Abstract
The apicomplexan parasite Cryptosporidium parvum is a major cause of serious diarrheal disease in both humans and animals. No efficacious chemo- or immunotherapies have been identified for cryptosporidiosis, but certain antibodies directed against zoite surface antigens and/or proteins shed by gliding zoites have been shown to neutralize infectivity in vitro and/or to passively protect against, or ameliorate, disease in vivo. We previously used monoclonal antibody 11A5 to identify a 15-kDa surface glycoprotein that was shed behind motile sporozoites and was recognized by several lectins that neutralized parasite infectivity for cultured epithelial cells. Here we report the cloning and sequence analysis of the gene encoding this 11A5 antigen. Surprisingly, the gene encoded a 330-amino-acid, mucin-like glycoprotein that was predicted to contain an N-terminal signal peptide, a homopolymeric tract of serine residues, 36 sites of O-linked glycosylation, and a hydrophobic C-terminal peptide specifying attachment of a glycosylphosphatidylinositol anchor. The single-copy gene lacked introns and was expressed during merogony to produce a 60-kDa precursor which was proteolytically cleaved to 15- and 45-kDa glycoprotein products that both localized to the surface of sporozoites and merozoites. The gp15/45/60 gene displayed a very high degree of sequence diversity among C. parvum isolates, and the numerous single-nucleotide and single-amino-acid polymorphisms defined five to six allelic classes, each characterized by additional intra-allelic sequence variation. The gp15/45/60 single-nucleotide polymorphisms will prove useful for haplotyping and fingerprinting isolates and for establishing meaningful relationships between C. parvum genotype and phenotype.
Cryptosporidium parvum, a protozoan parasite of the phylum Apicomplexa, is an enteric pathogen that infects humans and many animals and causes an acute diarrheal disease (30, 61). Cryptosporidiosis is self-limiting in immunocompetent individuals (22, 25, 116) but is often chronic and life-threatening in immunocompromised patients, such as those with AIDS (20, 22, 72, 76, 78) or in persons with weakened immune systems, for example, malnourished children and the elderly (18, 24, 27, 46, 89). Despite exhaustive attempts at chemotherapy with a wide variety of drugs, including many that are effective against related apicomplexan parasites, no efficacious treatment for cryptosporidiosis has been identified. Passive immunotherapy with hyperimmune bovine colostral immunoglobulin (HBC Ig) has, however, shown some ability to ameliorate the clinical symptoms of disease in humans (29, 62, 77, 88, 93, 108–110). HBC Ig inhibits sporozoite invasion in vitro and neutralizes sporozoite infectivity in animal models of cryptosporidiosis (5, 9, 10, 23, 69–71, 84–86, 107). Although the exact identities of the HBC Ig neutralization antigens are unknown, several candidate molecules have been identified (8, 39, 69, 84–86, 107). The majority are protein or glycoprotein antigens present on the zoite surface or resident in the secretory organelles of the zoite apical complex, and several are secreted during gliding motility and/or epithelial cell invasion.
Approximately 20 C. parvum proteins ranging in size from 11 to 900 kDa, including several recognized by HBC Ig, have been localized to the sporozoite plasmalemma by cell surface radioiodination experiments and are, therefore, candidate neutralization antigens (23, 84, 104). In addition, immunofluorescence microscopy experiments with a number of distinct monoclonal and monospecific, polyclonal anti-C. parvum antibodies localized seven antigens ranging in size from ∼15 to >1,200 kDa to the sporozoite and/or merozoite cell surfaces (31, 73, 84, 86, 87, 102, 105, 107). Many of these antigens were N- and/or O-glycosylated based on lectin binding profiles (45, 104), glycosidase reactivity patterns (73, 86), and periodate oxidation-glycotope ablation experiments (31, 86, 102, 105, 107), and at least three (gp15-17, p23-27, and gp900; the number represents size in kilodaltons) were present in the membranous and proteinaceous trails deposited by sporozoites during gliding locomotion (6, 26, 31, 73, 103). Moreover, the gp15-17 and p23-27 antigen families were strongly recognized by immunoglobulins present in human and animal infection and convalescent sera (48, 50, 54–56, 65, 67, 80–82), and high titers of these antibodies are believed to correlate with protection from clinical disease (56, 80, 83).
The few C. parvum neutralization antigens that have been identified are present on, or are secreted from, the apical complex and transit the zoite surface during gliding locomotion and/or host cell penetration. The antigens are deposited in trails behind gliding zoites and/or in “splashes” on the host cell surface at the site of parasite invasion. We previously used monoclonal antibody (MAb) 11A5 to identify a 15-kDa O-glycosylated protein that demonstrated these properties and hypothesized that it was a candidate sporozoite neutralization antigen (31). In this paper, we describe the cloning and characterization of a single-copy gene encoding the gp15 11A5 antigen and, surprisingly, a formerly unknown and complicated gp15/45/60 11A5 antigen family. Importantly, nucleic acid sequence analysis of the gp15/45/60 locus from 29 geographically diverse human and animal isolates of C. parvum showed that it was highly polymorphic, much more so than any Cryptosporidium locus examined to date. The locus manifests numerous single-nucleotide and single-amino-acid polymorphisms (SNPs and SAAPs), particularly among genotype I human isolates, and the amount of sequence variability is nearly sufficient to fingerprint individual genotype I isolates. The gp15/45/60 locus and associated SNPs will provide an important tool for investigating the relationships between parasite genotype and phenotype and will greatly facilitate studies designed to investigate the genotypic basis of parasite virulence and pathogenesis and determine the genetic population structure of the parasite.
MATERIALS AND METHODS
Parasites.
C. parvum oocysts (Iowa isolate) were purchased from P. Mason (Pleasant Hill Farms, Troy, Idaho). These oocysts were used to infect Madin-Darby canine kidney (MDCK; ATCC CCL 34) epithelial cells, to prepare parasite protein extracts, and to isolate RNA and genomic DNA (gDNA). Oocysts from human isolates 0542J, 0541L, 2064D, 2066K, and 0676I were provided by J. K. Griffiths (Tufts University School of Medicine, Boston, Mass.) and C. J. Fichtenbaum (University of Cincinnati School of Medicine, Cincinnati, Ohio) and were isolated from the stools of human immunodeficiency virus (HIV)-positive patients enrolled in the NIAID AIDS Clinical Trials Group (ACTG 336) phase II/III study of nitazoxanide (115). Oocysts from human isolates NT009 and Zaire were from J. H. Crabb (ImmuCell Corp., Portland, Maine); oocysts from human isolates CCPO1 (Texas), CCPO2 (Texas), and Peru were from C. L. Chappell and P. C. Okhuysen (Department of Medicine and School of Public Health, University of Texas Health Science Center, Houston); oocysts from human isolates 9877 and NEMC1 were from S. Tzipori and G. Widmer (Department of Infectious Diseases, Tufts University School of Veterinary Medicine, North Grafton, Mass.); oocysts from human isolates 6HMA1-10 (Milwaukee, 1993), HGM07 (Guatemala, 1997), and HGM10 (Guatemala, 1997) were from M. Arrowood (Centers for Disease Control and Prevention, Atlanta, Ga.); and oocysts from the human isolates SFGH1, SFGH3, SFGH4, SFGH6, and SFGH23 were purified from fecal samples obtained from the clinical laboratory at San Francisco General Hospital. Genomic DNA purified from a human Brazilian isolate was provided by C. Petersen (Agouron Pharmaceuticals, Inc., San Diego, Calif.) and is from the same sample studied earlier by Ortega et al. (64). Oocysts from animal isolates TAMU, UCP, and KSU were kindly provided by C. L. Chappell, J. H. Crabb, and S. J. Upton (Division of Biology, Kansas State University, Manhattan), respectively. Purified gDNA samples from calf isolates HS1 and HS2 were a gift from H. Schraft (Department of Food Science, University of Guelph, Ontario, Canada).
Cell culture.
MDCK cells were cultivated and infected as described previously (34). Briefly, cells were grown to confluency in eight-well chamber slides (Nalgene/Nunc) or T75 culture flasks (Corning) in RPMI 1640 (Gibco) containing 5% bovine calf serum (HyClone) at 37°C in a 5% CO2–95% air atmosphere. Cells were infected with bleach-sterilized oocysts or with purified sporozoites (33) in RPMI 1640 supplemented with 1% bovine calf serum and 0.048 U of insulin per ml.
Cloning of gp15/45/60.
Two libraries were used for cloning the gp15/45/60 gene: a NINC1 isolate gDNA expression library in λgt11 (43, 74), and a directionally cloned Iowa isolate sporozoite cDNA expression library in λZAP II XR (97). Approximately 200,000 PFU of the gDNA library was screened in Escherichia coli strain LE392, and 130,000 PFU of the cDNA library were screened in E. coli XL1 Blue MRF′ Kan. Recombinant protein expression was induced by overlaying plates with isopropyl-β-d-thiogalactopyranoside (IPTG)-impregnated nitrocellulose filters following standard methods (90). The libraries were screened in duplicate using a mixture of hybridoma culture supernatants containing monoclonal IgA antibodies CrA1 and CrA2; positive clones were detected using alkaline phosphatase-conjugated goat anti-mouse IgA secondary antibody (Southern Biotechnology Associates) followed by colorimetric development with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP).
Bacteriophage DNA from positive λgt11 clones was prepared using a Lambda Wizard Prep kit (Promega), digested with EcoRI, and fractionated by agarose gel electrophoresis. The recombinant DNA inserts were excised from the gel, purified using QIAEX II resin (Qiagen), ligated into EcoRI-cut phosphatase-treated pBlueScript II SK− plasmid (Stratagene), and transformed into E. coli JM109. Positive cDNA clones were derived from λZAP II XR bacteriophage by in vivo excision of the embedded pBlueScript II SK− cDNA plasmid using ExAssist M13 helper phage and E. coli SOLR as described by the manufacturer (Stratagene).
Nucleic acid preparation and analysis.
Total nucleic acids were isolated from purified oocysts (Iowa isolate) by several freeze-thaw cycles (liquid nitrogen, 65°C) in the presence of proteinase K followed by 55°C incubation for 2 h with additional proteinase K (43). Nucleic acids were digested with RNase A and phenol-chloroform extracted, and DNA was ethanol precipitated in the presence of 0.3 M sodium acetate (pH 5.2). Genomic DNA from the other isolates was prepared identically except proteolysis was extended to 6 h and additional proteinase K was added at 2-h intervals.
Total RNA was prepared from intracellular parasites by direct lysis of infected MDCK cell cultures with Trizol reagent (Gibco). Confluent monolayers growing in T75 culture flasks were infected with 8 × 107 sporozoites and incubated for various times at 37°C in a 95% air–5% CO2 atmosphere. Infected monolayers from single culture flasks were harvested at 30 min, 2 h, 4 h, 6 h, 9 h, and 11 h by Trizol lysis. Identical monolayers infected with 4 × 107 or 2 × 107 sporozoites were harvested at 24 and 48 h, respectively and a mock-infected control monolayer was harvested at 6 h. RNA preparations were treated with DNase I and extracted three times with phenol-chloroform and once with chloroform; the RNA was ethanol precipitated in the presence of 0.3 M sodium acetate (pH 5.2). Total RNA was also isolated from purified sporozoites using the Trizol procedure.
gp15/45/60 cDNA was prepared from sporozoite and intracellular parasite total RNA by reverse transcription followed by PCR amplification. Total cDNA was synthesized with a cDNA preamplification kit (Gibco) according to the manufacturer's instructions. Each reverse transcriptase (RT) reaction mixture contained 2 μg of DNase I-treated total RNA and the oligo(dT) primer provided with the kit. Following annealing of primer and RNA, reaction mixtures were divided into two aliquots; 200 U of Superscript II RT was added to one, and water was added to the other to control for gDNA contamination. After cDNA synthesis and before PCR, reaction mixtures were treated for 20 min at 37°C with 2 U of RNase H.
gp15/45/60 DNA fragments were PCR amplified using sense primer gp15ATG (5′-CGGGATCCATATGAGATTGTCGCTCATTATC) and antisense primer gp15STOP (5′-GGAATTCTTACAACACGAATAAGGCTG), which amplified a ca. 1-kb fragment extending from the translational start codon to the termination codon of the gp15/45/60 gene. The sense primer included upstream BamHI and NdeI restriction sites, and the antisense primer included a downstream EcoRI site for directional cloning of the amplified fragments. Amplifications were performed using Taq DNA polymerase and Q-solution (Qiagen) in a Perkin-Elmer model 9600 thermocycler for 35 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 45 s, and extension at 72°C for 60 s. A portion of the C. parvum 18S rDNA gene was amplified using primers CpBDIAGF (5′-AAGCTCGTAGTTGGATTTCTG) and CpBDIAGR (5′-TAAGGTGCTGAAGGAGTAAGG) as described elsewhere (41); reaction conditions were similar to those above except that annealing time was reduced to 30 s.
Southern blot analysis of C. parvum gDNA was performed by standard methods (90). Five-microgram aliquots of gDNA were digested with the enzymes EcoRI, BamHI, PstI, KpnI, or HindIII and size fractionated on a 0.8% agarose gel in 1× Tris-acetate-EDTA buffer. Following electrophoresis, alkaline denaturation, and neutralization, DNA was blotted overnight to a Qiabrane nylon plus membrane in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). A gp15/45/60 fluoresceinated DNA probe was prepared by random priming the gel-purified gp15/45/60 PCR fragment amplified from Iowa isolate gDNA, using a Tropix kit as described by the manufacturer. Blots were hybridized in ExpressHyb solution (Clontech) at 65°C for 1 h, washed several times in 2× SSC–1% SDS at room temperature, and given three 20-min washes in 0.2× SSC–1% SDS at 65°C. Following incubation of the blot with alkaline phosphatase-conjugated antifluorescein antibody and CDP-Star substrate (Southern Star kit; Tropix), the chemiluminescent hybridization signal was recorded on X-Omat AR film (Kodak).
DNA sequence analysis.
The gp15/45/60 cDNA and gDNA plasmid clone inserts were sequenced using the dideoxy-chain termination procedure with Sequenase T7 DNA polymerase and [35S]dATP (Amersham), and the reactions were fractionated on 5% Long Ranger acrylamide gels (FMC). The sequence of the NINC1 isolate gp15/45/60 insert was completely determined on both strands using primers spaced at ∼300-bp intervals, while several other clones identified in the screening were sequenced only on a single strand at their 5′ and 3′ ends.
All gp15/45/60 PCR and RT-PCR fragments were purified using a QiaQuick PCR Prep kit (Qiagen), and their sequences were determined by cycle sequencing using the same primers and either a dRhodamine dye terminator or a BigDye terminator cycle sequencing kit (Perkin-Elmer). Unincorporated dye terminators were removed by ethanol precipitation in the presence of 0.3 M sodium acetate (pH 5.2), and the nucleic acid pellet was dried under vacuum. Reactions were analyzed on a Perkin-Elmer model 377 automated fluorescence sequencer at the Iowa State University DNA Sequencing Facility, Ames. In a few instances, an additional primer was required to determine the complete sequence of one strand of the PCR fragment; these primers included sense primer gp15/45/60-2F (5′-ACTTCATTTRTHATGTGGTTCG) and antisense primer gp15/45/60-1R (5′-CCAAGTCTCCGTTCTCATTC).
