Abstract
Background
The gold standard treatment for locally advanced rectal cancer is total mesorectal excision after preoperative chemoradiotherapy. Response to chemoradiotherapy varies, with some patients completely responding to the treatment and some failing to respond at all. Identifying biomarkers of response to chemoradiotherapy could allow patients to avoid unnecessary treatment-associated morbidity rate. While previous studies have attempted to identify such biomarkers, none have reached clinical utility, which may be due to heterogeneity of the cancer. In this study, potential human gene and microbial biomarkers were explored in a cohort of rectal cancer patients who underwent chemoradiotherapy.
Methods
RNA sequencing was carried out on matched tumour and adjacent normal rectum biopsies from patients with rectal cancer with varying chemoradiotherapy responses treated between 2016 and 2019 at two institutions. Enriched genes and microbes from tumours of complete responders were compared with those from tumours of others with lesser response.
Results
In 39 patients analysed, enriched gene sets in complete responders indicate involvement of immune responses, including immunoglobulin production, B cell activation and response to bacteria (adjusted P values <0.050). Bacteria such as Ruminococcaceae bacterium and Bacteroides thetaiotaomicron were documented to be abundant in tumours of complete responders compared with all other patients (adjusted P value <0.100).
Conclusion
These results identify potential genetic and microbial biomarkers of response to chemoradiotherapy in rectal cancer, as well as suggesting a potential mechanism of complete response to chemoradiotherapy that may benefit further testing in the laboratory.
Rectal cancer patients respond differently to presurgical radiotherapy, such that biomarkers indicating efficacy of the treatment are vital to stratify patients to the most effective treatment regimen. Currently identified biomarkers have no clinical utility, possibly due to heterogeneity of the disease in patients. In this article we present a method that mitigates interpersonal variation between patients, allowing the identification of immune responses and microbes, and their possible contributions to the mechanisms of complete response to radiotherapy in rectal cancer.
Introduction
Locally advanced rectal cancer often requires neoadjuvant chemoradiotherapy (nCRT), aiming to downstage tumours and reduce rates of local recurrence, improving survival1,2. The response to nCRT varies: up to 20 per cent of patients achieve a pathological complete response (pCR), up to 60 per cent demonstrate partial response, and the remainder display resistance to nCRT1,3. These response rates could further be increased by total neoadjuvant therapy3.
A pCR is defined as the absence of residual viable tumour cells in the resected specimen and the reliance on pathological confirmation of a complete response fails to identify patients that may benefit from organ preservation. Habr-Gama pioneered the adoption of a clinical complete response (cCR) as a surrogate for pCR4. Patients who achieve a cCR may subsequently be managed using a ‘Watch and Wait’ approach, thereby avoiding the morbidity rate associated with major resectional surgery. Although up to 25 per cent of these patients show tumour regrowth by 2 years, the majority are amenable to salvage procedures5.
Even though extensive work has been carried out to identify clinical and biological markers of response to radiotherapy in rectal cancer1,6–8, no reliable biomarkers have been validated for clinical use. A robust biomarker that selects patients likely to achieve a pCR to nCRT would allow the accurate identification of responders and increase confidence in selecting patients amenable to non-operative management. Chemoradiotherapy (CRT) is associated with significant localized and systemic side-effects and has been demonstrated to negatively impact quality of life9. The identification of patients unlikely to benefit from conventional CRT would result in a reduction in CRT-associated side-effects, and targeting of patients for novel treatment strategies.
Due to the largely sporadic nature of colon and rectal cancer, environmental factors are likely to play a critical role in the development of the disease, and recent international data points to the importance of the microbiome in its development and progression10–12. Recent reports have also demonstrated that systemic effects of the gut microbiome may contribute to treatment response in other cancer types13,14 and could be predictors of a favourable response to immunotherapy. In addition, gut microbiota have been shown to locally influence the treatment efficacy of irinotecan for colorectal cancer (CRC)15. However, although some studies on the protective effect of the microbiome on radiotherapy-induced toxicity have been carried out16,17, very little is known about whether or how the gut microbiome may regulate the response of the tumour to radiotherapy.
This research aimed to analyse gene expression data from a unique cohort of pre-nCRT rectal cancer tumours and their matched normal mucosa samples to identify tumour and microbial genes and molecular pathways associated with response.
Methods
Patients
Two prospective, consecutively sampled cohorts of patients with rectal cancer treated at Christchurch Hospital, New Zealand (study period: 2018), and at the Peter MacCallum Cancer Centre, Melbourne, Australia (study period: August 2016 to February 2019) were analysed. Patients were selected for nCRT at multidisciplinary team meetings at their respective institutions. Pretreatment biopsies of tumour tissue and adjacent, macroscopic normal tissue (>10 cm from tumour) were taken at colonoscopy, before nCRT treatment. Patients who had received previous chemotherapy or radiation therapy for treatment of their rectal tumour were excluded from the study. Patient data, including clinical staging, treatment schemes, histology and follow-up (recurrence and metastases until March 2022), was collected. Response to long-course CRT (LCCRT) was assessed histologically from surgical resection specimens, and reported using Dworak grading (Christchurch cohort) or the American Joint Committee on Cancer (AJCC) grading (Melbourne cohort). Response groups were designated as complete responders (Dworak 4/AJCC 0), near-complete responders (Dworak 3/AJCC 1), incomplete responders (Dworak 2/AJCC 2) and non-responders (Dworak 1/AJCC 3). In addition, patients who developed progressive disease or died of disease, during the course of therapy, were also designated non-responders.
