Abstract
Porphyromonas gingivalis, a gram-negative bacterium, has been linked to the onset and progression of periodontitis, a chronic inflammatory disease of the supporting tissues of the teeth. A major virulence factor of P. gingivalis is an extracellular complex of Arg- and Lys-specific proteinases and adhesins designated the RgpA-Kgp complex (formerly the PrtR-PrtK complex). In this study we show that the RgpA-Kgp complex, when used as an immunogen with incomplete Freund adjuvant (IFA), protects against challenge with invasive and noninvasive strains of P. gingivalis in the murine lesion model. We identified a variety of peptide vaccine candidates from the RgpA and Kgp polyprotein sequences that involved the putative active site histidine of both proteinases and five repeat motifs in the adhesin domains of both polyproteins implicated in aggregation and binding to host substrates, designated adhesin-binding motif (ABM) peptides. These peptides were synthesized using standard, solid-phase protocols for 9-fluorenylmethoxy carbonyl chemistry with S-acetylmercaptoacetic acid (SAMA) as the N-terminal residue. The SAMA-peptides were then conjugated to diphtheria toxoid and used with IFA to immunize BALB/c mice. Both active-site peptides and three of the five ABM peptides gave protection (P < 0.005) against challenge with P. gingivalis in the murine lesion model. The three ABM peptide sequences that conferred protection exist within a 100-residue span in the RgpA44 and Kgp39 adhesins of the RgpA-Kgp complex. Protective anti-RgpA-Kgp complex mouse antisera recognized the RgpA27, Kgp39, and RgpA44 adhesins in an immunoblot. Epitope mapping of the RgpA27 adhesin using the protective anti-RgpA-Kgp antisera identified a major protective epitope that mapped immediately N terminal to one of the protective ABM peptides in the 100-residue span in RgpA44 and Kgp39. This identified protective epitope contains clusters of basic residues spatially surrounded by hydrophobic amino acids, a finding which is characteristic of a heparin binding motif.
Periodontitis is a chronic inflammatory disease of the supporting tissues of the teeth that is associated with the emergence of a consortium of gram-negative bacteria in subgingival plaque. Epidemiological surveys have indicated that periodontitis affects between 10 to 15% of dentate adults (15). Of the 300 or more bacteria found in subgingival plaque, three species, Porphyromonas gingivalis, Bacteroides forsythus, and Treponema denticola, have been strongly associated with disease (56). P. gingivalis has received considerable interest since it can represent up to 50% of the anaerobically cultivable bacteria from adult periodontal lesions (9, 52). An increase in the severity of periodontitis has also been associated with an increase in the subgingival level of P. gingivalis (34, 55), and resolution of the disease has been associated with a reduction in level of the bacterium (32, 61). Furthermore, subgingival implantation of P. gingivalis in mice, rats, and nonhuman primates has been reported to induce periodontal bone loss (1, 12, 19, 23). These studies in humans and animals implicate P. gingivalis as a major etiological agent in the development of periodontitis.
Several studies have reported that immunization with formalin- or heat-killed P. gingivalis cells, cell extracts, or outer membrane preparations can protect or minimize tissue destruction caused by a P. gingivalis challenge in the murine lesion model (3, 7, 8, 25). Furthermore, P. gingivalis-killed whole cells and sonicated cell surface extracts have been reported to reduce bone loss in the rat and nonhuman primate models of periodontitis (12, 43). Purified fimbrial protein from P. gingivalis strains ATCC 33277 and 381 have also been reported to confer protection in the murine lesion model and the rat periodontitis model (12, 40). Ogawa et al. (40) have reported that peptide immunogens derived from the fimbriae of P. gingivalis 381 reduced the lesion size by up to 88%. Furthermore, other fimbrial peptide epitopes from strain ATCC 33277 when used as a vaccine were reported to provide protection to 60% of mice against lethal P. gingivalis challenge (11).
P. gingivalis strains can be classified as invasive (e.g., strains W50 and A7A1-28) and noninvasive (e.g., strains ATCC 33277 and 381) based on their ability to cause ulcerative spreading lesions distant from the injection site or a localized abscess at the site of injection site, respectively (36, 62). P. gingivalis strains ATCC 33277 and 381 are fimbriated, whereas strain W50 is reported to be sparsely fimbriated with shorter fimbriae that have no hemagglutinating activity (57). Noninvasive and invasive P. gingivalis strains both induce periodontal bone loss in rats (12), although the comparative virulence of the different strains in the animal periodontitis models needs further elucidation. Virulence factors common to invasive and noninvasive strains of P. gingivalis are the Arg- and Lys-specific proteinases and their associated adhesins. A vaccine targeting this virulence factor may therefore provide protection against both invasive and noninvasive strains of P. gingivalis.
The extracellular Arg- and Lys-specific cysteine proteinases are believed to play a major role in the pathogenesis of periodontal disease (1, 26, 35, 53). Spontaneous P. gingivalis mutants with reduced Arg and Lys proteinase activity and wild-type cells treated with a protease inhibitor (TLCK [N-α-p-tosyl-l-lysine chloromethyl ketone]) have been reported to be avirulent in animal models (26). Furthermore, the Arg- and Lys-specific proteinases have been shown to degrade a variety of host proteins in vitro, e.g., fibrinogen, fibronectin, and laminin (30, 31, 44, 48, 60). Plasma host defense and regulatory proteinase inhibitors α-antitrypsin, α2-macroglobulin, antichymotrypsin, antithrombin III, and antiplasmin are also degraded by the P. gingivalis Arg and Lys proteinases (6). Inflammation at the site of infection may be enhanced by the Arg- and Lys-specific proteinases since the complement protein C5 has been reported to be hydrolyzed in vitro, releasing the biologically active C5a fragment (58). Cell lysis mediated by complement, however, may be avoided since the proteinases have been shown to degrade the other complement proteins (58, 65). Phagocytic and other functions of recruited neutrophils to the inflamed site may also be impaired, since the proteinases are capable of degrading cell surface receptors (20, 22). The host immune response may be further dysregulated by the proteinases since they can degrade immunoglobulin G (IgG), IgA, IgM, IgD, and IgE antibodies (14, 17).
These data collectively imply that the Arg- and Lys-specific proteinases of P. gingivalis are major virulence factors. We have previously characterized a cell-associated complex of noncovalently associated proteinases and adhesins purified from P. gingivalis W50 designated the RgpA-Kgp proteinase-adhesin complex, formerly the PrtR-PrtK proteinase-adhesin complex (2). This complex is composed of a 45-kDa Arg-specific, calcium-stabilized cysteine proteinase (RgpA45, formerly PrtR45); a 48-kDa Lys-specific cysteine proteinase (Kgp48, formerly PrtK48); and seven sequence-related adhesins designated RgpA44, RgpA15, RgpA17, RgpA27, Kgp39, Kgp15, and Kgp44 (formerly PrtR44, PrtR15, PrtR17, PrtR27, PrtK39, PrtK15, and PrtK44, respectively), encoded by the two genes rgpA (42) and kgp (41) (formerly prtR [50] and prtK [51] from P. gingivalis W50).
We report here the ability of the purified RgpA-Kgp proteinase-adhesin complex and peptides derived from the proteins within the complex to protect against P. gingivalis challenge in the murine lesion model.
MATERIALS AND METHODS
Synthesis of peptides derived from the RgpA-Kgp proteinase-adhesin complexes.
