Abstract
COVID-19 is an infectious disease caused by SARS-CoV-2 leading to the ongoing global pandemic. Infected patients developed a range of respiratory symptoms, including respiratory failure, as well as other extrapulmonary complications. Multiple comorbidities, including hypertension, diabetes, cardiovascular diseases, and chronic kidney diseases, are associated with the severity and increased mortality of COVID-19. SARS-CoV-2 infection also causes a range of cardiovascular complications, including myocarditis, myocardial injury, heart failure, arrhythmias, acute coronary syndrome, and venous thromboembolism. Although a variety of methods have been developed and many clinical trials have been launched for drug repositioning for COVID-19, treatments that consider cardiovascular manifestations and cardiovascular disease comorbidities specifically are limited. In this review, we summarize recent advances in drug repositioning for COVID-19, including experimental drug repositioning, high-throughput drug screening, omics data-based, and network medicine-based computational drug repositioning, with particular attention on those drug treatments that consider cardiovascular manifestations of COVID-19. We discuss prospective opportunities and potential methods for repurposing drugs to treat cardiovascular complications of COVID-19.
Keywords: cardiovascular disease, COVID-19, drug repositioning, heart failure, myocarditis, thromboembolism
COVID-19, caused by the SARS-CoV-2, spread rapidly from Wuhan, China in 2019 to over 200 countries and territories world-wide, leading to millions of infected individuals and numerous mortalities. Infected patients develop a range of symptoms, from mild to severe.1 Although some targeted therapies have been developed that show benefit, there is an urgent need for definitive therapies against SARS-CoV-2 from which patients would greatly benefit.2
Many clinical studies have reported that multiple comorbidities, including rheumatic diseases, hypertension, diabetes, cardiovascular diseases (CVD), and chronic kidney diseases, are associated with the severity and mortality of COVID-19.3 Systematic reviews and meta-analysis studies have shown that a high burden of cardiovascular events is significantly associated with mortality and intensive care among patients with COVID-19.4 The estimated mortality rate of COVID-19 in the general population is about 3.4%, whereas the comorbidity of CVD may result in a 10.5% mortality rate. A large-scale study that focused on independent clinical risk factors associated with a composite outcome of COVID-19 found that arterial hypertension was the most prevalent comorbidity (16.9%).5,6
Although the most common symptoms that COVID-19 causes are fever and dry cough, hospitalized patients can develop a range of cardiovascular complications, including myocarditis, heart failure, arrhythmia, acute coronary syndrome, and venous thromboembolism.7–10 A long-term observational study on a cohort of 153 760 individuals with COVID-19 indicated that the risk and 1-year burden of CVD in survivors are substantial (hazard ratio, 1.52, and burden, 4.03, for stroke; hazard ratio, 1.71 and burden, 10.74, for atrial fibrillation; hazard ratio, 1.72 and burden, 5.35, for acute coronary syndrome).11 Post-COVID-19 myocarditis may be caused by viral infection of the myocardium and associated cytokine storm; yet, the underlying pathophysiology of other COVID-19–associated cardiovascular complications has not been fully delineated.12
De novo development of new drugs for an infectious disease like COVID-19 in a short time frame is typically not realistic, for which reason patients would greatly benefit from the availability and repurposing of existing drugs. Drug repositioning or drug repurposing, which aims to find new indications for known drugs, can narrow drug candidates under consideration, avoid drug safety issues, and thereby shorten the development time, offering a promising alternative to de novo drug discovery. A variety of in vitro and in silico methods have been developed, and many resulting clinical trials have been launched for drug repositioning for COVID-19.13–16 Given the cardiovascular complications caused by SARS-CoV-2 infection, drug repurposing for COVID-19 should consider targeting the pathobiology underlying the cardiovascular manifestations and comorbidities of the infection. In this review, we summarize recent advances in repositioning drugs for COVID-19 and highlight prospective opportunities for repurposing drugs for the treatment of cardiovascular complications of COVID-19.
Epidemiology of COVID-19 and Cardiovascular Comorbidities
COVID-19 spread rapidly from Wuhan, China, in the late fall of 2019 to other countries and territories world-wide. The confirmed cases grew exponentially at the outset with an early estimated growth rate r of 0.1 to 0.2 per day. The outbreak reached an epidemic peak by late February 2020. By March 2020, a global SARS-CoV-2 pandemic had begun to unfold. Initial nonpharmaceutical interventions, such as lockdowns, social distancing, and the use of face masks, flattened the growth curve, which was, unfortunately, followed by subsequent waves due to evolution of the virus and changes in host immunity and behavior.17 By December, 2020, vaccines against SARS-CoV-2 had been developed and approved for emergency use; the durability of protection and new variant-specific protection, however, remain uncertain.
Many clinical studies have reported that multiple comorbidities are associated with the severity and mortality of COVID-19. For example, Guan et al5 conducted a large-scale study that analyzed independent clinical risk factors associated with a composite outcome (admission to intensive care, invasive ventilation, or death) using a Cox regression model on data from 1590 hospitalized patients with COVID-19, and found that the most prevalent comorbidity was arterial hypertension (16.9%), followed by diabetes (8.2%). Furthermore, analyses were performed to confirm the impact of cardiovascular comorbidities on the clinical outcomes of patients with COVID-19.6,18 A meta‐analysis of 22 studies based on a random-effects model revealed that the proportions of hypertension, diabetes, and cardiac disease in COVID‐19 was 17.1%, 8.5%, and 4.5%, respectively.19 In a retrospective study based on a cohort of 8438 patients with COVID-19, Kuno et al20 found significantly higher rates of mechanical ventilation and mortality in patients with COVID-19 with coronary artery disease (CAD), peripheral artery disease, heart failure, or cardiac injury.
Although patients with preexisting CVD have been shown to be at higher risk of adverse COVID-19 outcomes, it is unclear whether preexisting CVD are independently important predictors for severe COVID-19. In a nationwide Danish cohort of hospital-screened patients with COVID-19, Phelps et al21 investigated whether preexisting CVD predict the 30-day risk of composite outcome of severe COVID-19 and all-cause mortality or not using a Cox regression model, incorporating some covariates. The results show that pre-existing CVD does have modest effects on an increased risk of poor COVID-19 outcomes. A systematic meta-analysis of 51 studies with a total of 48 317 patients with COVID-19 demonstrated that the relative risk of developing severe COVID-19 or death was significantly higher in patients with risk factors for CVD,4 suggesting that CVD comorbidity was closely related to fatal outcomes in COVID-19 for patients across all ages.
