Abstract
Background
A recent study from Taiwan suggested that Clostridium innocuum may be an unrecognized cause of antibiotic-associated diarrhea (AAD) and clinically indistinguishable from Clostridioides difficile infection. Our objective was to compare C. innocuum prevalence and strain between those with AAD and asymptomatic controls.
Methods
In this cross-sectional study, we collected stool from 200 individuals with AAD and 100 asymptomatic controls. We evaluated the association between AAD and C. innocuum in stool using anaerobic culture and quantitative polymerase chain reaction (qPCR). To identify strain-specific associations with AAD, we performed whole-genome sequencing of C. innocuum isolates using Illumina MiSeq and constructed comparative genomics analyses.
Results
C. innocuum was isolated from stool of 126/300 (42%) subjects and more frequently from asymptomatic controls than AAD subjects (50/100 [50%] vs 76/200 [38%], respectively; P = .047). C. innocuum isolation frequency was not associated with AAD in either the adult or pediatric subgroups. C. innocuum and C. difficile were frequently co-prevalent in individuals with and without diarrhea. There were no phylogenetic differences or accessory genome associations between C. innocuum isolates from AAD subjects and asymptomatic controls.
Conclusions
C. innocuum was frequently isolated and at a greater frequency in asymptomatic controls than those with AAD. We did not identify strain lineages or accessory genomic elements associated with AAD. These data highlight that differentiating C. innocuum–associated diarrhea from asymptomatic colonization, and differentiating diarrhea caused by C. difficile from C. innocuum, are clinical microbiology challenges that require additional investigation to identify host-specific factors and/or biomarkers that distinguish these conditions.
Keywords: antibiotic-associated diarrhea, Clostridium innocuum, Clostridioides difficile, whole-genome sequencing, comparative genomics
In a cross-sectional study, we evaluated Clostridium innocuum as an emerging etiology of antibiotic-associated diarrhea. Using anaerobic culturing techniques and qPCR, we determined C. innocuum was frequently isolated at similar frequencies and with similar strain types irrespective of diarrheal symptoms.
Antibiotic-associated diarrhea (AAD) is a common complication of antibiotic use. Antibiotic treatment depletes the normal protective gut microbiota, resulting in an environment primed for invasive pathogenic species to colonize and proliferate. There are several known AAD etiologies. The most common cause of AAD is Clostridioides difficile, which is responsible for approximately 25% [1] of all AAD cases and is the most common healthcare-associated pathogen [2]. However, diagnosing C. difficile infection (CDI) as the cause of AAD is challenging because of the frequent use of highly sensitive nucleic acid amplification tests that do not adequately differentiate asymptomatic colonization from clinical infection [3]. Because asymptomatic colonization of C. difficile occurs in up to 20–30% of high-risk and/or hospitalized adult and pediatric populations, CDI may be frequently misdiagnosed and impair identification of other overlooked AAD etiologies [3].
Clostridium innocuum may be an emerging and overlooked cause of AAD. In a 2018 retrospective study in Taiwan, Chia et al. [4] isolated C. innocuum from stool from 5% of patients evaluated for CDI. These 103 adult and pediatric patients from whom C. innocuum was isolated were culture-negative for C. difficile despite exhibiting CDI-like symptoms, including severe and pseudomembranous colitis; the mortality rate was 14%. In addition, C. innocuum isolates were found to be multidrug resistant, including to vancomycin, cytotoxic to HT-29 cells, and pathogenic in a murine intestinal model of infection. Prior to this report, C. innocuum was generally considered clinically innocuous [5], only rarely causing opportunistic infections [6]. These novel observations were compelling and potentially paradigm-shifting for C. innocuum, and the findings raised several follow-up questions for further investigation [4]. For example, the prevalence of C. innocuum in individuals without diarrheal symptoms is unknown, and the genomic features of C. innocuum in patients with and without diarrhea have not been examined. Given the importance of these results, further investigation into C. innocuum as an etiology of AAD is needed.
The objective of this cross-sectional study was to characterize the clinical and molecular epidemiology of C. innocuum in a cohort of adults and children with and without AAD. Specifically, we aimed to measure the association between C. innocuum isolation from stool and AAD, and to compare strains associated with AAD and asymptomatic colonization through whole-genome sequencing. These findings further inform the clinical microbiology of C. innocuum and provide additional insight into the role of C. innocuum as an emerging diarrheal pathogen.