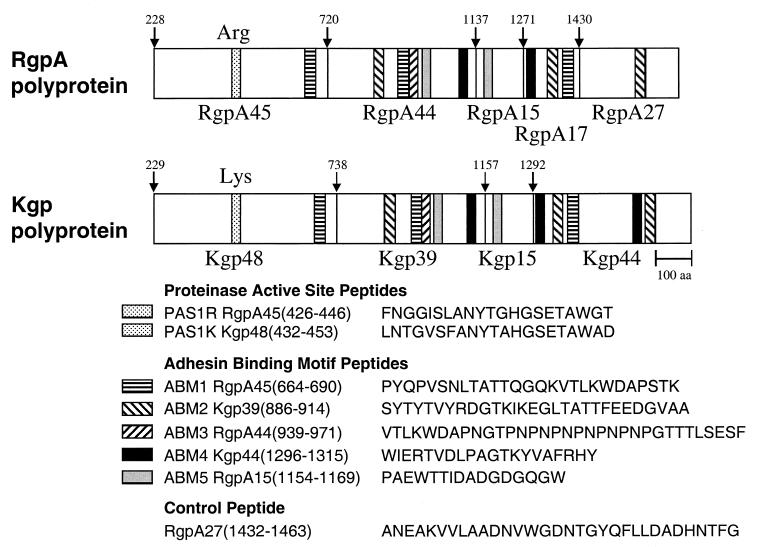
Analysis of the protein sequence of RgpA and Kgp polyproteins revealed that five sequences were repeated, with substitutions, throughout both the RgpA and Kgp adhesins (Fig. 1). Based on sequence similarity with known adhesins (50), as well as recent data showing involvement in aggregation and binding to host substrates (51; unpublished data), we have designated these repeat sequences adhesin-binding motifs (ABM) (Fig. 1). The putative active-site histidines of both the Arg- and Lys-specific proteinases have been identified previously (51). Peptides corresponding to the putative ABM sequences and the putative active sites of RgpA45 and Kgp48 (proteinase active site [PAS]) and a control peptide, ANE (occurring once in the RgpA and Kgp polyproteins), were selected for synthesis (Fig. 1).
FIG. 1.
Schematic representation of the RgpA and Kgp polyproteins showing the location and amino acid sequence of the synthesized peptides. Amino acid numbering is from the initial Met of the polyprotein. The downward arrows indicate the processing site for the proteinase and adhesin domains. The numbers above the arrows indicate the N-terminal residues of that domain. Similarly hatched areas at other locations correspond to repeat sequences (sometimes with substitutions) of the same motif.
Peptides were synthesized manually or using a 431A ABI peptide synthesizer. Standard solid-phase peptide synthesis protocols for Fmoc (9-fluorenylmethoxy carbonyl) chemistry were used throughout. Peptides were assembled as the carboxyamide form using Fmoc-Pal-Peg-PS resin (PerSeptive Biosystems, Inc., Framingham, Mass.). Coupling was accomplished with O-benzotriazole-N,N,N′,N′,-tetramethyl-uronium-hexafluorophosphate (HBTu) and N-hydroxybenzotriazole (HoBt) activation using 4 eq of Fmoc-amino acid and 6 eq of DIPEA. The Fmoc group was removed by 2% (vol/vol) DBU in DMF containing 2% (vol/vol) piperidine. Cleavage of peptides from the resin support was performed using trifluoroacetic acid (TFA)-phenol-triisopropylsilane (TIPS)-ethanedithiol (EDT)-water (92:2:2:2:2) cleavage cocktail for 2.5 or 4 h depending on the arginine content of the peptide. After cleavage the resin was removed by filtration and the filtrate was concentrated to approximately 1 ml under a stream of nitrogen. After the peptide products were precipitated in cold ether, they were centrifuged and washed three times. The peptide precipitates were dissolved in 5 to 10 ml of water containing 0.1% (vol/vol) TFA, and the insoluble residue was removed by centrifugation. Peptides were purified by reversed-phase high-pressure liquid chromatography as previously described (38).
Synthesis of SAMA peptides.
Resins bearing peptides were swollen in DMF, and the N-terminal Fmoc group removed by using 2% (vol/vol) DBU in DMF containing 2% (vol/vol) piperidine. The S-acetylmercaptoacetic acid (SAMA) group was coupled onto the N terminus by using 5 eq of SAMA-pentafluorophenylester (OPfp) and 5 eq of HOBt. The reaction was monitored by the trinitrobenzene sulfonic acid (TNBSA) test. Following a negative TNBSA test, the resin was washed with 5× DMF, 3× dichloromethane, and 3× diethyl ether. The resin was then dried under a vacuum, and the SAMA-peptides were cleaved from the resin support as described above.
Conjugation of SAMA-peptides to DT.
Diphtheria toxoid (DT) was kindly provided by I. Barr (CSL Pty., Ltd., Melbourne, Victoria, Australia) which contained nine equivalent amino groups per 62-kDa molecule. To a solution containing 10 mg of DT in phosphate-buffered saline (PBS; 0.1 M sodium phosphate, 0.9% NaCl; pH 7.4) per ml was added 0.1 ml of a 1% (wt/vol) solution of m-maleimido benzoyl-N-hydroxysuccinimide ester (MBS) in DMF. After 30 min the unreacted MBS was removed and MBS-modified DT was collected by gel filtration using a PD10 column (Pharmacia, Sydney, New South Wales, Australia) equilibrated in conjugation buffer (0.1 M sodium phosphate, 5 mM EDTA; pH 6.0). Purified SAMA-peptide (1.3 μmol) was dissolved in 200 μl of 6 M guanidine HCl containing 0.5 M Tris–2 mM EDTA (pH 6.0) and diluted with 800 μl of MilliQ water and then deprotected in situ by the addition of 25 μl of 2 M NH2OH (40 eq) dissolved in MilliQ water. The collected MBS-DT was immediately reacted with deprotected SAMA-peptide and stirred for 1 h at room temperature. The peptide-DT conjugate was separated from unreacted peptide by gel filtration using a PD10 column equilibrated in PBS (pH 7.4) and lyophilized. The reaction was monitored using the Ellman test (47). The conjugation yields of SAMA-peptides to MBS-DT ranged from 34 to 45%, indicating that three to four peptides were coupled per DT molecule.
Purification of the RgpA-Kgp proteinase-adhesin complex.
The growth of P. gingivalis W50 and the isolation and purification of the RgpA-Kgp proteinase-adhesin complex were done as previously described (2, 51).
Immunization and murine lesion model protocols.
BALB/c mice (6 to 8 weeks old) were immunized subcutaneously (s.c.) with either 50 μg of the peptide-DT conjugate, 50 μg of DT, 50 μg of the RgpA-Kgp proteinase-adhesin complex, 2 × 108 formalin-killed cells of P. gingivalis ATCC 33277 or W50, or 2 × 108 formalin-killed E. coli cells emulsified in incomplete Freund adjuvant (IFA). After 30 days the mice were injected s.c. with antigen emulsified in IFA and then bled from the retrobulbar plexus 12 days later. Mice were challenged with either 8 × 109 cells of P. gingivalis ATCC 33277 or 3 × 109 cells of P. gingivalis W50 (s.c.) in the abdomen, and the lesion sizes were measured over 14 days. Lesion sizes were statistically analyzed using the Kruskal-Wallis test and the Mann-Whitney U-Wilcoxon rank sum test with a Bonferroni correction (37).