The study was undertaken with ethical approval from the Health and Disability Ethics Committee of New Zealand (ethics approval number: 18/STH/40/AM01) and the Human Research Ethics Committees of Australia (ethics approval number: HREC 14/85). All participants provided written, informed consent before enrolment.
Outcomes of interest
The primary outcome of interest was to compare human gene expression and microbial taxa abundances in different groups of responders. To address this objective, complete responders were compared with all other responders grouped together, and secondly, non-responders were compared with all other response groups. In addition, the correlation between differentially expressed genes (DEGs) and differentially abundant bacteria was investigated, as well as microbial diversity.
RNA extraction, sequencing and processing
Tumour and normal tissue biopsies were taken at colonoscopy and immediately frozen in liquid nitrogen and stored at −80°C. RNA extraction was carried out as detailed previously18. Briefly, RNA was extracted from <20 mg of tissue using the RNEasy Plus Mini Kit (Qiagen, Hilden, Germany), including DNAse treatment, following tissue disruption using a Retsch Mixer Mill. Purified RNA was quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Asheville, NC, USA), and stored at −80°C. RNA sequencing was performed using an Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA) to produce 150 bp paired end reads, as previously described18. The Ribo-Zero™ Magnetic Kit (Human & Bacteria, Epicentre, Madison, WI, US) was used for ribosomal RNA depletion, and libraries were prepared using the NEBNext® Ultra™ RNA Library Prep Kit (New England BioLabs Inc.®, Ipswich, MA, USA). Approximately 50 million reads (15 Gb raw data) were produced per sample on an Illumina NovaSeq 6000 instrument. Raw sequencing reads were deposited at the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under BioProject ID PRJNA815861.
Raw sequencing data was parsed through the Metafunc pipeline19, which performs read preprocessing, host gene mapping and microbiome species identification. Further details of the computational pipeline may be found at https://gitlab.com/schmeierlab/workflows/metafunc, and complete analysis of this article is available at https://gitlab.com/alsulit08/uoc_response_rectalca. For the microbiome analysis part of the pipeline, no abundance filtering was performed at this stage of the analysis.
Microbiome data preprocessing
For the microbiome data set, raw counts of microbe taxonomies were gathered into a Phyloseq object20, with metadata information on their response and tumour or normal status. Prefiltering of the species was then performed before the analysis, only including those within the Bacterial Kingdom, and those with at least 10 reads in 20 per cent of the samples (Table S1).
Computational analyses and statistics
Expression levels for each human gene and sample were generated by the MetaFunc pipeline19, and differential human gene expression analysis (DGEA) using DESeq221 was used to detect DEGs. To detect DEGs that were significantly differentially expressed in the tumour relative to each participant’s normal tissue between groups of responders, the model fitted by DESeq2 included covariates for response (complete or other), tissue type (tumour or normal), response:participant (index) and response:tissue. Care was taken to ensure the model matrix was of full rank, the model converged and that modelling assumptions were met. The genes were considered differentially expressed if their adjusted P values were <0.100.
From the results of this DESeq2 comparison, a preranked list of all resulting genes based on P values and log2-fold change was generated, and these genes were used as input for gene set enrichment analysis (GSEA) using clusterProfiler22 with the C5 Ontology Gene Sets collection (version 7) from the molecular signatures database (MSigDB)23,24. Specifically, the genes were ranked using the formula:
This ranking places the genes with lowest P values and positive log2-fold change at the top of the list, and the genes with lowest P values and negative log2-fold change at the bottom of the list. Genes at the top of the list contribute to gene sets with positive enrichment scores and genes at the bottom contribute to gene sets with negative enrichment scores. Gene sets were significantly enriched in a responder group if their adjusted P values were <0.050.
For the differential metatranscriptome analysis of the microbiome data set, the same model for group-specific condition effects (see DGEA above) to obtain differentially abundant (DA) microbes in tumour samples compared with matched normal samples, specific to complete responders compared with other responders, was used. Differentially abundant bacteria were considered those species with adjusted P values <0.100.
The correlation between DEGs and differentially abundant bacteria in rectal cancer was then investigated. Using the rlog transformed values for gene expression and microbial abundance obtained from DESeq2 of 87 identified DEGs and 10 DA bacterial species, a Spearman correlation analysis between each gene and species was performed, correcting the final P values using Benjamini–Hochberg (BH) adjustment. Influential points (species rlog > 12) were removed for the Spearman correlation calculation and corresponding scatter plots.
