Abstract
Background
Metastasis is a major negative prognostic marker in clear cell renal cell carcinoma (ccRCC). Membrane palmitoylated proteins (MPPs) are a class of cell polarity-associated proteins that function in both cell-cell junction and adhesion. However, the relationship between MPP7 and the prognosis of ccRCC remains elusive. In this study, we aimed to investigate the associations between MPP7 expression with clinical prognosis of ccRCC using bioinformatics analyses.
Methods
The messenger RNA (mRNA) and protein expression patterns of MPP7 in different cancer types were examined using The Cancer Genome Atlas (TCGA) and Human Protein Atlas (HPA) databases, with key clinical characteristics (TNM and pathological stages, pathological grade, survival status) included. A nomogram model using MPP7 expressions and other clinical factors was built to predict the survival probability. The Kaplan-Meier plotter and Cox regression were employed to investigate the clinical significance and prognostic value of MPP7 in ccRCC. MPP7 expression-associated signaling pathways with were analyzed by the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) tools. The Tumor Immune Estimation Resource (TIMER) database was used to investigate the correlation between MPP7 and the infiltration patterns of immune cells.
Results
By analyzing TCGA-kidney renal clear cell carcinoma (TCGA-KIRC) and HPA databases, we found that MPP7 was differentially expressed in tumor tissues and adjacent normal tissues (P<0.001). The MPP7 expression patterns were associated with pathological stage (P<0.001), histological grade (P<0.01), and survival status (P<0.001). Using nomogram model, Cox regression and survival analysis, it showed that MPP7 expressions combined with key clinical factors could accurately predict the clinical prognosis. The promoter methylation patterns of MPP7 were correlated with the clinical factors of ccRCC patients. Furthermore, the KEGG and GO analyses demonstrated that MPP7 is associated with mitochondrial oxidative metabolism. MPP7 expression was associated with multiple types of immune cells and correlated with the enrichment of these cells.
Conclusions
MPP7 is a critical gene links with ccRCC prognosis and is associated with tumor immune status and metabolism. MPP7 could become a potential biomarker and important therapeutic target for ccRCC patients.
Keywords: Clear cell renal cell carcinoma (ccRCC), membrane palmitoylated protein-7 (MPP7), infiltrating immune cells, invasion, mitochondrial metabolism
Highlight box.
Key findings
• Using multi-dimensional bioinformatics analyses, we found that MPP7 is a critical gene whose expression level is significantly correlated with ccRCC prognosis, regulating tumor immune status and metabolism.
What is known and what is new?
• Metastasis is a major negative prognostic marker in ccRCC. MPPs are a class of cell polarity-associated proteins, which function in both cell junctions and adhesion.
• MPP7 is differentially expressed in tumor and paracancerous tissues and is correlated with the clinical prognosis. Moreover, MPP7 also correlates with mitochondrial metabolism and immune cell infiltration.
What is the implication, and what should change now?
• MPP7 could become a potential prognostic and therapeutic target for ccRCC, combined with other key factors. However, more in vivo experiments are still needed to validate the role of MPP7 in cancer.
Introduction
According to the latest statistics, renal cancer is the most common type of tumor in the urinary system, with an increasing incidence in both genders in the United States (1). Clear cell renal cell carcinoma (ccRCC) is an aggressive subtype of renal cancer, accounting for 90% of cases and the majority of deaths of renal cancer. Owing to the lack of typical clinical symptoms, approximately 25–30% of patients with ccRCC have already developed distant metastases upon diagnosis (2,3). In addition, improvement via surgical nephrectomy achieves a good prognosis for early-stage ccRCC patients; however, the 5-year survival rate of patients with advanced ccRCC is still less than 10% (4). Novel therapeutics such as antiangiogenic therapy, immune checkpoint inhibitors, tyrosine kinase inhibitors, and mammalian target of rapamycin (mTOR) inhibitors have brought hope to advanced ccRCC patients. Yet, the prognoses of metastatic ccRCC patients are still poor (5). Therefore, investigating the metastatic mechanisms of ccRCC is important for determining its pathogenesis and developing therapeutic options for ccRCC patients.
A previous study indicated that cell polarity is important for tumor pathogenesis and progression. Changes in the polarity-associated protein expression patterns have been shown to affect epithelial polarity, thereby regulating cell proliferation, migration, and tumorigenesis (6). The membrane-associated guanylate kinases (MAGUKs) are a class of scaffolding-associated proteins, which play key roles in cell adhesion, cell junctions, and cell polarity (7). Moreover, MAGUKs can be divided into several subfamilies according to their structural features, including the membrane palmitoylated protein (MPP) subfamily (8). It was reported that MPPs are a class of cell polarity-associated proteins, which function effectively in both cell junctions and adhesion (8). Recent studies have indicated that MPPs also function in tumorigenesis. For instance, MPP2 was reported to regulate the activity of the oncogene s-Src (9). Moreover, the downregulation of MPP6 is associated with ovarian cancer progression (10). Furthermore, MPP6 suppression ameliorates the anti-cancer activity of Saa3-knockout cancer-associated fibroblasts (11). MPP7 was also shown to function in both pancreatic ductal adenocarcinoma and breast cancer via the modulation of autophagy and activation of the epidermal growth factor receptor (EGFR)/protein kinase B (AKT) signaling, respectively (12,13). However, whether MPP7 functions in the pathogenesis and progression of ccRCC with the possible mechanisms are still not clear. Previous studies indicate that several biomarkers correlate with the clinical prognoses of ccRCC patients. For instance, PTP4A3, GPX1, TAZ were all reported with the clinical prognosis of renal cancer and function via different signaling pathways (14-16). However, the association between MPPs and ccRCC has not been reported before. Therefore, we intended to investigate the potential role of MPP7 in ccRCC by conducting bioinformatics-based analyses using a variety of databases and tools.
In this multi-dimensional bioinformatics-based study, we tried to analyze the associations between MPP7 and the clinical prognosis, while investigating the corresponding relations by data analyses. Data from The Cancer Genome Atlas-kidney renal clear cell carcinoma (TCGA-KIRC) were used to investigate the associations between the MPP7 expression and clinical prognoses [tumor-node-metastasis (TNM) stages, pathological grades, survival status, etc.] in ccRCC patients. Also, the MPP7 levels combined with other clinical factors were used to predict the clinical prognoses of ccRCC patients. Meanwhile, Kyoto Encyclopedia of Genes and Genome (KEGG) and Gene Ontology (GO) analyses were performed and the promoter methylation status of MPP7 was further analyzed. In addition, the correlations between MPP7 expressions and immune infiltration status were also investigated. Our study reveals the significance of MPP7 in ccRCC and its fundamental role in cancer metabolism, immunity, and tumorigenesis. We present the following article in accordance with the REMARK reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-166/rc).
