Abstract
Cryptosporidium parvum is a significant cause of diarrheal disease worldwide. The specific molecules that mediate C. parvum-host cell interactions and the molecular mechanisms involved in the pathogenesis of cryptosporidiosis are unknown. In this study we have shown that gp40, a mucin-like glycoprotein, is localized to the surface and apical region of invasive stages of the parasite and is shed from its surface. gp40-specific antibodies neutralize infection in vitro, and native gp40 binds specifically to host cells, implicating this glycoprotein in C. parvum attachment to and invasion of host cells. We have cloned and sequenced a gene designated Cpgp40/15 that encodes gp40 as well as gp15, an antigenically distinct, surface glycoprotein also implicated in C. parvum-host cell interactions. Analysis of the deduced amino acid sequence of the 981-bp Cpgp40/15 revealed the presence of an N-terminal signal peptide, a polyserine domain, multiple predicted O-glycosylation sites, a single potential N-glycosylation site, and a hydrophobic region at the C terminus, a finding consistent with what is required for the addition of a GPI anchor. There is a single copy of Cpgp40/15 in the C. parvum genome, and this gene does not contain introns. Our data indicate that the two Cpgp40/15-encoded proteins, gp40 and gp15, are products of proteolytic cleavage of a 49-kDa precursor protein which is expressed in intracellular stages of the parasite. The surface localization of gp40 and gp15 and their involvement in the host-parasite interaction suggest that either or both of these glycoproteins may serve as effective targets for specific preventive or therapeutic measures for cryptosporidiosis.
Cryptosporidium parvum, an intestinal apicomplexan parasite, is a significant cause of diarrheal disease worldwide (7, 36). Cryptosporidial infection is asymptomatic or self-limiting in immunocompetent hosts, but it may be chronic and life-threatening in immunocompromised patients such as those with AIDS. In children in developing countries, C. parvum infection has been reported to be associated with persistent diarrheal disease, which may result in subsequent growth impairment (8). Recently, this parasite has been implicated as the causative agent of numerous outbreaks of waterborne diarrheal disease (6). There is currently no specific therapy approved for the treatment of cryptosporidiosis.
Infection with C. parvum is initiated by ingestion of oocysts, which undergo excystation to release sporozoites. Sporozoites attach to and invade intestinal epithelial cells, where the parasite undergoes further intracellular development through asexual as well as sexual cycles. Merozoites released into the lumen during the asexual cycle can attach to and invade adjacent epithelial cells and thus maintain intracellular replication. The molecular mechanisms involved in the pathogenesis of cryptosporidiosis and the specific molecules that mediate C. parvum-host cell interactions are unknown. A number of C. parvum proteins have been implicated in attachment and invasion and are the subject of ongoing investigation (2, 9, 18, 21, 25, 29, 31, 33, 40).
We have previously identified a monoclonal antibody (MAb), 4E9, that neutralizes C. parvum infection and inhibits attachment in vitro (A. M. Cevallos, N. Bhat, R. Verdon, D. H. Hamer, B. Stein, S. Tzipori, M. E. A. Pereira, G. T. Keusch, and H. D. Ward, submitted for publication). 4E9 was found to recognize two glycoproteins, gp40 and GP900. Immunofluorescence (IF) studies using this MAb revealed that these glycoproteins are localized to the surface and apical region of sporozoites and merozoites, are shed in trails from sporozoites during gliding motility, and bind to intestinal epithelial cells. The epitope recognized by 4E9 contains α-N-acetylgalactosamine (αGalNAc) residues, which are present in a mucin-type O-glycosidic linkage. Lectins specific for these residues also block attachment in vitro. The gene encoding one of these glycoproteins, GP900, a high-molecular-weight mucin-like glycoprotein, has recently been cloned and sequenced (2).
In the present study, we have characterized gp40 and cloned, sequenced, and expressed the gene encoding it. The open reading frame (ORF) encoding gp40 also encodes a previously described (9) 15-kDa surface glycoprotein (gp15) (W. Strong, J. Gut, and R. Nelson, personal communication). We have investigated the relationship between gp40 and gp15 and present here evidence to suggest that these two glycoproteins are proteolytic fragments of a precursor protein.
MATERIALS AND METHODS
C. parvum oocysts, sporozoites, and shed proteins.
Oocysts of the GCH1 isolate (37) were treated with 1.75% (vol/vol) sodium hypochlorite for 10 min on ice and then washed with Dulbecco modified Eagle medium (DMEM; Life Technologies, Grand Island, N.Y.) containing 25 mM HEPES, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Oocysts were excysted for 1 h at 37°C in the presence of 0.75% taurocholic acid. Sporozoites were purified by filtration through a 2.0-μm (pore-size) Nucleopore polycarbonate filter (Costar Scientific Corp., Cambridge, Mass.).
Shed proteins were obtained by excystation of hypochlorite-treated oocysts in DMEM for 2 h at 37°C, followed by centrifugation at 5,000 × g at 4°C for 10 min. Protease inhibitors (final concentration, 2 mM phenylmethylsulfonyl fluoride [PMSF]–20 μM leupeptin–10 μM E64–2 mM EDTA) were added to the supernatant (containing shed proteins), which was concentrated 10-fold by ultrafiltration.
Cell culture.
Caco-2 (human intestinal epithelial) cells originally obtained from Hans Buller, Academic Medical Center, Amsterdam, The Netherlands (38), were designated Caco-2A and maintained by the GRASP Center Cell Culture Core facility. Cells were grown in 75-cm2 flasks in DMEM, supplemented with 10% fetal calf serum, 25 mM HEPES, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C and 5% CO2 as described earlier (19).
Isolation of native gp40 from shed proteins.
C. parvum shed proteins were fractionated by size exclusion ultrafiltration using Centricon filters (Millipore Corp., Bedford, Mass.) with molecular weight cutoffs of 10, 50, and 100 kDa. The 10- to 50-kDa fraction was enriched in gp40 as determined by silver staining and immunoblotting with MAb 4E9.
Lectin (HPA) affinity chromatography.
αGalNAc-containing glycoproteins were isolated from C. parvum oocysts as follows. Hypochlorite-treated oocysts (1 × 108 to 2 × 108/ml) in phosphate-buffered saline (PBS) containing protease inhibitors (described above) were lysed by five freeze-thaw cycles, followed by detergent extraction with 1% octylglucoside in PBS (OGS-PBS). Detergent-soluble material was incubated with Helix pomatia agglutinin (HPA)-agarose (EY Laboratories, Inc., San Mateo, Calif.) in 1% OGS-PBS for 18 h at 4°C. After an extensive washing, bound proteins were eluted with 0.1 M GalNAc in 0.05% OGS-PBS.
αGalNAc-containing glycoproteins were also isolated from C. parvum-infected Caco-2A monolayers as follows. Caco-2A cells were grown to confluence in 75-cm2 flasks and infected with 5 × 107 hypochlorite-treated oocysts for 12 or 18 h at 37°C. Infected cells were extracted with 1% OGS-PBS in the presence of protease inhibitors at 4°C. Detergent-soluble material was processed for HPA affinity chromatography as described above. Uninfected Caco-2A cells grown at the same time under identical conditions were processed in the same way.
Antibodies.
4E9, an immunoglobulin M (IgM) MAb was produced by immunization of mice with C. parvum sporozoites (Cevallos et al., submitted). Briefly, BALB/c mice were immunized intraperitoneally with C. parvum sporozoites, spleen cells fused with myeloma cells, and hybridomas cloned in liquid medium as previously described (41). Clones which reacted with the surface of sporozoites were identified using an IF assay. MAb CrA1, an IgA MAb against a C. parvum sporozoite surface protein (gp15) was obtained by oral immunization of mice with C. parvum sporozoites (X. Zhou, S. Tzipori, and M. R. Neutra, unpublished). Briefly, BALB/c mice were immunized intragastrically with a C. parvum sporozoite and oocyst mixture, and Peyer's patch cells were subsequently fused with mouse myeloma cells as previously described (42). C. parvum-specific IgA hybridomas were identified by enzyme-linked immunosorbent assay (ELISA) and cloned, and IgA antibodies were produced in serum-free medium.