Sequences were analyzed using DNA Strider 1.3 (47). The gp15/45/60 sequence was examined for similarity to sequences present in the GenBank nonredundant protein and nucleic acid databases, using the BLAST algorithm (2, 3). The N-terminal signal sequence and C-terminal glycosylphosphatidylinositol (GPI) anchor signal were predicted using the PSORT II program available over the World Wide Web (http://psort.nibb.ac.jp:8800/) (57). Potential GPI attachment sites were identified using the consensus GPI signal described by Coyne et al. (21). O-glycosylation sites were predicted using the NetOGlyc 2.0 server (http://130.225.67.199/services/NetOGlyc-2.0/) (35). All complete gp15/45/60 DNA and deduced protein sequences were compared in pairwise fashion to determine percentage sequence identity using the Genetics Computer Group GAP program (version 10.0, for Unix) with default settings; gp15/45/60 sequences were aligned using the ClustalW program, version 1.7 (101).
Parasite protein extracts.
Oocysts were excysted in phosphate-buffered saline (PBS) containing 2 mM sodium taurocholate; after the 3-min pulse at 37°C (33), the excystation mixture containing sporozoites and oocyst shells was made 1% in Triton X-100 and placed on ice for 10 min. The Triton X-100 lysate was adjusted to either 1× Complete mini protease inhibitor cocktail (Roche) or to 1× buffer A (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 1% Triton X-100, 1× Complete mini) and clarified by centrifugation at 14,000 × g at 4°C for 10 min. The Triton X-100-soluble supernatant was used for immunoblotting, immunoprecipitation, and lectin affinity chromatography.
Antibodies, immunoprecipitations, and immunoblotting.
MAb 11A5 has been described elsewhere (31). The anti-C. parvum IgA MAbs CrA1 and CrA2 were a gift from Marian R. Neutra (Children's Hospital, Harvard Medical School, Boston, Mass.). gp45 antisera were generated by intraperitoneal immunization of Sprague-Dawley rats with gel slices containing the HPA (Helix pomatia-derived) lectin affinity-purified and SDS-polyacrylamide gel electrophoresis (PAGE) size-fractionated, Coomassie blue-stained 45-kDa glycoprotein. Sera were collected after three additional immunizations with the same antigen.
Recombinant-eluted antibodies (REAs) recognizing proteins expressed by λgt11-gDNA or λZAP-cDNA bacteriophage clones were affinity purified from a polyspecific anti-oocyst/sporozoite rat ascites fluid on confluent plaque lifts of IPTG-induced phage clones, eluted with 0.1 M glycine (pH 2.5)–150 mM NaCl, and immediately neutralized as described elsewhere (74). Control REAs were prepared using wild-type λgt11 or λZAP II bacteriophage in the same manner.
CrA1/2 and 11A5 immunoprecipitations were performed by incubating a preformed ternary complex composed of primary MAb, biotin-conjugated secondary antibody (goat anti-mouse IgA or goat anti-mouse IgG), and streptavidin-agarose beads (EY Laboratories) with the extracts on a rotator for 1 h at room temperature. Immunoprecipitates were washed in PBS containing 1% Triton X-100, 0.01% bovine serum albumin, and 1× Complete mini protease inhibitors; antigens were eluted with 0.1 M glycine (pH 2.5)–150 mM NaCl, neutralized, boiled for 5 min in SDS-PAGE sample buffer, and electrophoretically separated on 16.5% (3% cross-linking) Tris-Tricine polyacrylamide gels (91). Gels were either stained with colloidal Coomassie (Serva Blue G) (60) or electrophoretically transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore). Blotted proteins were detected by chemiluminescence using a Western Star kit (Tropix) or by colorimetric methods using NBT and BCIP.
Immunofluorescence.
Sporozoite-infected MDCK cell monolayers in eight-well chamber slides were fixed at specified times postinfection with 4% formaldehyde–0.1% glutaraldehyde in PBS for 30 min and then washed with Tris-buffered saline. For detection of the 11A5, CrA1, or CrA2 antigen(s), air-dried cells were permeabilized with acetone, while for detection of the LOI antigen, a parasitophorous vacuole glycoprotein (14), cells were permeabilized with 1% Triton X-100 in PBS. After blocking with 0.25% bovine serum albumin in PBS, cells were sequentially incubated with primary antibody, biotin-conjugated secondary antibodies, and streptavidin-conjugated Cy3. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) in PBS. To identify antigens in gliding trials, sporozoites in PBS were allowed to glide on poly-l-lysine-coated slides for 5 min at 37°C (31); the parasites were fixed with 4% formaldehyde in PBS and immunostained, without permeabilization, as described above.
HPA lectin affinity chromatography.
A 1% Triton X-100-soluble protein extract in buffer A was applied to a column of HPA-agarose (EY Laboratories), and the column was washed extensively with buffer A lacking protease inhibitors. Alternatively, some experiments were carried out in a batch mode by incubating HPA-agarose resin with parasite lysates; in both cases, HPA-bound proteins were eluted with 0.2 M GalNAc.
HPA-binding proteins were also affinity purified from C. parvum-infected MDCK cells. Confluent monolayers (two T150 flasks) were infected with 108 oocysts/T150 for 2 h at 37°C in a 5% CO2–air atmosphere, washed several times with PBS to remove unexcysted oocysts and oocyst shells, and returned to the incubator for an additional 9 h. Infected monolayers were lysed in 3 ml of PBS containing 1% SDS, and the lysate was diluted with 10 volumes of buffer A containing 5 mM MgCl2, 200 μg of RNase A, 750 U of DNase I, and 200 μl of HPA-agarose. Samples were rotated for 1 h at room temperature; the HPA-resin was collected by centrifugation and washed extensively with buffer A without protease inhibitors, and bound proteins were eluted with 0.2 M GalNAc in wash buffer. The eluate was adjusted to 1× SDS-PAGE sample buffer and boiled for 5 min prior to electrophoretic fractionation. HPA-binding proteins were also affinity purified in the same manner from uninfected MDCK monolayers as a control.
N-terminal amino acid sequencing.
N-terminal sequencing was performed by automated Edman degradation at the University of California, Davis Protein Structure Laboratory. HPA-binding proteins were affinity purified from 109 oocysts as described above, size fractionated by SDS-PAGE (12% gel), and electrophoretically transferred to either an Immobilon PSQ (Millipore) or a Protoblot (Bio-Rad) polyvinylidene difluoride membrane in buffer containing 10 mM CAPS (3-(cyclohexylamino)-1-propanesulfonic acid; pH 11) and 10% methanol. Membranes were rinsed with water, stained for 1 min with 0.1% Coomassie brilliant blue R-250 in 50% methanol-water, destained for 5 min in several changes of 40% methanol–10% acetic acid, and finally rinsed with high-pressure liquid chromatography grade water. All stained protein bands were excised, dried, and submitted for N-terminal sequence analysis.
Nucleotide sequence accession numbers.
The complete C. parvum NINC1 isolate gp15/45/60 nucleotide sequence has been submitted to GenBank with accession number AF022929. The partial nucleotide sequences of gp15/45/60 genes from other isolates have been submitted to GenBank under accession numbers AF164487 to AF164505, AF164508, AF164509, AF178690 to AF178697, and AF224462 to AF224464. The partial 18S rDNA nucleotide sequences from different C. parvum isolates were submitted to GenBank with accession numbers AF178698 to AF178701 and AF224465.
RESULTS
MAbs 11A5, CrA1, and CrA2 recognize the same 15-kDa glycoprotein antigen.
We previously reported that MAb 11A5 recognized a carbohydrate or carbohydrate-dependent epitope on a 15-kDa glycoprotein antigen, gp15. This antigen was present on the surface of sporozoites and merozoites, in antigen trails deposited behind gliding sporozoites, and was shed from the sporozoite surface during invasion of MDCK epithelial cells (31). gp15 was shown to be O-glycosylated by lectin blotting and, based on its spectrum of lectin reactivity, to contain glycotopes similar or identical to the Thomsen-Freidenreich antigen [Gal(β1→3)GalNAcα1→Ser/Thr] and/or its precursor, the Tn antigen (GalNAcα1→Ser/Thr). The same lectins that identified the 11A5 antigen by lectin blotting also completely and irreversibly neutralized sporozoite infectivity for cultured MDCK cells (32). Together, these results suggested that gp15 might play a role in sporozoite motility and/or in host cell adhesion and invasion, and that it was a potential target of neutralizing antibodies and lectins. Thus, it was of interest to further characterize the 11A5 glycoprotein by cloning the corresponding gene; MAb 11A5 could not be used for this purpose, however, as it recognized a carbohydrate epitope. Serendipitously we learned of two anti-C. parvum IgA MAbs, CrA1 and CrA2, that recognized a 15-kDa surface antigen and provided partial passive protection against oral oocyst challenge of SCID mice when secreted from CrA1 or CrA2 hybridoma “backpack” tumors; both MAbs appeared to recognize protein epitopes as periodate oxidation of the antigen did not affect antibody reactivity (X. Zhou, S. Tzipori, and M. Neutra, personal communication).
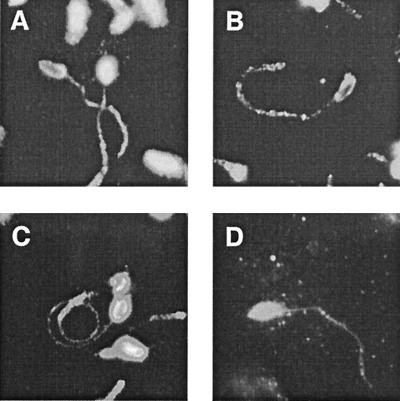
To compare the reactivity patterns of MAbs CrA1 and CrA2 (the kind gift of Marian R. Neutra, Children's Hospital, Harvard Medical School, Boston, Mass.), both with each other and with those of MAb 11A5 and the neutralizing H. pomatia α-GalNAc-specific lectin HPA, we microscopically examined the fluorescent staining patterns these reagents produced with formaldehyde-fixed sporozoites that had been gliding on poly-l-lysine-coated microscope slides prior to fixation. Each MAb and the lectin decorated sporozoites identically (Fig. 1), outlining the zoite surface and revealing antigen trails shed by gliding sporozoites, suggesting that the four reagents may identify the same glycoprotein. To determine whether the CrA1, CrA2, and 11A5 MAbs recognized the same antigen, each was used to precipitate a cognate antigen(s) from a Triton X-100-soluble sporozoite-oocyst extract, and the electrophoretically fractionated precipitates were blotted with each of the antibodies (Fig. 2; CrA1 results not shown). All three MAbs detected the 15-kDa antigen regardless of which antibody was used for immunoprecipitation, providing strong evidence that the antibodies all recognize gp15; a second antigen of ∼25 kDa was also recognized by all three MAbs. A third, ∼50-kDa antigen was detected in CrA2 precipitates blotted with CrA2 (Fig. 2B, lane 3) and in CrA2 immunoblots of the sporozoite-oocyst extract (Fig. 2B, lane 1) but was not identified by 11A5 (Fig. 2A, lanes 1 to 3) or CrA1 (not shown), suggesting that this ∼50-kDa antigen was unrelated to gp15. Since MAbs CrA1 and CrA2 both recognized protein epitopes of gp15, they were used to screen bacteriophage lambda expression libraries to identify and isolate the cognate gene as described below.
FIG. 1.
Anti-15-kDa antigen-specific MAbs and the HPA lectin decorate gliding sporozoites and identify shed antigen trails. Sporozoites locomoting on poly-l-lysine-coated slides were formaldehyde fixed, blocked, and incubated with MAb 11A5 (A), CrA1 (B), or CrA2 (C) or the biotin-conjugated lectin HPA (D). Reactivity profiles were visualized and documented by epifluorescence photomicroscopy following incubation of the washed slides with biotinylated secondary antibodies and/or Cy3-conjugated streptavidin.
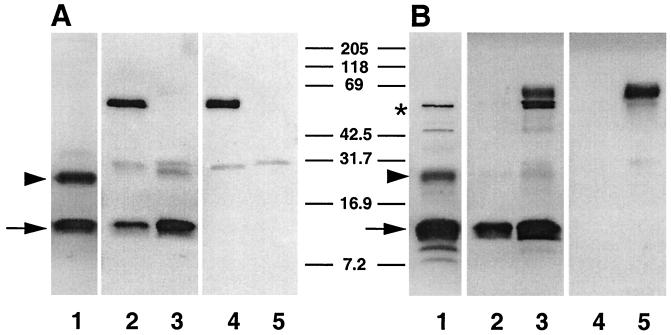
FIG. 2.
MAbs 11A5 and CrA2 recognize the same 15-kDa glycoprotein antigen. Triton X-100-soluble oocyst/sporozoite proteins were either directly fractionated and blotted following SDS-PAGE (A and B, lanes 1) or fractionated and blotted following immunoprecipitation with IgG MAb 11A5 (A and B, lanes 2 and 4) or IgA MAb CrA2 (A and B, lanes 3 and 5). The blots were probed with primary MAb 11A5 (A, lanes 1 to 3) or primary MAb CrA2 (B, lanes 1 to 3) followed by alkaline phosphatase-conjugated secondary anti-IgG (A) or anti-IgA (B) antibodies. The arrow indicates the position of the 15-kDa antigen, the arrowhead points to the 25-kDa antigen, the asterisk denotes the 50-kDa antigen, and lanes 4 and 5 in both panels are secondary antibody controls (lacking primary antibody) which reveal the position of the precipitating MAb 11A5 and CrA2 heavy and light chains.
Expression cloning of the gp15 gene.
A mixture of the CrA1 and CrA2 MAbs was used to screen 1.3 × 105 recombinant bacteriophage from a sporozoite cDNA expression library (Iowa isolate [97]) and 2 × 105 recombinant bacteriophage from a gDNA expression library (NINC1 isolate [43]). The antibody cocktail identified four cDNA and seven gDNA clones which were plaque purified and used to prepare REAs (74). REAs prepared using gDNA clones G7A1A and G4C1A identified an antigen of the same size as that recognized by MAbs 11A5, CrA1, and CrA2 in immunoblots of oocyst-sporozoite extracts (Fig. 3). To our surprise, however, none of the other REAs recognized a 15-kDa antigen. When plaque lifts containing recombinant proteins expressed from each of the clones were examined for reactivity with the CrA1 and CrA2 MAbs separately, only clones G7A1A and G4C1A were identified by both antibodies; the other nine clones reacted only with MAb CrA2. DNA sequence analyses of the inserts of several of the latter clones indicated that the CrA2-reactive peptide they encoded was, in fact, the C. parvum ortholog of protein translation elongation factor 1α (EF-1α), whose predicted size corresponds to the ∼50-kDa protein identified by MAb CrA2 in the immunoblot of Fig. 2. Remarkably, a C. parvum EF-1α cDNA was also recently isolated from a sporozoite cDNA expression library by Bonafonte et al. (12) using a different anti-gp15 MAb (MAb C6C1) that apparently also cross-reacts with an EF-1α epitope.
