Abstract
UBA1 is an X-linked gene and encodes an ubiquitin-activating enzyme. Three somatic mutations altering the alternative start codon (M41) in UBA1 in hematopoietic precursor cells have recently been described, resulting in a syndrome of severe inflammation, cytopenias, and the presence of intracellular vacuoles in hematopoietic precursors - termed VEXAS syndrome, a predominantly male disease. Here we present a patient with clinical features of VEXAS who harbored two novel somatic variants in UBA1 (I894S and N606I). To better understand the clinical relevance and biological consequences of non-M41 (UBA1non-M41) variants, we analyzed the whole genome and transcriptome data of 4168 patients with hematological malignancies and detected an additional 16 UBA1non-M41 putative somatic variants with a clear sex-bias in patients with myeloid malignancies. Patients diagnosed with myeloid malignancies carrying UBA1non-M41 putative somatic variants either had vacuoles or immunodysregulatory symptoms. Analysis of the transcriptome confirmed neutrophil activation in VEXAS patients compared to healthy controls but did not result in a specific transcriptomic signature of UBA1M41 patients in comparison with MDS patients. In summary, we have described multiple putative novel UBA1non-M41 variants in patients with various hematological malignancies expanding the genomic spectrum of VEXAS syndrome.
Subject terms: Haematological cancer, Genetics research, Clinical genetics
Introduction
UBA1 (Ubiquitin-like activating enzyme) is one of the two E1 enzymes of the ubiquitin-proteasome system (UPS) in humans and activates ubiquitins to transfer to E2 enzymes. UBA1 is located on the X chromosome in humans, but is a known X chromosome inactivation (XCI) escape gene [1, 2] and its total loss of function is considered embryonic lethal in males [3]. In late 2020, Beck et al. described somatic loss of function variants in UBA1 in the hematopoietic stem and progenitor cell (HSPC) compartment as a cause of a severe hematoinflammatory disease termed VEXAS (Vacuoles, E1 enzyme, X-linked, Autoinflammatory, Somatic) Syndrome [4]. VEXAS, observed exclusively in males, was characterized by treatment-refractory autoinflammatory symptoms, intracellular vacuoles in hematopoietic precursors and cytopenias. Half of the patients with VEXAS were observed to develop myelodysplastic syndromes (MDS) [5]. In the initial description, three causative variants were identified which altered the start codon of the cytoplasmic isoform in exon 3 (M41T, M41L, M41V). Sequencing of UBA1 exon 3 in more than 1000 patients identified one patient carrying a confirmed somatic non-synonymous variant (S56F), who had milder inflammatory symptoms, vacuoles, and a diagnosis of MDS [6, 7]. The S56F variant was shown to result in partial loss of function. This finding suggests that somatic UBA1 variants other than those at codon M41 (UBA1non-M41) may also be relevant in VEXAS syndrome pathogenesis. However, the precise frequency, clinical features and contribution to phenotype for non-M41 variants are unknown.
Herein, we report a patient presenting with the clinical features of VEXAS syndrome who harbored two novel somatic UBA1non-M41 variants (N606I and I894S) but no UBA1M41 variant. To further understand the clinicogenomic characteristics of somatic and germline UBA1non-M41 variants, we analyzed whole genome and transcriptome sequencing data from 4168 patients with hematological malignancies and describe five further UBA1non-M41 variants as potential novel causes of VEXAS.
Patients and methods
Patient cohort
A total of 4168 patients were recruited and analyzed as previously described [8]. The clinical diagnosis was based on standard procedures, following the 2017 WHO classification [9]. The cohort consisted of 774 patients with acute myeloid leukemia (AML), 120 patients with chronic myeloid leukemia (CML), 380 patients with myeloproliferative neoplasms (MPN), 397 patients with myelodysplastic/myeloproliferative neoplasm overlap syndromes (MDS/MPN), which included 78 patients with atypical CML (aCML), 222 patients with chronic myelomonocytic leukemia (CMML), 97 patients myelodysplastic/myeloproliferative neoplasm with ring sideroblasts (MDS/MPN-RS-T), 756 patients with myelodysplastic syndromes (MDS), 321 patients with B-lymphoblastic leukemia/lymphomas (B-ALL), 132 patients with T- lymphoblastic leukemia/lymphoma (T-ALL), 317 patients with chronic lymphocytic leukemia (CLL), 541 patients with mature B-cell neoplasms (labeled B-cell non-Hodgkin lymphoma; B-NHL), which included 97 patients with marginal zone lymphoma (MZL), 94 patients with mantle-cell lymphoma (MCL), 92 patients with hairy cell leukemia (HCL), 68 patients with follicular lymphoma (FL), and 190 patients with other mature B cell neoplasms, 148 patients with mature T- and NK-cell neoplasms (labeled T-cell non-Hodgkin lymphoma; T-NHL), 262 patients with plasma cell myeloma (labeled multiple myeloma; MM), and 20 patients with monoclonal gammopathy of undetermined significance (MGUS). The full details are given as part of Supplementary Table 1. 64 healthy controls were also used for comparison of the transcriptome (35 males and 29 females).
Whole genome sequencing (WGS) and whole transcriptome sequencing (WTS)
DNA and RNA were extracted from bone marrow and peripheral blood, and libraries were prepared by standard protocols as previously described [10].
WGS data processing and analysis
Samples were sequenced with an average coverage of 106x. Reads were aligned to the human reference genome (GRCh37, Ensembl annotation) using the Isaac aligner (v3.16.02.19) [11] through BaseSpace’s WGS app (v5, Illumina, San Diego, CA, USA) with default parameters. Tumor-unmatched normal variant calling was performed with a pool of sex-matched DNA (Promega, Madison, WI) using Strelka (v.2.4.7). Variant allele frequency (VAF) of over 2% and a minimum of 5 supporting reads were required for the variants to be reviewed. Variants were queried against the gnomAD database (v.2.1.1) to remove common germline calls (global population frequency >1%) and annotated with Ensembl VEP. Analysis was restricted to protein-altering and canonical splice-site variants. 74 genes and 64 genes were considered driver genes for myeloid and lymphoid malignancies, respectively, as defined in Supplementary Table 2. UBA1 variants with VAF < 10% were confirmed by targeted sequencing (Supplementary Table 3). For somatic copy number variations (CNV), GATK4 was used following the Broad Institute’s recommended best practices with a panel of normals (defined as patients with a normal karyotype according to conventional cytogenetics).
Classification of somatic versus germline origin of UBA1 variants
UBA1 variants were classified as follows; (i) putative germline – present in gnomAD plus VAF over 90% in males or between 40% and 60% in females (ii) putative somatic – not present in gnomAD (or present in gnomAD at low allele frequency with VAF supporting somatic origin) plus VAFs outside of the range described as putative germline (iii) unknown origin – not meeting criteria above. Frameshift variants not present in gnomAD were considered somatic regardless of their VAF. All putative somatic variants were reviewed for their X chromosome copy number status by standard cytogenetics to consider impact on VAF. When cytogenetic data were unavailable, WGS copy number variation call was referred to. The data necessary for the classification is provided in Supplementary Table 3.
Transcriptome analysis
Unsupervised clustering was done with uniform manifold approximation and projection (UMAP) using the top 5% most variable genes, excluding genes on the sex chromosomes. Differential gene expression analysis was performed with limma [12] and genes with an adjusted p-value < 0.05 and absolute logFC > 1.5 were considered significant. GO enrichment analysis was done by the web-based application Toppfun [13], from where GO gene sets were downloaded (accessed March 15, 2022). Single-sample gene set enrichment analysis (ssGSEA) was done through the GSVA R package [14] with parameter mx.diff = TRUE to suppose upregulation in all members of the gene set. The gene sets were downloaded from MSigDB [15, 16] or taken from references [17, 18] as listed in Supplementary Table 4. All statistical analyses were done using R.