Antisera to purified native gp40 were produced as follows. HPA-isolated glycoproteins (which are enriched in gp40) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After staining with Coomassie blue, the 40-kDa band (which was shown to represent gp40 by immunoblotting with MAb 4E9 of a parallel strip of gel transferred to nitrocellulose) was excised, lyophilized, and emulsified with complete Freund's adjuvant for the initial immunization and incomplete complete Freund's adjuvant for subsequent boosts. BALB/c mice were immunized at 3- to 4-week intervals with this material, and the presence of anti-gp40 antibodies in sera was monitored by immunoblotting of an oocyst-sporozoite antigen preparation.
Immunoblot analysis, immunoprecipitation, and immunofluorescence.
Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and then probed with MAbs 4E9 and CrA1 and anti-gp40 antisera. For MAb 4E9 and anti-gp40 antisera, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (heavy and light chain) antibody (Pierce Chemical Co., Rockford, Ill.) was used as secondary antibody. For MAb CrA1, HRP-conjugated goat anti-mouse IgA (α-chain-specific) antibody was used as secondary antibody (Southern Biotechnology Associates, Inc., Birmingham, Ala.). Immunoblots were developed with SuperSignal chemiluminescent substrate (Pierce). Perfect Protein markers, 10- to 225-kDa (Novagen, Madison, Wis.), were used as molecular weight standards and were detected by reactivity with HRP-conjugated S-Protein according to the manufacturer's protocol.
A mixture of sporozoites and oocysts (derived from excystation of 108 oocysts) in PBS containing protease inhibitors (described above) were lysed by five freeze-thaw cycles and detergent extraction with 1% Triton X-100. The lysate was centrifuged at 10,000 × g for 30 min. Detergent-soluble material was incubated with anti-gp40 antisera (or preimmune sera as controls) overnight, followed by incubation with protein G-Sepharose (Pharmacia Biotech, Inc., Piscataway, N.J.) for 2 h at 4°C. After an extensive washing with 20 mM phosphate–0.5 M NaCl–0.5% Triton X-100–0.1% SDS–0.1% deoxycholate, immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotting with MAb 4E9.
IF of C. parvum oocysts, sporozoites, and intracellular stages (present in Caco-2A monolayers grown to confluence in collagen-coated 16-well chamber slides, infected with oocysts for 24 h, and fixed and permeabilized with methanol) was performed as described earlier (10, 39). Anti-gp40 antisera and MAb CrA1 were used as primary antibodies and fluorescein isothiocyanate-conjugated goat anti-mouse IgG and IgA (Sigma Chemical Co., St. Louis, Mo.) were used as secondary antibodies.
Silver-stained SDS gels, immunoblots, and IF photomicrographs were scanned using an Agfa Arcus II scanner and Adobe Photoshop 4.0 software.
C. parvum in vitro infection assay.
The effect of anti-gp40 antisera (diluted 1:00) obtained from two different mice on C. parvum infection of Caco-2A cells was determined using an in vitro assay as described previously (39). Briefly, oocysts (104/well) preincubated with antisera for 30 min at room temperature were incubated with Caco-2A cells (2 × 104/well) grown in 96-well tissue culture plates for 24 h at 37°C and in 5% CO2. Cells were fixed and permeabilized with methanol for 10 min at room temperature and washed three times with Tris-buffered saline. Infection was quantified by ELISA as described earlier (39). Preimmune sera were used as controls.
gp40 binding assay.
Increasing concentrations of HPA-isolated glycoproteins or shed proteins were incubated with live or glutaraldehyde-fixed, Caco-2A cells (grown to confluence in 96-well plates) for 1 h at 4°C (live cells) or 2 h at 37°C (fixed cells). The amount of gp40 present in both preparations was quantified by comparison to known concentrations of Novagen Perfect Protein markers in silver-stained SDS gels by scanning densitometry using Quantity One software (Bio-Rad, Hercules, Calif.). Unbound protein was washed off and bound gp40 was detected by ELISA using anti-gp40 antisera and biotinylated horse anti-mouse IgG (Vector Laboratories, Burlingame, Calif.) as primary and secondary antibodies, respectively. Ovalbumin and β-galactosidase and antibodies specific for them (Sigma) were used as controls.
N-terminal and internal peptide amino acid sequence determination.
A gp40-enriched preparation obtained from shed proteins was subjected to SDS-PAGE, and transferred to a polyvinylidene difluoride membrane, which was stained with Coomassie blue. The 40-kDa band (identified as gp40 by probing an additional strip from the same membrane with MAb 4E9) was excised and processed for N-terminal amino acid microsequencing by automated Edman degradation using a Perkin-Elmer ABI 477A sequencer at the Tufts University Core Facility. The N-terminal sequence of the first five residues of the 40- and 13-kDa proteins isolated by HPA affinity chromatography was determined in a similar fashion.
To obtain amino acid sequence data from internal peptides, the 40-kDa band (which was shown to represent gp40 by immunoblotting with MAb 4E9 of a parallel strip of gel transferred to nitrocellulose) was excised from a Coomassie blue-stained SDS gel and submitted to the Harvard Microchemistry Facility, Cambridge, Mass. After in-gel tryptic digestion of the protein, sequence analysis of peptides was performed by microcapillary reverse-phase high-pressure liquid chromatography nanoelectrospray tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer. Two peptides, p70 and p81, were selected and analyzed by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS). These two peptides were then sequenced by Edman degradation. A high-confidence amino acid sequence (supported by the MALDI-TOF-MS results) was obtained (Table 1). The MS/MS spectra of peptides derived from native gp40 was correlated with the nucleotide and deduced amino acid sequence of Cpgp40/15 using the algorithm Sequest (4a) and programs developed at the Harvard Microchemistry Facility.
TABLE 1.
Amino acid sequence of gp40-derived peptides and synthetic oligonucleotides used for PCR and RT-PCR
| Primer (type)a | Amino acid sequenceb | Oligonucleotide |
|---|---|---|
| Primer 1 (F) | DVPVEG (N terminal) | 5′-GAYGTWCCWGTWGARGG-3′c |
| Primer 2 (F) | YISGEVTSVTFEK (p70) | 5′-GGWGARGTWACWTCWGTWACWTTYG-3′c |
| Primer 3 (R) | VNGQDFSTLSANSSSPTE (p81) | 5′-GTWGAAAAATCTTGWCCATTWAC-3′c |
| Primer 4 (R) | 5′-TAACTGTATTATCACTCTTTTCG-3′ | |
| Primer 5 (F) | 5′-GGCAAGAACTGGAGAAGACG-3′ | |
| Primer 6 (R) | 5′-GGTACCTTCTCCGAACCACA-3′ | |
| Primer 7 (F) | 5′-ATGCAAAAATACGTGGACTGGG-3′ | |
| Primer 8 (R) | 5′-TCGCACGAAAGATTTCCATTG-3′ | |
| Primer 9 (F) | 5′-TTACTCTCCGTTATAGTCTCCGCTG-3′ | |
| Primer 10 (R) | 5′-CGAATAAGGCTGCAAAGATTGC-3′ | |
| T7d | 5′-GTAATACGACTCACTATAGGGC-3′ | |
| T3d | 5′-AATTAACCCTCACTAAAGGG-3′ |
F, forward primer; R, reverse primer.
Underlining indicates the amino acids used for the design of degenerate oligonucleotides.
Degenerate primers. Ambiguous bases are abbreviated as follows: Y, C or T; W, A or T; R, S or G.
