Abstract
Haemophilus influenzae can utilize different protein-bound forms of heme for growth in vitro. A previous study (I. Maciver, J. L. Latimer, H. H. Liem, U. Muller-Eberhard, Z. Hrkal, and E. J. Hansen. Infect. Immun. 64:3703–3712, 1996) indicated that nontypeable H. influenzae (NTHI) strain TN106 expressed a protein that bound hemoglobin-haptoglobin and was encoded by an open reading frame (ORF) that contained a CCAA nucleotide repeat. Southern blot analysis revealed that several NTHI strains contained between three and five chromosomal DNA fragments that bound an oligonucleotide probe for CCAA repeats. Three ORFs containing CCAA repeats were identified in NTHI strain N182; two of these ORFs were arranged in tandem. The use of translational fusions involving these three ORFs and the β-lactamase gene from pBR322 revealed that these three ORFs, designated hgbA, hgbB, and hgbC, encoded proteins that could bind hemoglobin, hemoglobin-haptoglobin, or both compounds. Monoclonal antibodies (MAbs) specific for the HgbA, HgbB, and HgbC proteins were produced by immunizing mice with synthetic peptides unique to each protein. Both HgbA and HgbB were readily detected by Western blot analysis in N182 cells grown in the presence of hemoglobin as the sole source of heme, whereas expression of HgbC was found to be much less abundant than that of HgbA and HgbB. The use of these MAbs in a colony blot radioimmunoassay analysis revealed that expression of both HgbA and HgbB was subject to phase variation. PCR and nucleotide sequence analysis were used in conjunction with Western blot analyses to demonstrate that this phase variation involved the CCAA repeats in the hgbA and hgbB ORFs.
All Haemophilus influenzae strains have an absolute requirement for heme for aerobic growth because they are unable to convert δ-aminolevulinic acid to protoporphyrin IX, the immediate biosynthetic precursor of heme (13, 45). Analysis of the H. influenzae Rd genome (11) revealed the genetic basis for this growth requirement in that many of the genes encoding the relevant enzymes are missing in H. influenzae (42). Therefore, H. influenzae has evolved or acquired mechanisms for the binding and transport of exogenously supplied heme because aerobic growth and acquisition of heme by H. influenzae are absolutely codependent (10).
When H. influenzae is grown in vitro, free heme satisfies the porphyrin requirements (13) and, in part, the iron requirements (8) of this organism. For growth in vivo, however, H. influenzae faces a major impediment to heme acquisition. Free heme is toxic, and the human body possesses highly specific mechanisms for the complexing of this tetrapyrrole molecule (18). The abundant serum proteins albumin and hemopexin bind heme avidly, with Kd values of 10−8 and 10−13 M, respectively (17, 38). Under normal physiologic conditions, all circulating heme will be complexed to hemopexin because this glycoprotein has a much greater affinity for heme than does albumin (17). In addition, much of the body's heme is present in the form of hemoglobin. While free hemoglobin can be utilized readily by H. influenzae growing in vitro (40), the small amount of circulating free hemoglobin (i.e., that not present in erythrocytes) is tightly complexed (Kd, ∼10−23 M) by the serum protein haptoglobin (2).
A previous study from our laboratory identified a 115-kDa outer membrane protein (HhuA), expressed by nontypeable H. influenzae (NTHI) strain TN106, that was involved in the binding and utilization of hemoglobin-haptoglobin (23). The HhuA protein exhibited features typical of a TonB-dependent outer membrane receptor, having significant homology with other TonB-dependent proteins over the regions characteristic of these proteins (22). Elimination of expression of the hhuA gene product decreased but did not eliminate the ability of this NTHI strain to utilize hemoglobin-haptoglobin for aerobic growth, a finding which suggested the existence of an alternative mechanism or pathway for utilization of this heme-protein complex.
A striking feature of the hhuA gene is the presence of a four-nucleotide (CCAA) repeat motif near the start of this open reading frame (ORF). Inspection of the H. influenzae Rd genome (11) revealed that several other predicted ORFs shared this feature and encoded predicted proteins that were likely to be TonB dependent. The presence of these additional ORFs containing the CCAA repeat raised the possibility that there might exist a family of hemoglobin- or hemoglobin-haptoglobin-binding outer membrane proteins in H. influenzae. Moreover, expression of the encoded proteins might be affected by the recombination-independent slippage mechanism (i.e., slipped-strand mispairing) known to mediate the phase variation of surface antigens encoded by genes with homopolymeric or heteropolymeric nucleotide repeats. The latter include the PII protein of Neisseria gonorrhoeae (27), the enzymes that synthesize the lipooligosaccharide of H. influenzae (43), and hemoglobin-binding outer membrane proteins of both N. gonorrhoeae (3) and N. meningitidis (19, 34).
In the present study, we identified three genes in NTHI strain N182 that contain CCAA repeats and encode proteins very similar to HhuA. We used translational fusions to prove that these proteins can bind hemoglobin or hemoglobin-haptoglobin. Monoclonal antibodies (MAbs) were used to detect phase variation in the expression of these NTHI proteins.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type NTHI strain N182 and nine additional NTHI strains have been described previously (5). All NTHI isolates were cultured routinely in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) with NAD (10 μg/ml; Sigma Chemical Co., St. Louis, Mo.) (BHI) and hemin chloride (50 μg/ml; Sigma) (BHI-Hm). Isolates were also grown in BHI containing NAD and human hemoglobin (100 μg/ml; Sigma) (BHI-Hg). Addition of ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDA; Sigma) to a final concentration of 100 μM in broth media was used for iron chelation. All broth cultures were grown at 37°C with aeration; agar-solidified media were incubated at 37°C in an atmosphere of 95% air–5% CO2. Escherichia coli strain DH5α and recombinants derived from it were grown at 37 or 30°C on Luria-Bertani medium (36) supplemented with ampicillin (100 μg/ml) or tetracycline (15 μg/ml), as required.
Recombinant DNA methods.
Standard recombinant DNA methods, including restriction enzyme digestions, alkaline phosphatase reactions, ligation reactions, agarose gel electrophoresis, and plasmid purification, were performed as previously described (36) or in accordance with the manufacturer's instructions. Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). Shrimp alkaline phosphatase was purchased from USB (Cleveland, Ohio). T4 DNA ligase was purchased from GIBCO-BRL (Bethesda, Md.). Plasmid DNA was prepared with the Wizard Plus Miniprep DNA Purification System (Promega, Madison, Wis.) and the Qiagen Plasmid Midi Kit (Qiagen Inc., Valencia, Calif.). Chromosomal DNA was isolated by the method of Marmur (24).
Southern blot analysis.
NTHI chromosomal DNA was digested to completion with a variety of restriction enzymes and probed by Southern blot analysis with an oligonucleotide probe that consisted of five consecutive repeats of the tetranucleotide CCAA. This 20-mer was labeled by using the chemiluminescence-based Renaissance Oligonucleotide 3′ End-Labeling Kit (NEN, Boston, Mass.) as described by the manufacturer.
PCR.
