Abstract
The genetic basis for chronic persistence of Brucella abortus in lymphoid organs of mice, cows, and humans is currently unknown. We identified B. abortus genes involved in chronic infection, by assessing the ability of 178 signature-tagged mutants to establish and maintain persistent infection in mice. Each mutant was screened for its ability to colonize the spleens of mice at 2 and 8 weeks after inoculation. Comparison of the results from both time points identified two groups of mutants attenuated for chronic infection in mice. The first group was not recovered at either 2 or 8 weeks postinfection and was therefore defective in establishing infection. Mutants in this group carried transposon insertions in genes involved in lipopolysaccharide biosynthesis (wbkA), in aromatic amino acid biosynthesis, and in type IV secretion (virB1 and virB10). The second group, which was recovered at wild-type levels 2 weeks postinfection but not 8 weeks postinfection was able to establish infection but was unable to maintain chronic infection. One mutant in this group carried a transposon insertion in a gene with homology to gcvB of Mycobacterium tuberculosis, encoding glycine dehydrogenase, an enzyme whose activity is increased during the state of nonreplicating persistence. These results suggest that some mechanisms for long-term persistence may be shared among chronic intracellular pathogens. Furthermore, identification of two groups of genes, those required for initiating infection and those required only for long-term persistence, suggests that B. abortus uses distinct sets of virulence determinants to establish and maintain chronic infection in mice.
Bacteria causing chronic infections, such as Mycobacterium tuberculosis, Chlamydia trachomatis, and Brucella abortus are able to evade the host's immune system throughout the infection by colonizing an intracellular niche. This lifestyle may require adaptations other than the brief survival in phagocytic cells observed for well-characterized intracellular pathogens such as Salmonella serotypes which cause an acute infection. While M. tuberculosis and C. trachomatis are difficult to manipulate genetically, the genetic manipulation of the Brucella genome can be performed routinely, using tools such as plasmid vectors and systems for Tn5 mutagenesis (20–22). Thus, identification of the genes required by B. abortus to cause infection may reveal virulence mechanisms of chronic disease caused by other intracellular pathogens. A recent improvement in Tn5 mutagenesis, known as signature-tagged transposon mutagenesis (STM), has been developed for the in vivo selection of Tn5 mutants that are defective in colonization (18). This method uses experimentally infected animals to identify mutants that are attenuated in vivo from a large, mixed pool of mutants (29). Since Tn5 can be used in B. abortus, STM can be used to identify genes that are necessary for chronic intracellular infection.
Brucellosis is endemic in Mediterranean countries and Central and South America and is manifested as an undulant fever in humans that, if untreated, can develop into a chronic infection with symptoms persisting for several months (32). Chronic infections may result in infection of secondary tissues, including heart and brain, if the infection is left untreated. Symptoms may also recur years after the original infection. B. abortus infection is acquired by humans through contact with infected livestock and consumption of unpasteurized dairy products. Bacteria cause a systemic infection and localize preferentially to organs that are rich in elements of the reticuloendothelial system, such as liver, spleen, and lymph nodes, where they survive and multiply within host macrophages. B. abortus has been found to inhibit the bactericidal functions of phagocytes, including phagolysosomal fusion, neutrophil degranulation, and the oxidative burst (5); however, as with other intracellular pathogens which cause chronic infection, the genetic basis for the interaction of Brucella with phagocytic cells is still poorly understood. B. abortus virulence is conveniently studied in a mouse model that mimics the chronic infection observed in humans. Here bacteria are found intracellularly, within macrophages of infected organs (25). Experimental infection of BALB/c mice has shown that the infection has two phases: during the first 2 weeks, bacteria multiply rapidly. In the second phase, bacterial numbers stabilize over the next 5 to 6 weeks and then decrease slowly. Bacteria have been recovered from spleens of infected mice as late as 24 weeks postinfection (26, 28). The different phases of B. abortus infection in mice raise the question whether this pathogen uses different sets of virulence genes during the early and late stages of this disease.
To address this question, we have performed a random screen of the genome of B. abortus to identify genes required for infection at an early (2 weeks postinfection) and a late (8 weeks postinfection) time point postinoculation. Comparative analysis of these results provided new insight into the genetic basis for chronic intracellular infection and B. abortus pathogenesis. Furthermore, our results suggest that a better understanding of the mechanisms by which B. abortus is able to cause chronic intracellular infection may ultimately reveal strategies that are shared by other, less tractable pathogens.