λZapII-derived primers.
DNA libraries and synthetic oligonucleotides.
C. parvum genomic DNA libraries derived from the GCH1, SFGH-1, and NINC isolates, as well as a sporozoite cDNA library derived from the Iowa isolate, were generously provided by Giovanni Widmer, Tufts University School of Veterinary Medicine, North Grafton, Mass., and Richard Nelson, University of California at San Francisco. The oligonucleotide primers used for PCR are shown in Table 1. Degenerate primers (synthesized by Life Technologies) were designed on the basis of the amino acid sequences of the N-terminal (pN) and two internal peptides (p70 and p81) of gp40. The degeneracy of the primers was reduced by taking into account AT-rich codon usage in C. parvum (17). Nondegenerate primers were synthesized at the Tufts University Core Facility.
PCR and DNA sequencing.
DNA libraries or hypochlorite-treated, freeze-thawed oocysts were used as a source of template DNA for PCR. Conditions for PCR using degenerate primers were as follows: 95°C for 2 min; 95°C for 40 s, 37°C for 60 s, and 72°C for 50 s (5 cycles); 94°C for 40 s, 55°C for 50 s, and 72°C for 50 s (35 cycles); and 72°C for 5 min. The first five cycles were excluded for PCRs using nondegenerate primers. Reagents were used at the following final concentrations: deoxynucleoside triphosphates, 0.4 mM; degenerate primers, 1 μM; nondegenerate primers, 0.5 μM; MgCl2, 1.5 mM; and Taq DNA polymerase (Life Technologies), 50 U/ml. PCR products were cloned into the TA Cloning vectors, pCR2.1 or pCRII-TOPO (Invitrogen, Carlsbad, Calif.). Plasmids were purified using a Qiagen Miniprep Kit (Qiagen, Inc., Valencia, Calif.). DNA sequencing was performed by the dye-terminator method at the Tufts University Core Facility using a Perkin-Elmer ABI 377 sequencer.
The FASTA (28) and BLAST (1) algorithms were used to compare DNA and protein sequences to sequence databases. Analysis of nucleotide and amino acid sequences was performed with the MACVECTOR (Oxford Molecular Group, Oxford, United Kingdom) and PSORT II (15) programs. Predicted mucin-type O-glycosylation sites were identified using the NetOGlyc 2.0 program (11), and potential N-glycosylation sites were identified using the Motif program (43).
Expression in E. coli.
The following three fragments were cloned into the pET-32 Xa/LIC vector, which contains an internal S-tag, internal and C-terminal His tags, and N-terminal Trx tag sequences (Novagen): (i) an 891-bp fragment encoding amino acids 31 to 326 (corresponding to the entire Cpgp40/15 ORF, minus the putative signal peptide, designated pAMC40/15; (ii) a 315-bp fragment encoding amino acids 223 to 326 (corresponding to gp15, designated pAMC15); and (iii) a 576-bp fragment encoding amino acids 31 to 222 (corresponding to gp40, designated pAMC40). A control insert provided by Novagen was also cloned into the same vector. Overexpression in Escherichia coli AD494(DE3) was induced with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside). Fusion proteins encoded by pAMC40/15, pAMC40, and pAMC15 were designated pepAMC40/15, pepAMC40, and pepAMC15, respectively. pepAMC40 was toxic to E. coli; however, the transformation of E. coli AD494 was successful when performed in the presence of the plasmid pLysS (which encodes T7 lysozyme, a natural inhibitor of T7 RNA polymerase [Novagen]). The S-tag present in fusion proteins was identified in immunoblots by reactivity with HRP-conjugated S-Protein according to the manufacturer's instructions (Novagen).
Genomic Southern analysis.
Genomic DNA was extracted from a sporozoite-oocyst preparation using a G Nome DNA kit (Bio 101, Inc., Vista, Calif.). The restriction enzymes EcoRI and HindIII (New England Biolabs, Beverly, Mass.) alone or in combination were used to digest the genomic DNA. DNA fragments were separated on a 1% agarose gel, transferred to a nylon membrane (Duralon-UV; Stratagene, La Jolla, Calif.), and fixed by UV radiation using a UV Stratalinker 1800 (Stratagene). An 856-bp PCR product amplified from genomic DNA using primers 5 and 8 was used as a probe for this blot. The probe was labeled and hybridization was detected using the ECL direct nucleic acid labeling system and detection reagent (Amersham, Arlington Heights, Ill.) according to the manufacturer's instructions.
RT-PCR.
Caco-2A cells were grown to confluence in six-well plates and were infected with C. parvum oocysts (106) for 24 h. RNA was extracted from uninfected and infected monolayers using the RNeasy kit (Qiagen). Contaminating DNA was removed by RNase-free DNase treatment. Then, 0.5 μg of RNA was used in each reverse transcription (RT) reaction, which was performed using primers 9 and 10 (Table 1) and the Access RT-PCR System according to the manufacturer's instructions (Promega, Madison, Wis.). RNA from uninfected Caco-2A cells was used as a negative control. RT-PCR reactions without added reverse transcriptase were performed in parallel to confirm that the PCR products were not due to the amplification of contaminating DNA. The DNA sequence of the product obtained by RT-PCR was determined as described above.
The DNA sequence of Cpgp40/15 was deposited at GenBank under accession number AF155624.
RESULTS
gp40 is unrelated to GP900.
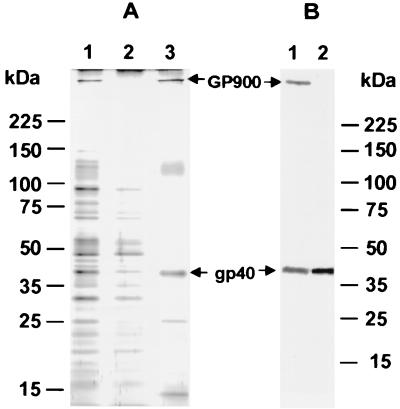
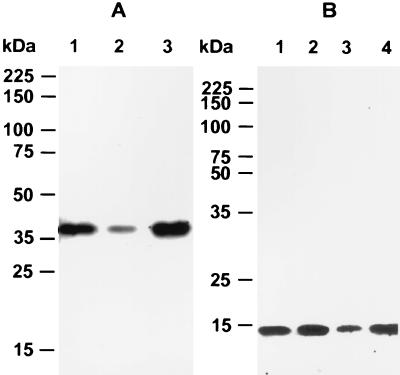
We previously identified gp40 using 4E9, an MAb which recognizes a carbohydrate epitope containing αGalNAc residues that are present on two glycoproteins, gp40 and GP900. The presence of these αGalNAc residues was exploited to isolate glycoproteins from C. parvum lysates by affinity chromatography using an αGalNAc-specific lectin, HPA. Analysis of this preparation by silver staining (Fig. 1A) and immunoblotting with MAb 4E9 (Fig. 1B, lane 1) revealed that a 40-kDa band recognized by MAb 4E9 was among the major proteins present (bands at >900, 128, 26, and 13 kDa were also present). In order to produce gp40-specific antibodies, native gp40 was purified by excision of the 40-kDa band from polyacrylamide-SDS gels and used to immunize mice. The purity of gp40 was verified by SDS-PAGE and silver staining of an aliquot of the protein (eluted from the excised gel with sample buffer), which revealed the presence of a single 40-kDa band (data not shown). Sera from these mice reacted exclusively with a 40-kDa band by immunoblotting, thus confirming the specificity of the antibody for gp40 (Fig. 1B, lane 2). The gp40-specific antisera immunoprecipitated a 40-kDa band from C. parvum sporozoite-oocyst lysates which was recognized by MAb 4E9 (data not shown), confirming that both antibodies recognized the same protein. However, the anti-gp40 antisera did not recognize GP900 (Fig. 1B, lane 2), suggesting that other than the presence of a common carbohydrate epitope, gp40 is unrelated to GP900.