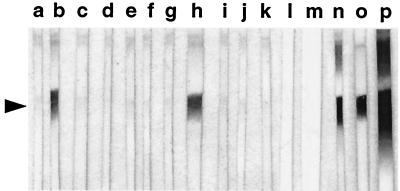
FIG. 3.
REAs affinity purified on plaque lifts of gDNA clones G4C1A and G7A1A recognize a 15-kDa C. parvum antigen. Oocyst lysates were fractionated by SDS-PAGE, blotted, and probed with REA prepared using gDNA (G) and cDNA (S) clones G9A1A (a), G7A1A (b), S10D1A (c), G4D1A (d), G15A1A (e), S9C1A (f), G12B1A (g), G4C1A (h), G1A1A (i), S5F1A (j), and S5C1A (k). The blots in lanes l and m were probed with REAs prepared on plaque lifts of the wild-type λZAP II and λgt11 phage vectors, respectively. Positive control blots were probed with MAbs 11A5 (n), CrA1 (o), and CrA2 (p); the position of the 15-kDa antigen is indicated by the arrowhead.
The gp15 gene is predicted to encode an acidic ∼34-kDa glycoprotein.
The inserts from the G4C1A and G7A1A λgt11 gDNA clones were subcloned into pBlueScript II SK− and sequenced from both ends using T3 and T7 universal primers. The partial sequences were identical with the exception that the G4C1A sequence extended 230 bp further 3′; therefore, this clone was selected for further analysis. The complete 1,355-bp G4C1A nucleotide sequence was determined on both strands. The sequence contained a single large open reading frame (ORF), beginning at nucleotide 289, that was predicted to encode an acidic (pI ∼4.4), 330-amino-acid, 34-kDa protein (Fig. 4). The nucleic acid and deduced protein sequences were used to search the GenBank nonredundant, dbEST and dbGSS databases using BLAST algorithms; one significant sequence similarity was discovered with C. parvum genome survey sequence CpG2126A, which corresponded to the 3′ end of the gp15 gene.
FIG. 4.
Nucleotide and deduced amino acid sequence of the NINC1 C. parvum gp15 gene. Locations of the gp15ATG and gp15STOP primers are indicated by arrows above and below the nucleotide sequence. The empirically determined N-terminal amino acid sequences of the HPA-affinity purified 15- and 45-kDa proteins are shown enclosed in boxes. The predicted N-terminal signal peptide cleavage site is indicated by the triangle, and the predicted GPI anchor attachment site is circled. Amino acid residues predicted to be O-glycosylated are subscripted by circles, and the single predicted N-glycosylated asparagine residue (Asn202) is boxed.
Despite its anomalous size, 34 kDa instead of 15 kDa, the deduced protein sequence had a number of interesting features, several of which were consistent with the cell surface localization and O-linked glycosylation of gp15. The amino terminus of the sequence contained a string of hydrophobic and hydroxylic amino acid residues that were predicted by the PSORT II program (57) to constitute an N-terminal signal sequence with a signal peptidase cleavage site between amino acids Ser19 and Ala20; similarly, the hydrophobic C terminus of the sequence was predicted by PSORT II to contain a C-terminal GPI anchor attachment sequence with proteolytic cleavage and anchor attachment occurring at residue Ser308 (Fig. 4). Adjacent to the signal sequence, the deduced protein sequence contained a homopolymeric serine tract comprised of 23 Ser residues, the majority of which, like many other Ser and Thr residues dispersed throughout the sequence, were predicted to be O-glycosylated (Fig. 4) by the NetOGlyc 2.0 program (35), an algorithm for prediction of mucin-type O-glycosylation sites. Last, the hypothetical protein was also predicted to contain a potential N-glycosylation site at Asn202 (Fig. 4); we have, however, no experimental data bearing on gp15 N-glycosylation.
The gp15 gene is single copy and lacks introns.
To examine gene copy number, C. parvum gDNA (Iowa isolate) was digested with three restriction enzymes that do not cleave within the gp15 sequence (EcoRI, PstI, and BamHI) and two that cleave once within the sequence (KpnI and HindIII). The digests were size fractionated and Southern blotted, and the blot was hybridized with a probe prepared by labeling the 1-kb PCR fragment amplified from the G4C1A plasmid clone with the gp15ATG and gp15STOP primers (Fig. 4). As expected for a single-copy gene, only a single hybridizing fragment was detected in the EcoRI, PstI, and BamHI restriction digests, while two hybridizing fragments were observed in digests with KpnI and HindIII (data not shown). Furthermore, to determine if the gene contained introns, we sequenced a gp15 cDNA fragment that was RT-PCR amplified from C. parvum-infected MDCK cell RNA with the same primers. The size and sequence of the amplified cDNA exactly matched those of the gp15 fragment amplified from genomic DNA, indicating that there were no introns within the coding sequence of the gene.
gp15 mRNA and protein are expressed late in merogony.
To examine the timing of gp15 transcription and translation during C. parvum intracellular development, we assayed the appearance of gp15 mRNA and antigen during a 48-h period following infection of MDCK epithelial cells with oocysts. gp15 mRNA expression was qualitatively assessed by RT-PCR using gp15ATG and gp15STOP primers (Fig. 5A) and gp15 antigen expression was monitored by indirect immunofluorescence with various anti-gp15 MAbs (Fig. 5B). Although gp15 is present on the sporozoite surface, it is shed as the sporozoite penetrates the host cell during invasion (31) and is absent from the earliest intracellular-stage parasites, single-nucleated trophozoites. As indicated by RT-PCR amplification of a 1-kb gp15 cDNA fragment from infected-host cell RNA, gp15 mRNA was first detected 4 h following infection (Fig. 5A, lane 5), a time when all parasites were still uninucleate. Even 2 h later, however, when gp15 mRNA expression was easily detectable (Fig. 5A, lane 6), little gp15 antigen was observed by immunofluorescence (Fig. 5B, images F, J, and N), although the control parasitophorous vacuole antigen LOI (14) was present (Fig. 5B, image B). Expression of gp15 mRNA increased and peaked between 9 and 24 h (Fig. 5A, lanes 7 to 9) before decreasing at 48 h (Fig. 5A, lane 10). By 11 h postinfection, many parasites had undergone three complete rounds of mitosis and nuclear division and existed as mature, eight-nucleated schizonts (Fig. 5B, images G, K, and O) containing fully developed merozoites. These merozoites contained significant amounts of the gp15 antigen, apparently localized to their surface membrane as judged by the pattern of reactivity with anti-gp15 MAbs, which outlined individual merozoites present within the schizont (Fig. 5B, images H, L, and P).
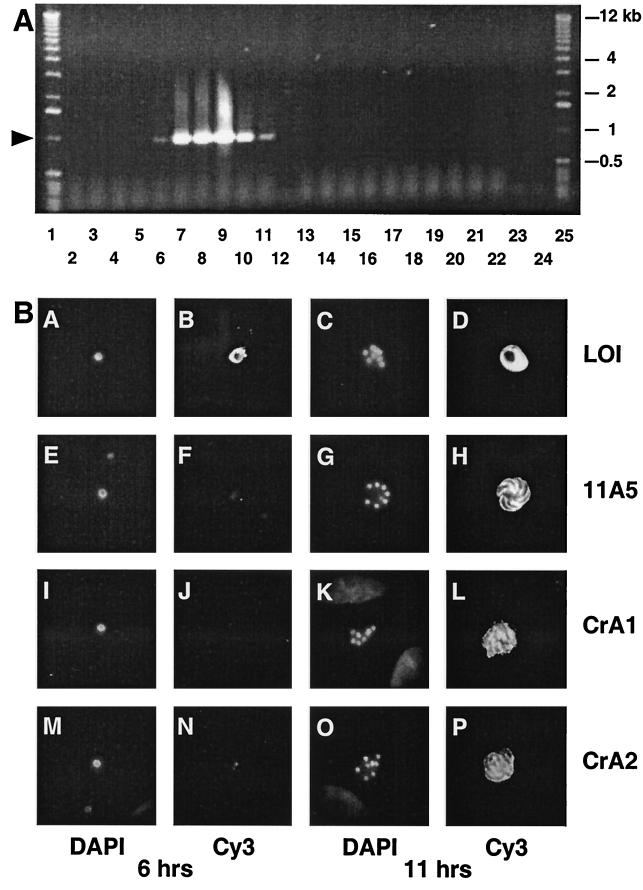
FIG. 5.
(A) Time course of gp15 mRNA expression in C. parvum-infected MDCK cell monolayers. Total RNA was isolated from infected MDCK cells at various times postinfection, DNase treated, and used as a template for RT-PCR (lanes 2 to 12) or control PCR (lanes 13 to 23), both primed with the gp15ATG and gp15STOP primers. The arrowhead indicates the position of the 1-kb gp15 amplicon. RNA template was isolated from infected MDCK cells at 0.5 h (lanes 3 and 14), 2 h (lanes 4 and 15), 4 h (lanes 5 and 16), 6 h (lanes 6 and 17), 9 h (lanes 7 and 18), 11 h (lanes 8 and 19), 24 h (lanes 9 and 20), and 48 h (lanes 10 and 21) postinfection; from uninfected MDCK cells (lanes 2 and 13); and directly from purified sporozoites (2 μg; lanes 11 and 22). Additional controls for the RT-PCR and PCRs lacked template nucleic acid (lanes 12 and 23 and 24, respectively). (B) Intracellular expression of gp15 protein occurs late in merogony. C. parvum-infected MDCK monolayers were fixed 6 and 11 h postinfection and incubated with control MAb LOI (B and D), which recognizes a parasitophorous vacuole antigen present throughout intracellular parasite development (14), or with anti-gp15 MAb 11A5 (F and H), CrA1 (J and L) or CrA2 (N and P) followed by biotinylated secondary antibodies and Cy3-conjugated streptavidin. Parasite nuclei in the same microscopic fields were stained with DAPI (lanes A, E, I, M, C, G, K, and O).
The “gp15 gene” actually encodes a 60-kDa glycoprotein that is processed during intracellular parasite development to produce gp15 and gp45 products.
Based on the dissimilar sizes of the 15-kDa 11A5-CrA1/2 antigen and the hypothetical ∼34-kDa glycoprotein encoded by the single large ORF present in the cloned gene, we hypothesized that gp15 was derived from the larger precursor by co- or posttranslational proteolytic processing. To investigate this possibility, we isolated oocyst/sporozoite O-linked glycoproteins by affinity chromatography of a Triton X-100-soluble parasite lysate on HPA-agarose, a lectin known to bind the 11A5 antigen (31). One of five major proteins (Fig. 6, lane c) specifically eluted with 0.2 M GalNAc was a 15-kDa glycoprotein recognized in Western blots by MAbs 11A5, CrA1, and CrA2, as well as by REA affinity purified on recombinant protein expressed by the G4C1A clone containing the ∼34-kDa ORF (Fig. 6, lanes 1 to 3 and 5). N-terminal sequence analysis of this 15-kDa glycoprotein produced the sequence NH2-ETXEAAAXVDLFAF, which corresponds to amino acids 227 through 240 of the deduced ∼34-kDa protein sequence (Fig. 4). The failure to identify the serine at position 229 and the threonine at position 234 suggested that these residues were most probably glycosylated. These results indicated that gp15 was derived from the C-terminal one-third of the ORF encoding the hypothetical ∼34-kDa glycoprotein, presumably by cleavage of a larger O-glycosylated precursor. Neither the other putative cleavage product, i.e., the N-terminal two-thirds of the ∼34-kDa ORF, nor the presumptive full-length precursor was obvious, however, as none of the anti-gp15 MAbs, nor the G4C1A REA, identified the larger proteins in immunoblots of the oocyst-sporozoite lysate or the HPA affinity-purified proteins.
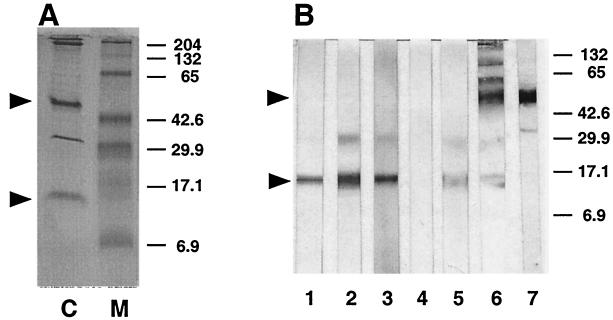
FIG. 6.
HPA affinity chromatography identifies 15- and 45-kDa glycoproteins. A Triton X-100-soluble, oocyst-sporozoite lysate was chromatographed on HPA-agarose, and bound proteins were eluted with GalNAc, fractionated by SDS-PAGE, and stained with Coomassie Serva blue G (lane C) or Western blotted (B) and probed with MAb CrA1 (lane 1), CrA2 (lane 2), 11A5 (lane 3), wild-type λgt11 REA (lane 4), G4C1A REA (lane 5), HPA-biotin (lane 6), or rat anti-gp45 polyclonal antibody (lane 7). The arrowheads indicate the positions of gp15 and gp45. Molecular weight markers are shown in lane M. Sizes are indicated in kilodaltons.
In an independent investigation Cevallos et al. (A. M. Cevallos, personal communication) cloned a gene encoding a ∼40-kDa HPA-binding glycoprotein (19) recognized by the sporozoite-neutralizing MAb 4E9 (A. M. Cevallos, N. Bhat, R. Verdon, D. H. Hamer, B. Stein, S. Tzipori, M. E. A. Pereira, G. T. Keusch, and H. D. Ward, submitted for publicaiton); in mutual discussions it became clear that the nucleic acid sequence of this gene was essentially identical to the sequence determined in this investigation. To determine if the ∼45-kDa glycoprotein (gp45) eluted from HPA-agarose with GalNAc in our experiments (Fig. 6, lane C) was the same as the ∼40-kDa protein identified by Cevallos et al. and was encoded by the ∼34 kDa ORF, we determined its N-terminal amino acid sequence. The sequence of the first six residues, NH2-DVPVEG, was identical to amino acid residues 31 through 36 of the hypothetical ∼34-kDa glycoprotein sequence (Fig. 4) beginning 12 residues downstream from the predicted signal peptide cleavage site. Since the CrA1, CrA2, and 11A5 MAbs failed to identify gp45 (Fig. 6, lanes 1 to 3), we believed it lacked the C-terminal gp15 sequence and was, in fact, the second proteolytic cleavage product corresponding to the N-terminal two-thirds of the ∼34-kDa ORF. This conjecture was supported by the observation that a rat antiserum prepared against HPA affinity-purified gp45 failed to detect gp15 on immunoblots (Fig. 6, lane 7). Thus, the evidence suggested that gp45 and gp15 were the respective N- and C-terminal proteolytic cleavage products of a larger precursor, i.e., the hypothetical ∼34-kDa glycoprotein, which was either not present or not abundant in the Triton X-100-soluble oocyst-sporozoite lysate.