ELISA.
Enzyme-linked immunosorbent assays (ELISAs) were performed in triplicate using a solution (1 μg/ml) of the RgpA-Kgp complex or a solution (5 μg/ml) of peptide antigen in 0.1 M PBS (pH 7.4) containing 0.1% (vol/vol) Tween 20 (PBST) and 0.1% (wt/vol) sodium azide to the coat wells of flat-bottom polyvinyl microtiter plates (Microtiter, Dynatech Laboratories, McLean, Va.) overnight at 4°C. After removal of the coating solution, 2% (wt/vol) skim milk powder in PBST was added to block the remaining uncoated plastic for 1 h at room temperature. After a washing (with 4× PBST), serial dilutions of antisera in PBST containing 1% (wt/vol) skim milk powder were added to each well and incubated for 3 h at 37°C. Bound antibody was detected, after washing with 6× PBST, by incubation with horseradish peroxidase-conjugated goat immunoglobulin directed against mouse immunoglobulin (1/4,000 dilution) (Sigma Chemical Co., Sydney, New South Wales, Australia) for 1.5 h at 37°C. The plates were then washed (6× PBST), and substrate [0.9 mM 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) in 80 mM citric acid (pH 4.0) buffer containing 0.005% (vol/vol) H2O2] was added. The optical density at 414 nm (OD414) was measured by using a Labsystems iEMS MF Ascent microplate reader (Pathtech).
To determine the IgG subclass antibody responses of mouse sera, microtiter plates were coated with the peptide antigen and incubated with peptide antisera as described above. After a washing (6× PBST), goat antisera to mouse IgM, IgG1, IgG2a, IgG2b, or IgG3 (Sigma) were added and allowed to bind for 3 h. Plates were again washed and a 1/4,000 dilution of horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin was added. After a washing in 6× PBST, the plates were developed as described above.
Immunoblotting.
Purified RgpA-Kgp complex was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE in 12.5% acrylamide gels (1 mm) by using the method of Laemmli (28) with a minigel system (Bio-Rad). Proteins were electrophoretically transferred onto polyvinylidene difluoride (PVDF) membrane by the method of Dashper et al. (10). After sectioning of the membrane, the molecular weight standards were stained with 0.1% (wt/vol) CBB R250. The remaining sections were blocked for 1 h at 20°C with 5% (wt/vol) nonfat skim milk powder in TN buffer (50 mM Tris-HCl, pH 7.4; 100 mM NaCl). Sections were subsequently incubated with mouse sera diluted 1:25 with TN buffer. After 5 h at 20°C the sections were washed (4× TN buffer containing 0.05% [vol/vol] Tween 20) and then incubated for an hour at 20°C with anti-mouse IgG horseradish peroxidase conjugate (Bio-Rad). After a washing with 4× TN buffer containing 0.05% (vol/vol) Tween 20, bound antibody was detected with 0.05% (wt/vol) 4-chloro-1-napthol in TN buffer containing 16.6% (vol/vol) methanol and 0.015% H2O2. Color development was stopped by rinsing the membranes with MilliQ water.
Epitope mapping analysis.
Epitope mapping of the first 148 residues of the N terminus of the RgpA27 adhesin of the RgpA-Kgp complex (2) was accomplished using 21 overlapping 13mer peptides (overlay by six and offset by seven residues). Peptides were synthesized by Chiron Technologies (Melbourne, Victoria, Australia) using the multipin peptide synthesis system. Epitope mapping of the pin-bound peptides was carried out by ELISA according to the instructions of the manufacturer using mouse sera at a dilution of 1:1,000 in 1% (wt/vol) nonfat skim milk powder in 0.1 M PBS (pH 7.4) containing 0.1% (vol/vol) Tween 20. The bound antibody was detected by incubating the pins with 0.9 mM 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) in 80 mM citric acid (pH 4.0) buffer containing 0.005% (vol/vol) H2O2. The OD414 was measured using a Labsystems iEMS MF Ascent microplate reader (Pathtech).
RESULTS
Immunization with the RgpA-Kgp proteinase-adhesin complex.
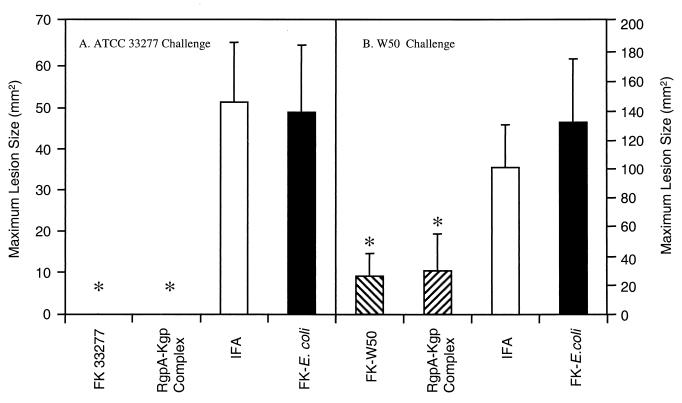
BALB/c mice were immunized with formalin-killed P. gingivalis strain ATCC 33277 or strain W50 (2 × 108 cells), formalin-killed E. coli cells (2 × 108), the RgpA-Kgp proteinase-adhesin complex (50 μg), or adjuvant alone (IFA). Formalin-killed E. coli cells were used as a gram-negative bacterial control. Twelve days after the second immunization mice were challenged s.c. with either P. gingivalis strain ATCC 33277 (7.5 × 109 viable cells) or strain W50 (3 × 109 viable cells). The mice were monitored for the next 14 days for the development of lesions. The maximum lesion size attained for each group is presented in Fig. 2. All of the mice immunized with adjuvant or formalin-killed E. coli cells developed lesions (reaching a maximum size by day 3 that receded over the next 11 days) when challenged with either P. gingivalis ATCC 33277 (Fig. 2A) or W50 (Fig. 2B), respectively. Animals challenged with strain W50 developed large, ulcerative lesions, whereas the challenge with strain ATCC 33277, at twice the inoculation dose, produced smaller localized abscesses. Mice immunized with either formalin-killed cells of strain ATCC 33277 or the RgpA-Kgp complex did not develop lesions when challenged with P. gingivalis strain ATCC 33277 (Fig. 2). Although all of the mice immunized with formalin-killed cells of strain W50 and the RgpA-Kgp complex developed lesions when challenged with W50, the lesions were significantly (P < 0.05) smaller than those of the control groups (Fig. 2).
FIG. 2.
Maximum lesion size of mice challenged with either P. gingivalis ATCC 33277 (A) or W50 (B). BALB/c mice were immunized s.c. with the RgpA-Kgp proteinase complex (50 μg), formalin-killed (FK) P. gingivalis cells (2 × 108 of either strain ATCC 33277 or strain W50), formalin-killed E. coli cells (2 × 108), or PBS administered in IFA for both the primary and secondary doses. All mice were challenged 12 days after the secondary immunization with either P. gingivalis strain ATCC 33277 (7.5 × 109 viable cells) or W50 (3 × 109 viable cells) and were weighed, and the lesion sizes were measured daily over 14 days. Lesion sizes were statistically analyzed using Mann-Whitney U-Wilcoxon rank sum test. ∗, Groups significantly different (P < 0.005) from the formalin-killed E. coli control group.