MATERIALS AND METHODS
Construction of mutants and growth conditions.
B. abortus strain 2308 (obtained from B. L. Deyoe, National Animal Disease Center, Ames, Iowa) was used as a host for STM. A bank comprising 240 signature-tagged B. abortus mutants (ST mutants) was constructed using a pool containing 10,000 uniquely tagged miniTn5Km2 derivatives carried on suicide plasmids, which was obtained from David Holden (18). The transposons were introduced into B. abortus S2308 by electroporation and selected on tryptic soy agar (TSA; Difco) containing 100 mg of kanamycin (Km) per liter (1). Approximately 20 mutants were taken from each of 10 individual electroporations in order to minimize the isolation of siblings. Mutants resistant to ampicillin were eliminated from the pool, since they carry the suicide vector inserted into the chromosome. For infection of mice, B. abortus strains were grown on potato infusion agar (PIA; Difco) for 48 h and resuspended at the appropriate concentration in phosphate-buffered saline (PBS) (2). For determination of auxotrophy, the chemically defined solid medium formulated by Gerhardt was used, which contains glycerol, glutamate, lactate, thiamine, nicotinic acid, biotin, calcium pantothenate, and inorganic salts (15). This basal medium was supplemented with 10 groups of amino acids to perform auxanography for attenuated mutants. All work with live B. abortus was performed in a biosafety level 3 containment facility following Centers for Disease Control-National Institutes of Health guidelines.
Infection of mice.
For the mutant screen, pools of 46 mutants were used to infect groups of six 6- to 8-week-old BALB/c mice intraperitoneally (i.p.) at a total dose of approximately 106 CFU. Groups of three mice were sacrificed at 2 and at 8 weeks postinfection, and bacteria were recovered from the spleens. Spleens were homogenized in 3 ml of PBS, and serial 10-fold dilutions were plated on TSA-Km.
Signature-tagged screen and hybridization analysis.
Bacteria recovered from murine spleens at 2 and 8 weeks postinfection were pooled to obtain one output pool from each mouse. Total chromosomal DNA was prepared from each output pool to serve as a template for generating probes containing labeled tags, as described before (18). Labeled tags used as probes were prepared by incorporation of [α-32P]dATP during PCR amplification of tags from total chromosomal DNA. Blots containing chromosomal DNA from individual mutants in each pool were prepared by transferring pools of B. abortus ST mutants from 96-well plates onto nylon membranes laid on top of TSA plates with a 48-prong replicator. Hybridization and washes were carried out under stringent conditions (4). To identify potentially attenuated mutants, hybridization signals on a blot probed with labeled tags prepared from the input pool (inoculum) were compared with three identical blots hybridized with tags prepared from the output pools recovered from the three mouse spleens at each time point. Mutants giving a hybridization signal with the input pool but no signal in the output pool of at least two mice were selected for further study. To confirm that each mutant carried a single insertion of mini-Tn5 in the chromosome, mutants were analyzed by Southern blot (4). Chromosomal DNA was prepared (4) and digested with EcoRI, which cuts once within the transposon, outside the kanamycin resistance gene. After agarose gel electrophoresis and transfer to nylon membranes, chromosomal DNA was analyzed by hybridization with a probe containing a 1.3-kb fragment of the Tn903 kanamycin resistance gene from plasmid pUC4KSAC (Pharmacia). Mutants whose chromosomal DNA gave a hybridization signal with a single band on the Southern blot were determined to have only one copy of the transposon inserted into the chromosome.
Competitive infection assay.
For competitive infection experiments, groups of four mice were inoculated i.p. with an approximately 1:1 mixture of mutant and wild-type B. abortus, at a total dose of approximately 105 CFU. Mice were sacrificed at 2 or 8 weeks postinfection, and bacteria were recovered from infected spleens. The CFU of mutant and wild-type B. abortus recovered from infected spleens were enumerated by serial dilution in PBS and plating in parallel on TSA and TSA-Km. Numbers of wild-type and mutant bacteria were calculated by subtracting the CFU recovered on TSA-Km, on which only the Tn5 mutants are able to grow, from the CFU recovered on TSA plates, representing the total number of bacteria recovered. Data were normalized by dividing the output ratio of (CFU [wild type]/CFU [mutant]) by the input ratio of (CFU [wild type]/CFU [mutant]). All data were then converted logarithmically for statistical analysis. A Student's t test was used to determine whether the wild-type/mutant ratio recovered from infected spleens was significantly different from the wild-type/mutant ratio present in the challenge inoculum.