In an attempt to identify the putative precursor containing both gp15 and gp45, we examined the C. parvum-infected epithelial cell cultures previously shown to actively transcribe the full-length, 1-kb gp15 mRNA and express gp15 antigenic determinants. Glycoproteins were HPA affinity purified from detergent lysates of infected and uninfected MDCK cell monolayers; GalNAc eluates were fractionated by Tris-Tricine SDS-PAGE and blotted with the anti-gp45 rat serum and MAb CrA2 (Fig. 7). Not only did anti-gp45 serum and MAb CrA2 individually recognize their respective antigens, gp45 and gp15, but both reagents also identified a larger, ∼60-kDa precursor (gp60). In addition, MAb CrA2 detected additional parasite glycoproteins of ∼17 and ∼25 kDa; the precise biochemical composition and origin of these two products are unknown, but their presence suggests that gp15/45/60 processing and maturation are complex.
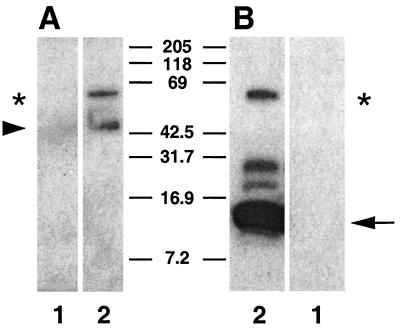
FIG. 7.
A 60-kDa precursor glycoprotein is synthesized by intracellular parasites and processed to produce gp15 and gp45 products. GalNAc-containing O-linked glycoproteins were affinity purified on HPA-agarose from uninfected (lane 1) and C. parvum-infected MDCK monolayers 11 h postinfection (lane 2), eluted with GalNAc, fractionated by SDS-PAGE, blotted, and probed with rat anti-gp45 polyclonal antiserum (A) or MAb CrA2 (B). The positions of the gp60 precursor (A and B, asterisk) and the presumptive gp15 (B, arrow) and gp45 (A, arrowhead) proteolytic products are indicated.
The nucleic acid and deduced amino acid sequences of gp15/45/60 are extremely polymorphic among human isolates of C. parvum.
Diverse human and animal isolates of C. parvum have been genotyped at a number of genetic loci, using comparative DNA sequence analysis to identify SNPs and SAAPs that distinguish alleles at a particular locus (7, 13, 15, 51, 52, 66, 68, 75, 94, 98, 99, 112, 113). The results of these investigations have been surprisingly uncomplicated and uniform. Essentially all genetic loci examined displayed dimorphic alleles that defined two prototype genotypes, one of these, commonly termed genotype I or the human (H) genotype, was found only among isolates derived from human infections; the other, termed genotype II or the calf (C) genotype, was found among both human and diverse animal isolates. Most of these genotyping studies characterized the SNP/SAAP patterns of housekeeping genes, although a few examined genetic loci that might be expected to be more polymorphic, e.g., the presumptive zoite surface antigens TRAP-C1 (95) and TRAP-C2 (68, 99) and a number of microsatellite simple sequence repeated sequences (1). The latter loci were, however, no more polymorphic than the former and offered no additional insight into the degree of genetic diversity within and between the two prototypal C. parvum genotypes.
Since gp15/45/60 is also a zoite surface antigen that might be under selective pressure, and since it contains a homopolymer serine tract encoded by a trinucleotide repeat, which are often hypervariable, we examined the SNPs and SAAPs at this locus in a number of animal and human isolates of C. parvum. The complete gp15/45/60 ORF was PCR amplified from gDNA isolated from 29 diverse isolates using the gp15ATG and gp15STOP primers; the amplicons from 23 isolates were completely sequenced, while six others were partially sequenced. Each complete DNA (and deduced protein) sequence was compared with each of the other complete sequences using the GAP alignment algorithm, and the percentages of identical nucleotide and amino acid residues were tabulated for each pairwise comparison (Table 1). Surprisingly, both comparisons clearly revealed that the gp15/45/60 sequences formed at least five, not two, distinct allelic groupings (Table 1). The nucleotide and deduced amino acid sequences of the gp15/45/60 amplicons derived from all of the animal isolates and four of the human isolates (Brazil, NT009, Peru, and Zaire) were identical or very nearly identical to each other and to the original NINC1 isolate sequence (Table 1). This homogeneous sequence family defined a single gp15/45/60 allele which, since it was found among both human and animal C. parvum isolates, is by definition a genotype II allele. The gp15/45/60 sequences derived from 16 other human isolates, however, displayed extensive polymorphism and conservatively defined at least four additional alleles which, since they were found only among human isolates, are by definition genotype I alleles (i.e., Ia, Ib, Ic and Id [Table 1]). The Ia allelic group contained sequences amplified from the SFGH1, SFGH3, SFGH4, SFGH6, and 2066K isolates, the Ib group contained sequences from the 0541L, 0542J, 2064D, CCPO2, NEMC1, and 9877 isolates; the Ic group contained sequences from the 0676I and SFGH23 isolates; and the Id group contained the complete sequence from the CCPO1 isolate and partial sequences from the HGM07 and HGM10 isolates. The gp15/45/60 sequences comprising any one particular allelic group exhibited 98 to 100% nucleotide and amino acid sequence identity, while the sequences from different allelic groupings shared only 77 to 88% nucleic acid sequence identity and even less amino acid sequence identity, 67 to 80% (Table 1). The two sequences grouped together as allele Ic were considerably less similar to one another (92% amino acid and 95% nucleotide sequence identity) than those defining the other type I and II alleles, and a case could be made for classifying these sequences as distinct alleles, thereby creating a fifth genotype I allelic group.
TABLE 1.
Sequence identitiesa define five distinct gp15/45/60 allelic groups: Ia, Ib, Ic, Id, and II
| Allelic group | Isolate | Identity (%)
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ia
|
Ib
|
Ic
|
Id
|
II
|
||||||||||||||||||||
| SFGH1 | SFGH3 | SFGH4 | SFGH6 | 2066K | 0541L | 0542J | 2064D | CCPO2 | NEMC1 | 9877 | 0676I | SFGH23 | CCPO1 | NINC1 | Brazil | Iowa | KSU1 | NT009 | Peru | TAMU | UCP | Zaire | ||
| Ia | SFGH1 | 100 | 99.1 | 99.1 | 99.1 | 99.0 | 70.5 | 69.7 | 70.5 | 71.6 | 71.8 | 71.8 | 74.5 | 70.9 | 70.3 | 72.3 | 73.2 | 77.0 | 77.0 | 77.0 | 77.0 | 77.0 | 76.9 | 73.5 |
| SFGH3 | 99.7 | 100 | 100 | 99.4 | 99.4 | 71.2 | 70.6 | 71.2 | 72.4 | 72.6 | 72.6 | 74.1 | 70.0 | 74.1 | 76.1 | 76.1 | 76.0 | 76.0 | 76.0 | 76.0 | 76.0 | 76.0 | 75.8 | |
| SFGH4 | 99.7 | 100 | 100 | 99.4 | 99.4 | 71.2 | 70.6 | 71.2 | 72.4 | 72.6 | 72.6 | 74.1 | 70.0 | 74.1 | 76.1 | 76.1 | 76.0 | 76.0 | 76.0 | 76.0 | 76.0 | 76.0 | 75.8 | |
| SFGH6 | 99.7 | 99.8 | 99.8 | 100 | 99.4 | 71.4 | 70.6 | 71.5 | 71.0 | 71.2 | 71.2 | 74.4 | 70.4 | 72.4 | 72.5 | 76.7 | 77.4 | 77.4 | 77.4 | 77.4 | 77.4 | 76.9 | 76.9 | |
| 2066K | 99.7 | 99.8 | 99.8 | 99.8 | 100 | 73.4 | 72.9 | 73.4 | 73.6 | 73.4 | 73.4 | 74.1 | 70.2 | 72.3 | 71.2 | 75.4 | 75.5 | 75.5 | 75.5 | 75.5 | 75.5 | 75.5 | 75.5 | |
| Ib | 0541L | 78.2 | 77.8 | 77.8 | 78.5 | 79.7 | 100 | 100 | 99.7 | 98.1 | 98.4 | 98.4 | 76.4 | 79.1 | 70.2 | 68.3 | 69.4 | 74.4 | 74.4 | 74.4 | 74.4 | 74.4 | 74.3 | 69.7 |
| 0542J | 78.2 | 77.4 | 77.4 | 78.0 | 79.5 | 100 | 100 | 99.7 | 98.4 | 98.4 | 98.4 | 75.7 | 78.8 | 69.6 | 67.3 | 68.8 | 73.6 | 73.6 | 73.6 | 73.6 | 73.6 | 73.6 | 68.9 | |
| 2064D | 78.2 | 77.7 | 77.7 | 78.4 | 79.6 | 99.9 | 99.9 | 100 | 97.8 | 98.1 | 98.1 | 76.4 | 79.0 | 69.9 | 68.3 | 69.5 | 74.4 | 74.4 | 74.4 | 74.4 | 74.4 | 74.4 | 69.8 | |
| CCPO2 | 77.7 | 77.0 | 77.0 | 77.8 | 79.8 | 98.7 | 99.2 | 98.7 | 100 | 99.7 | 99.7 | 75.7 | 79.0 | 70.4 | 69.3 | 69.3 | 74.0 | 74.0 | 74.0 | 74.0 | 74.0 | 73.9 | 69.6 | |
| NEMC1 | 78.1 | 77.4 | 77.4 | 78.2 | 79.9 | 99.2 | 99.3 | 99.2 | 99.6 | 100 | 100 | 75.8 | 79.1 | 70.6 | 69.6 | 69.5 | 74.3 | 74.3 | 74.3 | 74.3 | 74.3 | 74.1 | 69.8 | |
| 9877 | 78.1 | 77.4 | 77.4 | 78.2 | 79.9 | 99.2 | 99.3 | 99.2 | 99.6 | 100 | 100 | 75.8 | 79.1 | 70.6 | 69.6 | 69.5 | 74.3 | 74.3 | 74.3 | 74.3 | 74.3 | 74.1 | 69.8 | |
| Ic | 0676I | 79.6 | 79.3 | 79.3 | 79.5 | 79.0 | 85.3 | 85.0 | 85.3 | 85.0 | 85.3 | 85.3 | 100 | 92.2 | 73.0 | 76.9 | 76.9 | 77.3 | 77.3 | 77.3 | 77.3 | 77.3 | 77.2 | 76.6 |
| SFGH23 | 77.9 | 78.1 | 78.1 | 78.3 | 78.0 | 87.1 | 87.2 | 87.1 | 87.4 | 87.5 | 87.5 | 95.4 | 100 | 71.4 | 71.7 | 71.6 | 72.0 | 72.0 | 72.0 | 72.0 | 72.0 | 71.9 | 71.3 | |
| Id | CCPO1 | 80.9 | 79.7 | 79.7 | 82.4 | 82.8 | 80.1 | 79.7 | 80.0 | 79.5 | 79.9 | 79.9 | 79.9 | 78.9 | 100 | 79.5 | 79.9 | 80.2 | 80.2 | 80.2 | 80.2 | 80.2 | 80.2 | 80.2 |
| II | NINC1 | 80.8 | 84.6 | 84.6 | 82.5 | 79.7 | 78.6 | 78.1 | 78.7 | 77.5 | 77.9 | 77.9 | 83.1 | 80.3 | 80.9 | 100 | 99.7 | 99.7 | 99.7 | 99.7 | 99.7 | 99.7 | 99.7 | 99.4 |
| Brazil | 81.6 | 84.8 | 84.8 | 81.6 | 81.2 | 80.0 | 79.6 | 80.0 | 79.5 | 79.9 | 79.9 | 82.9 | 80.0 | 83.5 | 99.8 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.7 | |
| Iowa | 81.9 | 84.7 | 84.7 | 83.6 | 84.0 | 79.9 | 79.9 | 80.0 | 79.4 | 79.8 | 79.8 | 83.1 | 80.3 | 83.4 | 99.5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| KSU1 | 81.9 | 84.7 | 84.7 | 83.6 | 84.0 | 79.9 | 79.9 | 80.0 | 79.4 | 79.8 | 79.8 | 83.1 | 80.3 | 83.4 | 99.5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| NT009 | 81.9 | 84.7 | 84.7 | 83.6 | 84.0 | 79.9 | 79.9 | 80.0 | 79.4 | 79.8 | 79.8 | 83.1 | 80.3 | 83.4 | 99.5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| Peru | 81.9 | 84.7 | 84.7 | 83.6 | 84.0 | 79.9 | 79.9 | 80.0 | 79.4 | 79.8 | 79.8 | 83.1 | 80.3 | 83.4 | 99.5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| TAMU | 81.9 | 84.7 | 84.7 | 83.6 | 84.0 | 79.9 | 79.9 | 80.0 | 79.4 | 79.8 | 79.8 | 83.1 | 80.3 | 83.4 | 99.5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| UCP | 81.7 | 84.6 | 84.6 | 81.7 | 83.9 | 79.7 | 79.3 | 79.9 | 79.3 | 79.6 | 79.6 | 83.0 | 80.1 | 83.3 | 99.8 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 100 | 100 | |
| Zaire | 81.8 | 84.7 | 84.7 | 81.8 | 81.2 | 80.2 | 79.6 | 80.2 | 79.7 | 80.0 | 80.0 | 83.1 | 80.3 | 84.4 | 99.7 | 99.9 | 100 | 100 | 100 | 100 | 100 | 99.9 | 100 | |
Nucleotide (below diagonal) and amino acid (above diagonal). Intra-allelic sequence comparisons are in boldface.
A more detailed picture of the degree of sequence polymorphism and genetic diversity within and among gp15/45/60 alleles is provided by the ClustalW (101) alignment of the 15 different protein sequences shown in Fig. 8. The original NINC1 isolate gp15/45/60 sequence (Fig. 4) is the reference sequence to which all the others are compared, and sequences are grouped together by allele. The magnitude of sequence polymorphism observed at the gp15/45/60 locus is exceptional and is far greater than that detected at any other C. parvum locus examined to date, most of which manifest only a few SNPs and fewer or no SAAPs. For example, pairwise sequence comparisons of even the most highly conserved portions (3′ and C-terminal one-half) of the type I and type II gp15/45/60 allele sequences reveal 91 to 141 SNPs and 37 to 45 SAAPs, and similar levels of polymorphism are observed when the type I allele sequences are compared with one another.