FIG. 1.
Isolation of αGalNAc-containing glycoproteins by HPA affinity chromatography. (A) Aliquots of starting lysate (lane 1), effluent (lane 2), and GalNAc eluate (lane 3) were separated by 5 to 15% gradient SDS-PAGE and stained with silver. (B) Immunoblot of the GalNAc eluate separated by 5 to 15% gradient SDS-PAGE and probed with MAb 4E9 (lane 1) and anti-gp40 antisera (lane 2). The positions of gp40 and GP900 are indicated with arrows.
gp40-specific antisera neutralize C. parvum infection of intestinal epithelial cells.
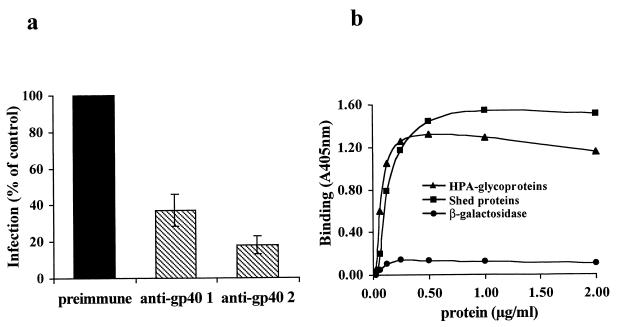
Our previous studies showed that MAb 4E9, which recognizes gp40 as well as GP900, neutralized infection and blocked attachment in vitro (Cevallos et al., submitted). In order to determine if gp40 is involved in mediating attachment and/or invasion, we assessed the effect of the gp40-specific antisera on C. parvum infection of intestinal epithelial cells by using an in vitro assay (39). Compared to their respective preimmune sera, gp40-specific antisera (from two different mice) significantly inhibited (by 82 and 68%, respectively) C. parvum infection of Caco-2A cells (Fig. 2a). This result implicates gp40 in C. parvum attachment to and/or invasion of host cells.
FIG. 2.
(a) Effect of gp40-specific antisera on C. parvum infection of Caco-2A cells. Oocysts were preincubated with preimmune sera or gp40-specific antisera obtained from two different mice and allowed to infect Caco-2A cells. Infection was quantified by ELISA. The results are expressed as a percentage of the control (respective preimmune sera from each of the mice). (b) Binding of gp40 to intestinal epithelial cells. Increasing concentrations of HPA-isolated glycoproteins and shed proteins containing the indicated concentrations of gp40 were incubated with live Caco-2A cells, and gp40 binding was quantified by ELISA using gp40-specific antisera. The results are expressed as the absorbance at 405 nm. β-Galactosidase was used as a control.
Native gp40 binds to intestinal epithelial cells.
Neutralization of infection by gp40-specific antisera suggested that gp40 is involved in C. parvum attachment to and/or invasion of host cells. In order to determine if gp40 bound to host cells, HPA-isolated glycoproteins or shed proteins (both of which are enriched in gp40) were incubated with live or glutaraldehyde-fixed Caco-2A cells, and gp40 binding was quantified by ELISA using gp40-specific antisera. As shown in Fig. 2b, binding of gp40 (present in both preparations) to Caco-2A cells occurred in a dose-dependent and saturable manner. There was no significant binding of the control proteins, β-galactosidase (Fig. 2b), or ovalbumin (not shown). Similar results were obtained with live as well as fixed cells (not shown). These results further implicate gp40 in sporozoite attachment to and/or invasion of host cells.
Cloning, sequencing, and analysis of the gene encoding gp40.
We set out to clone and sequence the gene encoding gp40. Our strategy was to ascertain the N-terminal and internal amino acid sequences from two internal tryptic peptides of native gp40 (purified from shed proteins by size exclusion ultrafiltration and gel isolation) and to design degenerate PCR primers based on these peptide sequences to amplify the gene encoding gp40. The sequences of the N-terminal six amino acids and two internal tryptic peptides, designated p70 and p81, are shown in Table 1. Comparison of these sequences with the deduced amino acid sequence of GP900 (2) revealed no significant similarity, confirming that these two proteins are unrelated. In addition, BLAST searches (1) with these sequences did not reveal significant similarity with known proteins in the databases. The sequence of the N-terminal five amino acids of gp40 isolated by HPA affinity chromatography was also determined and found to be identical to that isolated from shed proteins, confirming that both preparations contained the same protein.
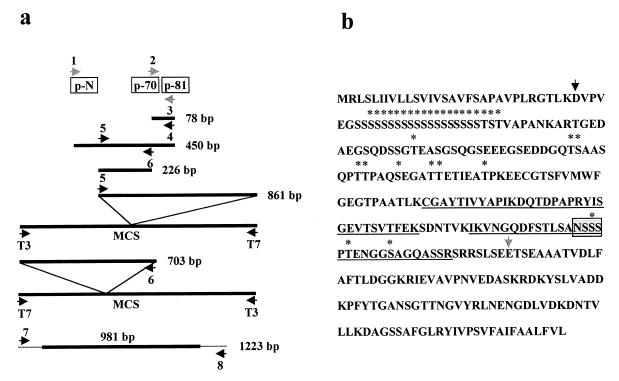
The multiple PCR steps that we used to clone the gene encoding gp40 are shown in Fig. 3a. Using degenerate primers 2 and 3 based on the amino acid sequence of internal peptides p70 and p81 (Table 1), a 78-bp fragment was amplified from oocyst genomic DNA, cloned into the pCR 2.1 vector, and sequenced. Using primer 4 (based on the nucleotide sequence of the 78-bp fragment) and degenerate primer 1 (based on the amino acid sequence of the N-terminal peptide) a 450-bp product was amplified from the same oocyst genomic DNA, cloned, and sequenced. The deduced amino acid sequences of the 78- and 450-bp fragments matched those of the known peptide sequences from the native protein. Primers 5 and 6 (based on the nucleotide sequence of the 450-bp fragment) were used to screen six C. parvum genomic and cDNA libraries by PCR. A product of the expected size (226 bp) was amplified from all of the libraries tested (data not shown). Anchored PCR of one of these libraries (GCH1; genomic DNA in λZapII) was used to obtain the sequences 5′ and 3′ of the 450-bp fragment. Thus, DNA from this library was used as template for PCR with primers based on the sequence of the T7 promoter of the λZapII phage vector (primer T7) and the 450-bp fragment (primer 5). The resulting 861-bp product amplified by PCR was sequenced. A similar approach with primer 6 was used to obtain a 703-bp product. Assembly and analysis of these overlapping fragments revealed a 981-bp ORF. Subsequently, to confirm our sequence, a 1,223-bp fragment was amplified by PCR from oocyst (GCH1) genomic DNA, using primers 7 and 8, along with a high-fidelity polymerase Pfu (Stratagene), and cloned into the Topo pCR II vector. Four independently derived clones were sequenced to correct potential PCR errors. BLAST comparison of the DNA and deduced amino acid sequences with protein and DNA sequence databases did not reveal significant similarity with any known genes or proteins.
FIG. 3.
(a) Sequential PCR steps used for cloning of Cpgp40/15 gene. Peptides pN, p-70, and p-81 and primers 1 to 8 are described in Table 1. MCS, multiple cloning site. The degenerate primers are indicated by stippled arrows. (b) Deduced amino acid sequence of Cpgp40/15 ORF. The N termini of gp40 (solid arrow) and gp15 (shaded arrow) are shown. The single potential N-glycosylation site is indicated with a stippled box. The predicted sites of mucin-type O-glycosylation are indicated with stars. The amino acid residues of contiguous peptides identified by MS/MS analysis are underlined.
