Abstract
The diagnosis of postacute sequelae of coronavirus disease 2019 (PASC) poses an ongoing medical challenge. To identify biomarkers associated with PASC we analyzed plasma samples collected from PASC and coronavirus disease 2019 patients to quantify viral antigens and inflammatory markers. We detect severe acute respiratory syndrome coronavirus 2 spike predominantly in PASC patients up to 12 months after diagnosis.
Keywords: COVID-19, post-acute sequelae of COVID-19 (PASC), long COVID, SARS-CoV-2 antigens, spike
Although symptoms resulting from infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) typically resolve within a few weeks, some individuals experience persistent symptoms following the acute phase of coronavirus disease (COVID-19). The associated syndrome, termed postacute sequelae of COVID-19 (PASC) or long COVID, encompasses a range of symptoms, affecting multiple organs [1]. Estimates vary as to the prevalence of PASC, but the World Health Organization reports around one-quarter of individuals with COVID-19 continue to experience symptoms 4 to 5 weeks after diagnosis and approximately 1 in 10 has continuing symptoms after 12 weeks [2].
Although recent studies provide some clues, the underlying causes of PASC remain elusive [3]. Heterogeneous patient recruitment and inconsistencies in defining PASC patients make it difficult to compare studies and validate current hypotheses. Disentangling the complex biology of PASC will rely on the identification of biomarkers that enable classification of patient phenotypes. Here, we measure SARS-CoV-2 antigen and cytokine levels in plasma samples collected from individuals infected with SARS-CoV-2, some of whom developed PASC.
METHODS
A retrospective pilot study was performed using plasma samples collected from 63 adults who developed acute COVID-19 or PASC. A full description of the patient cohort and sample collection is provided in the Supplementary Materials. SARS-CoV-2 antigens and a panel of 10 cytokines were measured in the collected samples, as described in the Supplementary Materials.
RESULTS
We analyzed plasma samples from a cohort of 63 individuals previously infected with SARS-CoV-2, 37 of whom were diagnosed with PASC. For most of the PASC patients (n = 31), blood samples were collected 2 or more times up to 12 months after their first positive result with a nasopharyngeal swab reverse transcriptase-polymerase chain reaction test. Blood was also collected from individuals who suffered from COVID-19 but were not diagnosed with PASC (hereafter referred to as COVID-19 patients), up to 5 months postdiagnosis. The majority of PASC patients were female (n = 30), reflecting the increased frequency with which women are affected by persistent symptoms following SARS-CoV-2 infection [4]. A complete overview of patient characteristics is provided in Supplementary Table 1 and their reported symptoms are shown in Supplementary Figure 1.
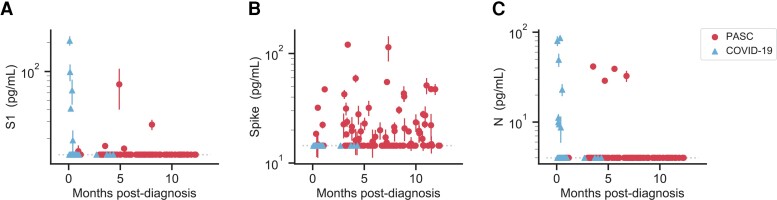
Using previously developed and optimized ultra-sensitive single molecule array assays (Supplementary Figure 2), we measured the concentration of SARS-CoV-2 antigens, including the S1 subunit of spike, full-length spike, and nucleocapsid (N), in the collected plasma samples (Figure 1) [5]. We detected either S1, spike, or N in approximately 65% of the patients diagnosed with PASC at any given time point, several months after SARS-CoV-2 infection. Of the 3 antigens, we detected spike most often in 60% of the PASC patients, whereas we did not detect spike in the COVID-19 patients. S1 was detected to a lesser degree in about one-fifth of the PASC patients and N was detected in a single patient at multiple time points. In line with our previous work [5], we detected S1 and N in COVID-19 patients, often those hospitalized with severe disease and within the first week after diagnosis.
Figure 1.