PCR was performed with either the GeneAmp XL PCR Kit (Perkin-Elmer Corp., Foster City, Calif.) or the Taq DNA Polymerase Kit (Promega). To amplify products from N182 genomic DNA, 1 μg of chromosomal DNA and 100 ng of each primer were used in a 100-μl reaction mixture. PCR products used for nucleotide sequence analysis were purified by agarose gel electrophoresis, followed by the use of the Wizard PCR DNA Purification System (Promega) in accordance with the manufacturer's directions. PCR products used for the construction of translational fusions were first digested with PstI and then subjected to gel purification as described above. The compositions of the oligonucleotide primers used to generate various PCR products are listed in Table 1.
TABLE 1.
Primers used to generate PCR products with N182 chromosomal DNA
| Primer | Sequencea |
|---|---|
| P1 | ACACGAAGCCAATCTGTGGG |
| P2 | ACAGGCGGATTTAAGAGCGG |
| P3 | CTTTCACAATATCATCGGG |
| P4 | AAAAAGGGATACGCTACG |
| P5 | AACTGCAGAACATTCCCTAACGTCGG |
| P6 | AACTGCAGCTACCGAAAATAGTGATTCG |
| P7 | AACTGCAGCGCATTAGTAGGAGACTGG |
| P8 | AACTGCAGTAGCGAAACAGCAGGCGTC |
| P9 | AACTGCAGCATTTTCGAGAAGCTCCG |
| P10 | AACTGCAGTTTCTGAACAACTAGAG |
| P11 | TTCAATGATATGGGCAGG |
| P12 | AACATTCCCTAACGTCGG |
| P13 | AGGGAACTGCAAATCCTG |
| P14 | TTTCACGGGTAGGACCAAC |
| P15 | CAAGAAATTGCGTTGCTG |
| P16 | TGCCAGACTCGTTCTATCC |
| P17 | TTCCACAACACTGTGACGC |
| P18 | AAATATCCAATGCAGGCG |
| P19 | CGGGATCCTCAAAATCCACACCGAAC |
| P20 | TGTTTTACCACGAGTCCC |
| P21 | GATCACGAGAATCAGACG |
| P22 | ATTTCACCCTCGCTACCAG |
The underlined sequence denotes a PstI site added to the primer.
Recombinant plasmid construction.
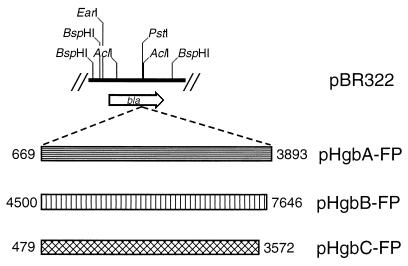
Plasmids p712-2, p661-1, and p661-8 were constructed by ligating DNA fragments, derived from the use of the GeneAmp XL PCR kit, into the TA Cloning Kit vector pCRII (Invitrogen, San Diego, Calif.). The 1.4-kb insert in p712-2 was generated from N182 chromosomal DNA by using the oligonucleotide primers P1 and P2. The two different 1.4-kb inserts in p661-1 and p661-8 were generated with primers P3 and P4. Translational fusions involving β-lactamase were generated by inserting PCR-derived partial ORFs of the hgbA, hgbB, and hgbC genes into the PstI site within the bla gene in pBR322. The 3,225-nucleotide (nt) insert in pHgbA-FP was obtained by PCR with the primers P5 and P6. Similarly, the 3,147-nt insert in pHgbB-FP and the 3,058-nt insert in pHgbC-FP were obtained by using primers P7-P8 and P9-P10, respectively.
Colony blot hybridization.
N182 chromosomal DNA that had been partially digested with Sau3AI was ligated into pBluescript II SK+ (Stratagene, La Jolla, Calif.) and used to transform E. coli DH5α. Total DNA from each transformant colony was hybridized with either the 1.4-kb insert from either p712-2 or p661-1 as previously described (36). The DNA probes were labeled by using [α-32P]dCTP and the Random Primed DNA Labeling Kit (Boehringer Mannheim, Indianapolis, Ind.) in accordance with the manufacturer's instructions. Two transformants were found to react with the p712-2-derived DNA probe, and 10 transformants were identified that bound with the p661-1-derived DNA probe.
Nucleotide sequence analysis.
Both strands of three overlapping PCR products generated from N182 chromosomal DNA with primers P11 and P12 (3.9 kb), primers P13 and P14 (3.4 kb), and primers P15 and P16 (4.1 kb) were sequenced in their entirety using a model 373A Automated DNA Sequencer (Applied Biosystems, Foster City, Calif.). These overlapping PCR products encompassed the complete sequence of the tandem hgbA and hgbB genes. The nucleotide sequence of the hgbC gene was derived from the 3.9-kb PCR product generated with primers P17 and P18. In each instance, at least three independent PCRs were performed to obtain the DNA segments which were then pooled for sequence analysis. PCR products used to determine the numbers of CCAA repeats in the various hgb genes were generated by using the oligonucleotide primers P19 and P20 for the hgbA gene, P15 and P21 for the hgbB gene, and P17 and P22 for the hgbC gene. Nucleotide sequence data were analyzed by using the MacVector analysis package (version 6.5; Oxford Molecular Group, Campbell, Calif.).
MAbs.
Spleens from mice individually immunized with three different keyhole limpet hemocyanin-conjugated synthetic peptides, derived from the HgbA, HgbB, and HgbC proteins, were fused with SP2/0-Ag14 plasmacytoma cells as previously described (35). These peptides included KEINNTTTPNSNSNKDKTYDFSKL from HgbA, KDSFNSQWTSMVERKEKQYTDITDIK from HgbB, and KFARIKDRKDKNNRDNRKIK from HgbC. The resultant lymphocyte hybridomas were screened in an enzyme-linked immunosorbent assay using an ovalbumin-conjugated form of the immunizing peptide as the antigen. Antibodies reactive in the enzyme-linked immunosorbent assay were then screened by Western blotting using a lysate of heme- and iron-starved NTHI N182 cells as the antigen. This approach resulted in the production of HgbA-specific MAb 17H3, HgbB-specific MAb 4B3, and HgbC-reactive MAb 12A2. All three of these MAbs were shown to readily bind their respective antigens (expressed as fusion proteins) by Western blot analysis. It should be noted that HgbC-reactive MAb 12A2 bound a doublet in a Western blot analysis of NTHI whole-cell lysates. The upper band of this doublet was HgbC; the identity of the lower band is not known. MAb 6B8 was used to detect the iron-regulated H. influenzae HitA protein (37).
SDS-PAGE, Western blotting, and colony blot RIA.
Whole-cell lysates of NTHI (29) were subjected to SDS-PAGE, followed by Coomassie blue staining or Western blot analysis as previously described (6). NTHI N182 colonies were examined for reactivity with HgbA-specific MAb 17H3 and HgbB-specific MAb 4B3 in the colony blot radioimmunoassay (RIA) (14). With MAb 17H3, this assay was performed as previously described (14). With MAb 4B3, the colonies were first lifted onto a nitrocellulose membrane filter (Schleicher & Schuell, Keene, N.H.), which was then laid atop a filter pad (Gel Blot Paper; Schleicher & Schuell), presoaked with 62.5 mM Tris-HCl (pH 6.8) containing 10% (wt/vol) SDS. The nitrocellulose was left on this filter pad for 10 min at room temperature prior to incubation with the blocking agent. HgbC-reactive MAb 12A2 did not function in colony blot RIA analysis.