Analysis of the deduced amino acid sequence revealed a 326-amino-acid protein with a calculated Mr of 33,600 (Fig. 3b). The amino acid sequence corresponding to the N terminus of the native protein was found 30 amino acids after the start codon. This N-terminal hydrophobic stretch of 30 amino acids is consistent with a signal peptide (26). Of note was a contiguous stretch of 19 serine residues in the N-terminal region of the mature protein. Hydropathicity analysis revealed hydrophobic regions at both the amino and carboxy termini (not shown). There was a short hydrophobic region at the C terminus, a finding consistent with that required for addition of a GPI anchor (23). A total of 32 threonine and serine residues in the deduced amino acid sequence of gp40 are predicted to be sites of mucin-type O-glycosylation (11). A single potential N-glycosylation site was identified (43).
The gene encoding gp40 is present in single copy and does not contain introns.
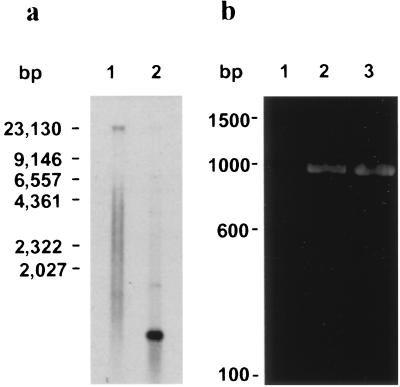
Southern blot analysis of genomic DNA isolated from GCH1 oocysts using an 856-bp Cpgp40/15 probe indicated that this sequence is present in single copy in the C. parvum genome. As seen in Fig. 4a, this probe hybridized to a single ∼800-bp EcoRI/HindIII restriction fragment of the expected size. When genomic DNA was digested with EcoRI alone, the probe hybridized to a single 23-kb fragment.
FIG. 4.
(a) Southern blot analysis of Cpgp40/15 locus. Genomic DNA was digested with EcoRI (lane 1) and HindIII and EcoRI (lane 2) and probed with an 856-bp Cpgp40/15 probe. (b) RT-PCR analysis of C. parvum RNA. RNA was extracted from Caco-2A cells (lane 1) or Caco-2A cells infected with C. parvum (lane 2) and analyzed by RT-PCR using gene-specific primers 9 and 10 (Table 1). Lane 3 shows the product obtained by PCR of genomic DNA using the same primers.
RT-PCR analysis using primers 9 and 10 (which span 949 bp of the Cpgp40/15 ORF) of RNA obtained from C. parvum intracellular stages revealed a single product of the same size as the product obtained by PCR of genomic DNA using the same primers (Fig. 4b). The DNA sequence of the RT-PCR product was identical to that of the corresponding genomic DNA sequence. This finding indicates that the gene encoding gp40 does not contain introns.
The gene encoding gp40 also encodes gp15.
In an independent study, IgA MAbs CrA1 and CrA2 which recognize a previously identified 15-kDa surface glycoprotein (9) were used to isolate a clone from a genomic DNA expression library (Strong et al., personal communication). Analysis of the DNA sequence of this clone revealed that it was almost identical to that of the gene identified in the present study. We therefore investigated the possibility that the gene encoding gp40 also encoded a 15-kDa protein (named gp15) as well. Previous studies indicated that gp15 bound the lectin HPA (9), suggesting that this protein may be present in the HPA-isolated glycoprotein preparation used to purify native gp40. Analysis of this preparation by SDS-PAGE and immunoblotting with MAb CrA1 revealed that a 13-kDa protein recognized by MAb CrA1 (indicating that this band represents gp15) was indeed present (Fig. 1A, lane 3; Fig. 5B, lane 4). We therefore obtained N-terminal amino acid sequence of the 13-kDa band from this preparation. The results indicated that the N-terminal residues ETSEA corresponded to amino acid residues 223 to 227 of the deduced amino acid sequence of the gene encoding gp40 (Fig. 3b). This finding, together with the work of Strong et al. (personal communication), indicates that gp15 is encoded by the same gene as gp40. Therefore, we have designated this gene Cpgp40/15.
FIG. 5.
Immunoblot of native gp40 and gp15. C. parvum oocysts (lanes 1), sporozoites (lanes 2), shed proteins (lanes 3), and HPA-isolated glycoproteins (panel B, lane 4) were separated by SDS-PAGE on a 5 to 15% gel, transferred to nitrocellulose, and probed with anti-gp40 antisera (A) or MAb CrA1 (B).
gp40 and gp15 are antigenically distinct proteins encoded by the same gene.
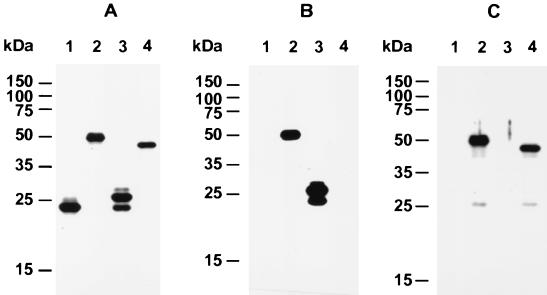
To characterize the relationship between gp40 and gp15, we determined whether antibodies to gp40 and gp15 cross-reacted with gp15 and gp40, respectively, by immunoblotting of various C. parvum preparations, including oocysts, sporozoites, and shed proteins. As shown in Fig. 5, in all of the antigen preparations tested, the anti-gp40 antisera only recognized gp40 and did not react with gp15 (confirming the specificity of the antibody for gp40). Similarly, MAb CrA1 only recognized gp15 and not gp40. In addition there was no difference in the Mr value of either protein under reducing or nonreducing conditions (data not shown). To further corroborate these findings, we overexpressed fragments of Cpgp40/15 encoding amino acids 31 to 326 (pepAMC40/15), amino acids 31 to 222 (pepAMC40), and amino acids 223 to 326 (pepAMC15) as thioredoxin fusion proteins in E. coli. These were probed with gp40- and gp15-specific antibodies. The results showed that both antibodies reacted with the fusion protein encoded by the entire ORF (Fig. 6B and C, lanes 2). However, the anti-gp40 antibody reacted only with pepAMC40 (Fig. 6C, lane 4), and MAb CrA1 reacted only with pepAMC15 (Fig. 6B, lane 3). These results confirm that gp40 and gp15 are antigenically distinct proteins encoded by the same gene Cpgp40/15.
FIG. 6.
Immunoblot of recombinant gp40 and gp15 fusion proteins. Control insert (lane 1), pAMC40/15 (lane 2), pAMC15 (lanes 3), and pAMC40 (lane 4) were cloned into pET-32 LIC/Xa and expressed in E. coli AD494(DE3). Bacterial lysates were separated by SDS–10% PAGE, transferred to nitrocellulose, and probed with S-Protein (A), MAb CrA1 (B), and anti-gp40 (C) antisera.
To determine whether peptides corresponding to gp15 were present in native gp40, the MS/MS spectra of six peptides obtained after tryptic digestion of gp40 were analyzed at the Harvard Microchemistry Facility. All six peptides (derived from gp40) analyzed on two separate occasions were represented in the portion of the deduced amino acid sequence of Cpgp40/15 present in the N-terminal 222 residues (Fig. 3B). In sharp contrast, none of the peptides corresponded to the amino acids present in residues 223 to 326 of gp15. This result further confirms that gp40 and gp15 are distinct polypeptides.
gp40 and gp15 are differentially localized in C. parvum sporozoites.