METHODS
Study Design, Setting, and Subjects
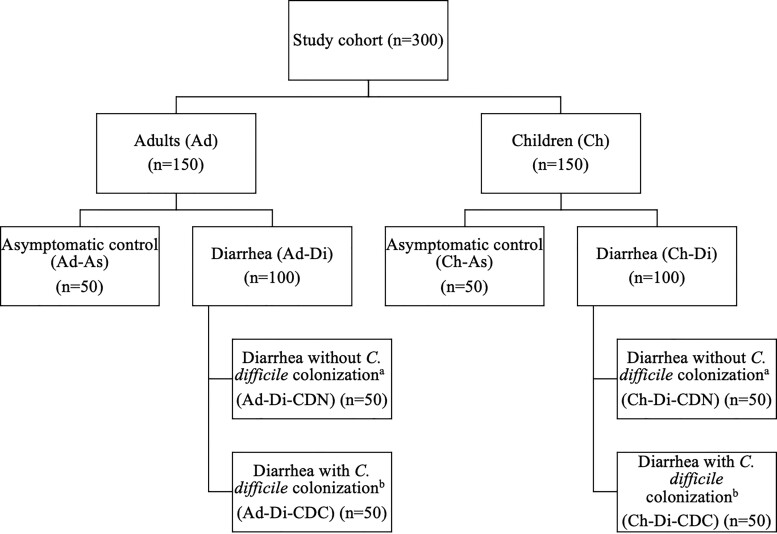
This cross-sectional study was performed in Chicago, Illinois, at the Ann & Robert H. Lurie Children’s Hospital of Chicago and Northwestern Memorial Hospital. The Institutional Review Boards from both Lurie Children’s and Northwestern University Feinberg School of Medicine approved this study. A cohort of 300 subjects (150 children age 2–18 years and 150 adults age >18 years) with and without diarrhea, and with and without C. difficile colonization, was assembled, as demonstrated in Figure 1.
Figure 1.
Flowchart of subjects included in this study. atcdB PCR negative, toxin EIA negative; btcdB PCR positive, toxin EIA negative. Abbreviations: EIA, enzyme immunoassay; PCR, polymerase chain reaction.
The clinical laboratories at each center saved specimens tested for C. difficile by tcdB polymerase chain reaction (PCR) (Xpert C. difficile; Cepheid, Sunnyvale, CA), irrespective of PCR result. Stools were refrigerated up to 7 days and then aliquoted and stored at −80°C until subject eligibility could be confirmed and the sample was ready for further processing. Patients with AAD were retrospectively identified through a series of steps by the research team. Antibiotic-associated diarrhea was defined as 3 or more unformed stools (ie, Bristol stool score of 5–7) in the preceding 24-hour period in those who had received systemic antibiotic therapy in the prior 30 days. To confirm eligibility, charts were manually reviewed for documentation of stool frequency and systemic antibiotic exposures within 24 hours and 30 days, respectively, preceding stool sample collection, as well as for laxative use and testing for other diarrheal pathogens. Subjects were excluded from the study if diarrhea was not clinically significant (ie, <3 stools in 24 hours), diarrhea was not antibiotic-associated (ie, no systemic antibiotic exposure in the previous 30 days), if the patient had another infectious etiology identified (ie, a positive clinical microbiology test for another pathogen), or had laxative use within the past 48 hours. For eligible subjects meeting the clinical definition of AAD, samples were thawed once and assigned a Bristol stool score by the research team; subjects whose Bristol stool score was 1–4 (ie, formed) upon visual inspection were excluded.