The α-diversity (microbial diversity within a sample or community) between different groups was compared using Observed (richness—number of observed taxa) and Shannon (richness and evenness—taking abundance of different taxa into consideration) measures. The sample set, as described in the microbiome preprocessing section above, was rarefied to 90 per cent of the smallest sample size in the data set and analysed in Phyloseq20. Observed and Shannon measures were calculated through the estimate_richness() function of the Phyloseq20 package, and compared between groups using Wilcoxon tests. Non-metric multidimensional scaling (NMDS) plots based on Bray–Curtis distances were used to visualize β-diversity between groups.
Analyses for DGEA, GSEA, DA, correlation and diversity were completed using R25,26 packages.
Results
Study population
This cohort comprised 40 patients (20 patients from each hospital) with diagnosed rectal cancer who were subsequently treated with CRT followed by surgical resection (Table 1). One patient was subsequently excluded, due to treatment cessation for palliative care. The majority (n = 36) were treated with LCCRT, with either capecitabine, FOLFIRI or 5FU. Two patients did not complete LCCRT due to the development of grade 3 toxicity. One patient received short-course radiotherapy, while the remaining two patients received sandwich CRT (FOLFOX). There were 12 females and 27 males, who ranged in age from 29 to 86 years (mean age: 62 years). There were five patients with complete response to LCCRT, five patients with near-complete response, 18 patients with incomplete response and eight patients who did not respond to LCCRT. In addition, three patients developed progressive disease or died of disease during the course of therapy, and these patients were also designated non-responders. Median follow-up was 42 months. Six patients died of the disease during a minimum follow-up period of 24 months.
Table 1.
Characteristics of the patients with rectal cancer cohort
| All (n = 39) | Complete response (n = 5) | Near-complete response (n = 5) | Incomplete response (n = 18) | No response (n = 11) | |
|---|---|---|---|---|---|
| Age (years), mean ± SD | 62.0 ± 14.5 | 66.2 ± 9.3 | 61.2 ± 4.8 | 62.4 ± 14.8 | 69.9 ± 19.3 |
| Sex | |||||
| Male | 26 (66.7) | 4 (80.0) | 4 (80.0) | 13 (72.2) | 5 (45.5) |
| Female | 13 (33.3) | 1 (20.0) | 1 (20.0) | 5 (27.8) | 6 (54.5) |
| Clinical stage | |||||
| 1 | 1 (2.6) | 0 (0.0) | 1 (20) | 0 (0.0) | 0 (0.0) |
| 2 | 4 (10.3) | 0 (0.0) | 0 (0.0) | 2 (11.1) | 2 (18.2) |
| 3 | 25 (64.1) | 4 (80.0) | 4 (80) | 11 (61.1) | 6 (54.5) |
| 4 | 9 (23.1) | 1 (20.0) | 0 (0.0) | 5 (27.8) | 3 (27.3) |
| Death due to disease progression | 5 (12.8) | 0 (0.0) | 0 (0.0) | 3 (16.7) | 2 (18.2) |
| Treatment | |||||
| LCCRT (no other data) | 4 (10.3) | 0 (0.0) | 0 (0.0) | 3 (16.7) | 1 (9.1) |
| LCCRT (capecitabine) | 26 (66.7) | 5 (100.0) | 3 (60.0) | 9 (50.0) | 9 (81.8) |
| LCCRT (5FU) | 3 (7.7) | 0 (0.0) | 0 (0.0) | 3 (16.7) | 0 (0.0) |
| LCCRT (FOLFIRI) | 2 (5.1) | 0 (0.0) | 0 (0.0) | 2 (11.1) | 0 (0.0) |
| Sandwich CRT | 2 (5.1) | 0 (0.0) | 2 (40.0) | 0 (0.0) | 0 (0.0) |
| SCRT | 1 (2.6) | 0 (0.0) | 0 (0.0) | 1 (5.6) | 0 (0.0) |
| Palliative | 1 (2.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (9.1) |
Values are n(%) unless otherwise stated. LCCRT, long-course chemoradiotherapy; FOLFIRI, folinic acid, fluorouracil and irinotecan; SCRT, short-course radiotherapy; 5FU, fluorouracil.