Cardiovascular Manifestations of COVID-19
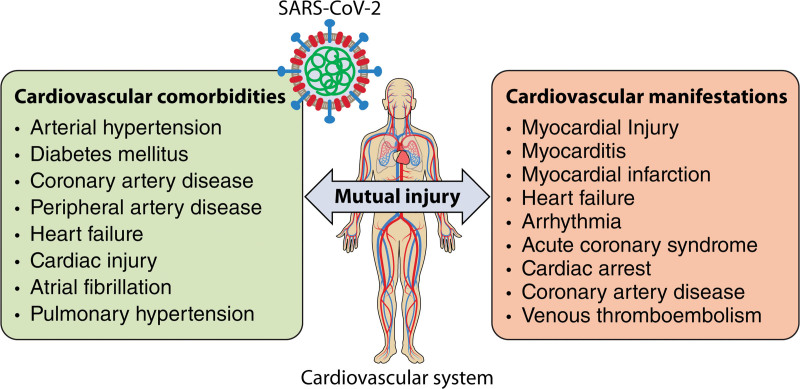
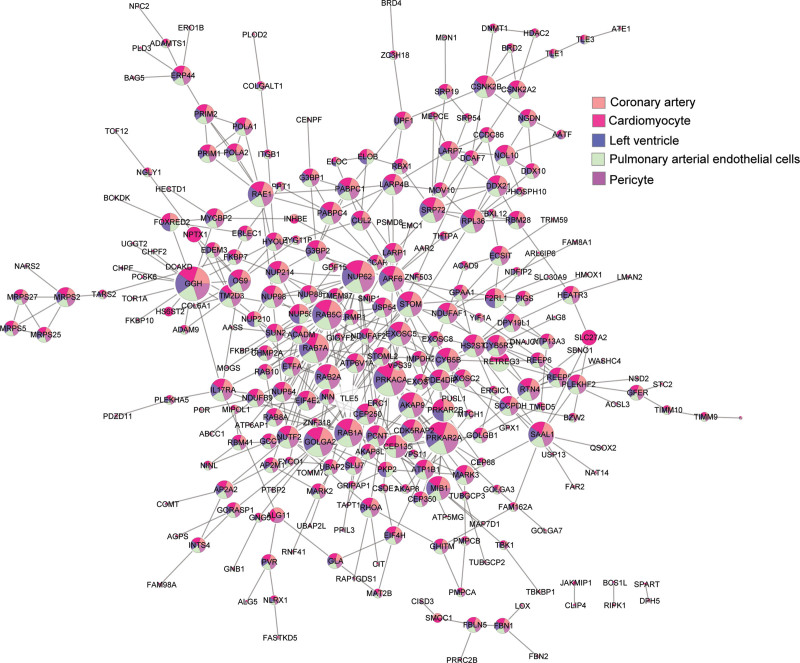
There is a mutual interaction between COVID-19 and the cardiovascular system: COVID-19 can also cause cardiovascular injury and a series of cardiovascular complications such as acute myocardial injury, myocarditis, arrhythmias, cardiac arrest, etc (Figure 1). SARS-CoV-2 binds the human membrane protein ACE2 (angiotensin-converting enzyme 2) receptor via its spike (S) protein (see below), providing a point of viral entry into the cell.8,9,22,23 Single-cell RNA sequencing showed that ACE2 expression in the adult human heart is higher than that in the lung,24 which may mediate SARS-CoV-2 entry into cardiomyocytes and contribute to cardiovascular complications. Most of the 332 host proteins from the first SARS-CoV-2 virus-host protein interaction map,25 which we denote SARS-CoV-2 targets, are connected with each other in the human protein-protein interactome (forming a distinct subnetwork or covidome) and are expressed in CVD-related cell types, such as cardiomyocytes, pulmonary arterial endothelial cells, and pericytes (Figure 2).
Figure 1.
The mutual interaction and injury between COVID-19 and the cardiovascular system. Cardiovascular comorbidities are significantly correlated with the severity and mortality of COVID-19; likewise, COVID-19 can cause cardiovascular injury and a series of cardiovascular complications. Illustration Credit: Sceyence Studios.
Figure 2.
The COVID-19 disease module. SARS-CoV-2 targets are not randomly scattered over the human interactome, but form a densely connected module (covidome). The majority of the SARS-CoV-2 targets are expressed in cardiovascular tissues, potentially serving as a basis for SARS-CoV-2 infection-induced cardiovascular manifestations.
Acute myocardial injury was observed even among early cases in China. In a study of 138 hospitalized patients with COVID-19, cardiac injury was present in 7.2% of patients and arrhythmias were present in 16.7% of patients, both with higher prevalence among patients requiring intensive care.26 A population-based, observational study indicated that COVID-19 infection accounted for ≈one-third of the increase in out of hospital cardiac arrest incidence during the pandemic.27 In a nation-wide Scotland cohort, Ho et al28 found that the risk of thromboembolism was significantly elevated over the whole risk period of COVID-19 but highest in the 7 days following a positive COVID-19 test; the risk of deep vein thrombosis and pulmonary embolism remained significantly elevated even 56 days following the test.
Long COVID or post-COVID refers to a condition in which patients continue to suffer from symptoms >3 months following acute infection. The cardiovascular manifestations of long COVID include stroke, chest pain, electrocardiographic abnormalities, postural tachycardia syndrome, atrial fibrillation, acute coronary syndrome, and chronic thromboembolic pulmonary hypertension.29,30 A long-term observational study of a cohort of 153 760 infected individuals showed that patients with COVID-19 are at increased risk for and 1-year burden of incident CVDs, including cerebrovascular disorders, arrhythmias, ischemic and nonischemic cardiac diseases, and thrombotic disorders.11 Thus, health care for those surviving acute COVID-19 should remain attentive to the potential for CVD.11 For more information on cardiovascular manifestations caused by SARS-CoV-2 infection, the reader is referred to recent comprehensive reviews.8–10,12,29–31
The underlying mechanisms of COVID-19–associated cardiovascular complications are yet to unfold. SARS-CoV-2 may directly infect human cardiomyocytes and exert cytotoxic effects, triggering severe cellular and organ-wide pathology and dysfunction.9 Hypoxia-induced myocardial injury due to hypoxic respiratory failure and hypoxemia, or acute right ventricle failure due to pulmonary embolism are indirect pathophysiological mechanisms. Cardiac injury may also stem from dysfunctional (excessively robust) inflammatory or immune responses.9 Post-COVID myocarditis may be caused by direct viral infection of the myocardium and associated cytokine storm.12 A recent study from our group uncovered common regulators and pathways common to COVID-19 and pulmonary arterial hypertension by integrating omics data and the human protein-protein interactome, viz., HIF1A (hypoxia inducible factor 1 subunit alpha), RUNX1 (RUNX family transcription factor 1), and JAK-STAT (Janus kinase-signal transducer and activator of transcription) pathways.32 Independent studies demonstrated that RUNX1 inhibition can reduce the expression of ACE2 which is critical for SARS-CoV-2 infection and may also be beneficial for pulmonary fibrosis and pulmonary hypertension.33,34 Identification of such overlapping pathways between COVID-19 and CVD is critical for designing treatments that consider cardiovascular manifestations of COVID-19.
Repurposing Drugs for Treating Cardiovascular Manifestations of COVID-19
Hospitalized patients with COVID-19 would greatly benefit from therapies against SARS-CoV-2 infection, some of which have been developed during the pandemic. Yet, definitive therapy and therapies that prevent long-term extrapulmonary and cardiovascular manifestations remain elusive. De novo development of new drugs for a rapidly spreading disease such as COVID-19 in a short period of time is not realistic. In contrast, patients may benefit from drug repositioning, which allows fast-tracking of existing drug candidates into human clinical trials for testing as COVID-19 therapies. Although many drug repurposing methods have been developed for COVID-19, treatments should consider targeting the pathobiological mechanisms underlying the cardiovascular manifestations and comorbidities. We highlight such prospective opportunities in this section.
Experimental Drug Repositioning for COVID-19
Understanding the cell entry mechanisms of SARS-CoV-2 is one key to deciphering a potential drug target and inhibiting infection. Cell entry of SARS-CoV-2 depends on binding of the viral spike (S) protein to its receptor human ACE2 and on S protein priming by the host cell protease TMPRSS2 (transmembrane serine protease 2).22 In addition to ACE2, this main protease of SARS-CoV-2 is an attractive drug target because of its central role in the SARS-CoV-2 life cycle and its conservation among different variants. A TMPRSS2 inhibitor camostat mesylate can block the entry of SARS-CoV-2 and might be a viable treatment option.23 Using a Fluorescence Resonance Energy Transfer (FRET)-based enzymatic assay, Ma et al35 discovered 4 compounds including boceprevir, GC-376, and calpain inhibitors II and XII that target the SARS-CoV-2 main protease and inhibit SARS-CoV-2 viral replication in cell culture. Using phosphoproteomics to study signaling changes in permissive human cells after SARS-CoV-2 infection, Klann et al36 found that inhibition of growth factor receptor downstream signaling by 5 compounds, pictilisib, omipalisib, RO5126766, lonafarnib, and sorafenib, can prevent SARS-CoV-2 replication in cells at clinically achievable concentrations. In addition, an orally bioavailable SARS-CoV-2 main protease inhibitor, PF-07321332 (Nirmatrelvir) was discovered and characterized as having in vitro pan-human coronavirus antiviral activity and excellent off-target selectivity and in vivo safety profiles.37 Recently, using the Ugi 4-component reaction, an α-ketoamide-containing main protease inhibitor Y180 with oral bioavailability was shown to protect against infection by wild-type SARS-CoV-2 and its variants including omicron.38
Although many studies concentrated on discovering SARS-CoV-2 inhibitors, few targeted the cardiovascular manifestations caused by COVID-19. This limitation is partly a consequence of our limited understanding of the underlying pathobiology. With the unfolding of the physiological mechanisms of COVID-19-associated cardiovascular complications,39,40 we envision there will be more research focused on this important topic.