Immunization with peptide-DT conjugates.
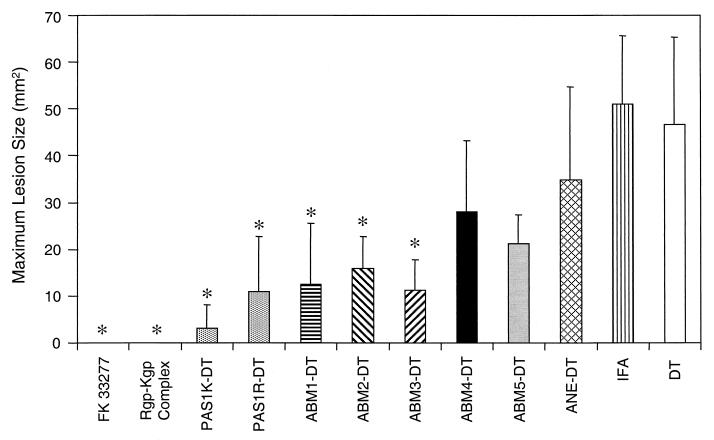
The amino acid sequence and location within the RgpA and the Kgp polyproteins of the synthesized PAS and ABM peptides are shown in Fig. 1. PAS and ABM peptide-DT conjugates were used to immunize BALB/c mice that were challenged with P. gingivalis ATCC 33277 or W50 12 days after the second immunization. Figure 3 shows the maximum lesion size for each immunized group challenged with strain ATCC 33277. Animals that were immunized with either formalin-killed P. gingivalis cells or the RgpA-Kgp complex (positive-control groups) did not develop lesions, whereas animals immunized with DT or the control peptide (ANE) conjugated to DT developed lesions that were not significantly smaller than those of the IFA negative control group (Fig. 3). However, animals immunized with ABM1, ABM2, ABM3, PAS1K, or PAS1R peptide-DT conjugates had significantly (P < 0.005) smaller lesions than that of the DT group. All of the animals immunized with ABM2, ABM4, and ABM5 developed lesions, whereas the percentages of animals immunized with either PAS1K, PAS1R, ABM1, or ABM3 which did not develop lesions were 70, 50, 40, and 30%, respectively. Animals immunized with the ABM1, ABM2, ABM3, PAS1K, and PAS1R peptide-DT conjugates and challenged with P. gingivalis W50 were also found to have significantly (P < 0.05) smaller lesions than animals immunized with DT alone (data not shown).
FIG. 3.
Maximum lesion size of mice immunized with peptide-DT conjugates. BALB/c mice were immunized s.c. with the peptide-DT conjugates (50 μg) administered in IFA for the primary and secondary doses and challenged s.c. 12 days after the second dose with P. gingivalis ATCC 33277 (7.5 × 109 viable cells). Animals were monitored over a 14-day period for weight loss and lesion size. Data were analyzed using the Mann-Whitney U-Wilcoxon rank sum test. ∗, Groups significantly different (P < 0.005) to the DT control group. FK, formalin killed.
Antibody specificity and antibody subclasses induced by the peptide-DT conjugates.
Table 1 shows the total IgG antibody and IgG subclass titers as measured by ELISA that were induced by the peptide-DT conjugates. Each peptide monomer was adsorbed on to microtiter plates and probed with peptide-DT conjugate antisera. All of the ABM peptide-DT sera induced a specific IgG response that only recognized the respective ABM peptide; however, antibodies generated toward PAS1K were also able to weakly recognize PAS1R and vice versa (data not shown). All of the peptide-DT conjugates induced a strong IgG1 response and weaker IgG3 and IgM responses (Table 1). PAS1K and the RgpA-Kgp complex also induced a weak IgG2a and IgG2b response, and ABM1 and ABM3 also induced a weak IgG2b response. Although all of the peptide-DT conjugate sera recognized the RgpA-Kgp complex, the antibody titers against the complex could be grouped into a strong response (>30,000 ELISA antibody titer) for peptide-DT conjugates that gave protection (PAS1K, PAS1R, ABM1, ABM2, and ABM3) and a weak response (<30,000 ELISA antibody titer) for the nonprotective peptide-DT conjugates (Table 1).
TABLE 1.
ELISA analysis of sera from mice immunized with peptide-DT conjugates and RgpA-Kgp complex
| Immunogenb | Mean ELISA antibody titera (103) ± SD
|
||||||
|---|---|---|---|---|---|---|---|
| Total IgGc | IgG1c | IgG2ac | IgG2bc | IgG3c | IgMc | RgpA-Kgp complexd | |
| PAS1K | 320 ± 7.1 | 256 ± 1.4 | 1.6 ± 0.2 | 0.8 ± 0.1 | 6.4 ± 0.2 | 0.6 ± 0.1 | 100 ± 1.6 |
| PAS1R | 384 ± 13.8 | 320 ± 38.4 | ND | ND | 2.8 ± 0.2 | 0.8 ± 0.1 | 44.7 ± 5.3 |
| ABM1 | 384 ± 1.5 | 320 ± 26.9 | ND | 1.2 ± 0.1 | 2.4 ± 0.1 | 1.0 ± 0.1 | 56.2 ± 7.1 |
| ABM2 | 256 ± 4.1 | 224 ± 20.6 | ND | ND | 1.2 ± 0.1 | 0.4 ± 0.1 | 33.1 ± 2.3 |
| ABM3 | 256 ± 17.4 | 256 ± 30.7 | ND | 1.2 ± 0.1 | 2.4 ± 0.1 | 0.4 ± 0.1 | 44.7 ± 5.4 |
| ABM4 | 512 ± 4.1 | 512 ± 8.1 | ND | ND | 6.4 ± 0.1 | 1.6 ± 0.1 | 12.6 ± 1.4 |
| ABM5 | 384 ± 3.1 | 384 ± 9.2 | ND | ND | 9.6 ± 1.9 | 1.2 ± 0.1 | 5.6 ± 0.6 |
| ANE | 384 ± 1.6 | 256 ± 3.1 | ND | ND | 0.4 ± 0.1 | 0.4 ± 0.1 | 15.8 ± 1.7 |
| RgpA-Kgp complex | 512 ± 18.4 | 512 ± 15.3 | 1.2 ± 0.3 | 1.2 ± 0.1 | 4.8 ± 0.2 | 0.8 ± 0.1 | 512 ± 18.4 |
Antibody titers are expressed as the dilution factor determined from sigmoidal dilution curves which produced an OD414 value fivefold greater than the background level in the ELISA. Each titer represents the mean ± the standard deviation of three values from sera from each animal in the immunogen group. ND, not detected.
All peptide epitopes were conjugated to DT, and 50 μg of the peptide-DT conjugate emulsified in IFA was used to immunize BALB/c mice.
Antibody isotype titer of antipeptide sera binding to the respective peptide monomer adsorbed (1 μg/ml) onto microtiter plates.
Antibody titer of antipeptide sera binding to the RgpA-Kgp complex adsorbed (1 μg/ml) onto microtiter plates.
Immunoblot analysis of the RgpA-Kgp complex.
