 copies/mL.
Table 2.
Linear Regression of Epigenetic Age Acceleration Markers With the Veterans Aging Cohort Study Index
| EAA Marker (per Year) | Median (Minimum, Maximum) | Mean (SD) | β Estimatea | (95% CI) | P Value |
|---|---|---|---|---|---|
| HorvathAA | 0.55 (−25.12, 22.94) | 0.61 (6.23) | .14 | (−.08 to .37) | .219 |
| HannumAA | −0.15 (−23.47, 29.92) | −0.15 (5.60) | .51 | (.26–.76) | <.001 |
| IEAA | 0.05 (−24.15, 19.49) | 0.08 (5.62) | −.14 | (−.40 to .11) | .276 |
| EEAA | 0.19 (−26.10, 36.00) | 0.27 (7.11) | .56 | (.37–.75) | <.001 |
| GrimAA | −0.06 (−12.23, 14.66) | 0.02 (4.88) | 1.18 | (.91–1.45) | <.001 |
| PhenoAA | 0.58 (−32.19, 30.04) | 0.59 (8.71) | .58 | (.42–.73) | <.001 |
Abbreviations: CI, confidence interval; EAA, epigenetic age acceleration; EEAA, extrinsic epigenetic age acceleration; GrimAA, Grim age acceleration; HannumAA, Hannum DNA methylation age acceleration; HorvathAA, Horvath DNA methylation age acceleration; IEAA, intrinsic epigenetic age acceleration; PhenoAA, Pheno age acceleration; SD, standard deviation.
Per point of Veterans Aging Cohort Study Index.
Figure 1.
Association of epigenetic age acceleration (EAA) markers with the Veterans Aging Cohort Study (VACS) Index. Mean and 95% confidence interval of the VACS Index score by age acceleration present (>0, squares) or absent (≤0, circles) for each EAA marker. The difference was tested by Student t test. *P < .001. Abbreviations: EAA, epigenetic age acceleration; EEAA, extrinsic epigenetic age acceleration; GrimAA, Grim age acceleration; HannumAA, Hannum DNA methylation age acceleration; HorvathAA, Horvath DNA methylation age acceleration; IEAA, intrinsic epigenetic age acceleration; PhenoAA, Pheno age acceleration; VACS, Veterans Aging Cohort Study.
Table 3.
Cox Regression of All-Cause Mortality for Each Measure of Epigenetic Age Acceleration and the Veterans Aging Cohort Study Index
| Predictors (per Year) | Unadjusted Models | Models Adjusted for VACS Index | Models Adjusted for Smoking Exposureb | ||||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P Value | C-Statistica (95% CI) | HR (95% CI) | P Value | HR (95% CI) | P Value | |
| HorvathAA | 1.00 (.98–1.03) | .843 | 0.48 (.43–.53) | 0.99 (.97–1.02) | .496 | 1.00 (.98–1.03) | .811 |
| HannumAA | 1.03 (1.00–1.06) | .038 | 0.56 (.51–.61) | 1.00 (.97–1.03) | .887 | 1.03 (1.00–1.06) | .039 |
| IEAA | 1.00 (.97–1.03) | .780 | 0.52 (.47–.57) | 1.00 (.97–1.03) | .874 | 1.00 (.97–1.03) | .774 |
| EEAA | 1.03 (1.01–1.05) | .014 | 0.57 (.52–.62) | 1.00 (.98–1.02) | .863 | 1.03 (1.00–1.05) | .019 |
| GrimAA | 1.10 (1.06–1.14) | <.001 | 0.62 (.57–.67) | 1.05 (1.01–1.09) | .016 | 1.08 (1.04–1.12) | <.001 |
| PhenoAA | 1.02 (1.00–1.04) | .028 | 0.56 (.51–.61) | 0.99 (.97–1.02) | .334 | 1.02 (.99–1.04) | .069 |
| VACS Indexc | 1.29 (1.22–1.35) | <.001 | 0.72 (.68–.77) | … | 1.28 (1.21–1.34) | <.001 | |
Abbreviations: CI, confidence interval; EEAA, extrinsic epigenetic age acceleration; GrimAA, Grim age acceleration; HannumAA, Hannum DNA methylation age acceleration; HorvathAA, Horvath DNA methylation age acceleration; HR, hazard ratio; IEAA, intrinsic epigenetic age acceleration; PhenoAA, Pheno age acceleration; VACS, Veterans Aging Cohort Study.