Differential gene expression and gene set enrichment between response groups
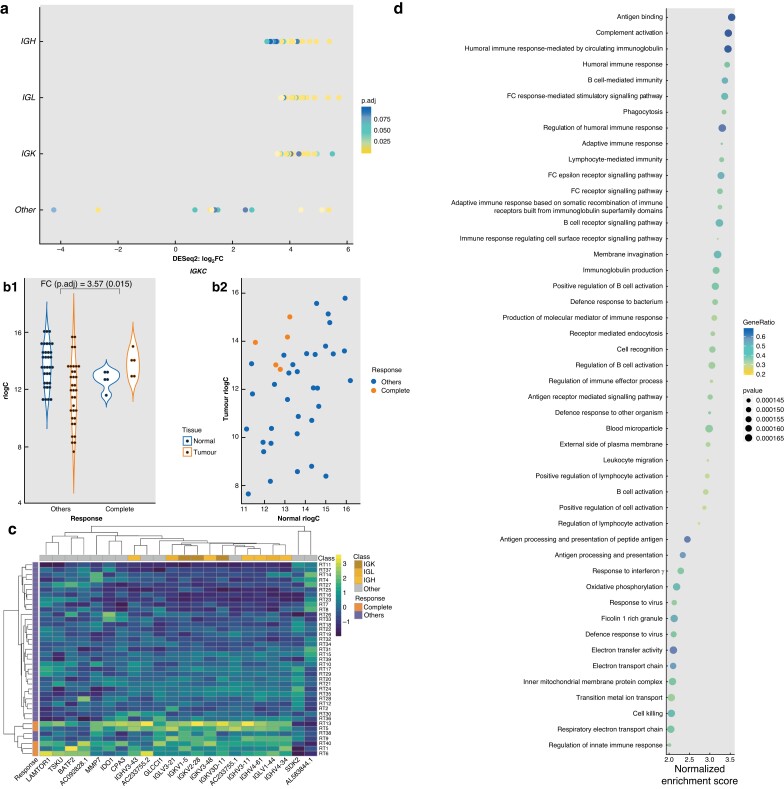
DEGs were analysed between matched pairs of tumour and normal tissues, to account for interpersonal variation in gene expression, and then these DEGs were compared between response groups. Eighty-seven genes were found to be differentially expressed between tumour and normal samples (adjusted P value <0.100), and only seen in complete responders. Interestingly, the majority of these genes are associated with immunoglobulin chains, with 75 of 87 genes having prefixes of IGH-, IGL- or IGK- (Fig. 1a), for immunoglobulin heavy, light, and kappa respectively. Tumour versus normal rlog was plotted as transformed counts per sample of representatives from these genes and showed that for most of these genes, complete responders cluster at high tumour–low normal values (upper left quadrant), indicating that in the complete responder group, these genes are more highly expressed in tumours compared with normal tissues (Fig. 1b and Fig. S1). When DGEA was carried out comparing non-responders to all other responders, no DEGs were identified between the two groups.
Fig. 1.
Host differential gene expression and gene set enrichment analysis
a Differentially expressed genes (DEGs) classified into immunoglobulin (Ig)-related genes and other non-Ig genes. p.adj, adjusted P value. b Transformed counts (rlog) of Tumour/Normal for complete responders compared with other responders of a representative DEG, IGKC. b1 Tumour versus Normal for each response category (FC, log2-fold change; p.adj, adjusted P value). b2 Tumour values and their corresponding normal values per sample by response. c Heatmap of the top 10 (by adjusted P values) DEGs, which are all Ig-related genes, and the other 12 non-Ig DEGs clustered by sample and gene. d Top 47 positively enriched gene sets (y axis) in Tumour versus Normal of Complete Responders. All have adjusted P values (Benjamini–Hochberg) of 0.00928. Gene ratio is the ratio of core enrichment genes to the gene set size. Gene sets coloured yellow are immune-related gene sets. Gene sets coloured red are gene sets that refer to a response to microbial input. All other gene sets are coloured black.
The majority of significant DEGs were upregulated in tumours compared with normal tissue. The top 10 DEGs (all immunoglobulin related), in addition to 12 non-immunoglobulin-related DEGs, could robustly distinguish complete responders from all other patients, as illustrated in Fig. 1c and Fig. S2.
Subsequently, Gene Set Enrichment Analysis (GSEA), which ascribes function to DEGs, was performed using genes ranked according to DESeq2 P value and log2-fold change (±). Gene sets relating to immune responses constitute the majority of the top 47 enriched gene sets in tumours of complete responders (Fig. 1d). Among the gene sets with the highest enrichment scores were Complement Activation and B Cell Mediated Immunity, consistent with the majority of the 87 DEGs in complete responders being related to immunoglobulins.
Diversity analysis of the microbiome of rectal tumours
No significant differences were found in Observed or Shannon measures of α-diversity (Fig. S3A–H). Any distinct group clustering by NMDS was not detected, indicating similar β-diversity between the response groups (Fig. S3I–L).
Differential abundance analysis of bacterial species between radiotherapy response groups
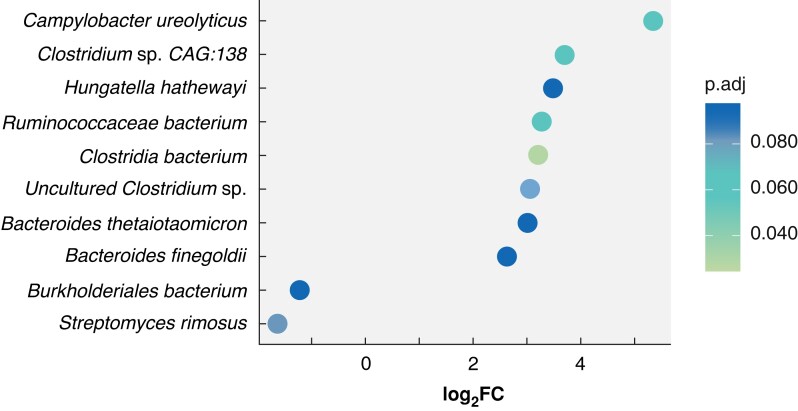
A similar approach to DGEA in the human data set was used to identify bacterial taxa that were differentially abundant in tumour versus matched normal samples, and specific to complete responders. Analysis of differences in the tumour microbiome between response groups identified 10 bacterial species that were differentially abundant (adjusted P values t <0.100) in tumour tissue compared with matched normal tissue in complete responders (Fig. 2). Bacterial species identified included Ruminococcaceae bacterium, Hungatella hathewayi, Bacteroides thetaiotaomicron and Clostridium species27–32. Plotting tumour versus normal rlog transformed counts per sample of these microbes showed that there is a separation between the complete responders compared with other responders, although not as pronounced as that seen in human DEGs (Fig. S4).