Results
Novel somatic UBA1 variants detected in a patient with clinical features of VEXAS syndrome
A 46-year-old previously healthy male presented with recurrent epistaxis, nodular erythema, increasing fatigue, night sweats and 10 kg of weight loss over several months. The patient denied drug/alcohol use or exposure to toxic substances. Family history was significant for lung cancer affecting his mother, who died at the age of 46. Physical examination revealed pale conjunctiva, jaundice, splenomegaly and erythema nodosum mainly in his trunk. Full blood examination revealed anemia (Hb 5.3 g/dL, reticulocytes 179.4 × 103/µL, MCV 90 fL) and thrombocytopenia (Plt 56 × 103/µL). Indirect bilirubin was high (T-Bil 3.9 mg/dL, D-Bil 0.4 mg/dL), but there were no other signs of hemolysis (LDH 199 U/L, haptoglobin 1.78 g/L). Both iron and ferritin values were low (serum iron 30 µg/dL, ferritin 24.1 µg/L, TS 7.2%), suggestive of iron-deficiency anemia. Hemorrhagic sources other than epistaxis and bleeding disorders besides thrombocytopenia were not identified. CT and gastrointestinal endoscopies showed no tumor or pathologically enlarged lymph nodes, other than enlarged spleen (>25 cm). A bone marrow biopsy was performed which revealed morphological dysplasia and excess blasts (5%), minimal fibrosis and no abnormal lymphoid infiltrate. A diagnosis of MDS-EB1 according to WHO 2017 [9] was made. Abundant vacuoles were observed in hematopoietic precursors and 3% ring sideroblasts were noted. Histological examination of the erythematous rash showed signs of small vessel vasculitis with histiocytes (CD31+, CD4+, MPO+, Lysozyme+, CD68+) and T-cell (CD3+) infiltration. Serological tests for rheumatological diseases including ANCA and ANA were negative. WGS was performed as part of the SIRIUS study (clinicalTrials.gov Identifier: NCT05046444) on a bone marrow aspirate sample which revealed two variants in UBA1 - I894S (VAF 56%) and N606I (VAF 9%). Neither of these variants was detected in CD3+ cells isolated from peripheral blood from the same patient consistent with their somatic origin. No typical MDS driver variants were detected.
Putative somatic UBA1non-M41 variants are detectable across hematological malignancies with a clear sex-bias in myeloid malignancies
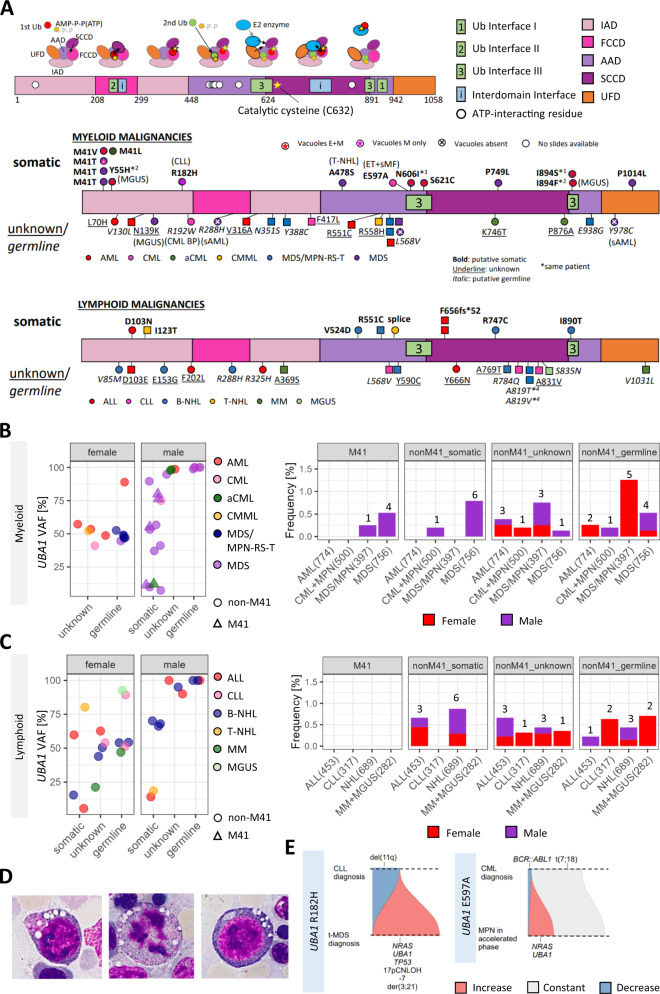
Given the identification of potential novel UBA1 variants causing VEXAS syndrome outside of the M41 codon, we went on to analyze WGTS of 4168 patients (see “Material and Methods”) with a broad range of myeloid and lymphoid malignancies. From this testing we identified 5 patients with UBA1M41 variants, 16 patients with putative somatic UBA1non-M41 variants, 20 patients with putative germline UBA1non-M41 variants and 16 patients where the origin of the UBA1 variant was not able to be definitively assigned. Three patients had longitudinal data available to confirm somatic status in two (R182H and E597A) and in one patients harboring a putative germline variant (R192W), no change in VAF (100%) was observed. Whilst identified variants were distributed over the whole coding region of UBA1, only putative somatic variants were observed in Ubiquitin interface III domain of the protein [19, 20] (Fig. 1A). The VAF ranged from 5% to 89% for the putative somatic variants, 21% to 100% for the unknown variants, and 44% to 100% for the putative germline variants, as certain variants with somatic VAFs are classified non-somatic due to their X chromosomal copy number status. Interestingly, a clear sex bias could be detected in myeloid malignancies, as potential somatic variants occurred only in males (Fig. 1B), whereas no sex difference was observed in occurring in lymphoid malignancies (Fig. 1C). The genetic and clinical characteristics of all the 12 patients with myeloid malignancies carrying the putative somatic variants are shown in Table 1.
Fig. 1. Distribution of variants in the gene UBA1 and their relevance in hematological malignancies.
A (Above) The gene UBA1 consists of five functional domains: two adenylation domains (active adenlyaltion domain [AAD] and inactive adenylation domain [IAD]), two catalytic half domains (first catalytic cysteine half domain [FCCD] and second catalytic cysteine half domain [SCCD]), and ubiquitin fold domain [UFD]. UBA1 IAD and AAD adenylate the first ubiquitin, and transfer ubiquitin to the catalytic cysteine domains to form a thioester bond. The second ubiquitin needs to be loaded to make the necessary conformational changes to transfer the charged ubiquitin to E2 enzymes. The regions constituting the interface contacting ubiquitin (Ub Interface I to III) defined in references [19] and [20] are labelled as boxes. Selected domains important in conformational changes are also represented in boxes. ATP-interacting residues mentioned in the Supplementary Fig. S12 of Beck et al. [4] are shown as circles. (Below) Loci of the variants are shown, separated by whether the patient was diagnosed with myeloid or lymphoid malignancies. *(number) denotes that the variants are carried by the same numbered patient. The putative somatic variants (bold) are shown above and putative germline (italic) and variants of unknown significance (underline) are shown below the gene. The presence of vacuoles are shown by signs inside the circle (male) or square (female). No signs means that bone marrow smear slides were not preserved at the facility. E + M: vacuoles both in erythroids and myeloids. M only: vacuoles in myeloids only. The diagnosis are color coded and the second diagnosis, if present, are denoted in brackets. AML (acute myeloid leukemia), CML (chronic myeloid leukemia), aCML (atypical chronic myeloid leukemia), CMML (chronic myelomonocytic leukemia), MDS/MPN-RS-T (myelodysplastic/myeloproliferative neoplasm with ring sideroblast and thrombocytosis). ALL (acute lymphoblastic leukemia), CLL (chronic lymphoblastic leukemia), B-NHL (B-cell non-Hodgkin lymphoma), T-NHL (T-cell non-Hodgkin lymphoma), MM (multiple myeloma), MGUS (monoclonal gammopathy of uncertain significance), CML BP (blast phase), sAML (secondary AML), ET (essential thrombocythemia), and sMF (secondary myelofibrosis). (B-C) Distribution of variant allele frequency by the lineage of the malignancies, and the frequency of each diagnostic categories and distribution of sexes are shown. The number of patients in the analyzed cohorts are shown in brackets. Further details are given in Supplementary Table 1. B Myeloid malignancies. C Lymphoid malignancies. D Characteristic vacuoles were present in myeloid precursors, even in mitotic phase, and in erythroid precursors from bone marrow aspirates (400x, oil) of the patient harboring variants Y55H and I894F. E Likely clonal history of two patients with longitudinal data available are shown.