Harrell C coefficient.
Smoking exposure classified as current/former and never.
Unit of VACS Index is per 5 points.
Figure 2.
Kaplan-Meier estimates of 10-year all-cause mortality for the predictors Veterans Aging Cohort Study (VACS) Index and Grim age acceleration (GrimAA). The number of participants at risk are below each graph. A, For the VACS Index, the median value of the score was used as the cutoff point to define comparison groups. B, For the GrimAA marker, the value of zero was used as the cutoff point to define accelerated aging (>0) or expected aging (≤0). Abbreviation: CI, confidence interval.
DISCUSSION
Several markers of EAA in PWH were associated with the VACS Index, a well-studied measure of physiologic frailty. PWH experiencing age acceleration, defined as EAA marker value >0, had significantly greater mean VACS Index (poorer prognosis) for HannumAA, EEAA, PhenoAA and GrimAA, but not HorvathAA and IEAA. These results were further supported by the predictive accuracy of the same EAA markers for all-cause mortality. In particular, GrimAA had the strongest predictive value, 10% increased risk of death per 1 year, and mortality discrimination (C-statistic, 0.62). While epigenetic biomarkers are quickly evolving, GrimAA may provide key insights into the molecular mechanisms mediating biological accelerated aging and physiologic frailty in PWH.
The VACS Index is a robust measure of physiologic frailty that encapsulates multiorgan function in combination with HIV progression but also has been validated in people without HIV [4]. Previous EWASs of PWH have leveraged the VACS Index as a prognostic indicator to identify CpG sites associated with illicit drug use and hepatitis C virus infection [34] and smoking [35]. Another EWAS identified 393 CpG sites located in genes controlling immune activation and cytokine receptor binding [36]. In contrast to this prior work, our objective was to determine the association of EAA markers with the VACS Index as a measure of physiologic frailty given our limited understanding of the interplay between frailty and accelerated aging in PWH. Shiau and colleagues assessed physical frailty with the Fried criteria in an epigenetic cross-sectional study of older African American adults with and without HIV [20]. Approximately one-third of participants in both groups were prefrail/frail, but none of the EAA markers (Horvath AA, IEAA, EEAA, GrimAA, and PhenoAA) were significantly different from the nonfrail participants. We found that EAA markers based on Hannum DNAm age (HannumAA and EEAA), DNAm Pheno age (PhenoAA), and DNAm Grim age (GrimAA) were significantly associated with the VACS Index. Intriguingly, the 2 EAA markers based on Horvath DNAm age (Horvath AA and IEAA) were not associated with the VACS Index. This finding was initially surprising given that Horvath DNAm age has been associated with 2 different indices of frailty in elderly adults without HIV [17, 37]. Yet more recent work found that GrimAA and PhenoAA, but not HorvathAA, were associated with the Fried frailty score [33], similar to our findings in PWH. Our results will require further investigation to determine potentially shared mechanisms that underlie the gene transcription captured by different epigenetic clocks in association with multiorgan decline captured by the VACS Index.
Studies investigating clinical outcomes support the use of EAA markers for HIV and aging research, yet also show different results across biomarkers [11]. Cross-sectional studies found evidence of age acceleration in PWH who are ART naive [22] or virally suppressed on ART [20] compared with people without HIV using the markers HorvathAA, EEAA, GrimAA, and PhenoAA, but not IEAA. A longitudinal study of ART-naive PWH found significantly greater age acceleration compared with age- and sex-matched controls for markers of HorvathAA, GrimAA, and PhenoAA, but not HannumAA [23]. Remarkably, after 2 years, all EAA markers were reduced, but only GrimAA and PhenoAA remained significantly higher than controls. In contrast, a small study of ART-naive PWH with age-matched adults without HIV showed that after 2 years of ART, Horvath AA and EEAA remained significantly greater in the HIV group, but there was no difference in GrimAA and PhenoAA between groups [21]. These differences in magnitude and change in EAA likely reflect clinical and environmental differences in study populations. Our results in the context of this literature support future research to determine the extent to which EAA markers reflect holistic versus specific organ system aging.