Methods
Gene expression profile analysis
This retrospective study analyzed the MPP7 levels and clinical prognoses of ccRCC patients. The messenger RNA (mRNA) expressions of MPP7 were analyzed by investigating the Tumor Immune Estimation Resource (TIMER) database according to the clinical data uploaded from TCGA (17). The TCGA-KIRC cohort consisted of 539 patients with ccRCC, including 539 tumor samples with 72 adjacent normal tissues, combined with the clinical prognoses and key information. Moreover, two Gene Expression Omnibus (GEO) datasets were analyzed to investigate the MPP7 expression patterns in the normal and tumor tissues. To further validate the expression profiles of MPP7, the Human Protein Atlas (HPA) database (18) was utilized to confirm the expression levels in ccRCC and adjacent tissues by immunohistochemical (IHC) staining. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Correlation analysis of MPP7 expression and clinical prognosis
The associations between MPP7 mRNA levels and the key clinicopathological characteristics, including T and N stages, M status, pathological stages, histological grades, gender, age, and survival status [overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI)], were determined using data from TCGA-KIRC cohort and the ggplot2 R package (R Foundation for Statistical Computing, Vienna, Austria).
Overall and disease-associated survival analyses
The Kaplan-Meier online database (https://kmplot.com/analysis/) was used to assess the associations between MPP7 and the cumulative survival rates of cancer patients based on data from TCGA-KIRC cohort. Specifically, the OS, DSS, and PFI were determined by classifying the ccRCC patients into two groups by the median MPP7 expression value. Log-rank tests with P values and hazard ratios (HRs) with 95% confidence intervals (CIs) were also analyzed. The time-dependent receiver operator characteristic (ROC) curves and a nomogram model graph were plotted using the timeROC, “Survival”, “survminer”, and “rms” R packages for prognosis prediction, based on data from TCGA-KIRC cohort. Moreover, prognosis-associated calibration analyses were conducted by incorporating the key prognosis-associated factors with the predicted 1-, 3-, and 5-year survival rates using the ‘rms’ R package.
Methylation analysis of MPP7 in ccRCC
A comprehensive analysis of the relationship between MPP7 methylation and clinical characteristics, including cancer type and stage, histological grade, gender, and age, was conducted using the University of Alabama Cancer Database (UALCAN) website. In addition, the significance of different methylation sites from TCGA cohort was explored using the online MethSurv tool (19). To determine the mechanisms of MPP7 dysregulations, mutation levels were also presented using the cBioPortal tool for cancer genomics based on TCGA PanCancer Atlas Studies.
Single-cell analysis of MPP7 in the functional states of ccRCC
The CancerSEA website (http://biocc.hrbmu.edu.cn/CancerSEA) was used to analyze the roles of specific genes in the cancer hallmarks of certain cancer types. These phenotypes include angiogenesis, cell apoptosis, cell cycle progression, cell differentiation, DNA damage/repair, epithelial-mesenchymal transition, hypoxia, inflammation status, cell invasion, metastatic status, cell proliferation, cell quiescence, and stemness properties. The results were based on certain single-cell sequencing study to provide more clinical information.
GO and KEGG analyses
To determine the associations between MPP7 and ccRCC, both the co-expressed genes and the differentially expressed genes (DEGs) were screened using the “limma” and “DESeq2” packages in R software (20). GO and KEGG analyses were performed using the “clusterProfiler” R package to explore biological functions and signal channels associated with MPP7. The GO analyses mainly included biological process (BP), cell composition (CC), and molecular function (MF) analyses (21). The STRING tool was used to investigate the functional association network of MPP7 with other proteins (22).
Correlations between MPP7 and immune-infiltrating cells
The TIMER database was adopted to further investigate the associations between the infiltration patterns of immune cells and MPP7 expressions (23). The CIBERSORT tool (24) was adopted to quantify the percentages of immune cells in ccRCC samples by classifying these participants according to their MPP7 expression levels. The “ggplot2”, “tidyverse”, and “reshape2” R packages were adopted. Meanwhile, the TIMER website and the Kaplan-Meier curves were used to investigate and visualize the correlations between infiltrating immune cells and the clinical prognoses of these patients.
Statistical analysis
SPSS software (version 20.0, SPSS Inc., Chicago, IL, USA) and R software (version 4.0.4, R Foundation for Statistical Computing, Vienna, Austria) were utilized for all statistical analyses in this study. The clinical data in this study are presented as the mean ± standard deviation (SD). Specifically, Student’s t-tests of both independent and paired samples were performed to analyze the MPP7 expression difference among different groups. The chi-squared test was used to analyze the MPP7 proportion difference in different groups in terms of the key clinical characteristic variables. The Cox proportional hazards regression models were constructed in both univariate and multivariate manners, adjusting for confounding factors. The OS, DSS, and PFI analyses were conducted using the Kaplan-Meier plots and log-rank tests. A two-tailed P<0.05 was considered to indicate a statistically significant difference.
Results
MPP7 expression is decreased in ccRCC compared with normal tissue
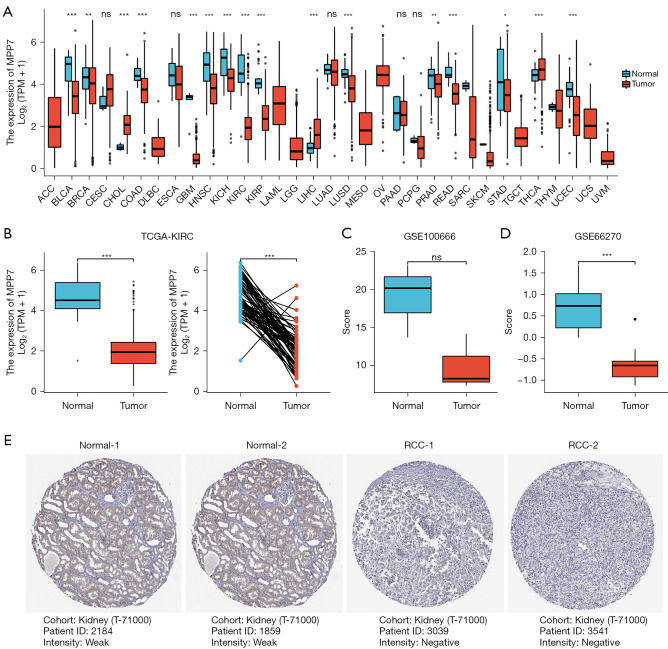
By analyzing the Pan-Cancer TCGA data, we found that MPP7 exhibited decreased expression patterns in several key cancer types (Figure 1A). Therefore, we mainly concentrated on the role of MPP7 in renal cancers, especially the ccRCC. More specifically, KIRC tissues exhibited decreased MPP7 expression using both the unpaired and paired data (P<0.001; Figure 1B). Furthermore, the expression patterns of MPP7 were further validated using the GEO datasets, GSE100666 (n=3 in each group) and GSE66270 (n=14 in each group), which revealed that MPP7 expression was lower in tumor tissues compared to the normal controls (Figure 1C,1D). The HPA database analysis showed via IHC staining that the MPP7 protein was weakly expressed in tumor tissues compared with adjacent normal tissues (Figure 1E).
Figure 1.