Cloning and sequence analysis.
Transposon-flanking DNA was cloned by inverse PCR as described previously (6) using RsaI for digestion of chromosomal DNA and the primer pair SIGN-10 (5′-GCCGAACTTGTGTATAAGAGTCAG-3′) and SIGN-11 (5′-AAAGGTAGCGTTGCCAATG-3′). The PCR products were ligated into cloning vector pCR II (Invitrogen), and plasmid DNA for sequencing was isolated from Escherichia coli strain DH5α (17) using ion-exchange columns from Qiagen. In addition, larger DNA fragments flanking the transposon insertions in several of the mutants were cloned by ligating PstI- or EcoRI-restricted genomic DNA into cloning vector pBluescript SK+ restricted with the appropriate enzyme and selection for the kanamycin resistance marker of mini-Tn5Km2 (12). Nucleotide sequences were analyzed using the MacVector 6.5 software package (Oxford Molecular Group). Sequence homology was determined using the BLAST2 search algorithm at the National Center for Biotechnology Information (NCBI) (3).
RESULTS
Generation of signature-tagged mutants of B. abortus.
B. abortus 2308 was mutagenized with a pool of signature-tagged mini-Tn5Km2 derivatives carried on plasmid pUT by electroporation, and mutants were selected on TSA-Km. These mutants were screened for susceptibility to ampicillin to eliminate strains (fewer than 2% of mutants) carrying cointegrates of the suicide vector pUT inserted in the chromosome. To confirm that mutants obtained from the same electroporation are not siblings but arise from independent transposition events, Southern hybridization was performed using the 1.3-kb EcoRI fragment of pUC4KSAC, which contains the Tn903 Kmr gene (12). Using this probe, the hybridization profiles of EcoRI-digested chromosomal DNA from 10 randomly chosen mutants from a single electroporation were compared. Since EcoRI cuts only once within miniTn5Km2, a mutant with a single transposon insertion should have only one band hybridizing with the probe. All of the mutants examined had a single band of unique size hybridizing with the probe, indicating that each mutant contained a single, unique insertion of the transposon (data not shown).
Signature-tagged screen of B. abortus mutants in mice.
To identify mutants defective in establishing chronic infection, we used the BALB/c mouse model of chronic brucellosis. Mice infected i.p. with B. abortus have been shown to harbor bacteria in the spleen for up to 24 weeks postinfection (5, 9). The persistence of B. abortus in mice is similar to chronic infection observed in other host species, including humans.
It has not been shown whether attenuated B. abortus mutants exhibit competitive infection defects in mice when coinoculated with a virulent strain. We therefore performed a preliminary competitive infection experiment using B. abortus 2308 and CA180, a Tn5 mutant defective in the synthesis of the O antigen of lipopolysaccharide (LPS), which was characterized previously (1). Three mice were infected i.p. with a mixture containing 6.4 × 104 CFU of B. abortus 2308 and 4.1 × 104 CFU of CA180. At 1 week postinfection, no mutant B. abortus could be recovered from the spleens of any infected mouse, while the number of wild-type B. abortus ranged from 6 × 103 to 2 × 106 CFU/spleen. This result showed that a mutant with a defect in a known B. abortus virulence factor, LPS, exhibits a competitive infection defect in the BALB/c mouse model of brucellosis. Furthermore, it suggested that STM, which utilizes competitive infection as the basis for screening pools of mutants, could be used to screen for mutants defective in chronic infection.