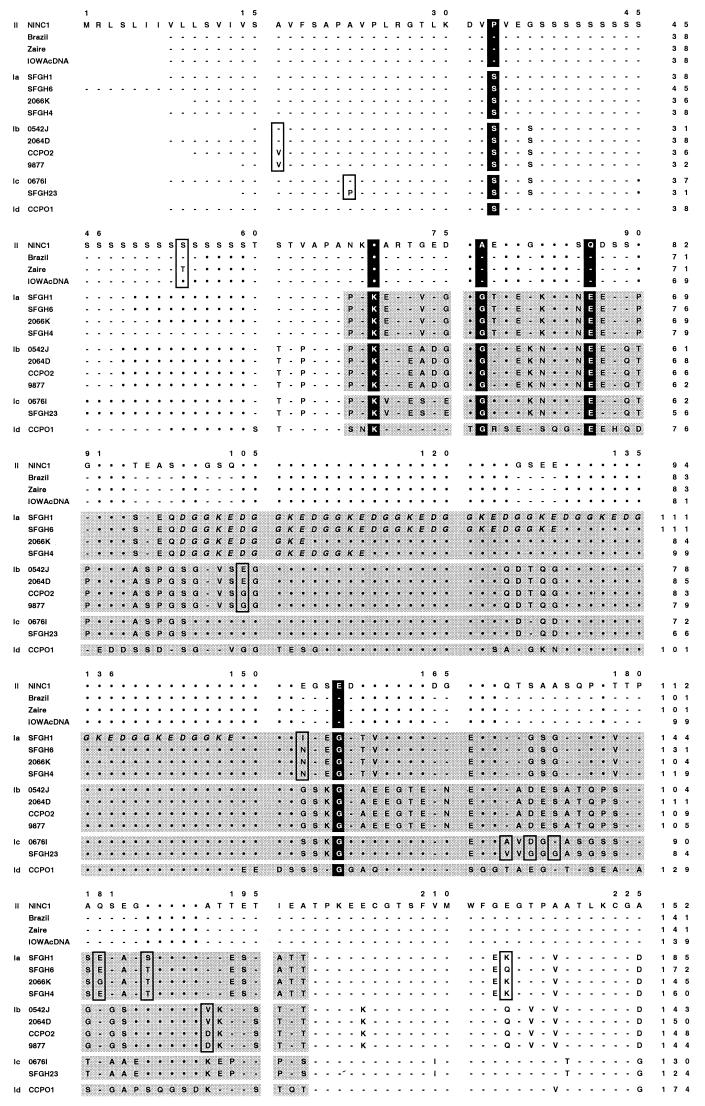
FIG. 8.
gp15/45/60 protein sequences are extremely polymorphic. The deduced gp15/45/60 amino acid sequences of 23 human and animal isolates were aligned using the ClustalW algorithm, and the sequence of the NINC1 animal isolate was used as a reference to which all others were aligned. The gp15/45/60 sequences from Iowa, NT009, KSU1, Peru, UCP, and TAMU were identical, and only the Iowa sequence is shown; likewise, only the first sequence from each of the identical pairs, 0542J and 0541L, SFGH4 and SFGH3, and 9877 and NEMC1, is presented. The five allelic groupings are explicitly indicated in the left margin by II, Ia, Ib, Ic, and Id. Hyphens in the alignment indicate identical residues, and filled circles indicate gaps; boxed residues depict SAAPs among sequences within a particular allelic grouping. The eight residues strictly conserved within either the allele I or allele II groupings are vertically highlighted throughout all sequences in black. The hypervariable region present within the gp45 sequence of human isolates (allelic groups Ia to Id) is indicated by shading. The tandemly repeated DGGKE sequence found within this region in all genotype Ia isolates is italicized. The two glutamic acid residues found in the putative protease cleavage site of the gp60 precursor are boxed throughout. The numbering above the sequences includes the gaps introduced for alignment purposes, while the numbers at the end of each line denote the amino acid position within that particular sequence.
Features of the gp15/45/60 sequences.
Further analysis of the deduced gp15/45/60 amino acid sequences revealed many conserved and polymorphic features of the protein that are presumably important to its structure and/or function. Eight amino acid residues were strictly conserved among the gp15/45/60 proteins of all human genotype I isolates, irrespective of allele, and distinguished these isolates from all animal genotype II isolates, which shared different conserved residues at each of these positions (Fig. 8). Second, the predicted N- and C-terminal peptides specifying the hypothetical signal sequence and GPI anchor attachment site were very highly conserved among all isolates, suggesting that both features are important. Third, the length of the putatively O-glycosylated polyserine region varied from 8 to 24 residues among isolates (Fig. 8 and Table 2), and the precise pattern of Ser codons encoding this repeat differed significantly among isolates (as an extreme example, the TCT Ser codon was observed only in the HGM10 isolate). Fourth, the sequence beginning just following the polyserine tract and ending at amino acid 125 (NINC1 numbering) was hypervariable among genotype I isolates but invariant among genotype II isolates; furthermore only proteins encoded by the Ia allelic group contained variable numbers of the peptide sequence DGGKE tandemly repeated within this hypervariable region (Fig. 8). Fifth, gp15/45/60 proteins encoded by alleles Ia and II contained the predicted N-linked glycosylation site ANSSS, while those encoded by allele Ic contained a different predicted N-linked glycosylation site, NNGST, eight residues further downstream, near the hypothetical C terminus of gp45; proteins encoded by alleles Ib and Id lacked potential N-glycosylation sites. Last and perhaps most significant, the putative proteolytic processing site apparently used to generate the mature gp15 and gp45 products from the gp15/45/60 precursor was conserved among all C. parvum isolates, irrespective of the specific gp15/45/60 allele they carried (Fig. 8).
TABLE 2.
Length polymorphisms in the gp15/45/60 polyserine tract
| Isolate | Origin | Genotype at gp15 locus | No. of serines in polyserine tract | Predicted N-glycosylation site |
|---|---|---|---|---|
| Ninc1 | Calf | Animal-II | 23 | ANSSS |
| Brazil | Humana | Animal-II | 19 | ANSSS |
| Zaire | Humana | Animal-II | 17 | ANSSS |
| Iowa gDNA | Calfa | Animal-II | 17 | ANSSS |
| Iowa cDNA | Calfa | Animal-II | 17 | ANSSS |
| Peru | Humana | Animal-II | 17 | ANSSS |
| UCP | Calfa | Animal-II | 17 | ANSSS |
| Tamu | Foala | Animal-II | 17 | ANSSS |
| KSU1 | Calfa | Animal-II | 17 | ANSSS |
| GCH1 | Humana | Animal-II | 19 | ANSSS |
| NT009 | Humana | Animal-II | 17 | ANSSS |
| HS1 | Calf | Animal-II | 19 | NDd |
| HS2 | Calf | Animal-II | 19 | ND |
| 6HMA1-10 | Human | Animal-II | 17 | ND |
| SFGH1 | Human | Human-Ia | 13 | ANSSS |
| SFGH6 | Human | Human-Ia | 13 | ANSSS |
| 2066K | Human | Human-Ia | 15 | ANSSS |
| SFGH4 | Human | Human-Ia | 23 | ANSSS |
| SFGH3 | Human | Human-Ia | 23 | ANSSS |
| 0542J | Human | Human-Ib | 12 | No site predicted |
| 0541L | Human | Human-Ib | 12 | No site predicted |
| 2064D | Human | Human-Ib | 12 | No site predicted |
| 9877 | Humanb | Human-Ib | 12 | No site predicted |
| NEMC1 | Humanb | Human-Ib | 12 | No site predicted |
| CCPO2 | Human | Human-Ib | 12 | No site predicted |
| 0676I | Human | Human-Ic | 8 | NNGST |
| SFGH23 | Human | Human-Ic | 8 | NNGST |
| CCPO1 | Human | Human-Id | 17 | No site predicted |
| HGM07 | Human | Human-Id | 24 | ND |
| HGM10c | Human | Human-Id | 17 | ND |
Oocysts have been passaged in calves.
Oocysts have been passaged in piglets.
Only sequence where TCT codon is found in polyserine tract; sequences from all other isolates use TCA and TCG.
ND, not determined.
SSU-rDNA genotyping confirms the species origin of type I gp15/45/60 alleles.
A recent small subunit ribosomal DNA (SSU-rDNA) genotyping investigation of 10 Cryptosporidium isolates obtained from HIV-infected persons identified two unusual alleles in addition to the usual dimorphic type I and II C. parvum SSU-rDNA alleles (75). Three isolates displayed an SSU-rDNA allele identical to that of the cat isolate, C. felis, and another had an rDNA allele identical to that of a canine Cryptosporidium sp. isolate. To determine if any of the diverse genotype I alleles observed at the gp15/45/60 locus might also be the result of human infections by Cryptosporidium species other than C. parvum, we determined the SSU-rDNA genotypes of one isolate from each of the genotype I allelic groups, viz., isolates SFGH3 (type Ia), 0542J (Ib), SFGH23 (Ic), and CCPO1 (Id). The SSU-rDNA variable region was amplified from isolated gDNA using primers CpBDIAGF and CpBDIAGR (41, 75), and the amplicons were sequenced; the SNP patterns of the SFGH3, 0542J, and CCPO1 isolates were identical to the canonical SSU-rDNA SNP pattern of human genotype I C. parvum isolates. The SFGH23 isolate (type Ic), although clearly a C. parvum isolate, displayed an unusual hybrid SNP pattern containing a type I SSU-rDNA SNP at one of the polymorphic positions and a type II SNP at the other (AF178700).
DISCUSSION
Proteins and glycoproteins expressed on the surface of the invasive C. parvum sporozoite and merozoite life cycle stages and shed in antigen trails by gliding zoites are thought to play essential roles in parasite motility and in parasite attachment to and invasion of host epithelial cells. Several such proteins have been identified as potential targets for active and/or passive immunoprophylaxsis and immunotherapy for cryptosporidiosis (5, 8, 10, 26, 39, 69, 84–86, 107). In particular, two antigen families, a 15-kDa group and a 27-kDa group, have consistently been identified by convalescent sera from infected humans and animals (48, 50, 54–56, 65, 67, 80, 82), and these specific humoral responses have been hypothesized to be associated with protection from subsequent infection and/or amelioration of disease (56). In this study, we have described the cloning, characterization, and sequence polymorphism analysis of a gp15/45/60 C. parvum gene family that encodes one of the 15-kDa group of antigens. This 15-kDa glycoprotein, the 11A5 antigen or gp15, is one of two presumptive proteolytic products of a 60-kDa precursor; the other glycoprotein product, gp45, also resides on the zoite surface and is a target recognized by neutralizing antibodies and lectins (Cevallos et al., submitted).
The 15-kDa antigen group has been shown to contain immunodominant, diagnostic, and perhaps protective antigen(s) and has been the subject of considerable study (38, 42, 54, 80, 102, 103, 105, 107). Several investigators have characterized C. parvum 15-kDa antigens, and the genes encoding two such proteins have been cloned and sequenced (37, 38, 42). Neither sequence is identical, nor similar, to the gp15/45/60 sequences determined in this study, suggesting, not surprisingly, that the parasite contains multiple 15-kDa antigens. On the other hand, in separate investigations Tilley et al. (102), Arrowood (4), and Zhou et al. (personal communication) produced MAbs that identified 15-kDa C. parvum sporozoite surface proteins; each of these MAbs recognizes the gp15 antigen encoded by the gp15/45/60 gene described herein (79; W. B. Strong et al., unpublished data), suggesting that it is, indeed, an immunodominant antigen (79). There have been only a few prior reports of C. parvum oocyst/sporozoite antigens in the size range of gp45 ((59, 74, 106); Cevallos et al., submitted), and the one gene sequenced to date (S2 antigen group (74); C. Petersen et al., unpublished data) has no similarity to the gp15/45/60 sequence. As described in Materials and Methods, the ∼40-kDa glycoprotein antigen (Cevallos et al., submitted) and corresponding gene that are the subjects of an accompanying paper (19) are essentially identical to gp45 and the gp15/45/60 gene described herein. Last, the “CP47” sporozoite antigen recently described by Nesterenko et al. (59) has surface labeling properties that suggest it may be related to or identical with gp45 (J. Gut et al., unpublished data).
The full-length protein encoded by the gp15/45/60 gene (NINC1 isolate) was predicted to contain many features characteristic of a membrane-bound surface glycoprotein including a N-terminal signal sequence, many O-glycosylated serine and threonine residues, one consensus N-glycosylation site, and a C-terminal GPI anchor attachment site. Cleavage of the gp15/45/60 N-terminal signal sequence was predicted to occur after Ser19, but the empirically determined N terminus of gp45 was found to be Asp31 instead. The N terminus of the second gp15/45/60 cleavage product, gp15, was determined to be Glu227. Assuming that the mature gp45 C terminus occurs at the preceding residue, Glu226, the predicted molecular masses of the gp45 and gp15 protein scaffolds are ∼20 and ∼11 kDa, respectively. Differences between the calculated and observed molecular masses are likely due to the presence of extensive mucin-like, O-linked glycosylation at multiple serine and threonine residues found throughout gp15/45/60 and presumptive N-glycosylation at the one asparagine residue present in the correct context near the C terminus of some of the predicted 45-kDa glycoproteins. Thirty-six of the 84 serine and threonine residues present in gp15/45/60 (NINC1 isolate) were predicted to be O-glycosylated (Fig. 4) by the NetOGlyc2.0 algorithm, but none of the predicted glycosylation sites were located in the gp15 portion of the molecule. However, as both gp15 and gp45 bound to the GalNAc-specific lectin HPA (and were also detected by HPA blotting), at least some of the O-glycosylation is likely to be the Tn antigen (GalNAcα1→Ser/Thr) and some of the serine and theorine residues present in gp15 must be O-glycosylated, contrary to the NetOGlyc2.0 prediction.
The Tn antigen is the precursor of almost all mammalian O-glycosylated glycoprotein core structures and is normally cryptic because it carries additional carbohydrate modification. However, when the Tn antigen is itself displayed on the surface of mammalian cells, e.g., on the aberrantly underglycosylated cell surface mucins synthesized by some tumors (44), it has been shown to play a role in malignant cell adhesion and metastasis (111). Thus, it is not unreasonable to hypothesize that the Tn-like antigenic determinants displayed by gp15 and gp45 on the zoite surface might also play an adhesive role in substrate-dependent zoite locomotion and in zoite adherence to, and invasion of, epithelial cells. Several other C. parvum sporozoite surface glycoproteins thought to play roles in zoite adhesion and host cell invasion also contain mucin-like domains that have been shown or predicted to carry Tn-like O-glycosylation (8, 92; Cevallos et al., submitted, and several lectins that recognize the Tn determinant have been shown to inhibit zoite attachment and invasion (32; Cevallos et al., submitted). Interestingly, a Gal/GalNAc-specific lectin has also been identified on the surface of sporozoites and implicated in adhesion to epithelial cells (40, 100). Thus, the zoite surface, like the host cell surface, contains both a Gal/GalNAc-specific lectin receptor as well as potential GalNAc-decorated glycoprotein ligands; whether the conceivable homotypic and heterotypic lectin-carbohydrate interactions function to organize the zoite cell surface, mediate cellular or cell-substrate adhesion, or both remains a subject for future investigation.