Fig. 2.
Differentially abundant bacteria in tumour samples compared with matched normal tissue, specific to complete responders
Plot of the 10 differentially abundant bacteria showing their log2-fold changes (x axis) and adjusted P (p.adj) values by colour.
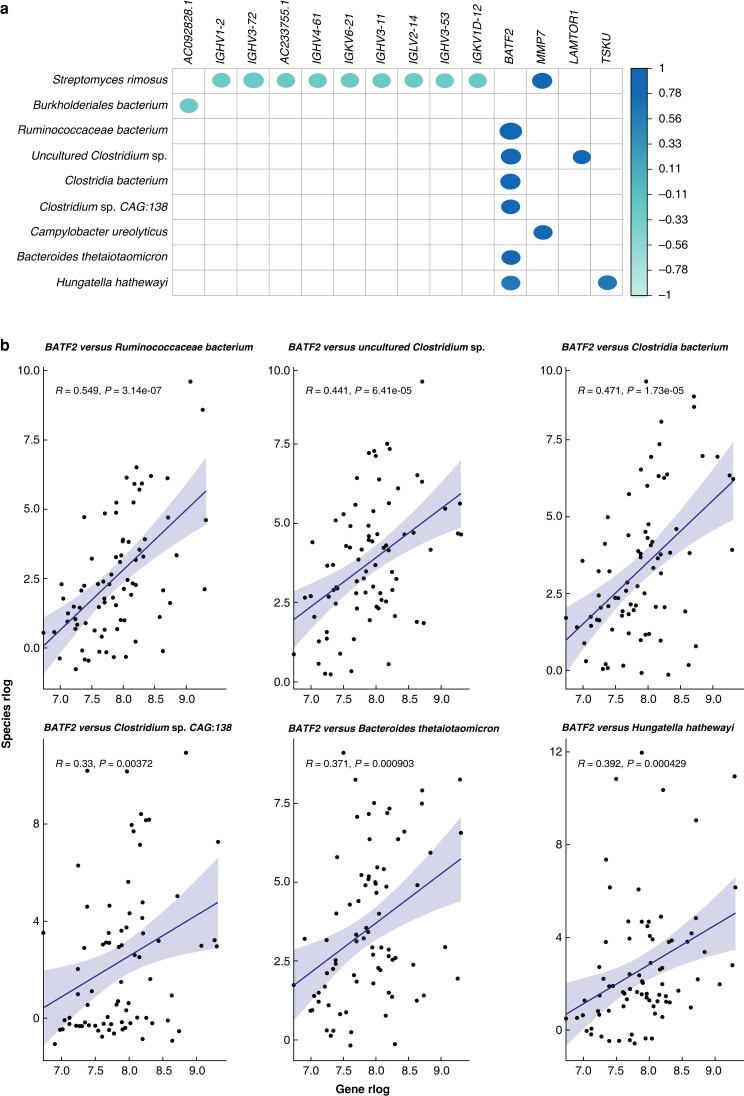
Correlation between host gene expression and microbial abundances
Significant correlations between DEGs and differentially abundant bacterial species were identified. Among DEGs that positively correlated with the microbial abundances, the BATF2 gene was notable. Positive correlations (Spearman coefficient: 0.355–0.549; BH-adjusted P value: 1.661 × 10−4−7.699 × 10−2) of this gene with several DA bacterial species, previously linked to CRC, including Ruminococacceae bacterium and Bacteroides thetaiotaomicron (Fig. 3), were found.
Fig. 3.
Correlations between differentially expressed genes and differentially abundant microbes
a Correlation plot showing bacteria–gene correlations with at least one significant correlation (adjusted P value <0.100). Spearman correlation coefficients (ρ) are represented by the colour bar; blank spaces represent correlations that are not significant. bBATF2 and its positive correlations with differentially abundant bacteria. R, Spearman coefficient; e, base number x10power.
Discussion
The identification of predictive biomarkers of response to CRT will result in improved survival, a reduction of morbidities rate related to unnecessary treatment, and a more targeted approach to treatment for patients with rectal cancer. Previous studies have attempted to identify markers of complete response to CRT, and while clinicopathological and radiological features have been identified, they are limited in sensitivity and specificity1. Intratumoural heterogeneity contributes to a lack of reproducibility between molecular biomarker studies, and as a result, no biomarker is currently in clinical use1,8. In order to identify a predictive biomarker of response for rectal tumours, a mechanistic link to the underlying tumour biology is also warranted. This study identifies potential biomarkers of complete response to CRT in patients with rectal cancer, in addition to uncovering novel links between the tumour microbiome and immune response in the rectal tumour microenvironment. Tumour tissues were firstly compared with their corresponding matched normal tissues, to mitigate interpersonal differences that are not due to tumour characteristics.