For the screen in mice, pools of 46 mutants were grown individually in tryptic soy broth (TSB) in 96-well plates for 48 h and then stamped with a 48-prong replicator onto PIA plates and grown for 48 h. Bacteria were resuspended from PIA plates, and the concentration was adjusted to approximately 107 CFU/ml with PBS. Of this suspension, 0.1 ml was injected i.p. into each of six BALB/c mice. In the pooled inoculum, the dose of each individual mutant was therefore approximately 104 CFU. At 2 and 8 weeks postinfection, groups of three mice were sacrificed. At necropsy, the only sign of disease evident was enlargement of the spleen. To recover bacteria, spleens were homogenized in PBS, and serial 10-fold dilutions were plated on TSA-Km. After incubation for 4 days, bacteria were resuspended from plates containing between 1,000 and 5,000 colonies, and chromosomal DNA was prepared from the recovered pool of mutants. Probes of input and output pools were generated by PCR amplification of tags from chromosomal DNA of pooled ST mutants as described before (18) and used for hybridization with dot blots containing individual mutants in the corresponding pool. For each time point, a fresh input pool probe was prepared and a blot was hybridized for comparison with the output pools recovered from the mice. We found that 39 mutants failed to give a reproducible hybridization signal when the input pool probe was prepared more than once from a chromosomal DNA preparation. These mutants were eliminated from the screen. Thus, a total of 178 mutants with consistently hybridizing tags were screened.
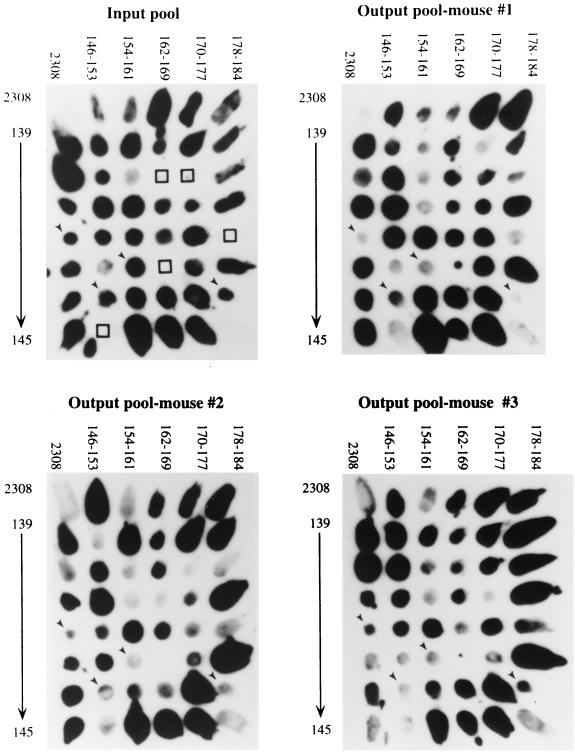
Mutants which gave weak or no hybridization signals on output pool blots from at least two of the three mice at each time point were identified as putatively attenuated by comparison with input pool blots (Fig. 1). At 2 weeks postinfection, 28 mutants exhibited a reduction in the hybridization signal, suggesting reduced recovery from the mouse spleens. Of these 28 mutants, 11 exhibited a reduced hybridization signal in output pools recovered at 8 weeks postinfection.
FIG. 1.
Colony blots showing hybridization of B. abortus ST mutants with probes containing tags amplified from the input pool and from output pools recovered from spleens of three mice at 8 weeks postinfection. At the upper left and lower right corner of each blot is the parent stain, B. abortus 2308. Arrows indicate mutants (BA142, BA152, BA159, and BA184 [left to right]) which gave weak signals in the output pools of at least two of the three mice inoculated and whose competitive infection defect was confirmed subsequently. Open squares indicate mutants that gave inconsistent hybridization results and were therefore eliminated from the screen.
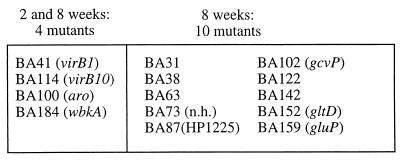
In addition to the 11 mutants reduced in colonization at both 2 and 8 weeks, we identified 16 mutants that were only defective for colonization at 8 weeks. While mutants identified from both the 2- and 8-week output pools may be unable to establish infection, those identified only from the 8-week output pools may be defective in sustaining chronic infection. The 17 mutants identified only from the 2-week output pools may represent mutants which are slow to colonize the spleen but are still able to persist. However, infection defects in these mutants were not characterized further, since these mutants were able to sustain chronic infection. Thus, two groups of mutants, those identified at both 2 and 8 weeks and those identified only at 8 weeks, were considered putatively attenuated for chronic infection and were chosen for further study (Table 1).
TABLE 1.