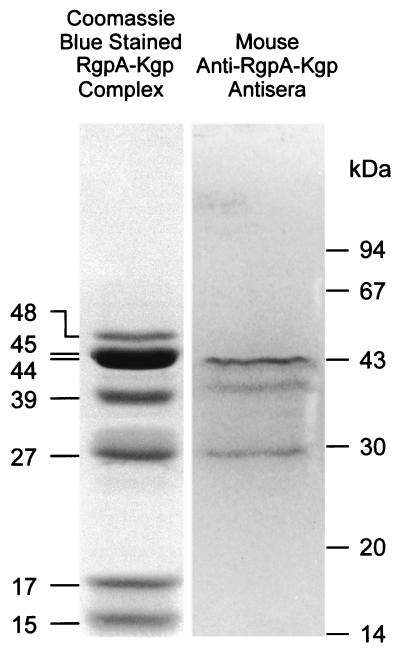
Immunoblot analysis results with the RgpA-Kgp proteinase complex using sera from mice immunized with the RgpA-Kgp proteinase complex are shown in Fig. 4. The anti-Rgp-Kgp complex mouse sera had an immunoreactive response to the 44-, 39-, and 27-kDa protein bands. These protein bands correspond to the RgpA44 or Kgp44, the Kgp39, and the RgpA27 adhesin proteins, respectively, from the complex (2). Antibodies directed to the Lys-specific proteinase (Kgp48), the Arg-specific proteinase (RgpA45), and the adhesins RgpA17, RgpA15, or Kgp15 were not detected with the anti-RgpA-Kgp complex sera.
FIG. 4.
Immunoblot analysis of the RgpA-Kgp complex using murine anti-RgpA-Kgp complex sera. The RgpA-Kgp complex was separated by SDS-PAGE and transferred onto PVDF membrane. The RgpA-Kgp complex was probed with sera (1:50 TN buffer) from mice immunized with the RgpA-Kgp proteinase complex. Molecular mass markers are indicated in kilodaltons.
Epitope mapping of the RgpA27 adhesin.
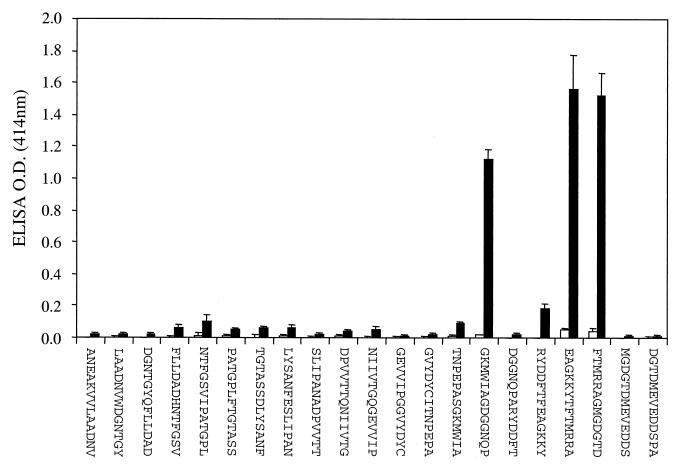
Analysis of the protein sequence of RgpA27 found that the N-terminal 148 residues are almost identical to the N-terminal 148 residues of the Kgp39 adhesin in the Kgp polyprotein. The C-terminal regions of RgpA27 and Kgp39 are also similar, but these sequences also share similarity with other adhesin proteins of the RgpA-Kgp complex that were not recognized by the protective antisera in the immunoblot. Therefore, 21 overlapping 13mer peptides representing the N-terminal 148 residues of RgpA27 were synthesized (offset, 7; overlap, 6) on PINS (Chiron Technologies). The peptide-PINS were probed with normal mouse sera and with sera from mice immunized with the RgpA-Kgp complex (Fig. 5). Two immunoreactive peptide epitopes were detected by all of the sera from mice immunized with the complex: 1542RYDDFTFEAGKKYTFTMRRAGMGDGTD1568 (EP1) and 1521TNPEPASGKMWIAGDGGNQP1540 (EP2).
FIG. 5.
Epitope mapping analysis of the RgpA27 adhesin protein of the RgpA-Kgp complex. Twenty-one overlapping 13mer peptides (overlay by six and offset by seven residues) were synthesized covering the N-terminal 148 residues of the RgpA27 by Chiron Technologies using the multipin peptide synthesis system. Epitope mapping of the pin-bound peptides was carried out by ELISA according to Chiron Technologies instructions. Pin-bound peptides were probed with sera from mice immunized with the RgpA-Kgp proteinase complex (■, n = 10) and normal mouse sera (□, n = 10).
DISCUSSION
The results presented here demonstrate that immunization of mice with the RgpA-Kgp proteinase-adhesin complex from P. gingivalis W50 protected against challenge with an invasive P. gingivalis strain (W50) and a noninvasive strain (ATCC 33277) in the murine lesion model. The fimbrial subunit FimA from P. gingivalis 381 (which is similar to strain ATCC 33277) has also been reported to protect rats and mice when challenged with the same strain (12, 40). However, the fimA gene is inactive in the virulent W50 strain, and thus the protein is not expressed (57); consequently, immunization with the FimA protein would not provide protection against this and other strains of P. gingivalis that do not express FimA.
Peptide sequences corresponding to the active site histidine of the RgpA and Kgp proteinases, as well as those repeated in the adhesin domains of RgpA and Kgp implicated in aggregation and host substrate binding, designated ABM peptides, were synthesized and conjugated to DT and used as vaccines in the murine lesion model. All of the peptide-DT conjugates induced a strong antibody response, and five of the eight peptide-DT conjugates protected mice against challenge with P. gingivalis. Although all the peptide-DT conjugates induced a strong peptide-specific antibody response, the ability to cross-react with the RgpA-Kgp complex varied. Anti-PAS1K antisera had the highest ELISA titer to the complex, and this peptide immunogen was the most protective, with 70% of the mice not developing lesions. The antibodies generated to other protective peptides (PAS1R and ABM1, -2, and -3) also exhibited strong cross-reactivity with the RgpA-Kgp complex. Peptides ABM4, ABM5, and ANE, which did not protect against challenge with P. gingivalis, generated antibodies that displayed a weak cross-reactivity with the RgpA-Kgp complex.
It is interesting to note that the protective ABM peptides ABM2, ABM1, and ABM3 are located clustered together in the RgpA44 and Kgp39 adhesin domains (Fig. 1). In fact, in both the RgpA44 and Kgp39 adhesin domains, ABM2, ABM1, and ABM3 are located within a 100-residue span, with ABM1 and ABM3 contiguous. In the RgpA44 adhesin this 100-residue span is from residues 865 to 965. Booth et al. (4) have demonstrated that passive immunization of humans with monoclonal antibody (MAb) 61BG1.3 restricted colonization of P. gingivalis in the oral cavity. The binding site of this MAb was mapped using recombinant RgpA44 to residues 907 to 931 (24), which is within the 100-residue span identified in this study. Part of the sequence recognized by MAb 61BG1.3 included the sequence PVXNLT, which is found in ABM1. This same sequence has also been identified using phage display epitope-mapping techniques as a hemagglutination-associated motif (49). Furthermore, the RgpA-Kgp complex has been shown to bind to the ABM1 peptide, indicating that it may be involved in the association of the RgpA and Kgp proteinases and adhesins into large complexes, thereby facilitating agglutination (51). Antibodies binding to ABM1 or proximal peptide epitopes ABM2 and ABM3 may therefore inhibit colonization, hemagglutination, and the formation of the proteinase-adhesin complex and thus reduce the virulence of P. gingivalis.