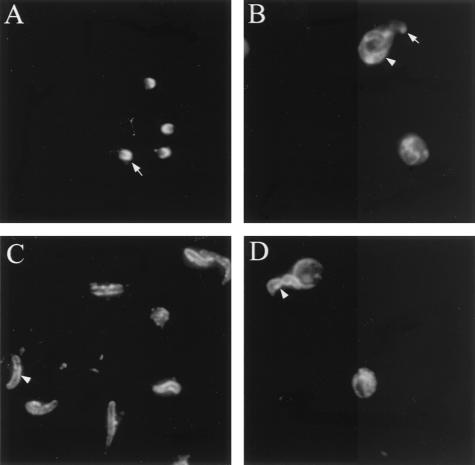
The localization of gp40 and gp15 in C. parvum developmental stages was determined by IF using antibodies specific for both proteins. The anti-gp40 antisera reacted mainly with the surface of the anterior portion of sporozoites, suggestive of an apical complex localization (Fig. 7A). In the intracellular stages, there was reactivity with the entire surface and apical region of merozoites (present within meronts) in infected Caco-2A cells (Fig. 7B). In contrast, gp15 was present on the entire surface of sporozoites (Fig. 7C) as well as merozoites (Fig. 7D). These results suggest that these two proteins are differentially localized in sporozoites.
FIG. 7.
Localization of gp40 and gp15 in C. parvum invasive stages by IF. The reactivity of anti-gp40 antisera (A and B) and MAb CrA1 (C and D) with purified sporozoites (A and C) and merozoites (B and D) in infected Caco-2A cells is shown. Apical and surface forms of localization are indicated with arrows and arrowheads, respectively.
gp40 and gp15 are products of proteolytic cleavage of a 49-kDa precursor protein.
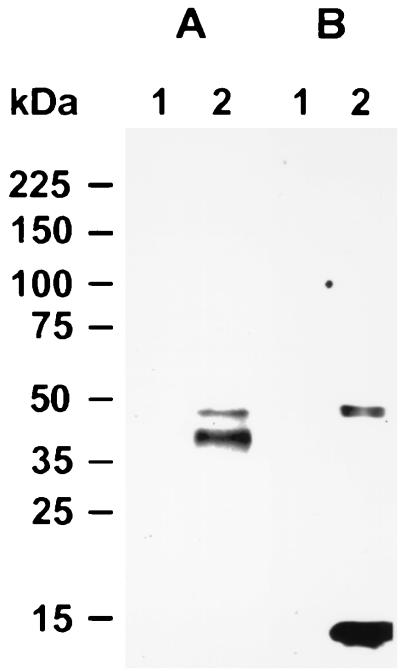
The finding that gp40 and gp15 are encoded by the same gene raised the possibility that they may be proteolytic fragments of a larger precursor protein. Immunoblotting of oocysts, sporozoites, shed proteins, and HPA-isolated glycoproteins with anti-gp40 antisera and MAb CrA1, however, did not reveal the presence of a putative precursor protein (Fig. 1B, lane 2, and Fig. 5). We therefore sought to determine whether a putative precursor protein was expressed by intracellular stages of the parasite. To do this, we infected Caco-2A cells with oocysts for 12 or 18 h, isolated αGalNAc-containing glycoproteins from them by HPA affinity chromatography, and analyzed them by immunoblotting with anti-gp40 antisera and MAb CrA1. This revealed the presence of a 49-kDa protein which was recognized by both antibodies (Fig. 8A and B, lanes 2) in addition to a 40-kDa band recognized only by anti-gp40 antisera (Fig. 8A, lane 2) and a ∼13-kDa band recognized only by MAb CrA1 (Fig. 8B, lane 2). Similar results were obtained at both time points (not shown). The 49-kDa band was not detected by either antibody in glycoproteins isolated in the same way from uninfected cells (Fig. 8A and B, lanes 1). These results implicate the 49-kDa protein recognized by both antibodies as the precursor of gp40 and gp15 and confirm that these two polypeptides are proteolytic fragments of the same protein.
FIG. 8.
Identification of gp40 and gp15 precursor in infected Caco-2A cells. αGalNAc-containing glycoproteins were first isolated from uninfected Caco-2A cells (lane 1) or Caco-2A cells infected with C. parvum for 18 h (lane 2), then separated by SDS-PAGE on a 5 to 15% gel, and finally transferred to nitrocellulose and probed with anti-gp40 antisera (A) and MAb CrA1 (B).
DISCUSSION
In this study we have cloned a C. parvum gene, Cpgp40/15, which encodes two antigenically distinct polypeptides, gp40 and gp15. These two glycoproteins are the products of cleavage of a 49-kDa precursor protein. gp40, a mucin-like glycoprotein was previously identified using a neutralizing MAb, 4E9, to a carbohydrate epitope present on gp40, as well as another glycoprotein (GP900). In the present study we have shown that other than the presence of a common carbohydrate epitope, gp40 is unrelated to GP900. gp40 is localized to the surface and apical region of invasive stages of the parasite and is shed from its surface. gp40-specific antisera neutralize infection in vitro, and native gp40 binds to host cells, implicating this protein in C. parvum attachment to and/or invasion of host cells. A recent study described the presence of a ∼47-kDa membrane-associated protein localized to the apical region of C. parvum sporozoites, which also bound to intestinal epithelial cells (25). The relationship of this protein to gp40 remains to be determined.
gp15 was identified by two groups of investigators. Gut and Nelson described a 15-kDa C. parvum glycoprotein using an MAb, 11A5, to a carbohydrate epitope (9). Interestingly, this protein is also present in sporozoites and merozoites, contains αGalNAc residues, and is shed in trails during the gliding movement of sporozoites. A 15-kDa protein was also identified using an IgA MAb, CrA1, which was obtained by oral immunization of mice with C. parvum (Zhou et al., unpublished). This MAb, which is directed against a protein epitope, is partially protective against C. parvum infection in a “backpack” tumor gamma interferon-preconditioned scid mouse model (Tzipori and Neutra, unpublished). Cross-immunoprecipitation studies revealed that these two MAbs recognized the same 15-kDa protein, (Strong et al., personal communication). Other investigators have also described 15-kDa proteins in C. parvum sporozoites (16, 17, 20, 34), two of which have been cloned and sequenced (16, 17, 20). However, there is no similarity in the deduced amino acid sequences of these proteins with that of gp15. The short hydrophobic stretch at the C-terminal region of gp15 is consistent with the signal required for the addition of a GPI anchor, suggesting that gp15 may be anchored in the membrane by this type of linkage. A number of parasite proteins, including those which are shed from the surface, are known to be GPI anchored (22, 23, 30, 35).
The molecular masses of the two Cpgp40/15-encoded proteins, as determined by SDS-PAGE and comparison to Novagen Perfect Protein markers, are 40 and 13 kDa. However, analysis of the deduced amino acid sequence of Cpgp40/15 predicts a mass of 20 kDa for gp40 (excluding the putative signal peptide) and a mass of 11 kDa for gp15. This discrepancy suggests that the additional mass present in the native proteins may be accounted for by glycosylation. This possibility is likely in view of several predicted mucin type O-glycosylation sites in the deduced amino acid sequence and is in keeping with our previous biochemical findings of terminal αGalNAc residues (which are present on oligosaccharides O-glycosidically linked to serine or threonine residues) in gp40 (Cevallos et al., submitted). Most of the predicted sites of O-glycosylation in gp40 are present in a polyserine domain in the N-terminal region of the deduced amino acid sequence. A serine-rich motif is also present in the sequence of TRAP C-1, another C. parvum protein which is implicated in attachment and invasion (33). Although TRAP C-1 has not been reported to be glycosylated, analysis of the deduced amino acid sequence using NetOGlyc 2.0 predicts several sites of mucin-type O-glycosylation. None of the serine or threonine residues in gp15 are predicted to be sites of mucin type O-glycosylation. However, lectin-binding studies suggest the presence of O-linked αGalNAc residues in this protein (9). We have previously also shown the presence of these αGalNAc residues in GP900 (Cevallos et al., submitted for publication), a C. parvum surface glycoprotein, which contains polythreonine mucin-like domains and has been shown to mediate invasion (2). Our previous findings that lectins specific for αGalNAc residues block attachment in vitro implicate these glycans as playing a role in adhesion. These glycans may mediate attachment by direct binding to host carbohydrate-binding proteins or by functioning as a “bridge” between host and parasite carbohydrate-binding proteins. The latter possibility may be likely in view of our earlier finding of a Gal/GalNAc-specific sporozoite lectin which may be involved in mediating attachment to host cells (18, 19). Mucin-like proteins in other protozoa, such as Trypanosoma cruzi, are also shed from the surface and are implicated in mediating attachment to host cells (30).