Confirmation of competitive defects of STM mutants by competitive infection of mice with B. abortus S2308a
| Mutant | Competitive index
|
Mutant | Competitive index in vivo at 8 wk | ||
|---|---|---|---|---|---|
| In vivo
|
In vitro | ||||
| 2 wk | 8 wk | ||||
| BA11 | 2.29* | 1.86 | BA18 | 0.064 | |
| BA41 | 4.74* | >29,512*** | 0.97 | BA27 | 2.24 |
| BA73 | 3.36 | 147.9* | 2.74 | BA31 | 3.63* |
| BA87 | 1.08 | 10.47** | 0.79 | BA38 | 7.76* |
| BA100 | 20.89*** | >758.6*** | 4.70 | BA63 | 1.86* |
| BA102 | 1.47 | 93.3*** | 1.80 | BA70 | 3.39 |
| BA109 | 0.92 | 3.21 | BA99 | 1.02 | |
| BA114 | 133.07** | >1,047.1*** | 0.91 | BA105 | 2.00 |
| BA157 | 0.74 | 1.58 | BA119 | 6.31 | |
| BA159 | 1.50 | 23.44* | 2.20 | BA116 | 2.51 |
| BA184 | 1,306.8*** | >2,168.2*** | 0.57 | BA122 | 95.50* |
| BA130 | 3.02 | ||||
| BA138 | 3.72 | ||||
| BA140 | 1.20 | ||||
| BA142 | 70.79*** | ||||
| BA152 | 72.44*** | ||||
Competitive index was calculated as [(CFUS2308/CFUmutant) recovered/(CFUS2308/CFUmutant) inoculated]. Significant differences between competitive indices of STM mutants and a control, nonattenuated mutant were compared using a Student t test. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.005.
Competitive infection of mice with mutants identified by STM.
Following our identification of mutants putatively attenuated for chronic infection, we performed a quantitative assay to confirm their colonization defect. To this end, each of the 27 mutants identified by the STM screen was inoculated i.p. at a 1:1 ratio with wild-type B. abortus at a total dose of approximately 105 CFU to groups of four mice (Table 1). As a control, mutant BA53, which was recovered at both 2 and 8 weeks postinfection from mice, was selected at random from the STM pool as a negative control and administered to mice in a competitive infection experiment as described above. During mixed-infection experiments, 15 mutants identified in the STM screen were found to have a significant defect in persistence in mouse spleens. One of these mutants, BA11, had a significant colonization defect only at 2 weeks and therefore was not characterized further. Thus, for 14 of 27 mutants identified in the preliminary STM screen, the putative chronic infection defect could be confirmed by a significant reduction in colonization during competitive-infection experiments. Mutants which did not have significant colonization defects in the competitive-infection assay either may have been false-positives in the STM screen or may only display competitive colonization defects when inoculated at a wild type-to-mutant ratio of greater than 1. However, these possibilities were not investigated further. Mutants with significant colonization defects during mixed infections fell into two groups: those reduced in colonization at both 2 and 8 weeks, and those reduced at only 8 weeks (Fig. 2). Subsequent mouse infection experiments showed that mutants with competitive colonization defects are also attenuated when administered alone to mice (data not shown). To determine whether colonization of the spleen was reduced due to a general growth defect, mutants displaying significant defects at both 2 and 8 weeks postinoculation were assayed for in vitro growth defects by inoculating TSB with a mixture of wild-type and ST mutant strains (Table 1). With the exception of BA100, which was outgrown almost fivefold by the parent strain, none of the mutants displayed a strong growth deficiency in laboratory medium.
FIG. 2.
Mutants with statistically significant colonization defects, as confirmed by individual competitive infections and statistical analysis. Homologues of disrupted genes are given in parentheses. nh, no homology.
Identification of inactivated genes.
Genes inactivated by mini-Tn5Km2 insertions were identified by cloning transposon-flanking DNA, sequence determination, and comparison with the GenBank database using the BLAST2 search program at NCBI. Transposon-flanking DNA was cloned from eight mutants.
Mutants with defects in establishing chronic infection included BA184, which carried a transposon insertion in a B. abortus homologue of Brucella melitensis wbkA, encoding a mannosyltransferase that functions in the biosynthesis of O antigen (16). LPS biosynthesis is required for virulence in Brucella species, and it has recently been shown that strains of B. abortus and B. melitensis carrying defined mutations in genes required for LPS biosynthesis are unable to establish infection in mice (1, 16, 24). Identification of this known virulence factor thus validated the STM screen.