Our understanding of the prognostic utility of epigenetic markers for PWH is limited. Esteban-Cantos and colleagues measured HorvathAA, PhenoAA, and GrimAA in blood samples collected at 2 time points in a longitudinal cohort of PWH on ART with viral suppression [26]. Baseline HorvathAA was significantly greater among those who had a serious clinical event during the 4-year follow-up (AIDS or non-AIDS-related event). However, none of the EAA markers at baseline predicted new clinical events in logistic regression models, with and without adjustment for potential confounders. Several smaller studies have shown associations of various EAA markers with cognitive function and HIV-associated neurocognitive disorder [20, 24, 25]. Zhang et al used machine learning to construct a DNAm Index in PWH based on 698 CpGs derived from smoking-associated CpGs (enriched for frailty) that strongly predicted mortality (HR, 1.46 [95% CI, 1.06–2.02]) [35]. Although this epigenetic marker was not based on predicted epigenetic age (eg, DNAm age), results provided the first evidence for predictive accuracy.
Our findings extend the epigenetic HIV literature by demonstrating that GrimAA, PhenoAA, HannumAA, and EEAA predicted all-cause death in a longitudinal cohort of PWH. Our negative result for EAA markers based on Horvath DNAm age (Horvath AA and IEAA) may reflect intrinsic cohort and/or environmental differences. In numerous HIV negative cohorts, Horvath DNAm age has predicted mortality using blood samples [15, 16] and further contributed to our understanding of accelerated aging in PWH, especially neurocognitive function [18]. The results our of survival analyses, combined with parallel relationships for the VACS Index, supports further evaluation of EAA markers to differentiate biologic mechanisms and environmental factors that drive accelerated aging due to multiorgan decline. The role of HIV disease progression and ART toxicity also needs to be considered in cohorts consisting of people with and without HIV. The cause of death, and thus underlying pathogenesis, may also be important for EAA markers but was not evaluated in our study. Wang and colleagues performed a survival analysis in 2 independent cohorts of >2000 individuals without HIV for the markers GrimAA, PhenoAA, EAAA, and IEAA [38]. Cardiovascular-related mortality (myocardial infarction and stroke) were predicted by GrimAA, PhenoAA, and EAAA, but not IEAA. None of the EAA markers predicted cancer-related mortality. Similar to our findings, GrimAA had the strongest association with all-cause mortality.
Among PWH, we found that a single-year increase in GrimAA predicted a 10% increased hazard for all-cause mortality (HR, 1.10 [95% CI, 1.06–1.14]; P < .001). Furthermore, GrimAA had the greatest mortality discrimination among EAA markers and remained an independent predictor of mortality after adjustment for the VACS Index. DNAm Grim age was derived from a combination of DNAm biomarkers of physiological stress correlated with plasma protein levels and a DNAm estimator of smoking pack-years [32]. The impact of smoking on DNA methylation is well-established in adults with and without HIV [37, 39, 40]. The higher mortality discrimination of GrimAA among the EAA markers we tested may have been driven by the large proportion of participants with a history of smoking (79%). Yet, in post hoc sensitivity analysis, GrimAA predicted death after adjustment for smoking (Table 3). Furthermore, our results are consistent with survival analyses in a larger cohort of older adults without HIV but a balanced distribution of smoking history [33]. In this study using the same EAA markers, McCrory et al found that only GrimAA predicted all-cause mortality in adjusted models. In PWH, the putative metabolic effects of ART also need to be considered. GrimAA correlated with visceral adipose tissue and was inversely related to skeletal muscle mass and plasma levels of micronutrients in a subsample of the Framingham Heart Study cohort [32]. Future research should evaluate if GrimAA differs by HIV strata defined by smoking history and other environmental factors. Given that the end-organ effects of lifestyle factors are difficult to measure, GrimAA may offer the opportunity to identify individuals with EAA for smoking cessation and other lifestyle interventions. Whether changes in GrimAA will reflect beneficial responses to interventions independent of VACS indices remains unclear.
The primary limitation of our study is the narrow demographic characteristics of the participants, mostly Black men. However, this is an understudied group in the methylation literature. Future research in PWH is needed to address the generalizability of our findings. In addition, the question remains if these relationships are comparatively different in people without HIV, and whether any differences are mediated by HIV, ART, or environmental determinants of health outcomes. Finally, we acknowledge that blood epigenetics alone does not capture the full breadth of pathology necessary for assessing biological aging in PWH [12].