In this study we have shown that Cpgp40/15 encodes two antigenically distinct polypeptides, gp40 and gp15. We have identified the regions of the gene encoding each protein by determining their respective N-terminal amino acid sequences. There is no similarity in the deduced amino acid sequences of gp40 and gp15, a finding which is consistent with the finding that antibodies specific for each protein reacted exclusively with the corresponding native or recombinant proteins. In addition, the recombinant fusion protein generated by in-frame expression of the entire Cpgp40/15 sequence is recognized by antibodies specific for both gp40 and gp15. However, fusion proteins encoded by sequences upstream and downstream of the experimentally determined N terminus of gp15 are recognized exclusively by antibodies specific for either gp40 or gp15, respectively. These data are consistent with the finding that gp40 and gp15 are products of proteolytic cleavage of a 49-kDa putative precursor protein which is recognized by antibodies to both gp40 and gp15.
The putative precursor was detected only in actively replicating intracellular stages of the parasite and not in the metabolically quiescent oocyst or sporozoite stages, suggesting that the protein is synthesized during the intracellular asexual cycle. It is not known at which stage in the life cycle cleavage of the precursor occurs. The differential localization of gp40 and gp15 in sporozoites by IF suggests that cleavage occurs in vivo in the parasite. The protease responsible for the cleavage of the precursor is also not known and may be of parasite or host origin. A serine protease, a cysteine protease, and an arginine aminopeptidase have been reported to be present in C. parvum (5, 24, 27). However, it remains to be determined whether any of these enzymes are involved in processing of the precursor protein.
Proteolytic cleavage of a precursor protein yielding polypeptides involved in adhesion or invasion has been described in other apicomplexan parasites. For example, Plasmodium merozoite surface proteins involved in the invasion of red cells are derived from proteolytic cleavage of MSP-1, a 200-kDa precursor protein that is synthesized during schizogony (3, 12–14). Posttranslational processing generates fragments of 83, 42, 38, and 28 to 30 kDa, which remain as a noncovalently linked complex on the surface of the merozoite. The 42-kDa protein is further cleaved to produce a 19-kDa C-terminal fragment at the time of invasion, and the other fragments are shed during this process. Toxoplasma rhoptry proteins secreted during the invasion process are also synthesized as pro-proteins, which are processed in the nascent rhoptries of dividing parasites (4, 32).
A number of findings suggest that gp40 and gp15 are involved in C. parvum-host cell interactions. Native gp40 binds to host cells and gp40-specific antisera neutralize infection in vitro. Previous studies have shown that a gp15-specific IgA MAb (CrA1) neutralizes infection in vivo (Tzipori and Neutra, unpublished). The apical and surface localization of gp40 and gp15 in invasive stages of the parasite and the finding that they are shed from the surface are consistent with a role in attachment and invasion. The likely involvement of these C. parvum proteins in the initial host-parasite interaction suggests a basis for devising strategies to inhibit this interaction. Either gp40, gp15, or both of these glycoproteins may serve as effective targets for specific preventive or therapeutic measures for cryptosporidiosis.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants U19 AI33384, P30 DK34928 (GRASP Digestive Diseases Center), and P30 DK34854 (Harvard Digestive Diseases Center).
We gratefully acknowledge Giovanni Widmer and Richard Nelson for providing DNA libraries and for helpful discussions; Bill Strong, Jiri Gut, and Richard Nelson for sharing data prior to publication; Mike Berne, Tufts University School of Medicine Core facility, for N-terminal amino acid sequencing, DNA sequencing, and oligonucleotide synthesis; Bill Lane, Harvard Microchemistry Facility, for internal peptide sequencing, MS/MS analysis, and helpful discussions; Anne Kane, GRASP Intestinal Microbiology Core, for providing microbiological media and for helpful discussions; Doug Jefferson, GRASP Cell Culture Core, for providing Caco-2A cells; and members of the Waldor laboratory, Shiv Pillai and Miercio Pereira, for helpful discussions.
ADDENDUM IN PROOF
Since the submission of this paper, Priest et al. (J. W. Priest, J. P. Kwon, M. J. Arrowood, and P. J. Laramie, Mol. Biochem. Parasitol. 106:261–271, 2000) have reported the cloning of a gene encoding the immunodominant 17-kDa antigen from Cryptosporidium parvum. The nucleotide sequence of this gene is very similar to that of Cpgp40/15.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes D A, Bonnin A, Huang J X, Gousset L, Wu J, Gut J, Doyle P, Dubremetz J F, Ward H, Petersen C. A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol Biochem Parasitol. 1998;96:93–110. doi: 10.1016/s0166-6851(98)00119-4. [DOI] [PubMed] [Google Scholar]
- 3.Blackman M, Holder A. Secondary processing of the Plasmodium falciparum merozoite surface protein (MSP-1) by a calcium-dependent membrane bound serine protease: shedding of MSP-133 as a non-covalently associated complex with other fragments of MSP-1. Mol Biochem Parasitol. 1992;50:307–316. doi: 10.1016/0166-6851(92)90228-c. [DOI] [PubMed] [Google Scholar]
- 4.Bradley P J, Boothroyd J C. Identification of the pro-mature processing site of Toxoplasma ROP1 by mass spectrometry. Mol Biochem Parasitol. 1999;100:103–109. doi: 10.1016/s0166-6851(99)00035-3. [DOI] [PubMed] [Google Scholar]
- 4a.Eng J, McCormack A L, Yates J R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 5.Forney J R, Yang S, Healey M C. Protease activity associated with excystation of Cryptosporidium parvum oocysts. J Parasitol. 1996;82:889–892. [PubMed] [Google Scholar]
- 6.Fricker C R, Crabb J H. Water-borne cryptosporidiosis: detection methods and treatment options. Adv Parasitol. 1998;40:241–278. doi: 10.1016/s0065-308x(08)60123-2. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths J K. Human cryptosporidiosis: epidemiology, transmission, clinical disease, treatment, and diagnosis. Adv Parasitol. 1998;40:37–85. doi: 10.1016/s0065-308x(08)60117-7. [DOI] [PubMed] [Google Scholar]
- 8.Guerrant R L. Cryptosporidiosis: an emerging, highly infectious threat. Emerg Infect Dis. 1997;3:51–57. doi: 10.3201/eid0301.970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gut J, Nelson R G. Cryptosporidium parvum sporozoites deposit trails of 11A5 antigen during gliding locomotion and shed 11A5 antigen during invasion of MDCK cells in vitro. J Eukaryot Microbiol. 1994;41:42S–43S. [PubMed] [Google Scholar]
- 10.Hamer D H, Ward H D, Tzipori S, Pereira M E, Alroy J P, Keusch G T. Attachment of Cryptosporidium parvum sporozoites to MDCK cells in vitro. Infect Immun. 1994;62:2208–2213. doi: 10.1128/iai.62.6.2208-2213.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen J E, Lund O, Tolstrup N, Gooley A A, Williams K L, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 12.Holder A A, Freeman R R. The three major antigens on the surface of Plasmodium falciparum merozoites are derived from a single high molecular weight precursor. J Exp Med. 1984;160:624–629. doi: 10.1084/jem.160.2.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holder A A, Lockyer M J, Odink K G, Sandhu J S, Riveros-Moreno V, Hillman S C N, Davi L S, Tizard M L V, Schwarz R T, Freeman R R. Primary structure of the precursor to the three major surface antigens of Plasmodium falciparum merozoites. Nature. 1985;317:270–273. doi: 10.1038/317270a0. [DOI] [PubMed] [Google Scholar]