Unformed stools whose Bristol stool score was 5–7 and had tested positive by tcdB PCR were thawed once and underwent C. difficile toxin enzyme immunoassay (EIA; C. difficile QUIK CHEK Complete; Techlab, Blacksburg, VA) to differentiate toxigenic C. difficile colonization (tcdB PCR-positive, toxin EIA-negative) and CDI (tcdB PCR-positive, toxin EIA-positive); patients with CDI (tcdB PCR-positive, toxin EIA-positive) were excluded from the primary analysis but included in a post hoc analysis described below. An equal number of subjects with (tcdB PCR-positive, toxin EIA-negative) and without (tcdB PCR-negative) C. difficile colonization were included in the AAD group; subjects were randomly chosen until sample size goals were met. Hospitalized children (2–18 years) and adults (>18 years) who reported consistently formed stools over the previous 48 hours were eligible for prospective enrollment into the asymptomatic control group. Control-group subjects were chosen randomly among patients admitted to a non–intensive care inpatient unit and who were anticipated to remain hospitalized for at least 24–48 hours after consent to permit time for stool sample collection. Control-group subjects with unformed stools upon visual inspection (ie, Bristol stool score of 5–7) after enrollment were excluded. Adults and children enrolled into the AAD group developed AAD between July 2019 and October 2020 and between February 2018 and April 2021, respectively. Adults and pediatric controls provided stool samples between September 2020 and December 2020 and between June 2016 and July 2020, respectively.
To estimate the co-prevalence of toxigenic C. difficile and C. innocuum, we conducted 2 post hoc analyses. First, we performed C. difficile tcdB endpoint PCR on stools from the 100 asymptomatic control subjects. Next, we performed culture and quantitative PCR (qPCR) for C. innocuum on stools from subjects with AAD caused by C. difficile who were omitted from the primary analysis above (ie, tcdB PCR-positive, toxin EIA-positive).
Laboratory Methods
Stool samples from subjects underwent anaerobic culture and qPCR for C. innocuum. Clostridium innocuum identification was confirmed by matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF). Clostridium innocuum isolates underwent whole-genome sequencing using the Illumina MiSeq. C. difficile tcdB endpoint PCR was performed on genomic DNA extracted from stools from asymptomatic control subjects. Laboratory, whole-genome sequencing, and bioinformatics analyses are detailed in the Supplementary Materials.
Statistical Analysis
A sample size of 300 subjects (200 AAD and 100 controls) was selected based on a priori estimates of anticipated C. innocuum positivity reported previously [4]. Wilcoxon rank-sum test was used to compare continuous data between 2 groups, and Kruskal-Wallis test followed by Dunn’s multiple comparisons test were used to compare continuous data among multiple groups. Two-sided P values less than .05 were considered statistically significant. When performing multiple pairwise comparisons of proportions among 3 groups, a Bonferroni-corrected P value less than .025 was considered statistically significant. Statistical analyses were performed using Prism9 software version 9.0.2 (GraphPad Software, Inc, La Jolla, CA) and Stata/IC 16.0 (StataCorp, College Station, TX).
RESULTS
Three-hundred subjects (150 children and 150 adults) were included in this study, 200 with AAD and 100 asymptomatic controls (Figure 1). Among the 200 subjects with AAD, 100 were determined to be colonized but not infected with C. difficile (tcdB PCR-positive, toxin EIA-negative) and the remaining 100 subjects with AAD were determined to have neither C. difficile colonization nor CDI (tcdB PCR-negative). Table 1 lists the subject demographic and clinical characteristics of those with AAD and asymptomatic controls, and Supplementary Table 1 lists the same data stratified by each subgroup.
Table 1.