Detection of hemoglobin- and hemoglobin-haptoglobin-binding activities.
NTHI strain N182 was screened for the ability to bind radioiodinated hemoglobin and hemoglobin-haptoglobin as previously described (23). (It must be noted that both the hemoglobin and haptoglobin in the hemoglobin-haptoglobin were radioiodinated.) To detect binding activity in the HgbA-FP, HgbB-FP, and HgbC-FP fusion proteins, total cell membranes were prepared from each recombinant E. coli strain using the Peripreps Periplasting Kit (Epicentre Technologies Corp., Madison, Wis.) and equal amounts of each fusion protein in membranes (as determined by SDS-PAGE and Coomassie blue staining) were loaded onto nitrocellulose membranes using a dot blot apparatus (Schleicher & Schuell) and tested for hemoglobin- and hemoglobin-haptoglobin-binding activities.
Detection of expression of Hgb proteins.
NTHI N182 cells were grown in BHI-Hm broth to mid-exponential phase, at which point the cells were serially diluted and plated onto both BHI-Hm and BHI-Hg agar plates. Approximately 900 of the resultant heme-grown colonies were tested for reactivity with HgbA-specific MAb 17H3 using the colony blot RIA; another 900 heme-grown colonies were tested for the ability to bind HgbB-specific MAb 4B3. Two sets of approximately 300 colonies each grown on BHI-Hg agar were tested for reactivity with these two MAbs. In a second experiment, N182 cells were grown to mid-exponential phase in both BHI-Hm and BHI-Hg broth and the resultant cells were serially diluted and plated onto agar plates of the homologous medium. Approximately 300 colonies from each set of plates were tested for reactivity with the two MAbs described above. From these plates, five MAb-reactive colonies and five non-MAb-reactive colonies were selected and streaked for the isolation of single colonies on the same medium. Each of the 40 isolates was then passaged two more times on the same medium. The final isolates were grown in the homologous broth medium for Western blot analysis.
Nucleotide sequence accession numbers.
The nucleotide sequences of the NTHI N182 hgbA, hgbB and hgbC genes were deposited in the GenBank database and assigned accession numbers AF221059 (for hgbA and hgbB) and AF221060 (for hgbC).
RESULTS
Determination of the number of NTHI genes bearing CCAA repeats.
We previously identified a gene (hhuA) in NTHI strain TN106 that contained a CCAA tetranucleotide repeat motif and encoded an outer membrane protein involved in the binding of hemoglobin-haptoglobin (23). However, an isogenic hhuA mutant was shown to still utilize both hemoglobin-haptoglobin and hemoglobin (23). Examination of the genome of H. influenzae strain Rd (11) revealed that it possessed four possible ORFs which encoded proteins homologous to HhuA and which also contained CCAA repeat motifs. We decided to investigate whether NTHI strains also possessed multiple genes with CCAA tetranucleotide repeats in their genomes.
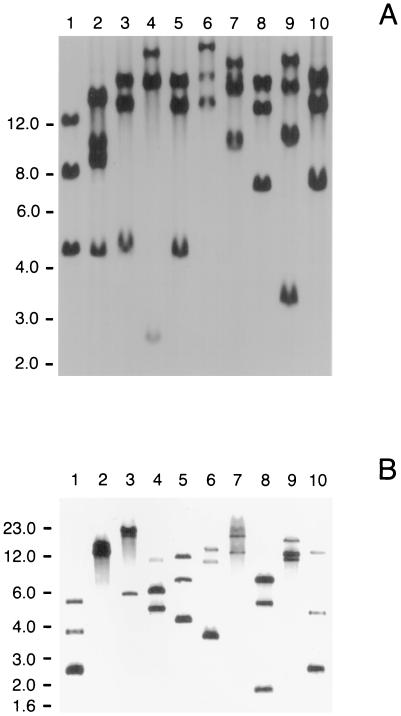
Chromosomal DNAs from 10 NTHI strains were digested with EcoRI and probed by Southern blot analysis with an oligonucleotide comprised of five consecutive repeats of CCAA. Each strain had at least three EcoRI fragments that bound this probe (Fig. 1A). Strain N182 (Fig. 1A, lane 1), which appeared to possess only three hybridizing bands, was chosen for further study. When N182 chromosomal DNA was digested to completion with 10 different restriction enzymes and probed with this same oligonucleotide, at most three different fragments in any single digest bound the CCAA probe (Fig. 1B).
FIG. 1.
Southern blot analysis of NTHI chromosomal DNA. The oligonucleotide probe was comprised of five consecutive repeats of the tetranucleotide CCAA. Panel A contains chromosomal DNA preparations from 10 NTHI strains that were digested to completion with EcoRI. Lanes: 1, N182; 2, TN106; 3, AAR200; 4, AAR203; 5, 152; 6, BF105; 7, KE2; 8, OC201; 9, BI102; 10, BO-2. Panel B contains NTHI strain N182 chromosomal DNA digested with 10 different restriction enzymes. Lanes: 1, AvaI; 2, BspHI; 3, Bsu36I; 4, EarI; 5, EcoRI; 6, EcoRV; 7, MluI; 8, AclI; 9, XbaI; 10, XhoI. Molecular size markers (in kilobases) are listed on the left side of each panel.
Identification of CCAA repeat-containing genes.
Oligonucleotide primers that flanked each of the four CCAA repeat-containing ORFs (HI661, HI635, HI712, and HI1566) found in the H. influenzae Rd genome (11) were used in a PCR with NTHI N182 chromosomal DNA, but no products were obtained. Four additional pairs of primers internal to these ORFs were designed, and two of these (P1 and P2 from the HI712 ORF and P3 and P4 from the HI661 ORF; Table 1) yielded 1.4-kb PCR products when used with N182 chromosomal DNA. These PCR products were cloned into the pCRII vector for nucleotide sequence analysis. The 1.4-kb fragment derived from the HI712-based primers contained a partial ORF that encoded a protein that was 82% identical to the predicted protein product of the H. influenzae Rd HI712 ORF. A single recombinant clone (p712-2) was chosen for further analysis. Sequence analysis of two recombinant clones (p661-1 and p661-8) generated from the HI661 internal primers showed two different incomplete ORFs that encoded protein products that were 84 and 40% identical, respectively, to the predicted protein product of the HI661 ORF in H. influenzae Rd and 39% identical to each other.
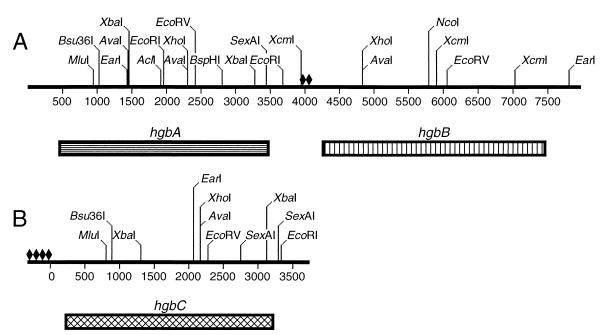
The remaining nucleotide sequences flanking these three partial ORFs were obtained by screening an N182 genomic library, constructed in E. coli as described in Materials and Methods, with the cloned partial ORFs described above. Compilation of nucleotide sequences derived from the various hybridization-positive recombinant clones revealed that the two complete ORFs containing the nucleotide sequences from the p661-8 and p712-2 DNA inserts were located in tandem in the N182 chromosome; these were designated (based on the fusion protein-based experiments discussed below) hemoglobin-binding proteins A (hgbA) and B (hgbB), respectively (Fig. 2). The complete ORF containing the nucleotide sequence from the p661-1 insert was not linked to these other two ORFs and was designated hgbC (Fig. 2). Immediately upstream from the hgbB ORF were tandem copies of 23-nt elements arranged as inverted repeats; these 23-nt elements were previously described as being associated with DNA duplications in the chromosome of H. influenzae (30). Immediately upstream from the hgbC ORF, there were at least four copies of this same 23-nt element (Fig. 2).