Table 1.
Genetic and clinical characteristics of the 12 patients with myeoid malignancies carrying putative somatic UBA1 variants.
| Patient_ID | Age | Sex | Diagnosis | UBA1 Variant | VAF [%] | Vacuole | Co-mutation | Karyotype | Hematological history (age) | Possible immunodysregulation (age) |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 77 | male | aCML | M41L | 12 | NA | NRAS (12%) | 46,XY[20] | clinically diagnosed with CML (65); CML diagnosis revised to aCML due to negative BCR::ABL1 [*](75); splenomegaly (circa 40 cm) | none recorded |
| P2 | 82 | male | MDS-MLD | M41T | 76 | NA | none | 46,XY[20] | ESA/transfusion-requring anemia | none recorded |
| P3 | 76 | male | MSD-RS-SLD | M41T | 11 | yes | SF3B1 (31%), JAK2 (14%), ZRSR2 (4%) | 46,XY[20] | thrombocytosis | none recorded |
| P4 | 50 | male | MDS-MLD | M41T | 80 | NA | none | 46,XY,t(2;11)(p21;q23)[13]/46,XY [7] | NA | unspecified autoimmune disease under immunosuppresion |
| P5 | 84 | male | MDS-MLD | M41V | 55 | yes | none | 46,XY,der(1)t(1;2)(p34;q24),der(2)t(1;2)(p36;q24)[7]/46,idem,del5(q14q34)[3]/46,XY[10] | transfusion-requring anemia, pancytopenia | polyneuropathy, latent hyperthyroidism |
| P6 | 76 | male | MDS-RS-MLD | Y55H | 41 | yes | SRSF2 (50%) | 46,XY[20] | [*]MGUS; pancytopenia; anemia treated with 30 mg prednisone/transfusion, successfully discontinued (79) | none recorded |
| I894F | 37 | |||||||||
| P7 | 66 | male | CLL | R182H | 0 | yes | 46,XY, del(11)(q21q23)[4]/46,XY[16] | [*]CLL treated with fludarabine, cyclophosphamide, rituximab (64-65) in CR (66); pancytopenia(65); treated with azacitidine (66) | Pneumocystis pneumonia (65/66), CMV reactivation (65/66) | |
| t-MDS/MDS-EB2 | 57 | NRAS (35%), TP53 (98%) | 44,XY,der(3;21)(q10;q10),-7[20] | |||||||
| P8 | 68 | male | MDS-MLD | A478S | 53 | NA | none | 45,X,-Y [18]/46,XY[2] | primary cutaneous large cell anaplastic T cell lymphoma in the head and neck treated with resection [*] and radiation (68-70); MDS no treatment until age 70 | hyperthyroidism |
| P9 | 69 | male | CML | E597A | 0 | NA | BCR::ABL1 (IS 0.034) | 46,XY,t(7;18)(q11,q23)[7]/46,XY[2] | ET (61) treated with anagrelide; CML [*1] treated with imatinib (67); anemia due to secondary MF (69); transfusion-dependent Coombs-positive anemia (69); 12% blasts with increased borderline monocyte/blasts in PB [*2] (69) | monoblast infiltration in the skin (69), steroid-resistant fever (69) |
| MPN | 75 | NRAS (50%), BCR::ABL1 (IS 0.006) | 46,XY,t(7;18)(q11,q23)[20] | |||||||
| P10 | 67 | male | MDS-RS-MLD | S621C | 90 | yes | FLT3-ITD (39%), RUNX1 (69%), SRSF2 (39%) | 47,XY, + 21[19]/46,XY[1] | steroid-treated hemolytic anemia[*] (67); 9 cycles of azacitidine against MDS (67); decitabine switch (68), 33% blasts in PB, suggesting secondary AML (68); transfusion-dependent anemia | azacitidine injection site reaction (67), antibiotic-resistant broncoscopy-negative pulmonary infiltrate post fungal infection (68) |
| P11 | 73 | male | MDS-MLD | P749L | 10 | NA | none | 47,XX, + X[20] | chronic hemolytic anemia requiring ESA/transfusion | hyperthyroidism treated with nuclear medicine (74) |
| P12 | 63 | male | MDS-RS-SLD | P1014L/canonical splice site | 7 | NA | SF3B1 (49%), TET2 (16%) | 46,XY[20] | autoimmune anemia treated with steroids (62-, tapered and reinitiated), azathioprine[*] (62–63), mycophenol-mofetil (63–74), and rituximab (63, 74), ESA/multiple transfusion (62-) | anti-parietal cell antibody positive gastritis with polyclonal hypergammaglobulinemia and elevated complements (56), IgG4-associated nephritis (58/histology 68), interstitial pneumotitis (62), leiomyosarcoma (71), diverse skin tumors (74) |
[*] indicates the time point the sequencing was performed.
MDS-MLD MDS with multi-lineage dysplasia, MDS-RS-MLD MDS with ring sideroblasts and multi-lineage dysplasia, CLL chronic lymphocytic leukemia, t-MDS therapy-related MDS, MDS-EB MDS with excess blast, CML chronic myeloid leukemia, MPN myeloproliferative neoplasms, MDS-RS-SLD MDS with ring sideroblasts and single-lineage dysplasia, aCML atypical CML, ESA Erythropoiesis-stimulating agent, NA not available, MGUS monoclonal gammopathy of undetermined significance, CR complete remission, ET essential thrombocythemia, MF myelofibrosis, PB peripheral blood, AML acute myeloid leukemia.
Consistent with previous descriptions [4], UBA1M41 variants occurred exclusively in males in our cohort and in myeloid malignancies (MDS n = 4, MDS/MPN [aCML] n = 1, Fig. 1B) but no UBA1M41 variant was detected in patients with AML. For 2/4 MDS patients with UBA1M41 variants, material was available for morphological assessment, which confirmed the presence of vacuoles in hematopoietic precursors.