Demographic and Clinical Characteristics of Included Subjects in Antibiotic-Associated Diarrhea and Asymptomatic Control Groups
| Study Cohort (N = 300) | AAD Group (n = 200) | Asymptomatic Control Group (n = 100) | P | |
|---|---|---|---|---|
| Age, years | ||||
| ȃ Median (IQR) | 20 (9–59) | 20 (11–59) | 19 (6–59) | .1 |
| ȃ Range | 2–94 | 2–94 | 2–85 | |
| Sex (male) | 155 (51.7%) | 105 (52.5%) | 50 (50%) | .68 |
| Race/ethnicity | .22 | |||
| ȃ Asian | 11 (3.7%) | 6 (3%) | 5 (5%) | |
| ȃ American Indian/Alaska Native | 0 (0%) | 0 (0%) | 0 (0%) | |
| ȃ Black | 64 (21.3%) | 36 (18%) | 28 (28%) | |
| ȃ Native Hawaiian/other Pacific Islander | 0 (0%) | 0 (0%) | 0 (0%) | |
| ȃ White | 169 (56.3%) | 119 (59.5%) | 50 (50%) | |
| Unknown | 6 (2%) | 1 (1%) | 5 (2.5%) | |
| ȃ Other | 50 (16.7%) | 16 (16%) | 34 (17%) | |
| Ethnicity | .09 | |||
| ȃ Hispanic or Latino | 57 (19%) | 42 (21%) | 15 (15%) | |
| ȃ Not Hispanic or Latino | 237 (79%) | 85 (85%) | 152 (76%) | |
| ȃ Unknown | 6 (2%) | 0 (0%) | 6 (3%) | |
| Comorbidities | ||||
| ȃ Any comorbid condition | 273 (91%) | 186 (93%) | 87 (87%) | .09 |
| ȃ Malignancy/stem cell transplant | 95 (31 7%) | 74 (37%) | 21 (21%) | .005* |
| ȃ IBD | 6 (2%) | 6 (3%) | 0 (0%) | .08 |
| ȃ Non-IBD gastrointestinal condition | 34 (11.3%) | 30 (15%) | 4 (4%) | .005* |
| ȃ Solid-organ transplant | 32 (10.7%) | 25 (12.5%) | 7 (7%) | .15 |
| ȃ Other immunocompromising condition | 21 (7%) | 18 (9%) | 3 (3%) | .06 |
| ȃ Cardiovascular | 99 (33%) | 60 (30%) | 39 (39%) | .12 |
| ȃ Liver | 7 (2.3%) | 5 (2.5%) | 2 (2%) | .79 |
| ȃ Kidney | 32 (10.7%) | 22 (11%) | 10 (10%) | .79 |
| ȃ Pulmonary | 20 (6 7%) | 6 (3%) | 14 (14%) | <.001* |
| ȃ Endocrine/diabetes | 34 (11 3%) | 21 (10.5%) | 13 (13%) | .52 |
| Antibiotic exposure (past 30 days) | ||||
| ȃ Any systemic antibiotic | 263 (87.7%) | 200 (100%) | 63 (63%) | <.001* |
| ȃ Third-/fourth-generation cephalosporin | 103 (34.3%) | 78 (39%) | 25 (25%) | .02* |
| ȃ Clindamycin | 16 (5.3%) | 8 (4%) | 8 (8%) | .15 |
| ȃ Fluoroquinolone | 45 (15%) | 39 (19.5%) | 6 (6%) | .002* |
Data are presented as n (%) unless otherwise indicated. *P < .05 (P value compares AAD group and asymptomatic control group).
Abbreviations: AAD, antibiotic-associated diarrhea; IBD, inflammatory bowel disease; IQR, interquartile range.
Prevalence of Clostridium innocuum
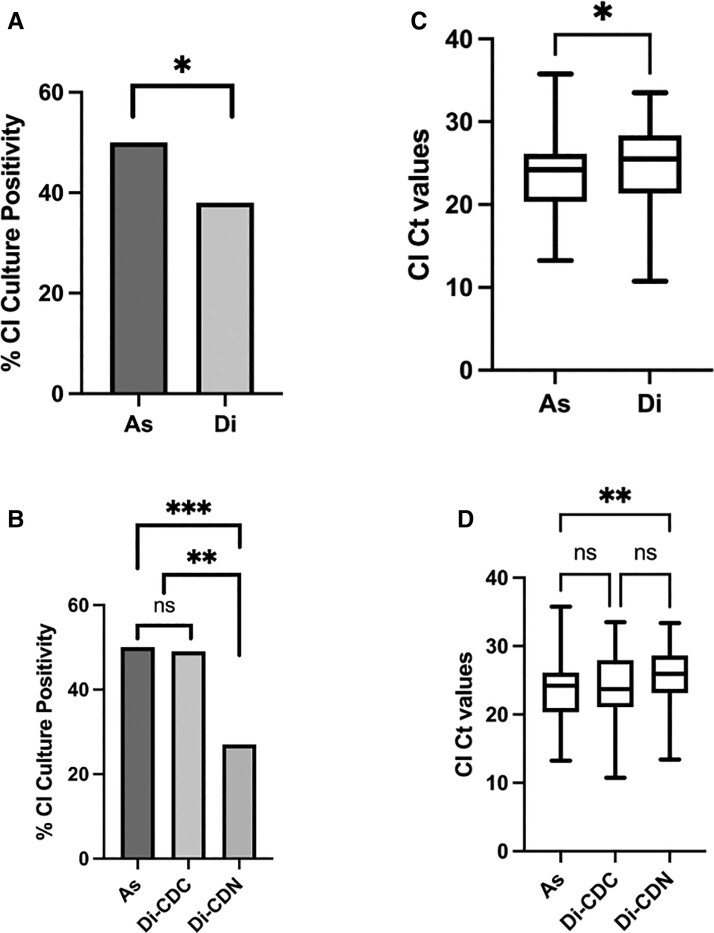