FIG. 2.
Partial restriction enzyme map of the NTHI N182 hgbA, hgbB, and hgbC genes. Panel A contains the tandem hgbA and hgbB genes. Panel B contains the hgbC gene. The boxes beneath the DNA fragments delineate the individuals ORFs. The diamonds indicate the locations of the 23-nt elements (30) described in Results.
Characteristics of the hgbA, hgbB, and hgbC genes and their encoded products.
The hgbA ORF contained 3,039 nt, and the encoded protein consisted of 1,009 amino acids (aa) with a calculated molecular weight of 115,811. The end of the hgbA ORF was separated from the beginning of the hgbB ORF by 855 nt. The 3,201-nt hgbB ORF encoded a predicted protein of 1,067 aa with a calculated molecular weight of 122,462. The hgbC ORF contained 2,979 nt, and its predicted protein had 993 aa and a calculated molecular weight of 113,608. All three ORFs possessed apparent transcriptional terminators located 14 to 27 nt 3′ from the translational stop codons and also putative consensus −35 and −10 promoter sequences located 34 to 41 nt upstream from the translation initiation codons. A dyad repeat sequence with weak homology to the Fur-binding consensus sequence (21) was located upstream from the translation initiation codons in both hgbA and hgbB; a similar dyad repeat sequence was not apparent upstream from the hgbC ORF (data not shown). Consecutive CCAA tetranucleotide repeats were found near the beginnings of all three ORFs, with the hgbA ORF containing 25 repeats, the hgbB ORF containing 19 repeats, and the hgbC ORF containing 9 repeats. (The possible number of CCAA repeats in each ORF varied [e.g., 23, 24, and 25 repeats in hgbA], as determined by nucleotide sequence analyses of PCR products derived independently from N182 chromosomal DNA—the selected numbers listed above for the hgbA, hgbB, and hgbC ORFs would allow full-length expression of each encoded protein as discussed below.)
Each protein appeared to possess a 24-aa leader peptide with a putative signal peptidase I cleavage site (Ala-Tyr-Ala) (Fig. 3). The repetitive amino acid sequence which resulted from the CCAA nucleotide repeats was QPTN and began immediately after the proposed N-terminal amino acid (alanine-25) of the mature protein. The three proteins were very homologous, with HgbA being 48% identical to HgbB and 83% identical to HgbC. HgbB was 44% identical to HgbC. All three proteins also contained a predicted TonB box (EQINVSGST/SEN) (16, 22) located 21 to 42 aa downstream from the proposed start of the mature protein (Fig. 3). Each of these proteins also contained six regions with homology to the six amino acid sequences common to TonB-dependent protein receptors (7, 16) (Fig. 3).
FIG. 3.
Comparison of the deduced amino acid sequences of the HgbA, HgbB, and HgbC proteins from NTHI strain N182. The asterisks indicate identical amino acids, and the dots indicate conservative amino acid substitutions. The arrowhead indicates the position of the putative signal peptidase I cleavage site. The bracket indicates the amino acids encoded by the CCAA repeats. The seven boxes enclose the amino acid sequences that have homology with other TonB-dependent proteins (16, 41).
BLAST analysis revealed that several bacterial outer membrane proteins reported to be involved in hemoglobin utilization possessed high levels of homology to the N182 HgbA, HgbB, and HgbC proteins. Not unexpectedly, those proteins with the greatest amount of homology to the N182 proteins were all from different strains of H. influenzae. The HgpB protein of H. influenzae type b strain HI689 (32) was 82% identical to the HgbA protein of NTHI strain N182 whereas the HgpC protein of this same H. influenzae type b strain (26) was 88% identical to the HgbB protein of N182. The predicted protein encoded by the HI661 ORF from H. influenzae Rd (11) was 83% identical to the HgbC protein of N182. None of these three NTHI N182 proteins was identical to the HhuA protein from NTHI strain TN106 (23); the HgbA and HgbC proteins were the most similar (50% identity) to the HhuA protein. All three of these N182 proteins had 43 to 45% identity with the H. ducreyi HupA (HgbA) outer membrane protein (9, 39) which is involved in the utilization of hemoglobin by this organism. Among non-Haemophilus species, the HpuB protein from N. meningitidis (20) was most similar (35 to 37% similarity) to these three NTHI N182 proteins.
Binding of hemoglobin and hemoglobin-haptoglobin by fusion proteins.
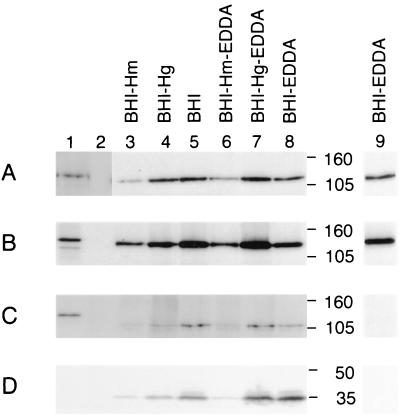
The ability of the HgbA, HgbB, and HgbC proteins to bind hemoglobin or hemoglobin-haptoglobin was examined in an E. coli background. Attempts to clone the intact NTHI N182 hgbA, hgbB, and hgbC genes into E. coli were unsuccessful, so slightly truncated versions of these three ORFs were fused to the β-lactamase gene in pBR322 to produce translational fusions (Fig. 4). Each ORF was truncated at the 5′ end to a point just beyond the end of the CCAA repeat region. Each of the resultant fusion proteins (HgbA-FP, HgbB-FP, or HgbC-FP) was shown to bind its homologous MAb by Western blot analysis (Fig. 5A to C, lane 1).
FIG. 4.
Partial enzyme restriction map of the bla gene in pBR322 depicting the insertion site for the construction of translational fusions involving the hgbA, hgbB, and hgbC genes. The three shaded bars denote the three PCR-derived DNA fragments inserted into the PstI site in pBR322 to construct pHgbA-FP, pHgbB-FP, and pHgbC-FP. The nucleotide numbers were derived from the restriction maps shown in Fig. 2.
FIG. 5.