Dysregulated MPP7 expression profiles between tumor and adjacent tissues of ccRCC. (A) The MPP7 expression levels in 26 types of cancer and normal tissues (TCGA cancer data vs. TCGA normal data). (B) The MPP7 expression levels in unpaired (n=72 in normal group and n=539 in tumor group) and paired (n=72) patients from the TCGA-KIRC cohort. (C,D) The MPP7 expression patterns of samples from the GSE100666 (n=3 in each group) and GSE66270 (n=14 in each group) datasets. (E) IHC staining (50×) was used to confirm the expression levels in ccRCC and adjacent tissues from the HPA database, with the cohort and patients’ ID listed. URL of each panel: normal-1 (patient ID: 2184) and normal-2 (patient ID: 1859): https://www.proteinatlas.org/ENSG00000150054-MPP7/tissue/kidney#img. RCC-1 (patient ID: 3039) and RCC-2 (patient ID: 3541): https://www.proteinatlas.org/ENSG00000150054-MPP7/pathology/renal+cancer#img. *P<0.05; **P<0.01; ***P<0.001; ns, P>0.05. MPP7, membrane palmitoylated protein-7; TPM, transcripts per million; TCGA, The Cancer Genome Atlas; KIRC, kidney renal clear cell carcinoma; RCC, renal cell carcinoma; ccRCC, clear cell renal cell carcinoma; IHC, immunohistochemical.
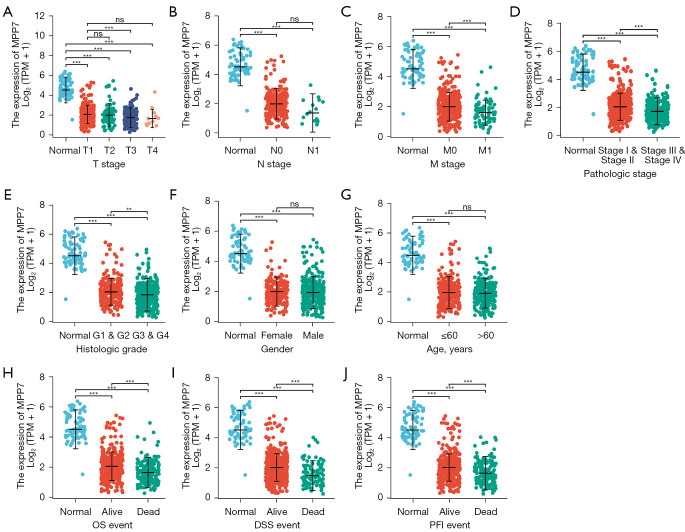
MPP7 expression is correlated with the clinical prognosis of ccRCC patients
The included ccRCC patients were further classified into high (n=270) and low (n=269) MPP7 groups in TCGA-KIRC cohort. Their baseline characteristics are presented in Table 1, which showed that MPP7 expression levels were correlated with T stages, M stages, pathologic stages, and histologic grades (P<0.001). Moreover, MPP7 expression was downregulated and the number of T stages was increased, with significant differences detected between patients with T1 and T4 stages (P<0.001; Figure 2A). Meanwhile, patients with N1 and N0 stages exhibited no difference of MPP7 expressions, while those with M1 stage had lower MPP7 expression levels than those with M0 stage (P<0.001; Figure 2B,2C).
Table 1. The baseline characteristics of patients with ccRCC according to the expression levels of MPP7 in TCGA-KIRC cohort.
| Characteristics | Low MPP7 | High MPP7 | P value |
|---|---|---|---|
| Sample size | 269 | 270 | – |
| T stage, n (%) | <0.001 | ||
| T1 | 118 (43.9) | 160 (59.3) | |
| T2 | 33 (12.3) | 38 (14.1) | |
| T3 | 110 (40.9) | 69 (25.6) | |
| T4 | 8 (3.0) | 3 (1.1) | |
| N stage, n (%) | 0.160 | ||
| N0 | 114 (42.4) | 127 (47.0) | |
| N1 | 11 (4.1) | 5 (1.9) | |
| M stage, n (%) | <0.001 | ||
| M0 | 196 (72.9) | 232 (85.9) | |
| M1 | 56 (20.8) | 22 (8.1) | |
| Pathologic stage, n (%) | <0.001 | ||
| Stage I | 114 (42.4) | 158 (58.5) | |
| Stage II | 25 (9.3) | 34 (12.6) | |
| Stage III | 70 (26.0) | 53 (19.6) | |
| Stage IV | 59 (21.9) | 23 (8.5) | |
| Histologic grade, n (%) | <0.001 | ||
| G1 | 7 (2.6) | 7 (2.6) | |
| G2 | 101 (37.5) | 134 (49.6) | |
| G3 | 104 (38.7) | 103 (38.1) | |
| G4 | 55 (20.4) | 20 (7.4) | |
| Gender, n (%) | 0.547 | ||
| Female | 89 (33.1) | 97 (35.9) | |
| Male | 180 (66.9) | 173 (64.1) | |
| Age (years), n (%) | 0.636 | ||
| ≤60 | 131 (48.7) | 138 (51.1) | |
| >60 | 138 (51.3) | 132 (48.9) |
ccRCC, clear cell renal cell carcinoma; MPP7, membrane palmitoylated protein-7; TCGA, The Cancer Genome Atlas; KIRC, kidney renal clear cell carcinoma.
Figure 2.
Relationship between the MPP7 expression and clinicopathological parameters of ccRCC from the TCGA-KIRC cohort. Correlations between the expression patterns of MPP7 and T stages (A), N stages (B), M status (C), pathological stages (D), histologic grades (E), gender (F), age (G), OS event (H), DSS event (I), and PFI event (J). **P<0.01; ***P<0.001; ns, P>0.05. MPP7, membrane palmitoylated protein-7; TPM, transcripts per million; OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval; ccRCC, clear cell renal cell carcinoma; TCGA, The Cancer Genome Atlas; KIRC, kidney renal clear cell carcinoma.
Moreover, ccRCC patients with a higher pathologic stage or histologic grade exhibited decreased MPP7 levels than those with a lower stage or grade (Figure 2D,2E). There were no significant differences in MPP7 levels between females and males or between elderly (>60 years) and younger (≤60 years) patients (Figure 2F,2G). Furthermore, according to the survival outcomes, dead patients exhibited reduced MPP7 levels compared to those in live patients (Figure 2H-2J).
Univariate Cox analysis revealed that except for gender, all of the key characteristics exhibited significant associations with the clinical prognosis, whereas only M stage, age, and MPP7 expression levels were included in the final model during multivariate analysis (Table 2). These results suggested that MPP7 expression patterns play vital roles in the clinical prognoses of ccRCC patients, combined with other key factors.
Table 2. Univariate and multivariate cox analyses of the key clinical characteristics in ccRCC.
| Characteristics | Total, n | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| T stage | 539 | |||||
| T1 & T2 | 349 | Reference | ||||
| T3 & T4 | 190 | 3.228 (2.382–4.374) | <0.001 | 1.453 (0.633–3.334) | 0.378 | |
| N stage | 257 | |||||
| N0 | 241 | Reference | ||||
| N1 | 16 | 3.453 (1.832–6.508) | <0.001 | 1.438 (0.715–2.894) | 0.309 | |
| M stage | 506 | |||||
| M0 | 428 | Reference | ||||
| M1 | 78 | 4.389 (3.212–5.999) | <0.001 | 2.299 (1.345–3.931) | 0.002 | |
| Pathologic stage | 536 | |||||
| Stage I & II | 331 | Reference | ||||
| Stage III & IV | 205 | 3.946 (2.872–5.423) | <0.001 | 1.348 (0.530–3.427) | 0.531 | |
| Histologic grade | 531 | |||||
| G1 & G2 | 249 | Reference | ||||
| G3 & G4 | 282 | 2.702 (1.918–3.807) | <0.001 | 1.586 (0.951–2.646) | 0.077 | |
| Gender | 539 | |||||
| Male | 353 | Reference | ||||
| Female | 186 | 1.075 (0.788–1.465) | 0.648 | |||
| Age (years) | 539 | |||||
| ≤60 | 269 | Reference | ||||
| >60 | 270 | 1.765 (1.298–2.398) | <0.001 | 1.712 (1.114–2.631) | 0.014 | |
| MPP7 | 539 | |||||
| Low | 269 | Reference | ||||
| High | 270 | 0.367 (0.266–0.505) | <0.001 | 0.423 (0.268–0.667) | <0.001 | |
ccRCC, clear cell renal cell carcinoma; HR, hazard ratio; CI, confidence interval; MPP7, membrane palmitoylated protein-7.