High-Throughput Drug Screening for COVID-19
The most critical step in drug repurposing is the development of in vitro assays that can evaluate the efficacy of drug candidates in an efficient way. Although individual experiments have been used to reposition drugs for COVID-19 through mechanistic exploration (as described above), a number of high-throughput screening assays have been developed to reposition approved and investigational drugs for SARS-CoV-2 infection in an unbiased, quantitative way, such as the ACE2 activity assay, 3CL protease activity assay, multitarget assays, spike-ACE2 protein–protein interaction assay, pseudotyped particle entry assay, and phenotypic antiviral efficacy assays.41 These high-throughput screens facilitate the rapid assessment of many drug candidates that may be repurposed for COVID-19.
A representative study is the high-throughput immunofluorescence-based screening of 6710 clinical and preclinical compounds from the Broad Institute’s drug repositioning repository that aimed to identify potent inhibitors of SARS-CoV-2 infection.42 Computational analysis of primary screen outcomes classified the compounds into 4 different categories (no-effect, strongly active, weakly active, and cytotoxic compounds), and prioritized the best candidates for follow-up in mechanistic assays. A cellular network integrating relationships between small molecule structure, dose-response antiviral activity, host target, and cell interactome was constructed, which provides important insights for SARS-CoV-2 infection. Some representative drugs were evaluated for mechanism of action in human cell models with SARS-CoV-2 variants. One promising candidate, obatoclax, was found to reduce the SARS-CoV-2 viral load significantly in the lungs of mice engineered to express the human ACE2 protein in their pulmonary epithelial cells. This work illustrates a rigorous approach for future pharmacological and computational identification of potential drug treatments for viral diseases in general, and for SARS-CoV-2 in particular.
Repurposing, Focused Rescue, and Accelerated Medchem, a best-in-class drug repurposing library consisting of 12 000 compounds,43 has been used widely in high-throughput drug screening for COVID-19. For example, the library was screened against 2 human cell lines (HeLa-ACE2 and Calu-3) using high-content imaging assays of SARS-CoV-2 infection.44 Among over 40 promising hits from each assay, the antivirals nelfinavir and the parent or prodrug molnupiravir were found to reduce SARS-CoV-2 replication in an orthogonal human differentiated primary cell model. The library was also screened for therapeutic agents to treat SARS-CoV-2 infection in Vero E6 cells45 and identified 21 drugs that inhibit viral replication of SARS-CoV-2 with dose–response relationships, including the kinase inhibitor apilimod. In a recent study, ≈18 000 drugs were screened for antiviral activity using live virus infection in human epithelial cells and 122 drugs were validated with antiviral activity and selectivity against SARS-CoV-2,46 among which were included 16 nucleoside analogues including the approved antivirals remdesivir and molnupiravir for use in COVID-19. In particular, this study identified a panel of host nucleoside biosynthesis inhibitors as effective antiviral agents which, when combined with antiviral nucleoside analogues, can synergistically inhibit SARS-CoV-2 infection in vitro and in vivo. In addition to high-throughput screening assays, virtual screening with predocking and postdocking pharmacophore filtering has also been used for drugs for COVID-19.47 Most patients with COVID-19 only develop no or mild symptoms, but some patients develop a severe inflammatory response that can be fatal. Based on this observation, a high-throughput screening assay using a 2560 small-molecule compound library was performed to identify Food and Drug Administration (FDA)-approved drugs that function as pan-inflammasome inhibitors.48 In vitro and in vivo experiments validated that niclosamide effectively inhibits both inflammasome activation and SARS-CoV-2 replication.
Using a lung organoid model based on human pluripotent stem cells, Han et al49 performed a high-throughput screen of FDA-approved drugs and identified inhibitors of SARS-CoV-2 infection, including imatinib, mycophenolic acid, and quinacrine dihydrochloride, demonstrating that this model infected with SARS-CoV-2 can serve as a relevant disease model for rapid screening. By screening ≈3000 compounds in hepatocyte Huh7.5 and respiratory Calu-3 cells against SARS-CoV-2, Dittmar et al50 found that SARS-CoV-2 enters different cell types through different entry pathways, and uncovered only 9 compounds that are active in both Huh7.5 cells and Calu-3 cells including cyclosporine and cyclophilin inhibitors. A pipeline for quantitative, high-throughput, image-based screening of SARS-CoV-2 infection was created through which 17 clinical drug candidates were identified, including lactoferrin, with in vitro antiviral activity across multiple cell lines from a library of 1425 FDA-approved drugs.51 In another study, a team of investigators developed a fluorogenic high-throughput in vitro screening assay through which a total of 6030 drug repurposing candidates from National Center for Advancing Translational Sciences (NCATS) annotated libraries were evaluated for inhibiting SARS-CoV-2 replication in Calu-3 cells.52 Next, a suite of subsequent assays, including a mass spectrometry-based detection assay and a cell-based SARS-CoV-2 pseudotyped particle entry assay, was developed to confirm and assess the hits from the biochemical screen, among which camostat, nafamostat, PCI-27483, otamixaban, and 2 peptidomimetic inhibitors of TMPRSS2 were identified.
Although many high-throughput screening assays have been developed for drug repositioning for SARS-CoV-2 infection, to the best of our knowledge, no assays have been developed in cardiovascular cells. We hypothesize that large-scale drug screening using human cardiomyocytes, in particular, may be critical for identifying drug candidates that may be used to treat the key cardiovascular manifestations of COVID-19.
Omics Data-Based Drug Repositioning for COVID-19
SARS-CoV-2 interacts with human host proteins and induces a series of molecular changes at different biological levels. Owing to the advances in high-throughput omics technologies, a large pool of SARS-CoV-2-induced transcriptomics, proteomics, and other omics data have been generated and accumulated. For example, a single-cell atlas of the peripheral immune response to severe COVID-19 was created based on single-cell RNA sequence profiling of peripheral blood mononuclear cells from 7 patients hospitalized for COVID-19.53 More recently, a molecular single-cell lung atlas of lethal COVID-19 was created based on single-nucleus RNA sequencing of the lungs of nineteen individuals who died of COVID-19.54 The SARS-CoV-2-induced transcriptome, proteome, ubiquitinome, and phosphoproteome of a lung-derived human cell line were integrated in a concurrent multi-omics study to define the pathogenic properties of SARS-CoV-2 at different levels.55 These omics atlases enable the dissection of lethal COVID-19 and inform our understanding of long-term complications of COVID-19 survivors, providing an important resource for therapeutic development in general as well as opportunities for therapeutic development for the cardiovascular complications of COVID-19 in particular.
In the integrated omics analysis of these data sets, SARS-CoV-2 targets represent causal layers, and differentially expressed genes or proteins caused by the infection represent the consequence layers. By integrating SARS-CoV-2 targets and differentially expressed genes into the human interactome and finding the shortest paths (hidden layer) between them, a SARS-CoV-2–induced protein network was constructed in a recent study.56 Key proteins and disease pathways were uncovered from the hidden layer and drugs that target key proteins were ranked. After using an unsupervised self-organized map to cluster the predicted drugs and uncover drug mechanisms of action, SARS-CoV-2–induced omics data have benefited drug repositioning for COVID-19. Two drugs, proguanil and sulfasalazine, were validated as inhibitors of viral replication in cell assays. This unbiased omics data analysis proves the power of multi-omics integration, which opens new avenues for the rapid repurposing of approved drugs.