Western blot-based detection of expression of the HgbA, HgbB, and HgbC proteins by recombinant E. coli strains and NTHI strain N182. Whole-cell lysates of the various strains were probed by Western blot analysis with the following MAbs: panel A, HgbA-specific MAb 17H3; panel B, HgbB-specific MAb 4B3; panel C, HgbC-reactive MAb 12A2; panel D, HitA protein-specific MAb 6B8. Lane 1 in panels A and D contains E. coli DH5α(pHgbA-FP). Lane 1 in panel B contains E. coli DH5α(pHgbB-FP). Lane 1 in panel C contains E. coli DH5α(pHgbC-FP). Lanes in all panels: 2, E. coli DH5α(pBR322); 3, NTHI N182 cells grown in BHI-Hm; 4, N182 cells grown in BHI-Hg; 5, N182 cells grown in BHI; 6, N182 cells grown in BHI-Hm with EDDA; 7, N182 cells grown in BHI-Hg with EDDA; 8, N182 cells grown in BHI with EDDA. Lanes 3 to 8 in panel C had to be exposed longer than the other three panels to detect HgbC. It should be noted that HgbC-reactive MAb 12A2 bound to a doublet in this Western blot; the upper band of the doublet is the HgbC protein. In panels A to C, lane 9 depicts the results obtained when the samples contained in lane 8 were all exposed to film for the same length of time. Molecular size markers (in kilodaltons) are between lanes 8 and 9.
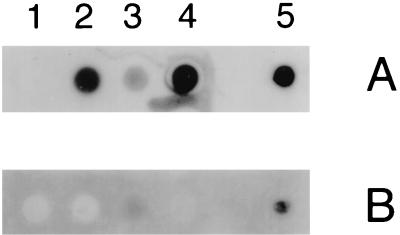
The total membrane fraction was isolated from the E. coli recombinant strains expressing the Hgb fusion proteins as described in Materials and Methods. Equivalent amounts of each fusion protein (in the total membrane fraction) were used in a dot blot assay for detection of hemoglobin- and hemoglobin-haptoglobin-binding activities. Whole cells of NTHI N182 were used as a positive control for hemoglobin and hemoglobin-haptoglobin binding (Fig. 6A and B, row 5). The total membrane fraction from E. coli DH5α(pBR322) was used as a negative control and did not bind either of the radiolabeled compounds (Fig. 6A and B, row 1). HgbA-FP bound the labeled hemoglobin (Fig. 6A, row 2) but did not appear to bind the labeled hemoglobin-haptoglobin (Fig. 6B, row 2). HgbB-FP bound hemoglobin (Fig. 6A, row 3), but to a lesser degree than did HgbA-FP. However, HgbB-FP was capable of binding hemoglobin-haptoglobin (Fig. 6B, row 3), albeit weakly. HgbC-FP bound hemoglobin (Fig. 6A, row 4) but did not detectably bind hemoglobin-haptoglobin (Fig. 6B, row 4).
FIG. 6.
Binding of hemoglobin and hemoglobin-haptoglobin by E. coli-derived fusion proteins. Equivalent amounts of the HgbA-FP, HgbB-FP, and HgbC-FP fusion proteins present in membrane fractions of recombinant E. coli strains were spotted onto nitrocellulose and probed with radioiodinated hemoglobin (A) and hemoglobin-haptoglobin (B). Rows: 1, E. coli DH5α(pBR322); 2, E. coli DH5α(pHgbA-FP);3, E. coli DH5α(pHgbB-FP); 4, E. coli DH5α(pHgbC-FP). Row 5 contains whole cells of NTHI N182 grown on BHI-Hm medium that were used as a control for binding of both radiolabeled compounds. Panels A and B were exposed to film for the same length of time.
Effects of heme and iron limitation on expression of the HgbA, HgbB, and HgbC proteins.
NTHI N182 was grown in a basal medium (BHI containing NAD) with high levels of heme (50 μg/ml), moderate levels of hemoglobin (100 μg/ml), or no added heme source. (H. influenzae can grow aerobically for four to five generations in the absence of exogenous heme—these latter conditions were used to induce heme starvation). N182 was also grown under these same conditions in the presence of the iron chelator EDDA. Western blot analysis revealed that when N182 was starved for both heme and iron, both HgbA (Fig. 5A, lane 9) and HgbB (Fig. 5B, lane 9) were readily detectable whereas HgbC was not apparent (Fig. 5C, lane 9). Long-term exposure of the autoradiogram revealed that HgbC was expressed (Fig. 5C, lane 8), albeit at relatively low levels.
In general, growth of N182 in the presence of a high level of heme resulted in the lowest apparent level of expression of all three proteins (Fig. 5A to C, lane 3). Growth in the presence of hemoglobin (Fig. 5A to C, lane 4) or in the absence of a heme source (Fig. 5A to C, lane 5) resulted in higher levels of expression of all three proteins. Iron limitation in the growth medium (as effected by EDDA) did not have an apparent effect on the expression of these proteins (Fig. 5A to C, lanes 6 to 8) but did affect the expression of the iron-regulated HitA protein (1, 37) (Fig. 5D). Detection of HitA expression was used to verify that the presence of EDDA in the growth medium had induced iron-limited growth conditions.
Expression of HgbA, HgbB, and HgbC by NTHI N182 cells grown with heme or hemoglobin.
The presence of the CCAA repeats in the hgbA, hgbB, and hgbC genes raised the possibilities that expression of these ORFs was regulated by the number of repeats and that phase variation of the encoded proteins could occur (27, 43). In a preliminary effort to ascertain whether HgbA and HgbB were consistently expressed by NTHI N182 cells, we first grew this strain in BHI-Hm broth and then plated these cells onto both BHI-Hm and BHI-Hg agar plates. The resultant individual colonies were evaluated for the ability to bind MAbs specific for the HgbA or HgbB protein in a colony blot RIA. (HgbC-reactive MAb 12A2 was not used in these experiments because it did not function in the colony blot RIA.) It was found that 96% of the colonies from the BHI-Hm agar plates bound the HgbA-specific MAb, while only 11% of the colonies reacted with the HgbB-specific MAb. Among the colonies that developed on the BHI-Hg plates, 98 and 8% bound the HgbA- and HgbB-specific MAbs, respectively. These results indicated that N182 cells, whether grown with heme or with hemoglobin, did not uniformly express HgbA and HgbB. (It should be noted that this experiment did not address the frequency of potential phase variation.)
To determine whether individual N182 isolates expressed all three proteins simultaneously, this NTHI strain was grown in BHI-Hm and in BHI-Hg broth, plated onto the homologous medium solidified with agar, and probed with the HgbA- and HgbB-specific MAbs. Five colonies that bound each MAb and five colonies that failed to bind each MAb were passaged by the single-colony isolation method three times on the homologous medium. The resultant 20 heme-grown and 20 hemoglobin-grown isolates were subjected to Western blot analysis to determine whether each isolate expressed the other two Hgb proteins.
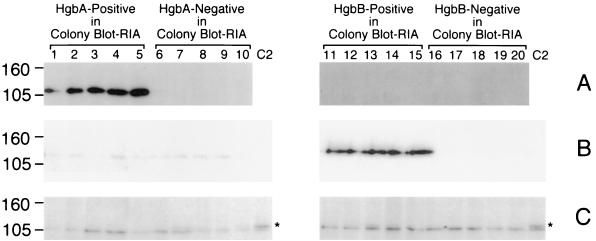
Heme-grown isolates originally identified in the colony blot RIA as reactive or unreactive with the HgbA MAb remained the same after serial passage on BHI-Hm agar (Fig. 7A, lanes 1 to 5 and 6 to 10, respectively). Similarly, isolates that bound the HgbB MAb or failed to bind this MAb in the colony blot RIA maintained these antigenic characteristics after in vitro passage (Fig. 7B, lanes 11 to 15 and 16 to 20, respectively). The 10 heme-grown isolates selected for their MAb reactivity (Fig. 7A, lanes 1 to 5, and B, lanes 11 to 15) expressed only the homologous Hgb protein (i.e., HgbA or HgbB); the other 10 MAb-unreactive isolates (Fig. 7, lanes 6 to 10 and 16 to 20) did not express detectable levels of any Hgb protein. No detectable HgbC protein was expressed by any of the 20 heme-grown isolates (Fig. 7C, lanes 1 to 10 and lanes 11 to 20).