In conclusion, epigenetic age acceleration markers combined with clinical and environmental data provide insight into molecular mechanisms mediating accelerated aging in PWH, and may, more generally, play an important role in precision medicine strategies for the prevention and treatment of disease.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Krisann K Oursler, Department of Internal Medicine, Virginia Tech Carilion School of Medicine and Veterans Affairs Salem Healthcare System, Roanoke, Virginia, USA.
Vincent C Marconi, Department of Medicine, Emory University School of Medicine and Rollins School of Public Health, Atlanta, Georgia, USA; Veterans Affairs Atlanta Healthcare System, Decatur, Georgia, USA.
Zeyuan Wang, Department of Epidemiology, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Ke Xu, Department of Psychiatry, Yale School of Medicine, West Haven, Connecticut, USA; Veterans Affairs Connecticut Healthcare System, West Haven, Connecticut, USA.
Monty Montano, Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Kaku So-Armah, Department of Medicine, Boston University School of Medicine, Boston, Massachusetts, USA.
Amy C Justice, Veterans Affairs Connecticut Healthcare System, West Haven, Connecticut, USA; Department of Medicine, Yale School of Medicine, West Haven, Connecticut, USA; Division of Health Policy, Yale School of Public Health, West Haven, Connecticut, USA.
Yan V Sun, Veterans Affairs Atlanta Healthcare System, Decatur, Georgia, USA; Department of Epidemiology, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Notes
Author contributions. K. K. O., V. C. M., and Y. V. S. designed the study. A. C. J. obtained the funding and specimens. Z. W., with input from K. K. O., Y. V. S., and A. C. J., conducted the statistical analysis. K. X. contributed to DNA methylation data. K. S. contributed to the phenotypic data. All authors have seen and approved the final version for publication.
Acknowledgments. The authors appreciate assistance from Chani Jain (tables and figures), Kim Birkett (references), and Dr Janet Tate (Veterans Aging Cohort Study Index).
Financial support. This work was supported by the US Department of Veterans Affairs (grant number I01 RX002790 to K. K. O. and V. C. M.) and the National Institutes of Health (NIH) (grant/award numbers P30 AI050409 to V. C. M. and Y. V. S.; R01DK125187 to Z. W. and Y. V. S.; R01DA047820, R01DA052846, R01DA042691, and R01DA047063 to K. X.; U24AA020794, U01AA020790, U24-AA022001, and U10 AA013566 to A. C. J.; K01HL134147 to K. S.; and P30AG031679 and P30AI060354 to M. M.).
References
- 1. Akgun KM, Gordon K, Pisani M, et al. . Risk factors for hospitalization and medical intensive care unit (MICU) admission among HIV-infected veterans. J Acquir Immune Defic Syndr 2013; 62:52–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Womack JA, Goulet JL, Gibert C, et al. . Physiologic frailty and fragility fracture in HIV-infected male veterans. Clin Infect Dis 2013; 56:1498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGinnis KA, Justice AC, Moore RD, et al. . Discrimination and calibration of the VACS Index 2.0 for predicting mortality among people with HIV in North America. Clin Infect Dis 2022; 75:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Justice AC, Tate JP. Strengths and limitations of the Veterans Aging Cohort Study Index as a measure of physiologic frailty. AIDS Res Hum Retroviruses 2019; 35:1023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tate JP, Justice AC, Hughes MD, et al. . An internationally generalizable risk index for mortality after one year of antiretroviral therapy. AIDS 2013; 27:563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bird A. Perceptions of epigenetics. Nature 2007; 447:396–8. [DOI] [PubMed] [Google Scholar]
- 7. Johansson A, Enroth S, Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS One 2013; 8:e67378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bollati V, Schwartz J, Wright R, et al. . Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 2009; 130:234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bocklandt S, Lin W, Sehl ME, et al. . Epigenetic predictor of age. PLoS One 2011; 6:e14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol 2015; 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Titanji BK, Gwinn M, Marconi VC, Sun Y V. Epigenome-wide epidemiologic studies of human immunodeficiency virus infection, treatment, and disease progression. Clin Epigenetics 2022; 14:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horvath S, Lin DTS, Kobor MS, et al. . HIV, pathology and epigenetic age acceleration in different human tissues. GeroScience 2022; 44:1609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nwanaji-Enwerem JC, Weisskopf MG, Baccarelli AA. Multi-tissue DNA methylation age: molecular relationships and perspectives for advancing biomarker utility. Ageing Res Rev 2018; 45:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013; 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics 2016; 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marioni