Clostridium innocuum was isolated by culture from stool from 126 of 300 (42%) subjects and more frequently among asymptomatic controls: 76 of 200 (38%) subjects with AAD and 50 of 100 (50%) asymptomatic controls (P = .047) (Figure 2A). By C. innocuum qPCR of the stool, median (interquartile range [IQR]) cycle threshold (Ct) values for subjects with AAD and asymptomatic controls were 25.5 (21.4–28.4) and 24.2 (20.4–26.1), respectively (P = .01) (Figure 2C). Among the subgroup of subjects with AAD who were with and without C. difficile colonization, C. innocuum was isolated less frequently from those without C. difficile colonization (49% vs 27%, P = .001) (Figure 2B); C. innocuum PCR Ct values did not significantly differ among those with or without C. difficile colonization (23.7 [21.1–28.0] vs 25.9 [23.2–28.6]; P = .06) (Figure 2D). All subjects with AAD, but only 63 (63%) asymptomatic controls, received antibiotics within the previous 30 days. Among asymptomatic controls with and without antibiotic exposure in the prior 30 days, C. innocuum stool culture positivity (44% vs 59%; P = .15) and median (IQR) C. innocuum PCR Ct values (24.1 [20.0–27.2] vs 24.3 [21.4–26.0]; P = .97) were similar between groups.
Figure 2.
Clostridium innocuum culture positivity rates and Ct values from subjects with AAD and asymptomatic controls. A, B, Percentage of CI culture positivity rates for a cohort of subjects with (A) diarrhea (Di) or asymptomatic controls (As), and (B) subjects with diarrhea who are either C. difficile colonized (Di-CDC) or C. difficile negative (Di-CDN). C, D, Box plots of CI Ct values as determined by qPCR of (C) As and Di subjects and (D) As, Di-CDC, and Di-CDN subjects. Boxes show the median and 25th and 75th percentiles. The whiskers represent the maximum and minimum data values. *P < .05 (or Bonferroni-corrected P < .25 for pairwise comparisons of multiple groups in panels B and D); **P < .01; ***P < .001. Abbreviations: AAD, antibiotic-associated diarrhea; CI, Clostridium innocuum; Ct, cycle threshold; ns, not significant; qPCR, quantitative polymerase chain reaction.
Stratified analyses of C. innocuum prevalence performed in adults (Supplementary Figure 1) and children (Supplementary Figure 2) were similar; C. innocuum was not isolated more frequently among those with AAD. Additional details of results from these adult and pediatric subgroup analyses are provided in the Supplementary Materials. To assess for confounding related to inclusion of subjects with cancer and gastrointestinal disease who often have multiple diarrheal risk factors, we measured C. innocuum stool positivity in subjects without underlying malignancy, stem cell transplant, or a gastrointestinal condition. The frequency of C. innocuum culture positivity in subjects with and without AAD was similar; C. innocuum was isolated by culture from stool from 56 of 138 (41%) subjects: 24 of 69 (35%) subjects with AAD and 32 of 69 (46%) asymptomatic controls (P = .17).