A recent paper by Genco et al. (16) reported that a 20-amino-acid synthetic peptide representing the N terminus of the RgpA catalytic domain, RgpA45 coupled to an oligolysine support (multiple antigenic peptide [59]) gave protection as a vaccine against P. gingivalis challenge in the murine chamber model. However, three other peptides representing (i) the proteinase active-site cysteine and flanking residues, (ii) ABM1, and (iii) a sequence which is between ABM2 and ABM1 in the RgpA44, Kgp39 protective ABM cluster, did not give protection in that study. Our study has shown that peptide sequences representing the proteinase active-site histidine and flanking residues and those in the ABM cluster region (ABM2, -1, and -3) conferred protection when used as a vaccine in the murine lesion model. The discrepancy between the two studies may be attributable to the availability of T-cell help. In the current study the synthetic peptides were coupled to DT as a source of T-cell help, whereas in the Genco et al. (16) study the peptides were coupled to an oligolysine support. Consequently, unless the peptides contained both B-cell and T-helper cell epitopes, a T-cell-dependent antibody response would not have been generated in the Genco et al. (16) study.
Using protective RgpA-Kgp mouse sera to probe the RgpA-Kgp complex by immunoblot, three bands corresponding to the RgpA44 or Kgp44, the Kgp39, and the RgpA27 peptides were detected. We have previously reported that sera from a group of healthy subjects that harbored P. gingivalis subgingivally as shown by DNA probe analysis also detected the 44-, 39-, and 27-kDa bands of the RgpA-Kgp complex in an immunoblot (39). However, sera from a group of age- and sex-matched diseased patients with periodontitis did not detect the Kgp39 and RgpA27 adhesins and only detected the 44-kDa band (39). The results from both human and mouse studies together suggest that epitopes within the RgpA27, Kgp39, and RgpA44 adhesins may induce protection. It is interesting to note that in this current mouse study and the previous human study (39) antibodies directed to the RgpA-Kgp complex did not recognize the RgpA45 and the Kgp48 proteinase domains of the complex. This may suggest that the proteinase domains are not readily accessible to the immune system as part of the complex since the adhesins of the complex may represent the major surface components and may be immunodominant. However, the recognition of the RgpA-Kgp complex by the antibodies directed against the proteinase active-site peptides demonstrates that at least the proteinase active sites are accessible to antibody binding when the proteinase domains are incorporated into the proteinase-adhesin complex.
We have previously shown that epitope mapping of RgpA27 with healthy subject sera identified two major and two minor epitopes that were recognized by IgG4 subclass antibodies (39). The epitopes recognized by the healthy subject sera in the previous study (39) are identical to the epitopes of RgpA27 identified in the current study with the protective mouse sera (Fig. 5). The major epitope of RgpA27, EP1, includes residues 1542 to 1568, which are also present in Kgp39, and a similar sequence is also present in the RgpA44 adhesin, as follows: epitope EP1, RgpA27 (residues 1544 to 1568), DDFTFEAGKKYTFTMRRAGMGDGTD; Kgp39 (residues 1083 to 1107), DDFTFEAGKKYTFTMRRAGMGDGTD; and RgpA44 (residues 831 to 855), DDYVFEAGKKYHFLMKKMGSGDGTE.
The presence of EP1-related epitopes in the RgpA27, Kgp39, and RgpA44 adhesins of the RgpA-Kgp complex is consistent with the protective mouse sera in the current study and the healthy human sera in the previous study recognizing the 27-, 39-, and 44-kDa proteins of the RgpA-Kgp complex in an immunoblot. The protective epitope EP1 in the 44-kDa adhesin, RgpA44 residues 831 to 855, is located immediately N terminal to the ABM cluster motif (ABM2, -1, and -3), residues 865 to 965, again supporting the importance of this region in protection. Each of the EP1 peptide sequences in RgpA27, Kgp39, and RgpA44 share the motif -X-B-B-X-X-X-X-X-B-B-X-, where B is a basic residue and X is predominantly a hydrophobic residue. Vyas et al. (64) have identified a heparin-binding motif -X-B-B-X-X-X-B-B-X- from snake cardiotoxin. Furthermore, heparin and heparan sulfate glycosaminoglycan binding motifs are characterized by clusters of basic residues separated by hydrophobic residues (5). Consensus sequences for heparin and heparan sulfate glycosaminoglycan recognition have been proposed to be -X-B-B-X-B-X, -X-B-B-B-X-X-B-X-, and -X-B-B-X2n−1-B-X- (5, 64). Hence, the sequence in EP1 may represent a heparin and/or heparan sulfate binding motif, suggesting that the entire span of residues from 831 to 965 in RgpA44 and the other adhesins (Fig. 1) may be involved in binding to host substrates.
The murine antibody subclass response generated by the RgpA-Kgp complex and the protective peptides was a high IgG1 response and a low IgG3 response. We have recently reported that in humans high IgG4 and low IgG2 antibody subclass responses to the RgpA-Kgp complex may be associated with protection against periodontitis (39). The Th2 cytokines interleukin-4 (IL-4) and IL-13 have been reported to induce murine γ1 and human γ4 germ line transcripts and direct B-cell switching to murine IgG1 or human IgG4, respectively (29, 33, 45, 63). Human IgG4 is reported not to bind complement or induce proinflammatory mediators (21). Murine IgG1 can be subdivided into non-complement-fixing and complement-fixing antibodies; however, antigen-bound IgG1 only weakly activates complement and like IgA only activates the alternative pathway (13, 27). Antigen-bound murine IgG1 has also been reported to induce Fc receptor-dependent cellular cytotoxicity and phagocytosis via the FcγRII and FcγRIII receptors (18, 46). Snapper and Paul (54) have reported that IL-4 prepares resting murine B-cells to secrete IgG1 upon subsequent stimulation with bacterial lipopolysaccharide. These results suggest that immunization with the RgpA-Kgp complex and the peptide-DT conjugates induced a Th2-dependent IgG1 response in mice. Subsequent challenge with P. gingivalis would be likely to enhance this response through the stimulation of antigen-primed B cells. Consequently, protection against P. gingivalis in this murine model may be mediated via Fc receptor-dependent phagocytosis of IgG1 opsonized RgpA-Kgp complex with either no induction or weak induction of proinflammatory mediators.
In conclusion, we have demonstrated that immunization with the RgpA-Kgp proteinase-adhesin complex protects against challenge with P. gingivalis in the murine lesion model. Further, we have shown that peptides conjugated to DT corresponding to the proteinase active sites and several adhesin-binding motifs also protect in this model when used as immunogens.
ACKNOWLEDGMENT
Support of the Australian National Health and Medical Research Council (project no. 990199) is acknowledged.