Sixteen putative somatic UBA1non-M41 variants were detected in 16 patients across both myeloid and lymphoid malignancies (Fig. 1B, C). UBA1non-M41 variants were observed exclusively in males in patients with myeloid malignancies (MDS n = 6, CML + MPN [ET + CML + sMF] n = 1), with 8 UBA1non-M41 variants observed in 7 patients (Table 1). Three patients with MDS and UBA1non-M41 variants (Y55H + I894F, R182H, S621C) had material available for morphological assessment of which all three showed vacuoles. Images of a representative patient (Y55H + I894F) are shown in Fig. 1D and Supplementary Fig. 1. In lymphoid malignancies, the distribution of putative somatic UBA1non-M41 across sexes was essentially equal with nine patients (5 male, 4 female) found to carry UBA1non-M41 putative somatic variants (B-ALL n = 3, B-NHL n = 4 [FL n = 3, HCL n = 1], T-NHL n = 2, Fig. 1A). Notably the same frameshift variant (F656SfsTer52) was identified in two female patients with ALL.
Concerning the UBA1 unknown origin and putative germline variants, the incidence showed a slight female predominance with M:F ratios of 1:1.3 and 1:1.6, respectively (total cohort M:F ratio 1:0.7). No patients carrying putative somatic variants were diagnosed with MDS/MPN-RS-T, however, 5 patients carrying putative germline variants were diagnosed with MDS/MPN-RS-T, all of whom were female (MDS/MPN-RS-T cohort male to female ratio = 1:1.1). Material was available for morphological assessment in 3 patients with putative germline origin UBA1 variants. None of these three patients (R288H, Y978C, and L568V) had vacuoles present.
Immunodysregulatory symptoms are common among patients with myeloid malignancies carrying UBA1non-M41 variants
If available, non-hematological co-diagnoses and history of patients, as provided by the referring physician, were extracted from the request form. The records of the 57 patients carrying UBA1 variants were examined, with particular attention to symptoms suggestive of VEXAS in the 16 patients carrying the putative somatic UBA1non-M41 variants. Whilst the records were incomplete, among the patients with putative somatic variants, 5 out of the 7 patients with myeloid malignancies had immunodysregulatory symptoms documented, whereas no unprovoked inflammatory symptoms were noted for the 9 patients with lymphoid malignancies. Table 1 summarizes the results of the 7 patients with myeloid malignancy carrying the putative somatic UBA1non-M41 variants along with the record of the 5 patients carrying the UBA1M41 variants. The documented clinical information included IgG4-associated nephritis/pneumonitis (P1014L, simultaneously affecting the canonical splice site), skin monoblast infiltration (E597A), hyperthyroidism (A478S and P749L), Coombs-positive hemolytic anemia (E597A), and sterile pulmonary infiltrate (S621C). Of interest, two patients developed asynchronous malignancies in the skin of the head and neck area after they were discovered to carry UBA1 likely somatic variants. The patient carrying the A478S variant was diagnosed with primary cutaneous anaplastic large-cell lymphoma of both ears, left temporal and right occipital regions recurring in a period of two years. The patient carrying the P1014L variant was under immunosuppressive treatment, when he developed basosquamous carcinoma (right nasolabial and cheek), squamous cell carcinoma (right retroauricular region), and Bowen’s disease (right cervical region) within a period of two months.
Among the patients with UBA1 variants of unknown origin or putative germline, Graves’ disease with recurrent thyroid nodules development after goiter operation was documented for one MDS patient (L568V) and there were two additional patients with thyroid involvement (hypothyroidism, R784Q, CLL; intrathyroidal parathyroid carcinoma, N139K, MDS). Polyarthritis (Y978C, MDS), childhood tonsillectomy (L568V, MDS/MPN-RS-T), steroid-treated sinusitis, pneumonitis, and non-malignant renal mass (Y388C, MDS/MPN-RS-T), and unclear polyneuropathy before chemotherapy (R551C, AML) were also documented.
The majority of patients with UBA1 putative somatic variants in myeloid malignancies have co-mutations in leukemic driver genes
In order to understand the broader genomic context of putative somatic UBA1 variants in myeloid malignancies, we searched for chromosomal aberrations and co-mutations in known leukemia driver genes (Table 1). Various co-mutations were detected in 7/12 (58%) patients, with NRAS (n = 3), SF3B1 (n = 2) and SRSF2 (n = 2) being the most frequent. The identified leukemogenic driver variants were present at higher VAFs than the X-chromosome adjusted VAF of UBA1 variants and, hence, these UBA1 clones are likely sub-clonal to the driver clones by pigeon-hole principle, assuming heterozygosity of the driver variants. Among the patients harboring NRAS variants, two carrying the UBA1 variants R182H (MDS) and E597A (CML + secondary myelofibrosis [MF] after essential thrombocythemia [ET] treatment) were known with a history of prior treatment against CLL (rituximab-fludarabine-cyclophosphamide; R-FC) and CML (imatinib), respectively. For both cases, WGS data were available at the pretreatment time point, which showed that the UBA1 and NRAS clones occurred only after treatment (Fig. 1E). In 2 patients (P4, M41T with t(2;11); P5, M41V with del(5q)), disease-defining chromosomal aberrations were noted to be at a smaller clone size than the UBA1 variant clones. In 3 patients (M41T, P749L, A478S) no apparent leukemogenic driver variants or disease-defining chromosomal aberrations were noted.
Whole transcriptome analysis of patients with UBA1non-M41 variants
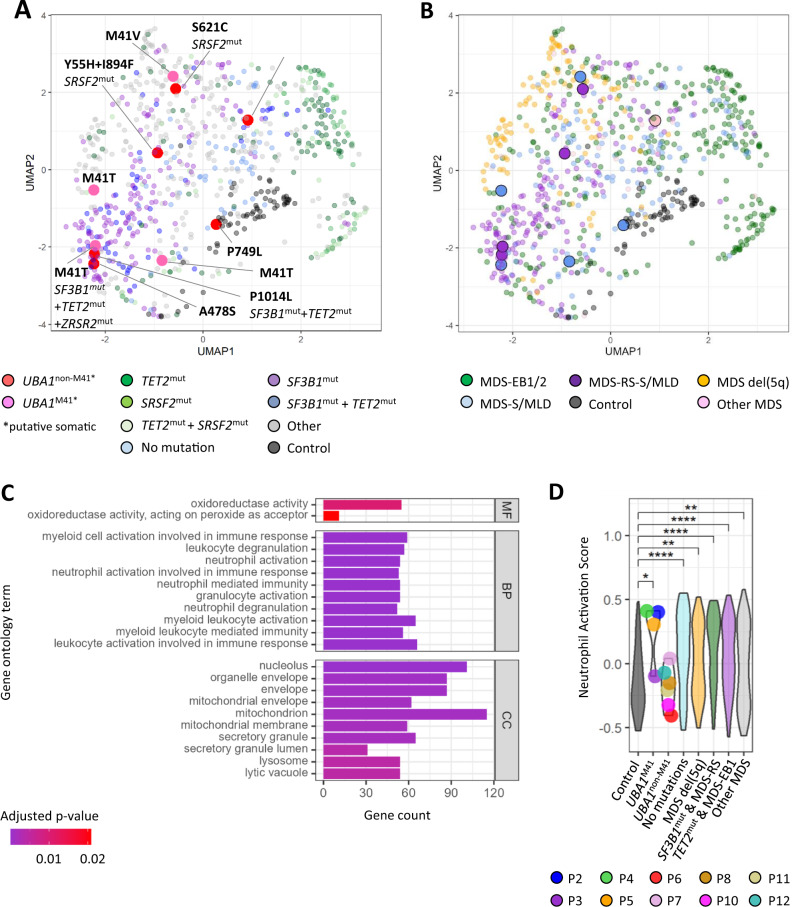
To better understand the biological effect and inflammogenicity of somatic UBA1non-M41 variants we analyzed the transcriptome from patients diagnosed with MDS. The samples were taken at the time of their first diagnosis of MDS, and the disease severity and treatment status of their co-existing non-hematological conditions, if any, were not uniformly available. An initial unsupervised clustering analysis based on the top 5% most variable genes (1012 genes) showed that samples with putative somatic UBA1 variants did not form a distinct group (Fig. 2A, B). As the top 5% most variable genes did not capture the characteristics of UBA1M41 samples, we subsequently performed supervised differential gene expression (DEG) analysis. Comparing the transcriptional profiles of UBA1M41 samples with healthy controls resulted in 1038 DEGs with 906 genes up-regulated and 132 genes down-regulated, respectively. GO enrichment analysis of up-regulated genes in UBA1M41 patients revealed an enrichment of myeloid cell/neutrophil activation and degranulation, as reported by Beck et al. [4] for VEXAS patients (Fig. 2C). To evaluate UBA1non-M41 variants individually in the context of VEXAS-upregulated neutrophil activation we analyzed the samples applying the single-sample gene set enrichment analysis method [14], which ranks and assigns an enrichment score of gene sets to each sample within the group. We created a group of MDS patients confirmed not to have any driver mutations (VAF > 2%) or chromosomal abnormalities to simulate background MDS controls as well as MDS subgroups reported with positive (del5q [21], TET2mut/MDS-EB1 [22, 23]) or negative (SF3B1mut/MDS-RS [22, 23]) association to inflammation. High neutrophil activation scores were not specific to UBA1M41 samples, and some MDS samples carrying no leukemia-driver mutations scored over top quantile (Fig. 2D). None of the UBA1non-M41 samples had reached over top quantile scores. Results of other myeloid cell/neutrophil activation and degranulation gene sets did not differ (data not shown).