Post Hoc Analyses: Co-prevalence of Toxigenic C. difficile and C. innocuum
With the higher-than-expected prevalence of C. innocuum in study subjects, we performed several post hoc analyses. First, we performed tcdB endpoint PCR on stools from asymptomatic controls. In total, 19 of 100 (19%) asymptomatic controls were positive for C. difficile tcdB PCR. There was no correlation between C. difficile tcdB PCR positivity and C. innocuum culture positivity among asymptomatic controls (Cramer’s V = 0.03) (Supplementary Table 2). We additionally performed culture and qPCR for C. innocuum on stools from subjects with AAD caused by C. difficile who were omitted from the primary analysis above (ie, tcdB PCR-positive, toxin EIA-positive). Among 46 individuals (23 children, 23 adults) with AAD whose stool was positive for both C. difficile tcdB PCR and toxin EIA, 13 (28%) were positive for C. innocuum by culture, which was similar to the 27% C. innocuum culture positivity rate among those with AAD who were colonized but not infected with C. difficile (ie, tcdB PCR-positive, toxin EIA-negative individuals) reported above (P = .87). Among the 46 individuals with CDI, 6 of 23 (26%) children and 7 of 23 (30%) adults with CDI were positive for C. innocuum. Median (IQR) C. innocuum PCR Ct values in individuals with CDI were 19.8 (16.4–24.0), which was significantly lower than C. innocuum Ct values among those with AAD who were colonized but not infected with C. difficile reported above (P < .0001).
Phylogenetic and Comparative Genomics Analyses
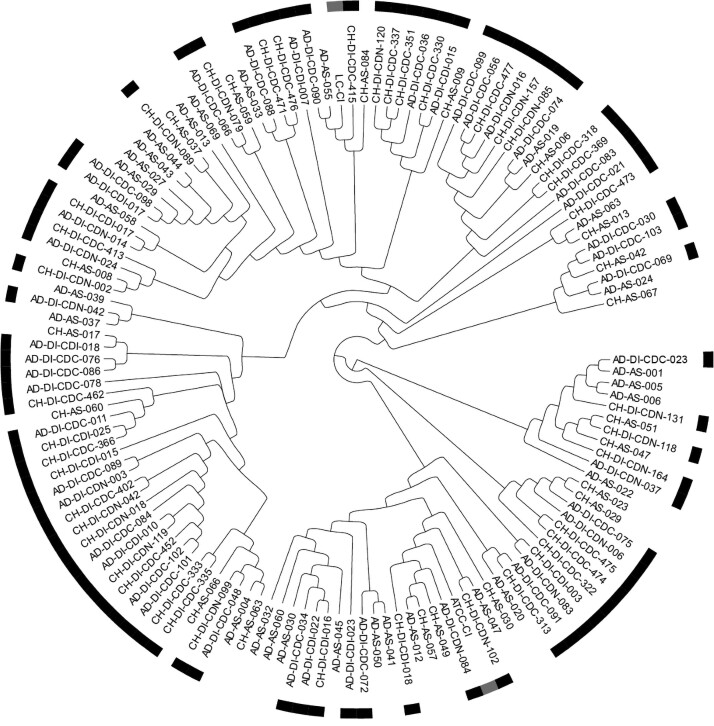
Of the 126 C. innocuum isolates, 119 (94%) were successfully sequenced. A cladogram was generated from the core genome alignment of the 2 reference sequences (ATCC [American Type Culture Collection] 14501 [7] and LC-CI [8]) and the 119 C. innocuum isolate sequences (Figure 3). Isolates from subjects with (n = 71) and without (n = 48) AAD were interspersed and coexistent in distinct tree lineages. Thus, we did not identify specific C. innocuum lineages associated with AAD. We performed a similar analysis stratified by age group (adults [n = 64] vs children [n = 55]) (Supplementary Figure 3). Similarly, we did not identify specific C. innocuum lineages associated with age group.
Figure 3.
Cladogram of C. innocuum isolates stratified by the presence of AAD showing 119 successfully sequenced isolates from adults (Ad) and children (Ch), 71 with AAD (Di), with and without C. difficile colonization (CDC and CDN, respectively), and 50 asymptomatic controls (As). Isolates from those with AAD are indicated by the black bars. The 2 reference strains, LC-CI [8] and CI-ATCC [7], are indicated by the gray bar bars. Abbreviations: AAD, antibiotic-associated diarrhea; ATCC, American Type Culture Collection; CI,Clostridium innocuum.