FIG. 7.
Western blot-based detection of the HgbA, HgbB, and HgbC proteins in heme-grown isolates of N182. Equivalent amounts of whole-cell lysates from the 20 BHI-Hm-grown isolates were probed with HgbA-specific MAb 17H3 (A); HgbB-specific MAb 4B3 (B); and HgbC-reactive MAb 12A2 (C). The BHI-Hm-grown isolates identified in the colony blot RIA as positive for HgbA expression are in lanes 1 to 5. The BHI-Hm-grown isolates that were negative for HgbA expression in the colony blot RIA are in lanes 6 to 10. The BHI-Hm-grown isolates positive for HgbB expression in the colony blot RIA are in lanes 11 to 15. The BHI-Hm-grown isolates that were negative for HgbB expression in the colony blot RIA are in lanes 16 to 20. Control lane C2 in panel C contains a cell lysate from N182 cells grown in BHI to allow detection of HgbC. The asterisk beside lane C2 denotes the location of the HgbC protein. It should be noted that the HgbC-reactive MAb bound to a doublet in this Western blot; the upper band of the doublet (marked with the asterisk) is the HgbC protein. Molecular size markers (in kilodaltons) are on the left.
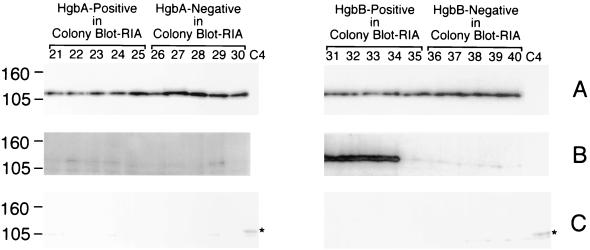
After in vitro passage on BHI-Hg medium, the HgbA protein was expressed by all 20 of the hemoglobin-grown isolates (Fig. 8A, lanes 21 to 30 and 31 to 40), regardless of whether the isolate was originally positive (Fig. 8A, lanes 21 to 25) or negative (Fig. 8A, lanes 26 to 30) with the HgbA MAb in the colony blot RIA. Four of the five isolates originally identified in the colony blot RIA as being reactive with the HgbB MAb remained reactive with this MAb after in vitro passage (Fig. 8B, lanes 31 to 34); these same five isolates also expressed HgbA (Fig. 8A, lanes 31 to 35). The five isolates originally negative for reactivity with the HgbB MAb remained unreactive with this MAb (Fig. 8B, lanes 36 to 40) but did express HgbA (Fig. 8A, lanes 36 to 40). None of these hemoglobin-grown isolates expressed HgbC (Fig. 8C, lanes 21 to 30 and 31 to 40).
FIG. 8.
Western blot-based detection of the HgbA, HgbB, and HgbC proteins in hemoglobin-grown isolates of N182. Equivalent amounts of whole-cell lysates from the 20 BHI-Hg-grown isolates were probed with HgbA-specific MAb 17H3 (A); HgbB-specific MAb 4B3 (B); and HgbC-reactive MAb 12A2 (C). The BHI-Hg-grown isolates identified in the colony blot RIA as positive for HgbA expression are in lanes 21 to 25. The BHI-Hg-grown isolates that were negative for HgbA expression in the colony blot RIA are in lanes 26 to 30. The BHI-Hg-grown isolates positive for HgbB expression in the colony blot RIA are in lanes 31 to 35. The BHI-Hg-grown isolates that were negative for HgbB expression in the colony blot RIA are in lanes 36 to 40. Control lane C4 in panel C contains a lysate from N182 cells grown in BHI to allow detection of HgbC. The asterisk beside lane C4 denotes the location of the HgbC protein. It should be noted that the HgbC-reactive MAb bound to a doublet in this Western blot; the upper band of the doublet (marked with the asterisk) is the HgbC protein. Molecular size markers (in kilodaltons) are on the left.
Effect of CCAA repeats on protein expression.
To determine whether the number of CCAA repeats in the hgbA, hgbB, and hgbC ORFs was involved in control of expression of the Hgb proteins, the region containing these repeats was amplified by PCR from several of the isolates included in Fig. 7 and 8. Two isolates from each group of five (e.g., 1 to 5, 6 to 10, 11 to 15, and 16 to 20 in Fig. 7) were randomly chosen for this analysis. Each heme-grown isolate positive for HgbA expression (4 and 5 in Fig. 7) contained 25 CCAA repeats in its hgbA ORF; this number of repeats would theoretically allow full-length expression of the encoded protein. The HgbA MAb-unreactive isolates grown on heme (9 to 12, 16, and 17 in Fig. 7) had 23, 26, or 27 repeats in their hgbA ORFs; all of these would result in premature translational termination codons in the ORF. The heme-grown HgbB-positive isolates (11 and 12 in Fig. 7) contained 19 repeats in their hgbB ORFs, consistent with the predicted expression of the entire encoded protein. The companion HgbB-negative isolates (4, 5, 9, 10, 16, and 17 in Fig. 7) possessed 20 or 21 CCAA repeats in their hgbB ORFs; these would cause premature translational termination.
All of the hemoglobin-grown isolates (21, 22, 26, 27, 31, 32, 36, and 37 in Fig. 8) contained 22 or 25 CCAA repeats in their hgbA ORFs; both of these numbers of repeats would allow full-length protein expression. The two hemoglobin-grown isolates that expressed the HgbB protein (31 and 32 in Fig. 7) possessed 19 and 22 repeats in their hgbB ORFs, consistent with the potential for full-length protein expression. The hemoglobin-grown HgbB-negative isolates (21, 22, 26, 27, 36, and 37 in Fig. 8) had either 20 or 21 repeats in their hgbB ORFs; either of these would cause premature translational termination. The HgbC protein was not expressed by any of the eight heme-grown or eight hemoglobin-grown isolates subjected to PCR and nucleotide sequence analysis; the presence of either 10 or 11 CCAA repeats in all of the hgbC ORFs examined in these 16 isolates would result in premature termination of translation. These results indicated that protein expression, as detected by MAb reactivity, could be correlated with predicted protein expression as determined by the number of CCAA repeats present in the different ORFs.
DISCUSSION
It is now apparent that H. influenzae strains have numerous mechanisms for the binding and uptake of heme, whether the heme is free or bound to a protein carrier (12, 15, 23, 31, 40). Considering the absolute dependence of H. influenzae on heme for aerobic growth, such redundancy in heme uptake systems is perhaps not surprising. Moreover, there are now data which indicate that some of the genes encoding these heme uptake systems are transcribed in the human body during the infectious process, at least in otitis media (44).