Fig. 2. Transcriptomic analysis of patients carrying putative somatic UBA1 variants in comparison to other MDS patients.
A UMAP of 816 (752 MDS patients, 64 healthy controls) samples color-coded based on selected co-mutations. B UMAP of 816 (752 MDS patients, 64 healthy controls) samples color-coded based on WHO sub-classification. MDS-EB (MDS with excess blast), MDS del5q (MDS with isolated del(5q)), MDS-RS (MDS with ring sideroblasts), MDS-MLD (MDS with multi-lineage dysplasia), MDS-SLD (MDS with single-lineage dysplasia) (C) GO analysis of upregulated genes of UBA1M41 patients compared to healthy controls. D ssGSEA (single-sample gene set enrichment analysis) scoring of neutrophil activation stratified by selected groups. Wilcoxon rank sum test was used with Benjamini & Hochberg correction. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To complement the functional analysis, we evaluated pathways that may be involved in VEXAS inflammation in a hypothesis-driven manner. Briefly, we selected pathways targeted by anti-inflammatory treatment given to VEXAS patients [24–28]. Furthermore we examined pathways related to autoinflammatory diseases due to loss of function variants in the proteasome genes [29–31], which are expected to provide mechanistic insights into VEXAS inflammation [32], as they both may induce interferon response via the unfolded protein response (UPR) pathway. UBA1M41 samples had significantly higher scores of the Interferon response gene set (IRG-6) [18], but did not differ to other MDS subgroups (Supplementary Fig. 2). Thus, UBA1M41 samples did not show any specific and sensitive inflammatory signature.
Discussion
UBA1 is a newly rediscovered gene in the field of hematology in the context of VEXAS syndrome, a prototypical hematoinflammatory disease. By analyzing comprehensive and unbiased WGTS data from 4168 patients we identified five UBA1M41 variants (total cohort 0.1%; myeloid malignancy cohort 0.2%; MDS cohort 0.5%), 16 UBA1non-M41 putative somatic variants (total cohort 0.4%), including eight UBA1non-M41 variants in seven patients with myeloid malignancies (myeloid malignancy cohort 0.3%; MDS cohort 0.8%), representing potential causative variants of VEXAS syndrome. In addition, nine UBA1non-M41 variants in patients with lymphoid malignancies (lymphoid malignancy cohort 0.5%) were detected. The incidence of UBA1 somatic variants in a community cohort has recently been reported to be approximately 1 in 4000 of older males [33]. The relatively high incidence of somatic UBA1 variants in our cohort of hematological malignancy suggests that UBA1 variants are not passenger variants. We further detected a clear sex bias in the occurrence of UBA1 somatic variants in myeloid malignancies, which is supportive of the potential clonal advantage conferred by UBA1 variants. In these male patients of myeloid malignancies, we observed three different scenarios: (i) UBA1 variants as the main clone, (ii) UBA1 variants as subclonal events in combination with known leukemic driver events, and (iii) UBA1 variants as a secondary major driver event following treatment. Scenario (iii) has been described in one case report of a patient initially diagnosed with ET treated with hydroxyurea subsequently developing MDS after acquiring a UBA1 variant [34]. Of note two of our confirmed somatic variants (R182H, E597A) co-emerged with a NRAS variant after treatment (R-FC for CLL + t-MDS R182H; imatinib for ET + CML + sMF E597A), which again suggests the contribution of UBA1 variants to clonal fitness. Among the variants we identified, P749L is a known partial loss of function mutation [35, 36]. Some of the identified somatic variants were also on functional residues (S478, R551, N606, S621, R747), one of which is a partial loss of function site (R747). The literature and prevalence in public databases on the identified putative somatic variants are summarized in Table 2 [7, 20, 33, 35–44]. Frameshift variants, which are likely total loss of function, were only discovered in females (F656SfsTer52), which may also be partial loss of function overall considering the two copies of the X chromosome. None of the identified variants in males were on total loss of function sites, but only proximal to such sites (V524D, E597A). Experimentally, partial loss of function mutations are known to result in a proliferative phenotype, whereas total loss of function mutations results in an apoptotic phenotype [35, 36]. Thus, it is possible that variants leading to partial loss of UBA1 function contribute to the development of cancers.
Table 2.
Literature review on the identified putative somatic missense variants in the hematological malignancy cohorts.