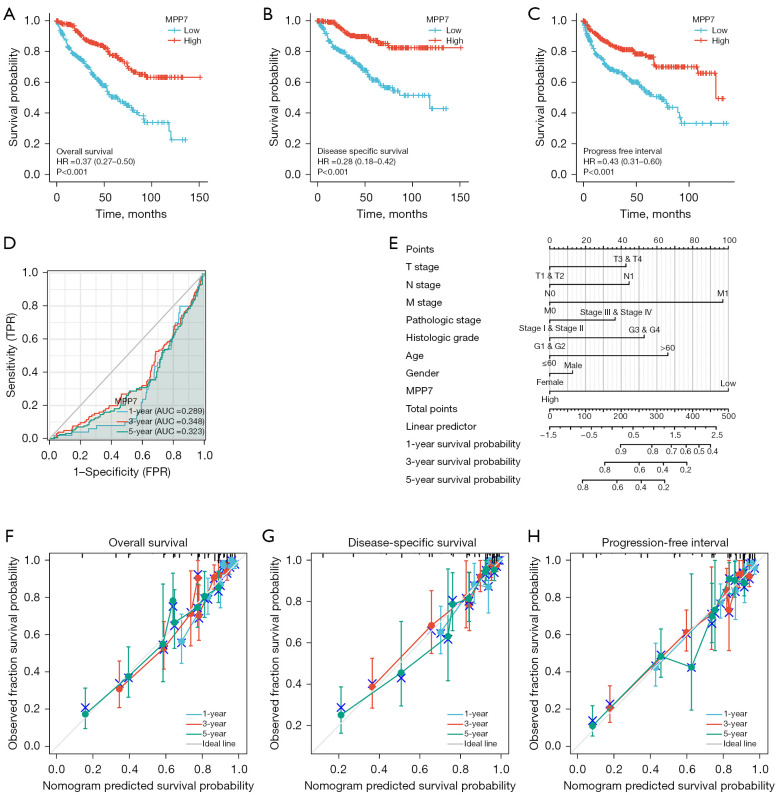
MPP7 is a prognostic factor that is associated with clinical outcomes in ccRCC patients
The prognostic value of MPP7 in ccRCC was assessed by Kaplan-Meier analyses and log-rank tests. Low MPP7 levels were associated with significantly decreased OS (HR =0.37, P<0.001; Figure 3A), DSS (HR =0.28, P<0.001; Figure 3B), and PFI (HR =0.43, P<0.001; Figure 3C), compared with higher MPP7 expression in the TCGA-KIRC cohort.
Figure 3.
Prognostic and predictive value of MPP7 expression in ccRCC patients from TCGA-KIRC cohort. (A-C) The OS, DSS, and PFI survival curves presenting patients with high (red) and low (blue) MPP7 expression in ccRCC. (D) Time-dependent curve analysis to predict the 1-, 3-, and 5-year survival rates. (E) Nomogram model integrating the clinicopathologic factors and MPP7 expression levels to predict the survival probability at 1, 3, and 5 years. (F-H) Prognosis-associated calibration analyses were conducted by incorporating the key prognosis-associated factors to predict the 1-, 3-, and 5-year OS, DSS, and PFI. MPP7, membrane palmitoylated protein-7; HR, hazard ratio; TPR, true positive rate; FPR, false positive rate; AUC, area under the curve; CI, confidence interval; ccRCC, clear cell renal cell carcinoma; TCGA, The Cancer Genome Atlas; KIRC, kidney renal clear cell carcinoma; OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval.
For the time-dependent ROC curves, time-dependent survival ROC curves of MPP7 were created to predict the 1-, 3-, and 5-year survival rates. However, all of these AUC values were lower than 0.6 (which was regarded as the threshold value, Figure 3D), indicating that MPP7 alone is not a suitable index to predict the time-dependent prognosis.
By combining the key clinicopathological factors (age, sex, T stage, N stage, M stage, and histological and pathological stages) and MPP7 expression, we built a nomogram model that can be used to predict the survival probabilities at 1-, 3-, and 5-year for patients in clinical practice (Figure 3E), indicating that the model combining MPP7 expression with other clinical factors is a dominant factor in predicting survival probability for ccRCC patients. Moreover, prognosis-associated calibration analyses were conducted by incorporating MPP7 expressions, M stages, and histologic grades, and demonstrated that these factors could perfectly predict the 1-, 3-, and 5-year survival probabilities (Figure 3F-3H).
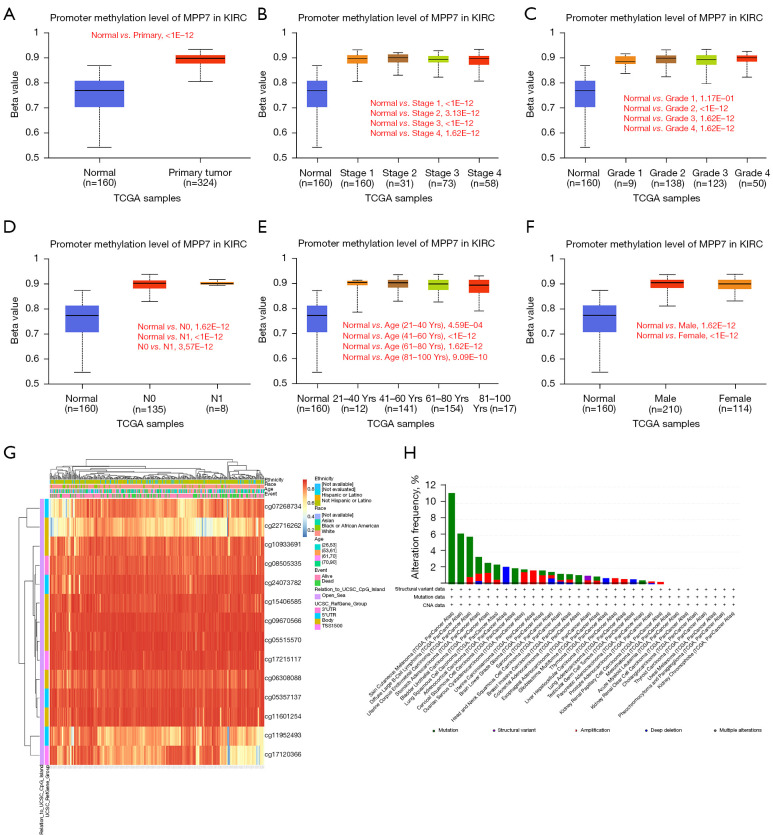
Association of MPP7 methylation and mutation status with clinical features in patients with ccRCC
The associations between promoter methylation levels of MPP7 and clinical features were analyzed, showing that ccRCC tumors exhibited increased methylation levels compared to those in normal tissues (P<1E-12; Figure 4A). Meanwhile, no significant differences were observed among different pathologic stages or histologic grades (Figure 4B,4C). A significant difference in methylation levels was observed between N0 and N1 (P<0.05; Figure 4D) but no significant differences were found between different age groups or genders (Figure 4E,4F).