SARS-CoV-2 antigen levels versus time. The concentration of S1 (A), spike (B), and N (C) measured in the plasma of individuals over time after diagnosis with PASC or COVID-19 following SARS-CoV-2 infection. Multiple data points may correspond to the same individual, where repeat sampling was available. Data points represent mean values ± SD (n = 2). Dashed lines indicate the LOD for each assay. Abbreviations: COVID-19, coronavirus disease; LOD, limit of detection; PASC, postacute sequelae of coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
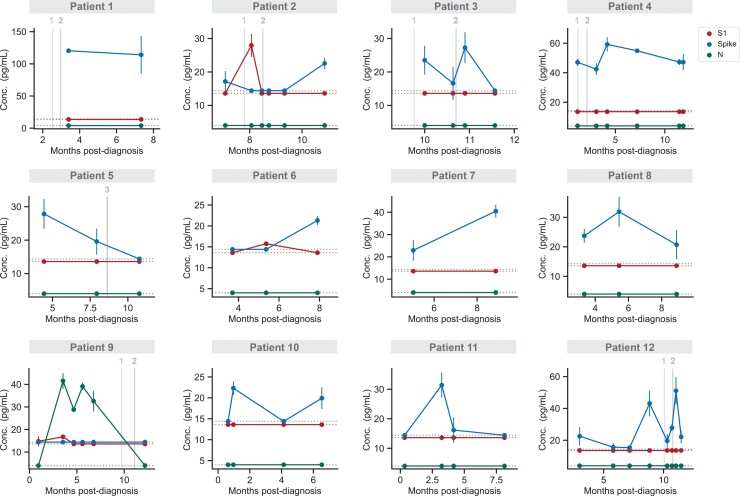
For a subset of PASC and COVID-19 patients, blood samples were collected at multiple times, allowing us to analyze temporal antigen profiles. Of the PASC patients for whom we had longitudinal samples, we detected antigen more than once in 12 patients (Figure 2). In only a few cases, the presence of S1 or spike may be correlated with vaccination; however, according to our previous findings, S1 is only detected soon after the first dose and spike is rarely detected [6]. Most significantly, we observe sustained spike levels over the course of several months in many patients. In other cases, we observe fluctuations in antigen detectability, indicating that the time of sampling is important. In contrast, temporal antigen profiles of 6 COVID-19 patients show high antigen levels soon after diagnosis, quickly dropping below the limit of detection (Supplementary Figure 3).
Figure 2.
Temporal profiling of SARS-CoV-2 antigens for individual PASC patients. S1, spike, and N levels versus time for 12 patients diagnosed with PASC. Data points represent mean values ± SD (n = 2). Vertical lines correspond to the times when each patient received either the first, second, or third dose of a COVID-19 vaccine. If an individual was not vaccinated within 1 month of the time frame shown, no vaccination information is shown. Dashed horizontal lines indicate the LOD for each assay. Abbreviations: COVID-19, coronavirus disease; LOD, limit of detection; PASC, postacute sequelae of coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Because PASC is known to affect multiple organ systems, we grouped reported persistent and acute symptoms according to the associated organ system and related those symptoms to whether S1, spike, or nucleocapsid was detected at any given time after infection (Supplementary Figures 4, 5). We detected antigen in > 70% of patients who reported ongoing cardiovascular, systemic, head-eye-ear-nose-throat, and musculoskeletal symptoms. On the other hand, we were more likely to detect antigen in patients who reported acute gastrointestinal and neuropsychiatric symptoms. Furthermore, the greater number of organ systems involved, the higher the probability a given patient was antigen positive.
We also measured the plasma concentrations of a panel of cytokines, including interferon-γ, interleukin 1β (IL-1β), IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-22, and tumor necrosis facto-α, but there were no apparent differences and levels mostly remained within normal ranges (Supplementary Figures 6, 7).
DISCUSSION
We measured the concentration of SARS-CoV-2 antigens and cytokines in plasma samples collected from PASC and COVID-19 patients. Most notably, we detected circulating SARS-CoV-2 spike in the majority of PASC patients. Although our sample size is small, the detection of spike at multiple time points 2 to 12 months after infection is an important finding. Temporal fluctuations in antigen levels for a subset of individuals indicates the importance of longitudinal sampling. Furthermore, the minority of our PASC cohort was hospitalized (n = 2), suggesting that our findings relate to SARS-CoV-2 infection rather than effects from severe illness and hospitalization. However, our COVID-19 cohort was more often hospitalized and demographically different, making it difficult to make concrete comparisons. In addition, samples were collected mostly from COVID-19 patients within the first month after diagnosis and not after 5 months. Future studies with better matched cohorts and sampling times will be necessary to validate these results.