| Patient ID | Cluster | Diagnosis | Variant | Cohort freq1 | Public freq2 | Proximal functional site | Methods | Function/mutant consequence/mutant phenotype | Reference | Organism |
|---|---|---|---|---|---|---|---|---|---|---|
| P6 | NA | MDS | Y55H | 1/756; 0.0013 | 0 | S56F | mutagenesis | temperature-sensitive partial loss of function | Poulter [7] | human |
| R57 | structural | ATP-contacting residue (gamma phosphate) | Lv [20] | human | ||||||
| R57K/A | mutagenesis | significantly reduced nitrate reductase activity in R515 (R73 in E. Coli) double mutants | Lake [40] | E. Coli (R14K/A) | ||||||
| P33 | NA | B-ALL | D103N | 1/321; 0.0031 | 0 | S140 | structural | SCCH/IAD/AAD interacting residue; substitution has minimal effect on thioester bond formation | Hann [41] | S. Pombe (Q105A) |
| P40 | NA | T-NHL | I123T | 1/148; 0.0068 | 0 | S140 | structural | SCCH/IAD/AAD interacting residue; substitution has minimal effect on thioester bond formation | Hann [41] | S. Pombe (Q105A) |
| P7 | NA | MDS | R182H | 1/756; 0.0013 | gnomAD (3/165293; 1.81e-5) | A189T* | mutagenesis | slightly less efficient ubiquitination, higher apoptotic rate than wild type | Lao [42] | mouse (A189T) |
| P8 | ATP-binding | MDS + T-NHL | A478S | 1/756; 0.0013 | 0 | A478 | structural | ATP-contacting residue (alpha phosphate); consists the oxyanion hole and GxGxxG ATP binding motif | Lv [20] | human |
| P37 | ATP-binding | B-NHL (FL) | V524D | 1/541; 0.0018 | 0 | K528N* | mutagenesis | mutant tissue apoptotic and enlarged wild type tissue (total loss of function) | Pfleger [39] | Drosophila(K663N) |
| K528A | structural; mutagenesis | ATP-contacting residue (beta phosphate); ATP binding attenuated and reduced Ub-adenylation | Tokgöz [43] | human | ||||||
| P39 | ATP-binding | B-NHL (HCL) | R551C | 1/541; 0.0018 | 0 | N550* | mutagenesis | 3-fold decrease in thioester bond formation | Hann [41] | S. Pombe (E511R) |
| R551 | structural | ATP-contacting residue | Lv [20] | human | ||||||
| P9 | Ub Interface III | CML + MPN | E597A | 1/500; 0.0020 | 0 | C588Y* | mutagenesis | mutant tissue apoptotic and enlarged wild type tissue (total loss of function) | Pfleger [39] | Drosophila (C723Y) |
| G599 | structural | ubiquitin-contacting residue | Lv [20] | human | ||||||
| case | Ub Interface III | MDS | N606I | 1/756; 0.0013 | 0 | N606 | structural | van der Waals contacts with Ub | Lv [20] | human |
| P10 | Ub Interface III | MDS | S621C | 1/756; 0.0013 | COSMIC MDS-RS (1/9; 0.11) | S621 | structural | hydrogen bond with Ub | Lv [20] | human |
| S621C | sequencing | somatic variant identified in a SF3B1-mut MDS patient (COSMIC-cited) | Papaemmanuil [44] | human | ||||||
| S621C | sequencing | somatic variant identified in a patient with vasculitis | Beck [36] | human | ||||||
| P36 | Interdomain Interface | B-NHL (FL) | R747C | 1/541; 0.0018 | COSMIC colorectal carcinoma (1/619; 0.0016) | R747E* | mutagenesis | 4-fold decrease in thioester bond formation | Hann [41] | S. Pomne (R707E) |
| R747 | structural | salt bridge between E252 of the FCCH domain | Lv [20] | human | ||||||
| P747C | sequencing | somatic variant identified in a female patient with large intestine adenocarcinoma (COSMIC-cited) | Giannakis () | human | ||||||
| P11 | Interdomain Interface | MDS | P749L | 1/756; 0.0013 | 0 | P749L* | mutagenesis | enlarged mutant tissue (temperature-sensitive partial loss of function) | Pfleger [39], Lee [38] | Drosophila (P884L) |
| P38 | Ub Interface III | B-NHL (FL) | I890T | 1/541; 0.0018 | 0 | I891 | structural | van der Waals contacts with Ub | Lv [20] | human |
| P6 | Ub Interface III | MDS | I894F | 1/756; 0.0013 | 0 | I891 | structural | van der Waals contacts with Ub | Lv [20] | human |
| case | Ub Interface III | MDS | I894S | 1/756; 0.0013 | 0 | I891 | structural | van der Waals contacts with Ub | Lv [20] | human |
| P12 | NA | MDS | P1014L/splice | 1/756; 0.0013 | 0 | R1010 | structural | E2 enzyme (Ubc4) contact site | Olsen [45] | S. Pombe (R965) |
| del(923-1058) | mutagenesis | IRF3 binding lost | Chen [46] | human/zebrafish |
[*] indicates that the site is a corresponding conserved residue in human.
1: The frequency in the respective subcohort.
2: The frequency reported in gnomAD or COSMIC databases. In case of COSMIC the cancer name is given and the frequency is in the brackets.
Subject Ontology:UBA1, VEXAS, MDS, inflammation, cytopenia, leukemia, lymphoma, sex difference.
Whilst there are no official diagnostic criteria for VEXAS syndrome, VEXAS is characterized by the co-existence of acquired inflammatory and hematological symptoms as well as the presence of vacuoles in hematopoietic stem and progenitor cells and somatic UBA1 mutations [4, 24]. Our index case showed all the clinical features of VEXAS but carried two novel UBA1non-M41 variants (N606I VAF 9%, I894S VAF 56%) instead of the canonical UBA1M41 variant, one of which variant locus was identified in another patient with double mutant (Y55H VAF 41%, I894F VAF 37%) in our cohort. The repeated appearance of the I894 loci and the high VAF suggests I894 as a potential disease-causative loci of VEXAS. Interestingly, residues N606 and I894 face each other at the ubiquitin interface III, and the likely later acquisition of N606 with a lower VAF may have had an advantageous effect for clonal survival. We additionally report that 5 of the 7 patients diagnosed with myeloid malignancies in the retrospective cohort carrying somatic UBA1non-M41 variants (A478S, E597, S621C, P749L, P1014L) showed immunodysregulatory symptoms (Table 1), although a clinical rheumatologic diagnosis was recorded in only one variant (P1014L). Undifferentiated inflammatory symptoms are frequently observed in patients harboring UBA1M41 mutations [6, 28, 45, 46], and our records on patients carrying UBA1non-M41 variants would contribute to the shaping of VEXAS Syndrome as a spectrum of inflammatory manifestations.
As the uniqueness and pathogenesis of UBA1M41 somatic variants in hematopoietic precursors pertains to its inflammogenicity, we analyzed the transcriptome with respect to inflammation-related signatures. However, no UBA1M41-specific signature could be identified. Furthermore, activation of inflammatory pathways relevant in VEXAS syndrome were neither specific nor sensitive in comparison to other patients with MDS. The retrospective nature of the dataset did not allow us to assign the state of active inflammation or reception of anti-inflammatory treatment, leaving the possibility that specific VEXAS inflammation is detectable when patients at disease onset are compared. However, 10–20% of patients with MDS show treatment-refractory systemic autoimmune and inflammatory diseases [22] with UBA1M41 variants only identifiable in the minority [47, 48]. Moreover, since not only autoinflammatory (dysregulation of innate immunity) but also autoimmune diseases (dysregulation of acquired immunity/antibody-mediated) can be part of VEXAS presentation [49], it may be that a transcriptomic signature specific for the inflammation seen in VEXAS does not exist. This necessitates the emphasis of UBA1 somatic mutations in the definition of VEXAS rather than defining the disease based on the co-existence of inflammatory symptoms and cytopenias, which can be observed in patients with MDS harboring other mutations [23, 50, 51] and chromosomal abnormalities [52, 53].
Although only anecdotal, an intriguing finding in our cohort are the described recurrent malignancies in the skin for two MDS patients carrying putative UBA1non-M41 somatic variants. Skin cancer (Merkel cell carcinoma) has recently been reported in one CMML patient carrying a S56F variant [48]. In addition to being apoptotic, UBA1 total loss of function variants are known to para-clonally cause an overgrowth of the surrounding wild type tissues [35, 36]. Moreover, UBA1M41 variants were reported to lead to para-clonal cutaneous involvements in VEXAS syndrome [54]. Thus, non-hematological co-malignancies may also have relevance to the presence of UBA1 variants by para-clonal effect.