We also performed comparative genomic analyses of the 119 successfully sequenced C. innocuum isolates from our cohort. Among these 119 isolates, we delineated the core and accessory genomes. Within the accessory genomes of the 119 isolates, we identified 11 165 unique accessory genomic elements (AGEs). We compared the prevalence of each AGE between the AAD and asymptomatic control-group isolates. None of the AGEs were strongly correlated (Cramer’s V >0.8) with AAD; Cramer’s V of each AGE ranged from 0.001 to 0.41.
DISCUSSION
In a recent study by Chia et al [4] in Taiwan, C. innocuum was identified as a potential pathogen causing a CDI-like AAD illness. Subsequently, they identified C. innocuum as associated with high AAD mortality (15%) and more likely to be associated with extraintestinal infection compared with CDI [9]. In our cross-sectional study of children and adults, we aimed to evaluate the association between C. innocuum and AAD. We measured the prevalence of C. innocuum in stool of 300 adults and children with or without AAD. We hypothesized that, if C. innocuum was a prominent cause of AAD, we would isolate C. innocuum more frequently and/or more abundantly in patients with AAD. Instead, we isolated C. innocuum more frequently among asymptomatic controls than those with AAD, and stool C. innocuum bacterial loads were not greater among those with AAD. Upon subgroup analyses, AAD was not associated with C. innocuum isolation, irrespective of age group (ie, adults vs children) or C. difficile colonization status (ie, C. difficile PCR-negative vs C. difficile PCR-positive, toxin EIA-negative). Additionally, C. innocuum and toxigenic C. difficile were frequently co-prevalent in stool of children with and without diarrhea.
Our findings may suggest that there could be geographic differences in strain types and pathogenicity between the Taiwan cohort and our 2-center US cohort. However, our findings also highlight significant clinical microbiological challenges for discerning C. innocuum–associated diarrhea and C. innocuum colonization, similar to well-described challenges with CDI diagnosis [3]. Additionally, because C. innocuum and toxigenic C. difficile are often concomitantly present in stool of subjects with and without diarrhea, differentiating between these 2 pathogens and their relative contribution to diarrhea is also a significant challenge. For CDI diagnosis, clinical microbiology laboratories differentiate toxigenic from nontoxigenic C. difficile by PCR, EIA, and/or toxigenic stool culture [10]. However, at the present time, relative pathogenicity among C. innocuum strains is unknown and a potential C. innocuum toxin has not been identified. Although Chia et al [4] demonstrated C. innocuum pathogenicity in murine and cell culture models, they were unable to discern a pathogenetic mechanism for C. innocuum. In our prior study [11], we identified 2 proteins in C. innocuum supernatant that cross-react with commercial antibodies against C. difficile toxins A and B. While these proteins are ubiquitously present in genomes of all C. innocuum strains our laboratory has isolated, including those in this present study (data not shown), these proteins lacked toxin activity in a cell culture model. Because strain-specific differences in pathogenicity and clinical significance have been identified in other diarrheal pathogens, including C. difficile and Escherichia coli, we additionally performed comparative genomics analyses of our C. innocuum isolates. We did not identify strain lineages or AGEs associated with AAD. Thus, identification of the pathogenetic mechanism of C. innocuum–associated diarrhea requires further investigation. Identification of pathogenic and/or toxigenic strain types may inform clinical microbiology diagnostic strategies.
Our study has several limitations. As a study performed at 2 centers in the same city, our findings may not be externally generalizable. We used culturing methods previously optimized for isolation of C. difficile. It is possible that our culturing methods may not be optimized for C. innocuum. However, our culture yield was robust and more frequent than the prior study by Chia et al [4]. We included patients with AAD with and without C. difficile colonization, having defined colonization as a patient whose stool tested positive by C. difficile PCR and negative by toxin EIA. Although in prior studies [12, 13] this definition suggests C. difficile colonization, rather than CDI, this remains controversial [3, 14]. Nonetheless, our findings of a lack of association of C. innocuum with AAD did not change when only comparing controls with those without toxigenic C. difficile in stool. As a cross-sectional study, we did not assess outcomes of AAD. Given the frequent concomitant presence of C. difficile and C. innocuum in stool, comparison of diarrheal illness outcomes among patients with only 1 pathogen versus both pathogens is an important area of future investigation.