Our data indicate that the gp15/45/60 mRNA is transcribed and translated, and the resulting protein product is glycosylated, to produce an ∼60-kDa glycoprotein precursor during the intracellular stages of the parasite life cycle. This precursor is apparently proteolytically processed shortly after synthesis to generate the mature 15- and 45-kDa glycoproteins that associate with the cell surfaces of developing merozoites and sporozoites during late stages of merogony and, presumably, sporogony, respectively. Furthermore, since only the deduced gp15 protein sequence contained features predicting membrane attachment, i.e., a GPI anchor, gp45 would appear to be a peripheral membrane protein that must associate with the merozoite and sporozoite surfaces through interaction(s) with other surface molecules, perhaps gp15 or the previously identified Gal/GalNAc sporozoite-surface lectin (40, 100). Since gp15 and gp45 are clearly the predominant forms of gp15/45/60 present in intracellular meronts and since none of the gp15/45/60 precursor was found in either oocysts or sporozoites, it appears likely that gp15/45/60 processing is completed considerably prior to zoite attachment to and invasion of host cells, probably even before the zoites are released from schizonts or oocysts. This contrasts with the processing that some malaria and Toxoplasma zoite surface proteins undergo just before and during attachment and invasion. For example, the 200-kDa Plasmodium falciparum merozoite surface protein 1 undergoes two processing events; the first generates four fragments ranging from 42 to 83 kDa, and the second cleaves the GPI-anchored 42-kDa protein into a 33-kDa peptide and a 19-kDa GPI-anchored domain. The second proteolytic cleavage occurs immediately prior to host cell invasion and is obligatory for successful merozoite invasion of erythrocytes (11, 36). Similarly, following its secretion to the tachyzoite surface, the T. gondii microneme protein MIC2 undergoes N-terminal proteolytic processing that exposes an integrin-like adhesive domain and C-terminal processing that releases the transmembrane protein from the parasite surface during host cell penetration (17).
No information is available about which specific proteinase, or which generic class of proteinase, is responsible for processing the gp15/45/60 precursor. Interestingly, however, hydropathy plots of the diverse gp15/45/60 protein sequences deduced in this study shared similar profiles, viz., hydrophobic N and C termini and three central hydrophilic regions each separated by a hydrophobic stretch of ∼20 amino acid residues. In each case, the predicted proteolytic cleavage site containing adjacent glutamic acid residues occurred at the end of the second hydrophilic region, suggesting that despite significant primary sequence variability, overall protein structure and cleavage site accessibility are probably similar and important for gp15/45/60 processing. To date, a cysteine proteinase and a serine protease have been identified on the surface of C. parvum sporozoites (28, 58), but in neither case have the endogenous substrates been determined.
In this paper we have reported the nucleic acid and deduced protein sequences of the single-copy gp15/45/60 genes of 29 temporally and geographically diverse C. parvum isolates. The gp15/45/60 locus is highly polymorphic, far more so than any other C. parvum protein-coding locus examined to date. The gp15/45/60 sequence comparisons and alignments revealed at least five allelic groups which we have termed Ia, Ib, Ic, Id, and II. Although the deduced protein sequences in any one group were not all identical, they were considerably more similar to one another (92 to 100% amino acid sequence identity) than they were to the sequences in other gp15/45/60 allelic groups (67 to 80% identity). All of the animal C. parvum isolates carried the gp15/45/60 type II allele which was also found in a subset of human isolates; the remainder of the human isolates displayed one of four type I alleles. Although the type I alleles had no more (and in some cases had less) overall sequence similarity to each other than they had to the type II allele, the encoded proteins shared invariant amino acids at eight positions within the sequence that were distinct from the invariant residues present at these (and many other) positions in proteins encoded by members of the type II allelic class. Thus, the dimorphic SAAP patterns allowed assignment of the corresponding gp15/45/60 alleles to one of the two prototypical genotypes found to characterize all other C. parvum genetic loci (7, 13, 15, 51, 52, 63, 66, 68, 75, 94, 98, 99, 112, 113). Namely, alleles Ia, Ib, Ic, and Id shared one eight-residue SAAP pattern, were found only among human C. parvum isolates, and therefore, by convention, were grouped together as gp15/45/60 genotype I alleles. Conversely, the allele II sequences were considerably more similar (99 to 100% identity), shared the second eight-residue SAAP pattern, were found among both human and animal C. parvum isolates, and therefore, again by convention, were assigned to genotype II.
The high degree of nucleic acid sequence polymorphism displayed by gp15/45/60 alleles is surprising and unprecedented; not only are the various genotype I allele nucleotide sequences very different from the genotype II sequences (78 to 85% sequence identity), they are also very different from one another (77 to 88% identity). This observation contrasts with previous DNA sequencing-based genotyping studies at many other C. parvum loci that have most often identified a dimorphic pair of alleles differing by only a few (15, 63, 98, 99) or, in one case, up to 38 SNPs (112). At some loci, however, an additional variant allele(s) whose sequence differed only slightly (e.g., by one SNP) from that of the prototypical type I or type II allele was detected (1, 68, 99, 113; D. Champliaud, W. B. Strong, J. Lopez, P. Gobet, F. Dalle, G. Dautin, M. Naciri, M. L. Dardé, L. Favennec, A. Datry, G. Harly, R. G. Nelson, and A. Bonnin, submitted for publication; W. B. Strong, J. Gut, M. Gemeniano, A. Bonnin, and R. G. Nelson, submitted for publication); in one study of the β-tubulin intron locus (114), but not in others (15, 98), considerably more sequence polymorphism, and two additional alleles were observed, albeit only in one isolate each. The investigators hypothesized that one of these atypical β-tubulin intron alleles (observed in C. parvum isolate 0676I) was due to genetic recombination between genotype I and II parasites because the allele sequence displayed a hybrid pattern of both type I- and type II-specific SNPs (114).
Many of the gp15/45/60 genotype I isolates described in this paper have also been genotyped at other loci, either in this study or in the above-referenced investigations. As described in Results, isolates SFGH3, 0542J, SFGH23, and CCPO1, representing the gp15/45/60 Ia, Ib, Ic, and Id allelic groups, respectively, were genotyped by DNA sequencing at the SSU-rDNA locus. The SFGH3, 0542J, and CCPO1 isolates displayed identical C. parvum SSU-rDNA genotype I SNP patterns (41, 75), but the SFGH23 isolate displayed a hybrid SNP pattern containing both SSU-rDNA genotype I and genotype II-specific SNPs. We also genotyped isolates SFGH3; 2064D, CCPO2, and 0542J; and CCPO1, representing the gp15/45/60 Ia, Ib, and Id allelic groups respectively, by DNA sequencing at the polymorphic DHFR-TS locus (Strong et al., submitted) and found that all carried the previously described (112) genotype I allele. Sulaiman et al. genotyped isolates HGM07 and HGM10, both of which carry the gp15/45/60 Id allele, by DNA sequencing at the TRAP-C2 (99) and the β-tubulin intron (98) loci. The two isolates displayed identical genotype I alleles at both loci, although the TRAP-C2 allele was the less common type I variant carrying the T→C transition (68, 99) at one of the five polymorphic positions. Last, Widmer et al. (114, 115) genotyped isolate 2066K and isolates 0541L, 0542J and 2064D, representing gp15/45/60 allelic groups Ia and Ib, respectively, by PCR-restriction fragment length polymorphism (RFLP) and/or DNA sequencing at one or more genetic loci including the polythreonine locus (16), the COWP locus (96), the RNR locus (115), and the β-tubulin intron locus (114). Each isolate displayed the genotype I allele at each locus examined (three, four, three, and one locus, respectively). These and similar results observed in recent bi- and multilocus C. parvum genotyping investigations (15, 49, 52, 63, 94, 95; Champliaud et al., submitted) have demonstrated extensive linkage disequilibrium between unlinked genetic loci. In the vast majority of cases examined, genotype I alleles exclusively cosegregated with other genotype I alleles and genotype II alleles exclusively cosegregated with other genotype II alleles. These observations strongly suggest that the C. parvum genetic population structure is clonal and made up of two prominent lineages that manifest either genotype I or genotype II alleles at all genomic loci, i.e., C. parvum haplotypes I and II. It is also clear, however, that both parasite haplotypes undergo intraallelic variation as both minor (1, 68, 99, 113; Champliaud et al., submitted; Strong et al., submitted) and major (e.g., at the gp15/45/60 locus) alterations have been observed in the prototypical SNP patterns at several loci.
Due to a lack of compelling evidence for sexual recombination between the clonal lineages, some investigators have questioned whether the two C. parvum clones are, in fact, distinct Cryptosporidium species (53). We believe that some of the gp15/45/60 genotyping data obtained in this study, together with some of the previous results reported by Widmer and colleagues (114, 115), present a reasonable case for the existence of rare recombinant C. parvum haplotypes and therefore for the occurrence of occasional biparental mating and sexual recombination both within and between the prototypal clonal lineages. These data, described in more detail in what follows, support the current taxonomic classification that assigns both lineages to the same Cryptosporidium species, C. parvum.
As described in the results, the two sequences (SFGH23 and 0676I) making up gp15/45/60 allelic class Ic were considerably less similar to one another than were the gp15/45/60 sequences that defined all other allelic classes. Both a pairwise DNA sequence alignment (not shown) and the protein sequence alignment shown in Fig. 8 reveal that the vast majority of differences between the SFGH23 and 0676I sequences occur in the 3′ one-third and C-terminal region of the sequences (Fig. 8). Specifically, this region contains 36 of 40 SNPs and 19 of 23 SAAPs that distinguish the SFGH23 and 0676I DNA and protein sequences, respectively. Thus, although the two allele Ic sequences are nearly identical over their 5′ two-thirds, they differ considerably at their 3′ ends, and over this region they are, in fact, much more similar to other gp15/45/60 alleles than they are to each other. For example, this region of the SFGH23 sequence is nearly identical to the same portion of the 9877 allele Ib sequence (the sequences share 29 of the 36 3′ SNPs and 16 of the 19 C-terminal SAAPs mentioned above), while the same region of the 0676I sequence is very similar to the NINC1 and other allele II sequences (they share 27 of 36 3′ SNPs and 15 of 19 C-terminal SAAPs). These observations strongly suggest that progenitors of the SFGH23 and 0676I gp15/45/60 alleles were derived by intragenic recombination between parasites carrying a prototypal Ic allele, which donated the 5′ two-thirds of both sequences, and isolates carrying either allele Ib or allele II, which provided the respective 3′ ends of the two sequences. Some additional evidence supporting this hypothesis comes from other genotyping investigations of isolate 0676I (114, 115). These studies showed that (i) the β-tubulin intron allele also contained a recombinant SNP pattern intermediate between those manifested by prototypical genotype I and II alleles, (ii) the RNR, COWP, and polythreonine alleles each displayed RFLPs characteristic of genotype II isolates, and (iii) isolate 0676I was not infective for neonatal ICR mice, a phenotype characteristic of genotype I, but not genotype II parasites. Thus, the 0676I parasite isolate has genetic features of both clonal lineages and a recombinant haplotype presumably derived by sexual recombination between them, while the SFGH23 parasite isolate, which also carries a recombinant gp15/45/60 allele, appears to have been derived by recombination between distinct genetic subpopulations within the genotype I lineage.
Finally, gp45 (19) and the possibly related or identical protein, CP47 (59), have been shown to bind epithelial cell lines and enterocytes and have been suggested to function as adhesion ligands mediating zoite attachment to host epithelium. This is somewhat curious as gp45, lacking both a transmembrane domain and a GPI anchor, is presumably a peripheral membrane protein associated with the zoite plasmalemma only through its interaction with an as yet unidentified molecule. Furthermore, at least within parasites of the genotype I lineage, gp45 contains the most hypervariable region of the entire gp15/45/60 protein sequence. Thus, it is unclear whether the diverse gp45 molecules expressed by various parasite isolates serve as adhesins recognized by the same epithelial cell receptor or whether they mediate adhesion to distinct receptors. An argument could be made for either possibility based on (i) the predicted conservation of multiple Tn glycotopes within the conserved gp45 polyserine tract of all isolates or (ii) the extreme variation of the gp45 amino acid sequence among parasite isolates. It is also quite possible that these two features function together in more novel ways. For example, multiple, adjacent Tn glycotopes in different primary, secondary, and tertiary structural environments might either produce a multiplicity of adhesive phenotypes or, instead, camouflage a conserved adhesive glycotope within an antigenically diverse gp45 protein superstructure. The possibilities are not mutually exclusive and both would be advantageous to individual parasites and the parasite genetic population. The former would permit parasites to specifically adhere to diverse adhesion receptors (e.g., present in different host species, different host genotypes, or different host tissues), thereby providing redundancy for the biologically crucial process of zoite-host cell attachment and invasion. The latter would allow genetically distinct parasites to utilize a common, albeit disguised, ligand for host cell attachment and thereby evade the host's immune response to previously encountered gp45 antigenic types. A more complete and less speculative understanding of gp15/45/60 allelic diversity and the role it plays in the antigenic and functional diversity of the gp15 and gp45 glycoproteins clearly requires further investigation.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI40319 and AI42565 to R.G.N.
We thankfully acknowledge Marian R. Neutra (Children's Hospital, Harvard Medical School, Boston, Mass.) for her kind gift of the CrA1 and CrA2 MAbs and Saul Tzipori (Department of Infectious Diseases, Tufts University School of Veterinary Medicine, North Grafton, Mass.) for his agreement and help in obtaining these antibodies. We especially acknowledge Honorine D. Ward and Ana Maria Cevallos for their extensive patience and for unselfishly sharing a large amount of unpublished data. We also gratefully acknowledge many of our colleagues for supplying other MAbs, sharing unpublished data, and/or providing isolates of C. parvum and other Cryptosporidium spp., including Alain Bonnin (Université de Bourgogne, Faculté de Médecine de Dijon, Dijon, France), Michael J. Arrowood, Norman J. Pieniazek, Jeffrey W. Priest, and Altaf A. Lal (Division of Parasitic Diseases, CDC, Atlanta, Ga.), Saul Tzipori, Giovanni Widmer, and Donna Akiyoshi (Department of Infectious Diseases, Tufts University School of Veterinary Medicine), Joe H. Crabb (ImmuCell Corporation, Portland, Maine), Jeffrey K. Griffiths (Tufts University School of Medicine, Boston, Mass.), Carl J. Fichtenbaum (University of Cincinnati School of Medicine, Cincinnati, Ohio), Cynthia L. Chappell and Pablo C. Okhuysen (Department of Medicine and School of Public Health, University of Texas Health Science Center, Houston, Tex.), Carolyn Petersen (Agouron Pharmaceuticals, Inc., San Diego, Calif.), Steve J. Upton (Division of Biology, Kansas State University, Manhattan, Kans.), Heidi Schraft (Department of Food Science, University of Guelph, Guelph, Ontario, Canada), and Byron Blagburn (Department of Pathobiology, College of Veterinary Medicine, Auburn University, Auburn, Ala.). Last, we thank C. J. Hackbarth (Versicor Inc., Fremont, Calif.) for help with editing the manuscript.