In a recent study, multi-omics profiling of peripheral blood mononuclear cells from patients hospitalized with severe COVID-19 was performed, including transcriptomic profiling by single-cell RNA sequence and epigenetic profiling by an assay for transposase-accessible chromatin with high-throughput sequencing and whole-genome bisulfite sequencing.57 Integration of the single-cell transcriptomes and epigenomes revealed significant differential monocyte transcriptomic and epigenetic attributes between those who would survive and those who would die from COVID-19. The study also showed that peripheral blood mononuclear cell omics composition is strongly correlated with disease progression. A significant reduction of T cells and a significant increase of monocytes were observed in critically ill patients. Through data integration and Ingenuity pathway analysis, several promising drug candidates, including dexamethasone, baricitinib, tacrolimus, zotatifin, and nintedanib, have been identified for further study. Nintedahib could be potentially useful for the treatment of post-COVID-19 pulmonary fibrosis.58,59
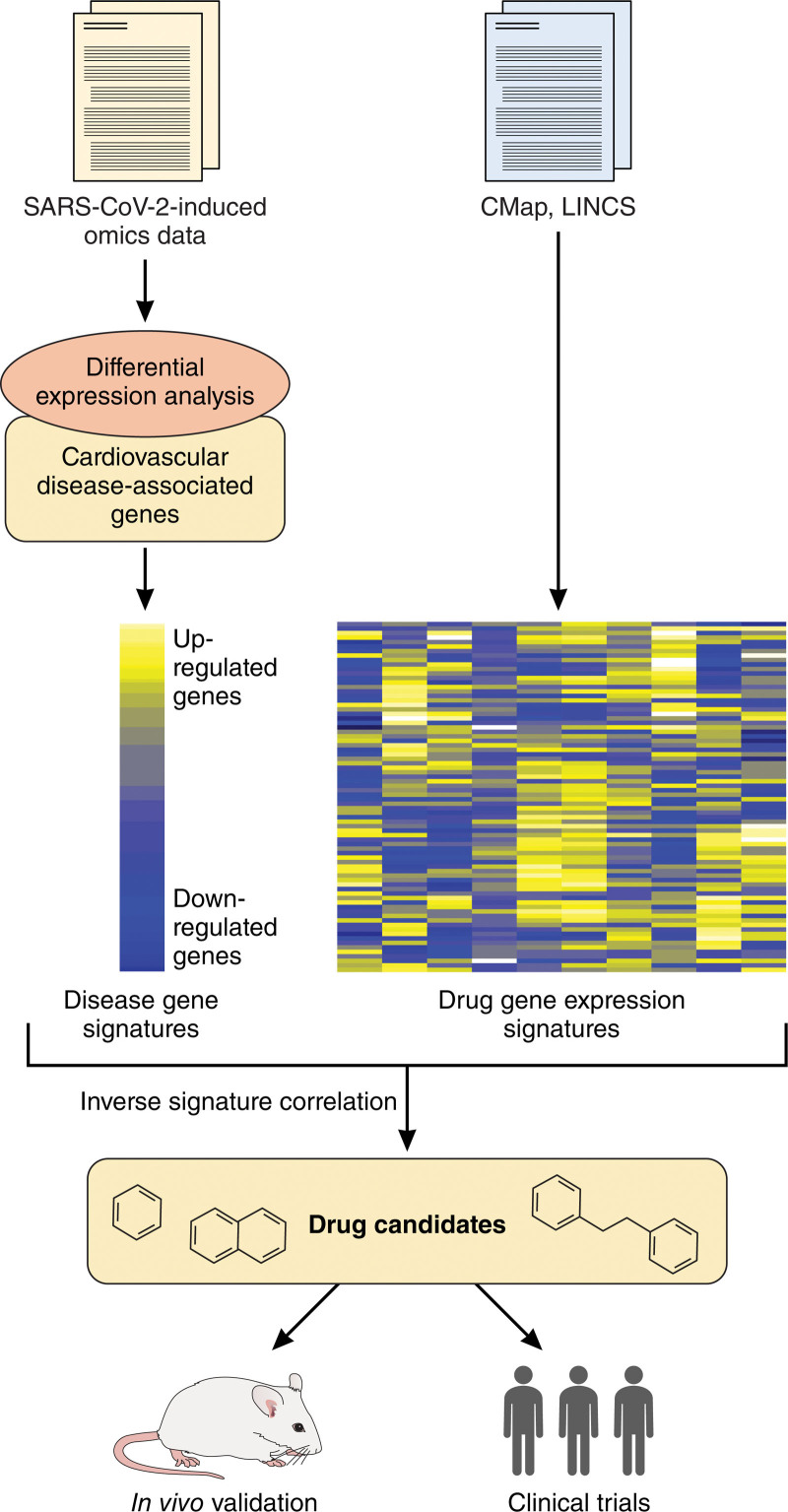
Although there have not been methods proposed specifically designed for drug repurposing in the treatment of cardiovascular manifestations of COVID-19, SARS-COV-2 infection-induced omics data combined with CVD gene signatures can potentially identify drug candidates for COVID-19 with cardiovascular complication (Figure 3). For example, the Connectivity Map and the Library of Integrated Network-Based Cellular Signatures Program provide a data repository that contains almost 2 million gene expression profiles covering 72 human cell lines for thousands of small molecules and drugs.60,61 Differentially expressed genes can be obtained from SARS-CoV-2-induced omics data. Overlapping analysis between CVD-associated genes and SARS-CoV-2 differentially expressed genes leads to disease gene signatures representing the underlying pathobiology of COVID-19-associated cardiovascular complications. Inverse signature correction between disease gene signatures and drug gene expression signatures from the Connectivity Map and Library of Integrated Network-Based Cellular Signatures can identify potential drug candidates for targeting cardiovascular manifestations for COVID-19. In addition, some strategies for cardiovascular drug repositioning can be used for treating cardiovascular manifestation of COVID-19, as well.62
Figure 3.
A potential scheme for repositioning drug candidates to target cardiovascular manifestations of COVID-19. Differentially expressed genes can be obtained from SARS-CoV-2-induced omics data. Overlapping analysis between cardiovascular disease-associated genes and SARS-CoV-2 differentially expressed genes leads to disease gene signatures. Inverse signature correction between disease gene signatures and drug gene expression signatures from the Connectivity Map (CMap) and Library of Integrated Network-Based Cellular Signatures (LINCS) can identify potential drug candidates for targeting cardiovascular manifestation for COVID-19. The drug candidates are then subject to in vitro and in vivo validation through animal models and clinical trials in patients with COVID-19. Illustration Credit: Sceyence Studios.
Network Medicine-Based Drug-Repurposing for COVID-19
Network medicine is an emerging interdisciplinary field that integrates systems biology and network science, focuses on the interaction between biological components, and applies concepts and tools from network science to understanding, preventing, and treating diseases.63 Network medicine approaches have been successfully applied to the identification of disease pathways and molecular relationships between different pathophenotypes. They also offer important insights into the relationships between drugs, their targets, and diseases, including network-based drug repositioning.64 The first SARS-CoV-2 protein interaction map consisting of 332 high-confidence protein–protein interactions between SARS-CoV-2 and human proteins provided opportunities for revealing therapeutic targets for drug repositioning.25 In fact, drug repositioning for CVDs and COVID-19 has benefited from network medicine approaches, as well, which paves the way toward the development of preventive and therapeutic solutions for COVID-19 with particular consideration of cardiovascular complications and cardiovascular comorbidities.
In our research group, we have developed a network module-based method for illuminating the mechanisms of drug action for myocardial infarction by constructing drug-target-disease modules,65 which suggested a list of potential drug repurposing candidates for myocardial infarction, for example, valproic acid, minocycline, and tetracycline. A similar approach was adopted for drug repositioning for pulmonary arterial hypertension and identified a number of drug candidates that were either verified by the curated literature or included in (ongoing) clinical trials.66 Unlike machine learning-based black box methods, such network module-based approaches can provide insights into the mechanisms of action of drugs. This method could further be expanded for drug repositioning for treating cardiovascular manifestations of COVID-19 by integrating SARS-CoV2 targets (covidome), CVD genes, and drug targets into the interactome, constructing network modules consisting of 3 types of nodes and identifying drugs that target both the covidome and CVD modules.