Two additional mutants with defects in establishing persistent infection, BA41 and BA114, carried transposon insertions in homologues (virB1 and virB10, respectively) of the Brucella suis virB locus (27). B. suis mutants carrying disruptions at this locus show a reduced ability to multiply in vitro within macrophages and HeLa cells. Our data suggest that this locus is required for establishing chronic infection in organs of the mouse.
We could obtain no clone from DNA flanking the transposon insertion of the fourth mutant (BA100) defective in establishing chronic infection. However, BA100 was unable to grow on Brucella minimal medium unless supplemented with a combination of aromatic amino acids. These auxanography data indicated that this mutant was defective in an early step of the aromatic amino acid biosynthesis pathway.
Sequences obtained from the transposon insertion site in three mutants defective in sustaining chronic infection showed homology to metabolic genes. BA159 was interrupted at the gluP locus, encoding a putative transporter for glucose and galactose (14). Mutant BA152 carried a transposon insertion in a homologue of Rhizobium etli gltD, encoding the small subunit of glutamate synthase (10). The identification of these two mutants in the STM screen suggests that glucose, galactose, or glutamate may serve as carbon and/or nitrogen sources during growth of B. abortus in the host. BA102 carried an insertion in a putative glycine dehydrogenase or glycine cleavage system. Since BA102 did not have a competitive growth defect in TSA and was able to grow on minimal medium without glycine, the virulence defect of BA102 is likely not due to auxotrophy. Interestingly, the activity of glycine dehydrogenase has been shown to increase 10-fold upon entry of M. tuberculosis into a state of nonreplicating persistence (30, 31). Since B. abortus, like M. tuberculosis, is able to persist chronically in infected hosts, glycine dehydrogenase may play a similar role in the entry of these pathogens into a latent state in the host.
Finally, DNA flanking two insertions in mutants defective for maintaining chronic infection showed either no homology to genes in the database (BA73) or homology to an open reading frame of unknown function (BA87) (Table 2).
TABLE 2.
Characterization of B. abortus genes identified by STM
| Mutant | Closest homologue (organism) | GenBank accession no. | Inferred function |
|---|---|---|---|
| BA41 | virB1 (B. suis) | AF141604 | Type IV secretion |
| BA114 | virB10 (B. suis) | AF141604 | Type IV secretion |
| BA184 | wbkA (B. melitensis) | AF047478 | O antigen biosynthesis |
| BA159 | gluP (B. abortus) | U43785 | Uptake of glucose and galactose |
| BA102 | P protein (Synechocystis sp.) | D90914 | Glycine dehydrogenase |
| gcvB (M. tuberculosis) | Q50601 | ||
| BA152 | gltD (R. etli) | AF107264 | Glutamate synthase |
| BA87 | HP1225 (Helicobacter pylori) | AE000628 | Conserved inner membrane protein |
| BA73 | No homology | ? |
DISCUSSION
The goal of the STM screen was to identify and compare B. abortus genes required for establishment and maintenance of chronic persistence in the mouse. Since fewer than 200 genes have been sequenced in Brucella species, we reasoned that screening a small number of mutants would be sufficient to identify new B. abortus genes required for chronic infection. The STM screen identified 27 mutants putatively attenuated for chronic infection. A statistically significant competitive-infection defect could be detected in 14 of these mutants. Thus, statistically significant evidence for attenuation was obtained for 8% (14 of 178) of the mutants screened. Assuming that our mutagenesis was random and that the coding density of the 3,200-kb B. abortus genome is similar to that of the E. coli genome, our data suggest that an estimated 257 genes may be required for establishing and maintaining chronic persistence in mice after i.p. infection. In contrast, i.p. infection of mice with an STM bank of Salmonella enterica Typhimurium revealed that only 3% of its 4,400-kb genome, or an estimated 153 genes, is required for the acute infection caused by this intracellular pathogen (18). The greater number of virulence genes required for chronic infection versus acute disease may reflect the requirement for additional adaptations to ensure long-term persistence, such as those which prevent clearance of B. abortus by the host immune system.