Rather than a continuum of gene expression changes from complete responders to non-responders, a tight clustering of complete responders based on gene expression of mainly immune-related genes, compared with all other patients, was observed. This suggests the presence of a distinct tumour microenvironment in a group of patients that predisposes them to a complete response to radiotherapy. In these cohorts, the patients showed significantly higher expression of genes responsible for complement activation and B cell-related functions in their tumour tissue compared with adjacent normal tissue, supporting previously published reports that immunoglobulins may recognize radiotherapy-induced neoantigens resulting in complement activation and CD8+ T cell responses33.
Enriched gene sets related to antigen presentation in the cohort of complete responders were also observed. Ionizing radiation can cause DNA mutations, which, when translated into peptides, are presented on MHC-1 molecules, eliciting a cytotoxic immune response. This has been observed in studies of other cancer types, where radiation has been shown to recruit neutrophils and monocytes, as well as promote maturation of antigen presenting cells34. Radiotherapy also results in mimicry of viral infections, where cytosolic DNA induces the release of type 1 interferons that recruit dendritic cells specialized in antigen presentation to CD8+ T cells35. Our findings indicate that enhanced antigen presentation in pretreatment tumours predisposes them to a successful response to CRT.
A significant enrichment of the BATF2 gene was found in patients who went on to have a complete response to CRT, which is consistent with findings that BATF2 depletion in tumour compared with normal tissue is correlated with poor prognosis in CRC36. BATF2 is thought to be induced by type 1 interferons and play a role in viral infections37 and may, as a result, contribute to radiotherapy-induced responses that mimic viral infection. Type-1 interferons are important players in antiviral immune responses, recruiting dendritic cells specialized in antigen presentation to CD8+ T cells35. This is again consistent with viral infection and antigen presentation-enriched gene sets identified in complete responders in this cohort. Furthermore, BATF2 may also induce antitumour effects through the induction of CD8+ T cells38.
The enrichment of immune response-related genes and gene sets in tumours of complete responders to CRT in our cohort is consistent with the findings of a previous study39. In that study, they identified antigen presentation, interferon (IFN) activity and B cell activity to be enriched in good responders to preoperative CRT in locally advanced rectal cancer (LARC) patients, and that these factors may relate to good responses to the treatment via activation of an antiviral-like response and CD8+ T cell recruitment. While their study largely used microarray gene expression and targeted sequencing of CRC genes, this study mainly used whole RNA sequencing, which also allowed investigation of possible contributions of the microbiome.
A key finding of this study is the enrichment of gene sets relating to response to bacteria in complete responders. The role of the microbiome in therapy response has recently become a focal point for studies of different types of treatments in a variety of cancer types. Early studies of the microbiome in CRC had suggested a decrease in microbial diversity associated with tumours compared with healthy controls10,11,40. However, more recent studies refute this finding, where CRC samples have increased richness41 or are no different27 compared with controls. Consistent with these more recent studies42, here, no difference was found in microbiome diversity metrics between the response groups. However, a number of bacterial species that were differentially abundant in tumours of complete responders were identified, several of which had previously been implicated in CRC carcinogenesis and prognosis. Hungatella hathewayi has been reported to be differentially abundant in CRC27, and has been reported to drive methylation of tumour suppressor genes28 and was also found to be significantly more abundant in complete responders in the present study. Ruminococcaceae, a putative commensal genus, has been linked to the expression of T cell recruiting chemokines31, indicating a role in tumour-killing immune activation, and a low abundance has been associated with CRC29,30. High levels of this genus may indicate a beneficial role in driving response to CRT in the tumours of complete responders through enhanced T cell activation. Similarly, Clostridium species have been associated with a protective effect against CRC, due to their ability to synthesize short-chain fatty acids, notably butyrate, which can promote CRC cell apoptosis and inhibit carcinogenesis32.
The importance of the gut microbiome in response to immunotherapy has garnered much attention in recent years, where species of microbes have been associated with good response to immunotherapy treatments13,14. Given the similarities with the known immune mechanisms at play in radiotherapy response, for example CD8+ T cell activation, an overlap between the established systemic effect of the microbiome on immunotherapy efficacy and that of radiotherapy is likely. Ruminococcaceae and Bacteroides thetaiotaomicron, both enriched in complete responders in our cohort, have been reported to enhance immunotherapy effects13,43, and this mechanism may also contribute to radiotherapy-induced immune clearance of cancer cells. Indeed, a previous study13 showed that there is a correlation between CD8+ T cells and abundance of the Ruminococcaceae family, and that there are statistically more CD8+ T cells in responders versus non-responders to immunotherapy in melanoma patients. Taken together with the associations between differentially abundant bacteria and BATF2 expression, and the increased abundance of commensal bacteria, such as Clostridium spp., the activation of immune pathways in the tumour microenvironment indicates a potential role of the tumour microbiota in the radiotherapy response in rectal cancer. These findings also suggest that there may be value in changing the gut microenvironment (for example through faecal microbiota transplants) as a means of improving the efficacy of CRT, in a similar manner to that reported for immunotherapy. Nevertheless, future studies to assess the clinical utility of incorporating immune and bacterial gene markers to stratify patients for targeted therapy are necessary in larger prospective cohorts.