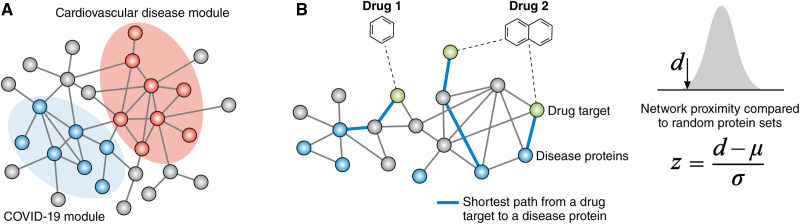
Previous studies have demonstrated that disease proteins tend to cluster in the same network neighborhood within the human protein–protein interactome and form a subnetwork called the disease module.67 For drugs to have on-target therapeutic effects on a disease or off-target adverse effects, their targets have to be in the vicinity of disease modules in the human interactome.68 Based on this principle, network proximity has been demonstrated as a powerful tool for predicting drug efficacy. This method quantifies the closeness relationship between disease modules and drug targets in the human interactome (Figure 4). For example, we used a network proximity approach to systematically repositioning FDA-approved drugs for many diseases and hundreds of new drug-disease associations have been identified as a result.69 In particular, CVDs were chosen as a test case of this principle owing to their high prevalence, morbidity, and mortality in the population. Nine hundred eighty-four FDA-approved drugs (including 177 FDA-approved cardiovascular drugs defined by Anatomical Therapeutic Chemical (ATC) classification codes) and 22 types of CVDs were investigated. In this work, we demonstrated a population-based validation approach using electronic medical records from large administrative healthcare databases to test the predicted drug-disease relationships. Two drugs, carbamazepine and hydroxychloroquine, were validated using propensity score matching in the patient-level data: carbamazepine is associated with an increased risk of CAD and hydroxychloroquine is associated with a decreased risk of CAD. Of course, this validation only verifies drug-disease associations, not real causal relationships. In vitro and in vivo experimental validation is necessary to confirm the roles of these drugs in CVDs. These studies demonstrate that a unique integration of human interactome and large-scale patient-level data complemented by mechanistic in vitro studies can facilitate drug repurposing, which can be adapted for COVID-19 drug repositioning with consideration of CVD comorbidity and complications, as well. The drugs whose targets are significantly proximate to both the covidome and CVD modules may be good candidates for experimental validation.
Figure 4.
A network proximity approach for repositioning drugs to treat cardiovascular manifestations of COVID-19. A, COVID-19 disease module and cardiovascular disease module in the human interactome. B, For each target of a drug, a disease protein in the module closest to the target is identified and shortest path length is calculated. The average shortest path length from the targets of a drug to a disease module defines the network proximity d from the drug to the disease module. The significance is evaluated by the null distribution generated through a defined number of random modules. Illustration Credit: Sceyence Studios.
In a recent study, we created a human calcification endophenotype module by mapping vascular calcification proteins to the human vascular smooth muscle-specific protein-protein interactome.70 Network proximity analysis demonstrated that the cardiovascular calcification module significantly overlaps with endophenotype modules governing inflammation, thrombosis, and fibrosis in the human interactome and identified 3 drugs, everolimus, temsirolimus, and pomalidomide, targeting the module. The efficacy of these drugs in reducing vascular calcification was confirmed experimentally by treating human coronary artery smooth muscle cells in an in vitro calcification assay, demonstrating that the integrative network analytical approach followed by experimental validation has broad applicability to other diseases.
By incorporating SARS-CoV-2 virus-host protein-protein interactions, transcriptomics, and proteomics into the human interactome, Zhou et al71 used a network proximity metric to reveal the underlying pathogenesis for broad COVID-19-associated disease manifestations and explored a potential treatment, melatonin, for COVID-19 by combining network-based predictions and a propensity score matching observational study of 26 779 individuals from a COVID-19 registry. They also used network proximity analyses of drug targets and human coronaviruses host interactions in the human interactome to prioritize potential anti-human coronaviruses drugs.72 Pulmonary fibrosis is one of the long-term complications that appears in some COVID-19 survivors. In a recent study, novel gene signatures of post-COVID pulmonary fibrosis were revealed through network analyses combined with the SARS-CoV-2-induced transcriptome at single-cell resolution. Several potential drug candidates have been proposed for targeting pulmonary fibrosis caused by COVID-19 by quantifying the network proximity between the drug targets and the pulmonary fibrosis signatures in the human interactome.73
A recent breakthrough in discovering potential host therapeutic targets for SARS-CoV-2 infection was made through generating a comprehensive SARS-CoV-2–human protein–protein network of 739 high-confidence interactions using a high-throughput yeast 2-hybrid screening experiment.74 In this network, many known SARS-CoV-2 host factors were validated and 361 novel ones were revealed. Network-based screens of FDA-approved or investigational drugs using network proximity analysis identified 23 candidates for COVID-19. One of these drugs, carvedilol, was validated to show clinical benefits for patients with COVID-19 in an electronic health records analysis and antiviral properties in a human lung cell line infected with SARS-CoV-2. Similarly, Kim et al75 created another proteome-scale map of the SARS-CoV-2–human interactome through which the human genetic architecture for COVID-19 severity was revealed, offering an important data resource for discovering potential therapeutic targets. Although SARS-CoV-2-host interactomes are great resources for understanding viral replication, knowledge of downstream pathways, including all potential viral receptors, host cell proteases, and cofactors, is necessary for the validation of critical host machineries. For this purpose, Liu et al76 applied both affinity purification mass spectrometry and the complementary proximity-based labeling mass spectrometry method on 29 viral open reading frames and 18 host proteins with potential roles in viral replication to map the interactions relevant to viral processing. Some drug candidates identified through this approach, including methotrexate, were validated to have antiviral effects using an image-based drug screen assay.
A potential limitation of network proximity is that it cannot distinguish on-target therapeutic effects from off-target adverse effects until further experimental validation is performed. Nevertheless, it can be empowered by inclusion of other methods such as machine learning. In Gysi et al,77 we assembled algorithms relying on graph convolutional network-based machine learning, network diffusion, and network proximity to rank 6340 drugs for their expected efficacy against SARS-CoV-2 infection. We used as ground truth 918 drugs experimentally screened in VeroE6 cells to test the predictions. Importantly, no single predictive algorithm offers consistently reliable outcomes across all data sets and metrics, suggesting the complexity and challenge of the drug repositioning task and the necessity of integrating different methods for optimization. This work offers a useful methodological pathway to identify repurposable drugs for other infectious diseases or treating other disease manifestations of COVID-19, including cardiovascular manifestations.
Despite different methods that have been developed for drug repositioning, few of them consider potential side effects that could worsen the condition of patients. Although FDA-approved drugs have passed safety assessment, minimizing the side effects of repurposed drugs for a disease is still beneficial. This issue is important to consider because unique drug-by-disease interactions may occur that would not have been predicted (or excluded) based on prior clinical trials in which the drug was used to treat a different illness. Focusing on 9 cardiovascular disorders, we have developed an adjusted network-based similarity measure implemented by the algorithm originally applied to COVID-1978 to reposition drugs for CVDs while, at the same time, considering 2 side effects of drug candidates—long QT syndrome and drug-induced asthma.79 This method formulated both disease disorders and side effects as network modules in the human interactome and considered those drug candidates that are proximal to disease modules but far from side-effects modules as ideal. A list of drug candidates for CVDs that are unlikely to produce common, adverse side-effects was provided in this work. This study demonstrated the possibility of incorporating side effects in drug repositioning and paved a way towards identifying drug treatments with lower risks of adverse effects. This approach could be adopted to the identification of drugs repurposed that have therapeutic effects for COVID-19 with minimal adverse effects.
Concluding Remarks and Future Perspectives
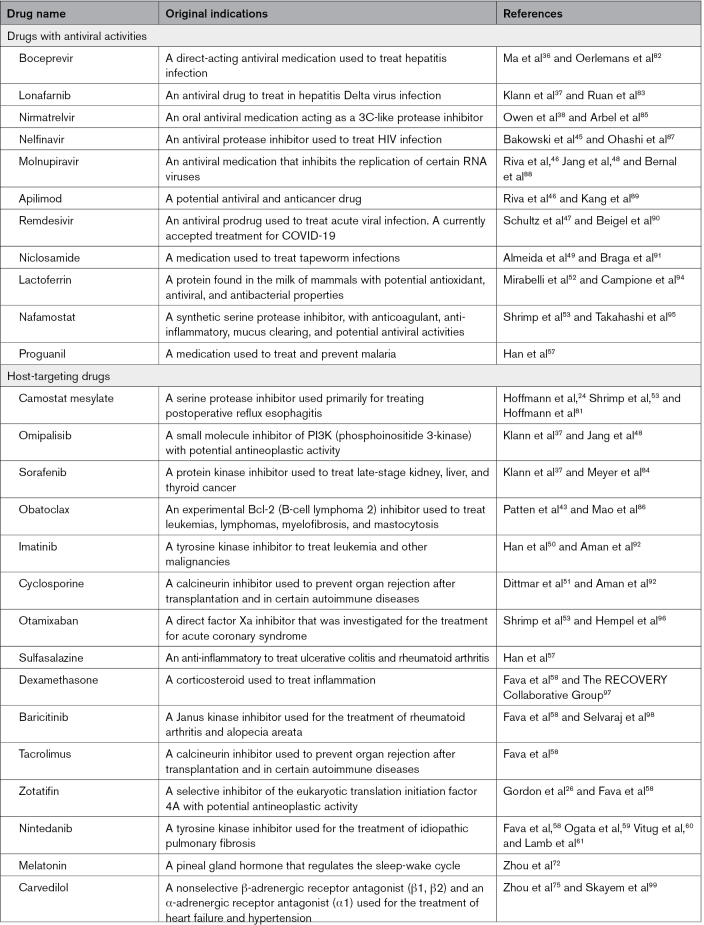
The COVID-19 pandemic has been challenging to address using conventional drug development methods. For this reason, drug repositioning, or the application of approved drugs for new purposes, has been used to accelerate drug discovery. Experimental approaches based on cell entry mechanisms of SARS-CoV-2 provide drug candidates that could potentially inhibit viral infection and replication. High-throughput screening assays offer an unbiased, efficient way to identify approved and investigational drugs for COVID-19. High-throughput omics technologies have generated significant of SARS-CoV-2-induced transcriptomics, proteomics, and other omics data, which facilitate drug repositioning for COVID-19, as well. Using a combination of rigorous computational and network medicine approaches that rely on drug-target interactions and are guided by molecular interaction networks, especially the protein-protein interaction network, potential therapies have been identified that are undergoing clinical testing. Table lists some repositioned drug candidates we discussed in this review which are now used for COVID-19 or with promising potential for COVID-19. Noted that some of these candidates do not have consistent anti-COVID-19 therapeutic effects across different studies and need further large-population-based validations.