The present study extends our earlier finding that an NTHI outer membrane protein encoded by an ORF containing a CCAA repeat motif was involved in the binding and utilization of hemoglobin-haptoglobin (23). In NTHI strain N182, there appears to be a family of related proteins which can bind hemoglobin or hemoglobin-haptoglobin and which have in common the presence of a CCAA repeat in their respective ORFs. Work by Stull and colleagues (26) that was reported while the present study was in progress indicates that H. influenzae type b strain HI689 possesses three ORFs (i.e., hgpA, hgpB, and hgpC) that contain CCAA repeats and encode hemoglobin- or hemoglobin-haptoglobin-binding proteins, a finding which raises the possibility that this type of protein family is common to both NTHI and H. influenzae type b strains. The presence of CCAA repeats in the other NTHI strains described in the present study, as well as in other H. influenzae strains described independently by Morton and Stull (25), indicates that these genes are ubiquitous among H. influenzae strains.
Using the HgbA- and HgbB-specific MAbs in the colony blot RIA, we were able to detect the expression of these proteins individually and then use PCR to determine the number of CCAA repeats present in the relevant ORF. These data indicate that expression of HgbA or HgbB by a given isolate could be directly correlated with the presence of an appropriate number of CCAA repeats in the selected ORF (i.e., that which would be predicted to allow full-length protein expression). We were also able to use PCR to amplify the CCAA repeat-containing regions from the other two, unselected hgb ORFs in each isolate. In every case, when an isolate did not express a protein reactive with a given MAb, the number of CCAA repeats in that particular ORF was consistent with premature translational termination. These data, based on detection of protein expression with MAbs, are complemented by those of Ren et al. (33), who used a CCAA-containing gene from H. influenzae type b in a lacZ-based translational gene fusion to correlate phase-variable expression of LacZ with alterations in the number of CCAA repeats.
We were able to readily detect individual N182 isolates that expressed either HgbA or HgbB or both simultaneously (Fig. 7 and 8). In contrast, we did not detect any individual N182 isolates (Fig. 7 and 8) that readily expressed HgbC even though we could detect some HgbC expression in a population of N182 cells that had been starved for both heme and iron (Fig. 5C, lane 8). Again, nucleotide sequence analysis of the 5′ end of the hgbC ORF in the 16 individual isolates that did not express HgbC indicated that, in every case, the number of CCAA repeats in the hgbC ORF would have resulted in premature termination of translation. The fact that HgbC-reactive MAb 12A2 did not function in a colony blot RIA analysis precluded direct identification of individual N182 isolates that expressed HgbC.
Slipped-strand mispairing resulting in phase-variable expression of an outer membrane protein was first reported by Cannon and colleagues with the PII (Opa) protein of N. gonorrhoeae, where a pentanucleotide repeat was present near the beginning of the ORF encoding this protein (27). Subsequently, the occurrence of slipped-strand mispairing has been described for several ORFs that encode proteins involved in the uptake of hemoglobin in both N. gonorrhoeae (3) and N. meningitidis (19, 34). Polyguanine tracts within the ORFs encoding the HpuA protein of N. gonorrhoeae (3) and the HpuA and HmbR proteins of N. meningitidis (19, 34) have been shown to be involved in phase variation that is controlled by slipped-strand mispairing.
The frequency of phase variation in the gonococcal hemoglobin utilization system described above has been reported to be approximately 10−3 (4), whereas different serotypes of meningococci exhibited phase variation in hemoglobin utilization at frequencies as high as 10−2 and as low as 10−6 (19, 34). We observed that when a heme-grown isolate of N182 (Fig. 7, lane 7) that did not express HgbA, HgbB, or HgbC was grown in BHI-Hm medium and then plated on BHI-Hm plates, colonies that bound either the HgbA-specific MAb or the HgbB-specific MAb arose at a frequency of 10−2 to 10−3 (data not shown).
While a number of different genes encoding proteins involved in heme acquisition have now been described in H. influenzae, the hgbA, hgbB, and hgbC genes in NTHI N182 and the very similar hgpA, hgpB, and hgpC genes in H. influenzae type b strain HI689 represent the first descriptions of genes that may have been derived from duplication events. The likelihood of this possibility is reinforced by our detection of large inverted repeats, proposed to be associated with gene duplications (30), immediately upstream from both the hgbB and hgbC ORFs in NTHI N182. Why other H. influenzae genes involved in heme acquisition (e.g., hxuCBA) (6) have not been duplicated in the H. influenzae chromosome is not known. It is possible that this redundancy in genes that express hemoglobin- or hemoglobin-haptoglobin-binding proteins reflects a greater functional significance of these particular protein-bound sources of heme to H. influenzae. We also cannot formally exclude the possibility that the presence of these related genes in NTHI strain N182 is the result of horizontal genetic exchange, especially in view of the fact that multiple strains of NTHI can coexist simultaneously in the human respiratory tract (28).
Our data on the expression of HgbA, HgbB, and HgbC by individual N182 isolates (Fig. 8, lanes 21 to 40) indicate that repeated passage of these cells with hemoglobin as the sole source of heme resulted in expression of HgbA by all of the individual isolates regardless of whether the original isolate (as identified by colony blot RIA) expressed this protein. This finding is likely the result of the selection of a population of HgbA-expressing cells. Whether HgbA functions more or less effectively than HgbB and HgbC in the binding and utilization of hemoglobin by strain N182 remains to be determined. A recent report by Stull and colleagues (26) indicates that a hgpA hgpB hgpC mutant of H. influenzae type b HI689 is still able to utilize hemoglobin as its sole source of heme for growth. This finding indicates that, at least in H. influenzae type b HI689, a protein encoded by a gene that does not contain CCAA repeats will function to allow this H. influenzae strain to acquire heme from hemoglobin. The identity of this gene product and its possible existence in NTHI strains remain to be determined.
ACKNOWLEDGMENTS
This study was supported by U.S. Public Health Service grant AI17621 to E.J.H.
We thank Jo Latimer, Sheryl Lumbley, Sharon Thomas, and Yufan Zhu for technical assistance. We also thank Kathryn Edwards, Janet Gilsdorf, Timothy Murphy, and Peter Rice for providing many of the NTHI isolates used in this study.