Figure 4.
DNA methylation levels of MPP7 with its prognostic value in ccRCC. (A) Promoter methylation patterns of MPP7 in both normal and ccRCC tissues using the UALCAN database. (B-F) Promoter methylation level of MPP7 in ccRCC cancer tissues of various tumor stages, tumor grades, nodal metastasis status, ages, and genders using the UALCAN database. (G) The heat map of DNA methylation at CpG sites in the MPP7 gene by the MethSurv database. (H) The structural and mutation data of all cancer types from TCGA PanCancer data. MPP7, membrane palmitoylated protein-7; KIRC, kidney renal clear cell carcinoma; TCGA, The Cancer Genome Atlas; UCSC, University of California Santa Cruz; CNA, copy number alteration; ccRCC, clear cell renal cell carcinoma; UALCAN, University of Alabama Cancer Database; CpG, cytosine-phospho-guanine.
Furthermore, the DNA methylation patterns of MPP7 were assessed according to the prognostic value of each single cytosine-phospho-guanine (CpG) using the MethSurv tool. The MethSurv database results suggested 14 methylation CpG sites, among which most sites had a high level of DNA methylation (Figure 4G). The methylation levels of five CpG sites were correlated with prognosis: cg06308088, cg08505335, cg11601254, cg11952493, and cg17120366 (P<0.05; Table 3). Until now, no significant MPP7 gene mutation data have been observed in ccRCC (Figure 4H).
Table 3. The significant prognostic values of CpG in the MPP7 promoter.
| Gene symbol | CpG name | HR | 95% CI | LR test P value | UCSC Ref gene group | Relation to UCSC CpG island |
|---|---|---|---|---|---|---|
| MPP7 | cg05357137 | 0.652 | 0.424–1.003 | 0.059628 | 5'UTR | Open_Sea |
| cg05515570 | 1.340 | 0.806–2.228 | 0.244227 | Body | Open_Sea | |
| cg06308088 | 0.583 | 0.396–0.857 | 0.007021 | Body | Open_Sea | |
| cg07268734 | 1.228 | 0.772–1.952 | 0.376782 | 5'UTR | Open_Sea | |
| cg08505335 | 1.737 | 1.033–2.923 | 0.027180 | 3'UTR | Open_Sea | |
| cg09670566 | 1.457 | 0.987–2.150 | 0.056424 | Body | Open_Sea | |
| cg10933691 | 1.270 | 0.864–1.867 | 0.223664 | Body | Open_Sea | |
| cg11601254 | 0.487 | 0.324–0.733 | 0.000981 | Body | Open_Sea | |
| cg11952493 | 0.505 | 0.302–0.844 | 0.005223 | 5'UTR | Open_Sea | |
| cg15406585 | 0.640 | 0.398–1.028 | 0.054910 | Body | Open_Sea | |
| cg17120366 | 0.400 | 0.265–0.604 | 5.86E-06 | TSS1500 | Open_Sea | |
| cg17215117 | 1.284 | 0.801–2.056 | 0.287787 | TSS1500 | Open_Sea | |
| cg22716262 | 0.705 | 0.469–1.058 | 0.098402 | Body | Open_Sea | |
| cg24073782 | 1.295 | 0.847–1.980 | 0.242171 | 5'UTR | Open_Sea |
CpG, cytosine-phospho-guanine; MPP7, membrane palmitoylated protein-7; HR, hazard ratio; CI, confidence interval; LR, likelihood ratio; UCSC, University of California Santa Cruz; UTR, untranslated region.
Single-cell sequencing analysis revealed the role of MPP7 in ccRCC tissues
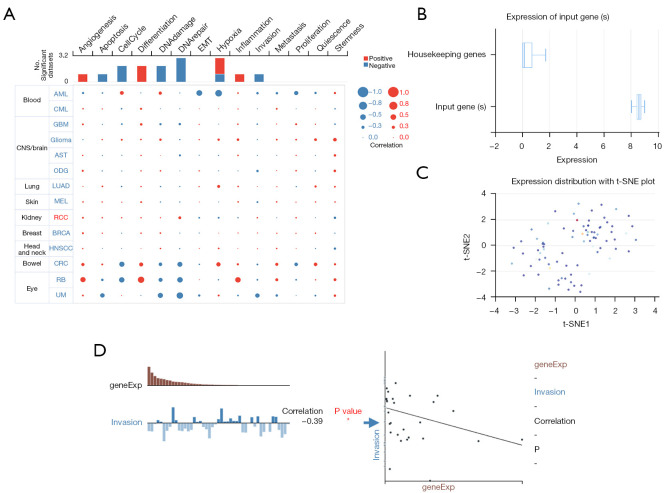
Based on data from the CancerSEA website, we found that no significant correlation existed between MPP7 expression and the hallmarks of ccRCC (Figure 5A). A single-cell sequencing study on ccRCC (25) obtained a total of 83 cells, and the expression distribution was examined with a t-distributed stochastic neighbor embedding (t-SNE) plot (Figure 5B,5C). Moreover, MPP7 expression was correlated with cell invasion in ccRCC, with a correlation of −0.39 and P<0.05 (Figure 5D).
Figure 5.
Single-cell sequencing analysis of MPP7 in the functional states of ccRCC. (A) The CancerSEA database was used to reveal the associations between cancer hallmarks and MPP7 expression. (B-D) A specific single-cell sequencing study concentrated on the role of MPP7 in ccRCC. *P<0.05. EMT, epithelial-to-mesenchymal transition; CNS, central nervous system; t-SNE, t-distributed stochastic neighbor embedding; MPP7, membrane palmitoylated protein-7; ccRCC, clear cell renal cell carcinoma. AML, acute myelocytic leukemia; CML, chronic myelocytic leukemia; GBM, glioblastoma multiforme; AST, astrocytes; ODG, oligodendrogliomas; LUAD, lung adenocarcinoma; MEL, melanoma; BRCA, breast cancer; HNSCC, head and neck squamous cell carcinoma; CRC, colorectal cancer; RB, retinoblastoma; UM, uveal melanoma.