The presence of circulating spike supports the hypothesis that a reservoir of active virus persists in the body. Another preliminary postmortem tissue study found SARS-CoV-2 RNA in multiple anatomic sites up to 7 months after symptom onset, corroborating viral antigen persistence [7]. Furthermore, we previously detected elevated levels of spike in children who develop multisystem inflammatory syndrome weeks after SARS-CoV-2 infection resulting from leakage from the gut into the blood stream [8]. Additionally, a recent study detected SARS-CoV-2 RNA shed in the feces up to 7 months after initial infection [9]. Although the detection of spike in PASC patients months after diagnosis suggests the presence of replicating viral reservoirs, further analyses are needed to confirm this hypothesis.
If viral reservoirs persist in the body of PASC patients, why do we not also detect N in more patients? It is possible that N is preferentially hydrolyzed, whereas spike may be more efficiently transported into the bloodstream, evading degradation. For instance, SARS-CoV-2 spike was shown to be incorporated in extracellular vesicles circulating in patient blood [10]. Alternatively, circulating anti-N antibodies may be more effective at clearing N compared with the anti-spike antibodies produced. However, after determining the neutralization capacity for anti-spike antibodies in many spike positive samples, we found that spike could be detected even if neutralization levels were high (Supplementary Figure 8). Furthermore, PASC is a heterogeneous syndrome, possibly dependent on the tissue in which the viral reservoir persists. This heterogeneity may influence whether spike or N is detected; future investigations are necessary to resolve these unanswered questions.
Similarly, we detect S1 at a lower frequency compared with full spike. In principle, we should detect S1 as part of spike; however, our results indicate that both S1 epitopes may not be accessible depending on the conformation of circulating spike. We assume that spike will be incorporated in a membrane, either in viral particles, extracellular vesicles, or remnants of dead infected cells. Given that we calibrate our assay with the spike ectodomain, which is missing the transmembrane domain, we cannot confirm that our S1 assay should bind spike in its natural conformation.
Active viral reservoirs could cause PASC symptoms, but circulating spike may also give rise to symptoms. Similar to bacterial superantigens, SARS-CoV-2 spike contains structural motifs that skew the T-cell receptor repertoire, possibly accounting for the hyperinflammatory response observed in severe COVID-19 and multisystem inflammatory syndrome in children [11]. Although spike does not instigate a cytokine storm in PASC patients, spike alone has been shown to induce dysfunction in pericytes, vascular endothelial cells, and the blood–brain barrier [12–14]. Alternatively, PASC may manifest as cellular exhaustion or the induction of refractory inflammatory response—but with persistent tissue localized dysfunction. Confined inflammation may also explain why changes in circulating cytokines may be subtle when measured in blood. Other work indicates a trend toward elevated IL-6 and tumor necrosis factor-α in PASC patients [15] or that the combination of inflammatory mediators is more predictive [16]; therefore, despite our results, monitoring cytokines over time in a larger cohort warrants further investigation.
In conclusion, the presence of circulating spike in PASC patients up to 12 months after diagnosis suggests that SARS-CoV-2 viral reservoirs may persist in the body. There are likely many overlapping immunological and inflammatory phenomena contributing to PASC and the detection of spike cannot alone confirm the presence of active viral reservoirs. However, if our finding can be validated in a larger cohort, it would provide strong support for the use of spike as a biomarker for PASC, making it easier to identify patients and assess treatment strategies.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Zoe Swank, Harvard Medical School, Boston, Massachusetts, USA; Department of Pathology, Brigham and Women's Hospital, Boston, Massachusetts, USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA.
Yasmeen Senussi, Harvard Medical School, Boston, Massachusetts, USA; Department of Pathology, Brigham and Women's Hospital, Boston, Massachusetts, USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA.
Zachary Manickas-Hill, Ragon Institute of MGH, MIT and Harvard, Cambridge, Massachusetts, USA.