Table.
Select Drugs That Have Been or Can Potentially be Repositioned for COVID-19
Despite these advances, designing an optimal, generalizable algorithm for drug repurposing in general and for SARS-CoV-2 and its cardiovascular manifestations in particular remains a challenge. More recently, attention has turned to long-term post-COVID complications, including cardiovascular complications and lung diseases,11,99 with drug treatments suggested for COVID-19–induced pulmonary fibrosis,53,54,67 as well. With more evolving knowledge of the pathophysiological mechanisms underlying post-COVID cardiovascular complications, the methodological approaches available to surmount this challenge are expanding with a promise of direct clinical application advancing rapidly. Coupled with a computational strategy for identifying potential adverse effects of repurposed drugs, this general approach to drug development offers a clear path forward for this and future epidemics requiring timely effective response.
Article Information
Sources of Funding
This work is supported, in part, by NIH grants U01 GH 007691, R01 HL155097, and R01 HL155106; AHA grant 957729, and EU Horizon Health 2021 grant 10157619.
Disclosures
J.L. is scientific co-founder of Scipher Medicine, Inc., a network medicine-based therapeutics company. R.-S.W. has no conflicts of interest to declare.
Nonstandard Abbreviations and Acronyms
- ACE2
- angiotensin-converting enzyme 2
- CAD
- coronary artery disease
- CVD
- cardiovascular diseases
- FDA
- Food and Drug Administration
- HIF1A
- hypoxia inducible factor 1 subunit alpha
- JAK-STAT
- janus kinase-signal transducer and activator of transcription
- RUNX1
- RUNX family transcription factor 1
- TMPRSS2
- transmembrane serine protease 2
For Sources of Funding and Disclosures, see page 1384.
References
- 1.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3 [DOI] [PubMed] [Google Scholar]
- 2.Murakami N, Hayden R, Hills T, Al-Samkari H, Casey J, Sorbo LD, Lawler PR, Sise ME, Leaf DE. Therapeutic advances in COVID-19. Nat Rev Nephrol. 2023;19:38–52. doi: 10.1038/s41581-022-00642-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida-Pititto B, Dualibb PM, Zajdenverg L, Dantas JR, Souza FD, Rodacki M, Bertoluci MC. Severity and mortality of COVID 19 in patients with diabetes, hypertension and cardiovascular disease: a meta-analysis. Diabetology & Metabolic Syndrome. 2020;12:75. doi: 10.1186/s13098-020-00586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae SA, Kim SR, Kim MN, Shim WJ, Park SM. Impact of cardiovascular disease and risk factors on fatal outcomes in patients with COVID-19 according to age: a systematic review and meta-analysis. Heart. 2021;107:373–380. doi: 10.1136/heartjnl-2020-317901 [DOI] [PubMed] [Google Scholar]
- 5.Guan WJ, Liang WH, Zhao Y, Liang H-R, Chen Z-S, Li Y-M, Liu X-Q, Chen R-C, Tang C-L, Wang T, et al. ; China Medical Treatment Expert Group for COVID-19. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020;55:2000547. doi: 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan WJ, Liang WH, He JX, Zhong NS. Cardiovascular comorbidity and its impact on patients with COVID-19. Eur Respir J. 2020;55:2001227. doi: 10.1183/13993003.01227-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hessami A, Shamshirian A, Heydari K, Pourali F, Alizadeh-Navaei R, Moosazadeh M, Abrotan S, Shojaie L, Sedighi S, Shamshirian D, et al. Cardiovascular diseases burden in COVID-19: systematic review and meta-analysis. Am J Emerg Med. 2021;46:382–391. doi: 10.1016/j.ajem.2020.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clerkin KJ, Fried JA, Raikhelkar J, Sayer G, Griffin JM, Masoumi A, Jain SS, Burkhoff D, Kumaraiah D, Rabbani L, et al. COVID-19 and cardiovascular disease. Circulation. 2020;141:1648–1655. doi: 10.1161/CIRCULATIONAHA.120.046941 [DOI] [PubMed] [Google Scholar]
- 9.Chung MK, Zidar DA, Bristow MR, Cameron SJ, Chan T, Harding CV, III, Kwon DH, Singh T, Tilton JC, Tsai TJ, et al. COVID-19 and cardiovascular disease from bench to bedside. Circ Res. 2021;128:1214–1236. doi: 10.1161/CIRCRESAHA.121.317997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishiga M, Wang DW, Han Y, Lewis DB, Wu JC. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat Rev Cardiol. 2020;17:543–558. doi: 10.1038/s41569-020-0413-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y, Xu E, Bowe B, Al-Aly Z. Long-term cardiovascular outcomes of COVID-19. Nat Med. 2022;28:583–590. doi: 10.1038/s41591-022-01689-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farshidfar F, Koleini N, Ardehali H. Cardiovascular complications of COVID-19. JCI Insight. 2021;6:e148980. doi: 10.1172/jci.insight.148980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Wang F, Tang J, Nussinov R, Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit Health. 2020;2:e667–e676. doi: 10.1016/S2589-7500(20)30192-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guy RK, DiPaola RS, Romanelli F, Dutch RE. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829–830. doi: 10.1126/science.abb9332 [DOI] [PubMed] [Google Scholar]
- 15.Galindez G, Matschinske J, Rose TD, Sadegh S, Salgado-Albarrán M, Späth J, Baumbach J, Pauling JK. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nature Computational Science. 2021;1:33–41. doi: 10.1038/s43588-020-00007-6 [DOI] [PubMed] [Google Scholar]
- 16.Dotolo S, Marabotti A, Facchiano A, Tagliaferri P. A review on drug repurposing applicable to COVID-19. Brief Bioinform. 2021;22:726–741. doi: 10.1093/bib/bbaa288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koelle K, Martin MA, Antia R, Lopman B, Dean NE. The changing epidemiology of SARS-CoV-2. Science. 2022;375:1116–1121. doi: 10.1126/science.abm4915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sisnieguez CEL, Espeche WG, Salazar MR. Arterial hypertension and the risk of severity and mortality of COVID-19. Eur Respir J. 2020;55:2001148. doi: 10.1183/13993003.01148-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Wu S, Qin M, Jiang W, Liu X. Prevalence of cardiovascular comorbidities in coronavirus disease 2019, severe acute respiratory syndrome, and Middle East respiratory syndrome: pooled analysis of published data. J Am Heart Assoc. 2020;9:e016812. doi: 10.1161/JAHA.120.016812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuno T, Takahashi M, Obata R, Maeda T. Cardiovascular comorbidities, cardiac injury, and prognosis of COVID-19 in New York City. Am Heart J. 2020;226:24–25. doi: 10.1016/j.ahj.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelps M, Christensen DM, Gerds T, Fosbøl E, Torp-Pedersen C, Schou M, Køber L, Kragholm K, Andersson C, Biering-Sørensen T, et al. Cardiovascular comorbidities as predictors for severe COVID-19 infection or death. Eur Heart J Qual Care Clin Outcomes. 2021;7:172–180. doi: 10.1093/ehjqcco/qcaa081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Fang Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Gai S, Wang X, Zeng J, Sun C, Zhao Y, Zheng Z. Single-cell analysis of SARS-CoV-2 receptor ACE2 and spike protein priming expression of proteases in the human heart. Cardiovasc Res. 2020;116:1733–1741. doi: 10.1093/cvr/cvaa191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marijon E, Karam N, Jost D, Perrot D, Frattini B, Derkenne C, Sharifzadehgan A, Waldmann V, Beganton F, Narayanan K, et al. Out-of-hospital cardiac arrest during the COVID-19 pandemic in Paris, France: a population-based, observational study. Lancet Public Health. 2020;5:e437–e443. doi: 10.1016/S2468-2667(20)30117-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho FK, Man KKC, Toshner M, Church C, Celis-Morales C, Wong ICK, Berry C, Sattar N, Pell JP. Thromboembolic risk in hospitalized and nonhospitalized COVID-19 patients: a self-controlled case series analysis of a nationwide cohort. Mayo Clin Proc. 2021;96:2587–2597. doi: 10.1016/j.mayocp.2021.