GO and KEGG pathway analyses of MPP7
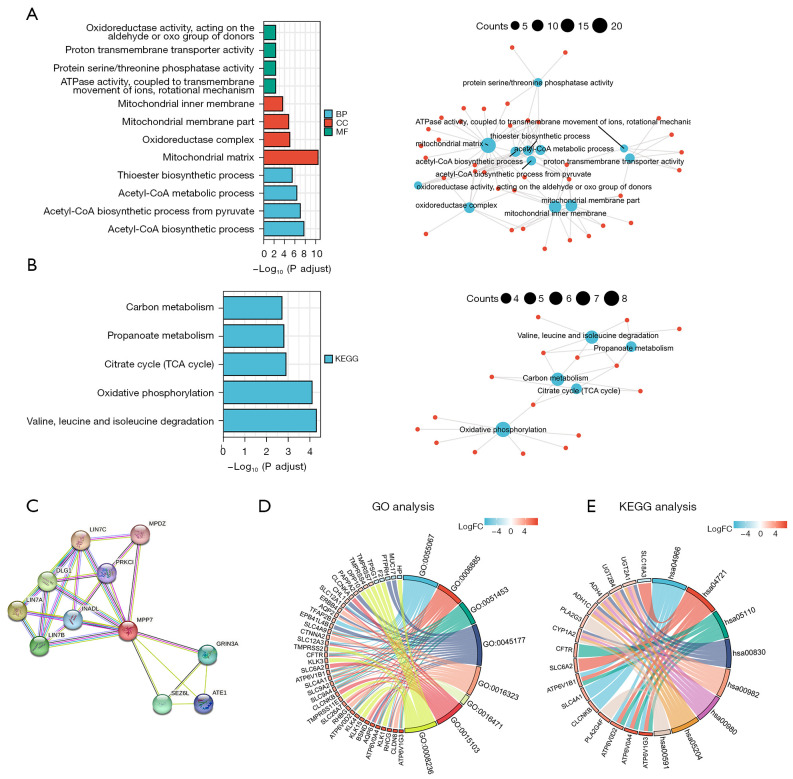
The top 100-most correlated genes with MPP7 in the GO and KEGG enrichment analyses were determined using the “clusterProfiler” R package. Previous studies indicated that MPP7 functions in the regulation of cell polarity and cell junctions. The GO analysis showed that most of the genes were associated with adenosine triphosphatase (ATPase) activity, proton transport, mitochondrial matrix, and the acetyl-CoA metabolic process (Figure 6A). The KEGG data suggested that oxidative phosphorylation and citrate cycle were related to the function of MPP7 (Figure 6B). Moreover, MPP7-binding proteins were investigated using the STRING network (Figure 6C) and were combined with key genes on autophagy, metabolism, cell junction, etc.
Figure 6.
Functional and pathway enrichment analysis of MPP7 in ccRCC. (A) Significant GO terms (including BP, MF, and CC) of the top 100 genes most associated with MPP7. (B) Significant KEGG pathway of the top 100 genes most associated with MPP7. (C) MPP7-binding proteins obtained using the STRING tool. (D,E) GO and KEGG analyses of DEGs associated with MPP7 using TCGA-KIRC data. BP, biological process; CC, cell composition; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genome; TCA, tricarboxylic acid; GO, Gene Ontology; MPP7, membrane palmitoylated protein-7; ccRCC, clear cell renal cell carcinoma; DEGs, differentially expressed genes; TCGA, The Cancer Genome Atlas; KIRC, kidney renal clear cell carcinoma.
To further investigate the signaling pathways associated with MPP7, DEGs exhibiting P<0.05 and log2fold change >2 were collected. Both GO and KEGG analyses were performed using these DEGs, which showed that these DEGs were mainly associated with ion balance, transmembrane transport, and cell polarity by GO analysis, and the vesicle cycle, drug metabolism, and carcinogenesis processes by KEGG analysis (Figure 6D,6E).
MPP7 was significantly correlated with tumor-infiltrating immune cells in ccRCC
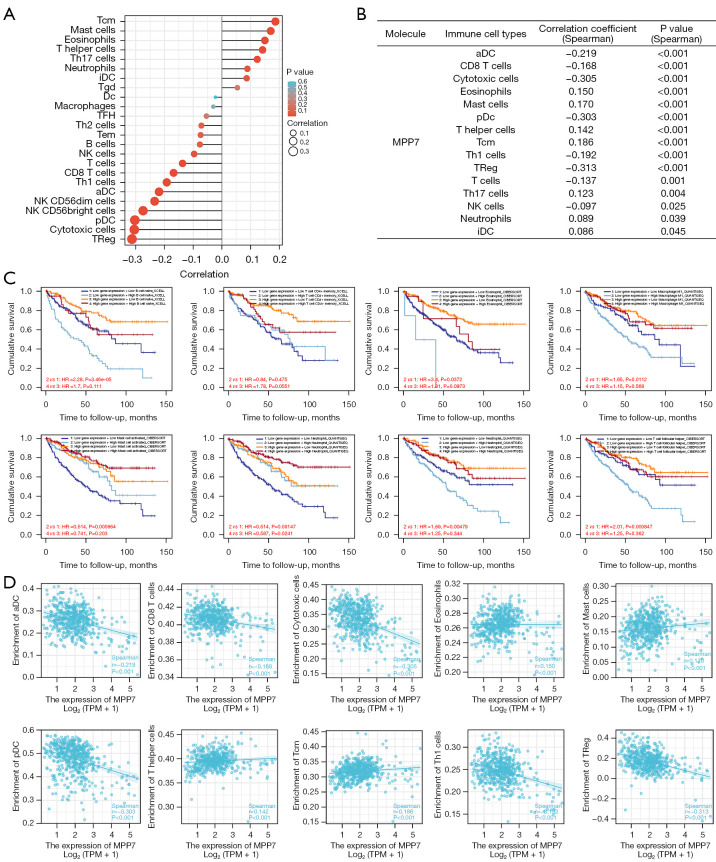
To better reveal the relationship between MPP7 and immune status of ccRCC, we investigatde the potential association between MPP7 expression and infiltration status of 24 immune cell types using R software. MPP7 expression was significantly correlated with dendritic cells (DCs), CD8+ T cells, cytotoxic cells, eosinophils, mast cells, T helper (Th) cells, macrophages, neutrophils, natural killer (NK) cells, central memory T cells (TCMs), Th1 cells, Th17 cells, and regulatory T cells (Tregs) (Figure 7A,7B). Using the TIMER tool, we further investigated the effects of MPP7 and infiltrating cells on the survival rates of patients with ccRCC (Figure 7C). The results showed that naïve B cells, CD4+ memory T cells, eosinophils, M1 macrophages, activated mast cells, neutrophils, activated NK cells, and T follicular helper cells (TFHs) were correlated with survival rates of ccRCC patients.
Figure 7.
Associations between MPP7 expression and immune cell infiltration status of ccRCC. (A) Lollipop chart of the MPP7 expression levels in 24 immune cells. (B) The immune cell infiltration status associated with MPP7 expression with statistical significance. (C) The effects of immune cell infiltration on the prognoses in ccRCC patients. (D) Correlation between MPP7 expression and immune cell infiltration levels, including the aDCs, CD8 T cells, cytotoxic cells, eosinophils, mast cells, pDCs, Th cells, TCMs, Th1 cells, Tregs. TCMs, central memory T cells; Th, T helper; iDCs, immature DCs; DCs, dendritic cells; Tgd, T gamma delta; TFHs, T follicular helper cells; NK, natural killer; aDCs, activated DCs; pDCs, plasmacytoid DCs; Tregs, regulatory T cells; HR, hazard ratio; MPP7, membrane palmitoylated protein-7; TPM, transcripts per million; ccRCC, clear cell renal cell carcinoma; MPP7, membrane palmitoylated protein-7.