RE, Shah S, McRae AF, et al. . DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol 2015; 16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Breitling LP, Saum KU, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics 2016; 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horvath S, Levine AJ. HIV-1 Infection accelerates age according to the epigenetic clock. J Infect Dis 2015; 212:1563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nelson KN, Hui Q, Rimland D, et al. . Identification of HIV infection-related DNA methylation sites and advanced epigenetic aging in HIV-positive, treatment-naive U.S. veterans. AIDS 2017; 31:571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shiau S, Arpadi SM, Shen Y, et al. . Epigenetic aging biomarkers associated with cognitive impairment in older African American adults with human immunodeficiency virus (HIV). Clin Infect Dis 2021; 73:1982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sehl ME, Rickabaugh TM, Shih R, et al. . The effects of anti-retroviral therapy on epigenetic age acceleration observed in HIV-1–infected adults. Pathog Immun 2020; 5:291–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang CX, Schon E, Obeidat M, et al. . Occurrence of accelerated epigenetic aging and methylation disruptions in human immunodeficiency virus infection before antiretroviral therapy. J Infect Dis 2021; 223:1681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Esteban-Cantos A, Rodríguez-Centeno J, Barruz P, et al. . Epigenetic age acceleration changes 2 years after antiretroviral therapy initiation in adults with HIV: a substudy of the NEAT001/ANRS143 randomised trial. Lancet HIV 2021; 8:e197–205. [DOI] [PubMed] [Google Scholar]
- 24. Levine AJ, Quach A, Moore DJ, et al. . Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol 2016; 22:366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shiau S, Cantos A, Ramon C V, et al. . Epigenetic age in young African American adults with perinatally acquired HIV. J Acquir Immune Defic Syndr 2021; 87:1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Esteban-Cantos A, Montejano R, Rodríguez-Centeno J, et al. . Longitudinal changes in epigenetic age acceleration in aviremic human immunodeficiency virus–infected recipients of long-term antiretroviral treatment. J Infect Dis 2022; 225:287–94. [DOI] [PubMed] [Google Scholar]
- 27. Justice AC, Dombrowski E, Conigliaro J, et al. . Veterans Aging Cohort Study (VACS): overview and description. Med Care 2006; 44:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tate JP, Sterne JAC, Justice AC. Albumin, white blood cell count, and body mass index improve discrimination of mortality in HIV-positive individuals. AIDS 2019; 33:903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mathur R, Hui Q, Huang Y, et al. . DNA methylation markers of type 2 diabetes mellitus among male veterans with or without human immunodeficiency virus infection. J Infect Dis 2019; 219:1959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 2014; 30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levine ME, Lu AT, Quach A, et al. . An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018; 10:573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu AT, Quach A, Wilson JG, et al. . DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019; 11:303–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCrory C, Fiorito G, Hernandez B, et al. . GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J Gerontol A Biol Sci Med Sci 2021; 76:741–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X, Hu Y, Justice AC, et al. . DNA methylation signatures of illicit drug injection and hepatitis C are associated with HIV frailty. Nat Commun 2017; 8:2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X, Hu Y, Aouizerat BE, et al. . Machine learning selected smoking-associated DNA methylation signatures that predict HIV prognosis and mortality. Clin Epigenetics 2018; 10:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shu C, Justice AC, Zhang X, et al. . DNA methylation biomarker selected by an ensemble machine learning approach predicts mortality risk in an HIV-positive veteran population. Epigenetics 2021; 16:741–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao X, Zhang Y, Saum KU, Schottker B, Breitling LP, Brenner H. Tobacco smoking and smoking-related DNA methylation are associated with the development of frailty among older adults. Epigenetics 2017; 12:149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang C, Ni W, Yao Y, et al. . DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: the NAS, and KORA F4. EBioMedicine 2021; 63:103151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Yang R, Burwinkel B, Breitling LP, Brenner H. F2RL3 methylation as a biomarker of current and lifetime smoking exposures. Environ Health Perspect 2014; 122:131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun YV, Smith AK, Conneely KN, et al. . Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum Genet 2013; 132:1027–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.