REFERENCES
- 1.Baker P J, Evans R T, Roopenian D C. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch Oral Biol. 1994;39:1035–1040. doi: 10.1016/0003-9969(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 2.Bhogal P S, Slakeski N, Reynolds E C. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143:2485–2495. doi: 10.1099/00221287-143-7-2485. [DOI] [PubMed] [Google Scholar]
- 3.Bird P S, Gemmell E, Polak B, Paton R G, Sosroseno W, Seymour G J. Protective immunity to Porphyromonas gingivalis infection in a murine model. J Periodontol. 1995;66:351–362. doi: 10.1902/jop.1995.66.5.351. [DOI] [PubMed] [Google Scholar]
- 4.Booth V, Ashley F P, Lehner T. Passive immunization with monoclonal antibodies against Porphyromonas gingivalis in patients with periodontitis. Infect Immun. 1996;64:422–427. doi: 10.1128/iai.64.2.422-427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardin A D, Weintraub H J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson J, Herrmann B F, Hofling J F, Sundqvist G K. Degradation of the human proteinase inhibitors alpha-1-antitrypsin and alpha-2-macroglobulin by Bacteroides gingivalis. Infect Immun. 1984;43:644–648. doi: 10.1128/iai.43.2.644-648.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P B, Neiders M E, Millar S J, Reynolds H S, Zambon J J. Effect of immunization on experimental Bacteroideis gingivalis infection in a murine model. Infect Immun. 1987;55:2534–2537. doi: 10.1128/iai.55.10.2534-2537.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P B, Davern L B, Schifferle R, Zambon J. Protective immunization against experimental Bacteroides (Porphyromonas) gingivalis infection. Infect Immun. 1990;58:3394–3400. doi: 10.1128/iai.58.10.3394-3400.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christersson L A, Zambon J J, Dunford R G, Grossi S G, Genco R J. Specific subgingival bacteria and diagnosis of gingivitis and periodontitis. J Dent Res. 1989;68:1633–1639. [Google Scholar]
- 10.Dashper S G, O'Brien-Simpson N M, Bhogal P S, Franzmann A D, Reynolds E C. Purification and characterization of a putative fimbrial protein/receptor of Porphyromonas gingivalis. Aust Dent J. 1998;43:99–104. [PubMed] [Google Scholar]
- 11.Deslauriers M, Haque S, Flood P M. Identification of murine protective epitopes on the Porphyromonas gingivalis fimbrillin molecule. Infect Immun. 1996;64:434–440. doi: 10.1128/iai.64.2.434-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans R T, Klausen B, Sojar H T, Bedi G S, Sfintescu C, Ramamurthy N S, Golub L M, Genco R J. Immunization with Porphyromonas (Bacteroides) gingivalis fimbriae protects against periodontal destruction. Infect Immun. 1992;60:2926–2935. doi: 10.1128/iai.60.7.2926-2935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ey P L, Russell Jones G J, Jenkin C R. Isotypes of mouse IgG. I. Evidence for ‘non-complement-fixing’ IgG1 antibodies and characterization of their capacity to interfere with IgG2 sensitization of target red blood cells for lysis by complement. Mol Immunol. 1980;17:699–710. doi: 10.1016/0161-5890(80)90139-x. [DOI] [PubMed] [Google Scholar]
- 14.Fishburn C S, Slaney J M, Carman R J, Curtis M A. Degradation of plasma proteins by the trypsin-like enzyme of Porphyromonas gingivalis and inhibition of protease activity by a serine protease inhibitor of human plasma. Oral Microbiol Immunol. 1991;6:209–215. doi: 10.1111/j.1399-302x.1991.tb00479.x. [DOI] [PubMed] [Google Scholar]
- 15.Fox C H. New considerations in the prevalence of periodontal disease. Curr Opin Dent. 1992;2:5–11. [PubMed] [Google Scholar]
- 16.Genco C A, Odusanya B M, Potempa J, Mikolajczyk Pawlinska J, Travis J. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalis infection in mice. Infect Immun. 1998;66:4108–4114. doi: 10.1128/iai.66.9.4108-4114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory R L, Kim D E, Kindle J C, Hobbs L C, Lloyd D R. Immunoglobulin-degrading enzymes in localized juvenile periodontitis. J Periodontal Res. 1992;27:176–183. doi: 10.1111/j.1600-0765.1992.tb01666.x. [DOI] [PubMed] [Google Scholar]
- 18.Hazenbos W L, Heijnen I A, Meyer D, Hofhuis F M, Renardel de Lavalette C R, Schmidt R E, Capel P J, van de Winkel J G, Gessner J E, van den Berg T K, Verbeek J S. Murine IgG1 complexes trigger immune effector functions predominantly via Fc gamma RIII (CD16) J Immunol. 1998;161:3026–3032. [PubMed] [Google Scholar]
- 19.Holt S C, Ebersole J, Felton M, Brunsvold M, Korman K S. Implantation of Bacteroides gingivalis in non-human primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- 20.Jagels M A, Travis J, Potempa J, Pike R, Hugli T E. Proteolytic inactivation of the leukocyte C5a receptor by proteinases derived from Porphyromonas gingivalis. Infect Immun. 1996;64:1984–1991. doi: 10.1128/iai.64.6.1984-1991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jefferis R, Pound J, Lund J, Goodall M. Effector mechanisms activated by human IgG subclass antibodies: clinical and molecular aspects. Review article. Ann Biol Clin Paris. 1994;52:57–65. [PubMed] [Google Scholar]
- 22.Kadowaki T, Yoneda M, Okamoto K, Maeda K, Yamamoto K. Purification and characterization of a novel arginine-specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J Biol Chem. 1994;269:21371–21378. [PubMed] [Google Scholar]
- 23.Katz J, Ward D C, Michalek S M. Effect of host response on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;5:309–318. doi: 10.1111/j.1399-302x.1996.tb00187.x. [DOI] [PubMed] [Google Scholar]
- 24.Kelly C G, Booth H, Kendal J M, Slaney M A, Curtis M A, Lehner T. The relationship between colonisation and haemagglutination inhibiting and B cell epitopes of P. gingivalis. Clin Exp Immunol. 1997;110:285–291. doi: 10.1111/j.1365-2249.1997.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kesavalu L, Ebersole J L, Machen R L, Holt S C. Porphyromonas gingivalis virulence in mice: induction of immunity to bacterial components. Infect Immun. 1992;60:1455–1484. doi: 10.1128/iai.60.4.1455-1464.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesavalu L, Holt S C, Ebersole J L. Porphyromonas gingivalis virulence in a murine lesion model: effects of immune alterations. Microb Pathog. 1997;23:317–326. doi: 10.1006/mpat.1997.0161. [DOI] [PubMed] [Google Scholar]
- 27.Klaus G G, Pepys M B, Kitajima K, Askonas B A. Activation of mouse complement by different classes of mouse antibody. Immunology. 1979;38:687–695. [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lai Y H, Mosmann T R. Mouse IL-13 enhances antibody production in vivo and acts directly on B cells in vitro to increase survival and hence antibody production. J Immunol. 1999;162:78–87. [PubMed] [Google Scholar]
- 30.Lantz M S, Rowland R W, Switalski L M, Hook M. Interactions of Bacteroides gingivalis with fibrinogen. Infect Immun. 1986;54:654–658. doi: 10.1128/iai.54.3.654-658.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larjava H, Uitto V J, Haapasalo M, Heino J, Vuento M. Fibronectin fragmentation induced by dental plaque and Bacteroides gingivalis. Scand J Dent Res. 1987;95:308–314. doi: 10.1111/j.1600-0722.1987.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 32.Loesche W J, Syed S A, Morrison E C, Laughon B, Grossman N S. Treatment of periodontal infections due to anaerobic bacteria with short-term treatment with metronidazole. J Clin Periodont. 1981;8:29–44. doi: 10.1111/j.1600-051x.1981.tb02022.x. [DOI] [PubMed] [Google Scholar]