Because of the retrospective nature of our study and the associated incompleteness of the clinical records, our work can only provide a limited assessment of the clinical relevance of UBA1non-M41 variants. In addition, we did not have the option to investigate matched germline material to definitively assign somatic and germline status. As VEXAS pertains to somatic variants, we adopted a conservative threshold of 90% for males to assign somatic status. However UBA1M41 somatic variants in VEXAS patients may show exceedingly high VAF (>90%) [55, 56], and the UBA1non-M41 variants classified as unknown and putative germlines may well have been somatic. Furthermore, it should be noted that rare germline variants are sometimes found as somatic variants in MDS/AML patients [57, 58], and the assignment of germline origin does not exclude its potential pathogenicity or predisposition to the disease. One of the identified variants (R551C) was classified as putative somatic in a patient with T-NHL, whereas it was classified as unknown in a patient with AML, and the classification approach is limited in understanding the effect of the variant on the function of the gene. Finally, the transcriptomic analysis was limited because of the relatively small number of patients with UBA1 putative somatic variants in our cohort.
In summary, by applying comprehensive WGTS data analyses we identified several potentially clinically relevant UBA1non-M41 variants that contribute to a more detailed and exhaustive description of the landscape of UBA1 variants in hematologic malignancies. Further functional studies of these novel UBA1 variants are warranted to understand their contribution to the VEXAS phenotype. However, given the broad spectrum of identified variants, we recommend that future VEXAS studies should include the entire UBA1 gene for analysis in order to better understand the variants causing this rare inflammatory syndrome.
Supplementary information
Acknowledgements
The authors thank all coworkers at the MLL Munich Leukemia Laboratory for their dedicated work and all the physicians who provided samples, cared for patients, and collected data.
Author contributions
MS, PB, WW and TH designed the study; MS, WW and MM were responsible for bioinformatic analyses, MM, CH, WK, and TH for diagnostics, MS, ML, UMM and PB for clinical data interpretation; MS drafted the manuscript and all authors read and contributed to the final version of the manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 953407. PB received grant support by Torsten Haferlach Leukämiediagnostik Stiftung.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
CH, WK, and TH declare part ownership of Munich Leukemia Laboratory (MLL). MS, MM and WW are employed by the MLL.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-023-01857-5.
References
- 1.Loda A, Brandsma JH, Vassilev I, Servant N, Loos F, Amirnasr A, et al. Genetic and epigenetic features direct differential efficiency of Xist-mediated silencing at X-chromosomal and autosomal locations. Nat Commun. 2017;8:690. doi: 10.1038/s41467-017-00528-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haupt S, Caramia F, Herschtal A, Soussi T, Lozano G, Chen H, et al. Identification of cancer sex-disparity in the functional integrity of p53 and its X chromosome network. Nat Commun. 2019;10:5385. doi: 10.1038/s41467-019-13266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulkarni M, Smith HE. E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 2008;4:e1000131. doi: 10.1371/journal.pgen.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck DB, Ferrada MA, Sikora KA, Ombrello AK, Collins JC, Pei W, et al. Somatic mutations in UBA1 and severe adult-onset autoinflammatory disease. N. Engl J Med. 2020;383:2628–38. doi: 10.1056/NEJMoa2026834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgin-Lavialle S, Terrier B, Guedon AF, Heiblig M, Comont T, Lazaro E, et al. Further characterization of clinical and laboratory features in VEXAS syndrome: large-scale analysis of a multicentre case series of 116 French patients. Br J Dermatol. 2022;186:564–74. doi: 10.1111/bjd.20805. [DOI] [PubMed] [Google Scholar]
- 6.Poulter J, Consortium UV, Morgan A, Cargo C, Savic S. A high-throughput amplicon screen for somatic UBA1 variants in cytopenic and giant cell arteritis cohorts. J Clin Immunol. 2022;42:947–51. doi: 10.1007/s10875-022-01258-w. [DOI] [PubMed] [Google Scholar]
- 7.Poulter JA, Collins JC, Cargo C, De Tute RM, Evans P, Ospina Cardona D, et al. Novel somatic mutations in UBA1 as a cause of VEXAS syndrome. Blood. 2021;137:3676–81. doi: 10.1182/blood.2020010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palomo L, Meggendorfer M, Hutter S, Twardziok S, Adema V, Fuhrmann I, et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood. 2020;136:1851–62. doi: 10.1182/blood.2019004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swerdlow SHCE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: International Agency for Research on Cancer, 2017; 2017.
- 10.Stengel A, Shahswar R, Haferlach T, Walter W, Hutter S, Meggendorfer M, et al. Whole transcriptome sequencing detects a large number of novel fusion transcripts in patients with AML and MDS. Blood Adv. 2020;4:5393–401. doi: 10.1182/bloodadvances.2020003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raczy C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH, et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 2013;29:2041–3. doi: 10.1093/bioinformatics/btt314. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, de Jesus AA, Brooks SR, Liu Y, Huang Y, VanTries R, et al. Development of a validated interferon score using nanostring technology. J Interferon Cytokine Res. 2018;38:171–85. doi: 10.1089/jir.2017.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice GI, Melki I, Fremond ML, Briggs TA, Rodero MP, Kitabayashi N, et al. Assessment of type I interferon signaling in pediatric inflammatory disease. J Clin Immunol. 2017;37:123–32. doi: 10.1007/s10875-016-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–78. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 20.Lv Z, Williams KM, Yuan L, Atkison JH, Olsen SK. Crystal structure of a human ubiquitin E1-ubiquitin complex reveals conserved functional elements essential for activity. J Biol Chem. 2018;293:18337–52. doi: 10.1074/jbc.RA118.003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang J, Barker B, Bolanos L, Liu X, Jerez A, Makishima H, et al. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-kappaB gene network. Cell Rep. 2014;8:1328–38. doi: 10.1016/j.celrep.2014.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mekinian A, Grignano E, Braun T, Decaux O, Liozon E, Costedoat-Chalumeau N, et al. Systemic inflammatory and autoimmune manifestations associated with myelodysplastic syndromes and chronic myelomonocytic leukaemia: a French multicentre retrospective study. Rheumatol (Oxf) 2016;55:291–300. doi: 10.1093/rheumatology/kev294. [DOI] [PubMed] [Google Scholar]
- 23.Zhao LP, Boy M, Azoulay C, Clappier E, Sebert M, Amable L, et al. Genomic landscape of MDS/CMML associated with systemic inflammatory and autoimmune disease. Leukemia. 2021;35:2720–4. doi: 10.1038/s41375-021-01152-1. [DOI] [PubMed] [Google Scholar]
- 24.Bourbon E, Heiblig M, Gerfaud Valentin M, Barba T, Durel CA, Lega JC, et al. Therapeutic options in VEXAS syndrome: insights from a retrospective series. Blood. 2021;137:3682–4. doi: 10.1182/blood.2020010177. [DOI] [PubMed] [Google Scholar]
- 25.Heiblig M, Patel BA, Groarke EM, Bourbon E, Sujobert P. Toward a pathophysiology inspired treatment of VEXAS syndrome. Semin Hematol. 2021;58:239–46. doi: 10.1053/j.seminhematol.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Heiblig M, Ferrada MA, Koster MT, Barba T, Gerfaud-Valentin M, Mekinian A, et al. Ruxolitinib is more effective than other JAK inhibitors to treat VEXAS syndrome: a retrospective multicenter study. Blood. 2022;140:927–31. doi: 10.1182/blood.2022016642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirino Y, Takase-Minegishi K, Tsuchida N, Hirahara L, Kunishita Y, Yoshimi R, et al. Tocilizumab in VEXAS relapsing polychondritis: a single-center pilot study in Japan. Ann Rheum Dis. 2021;80:1501–2. doi: 10.1136/annrheumdis-2021-220876. [DOI] [PubMed] [Google Scholar]