This study has some limitations: one is the possible selection bias this study has in collecting from two different centres (Christchurch Hospital in New Zealand and the Peter MacCallum Cancer Centre in Melbourne), although a mixture of responses come from both study groups. In addition, this study is computational and further laboratory testing of identified genes, microbes and mechanisms in experimental procedures is highly advocated.
Finally, the results reported here suggest that an increase in the expression of genes contributing to immune activation in tumours compared with normal samples contributes to radiosensitivity. The main hypothesis was that this primes a microenvironment that activates antitumour responses during radiotherapy. Furthermore, bacteria enriched in the tumours of complete responders in this study have previously been associated with CRC and improved efficacy of immunotherapy, and could possibly contribute to the immune activation taking place in complete response to radiotherapy. These data provide future targets for biomarker validation and provide direction to investigate the mechanisms of radiotherapy response in rectal cancer.
Supplementary Material
Acknowledgements
A.K.S and K.W. contributed equally to this study.
Contributor Information
Arielle Kae Sulit, School of Natural Sciences, Massey University, Auckland, New Zealand; Department of Surgery, University of Otago, Christchurch, New Zealand.
Kasmira Wilson, Department of Surgical Oncology, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia; Differentiation and Transcription Laboratory, Sir Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia; Sir Peter MacCallum Department of Oncology, University of Melbourne, Melbourne, Victoria, Australia.
John Pearson, Biostatistics and Computational Biology Unit, University of Otago, Christchurch, New Zealand.
Olin K Silander, School of Natural Sciences, Massey University, Auckland, New Zealand.
Shienny Sampurno, Differentiation and Transcription Laboratory, Sir Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia.
Michael Michael, Sir Peter MacCallum Department of Oncology, University of Melbourne, Melbourne, Victoria, Australia; Department of Medical Oncology, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia.
Robert Ramsay, Department of Surgical Oncology, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia; Differentiation and Transcription Laboratory, Sir Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia.
Alexander Heriot, Department of Surgical Oncology, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia; Sir Peter MacCallum Department of Oncology, University of Melbourne, Melbourne, Victoria, Australia.
Frank Frizelle, Department of Surgery, University of Otago, Christchurch, New Zealand.
Rachel Violet Purcell, Department of Surgery, University of Otago, Christchurch, New Zealand.
Funding
The Health Research Council of New Zealand (Sir Charles Hercus Fellowship), Cancer Society of New Zealand.
Disclosure
The authors declare no conflict of interest.
Supplementary material
Supplementary material is available at BJS Open online.
Data availability
Sequencing data can be found under Bioproject ID PRJNA815861 in the NCBI SRA database. Details for bioinformatics and statistical analyses may be found at https://gitlab.com/alsulit08/uoc_response_rectalca. Limited patient metadata may be provided upon reasonable request to the authors.
References
- 1. Dayde D, Tanaka I, Jain R, Tai MC, Taguchi A. Predictive and prognostic molecular biomarkers for response to neoadjuvant chemoradiation in rectal cancer. Int J Mol Sci 2017;18:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feeney G, Sehgal R, Sheehan M, Hogan A, Regan M, Joyce Met al. Neoadjuvant radiotherapy for rectal cancer management. World J Gastroenterol 2019;25:4850–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fischer J, Eglinton TW, Richards SJ, Frizelle FA. Predicting pathological response to chemoradiotherapy for rectal cancer: a systematic review. Expert Rev Anticancer Ther 2021;21:489–500 [DOI] [PubMed] [Google Scholar]
- 4. Habr-Gama A, Perez RO, Nadalin W, Sabbaga J, Ribeiro U, Silva e Sousa AHet al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy. Ann Surg 2004;240:711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van der Valk MJM, Hilling DE, Bastiaannet E, Meershoek-Klein Kranenbarg E, Beets GL, Figueiredo NLet al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): an international multicentre registry study. Lancet 2018;391:2537–2545 [DOI] [PubMed] [Google Scholar]
- 6. Fischer J, Eglinton TW, Frizelle FA. Clinical predictors of response to chemoradiotherapy for rectal cancer as an aid to organ preservation. ANZ J Surg 2021;91:1190–1195 [DOI] [PubMed] [Google Scholar]
- 7. Garland ML, Vather R, Bunkley N, Pearse M, Bissett IP. Clinical tumour size and nodal status predict pathologic complete response following neoadjuvant chemoradiotherapy for rectal cancer. Int J Colorectal Dis 2014;29:301–307 [DOI] [PubMed] [Google Scholar]
- 8. Ryan JE, Warrier SK, Lynch AC, Ramsay RG, Phillips WA, Heriot AG. Predicting pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a systematic review. Colorectal Dis 2016;18:234–246 [DOI] [PubMed] [Google Scholar]