In summary, our findings confirm that C. innocuum is commonly present in the stool of adults and children with AAD, including AAD caused by C. difficile, but even more common in stool from asymptomatic adults and children. We did not identify strain lineages or AGEs associated with AAD. These findings highlight that differentiating C. innocuum–associated diarrhea from asymptomatic colonization, and differentiating C. innocuum–associated diarrhea from CDI, are important clinical microbiology challenges that require additional investigation. Identification of host-specific factors and/or biomarkers that distinguish these conditions will be essential to optimize evaluation and treatment for AAD.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The funders had no role in the study design, data collection, and interpretation, or the decision to submit the work for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by grants from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (grant number R21 AI144549 to L. K. K. and T32 916225 to K. E. C.). Research reported in this publication was supported, in part, by the National Institutes of Health’s National Center for Advancing Translational Sciences (grant number UL1TR001422). This research was also supported in part through the computational resources and staff contributions provided for the Quest high-performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
Supplementary Material
Contributor Information
Kathryn E Cherny, Northwestern University, Feinberg School of Medicine, Chicago, Illinois, USA.
Emily B Muscat, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, Illinois, USA.
Aakash Balaji, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, Illinois, USA.
Jayabrata Mukherjee, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, Illinois, USA.
Egon A Ozer, Northwestern University, Feinberg School of Medicine, Chicago, Illinois, USA.
Michael P Angarone, Northwestern University, Feinberg School of Medicine, Chicago, Illinois, USA.
Alan R Hauser, Northwestern University, Feinberg School of Medicine, Chicago, Illinois, USA.
Joseph S Sichel, Northwestern University, Feinberg School of Medicine, Chicago, Illinois, USA.
Emmanuel Amponsah, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, Illinois, USA.
Larry K Kociolek, Northwestern University, Feinberg School of Medicine, Chicago, Illinois, USA; Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, Illinois, USA.
References
- 1. Nasiri MJ, Goudarzi M, Hajikhani B, Ghazi M, Goudarzi H, Pouriran R. Clostridioides (Clostridium) difficile infection in hospitalized patients with antibiotic-associated diarrhea: a systematic review and meta-analysis. Anaerobe 2018; 50:32–7. [DOI] [PubMed] [Google Scholar]
- 2. Magill SS, Edwards JR, Bamberg W, et al. . Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kociolek LK. Strategies for optimizing the diagnostic predictive value of Clostridium difficile molecular diagnostics. J Clin Microbiol 2017; 55:1244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chia J-H, Wu T-S, Wu T-L, et al. . Clostridium innocuum is a vancomycin-resistant pathogen that may cause antibiotic-associated diarrhoea. Clin Microbiol Infect 2018; 24:1195–9. [DOI] [PubMed] [Google Scholar]
- 5. Smith LD, King E. Clostridium innocuum, sp. n., a sporeforming anaerobe isolated from human infections. J Bacteriol 1962; 83:938–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cherny KE, Muscat EB, Reyna ME, Kociolek LK. Clostridium innocuum: microbiological and clinical characteristics of a potential emerging pathogen. Anaerobe 2021; 71:102418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cherny KE, Ozer EA, Kochan TJ, Kociolek LK. Complete genome sequence of Clostridium innocuum strain ATCC 14501. Microbiol Resour Announc 2020; 9:e00452-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cherny KE, Ozer EA, Kochan TJ, Johnson S, Kociolek LK. Complete genome sequence of Clostridium innocuum strain LC-LUMC-CI-001, isolated from a patient with recurrent antibiotic-associated diarrhea. Microbiol Resour Announc 2020; 9:e00365-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y-C, Kuo Y-C, Chen M-C, et al. . Case-control study of Clostridium innocuum infection, Taiwan. Emerg Infect Dis 2022; 28:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burnham CA, Carroll KC. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev 2013; 26:604–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherny KE, Balaji A, Mukherjee J, et al. . Identification of Clostridium innocuum hypothetical protein that is cross-reactive with C. difficile anti-toxin antibodies. Anaerobe 2022; 75:102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Planche TD, Davies KA, Coen PG, et al. . Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis 2013; 13:936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Polage CR, Gyorke CE, Kennedy MA, et al. . Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 2015; 175:1792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McDonald LC, Gerding DN, Johnson S, et al. . Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66:e1–e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.