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyöngyösi M, Alcaide P, Asselbergs FW, et al. Long COVID and the cardiovascular system - elucidating causes and cellular mechanisms in order to develop targeted diagnostic and therapeutic strategies: a joint scientific statement of the ESC working groups on cellular biology of the heart and myocardial & pericardial diseases. Cardiovasc Res. 2022;cvac115. doi: 10.1093/cvr/cvac115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman B, Bluemke DA, Lüscher TF, Neubauer S. Long COVID: post-acute sequelae of COVID-19 with a cardiovascular focus. Eur Heart J. 2022;43:1157–1172. doi: 10.1093/eurheartj/ehac031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jone PN, John A, Oster ME, Allen K, Tremoulet AH, Saarel EV, Lambert LM, Miyamoto SD, de Ferranti SD, et al.; American Heart Association Leadership Committee and Congenital Cardiac Defects Committee of the Council on Lifelong Congenital Heart Disease and Heart Health in the Young; Council on Hypertension, and Council on Peripheral Vascular Disease. SARS-CoV-2 infection and associated cardiovascular manifestations and complications in children and young adults: a Scientific Statement from the American Heart Association. Circulation. 2022;145:e1037–e1052. doi: 10.1161/CIR.0000000000001064 [DOI] [PubMed] [Google Scholar]
- 32.Wang RS, Loscalzo J. Uncovering common pathobiological processes between COVID-19 and pulmonary arterial hypertension by integrating omics data. Pulm Circ. 2023;13:e12191. doi: 10.1002/pul2.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Hare M, Amarnani D, Whitmore HAB, An M, Marino C, Ramos L, Delgado-Tirado S, Hu X, Chmielewska N, Chandrahas A, et al. Targeting runt-related transcription factor 1 prevents pulmonary fibrosis and reduces expression of severe acute respiratory syndrome coronavirus 2 host mediators. Am J Pathol. 2021;191:1193–1208. doi: 10.1016/j.ajpath.2021.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong EM, Pereira M, So EY, Wu KQ, Del Tatto M, Wen S, Dooner MS, Dubielecka PM, Reginato AM, Ventetuolo CE, et al. Targeting RUNX1 as a novel treatment modality for pulmonary arterial hypertension. Cardiovasc Res. 2022;118:3211–3224. doi: 10.1093/cvr/cvac001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T, Zhang X, Tarbet B, Marty MT, Chen Y, et al. Boceprevir, gc-376, and calpain inhibitors ii, xii inhibit sars-cov-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klann K, Bojkova D, Tascher G, Ciesek S, Münch C, Cinat J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol Cell. 2020;80:164–174.e4. doi: 10.1016/j.molcel.2020.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784 [DOI] [PubMed] [Google Scholar]
- 38.Quan BX, Shuai H, Xia AJ, Hou Y, Zeng R, Liu XL, Lin GF, Qiao JX, Li WP, Wang FL, et al. An orally available Mpro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron. Nat Microbiol. 2022;7:716–725. doi: 10.1038/s41564-022-01119-7 [DOI] [PubMed] [Google Scholar]
- 39.Kang Y., ChenT, Mui D, Ferrari V, Jagasia D, Scherrer-Crosbie M, Chen Y, Han Y. Cardiovascular manifestations and treatment considerations in COVID-19. Heart. 2020;106:1132–1141. doi: 10.1136/heartjnl-2020-317056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basu D, Chavda VP, Mehta AA. Therapeutics for COVID-19 and post COVID-19 complications: an update. Curr Res Pharmacol Drug Discov. 2022;3:100086. doi: 10.1016/j.crphar.2022.100086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu T, Zheng W, Huang R. High-throughput screening assays for SARS-CoV-2 drug development: current status and future directions. Drug Discov Today. 2021;26:2439–2444. doi: 10.1016/j.drudis.2021.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patten JJ, Keiser PT, Morselli-Gysi D, Menichetti G, Mori H, Donahue CJ, Gan X, Valle I, Geoghegan-Barek K, Anantpadma M, et al. Identification of potent inhibitors of SARS-CoV-2 infection by combined pharmacological evaluation and cellular network prioritization. iScience. 2022;25:104925. doi: 10.1016/j.isci.2022.104925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janes J, Young ME, Chen E, Rogers NH, Burgstaller-Muehlbacher S, Hughes LD, Love MS, Hull MV, Kuhen KL, Woods AL, et al. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc Natl Acad Sci USA. 2018;115:10750–10755. doi: 10.1073/pnas.1810137115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakowski MA, Beutler N, Wolff KC, Kirkpatrick MG, Chen E, Nguyen TTH, Riva L, Shaabani N, Parren M, Ricketts J, et al. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat Commun. 3309;12:2021. doi: 10.1038/s41467-021-23328-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, Burgstaller-Muehlbacher S, Jesus PDD, Teriete P, Hull MV, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz DC, Johnson RM, Ayyanathan K, Miller J, Whig K, Kamalia B, Dittmar M, Weston S, Hammond HL, Dillen C, et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature. 2022;604:134–140. doi: 10.1038/s41586-022-04482-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang WD, Jeon S, Kim S, Lee SY. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc Natl Acad Sci USA. 2021;118:e2024302118. doi: 10.1073/pnas.2024302118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almeida L, da Silva ALN, Rodrigues TS, Oliveira S, Ishimoto AY, Seribelli AA, Becerra A, Andrade WA, Marco A Ataide MA, Caetano CCS, et al. Identification of immunomodulatory drugs that inhibit multiple inflammasomes and impair SARS-CoV-2 infection. Sci Adv. 2022;8:eabo5400. doi: 10.1126/sciadv.abo5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uh S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dittmar M, Lee JS, Whig K, Segrist E, Li M, Kamalia B, Castellana L, Ayyanathan K, Cardenas-Diaz FL, Morrisey EE, et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-CoV-2. Cell Rep. 2021;35:108959. doi: 10.1016/j.celrep.2021.108959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mirabelli C, Wotring JW, Zhang CJ, McCarty SM, Fursmidt R, Pretto CD, Qiao Y, Zhang Y, Frum T, Kadambi NS, et al. Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19. Proc Natl Acad Sci USA. 2021;118:e2105815118. doi: 10.1073/pnas.2105815118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shrimp JH, Janiszewski J, Chen CZ, Xu M, Wilson KM, Kales SC, Sanderson PE, Shinn P, Schneider R, et al. Suite of TMPRSS2 assays for screening drug repurposing candidates as potential treatments of COVID-19. ACS Infect Dis. 2022;8:1191–1203. doi: 10.1021/acsinfecdis.2c00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melms JC, Biermann J, Huang H, Wang Y, Nair A, Tagore S, Katsyv I, Rendeiro AF, Amin AD, Schapiro D, et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595:114–119. doi: 10.1038/s41586-021-03569-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C, Haas DA, Huang Y, Oubraham L, Wang A, et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4 [DOI] [PubMed] [Google Scholar]
- 56.Han N, Hwang W, Tzelepis K, Schmerer P, Yankova E, MacMahon M, Lei W, Katritsis NM, Liu A, Felgenhauer U. Identification of SARS-CoV-2–induced pathways reveals drug repurposing strategies. Sci Adv. 2021;7:eabh3032. doi: 10.1126/sciadv.abh3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fava VM, Bourgey M, Nawarathna PM, Orlova M, Cassart P, Vinh DC, Cheng MP, Bourque G, Schurr E, Langlais D. A systems biology approach identifies candidate drugs to reduce mortality in severely ill patients with COVID-19. Sci Adv. 2022;8:eabm2510. doi: 10.1126/sciadv.abm2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogata H, Nakagawa T, Sakoda S, Ishimatsu A, Taguchi K, Kadowaki M, Moriwaki A, Yoshida M. Nintedanib treatment for pulmonary fibrosis after coronavirus disease 2019. Respirol Case Rep. 2021;9:e00744. doi: 10.1002/rcr2.744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vitug LC, Santiaguel J. Nintedanib as an adjunct treatment in improving lung function of post-COVID-19 pulmonary fibrosis in an elderly patient: a case report. Chest. 2021;160:A2166. doi: 10.1016/j.chest.2021.07.1914 [Google Scholar]