REFERENCES
- 1.Aiello A E, Xiao L, Limor J R, Liu C, Abrahamsen M S, Lal A A. Microsatellite analysis of the human and bovine genotypes of Cryptosporidium parvum. J Eukaryot Microbiol. 1999;46:46S–47S. [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrowood M. Ph.D. thesis. Tuscon: University of Arizona; 1988. [Google Scholar]
- 5.Arrowood M J, Mead J R, Mahrt J L, Sterling C R. Effects of immune colostrum and orally administered antisporozoite monoclonal antibodies on the outcome of Cryptosporidium parvum infections in neonatal mice. Infect Immun. 1989;57:2283–2288. doi: 10.1128/iai.57.8.2283-2288.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrowood M J, Sterling C R, Healey M C. Immunofluorescent microscopical visualization of trails left by gliding Cryptosporidium parvum sporozoites. J Parasitol. 1991;77:315–317. [PubMed] [Google Scholar]
- 7.Awad-el-Kariem F M, Robinson H A, Dyson D A, Evans D, Wright S, Fox M T, McDonald V. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology. 1995;110:129–132. doi: 10.1017/s0031182000063885. [DOI] [PubMed] [Google Scholar]
- 8.Barnes D A, Bonnin A, Huang J X, Gousset L, Wu J, Gut J, Doyle P, Dubremetz J F, Ward H, Petersen C. A novel multi-domain mucin-like glycoprotein of Cryptosporidium parvum mediates invasion. Mol Biochem Parasitol. 1998;96:93–110. doi: 10.1016/s0166-6851(98)00119-4. [DOI] [PubMed] [Google Scholar]
- 9.Bjorneby J M, Hunsaker B D, Riggs M W, Perryman L E. Monoclonal antibody immunotherapy in nude mice persistently infected with Cryptosporidium parvum. Infect Immun. 1991;59:1172–1176. doi: 10.1128/iai.59.3.1172-1176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjorneby J M, Riggs M W, Perryman L E. Cryptosporidium parvum merozoites share neutralization-sensitive epitopes with sporozoites. J Immunol. 1990;145:298–304. [PubMed] [Google Scholar]
- 11.Blackman M J, Holder A A. Secondary processing of the Plasmodium falciparum merozoite surface protein-1 (MSP1) by a calcium-dependent membrane-bound serine protease: shedding of MSP133 as a noncovalently associated complex with other fragments of the MSP1. Mol Biochem Parasitol. 1992;50:307–315. doi: 10.1016/0166-6851(92)90228-c. [DOI] [PubMed] [Google Scholar]
- 12.Bonafonte M T, Priest J W, Garmon D, Arrowood M J, Mead J R. Isolation of the gene coding for elongation factor-1alpha in Cryptosporidium parvum. Biochim Biophys Acta. 1997;1351:256–260. doi: 10.1016/s0167-4781(97)00013-4. [DOI] [PubMed] [Google Scholar]
- 13.Bonnin A, Fourmaux M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygauthier-Toubas D, Gabriel-Pospisil G, Naciri M, Camerlynck P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 14.Bonnin A, Gut J, Dubremetz J F, Nelson R G, Camerlynck P. Monoclonal antibodies identify a subset of dense granules in Cryptosporidium parvum zoites and gamonts. J Eukaryot Microbiol. 1995;42:395–401. doi: 10.1111/j.1550-7408.1995.tb01601.x. [DOI] [PubMed] [Google Scholar]
- 15.Caccio S, Homan W, van Dijk K, Pozio E. Genetic polymorphism at the beta-tubulin locus among human and animal isolates of Cryptosporidium parvum. FEMS Microbiol Lett. 1999;170:173–179. doi: 10.1111/j.1574-6968.1999.tb13371.x. [DOI] [PubMed] [Google Scholar]
- 16.Carraway M, Tzipori S, Widmer G. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carruthers V B, Sherman G D, Sibley L D. The Toxoplasma adhesive protein MIC2 is proteolytically processed at multiple sites by two parasite-derived proteases. J Biol Chem. 2000;275:14346–14353. doi: 10.1074/jbc.275.19.14346. [DOI] [PubMed] [Google Scholar]
- 18.Cegielski J P, Ortega Y R, McKee S, Madden J F, Gaido L, Schwartz D A, Manji K, Jorgensen A F, Miller S E, Pulipaka U P, Msengi A E, Mwakyusa D H, Sterling C R, Reller L B. Cryptosporidium, enterocytozoon, and cyclospora infections in pediatric and adult patients with diarrhea in Tanzania. Clin Infect Dis. 1999;28:314–321. doi: 10.1086/515131. [DOI] [PubMed] [Google Scholar]
- 19.Cevallos A M, Zhang X, Waldor M K, Jaison S, Zhou X, Tzipori S, Neutra M R, Ward H D. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect Immun. 2000;68:4108–4116. doi: 10.1128/iai.68.7.4108-4116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colford J M, Jr, Tager I B, Hirozawa A M, Lemp G F, Aragon T, Petersen C. Cryptosporidiosis among patients infected with human immunodeficiency virus. Factors related to symptomatic infection and survival. Am J Epidemiol. 1996;144:807–816. doi: 10.1093/oxfordjournals.aje.a009015. [DOI] [PubMed] [Google Scholar]
- 21.Coyne K E, Crisci A, Lublin D M. Construction of synthetic signals for glycosyl-phosphatidylinositol anchor attachment. Analysis of amino acid sequence requirements for anchoring. J Biol Chem. 1993;268:6689–6693. [PubMed] [Google Scholar]
- 22.Current W L, Reese N C, Ernst J V, Bailey W S, Heyman M B, Weinstein W M. Human cryptosporidiosis in immunocompetent and immunodeficient persons. Studies of an outbreak and experimental transmission. N Engl J Med. 1983;308:1252–1257. doi: 10.1056/NEJM198305263082102. [DOI] [PubMed] [Google Scholar]
- 23.Doyle P S, Crabb J, Petersen C. Anti-Cryptosporidium parvum antibodies inhibit infectivity in vitro and in vivo. Infect Immun. 1993;61:4079–4084. doi: 10.1128/iai.61.10.4079-4084.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duong T H, Dufillot D, Koko J, Nze-Eyo'o R, Thuilliez V, Richard-Lenoble D, Kombila M. Digestive cryptosporidiosis in young children in an urban area in Gabon. Sante. 1995;5:185–188. [PubMed] [Google Scholar]
- 25.DuPont H L, Chappell C L, Sterling C R, Okhuysen P C, Rose J B, Jakubowski W. The infectivity of Cryptosporidium parvum in healthy volunteers. N Engl J Med. 1995;332:855–859. doi: 10.1056/NEJM199503303321304. [DOI] [PubMed] [Google Scholar]
- 26.Enriquez F J, Riggs M W. Role of immunoglobulin A monoclonal antibodies against P23 in controlling murine Cryptosporidium parvum infection. Infect Immun. 1998;66:4469–4473. doi: 10.1128/iai.66.9.4469-4473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enriquez J F, Avila C R, Ignacio Santos J, Tanaka-Kido J, Vallejo O, Sterling C R. Cryptosporidium infections in Mexican children: clinical, nutritional, enteropathogenic, and diagnostic evaluations. Am J Trop Med Hyg. 1997;56:254–257. doi: 10.4269/ajtmh.1997.56.254. [DOI] [PubMed] [Google Scholar]
- 28.Forney J R, Yang S, Healey M C. Interaction of the human serine protease inhibitor alpha-1-antitrypsin with Cryptosporidium parvum. J Parasitol. 1996;82:496–502. [PubMed] [Google Scholar]
- 29.Greenberg P D, Koch J, Cello J P. Diagnosis of Cryptosporidium parvum in patients with severe diarrhea and AIDS. Dig Dis Sci. 1996;41:2286–2290. doi: 10.1007/BF02071413. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths J K. Human cryptosporidiosis: epidemiology, transmission, clinical diagnosis, treatment, and diagnosis. Adv Parasitol. 1998;40:37–85. doi: 10.1016/s0065-308x(08)60117-7. [DOI] [PubMed] [Google Scholar]
- 31.Gut J, Nelson R G. Cryptosporidium parvum sporozoites deposit trails of 11A5 antigen during gliding locomotion and shed 11A5 antigen during invasion of MDCK cells in vitro. J Eukaryot Microbiol. 1994;41:42S–43S. [PubMed] [Google Scholar]
- 32.Gut J, Nelson R G. Cryptosporidium parvum: lectins mediate irreversible inhibition of sporozoite infectivity in vitro. J Eukaryot Microbiol. 1999;46:48S–49S. [PubMed] [Google Scholar]
- 33.Gut J, Nelson R G. Cryptosporidium parvum: synchronized excystation in vitro and evaluation of sporozoite infectivity with a new lectin-based assay. J Eukaryot Microbiol. 1999;46:56S–57S. [PubMed] [Google Scholar]
- 34.Gut J, Petersen C, Nelson R, Leech J. Cryptosporidium parvum: in vitro cultivation in Madin-Darby canine kidney cells. J Protozool. 1991;38:72S–73S. [PubMed] [Google Scholar]
- 35.Hansen J E, Lund O, Tolstrup N, Gooley A A, Williams K L, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 36.Holder A A. Proteins on the surface of the malaria parasite and cell invasion. Parasitology. 1994;108(Suppl.):S5–S18. doi: 10.1017/s0031182000075673. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins M C, Fayer R. Cloning and expression of cDNA encoding an antigenic Cryptosporidium parvum protein. Mol Biochem Parasitol. 1995;71:149–152. doi: 10.1016/0166-6851(95)00050-b. [DOI] [PubMed] [Google Scholar]
- 38.Jenkins M C, Fayer R, Tilley M, Upton S J. Cloning and expression of a cDNA encoding epitopes shared by 15- and 60-kilodalton proteins of Cryptosporidium parvum sporozoites. Infect Immun. 1993;61:2377–2382. doi: 10.1128/iai.61.6.2377-2382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenkins M C, O'Brien C, Trout J, Guidry A, Fayer R. Hyperimmune bovine colostrum specific for recombinant Cryptosporidium parvum antigen confers partial protection against cryptosporidiosis in immunosuppressed adult mice. Vaccine. 1999;17:2453–2460. doi: 10.1016/s0264-410x(98)00369-7. [DOI] [PubMed] [Google Scholar]
- 40.Joe A, Hamer D H, Kelley M A, Pereira M E, Keusch G T, Tzipori S, Ward H D. Role of a Gal/GalNAc-specific sporozoite surface lectin in Cryptosporidium parvum-host cell interaction. J Eukaryot Microbiol. 1994;41:44S. [PubMed] [Google Scholar]
- 41.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khramtsov N V, Oppert B, Montelone B A, Upton S J. Sequencing, analysis and expression in Escherichia coli of a gene encoding a 15 kDa Cryptosporidium parvum protein. Biochem Biophys Res Commun. 1997;230:164–166. doi: 10.1006/bbrc.1996.5877. [DOI] [PubMed] [Google Scholar]
- 43.Kim K, Gooze L, Petersen C, Gut J, Nelson R G. Isolation, sequence and molecular karyotype analysis of the actin gene of Cryptosporidium parvum. Mol Biochem Parasitol. 1992;50:105–113. doi: 10.1016/0166-6851(92)90248-i. [DOI] [PubMed] [Google Scholar]
- 44.Kim Y S, Gum J, Jr, Brockhausen I. Mucin glycoproteins in neoplasia. Glycoconj J. 1996;13:693–707. doi: 10.1007/BF00702333. [DOI] [PubMed] [Google Scholar]
- 45.Luft B J, Payne D, Woodmansee D, Kim C W. Characterization of the Cryptosporidium antigens from sporulated oocysts of Cryptosporidium parvum. Infect Immun. 1987;55:2436–2441. doi: 10.1128/iai.55.10.2436-2441.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macfarlane D E, Horner-Bryce J. Cryptosporidiosis in well-nourished and malnourished children. Acta Paediatr Scand. 1987;76:474–477. doi: 10.1111/j.1651-2227.1987.tb10502.x. [DOI] [PubMed] [Google Scholar]
- 47.Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple MacIntosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLauchlin J, Casemore D P, Moran S, Patel S. The epidemiology of cryptosporidiosis: application of experimental sub-typing and antibody detection systems to the investigation of water-borne outbreaks. Folia Parasitol (Praha) 1998;45:83–92. [PubMed] [Google Scholar]
- 49.McLauchlin J, Pedraza-Diaz S, Amar-Hoetzeneder C, Nichols G L. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J Clin Microbiol. 1999;37:3153–3158. doi: 10.1128/jcm.37.10.3153-3158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mead J R, Arrowood M J, Sterling C R. Antigens of Cryptosporidium sporozoites recognized by immune sera of infected animals and humans. J Parasitol. 1988;74:135–143. [PubMed] [Google Scholar]
- 51.Morgan U M, Constantine C C, O'Donoghue P, Meloni B P, O'Brien P A, Thompson R C. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 52.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertzberg H, Elliot A, Thompson R C. Molecular characterization of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- 53.Morgan U M, Xiao L, Fayer R, Lal A A, Thompson R C. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol. 1999;29:1733–1751. doi: 10.1016/s0020-7519(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 54.Moss D M, Bennett S N, Arrowood M J, Hurd M R, Lammie P J, Wahlquist S P, Addiss D G. Kinetic and isotypic analysis of specific immunoglobulins from crew members with cryptosporidiosis on a U.S. Coast Guard cutter. J Eukaryot Microbiol. 1994;41:52S–55S. [PubMed] [Google Scholar]
- 55.Moss D M, Bennett S N, Arrowood M J, Wahlquist S P, Lammie P J. Enzyme-linked immunoelectrotransfer blot analysis of a cryptosporidiosis outbreak on a United States Coast Guard cutter. Am J Trop Med Hyg. 1998;58:110–118. doi: 10.4269/ajtmh.1998.58.110. [DOI] [PubMed] [Google Scholar]
- 56.Moss D M, Chappell C L, Okhuysen P C, DuPont H L, Arrowood M J, Hightower A W, Lammie P J. The antibody response to 27-, 17-, and 15-kDa Cryptosporidium antigens following experimental infection in humans. J Infect Dis. 1998;178:827–833. doi: 10.1086/515377. [DOI] [PubMed] [Google Scholar]
- 57.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 58.Nesterenko M V, Tilley M, Upton S J. A metallo-dependent cysteine proteinase of Cryptosporidium parvum associated with the surface of sporozoites. Microbios. 1995;83:77–88. [PubMed] [Google Scholar]
- 59.Nesterenko M V, Woods K, Upton S J. Receptor/ligand interactions between Cryptosporidium parvum and the surface of the host cell. Biochim Biophys Acta. 1999;1454:165–173. doi: 10.1016/s0925-4439(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 60.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 61.O'Donoghue P J. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. [DOI] [PubMed] [Google Scholar]