The working hypothesis of this study was that different sets of genes may be required for the initial steps or the establishment of chronic infection, which is characterized by rapid bacterial growth, than for maintenance of chronic infection, in which little or no growth is observed (5, 9). Indeed, the 14 attenuated mutants identified in this study fell into two classes (Fig. 2). Four mutants were unable to establish infection by either 2 or 8 weeks postinfection. In contrast, the remaining 10 mutants were able to establish infection at 2 weeks postinfection but displayed a defect in chronic persistence at 8 weeks postinfection. The first class included mutants with transposon insertions in genes required for O antigen biosynthesis (BA184), type IV secretion (BA41 and BA114), and biosynthesis of aromatic amino acids (BA100). The genes inactivated in these mutants are predicted to play a role early during infection. For example, BA184, a wbkA mutant, was the most highly attenuated and was outcompeted by the wild type by 1,000-fold at 2 weeks postinfection, suggesting that it was eliminated early in the infection process. Since rough mutants are sensitive to the bactericidal action of complement, it is possible that BA184 is cleared by complement-mediated lysis and may reach the spleen only in small numbers (1, 11, 13). The transposon insertion in BA100 rendered this mutant defective in the biosynthesis of aromatic amino acids, as determined by auxanography. This biosynthesis pathway is also required for the biosynthesis of 2,3-dihydroxybenzoic acid, the only siderophore known to be produced by B. abortus (23). However, since this siderophore has been shown to be dispensable for growth in mice (7), it is more likely that attenuation of this mutant is the result of its inability to acquire aromatic amino acids in the host. Salmonella aro mutants are unable to survive and replicate within macrophages and are attenuated for virulence (19). Thus, some of the virulence genes required in an early phase during chronic B. abortus infection may be similar to those used by intracellular pathogens, such as S. enterica serovar Typhimurium, which cause an acute infection. Two mutants (BA114 and BA41) defective for initiation of chronic infection carried insertions in a putative type IV secretion system of B. abortus, encoded by the virB genes (27). Mutant BA114 (virB10) displayed a greater competitive colonization defect in murine spleens at 2 weeks postinfection than BA41 (virB1) (Table 1). A similar effect has been described for the homologues of the B. abortus virB10 and virB1 genes present in the genome of the plant pathogen Agrobacterium tumefaciens. Inactivation of virB1 in A. tumefaciens causes a lower degree of attenuation than a mutation in virB10 (8). Mutations in the virB locus render B. suis unable to multiply in HeLa cells or macrophage cell lines in vitro (27). These data, together with our findings that B. abortus virB1 and virB10 mutants are unable to persist in mouse spleens after i.p. inoculation, suggest that attenuation in the animal model is due to an inability of these strains to grow intracellularly.
The second class of mutants, which were unable to maintain chronic infection, included strains defective in production of glutamate synthase (BA152), glycine cleavage (BA102), nutrient uptake (BA159), and several unknown functions (BA31, BA38, BA73, BA87, BA63, BA122, and BA142). While the inactivated genes in these mutants were not required for initiation of infection, they were required for chronic persistence. Mutant BA102 carried a transposon insertion in a gene with homology to gcvB, encoding glycine dehydrogenase, from M. tuberculosis. The activity of this enzyme has been found to increase 10-fold upon entry of M. tuberculosis into a state of nonreplicating persistence in vitro (31). The finding that glycine dehydrogenase is required for persistence of B. abortus in the mouse spleen suggests that M. tuberculosis and B. abortus may depend on similar metabolic pathways for chronic persistence in the host. This result underscores the potential for research on host pathogen interactions of B. abortus to elucidate mechanisms of intracellular persistence which are shared by other chronic intracellular pathogens.
ACKNOWLEDGMENTS
We thank Andreas Bäumler for critical comments on the manuscript and Garry Adams for helpful discussions throughout the course of this work.
Work in T.A.F.'s laboratory is supported by USDA/BARD Project No. US-2781-96 and Animal Formula Health AH8675. P.C.H. was supported by Food and Agriculture Science Fellowship 96-38420-3059 from USDA. R.T. was supported by National Research Service Award AI10050 from NIH/NIAID.