Further research showed that MPP7 expression was positively correlated with the infiltration levels of eosinophils (r=0.150, P<0.001), mast cells (r=0.170, P<0.001), Th cells (r=0.142, P<0.001), and TCMs (r=0.186, P<0.001). In contrast, MPP7 expression was negatively correlated with activated DCs (aDCs) (r=−0.219, P<0.001), CD8 T cells (r=−0.168, P<0.001), cytotoxic cells (r=−0.305, P<0.001), plasmacytoid DCs (pDCs) (r=−0.303, P<0.001), Th1 cells (r=−0.192, P<0.001), and Tregs (r=−0.313, P<0.001) (Figure 7D). These results indicated that MPP7 participates in immune cell infiltration and immunity-associated clinical prognosis in ccRCC.
Discussion
Recent advances in understanding the molecular background of ccRCC have led to unprecedented progress in the therapeutic strategies of ccRCC (26). Also, developments have been made in molecularly targeted drugs in advanced ccRCC (27). However, ccRCC is a highly vascularized tumor and prone to distant metastasis. In this study, we mainly focused on MPP7, a critical polarity-related protein, which plays important roles in cell-cell junction and adhesion. However, its role in ccRCC is still unknown. Using TCGA, GEO, and HPA databases, we confirmed that MPP7 expression is decreased in ccRCC tissues compared to that in adjacent tissues. Dysregulated promoter methylation status contribute to the aberrant expression profile. MPP7 expression was markedly correlated with key clinicopathological factors (M stage, pathological stage, histological grade, and survival status). Moreover, MPP7 correlated with dysregulated immune filtration status of several cell types which was associated with clinical prognosis. Furthermore, KEGG and GO analyses revealed that MPP7 functions in mitochondrial oxidative metabolism and ion balance. Therefore, MPP7 could become a potential biomarker for prognostic prediction, combined with other key factors, while could function in the pathogenesis and progression of ccRCC in relation to mitochondrial or immune functions regulation.
A previous report has shown that changes in polarity-associated proteins affect epithelial polarity, thereby regulating cell proliferation, migration, and tumorigenesis (6). MPPs are a class of cell polarity-related proteins comprising a total of seven members (MPP1 to MPP7), which play important roles in cell junctions and adhesion (8). Generally, the protein structure of MPP members contains a highly conserved core element composing of three sequentially-arranged domains. These three domains include a post-synaptic density protein 95/disc large/zonula occludens-1 (PDZ) domain, a Src homology 3 (SH3) domain and a catalytically inactive guanylate kinase (GUK) domain (8). This multidomain architecture and structure allow MPPs to form various protein-protein complexes, facilitating its function in signal transduction, secretion and maintaining cell polarity or cell-cell adhesion (28-30). A previous study by Bohl et al. demonstrated that MPP7 forms a tripartite complex with Discs large homolog 1 (DLG1) and Lin-7 homolog A (LIN7A) or Lin-7 homolog C (LIN7C), which regulates the stability and localization of DLG1 to cell junctions (28). Therefore, it could be speculated that MPPs may participate in tumorigenesis and metastasis. Furthermore, it has also been shown that MPP6 downregulation is associated with ovarian cancer progression (10). The inhibition of MPP6 can reverse the antitumor activity of Saa3-null cancer-associated fibroblasts (11). Moreover, MPP7 functions in both pancreatic ductal adenocarcinoma and breast cancer by modulating autophagy and activating the EGFR/AKT signaling, respectively (12,13). In this study, we systematically analyzed the role of MPP7 in the clinical outcomes of ccRCC; our results showed that MPP7 was correlated with metastasis stage, pathological stage, histological grade, and survival status. More importantly, MPP7 functions in immune infiltration, which may be attributed to the complex protein interactions between the tumor and immune cells. Therefore, MPP7 could be a useful biomarker and a potential therapeutic target for ccRCC patients. However, the effects of MPP7 on tumor proliferation, invasion and metastasis should be validated using both cellular and animal experiments with strict controls.
The tumor microenvironment (TME) comprises both tumor cells and various types of immune cells, and functions in tumor progression, metastasis, and treatment resistance (31). Given that MPP7 is a critical gene in cell junctions and adhesion, we speculated that MPP7 affects the TME by modulating the interactions and infiltration of immune cells and tumor cells. In the present study, we systematically analyzed the correlations between MPP7 and immune cell infiltration as well as the associated clinical prognosis. Our findings showed that MPP7 expression was positively correlated with the infiltration levels of eosinophils, mast cells, Th cells, and TCMs, but was negatively correlated with aDCs, CD8 T cells, cytotoxic cells, pDCs, Th1 cells, and Tregs. Also, the TME comprises a variety of immune cell types that influence tumor cell survival and clinical prognosis. Therefore, we can speculate that MPP7 functions in TME by affecting the immune status of TME. Future studies are needed to further explore the relationship between MPP7 and the functional regulation of these cell types.
To determine the pathways in which MPP7 is involved, a functional analysis of both co-expressed genes and DEGs was conducted using GO/KEGG. Interestingly, we found that most of the MPP7-related genes were associated with the mitochondrial membrane, citrate cycle, and oxidative phosphorylation, instead of cell junction proteins, indicating that MPP7 functions in mitochondrial metabolism and redox regulation. However, previous studies did not report on the relationship between MPPs and mitochondrial functions. Therefore, in addition to its role in cell adhesion, MPP7 might become an important target in mitochondrial metabolism. However, the relationship between MPP7 and mitochondrial functions is still not clear and further studies are needed to explore the role of MPP7 on redox status, energy metabolism, mitochondrial dynamics and cell death.
Energy and mitochondrial metabolism have been shown to play important roles in cancer pathogenesis and progression. Metabolic demands alterations frequently occur during tumor initiation, progression, and metastasis, challenging our understanding of tumor biology and resulting in the development of novel therapeutics. In particular, RCC is considered to be more prone to metabolic disease, with unique glucose and lipid metabolism patterns (32). Wu et al. reported that redox-related genes could accurately predict ccRCC prognosis (33). Xing et al. revealed that glycolysis-related genes could predict OS in ccRCC (34). Meanwhile, tumor-associated immune cells function in the carcinogenesis and treatment of ccRCC (35). Therefore, MPP7 may become an important connection between energy metabolism and immune status in both cancer pathogenesis and treatment.
Although our study has systematically explored the relationship between MPP7 expression and ccRCC in bioinformatic manner, it has some limitations that should be noted. Firstly, most of the data analyzed by bioinformatics methods were downloaded directly from public databases, and our findings require systematical experimental investigations. Secondly, a longer follow-up is required to validate MPP7 as an accurate prognostic predictor for ccRCC patients.
Conclusions
In this study, we report the role of MPP7 in the prognostic prediction of ccRCC for the first time, combining with other key factors. MPP7 was associated with mitochondrial metabolism, immune infiltration, and ion balance, which may be an important link between energy metabolism and immunity of cancer. Therefore, MPP7 could be a potential prognostic biomarker and therapeutic target for ccRCC, combined with other key factors. Further mechanistic studies are needed to validate our findings.
Supplementary
The article’s supplementary files as
Acknowledgments
We acknowledge TCGA, GEO, and HPA databases for providing their platforms and contributors for uploading their meaningful datasets. The results published here are in part based on data generated by TCGA Research Network: https://www.cancer.gov/tcga.