- 28.Sakuma M, Tanimura A, Yasui S, Ishiguro K, Kobayashi T, Ohshiro Y, et al. A Case of polychondritis-onset refractory organizing pneumonia with cytopaenia diagnosed as VEXAS syndrome: the disease course of 7 years. Rheumatol (Oxf) 2021;60:e356–e9. doi: 10.1093/rheumatology/keab349. [DOI] [PubMed] [Google Scholar]
- 29.Arima K, Kinoshita A, Mishima H, Kanazawa N, Kaneko T, Mizushima T, et al. Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc Natl Acad Sci USA. 2011;108:14914–9. doi: 10.1073/pnas.1106015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brehm A, Liu Y, Sheikh A, Marrero B, Omoyinmi E, Zhou Q, et al. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J Clin Invest. 2015;125:4196–211. doi: 10.1172/JCI81260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jesus AA, Hou Y, Brooks S, Malle L, Biancotto A, Huang Y, et al. Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J Clin Invest. 2020;130:1669–82. doi: 10.1172/JCI129301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck DB, Werner A, Kastner DL, Aksentijevich I. Disorders of ubiquitylation: unchained inflammation. Nat Rev Rheumatol. 2022;18:435–47. doi: 10.1038/s41584-022-00778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck DB, Bodian DL, Shah V, Mirshahi UL, Kim J, Ding Y, et al. Estimated prevalence and clinical manifestations of UBA1 variants associated with VEXAS syndrome in a clinical population. JAMA. 2023;329:318–24. doi: 10.1001/jama.2022.24836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hage-Sleiman M, Lalevee S, Guermouche H, Favale F, Chaquin M, Battistella M, et al. Dominance of an UBA1 mutant clone over a CALR mutant clone: from essential thrombocytemia to VEXAS. Haematologica. 2021;106:3245–8. doi: 10.3324/haematol.2021.279418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, Lackey M, et al. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development. 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfleger CM, Harvey KF, Yan H, Hariharan IK. Mutation of the gene encoding the ubiquitin activating enzyme ubal causes tissue overgrowth in Drosophila. Fly (Austin) 2007;1:95–105. doi: 10.4161/fly.4285. [DOI] [PubMed] [Google Scholar]
- 37.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–9. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 38.Hann ZS, Ji C, Olsen SK, Lu X, Lux MC, Tan DS, et al. Structural basis for adenylation and thioester bond formation in the ubiquitin E1. Proc Natl Acad Sci USA. 2019;116:15475–84. doi: 10.1073/pnas.1905488116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lao T, Chen S, Sang N. Two mutations impair the stability and function of ubiquitin-activating enzyme (E1) J Cell Physiol. 2012;227:1561–8. doi: 10.1002/jcp.22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokgoz Z, Bohnsack RN, Haas AL. Pleiotropic effects of ATP.Mg2+ binding in the catalytic cycle of ubiquitin-activating enzyme. J Biol Chem. 2006;281:14729–37. doi: 10.1074/jbc.M513562200. [DOI] [PubMed] [Google Scholar]
- 41.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl J Med. 2011;365:1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen SK, Lima CD. Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer. Mol Cell. 2013;49:884–96. doi: 10.1016/j.molcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen DD, Jiang JY, Lu LF, Zhang C, Zhou XY, Li ZC, et al. Zebrafish Uba1 degrades IRF3 through K48-linked ubiquitination to inhibit IFN production. J Immunol. 2021;207:512–22. doi: 10.4049/jimmunol.2100125. [DOI] [PubMed] [Google Scholar]
- 44.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;17:1206. doi: 10.1016/j.celrep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oganesyan A, Hakobyan Y, Terrier B, Georgin-Lavialle S, Mekinian A. Looking beyond VEXAS: coexistence of undifferentiated systemic autoinflammatory disease and myelodysplastic syndrome. Semin Hematol. 2021;58:247–53. doi: 10.1053/j.seminhematol.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Ferrada MA, Savic S, Ospina Cardona D, Collins JC, Alessi H, Gutierrez-Rodrigues F, et al. Translation of cytoplasmic UBA1 contributes to VEXAS syndrome pathogenesis. Blood. 2022;140:1496–506. doi: 10.1182/blood.2022016985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao LP, Schell B, Sebert M, Kim R, Lemaire P, Boy M, et al. Prevalence of UBA1 mutations in MDS/CMML patients with systemic inflammatory and auto-immune disease. Leukemia. 2021;35:2731–3. doi: 10.1038/s41375-021-01353-8. [DOI] [PubMed] [Google Scholar]
- 48.Gurnari C, Mannion P, Pandit I, Pagliuca S, Voso MT, Maciejewski JP, et al. UBA1 screening in sweet syndrome with hematological neoplasms reveals a novel association between VEXAS and chronic myelomonocytic leukemia. Hemasphere. 2022;6:e775. doi: 10.1097/HS9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A, Naidu G, Deo P, Beck DB. VEXAS syndrome with systemic lupus erythematosus: expanding the spectrum of associated conditions. Arthritis Rheumatol. 2022;74:369–71. doi: 10.1002/art.41957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh YJ, Shin DY, Hwang SM, Kim SM, Im K, Park HS, et al. Mutation of ten-eleven translocation-2 is associated with increased risk of autoimmune disease in patients with myelodysplastic syndrome. Korean J Intern Med. 2020;35:457–64. doi: 10.3904/kjim.2018.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watad A, Kacar M, Bragazzi NL, Zhou Q, Jassam M, Taylor J, et al. Somatic mutations and the risk of undifferentiated autoinflammatory disease in MDS: an under-recognized but prognostically important complication. Front Immunol. 2021;12:610019. doi: 10.3389/fimmu.2021.610019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wesner N, Drevon L, Guedon A, Fraison JB, Trad S, Kahn JE, et al. Inflammatory disorders associated with trisomy 8-myelodysplastic syndromes: French retrospective case-control study. Eur J Haematol. 2019;102:63–9. doi: 10.1111/ejh.13174. [DOI] [PubMed] [Google Scholar]
- 53.Nanah R, Zblewski D, Patnaik MS, Begna K, Ketterling R, Iyer VN, et al. Deletion 5q is frequent in myelodysplastic syndrome (MDS) patients diagnosed with interstitial lung diseases (ILD): Mayo Clinic experience. Leuk Res. 2016;50:112–5. doi: 10.1016/j.leukres.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Lacombe V, Beucher A, Urbanski G, Le Corre Y, Cottin L, Croue A, et al. Distinction between clonal and paraclonal cutaneous involvements in VEXAS syndrome. Exp Hematol Oncol. 2022;11:6. doi: 10.1186/s40164-022-00262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuchida N, Kunishita Y, Uchiyama Y, Kirino Y, Enaka M, Yamaguchi Y, et al. Pathogenic UBA1 variants associated with VEXAS syndrome in Japanese patients with relapsing polychondritis. Ann Rheum Dis. 2021;80:1057–61. doi: 10.1136/annrheumdis-2021-220089. [DOI] [PubMed] [Google Scholar]
- 56.van der Made CI, Potjewijd J, Hoogstins A, Willems HPJ, Kwakernaak AJ, de Sevaux RGL, et al. Adult-onset autoinflammation caused by somatic mutations in UBA1: A Dutch case series of patients with VEXAS. J Allergy Clin Immunol. 2022;149:432–9. doi: 10.1016/j.jaci.2021.05.014. [DOI] [PubMed] [Google Scholar]
- 57.Cheah JJC, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germline DDX41 mutation. Int J Hematol. 2017;106:163–74. doi: 10.1007/s12185-017-2260-y. [DOI] [PubMed] [Google Scholar]
- 58.Lewinsohn M, Brown AL, Weinel LM, Phung C, Rafidi G, Lee MK, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127:1017–23. doi: 10.1182/blood-2015-10-676098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.