REFERENCES
- 1.Allen C A, Adams L G, Ficht T A. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect Immun. 1998;66:1008–1016. doi: 10.1128/iai.66.3.1008-1016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alton G G, Jones L M, Pietz D E. Laboratory techniques in brucellosis. 2nd ed. Geneva, Switzerland: World Health Organization; 1975. [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. J. New York, N.Y: Wiley & Sons; 1994. [Google Scholar]
- 5.Baldwin C L, Winter A J. Macrophages and brucella. Immunol Ser. 1994;60:363–380. [PubMed] [Google Scholar]
- 6.Bäumler A J, Kusters J G, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellaire B H, Elzer P H, Baldwin C L, Roop R M., 2nd The siderophore 2,3-dihydroxybenzoic acid is not required for virulence of Brucella abortus in BALB/c mice. Infect Immun. 1999;67:2615–2618. doi: 10.1128/iai.67.5.2615-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birmingham J R, Jeska E L. Characterization of macrophage functions in mice infected with Brucella abortus. Infect Immun. 1981;32:1079–1083. doi: 10.1128/iai.32.3.1079-1083.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo A, Taboada H, Mendoza A, Valderrama B, Encarnacion S, Mora J. GenBank accession number AF107264 Rhizobium etli glutamate synthase large subunit (gltB) and glutamate synthase small subunit (gltD) genes, complete cds. 1998. [Google Scholar]
- 11.Corbeil L B, Blau K, Inzana T J, Nielsen K H, Jacobson R H, Corbeil R R, Winter A J. Killing of Brucella abortus by bovine serum. Infect Immun. 1988;56:3251–3261. doi: 10.1128/iai.56.12.3251-3261.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.deLorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenschenk F C, Houle J J, Hoffmann E M. Serum sensitivity of field isolates and laboratory strains of Brucella abortus. Am J Vet Res. 1995;56:1592–1598. [PubMed] [Google Scholar]
- 14.Essenberg R C, Candler C, Nida S K. Brucella abortus strain 2308 putative glucose and galactose transporter gene: cloning and characterization. Microbiology. 1997;143:1549–1555. doi: 10.1099/00221287-143-5-1549. [DOI] [PubMed] [Google Scholar]
- 15.Gerhardt P. The nutrition of brucellae. Microbiol Rev. 1958;22:81–98. doi: 10.1128/br.22.2.81-98.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godfroid F, Taminiau B, Danese I, Denoel P, Tibor A, Weynants V, Cloeckaert A, Godfroid J, Letesson J J. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect Immun. 1998;66:5485–5493. doi: 10.1128/iai.66.11.5485-5493.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant S G N, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 19.Hoiseth S K, Stocker B A D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live oral vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 20.Kagaya K, Watanabe K, Fukazawa Y. Capacity of recombinant gamma interferon to activate macrophages for Salmonella-killing activity. Infect Immun. 1989;57:609–615. doi: 10.1128/iai.57.2.609-615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovach M E, Phillips R W, Elzer P H, Roop R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–801. [PubMed] [Google Scholar]
- 22.Lai F, Schurig G G, Boyle S M. Electroporation of a suicide plasmid bearing a transposon into Brucella abortus. Microb Pathog. 1990;9:363–368. doi: 10.1016/0882-4010(90)90070-7. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Goni I, Moriyon I, Neilands J B. Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect Immun. 1992;60:4496–4503. doi: 10.1128/iai.60.11.4496-4503.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McQuiston J R, Vemulapalli R, Inzana T J, Schurig G G, Sriranganathan N, Fritzinger D, Hadfield T L, Warren R A, Snellings N, Hoover D, Halling S M, Boyle S M. Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect Immun. 1999;67:3830–3835. doi: 10.1128/iai.67.8.3830-3835.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meador V P, Tabatabai L B, Hagemoser W A, Deyoe B L. Identification of Brucella abortus in formalin-fixed, paraffin-embedded tissues of cows, goats, and mice with an avidin-biotin-peroxidase complex immunoenzymatic staining technique. Am J Vet Res. 1986;47:2147–2150. [PubMed] [Google Scholar]
- 26.Montaraz J A, Winter A J. Comparison of living and nonliving vaccines for Brucella abortus in BALB/c mice. Infect Immun. 1986;53:245–251. doi: 10.1128/iai.53.2.245-251.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 28.Plommet M, Plommet A M. Évolution de l'infection splénique de souris de quatre lignées, inoculées par voie veneuse, par trois doses de Brucella abortus. Ann Rech Vet. 1981;12:345–351. [PubMed] [Google Scholar]
- 29.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wayne L G. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 31.Wayne L G, Lin K Y. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young E J. An overview of human brucellosis. Clin Infect Dis. 1995;21:283–289. doi: 10.1093/clinids/21.2.283. ; quiz, 290. [DOI] [PubMed] [Google Scholar]