Funding: This study was funded by the Specialized Scientific Research Project of Military Health Care (No. 21BJZ37), the Hainan Provincial Natural Science Foundation of China (No. 821QN383), and the National Natural Science Foundation of China (No. 82270769).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-166/rc
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-166/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-166/coif). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Miller KD, Nogueira L, Devasia T, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin 2022;72:409-36. 10.3322/caac.21731 [DOI] [PubMed] [Google Scholar]
- 2.Grimaldo-Roque HJ, Martinez-Castaneda EA, Morales-Garcia MG, et al. Impact of the Discordance Between Scales of Memorial Sloan-Kettering Cancer Center and International Metastatic Renal Cell Carcinoma Database Consortium in Patients' Prognosis With Metastatic Renal Cancer. World J Oncol 2022;13:53-8. 10.14740/wjon1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur Urol 2022;82:399-410. 10.1016/j.eururo.2022.03.006 [DOI] [PubMed] [Google Scholar]
- 4.Larroquette M, Lefort F, Heraudet L, et al. Therapeutic Management of Metastatic Clear Cell Renal Cell Carcinoma: A Revolution in Every Decade. Cancers (Basel) 2022;14:6230. 10.3390/cancers14246230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choueiri TK, Halabi S, Sanford BL, et al. Cabozantinib Versus Sunitinib As Initial Targeted Therapy for Patients With Metastatic Renal Cell Carcinoma of Poor or Intermediate Risk: The Alliance A031203 CABOSUN Trial. J Clin Oncol 2017;35:591-7. 10.1200/JCO.2016.70.7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandalovičová A, Vomastek T, Rosel D, et al. Cell polarity signaling in the plasticity of cancer cell invasiveness. Oncotarget 2016;7:25022-49. 10.18632/oncotarget.7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye F, Zeng M, Zhang M. Mechanisms of MAGUK-mediated cellular junctional complex organization. Curr Opin Struct Biol 2018;48:6-15. 10.1016/j.sbi.2017.08.006 [DOI] [PubMed] [Google Scholar]
- 8.Chytła A, Gajdzik-Nowak W, Olszewska P, et al. Not Just Another Scaffolding Protein Family: The Multifaceted MPPs. Molecules 2020;25:4954. 10.3390/molecules25214954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumgartner M, Weiss A, Fritzius T, et al. The PDZ protein MPP2 interacts with c-Src in epithelial cells. Exp Cell Res 2009;315:2888-98. 10.1016/j.yexcr.2009.07.028 [DOI] [PubMed] [Google Scholar]
- 10.Xu F, Si X, Du J, et al. Downregulating SynCAM and MPP6 expression is associated with ovarian cancer progression. Oncol Lett 2019;18:2477-83. 10.3892/ol.2019.10542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djurec M, Graña O, Lee A, et al. Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci U S A 2018;115:E1147-56. 10.1073/pnas.1717802115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao W, Fan L, Li M, et al. MPP7 promotes the migration and invasion of breast cancer cells via EGFR/AKT signaling. Cell Biol Int 2021;45:948-56. 10.1002/cbin.11538 [DOI] [PubMed] [Google Scholar]
- 13.New M, Van Acker T, Sakamaki JI, et al. MDH1 and MPP7 Regulate Autophagy in Pancreatic Ductal Adenocarcinoma. Cancer Res 2019;79:1884-98. 10.1158/0008-5472.CAN-18-2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y, Xu T, Li S, et al. GPX1, a biomarker for the diagnosis and prognosis of kidney cancer, promotes the progression of kidney cancer. Aging (Albany NY) 2019;11:12165-76. 10.18632/aging.102555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan H, Bao L, Song Z, et al. High expression of TAZ serves as a novel prognostic biomarker and drives cancer progression in renal cancer. Exp Cell Res 2019;376:181-91. 10.1016/j.yexcr.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 16.Song Q, Zheng Y, Wu J, et al. PTP4A3 Is a Prognostic Biomarker Correlated With Immune Infiltrates in Papillary Renal Cell Carcinoma. Front Immunol 2021;12:717688. 10.3389/fimmu.2021.717688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Li Z, Zhou S, et al. NCAPG2 could be an immunological and prognostic biomarker: From pan-cancer analysis to pancreatic cancer validation. Front Immunol 2023;14:1097403. 10.3389/fimmu.2023.1097403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colwill K, Renewable Protein Binder Working Group ; Gräslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods 2011;8:551-8. 10.1038/nmeth.1607 [DOI] [PubMed] [Google Scholar]
- 19.Modhukur V, Iljasenko T, Metsalu T, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018;10:277-88. 10.2217/epi-2017-0118 [DOI] [PubMed] [Google Scholar]
- 20.Shang BB, Chen J, Wang ZG, et al. Significant correlation between HSPA4 and prognosis and immune regulation in hepatocellular carcinoma. PeerJ 2021;9:e12315. 10.7717/peerj.12315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Q, Hou Z, Zuo S, et al. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52. Cancer Sci 2019;110:1194-207. 10.1111/cas.13951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim KT, Lee HW, Lee HO, et al. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol 2016;17:80. 10.1186/s13059-016-0945-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801-6. 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava A, Doppalapudi SK, Patel HV, et al. The roaring 2020s: a new decade of systemic therapy for renal cell carcinoma. Curr Opin Oncol 2022;34:234-42. 10.1097/CCO.0000000000000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohl J, Brimer N, Lyons C, et al. The stardust family protein MPP7 forms a tripartite complex with LIN7 and DLG1 that regulates the stability and localization of DLG1 to cell junctions. J Biol Chem 2007;282:9392-400. 10.1074/jbc.M610002200 [DOI] [PubMed] [Google Scholar]
- 29.Stucke VM, Timmerman E, Vandekerckhove J, et al. The MAGUK protein MPP7 binds to the polarity protein hDlg1 and facilitates epithelial tight junction formation. Mol Biol Cell 2007;18:1744-55. 10.1091/mbc.e06-11-0980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podkalicka J, Biernatowska A, Majkowski M, et al. MPP1 as a Factor Regulating Phase Separation in Giant Plasma Membrane-Derived Vesicles. Biophys J 2015;108:2201-11. 10.1016/j.bpj.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MC, Jin Z, Kolb R, et al. Updates on Immunotherapy and Immune Landscape in Renal Clear Cell Carcinoma. Cancers (Basel) 2021;13:5856. 10.3390/cancers13225856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi X, Li Q, Che X, et al. The Uniqueness of Clear Cell Renal Cell Carcinoma: Summary of the Process and Abnormality of Glucose Metabolism and Lipid Metabolism in ccRCC. Front Oncol 2021;11:727778. 10.3389/fonc.2021.727778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Wei X, Feng H, et al. Integrated Analysis to Identify a Redox-Related Prognostic Signature for Clear Cell Renal Cell Carcinoma. Oxid Med Cell Longev 2021;2021:6648093. 10.1155/2021/6648093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing Q, Zeng T, Liu S, et al. A novel 10 glycolysis-related genes signature could predict overall survival for clear cell renal cell carcinoma. BMC Cancer 2021;21:381. 10.1186/s12885-021-08111-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarrabi K, Walzer E, Zibelman M. Immune Checkpoint Inhibition in Advanced Non-Clear Cell Renal Cell Carcinoma: Leveraging Success from Clear Cell Histology into New Opportunities. Cancers (Basel) 2021;13:3652. 10.3390/cancers13153652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as