Xu G Yu, Harvard Medical School, Boston, Massachusetts, USA; Ragon Institute of MGH, MIT and Harvard, Cambridge, Massachusetts, USA; Division of Infectious Diseases, Brigham and Women's Hospital, Boston, Massachusetts, USA.
Jonathan Z Li, Harvard Medical School, Boston, Massachusetts, USA; Division of Infectious Diseases, Brigham and Women's Hospital, Boston, Massachusetts, USA.
Galit Alter, Ragon Institute of MGH, MIT and Harvard, Cambridge, Massachusetts, USA; Division of Infectious Diseases, Massachusetts General Hospital, Boston, Massachusetts, USA.
David R Walt, Harvard Medical School, Boston, Massachusetts, USA; Department of Pathology, Brigham and Women's Hospital, Boston, Massachusetts, USA; Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA.
Notes
Acknowledgments. This work was supported by a generous donation from Barbara and Amos Hostetter. We thank the MassCPR PASC consortium, the Ragon Institute/MassCPR Biobank team, and the Mass General Brigham Biobank for financial and sample processing support.
Financial support. G. A. reports support from NIH, Moderna, Inc., Gates Foundation, Pfizer, Sanofi, Janssen, Abbvie, Medicago, Inc., Gilead, and GlaxoSmithKline. D. W. reports support from Mass Consortium for Pathogen Readiness and Amos and Barbara Hostetter.
References
- 1. Nalbandian A, Sehgal K, Gupta A, et al. Post-acute COVID-19 syndrome. Nat Med 2021; 27:601–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization (WHO) . In the wake of the pandemic: preparing for Long COVID. 2021. Available at: https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/publications-and-technical-guidance/2021/in-the-wake-of-the-pandemic-preparing-for-long-covid-2021. Accessed 11 May 2022
- 3. Peluso MJ, Deeks SG. Early clues regarding the pathogenesis of long-COVID. Trends Immunol 2022; 43:268–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ganesh R, Grach SL, Ghosh AK, et al. The female-predominant persistent immune dysregulation of the post-COVID syndrome. Mayo Clin Proc 2022; 97:454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ogata AF, Maley AM, Wu C, et al. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in COVID-19 patients with severe disease. Clin Chem 2020; 66:1562–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ogata AF, Cheng CA, Desjardins M, et al. Circulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccine antigen detected in the plasma of mRNA-1273 vaccine recipients. Clin Infect Dis 2022; 74:715–8. 10.1093/cid/ciab465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chertow D, Stein S, Ramelli A, et al. SARS-CoV-2 infection and persistence throughout the human body and brain. Published online December 20, 2021. 10.21203/rs.3.rs-1139035/v1 [DOI] [Google Scholar]
- 8. Yonker LM, Gilboa T, Ogata AF, et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J Clin Invest. Published online May 2021; 131. 10.1172/JCI149633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Natarajan A, Zlitni S, Brooks EF, et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med 2022; 3:371–387.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Troyer Z, Alhusaini N, Tabler CO, et al. Extracellular vesicles carry SARS-CoV-2 spike protein and serve as decoys for neutralizing antibodies. J Extracell Vesicles 2021; 10:e12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng MH, Zhang S, Porritt RA, et al. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc Natl Acad Sci 2020; 117:25254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avolio E, Carrabba M, Milligan R, et al. The SARS-CoV-2 spike protein disrupts human cardiac pericytes function through CD147 receptor-mediated signalling: a potential non-infective mechanism of COVID-19 microvascular disease. Clin Sci 2021; 135:2667–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lei Y, Zhang J, Schiavon CR, et al. SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ Res 2021; 128:1323–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeOre BJ, Tran KA, Andrews AM, Ramirez SH, Galie PA. SARS-CoV-2 spike protein disrupts blood–brain barrier integrity via RhoA activation. J Neuroimmune Pharmacol 2021; 16:722–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peluso MJ, Lu S, Tang AF, et al. Markers of immune activation and inflammation in individuals with postacute sequelae of severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis 2021; 224:1839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phetsouphanh C, Darley DR, Wilson DB, et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat Immunol 2022; 23:210–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.