REFERENCES
- 1.Adhikari P, Kirby S D, Nowalk A J, Veraldi K L, Schryvers A B, Mietzner T A. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J Biol Chem. 1995;270:25142–25149. doi: 10.1074/jbc.270.42.25142. [DOI] [PubMed] [Google Scholar]
- 2.Bowman B H, Barnett D R, Lum J B, Yang F. Haptoglobin. Methods Enzymol. 1988;163:452–474. doi: 10.1016/0076-6879(88)63043-6. [DOI] [PubMed] [Google Scholar]
- 3.Chen C-J, Elkins C, Sparling P F. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect Immun. 1998;66:987–993. doi: 10.1128/iai.66.3.987-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C-J, Sparling P F, Lewis L A, Dyer D W, Elkins C. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect Immun. 1996;64:5008–5014. doi: 10.1128/iai.64.12.5008-5014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cope L D, Thomas S E, Latimer J L, Slaughter C A, Muller-Eberhard U, Hansen E J. The 100 kDa heme:hemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol Microbiol. 1994;13:863–873. doi: 10.1111/j.1365-2958.1994.tb00478.x. [DOI] [PubMed] [Google Scholar]
- 6.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelissen C N, Biswas G D, Tsai J, Paruchuri D K, Thompson S A, Sparling P F. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J Bacteriol. 1992;174:5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulton J W, Pang J C S. Transport of hemin by Haemophilus influenzae type b. Curr Microbiol. 1983;9:93–98. [Google Scholar]
- 9.Elkins C, Chen C-J, Thomas C E. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans N M, Smith D D, Wicken A J. Haemin and nicotinamide adenine dinucleotide requirements of Haemophilus influenzae and Haemophilus parainfluenzae. J Med Microbiol. 1974;7:359–365. doi: 10.1099/00222615-7-3-359. [DOI] [PubMed] [Google Scholar]
- 11.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback R C, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Frangipane M E, Morton D J, Wooten J A, Pozsgay J M, Stull T L. Binding of human hemoglobin by Haemophilus influenzae. FEMS Microbiol Lett. 1994;118:243–248. doi: 10.1111/j.1574-6968.1994.tb06835.x. [DOI] [PubMed] [Google Scholar]
- 13.Granick S, Gilder H. The porphyrin requirements of Haemophilus influenzae and some functions of the vinyl and propionic acid side chains of heme. J Gen Physiol. 1946;30:1–13. [PMC free article] [PubMed] [Google Scholar]
- 14.Gulig P A, McCracken G H, Jr, Holmans P L, Hansen E J. Immunogenic proteins in cell-free culture supernatants of Haemophilus influenzae type b. Infect Immun. 1984;44:41–48. doi: 10.1128/iai.44.1.41-48.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin H, Ren Z, Pozsgay J M, Elkins C, Whitby P W, Morton D J, Stull T L. Cloning of a DNA fragment encoding a heme-repressible hemoglobin-binding outer membrane protein from Haemophilus influenzae. Infect Immun. 1996;64:3134–3141. doi: 10.1128/iai.64.8.3134-3141.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadner R J. Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol Microbiol. 1990;4:2027–2033. doi: 10.1111/j.1365-2958.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 17.Koskelo P, Muller-Eberhard U. Interaction of porphyrins with proteins. Semin Hematol. 1977;14:253–262. [PubMed] [Google Scholar]
- 18.Lee B C. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 19.Lewis L A, Gipson M, Hartman K, Ownbey T, Vaughn J, Dyer D W. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol Microbiol. 1999;32:977–989. doi: 10.1046/j.1365-2958.1999.01409.x. [DOI] [PubMed] [Google Scholar]
- 20.Lewis L A, Gray E, Wang Y-P, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 21.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundrigan M D, Kadner R J. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli: homology among outer membrane receptors that interact with TonB. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 23.Maciver I, Latimer J L, Liem H H, Muller-Eberhard U, Hrkal Z, Hansen E J. Identification of an outer membrane protein involved in utilization of hemoglobin-haptoglobin complexes by nontypeable Haemophilus influenzae. Infect Immun. 1996;64:3703–3712. doi: 10.1128/iai.64.9.3703-3712.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 25.Morton D J, Stull T L. Distribution of a family of Haemophilus influenzae genes containing CCAA nucleotide repeating units. FEMS Microbiol Lett. 1999;174:303–309. doi: 10.1111/j.1574-6968.1999.tb13583.x. [DOI] [PubMed] [Google Scholar]
- 26.Morton D J, Whitby P W, Jin H, Ren Z, Stull T L. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type B. Infect Immun. 1999;67:2729–2739. doi: 10.1128/iai.67.6.2729-2739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy G L, Connell T D, Barritt D S, Koomey M, Cannon J G. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989;56:539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- 28.Murphy T F, Sethi S, Klingman K L, Brueggemann A B, Doern G V. Simultaneous respiratory tract colonization by multiple strains of nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J Infect Dis. 1999;180:404–409. doi: 10.1086/314870. [DOI] [PubMed] [Google Scholar]
- 29.Patrick C C, Kimura A, Jackson M A, Hermanstorfer L, Hood A, McCracken G H, Jr, Hansen E J. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypeable Haemophilus influenzae. Infect Immun. 1987;55:2902–2911. doi: 10.1128/iai.55.12.2902-2911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Read T D, Farley M M. Conserved extragenic DNA elements in Haemophilus influenzae. Mol Microbiol. 1997;23:627–628. doi: 10.1046/j.1365-2958.1997.d01-1862.x. [DOI] [PubMed] [Google Scholar]
- 31.Reidl J, Mekalanos J J. Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J Exp Med. 1996;183:621–629. doi: 10.1084/jem.183.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren Z, Jin H, Morton D J, Stull T L. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect Immun. 1998;66:4733–4741. doi: 10.1128/iai.66.10.4733-4741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren Z, Jin H, Whitby P W, Morton D J, Stull T L. Role of CCAA nucleotide repeats in regulation of hemoglobin and hemoglobin-haptoglobin binding protein genes of Haemophilus influenzae. J Bacteriol. 1999;181:5865–5870. doi: 10.1128/jb.181.18.5865-5870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson A R, Stojiljkovic I. HmbR, a hemoglobin-binding outer membrane protein of Neisseria meningitidis, undergoes phase variation. J Bacteriol. 1999;181:2067–2074. doi: 10.1128/jb.181.7.2067-2074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson S M, Frisch C F, Gulig P A, Kettman J R, Johnston K H, Hansen E J. Monoclonal antibodies directed against a cell surface-exposed outer membrane protein of Haemophilus influenzae type b. Infect Immun. 1982;36:80–88. doi: 10.1128/iai.36.1.80-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanders J D, Cope L D, Hansen E J. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect Immun. 1994;62:4515–4525. doi: 10.1128/iai.62.10.4515-4525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seery V L, Muller-Eberhard U. Binding of porphyrins to rabbit hemopexin and albumin. J Biol Chem. 1973;248:3796–3800. [PubMed] [Google Scholar]
- 39.Stevens M K, Porcella S, Klesney-Tait J, Lumbley S R, Thomas S E, Norgard M V, Radolf J D, Hansen E J. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect Immun. 1996;64:1724–1735. doi: 10.1128/iai.64.5.1724-1735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stull T L. Protein sources of heme for Haemophilus influenzae. Infect Immun. 1987;55:148–153. doi: 10.1128/iai.55.1.148-153.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stuy J H, Walter R B. Cloning, characterization, and DNA base sequence of the high-level streptomycin resistance gene strA1 of Haemophilus influenzae Rd. J Bacteriol. 1992;174:5604–5608. doi: 10.1128/jb.174.17.5604-5608.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatusov R L, Mushegian A R, Bork P, Brown N P, Hayes W S, Borodovsky M, Rudd K E, Koonin E V. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- 43.Weiser J N, Love J M, Moxon E R. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 44.Whitby P W, Sim K E, Morton D J, Patel J A, Stull T L. Transcription of genes encoding iron and heme acquisition proteins of Haemophilus influenzae during acute otitis media. Infect Immun. 1997;65:4696–4700. doi: 10.1128/iai.65.11.4696-4700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White D C, Granick S. Hemin biosynthesis in Haemophilus. J Bacteriol. 1963;85:842–850. doi: 10.1128/jb.85.4.842-850.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]