- 9. Herman JM, Narang AK, Griffith KA, Zalupski MM, Reese JB, Gearhart SLet al. The quality-of-life effects of neoadjuvant chemoradiation in locally advanced rectal cancer. Int J Radiat Oncol 2013;85:e15–e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi Jet al. Human gut microbiome and risk for colorectal cancer. JNCI J Natl Cancer Inst 2013;105:1907–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol 2015;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marchesi JR, Dutilh BE, Hall N, Peters WHM, Roelofs R, Boleji Aet al. Towards the human colorectal cancer microbiome. PLoS One 2011;6:e20447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TVet al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Routy B, Chatelier EL, Derosa L, Duong CPM, Alou MT, Daillère Ret al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–97 [DOI] [PubMed] [Google Scholar]
- 15. Guthrie L, Gupta S, Daily J, Kelly L. Human microbiome signatures of differential colorectal cancer drug metabolism. NPJ Biofilms Microbiomes 2017;3:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Touchefeu Y, Montassier E, Nieman K, Gastinne T, Potel G, Bruley des Varannes Set al. Systematic review: the role of the gut microbiota in chemotherapy- or radiation-induced gastrointestinal mucositis—current evidence and potential clinical applications. Aliment Pharmacol Ther 2014;40:409–421. [DOI] [PubMed] [Google Scholar]
- 17. Vanhoecke BW, De Ryck TR, De boel K, Wiles S, Boterberg T, Van de Wiele Tet al. Low-dose irradiation affects the functional behavior of oral microbiota in the context of mucositis. Exp Biol Med 2016;241:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep 2017;7:11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sulit AK, Kolisnik T, Frizelle FA, Purcell R, Schmeier S. Metafunc: taxonomic and functional analyses of high throughput sequencing for microbiomes. Gut Microbiome 2023;4:e4. [Google Scholar]
- 20. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu G, Wang L-G, Han Y, He Q-Y. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol 2012;16:284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MAet al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. R Core Team . R: A language and environment for statistical computing. https://www.r-project.org/; 2019. (accessed 18 March 2021)
- 26. R Core Team . R: A language and environment for statistical computing.https://www.r-project.org/; 2022
- 27. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese Aet al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 2019;25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xia X, Wu WKK, Wong SH, Liu D, Kwong TNY, Nakatsu Get al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome 2020;8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med 2015;7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu LC-H, Wei S-C, Ni Y-H. Impact of microbiota in colorectal carcinogenesis: lessons from experimental models. Intest Res 2018;16:346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MGet al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 2018;67:1984–1994 [DOI] [PubMed] [Google Scholar]
- 32. Zou S, Fang L, Lee M-H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol Rep 2018;6:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Surace L, Lysenko V, Fontana AO, Cecconi V, Janssen H, Bicvic Aet al. Complement is a central mediator of radiotherapy-induced tumor-specific immunity and clinical response. Immunity 2015;42:767–777 [DOI] [PubMed] [Google Scholar]
- 34. Krombach J, Hennel R, Brix N, Orth M, Schoetz U, Ernst Aet al. Priming anti-tumor immunity by radiotherapy: dying tumor cell-derived DAMPs trigger endothelial cell activation and recruitment of myeloid cells. OncoImmunology 2019;8:e1523097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lhuillier C, Rudqvist N-P, Elemento O, Formenti SC, Demaria S. Radiation therapy and anti-tumor immunity: exposing immunogenic mutations to the immune system. Genome Med 2019;11:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Z, Wei P, Yang Y, Cui W, Cao B, Tan Cet al. BATF2 deficiency promotes progression in human colorectal cancer via activation of HGF/MET signaling: a potential rationale for combining MET inhibitors with IFNs. Clin Cancer Res 2015;21:1752–1763 [DOI] [PubMed] [Google Scholar]
- 37. Guler R, Roy S, Suzuki H, Brombacher F. Targeting Batf2 for infectious diseases and cancer. Oncotarget 2015;6:26575–26582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kanemaru H, Yamane F, Fukushima K, Matsuki T, Kawasaki T, Ebina Iet al. Antitumor effect of Batf2 through IL-12 p40 up-regulation in tumor-associated macrophages. Proc Natl Acad Sci USA 2017;114:E7331–E7340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sendoya JM, Iseas S, Coraglio M, Golubicki M, Robbio J, Salanova Ret al. Pre-existing tumoral B cell infiltration and impaired genome maintenance correlate with response to chemoradiotherapy in locally advanced rectal cancer. Cancers (Basel) 2020;12:E2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu Wet al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 2017;153:1621–1633.e6 [DOI] [PubMed] [Google Scholar]
- 41. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo Met al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 2019;25:667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi W, Shen L, Zou W, Wang Y, Yang J, Wang Yet al. The gut microbiome is associated with therapeutic responses and toxicities of neoadjuvant chemoradiotherapy in rectal cancer patients—a pilot study. Front Cell Infect Microbiol 2020;10:562463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament Cet al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data can be found under Bioproject ID PRJNA815861 in the NCBI SRA database. Details for bioinformatics and statistical analyses may be found at https://gitlab.com/alsulit08/uoc_response_rectalca. Limited patient metadata may be provided upon reasonable request to the authors.