- 62.Okhuysen P C, Chappell C L, Crabb J, Valdez L M, Douglass E T, DuPont H L. Prophylactic effect of bovine anti-Cryptosporidium hyperimmune colostrum immunoglobulin in healthy volunteers challenged with Cryptosporidium parvum. Clin Infect Dis. 1998;26:1324–1329. doi: 10.1086/516374. [DOI] [PubMed] [Google Scholar]
- 63.Ong C S, Eisler D L, Goh S H, Tomblin J, Awad-El-Kariem F M, Beard C B, Xiao L, Sulaiman I, Lal A, Fyfe M, King A, Bowie W R, Isaac-Renton J L. Molecular epidemiology of cryptosporidiosis outbreaks and transmission in British Columbia, Canada. Am J Trop Med Hyg. 1999;61:63–69. doi: 10.4269/ajtmh.1999.61.63. [DOI] [PubMed] [Google Scholar]
- 64.Ortega Y R, Sheehy R R, Cama V A, Oishi K K, Sterling C R. Restriction fragment length polymorphism analysis of Cryptosporidium parvum isolates of bovine and human origin. J Protozool. 1991;38:40S–41S. [PubMed] [Google Scholar]
- 65.Ortega-Mora L M, Troncoso J M, Rojo-Vazquez F A, Gomez-Bautista M. Identification of Cryptosporidium parvum oocyst/sporozoite antigens recognized by infected and hyperimmune lambs. Vet Parasitol. 1994;53:159–166. doi: 10.1016/0304-4017(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 66.Patel S, Pedraza-Diaz S, McLauchlin J, Casemore D P. Molecular characterisation of Cryptosporidium parvum from two large suspected waterborne outbreaks. Outbreak Control Team South and West Devon 1995, Incident Management Team and Further Epidemiological and Microbiological Studies Subgroup North Thames 1997. Commun Dis Public Health. 1998;1:231–233. [PubMed] [Google Scholar]
- 67.Peeters J E, Villacorta I, Vanopdenbosch E, Vandergheynst D, Naciri M, Ares-Mazas E, Yvore P. Cryptosporidium parvum in calves: kinetics and immunoblot analysis of specific serum and local antibody responses (IgA, IgG, and IgM) after natural and experimental infections. Infect Immun. 1992;60:2309–2316. doi: 10.1128/iai.60.6.2309-2316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perryman L E, Jasmer D P, Riggs M W, Bohnet S G, McGuire T C, Arrowood M J. A cloned gene of Cryptosporidium parvum encodes neutralization-sensitive epitopes. Mol Biochem Parasitol. 1996;80:137–147. doi: 10.1016/0166-6851(96)02681-3. [DOI] [PubMed] [Google Scholar]
- 70.Perryman L E, Kegerris K A, Mason P H. Effect of orally administered monoclonal antibody on persistent Cryptosporidium parvum infection in scid mice. Infect Immun. 1993;61:4906–4908. doi: 10.1128/iai.61.11.4906-4908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perryman L E, Riggs M W, Mason P H, Fayer R. Kinetics of Cryptosporidium parvum sporozoite neutralization by monoclonal antibodies, immune bovine serum, and immune bovine colostrum. Infect Immun. 1990;58(1):257–259. doi: 10.1128/iai.58.1.257-259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petersen C. Cryptosporidiosis in patients infected with the human immunodeficiency virus. Clin Infect Dis. 1992;15:903–909. doi: 10.1093/clind/15.6.903. [DOI] [PubMed] [Google Scholar]
- 73.Petersen C, Gut J, Doyle P S, Crabb J H, Nelson R G, Leech J H. Characterization of a >900,000-MrCryptosporidium parvum sporozoite glycoprotein recognized by protective hyperimmune bovine colostral immunoglobulin. Infect Immun. 1992;60:5132–5138. doi: 10.1128/iai.60.12.5132-5138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petersen C, Gut J, Leech J H, Nelson R G. Identification and initial characterization of five Cryptosporidium parvum sporozoite antigen genes. Infect Immun. 1992;60:2343–2348. doi: 10.1128/iai.60.6.2343-2348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pieniazek N J, Bornay-Llinares F J, Slemenda S B, da Silva A J, Moura I N, Arrowood M J, Ditrich O, Addiss D G. New Cryptosporidium genotypes in HIV-infected persons. Emerg Infect Dis. 1999;5:444–449. doi: 10.3201/eid0503.990318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pitlik S D, Fainstein V, Garza D, Guarda L, Bolivar R, Rios A, Hopfer R L, Mansell P A. Human cryptosporidiosis: spectrum of disease. Report of six cases and review of the literature. Arch Intern Med. 1983;143:2269–2275. doi: 10.1001/archinte.1983.00350120059015. [DOI] [PubMed] [Google Scholar]
- 77.Plettenberg A, Stoehr A, Stellbrink H J, Albrecht H, Meigel W. A preparation from bovine colostrum in the treatment of HIV-positive patients with chronic diarrhea. Clin Investig. 1993;71:42–45. doi: 10.1007/BF00210962. [DOI] [PubMed] [Google Scholar]
- 78.Pozio E, Rezza G, Boschini A, Pezzotti P, Tamburrini A, Rossi P, Di Fine M, Smacchia C, Schiesari A, Gattei E, Zucconi R, Ballarini P. Clinical cryptosporidiosis and human immunodeficiency virus (HIV)-induced immunosuppression: findings from a longitudinal study of HIV-positive and HIV-negative former injection drug users. J Infect Dis. 1997;176:969–975. doi: 10.1086/516498. [DOI] [PubMed] [Google Scholar]
- 79.Priest J W, Kwon J P, Arrowood M J, Lammie P J. Cloning of the immunodominant 17-kilodalton antigen from Cryptosporidium parvum. Mol Biochem Parasitol. 2000;106:261–271. doi: 10.1016/s0166-6851(99)00223-6. [DOI] [PubMed] [Google Scholar]
- 80.Priest J W, Kwon J P, Moss D M, Roberts J M, Arrowood M J, Dworkin M S, Juranek D D, Lammie P J. Detection by enzyme immunoassay of serum immunoglobulin G antibodies that recognize specific Cryptosporidium parvum antigens. J Clin Microbiol. 1999;37:1385–1392. doi: 10.1128/jcm.37.5.1385-1392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reperant J M, Naciri M, Chardes T, Bout D T. Immunological characterization of a 17-kDa antigen from Cryptosporidium parvum recognized early by mucosal IgA antibodies. FEMS Microbiol Lett. 1992;78:7–14. doi: 10.1016/0378-1097(92)90280-2. . (Erratum, 10:349, 1993.) [DOI] [PubMed] [Google Scholar]
- 82.Reperant J M, Naciri M, Iochmann S, Tilley M, Bout D T. Major antigens of Cryptosporidium parvum recognised by serum antibodies from different infected animal species and man. Vet Parasitol. 1994;55:1–13. doi: 10.1016/0304-4017(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 83.Riggs M W. Immunology: host response and development of passive immunotherapy and vaccines. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. New York, N.Y: CRC Press; 1997. pp. 129–162. [Google Scholar]
- 84.Riggs M W, McGuire T C, Mason P H, Perryman L E. Neutralization-sensitive epitopes are exposed on the surface of infectious Cryptosporidium parvum sporozoites. J Immunol. 1989;143:1340–1345. [PubMed] [Google Scholar]
- 85.Riggs M W, McNeil M R, Perryman L E, Stone A L, Scherman M S, O'Connor R M. Cryptosporidium parvum sporozoite pellicle antigen recognized by a neutralizing monoclonal antibody is a beta-mannosylated glycolipid. Infect Immun. 1999;67:1317–1322. doi: 10.1128/iai.67.3.1317-1322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riggs M W, Stone A L, Yount P A, Langer R C, Arrowood M J, Bentley D L. Protective monoclonal antibody defines a circumsporozoite-like glycoprotein exoantigen of Cryptosporidium parvum sporozoites and merozoites. J Immunol. 1997;158:1787–1795. [PubMed] [Google Scholar]
- 87.Robert B, Antoine H, Dreze F, Coppe P, Collard A. Characterization of a high molecular weight antigen of Cryptosporidium parvum micronemes possessing epitopes that are cross-reactive with all parasitic life cycle stages. Vet Res. 1994;25:384–398. [PubMed] [Google Scholar]
- 88.Rump J A, Arndt R, Arnold A, Bendick C, Dichtelmuller H, Franke M, Helm E B, Jager H, Kampmann B, Kolb P, Kreuz W. Treatment of diarrhoea in human immunodeficiency virus-infected patients with immunoglobulins from bovine colostrum. Clin Investig. 1992;70:588–594. doi: 10.1007/BF00184800. [DOI] [PubMed] [Google Scholar]
- 89.Sallon S, Deckelbaum R J, Schmid I I, Harlap S, Baras M, Spira D T. Cryptosporidium, malnutrition, and chronic diarrhea in children. Am J Dis Child. 1988;142:312–315. doi: 10.1001/archpedi.1988.02150030086027. [DOI] [PubMed] [Google Scholar]
- 90.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 91.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 92.Schroeder A A, Lawrence C E, Abrahamsen M S. Differential mRNA display cloning and characterization of a Cryptosporidium parvum gene expressed during intracellular development. J Parasitol. 1999;85:213–220. [PubMed] [Google Scholar]
- 93.Shield J, Melville C, Novelli V, Anderson G, Scheimberg I, Gibb D, Milla P. Bovine colostrum immunoglobulin concentrate for cryptosporidiosis in AIDS. Arch Dis Child. 1993;69:451–453. doi: 10.1136/adc.69.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blancq S M, Tchack L, Tzipori S, Widmer G. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spano F, Putignani L, Guida S, Crisanti A. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp Parasitol. 1998;90:195–198. doi: 10.1006/expr.1998.4324. [DOI] [PubMed] [Google Scholar]
- 96.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 97.Strong W B, Nelson R G. Preliminary profile of the Cryptosporidium parvum genome: an expressed sequence tag and genome survey sequence analysis. Mol Biochem Parasitol. 2000;107:1–32. doi: 10.1016/s0166-6851(99)00225-x. [DOI] [PubMed] [Google Scholar]
- 98.Sulaiman I M, Lal A A, Arrowood M J, Xiao L. Biallelic polymorphism in the intron region of beta-tubulin gene of Cryptosporidium parasites. J Parasitol. 1999;85:154–157. [PubMed] [Google Scholar]
- 99.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thea D M, Pereira M E, Kotler D, Sterling C R, Keusch G T. Identification and partial purification of a lectin on the surface of the sporozoite of Cryptosporidium parvum. J Parasitol. 1992;78:886–893. [PubMed] [Google Scholar]
- 101.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tilley M, Eggleston M T, Upton S J. Multiple oral inoculations with Cryptosporidium parvum as a means of immunization for production of monoclonal antibodies. FEMS Microbiol Lett. 1993;113(2):235–240. doi: 10.1111/j.1574-6968.1993.tb06520.x. [DOI] [PubMed] [Google Scholar]
- 103.Tilley M, Upton S J. Both CP15 and CP25 are left as trails behind gliding sporozoites of Cryptosporidium parvum (Apicomplexa) FEMS Microbiol Lett. 1994;120:275–278. doi: 10.1111/j.1574-6968.1994.tb07045.x. [DOI] [PubMed] [Google Scholar]
- 104.Tilley M, Upton S J. Electrophoretic characterization of Cryptosporidium parvum (KSU-1 isolate) (Apicomplexa: Cryptosporidiidae) Can J Zool. 1990;68:1513–1519. [Google Scholar]
- 105.Tilley M, Upton S J. Sporozoites and merozoites of Cryptosporidium parvum share a common epitope recognized by a monoclonal antibody and two-dimensional electrophoresis. J Protozool. 1991;38:48S–49S. [PubMed] [Google Scholar]
- 106.Tilley M, Upton S J, Blagburn B L, Anderson B C. Identification of outer oocyst wall proteins of three Cryptosporidium (Apicomplexa: Cryptosporidiidae) species by 125I surface labeling. Infect Immun. 1990;58:252–253. doi: 10.1128/iai.58.1.252-253.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tilley M, Upton S J, Fayer R, Barta J R, Chrisp C E, Freed P S, Blagburn B L, Anderson B C, Barnard S M. Identification of a 15-kilodalton surface glycoprotein on sporozoites of Cryptosporidium parvum. Infect Immun. 1991;59:1002–1007. doi: 10.1128/iai.59.3.1002-1007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tzipori S, Roberton D, Chapman C. Remission of diarrhoea due to cryptosporidiosis in an immunodeficient child treated with hyperimmune bovine colostrum. Br Med J. 1986;293:1276–1277. doi: 10.1136/bmj.293.6557.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tzipori S, Roberton D, Cooper D A, White L. Chronic cryptosporidial diarrhoea and hyperimmune cow colostrum. Lancet. 1987;2:344–345. doi: 10.1016/s0140-6736(87)90944-5. [DOI] [PubMed] [Google Scholar]
- 110.Ungar B L, Ward D J, Fayer R, Quinn C A. Cessation of Cryptosporidium-associated diarrhea in an acquired immunodeficiency syndrome patient after treatment with hyperimmune bovine colostrum. Gastroenterology. 1990;98:486–489. doi: 10.1016/0016-5085(90)90842-o. [DOI] [PubMed] [Google Scholar]
- 111.Van Klinken B J-W, Dekker J, Büller H A, Einerhand A W C. Mucin gene structure and expression: protection vs. adhesion. Am J Physiol. 1995;269:G613–G627. doi: 10.1152/ajpgi.1995.269.5.G613. [DOI] [PubMed] [Google Scholar]
- 112.Vasquez J R, Gooze L, Kim K, Gut J, Petersen C, Nelson R G. Potential antifolate resistance determinants and genotypic variation in the bifunctional dihydrofolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol Biochem Parasitol. 1996;79:153–165. doi: 10.1016/0166-6851(96)02647-3. [DOI] [PubMed] [Google Scholar]
- 113.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:223–239. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 114.Widmer G, Tchack L, Chappell C L, Tzipori S. Sequence polymorphism in the beta-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl Environ Microbiol. 1998;64:4477–4481. doi: 10.1128/aem.64.11.4477-4481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]
- 116.Wolfson J S, Richter J M, Waldron M A, Weber D J, McCarthy D M, Hopkins C C. Cryptosporidiosis in immunocompetent patients. N Engl J Med. 1985;312:1278–1282. doi: 10.1056/NEJM198505163122002. [DOI] [PubMed] [Google Scholar]