- 33.Lundgren M, Persson U, Larsson P, Magnusson C, Smith C I, Hammarstrom L, Severinson E. Interleukin 4 induces synthesis of IgE and IgG4 in human B cells. Eur J Immunol. 1989;19:1311–1315. doi: 10.1002/eji.1830190724. [DOI] [PubMed] [Google Scholar]
- 34.Mayrand D, Holt S C. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol Rev. 1988;52:134–152. doi: 10.1128/mr.52.1.134-152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 36.Neiders M E, Chen P B, Suido H, Reynolds H S, Zambon J J, Shlossman M, Genco R J. Heterogeneity of virulence of Bacteroides gingivalis. J Periodont Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 37.Norusis M J. SPPS for Windows: base systems user's guide, release 6.0. Chicago, Ill: SPSS, Inc.; 1993. [Google Scholar]
- 38.O'Brien-Simpson N M, Daspher S G, Reynolds E C. Histatin 5 is a substrate and not an inhibitor of the Arg- and Lys-specific proteinases of P. gingivalis. Biochem Biophys Res Commun. 1998;254:474–478. doi: 10.1006/bbrc.1998.9318. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien-Simpson, N. M., C. L. Black, P. S. Bhogal, S. M. Cleal, N. Slakeski, T. J. Higgins, and E. C. Reynolds. Serum IgG and IgG subclass responses to the RgpA-Kgp proteinase-adhesin complex of Porphyromonas gingivalis in adult periodontitis. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 40.Ogawa T, Uchida H, Hamada S. Porphyromonas gingivalis fimbriae and their synthetic peptides induce proinflammatory cytokines in human peripheral blood monocyte cultures. FEMS Microbiol Lett. 1994;116:237–242. doi: 10.1111/j.1574-6968.1994.tb06707.x. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine protease (lys-gingipain) in Porphyromonas gingivalis: structural relationship with arginine-specific cysteine protease (arg-gingipain) J Biochem. 1996;120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 42.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 43.Persson G R, Engel D, Whitney G, Darveau R, Weinberg A, Brunsvold M, Page R C. Immunization against Porphyromonas gingivalis inhibits progression of experimental periodontitis in non-human primates. Infect Immun. 1994;62:1026–1031. doi: 10.1128/iai.62.3.1026-1031.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pike R N, Potempa J, McGraw W, Coetzer T H, Travis J. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J Bacteriol. 1996;178:2876–2882. doi: 10.1128/jb.178.10.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punnonen J, Aversa G, Cocks B G, McKenzie A N, Menon S, Zurawski G, de Waal Malefyt R, de Vries J E. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci USA. 1993;90:3730–3734. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravetch J V, Kinet J P. Fc receptors. Annu Rev Immunol. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- 47.Riddles P W, Blakeley R L, Zerner B. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid)—a reexamination. Anal Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 48.Scott C F, Whitaker E J, Hammond B F, Colman R W. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogen. J Biol Chem. 1993;268:7935–7942. [PubMed] [Google Scholar]
- 49.Shibata Y, Hayakawa M, Takiguchi H, Shiroza T, Abiko Y. Determination and characterisation of the hemagglutinin-associated short motifs found in P. gingivalis multiple gene products. J Biol Chem. 1999;274:5012–5020. doi: 10.1074/jbc.274.8.5012. [DOI] [PubMed] [Google Scholar]
- 50.Slakeski N, Cleal S M, Reynolds E C. Characterization of a Porphyromonas gingivalis gene prtR that encodes an arginine-specific thiol proteinase and multiple adhesins. Biochem Biophys Res Commun. 1996;224:605–610. doi: 10.1006/bbrc.1996.1073. [DOI] [PubMed] [Google Scholar]
- 51.Slakeski N, Bhogal P S, O'Brien-Simpson N M, Reynolds E C. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin binding motif involved in association of the PrtR and PrtK proteinases and adhesins into large complexes. Microbiology. 1998;144:1583–1592. doi: 10.1099/00221287-144-6-1583. [DOI] [PubMed] [Google Scholar]
- 52.Slots J. Importance of black-pigmented Bacteroides in human periodontal disease. In: Genco R J, Merganhagan S E, editors. Host-parasite interaction in periodontal disease. Washington, D.C.: American Society for Microbiology; 1982. pp. 27–45. [Google Scholar]
- 53.Smalley J W, Birss A J, Kay H M, McKee A S, Marsh P D. The distribution of trypsin-like enzyme activity in cultures of a virulent and an avirulent strain of Bacteroides gingivalis W50. Oral Microbiol Immunol. 1989;4:178–181. doi: 10.1111/j.1399-302x.1989.tb00249.x. [DOI] [PubMed] [Google Scholar]
- 54.Snapper C M, Paul W E. B cell stimulatory factor-1 (interleukin 4) prepares resting murine B cells to secrete IgG1 upon subsequent stimulation with bacterial lipopolysaccharide. J Immunol. 1987;139:10–17. [PubMed] [Google Scholar]
- 55.Socransky S S, Haffajee A D, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol. 1991;18:766–775. doi: 10.1111/j.1600-051x.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 56.Socransky S S, Haffajee A D, Cugini M A, Smith C, Kent R L., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 57.Sojar H T, Hamada N, Genco R J. Isolation and characterization of fimbriae from a sparsely fimbriated strain of Porphyromonas gingivalis. Appl Environ Microbiol. 1997;63:2318–2323. doi: 10.1128/aem.63.6.2318-2323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundqvist G, Bengston A, Carlsson J. Generation and degradation of the complement fragment C5a in human serum by Bacteroides gingivalis. Oral Microbiol Immunol. 1988;3:103–107. doi: 10.1111/j.1399-302x.1988.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 59.Tam J P. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uitto V J, Haapasalo M, Laakso T, Salo T. Degradation of basement membrane collagen by proteases from some anaerobic oral microorganisms. Oral Microbiol Immunol. 1988;3:97–102. doi: 10.1111/j.1399-302x.1988.tb00092.x. [DOI] [PubMed] [Google Scholar]
- 61.Van Dyke T E, Offenbacher S, Place D, Dowell V R, Jones J. Refractory periodontitis: mixed infection with Bacteroides gingivalis and other unusual Bacteroides species. J Periodont. 1988;59:184–189. doi: 10.1902/jop.1988.59.3.184. [DOI] [PubMed] [Google Scholar]
- 62.van Steenbergen T J, Kastelein P, Touw J J, de Graaff J. Virulence of black-pigmented Bacteroides strains from periodontal pockets and other sites in experimentally induced skin lesions in mice. J Periodont Res. 1982;17:41–49. doi: 10.1111/j.1600-0765.1982.tb01129.x. [DOI] [PubMed] [Google Scholar]
- 63.Vitetta E S, Ohara J, Myers C D, Layton J E, Krammer P H, Paul W E. Serological, biochemical, and functional identity of B cell-stimulatory factor 1 and B cell differentiation factor for IgG1. J Exp Med. 1985;162:1726–1731. doi: 10.1084/jem.162.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vyas A A, Pan J J, Patel H V, Vyas K A, Chiang C M, Sheu Y C, Hwang J K, Wu W. Analysis of binding of cobra cardiotoxins to heparin reveals a new beta-sheet heparin-binding structural motif. J Biol Chem. 1997;272:9661–9670. doi: 10.1074/jbc.272.15.9661. [DOI] [PubMed] [Google Scholar]
- 65.Wingrove J A, DiScipio R G, Chen Z, Potempa J, Travis J, Hugli T E. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J Biol Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]