- 14.Holder A A, Sandhu J S, Hillman Y, Davey L S, Nicholls S C, Cooper H, Lockyer M J. Processing of the precursor to the major merozoite surface antigens of Plasmodium falciparum. Parasitology. 1987;94:199–208. doi: 10.1017/s0031182000053889. [DOI] [PubMed] [Google Scholar]
- 15.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Ismb. 1997;5:147–152. [PubMed] [Google Scholar]
- 16.Jenkins M C, Fayer R. Cloning and expression of cDNA encoding an antigenic Cryptosporidium parvum protein. Mol Biochem Parasitol. 1995;71:149–152. doi: 10.1016/0166-6851(95)00050-b. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins M C, Petersen C. Molecular biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 225–232. [Google Scholar]
- 18.Joe A, Hamer D H, Kelley M A, Pereira M E, Keusch G T, Tzipori S, Ward H D. Role of a Gal/GalNAc-specific sporozoite surface lectin in Cryptosporidium parvum-host cell interaction. J Eukaryot Microbiol. 1994;41:44S. [PubMed] [Google Scholar]
- 19.Joe A, Verdon R, Tizpori S, Keusch G T, Ward H D. Attachment of Cryptosporidium parvum sporozoites to human intestinal epithelial cells. Infect Immun. 1998;66:3429–3432. doi: 10.1128/iai.66.7.3429-3432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khramtsov N V, Oppert B, Montelone B A, Upton S J. Sequencing, analysis and expression in Escherichia coli of a gene encoding a 15-kDa Cryptosporidium parvum protein. Biochem Biophys Res Commun. 1997;230:164–166. doi: 10.1006/bbrc.1996.5877. [DOI] [PubMed] [Google Scholar]
- 21.Langer R, Riggs M. Cryptosporidium parvum apical complex glycoprotein CSL contains a sporozoite ligand for intestinal epithelial cells. Infect Immun. 1999;67:5282–5291. doi: 10.1128/iai.67.10.5282-5291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manger I D, Hehl A B, Boothroyd J C. The surface of Toxoplasma tachyzoites is dominated by a family of glycosylphosphatidylinositol-anchored antigens related to SAG1. Infect Immun. 1998;66:2237–2244. doi: 10.1128/iai.66.5.2237-2244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McConville M J, Ferguson A J. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J. 1993;249:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nesterenko M V, Tilley M, Upton S J. A metallo-dependent cysteine proteinase of Cryptosporidium parvum associated with the surface of sporozoites. Microbios. 1995;83:77–88. [PubMed] [Google Scholar]
- 25.Nesterenko M V, Woods K, Upton S J. Receptor/ligand interactions between Cryptosporidium parvum and the surface of the host cell. Biochim Biophys Acta. 1999;1454:165–173. doi: 10.1016/s0925-4439(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Okhuysen P C, DuPont H L, Sterling C R, Chappell C L. Arginine aminopeptidase, an integral membrane protein of the Cryptosporidium parvum sporozoite. Infect Immun. 1994;62:4667–4670. doi: 10.1128/iai.62.10.4667-4670.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 29.Riggs M W, Stone A L, Yount P A, Langer R C, Arrowood M J, Bentley D L. Protective monoclonal antibody defines a circumsporozoite-like glycoprotein exoantigen of Cryptosporidium parvum sporozoites and merozoites. J Immunol. 1997;158:1787–1795. [PubMed] [Google Scholar]
- 30.Schenkman S, Ferguson M A, Heise N, de Almeida M L, Mortara R A, Yoshida N. Mucin-like glycoproteins linked to the membrane by glycosylphosphatidylinositol anchor are the major acceptors of sialic acid in a reaction catalyzed by trans-sialidase in metacyclic forms of Trypanosoma cruzi. Mol Biochem Parasitol. 1993;59:293–303. doi: 10.1016/0166-6851(93)90227-o. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder A A, Lawrence C E, Abrahamsen M S. Differential mRNA display cloning and characterization of a Cryptosporidium parvum gene expressed during intracellular development. J Parasitol. 1999;85:213–220. [PubMed] [Google Scholar]
- 32.Soldati D, Lassen A, Dubremetz J F, Boothroyd J C. Processing of Toxoplasma ROP1 protein in nascent rhoptries. Mol Biochem Parasitol. 1998;96:37–48. doi: 10.1016/s0166-6851(98)00090-5. [DOI] [PubMed] [Google Scholar]
- 33.Spano F, Putignani L, Naitza S, Puri C, Wright S, Crisanti A. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol Biochem Parasitol. 1998;92:147–162. doi: 10.1016/s0166-6851(97)00243-0. [DOI] [PubMed] [Google Scholar]
- 34.Tilley M, Upton S J, Fayer R, Barta J R, Chrisp C E, Freed P S, Blagburn B L, Anderson B C, Barnard S M. Identification of a 15-kilodalton surface glycoprotein on sporozoites of Cryptosporidium parvum. Infect Immun. 1991;59:1002–1007. doi: 10.1128/iai.59.3.1002-1007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomavo S, Schwarz R T, Dubremetz J F. Evidence for glycosyl-phosphatidylinositol anchoring of Toxoplasma gondii major surface antigens. Mol Cell Biol. 1989;9:4576–4580. doi: 10.1128/mcb.9.10.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzipori S, Griffiths J K. Natural history and biology of Cryptosporidium parvum. Adv Parasitol. 1998;40:6–36. doi: 10.1016/s0065-308x(08)60116-5. [DOI] [PubMed] [Google Scholar]
- 37.Tzipori S, Rand W, Griffiths J, Widmer G, Crabb J. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin Diagn Lab Immunol. 1994;1:450–463. doi: 10.1128/cdli.1.4.450-463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Beers E H, Al R H, Rings E H, Einerhand A W, Dekker J, Buller H A. Lactase and sucrase-isomaltase gene expression during Caco-2 cell differentiation. Biochem J. 1995;308:769–775. doi: 10.1042/bj3080769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verdon R, Keusch G T, Tzipori S, Grubman S A, Jefferson D M, Ward H D. An in vitro model of infection of human biliary epithelial cells by Cryptosporidium parvum. J Infect Dis. 1997;175:1268–1272. doi: 10.1086/593695. [DOI] [PubMed] [Google Scholar]
- 40.Ward H D, Cevallos A M. Cryptosporidium: molecular basis of host-parasite interaction. Adv Parasitol. 1998;40:151–185. doi: 10.1016/s0065-308x(08)60120-7. [DOI] [PubMed] [Google Scholar]
- 41.Ward H D, Kane A V, Ortega-Barria E, Keusch G T, Pereira M E. Identification of developmentally regulated Giardia lamblia cyst antigens using GCSA-1, a cyst- specific monoclonal antibody. Mol Microbiol. 1990;4:2095–2102. doi: 10.1111/j.1365-2958.1990.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 42.Weltzin R L, Lucia-Zandris P, Michetti P, Fields B N, Kraehnbuhl J P, Neutra M R. Binding and transepithelial transport of immunoglobulins by intestinal M cells: demonstration using monoclonal IgA against enteric viral proteins. J Cell Biol. 1989;108:1673–1685. doi: 10.1083/jcb.108.5.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yada T, Totoki Y, Ishikawa M, Asai K, Nakai K. Automatic extraction of motifs represented in the hidden Markov model from a number of DNA sequences. Bioinformatics. 1998;14:317–325. doi: 10.1093/bioinformatics/14.4.317. [DOI] [PubMed] [Google Scholar]