- 60.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939 [DOI] [PubMed] [Google Scholar]
- 61.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdelsayed M, Kort EJ, Jovinge S, Mercola M. Repurposing drugs to treat cardiovascular disease in the era of precision medicine. Nat Rev Cardiol. 2022;19:751–764. doi: 10.1038/s41569-022-00717-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seth IB, Ravi I. Network analyses in systems pharmacology. Bioinformatics. 2009;25:2466–2472. doi: 10.1093/bioinformatics/btp465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang RS, Loscalzo J. Illuminating drug action by network integration of disease genes: a case study of myocardial infarction. Mol Biosyst. 2016;12:1653–1666. doi: 10.1039/c6mb00052e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang RS, Loscalzo J. Network module-based drug repositioning for pulmonary arterial hypertension. CPT: Pharmacometrics Syst Pharmacol. 2021;10:994–1005. doi: 10.1002/psp4.12670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabási AL. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:12576011257601–125760111257601. doi: 10.1126/science.1257601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guney E, Menche J, Vidal M, Barábasi AL. Network-based in silico drug efficacy screening. Nat Commun. 2016;7:10331. doi: 10.1038/ncomms10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng F, Desai RJ, Handy DE, Wang RS, Sebastian Schneeweiss S, Barabási AL, Loscalzo J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nature Communications. 2018;9:2691. doi: 10.1038/s41467-018-05116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song JS, Wang RS, Leopold JA, Loscalzo J. Network determinants of cardiovascular calcification and repositioned drug treatments. FASEB J. 2020;34:11087–11100. doi: 10.1096/fj.202001062R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Y, Hou Y, Shen J, Mehra R, Kallianpur A, Culver DA, Gack MU, Farha S, Zein J, Comhair S, et al. A network medicine approach to investigation and population-based validation of disease manifestations and drug repurposing for COVID-19. PLoS Biol. 2020;18:e3000970. doi: 10.1371/journal.pbio.3000970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020;6:14. doi: 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li A, Chen JY, Hsu CL, Oyang YJ, Huang HC, Juan HF. A single-cell network-based drug repositioning strategy for post-COVID-19 pulmonary fibrosis. Pharmaceutics. 2022;14:971. doi: 10.3390/pharmaceutics14050971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Y, Liu Y, Gupta S, Paramo MI, Hou Y, Mao C, Luo Y, Judd J, Wierbowski S, et al. A comprehensive SARS-CoV-2–human protein–protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets. Nat Biotechnol. 2022;41:128–139. doi: 10.1038/s41587-022-01474-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim DK, Weller B, Lin CW, Sheykhkarimli D, Knapp JJ, Dugied G, Zanzoni A, Pons C, Tofaute MJ, Maseko SB, et al. A proteome-scale map of the SARS-CoV-2–human contactome. Nat Biotechnol. 2022;41:140–149. doi: 10.1038/s41587-022-01475-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X, Huuskonen S, Laitinen T, Redchuk T, Bogacheva M, Salokas K, Pöhner I, Öhman T, Tonduru AK, Hassinen A, et al. SARS-CoV-2-host proteome interactions for antiviral drug discovery. Mol Syst Biol. 2021;17:e10396. doi: 10.15252/msb.202110396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gysi DM, do Valle I, Zitnik M, Ameli A, Gan X, Varol O, Ghiassian SD, Patten JJ, Davey RA, Loscalzo J, et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc Natl Acad Sci USA. 2021;118:e2025581118. doi: 10.1073/pnas.2025581118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fiscon G, Conte F, Farina L, Paci P. SAveRUNNER. A network-based algorithm for drug repurposing and its application to COVID-19. PLoS Comput Biol. 2021;17:e1008686. doi: 10.1371/journal.pcbi.1008686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paci P, Fiscon G, Conte F, Wang RS, Handy DE, Farina L, Loscalzo J. Comprehensive network medicine-based drug repositioning via integration of therapeutic efficacy and side effects. npj Syst Biol Appl. 2022;8:1–12. doi: 10.1038/s41540-022-00221-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoffmann M, Hofmann-Winkler H, Smith JC, Krüger N, Arora P, Sørensen LK, Søgaard OS, Hasselstrøm JB, Winkler M, Hempel T, et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65:103255. doi: 10.1016/j.ebiom.2021.103255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oerlemans R, Ruiz-Moreno AJ, Cong Y, Kumar ND, Velasco-Velazquez MA, Neochoritis CG, Smith J, Reggiori F, Groves MR, Dömling A. Repurposing the HCV NS3–4A protease drug boceprevir as COVID-19 therapeutics. RSC Med Chem. 2021;12:370–379. doi: 10.1039/d0md00367k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ruan Z, Liu C, Guo Y, He Z, Huang X, Jia X, Yang T. SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12). J Med Virol. 2021;93:389–400. doi: 10.1002/jmv.26222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meyer B, Chiaravalli J, Gellenoncourt S, Brownridge P, Bryne DP, Daly LA, Grauslys A, Walter M, Agou F, Chakrabarti LA, et al. Characterizing proteolysis during SARS-CoV-2 infection identifies viral cleavage sites and cellular targets with therapeutic potential. Nat Commun. 5553;12:2021. doi: 10.1038/s41467-021-25796-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arbel R, Sagy YW, Hoshen M, Battat E, Lavie G, Sergienko R, Friger M, Jacob G., Waxman JG, et al. Nirmatrelvir use and severe Covid-19 outcomes during the omicron surge. N Engl J Med. 2022; 387:790–798. doi: 10.1056/NEJMoa2204919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mao B, Le-Trilling VTK, Wang K, Mennerich D, Hu J, Zhao Z, Zheng J, Deng Y, Katschinski B, Xu S, et al. Obatoclax inhibits SARS-CoV-2 entry by altered endosomal acidification and impaired cathepsin and furin activity in vitro. Emerg Microbes Infect. 2022;11:483–497. doi: 10.1080/22221751.2022.2026739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohashi H, Watashi K, Saso W, Shionoya K, Iwanami S, Hirokawa T, Shirai T, Kanaya S, Ito Y, Kim KS, et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience. 2021;24:102367. doi: 10.1016/j.isci.2021.102367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernal AJ, da Silva MMG, Musungaie DB, Kovalchuk E, Gonzalez A, Reyes VD, Martín-Quirós A, Caraco Y, Williams-Diaz A, Brown ML, et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized Patients. N Engl J Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang YL, Chou YY, Rothlauf PW, Liu Z, Soh TK, Cureton D, Case JB, Chen RE, Diamond MS, Whelan SPJ, et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, et al. ; ACTT-1 Study Group Members. Remdesivir for the treatment of Covid-19 - final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Braga L, Ali H, Secco I, Chiavacci E, Neves G, Goldhill D, Penn R, Jimenez-Guardeño JM, Ortega-Prieto AM, Bussani R, et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aman J, Duijvelaar E, Botros L, Kianzad A, Schippers JR, Smeele PJ, Azhang S, Bartelink IH, Bayoumy AA, Bet PM, et al. Imatinib in patients with severe COVID-19: a randomised, double-blind, placebo-controlled, clinical trial. Lancet Respir Med. 2021;9:957–968. doi: 10.1016/S2213-2600(21)00237-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sauerhering L, Kuznetsova I, Kupke A, Meier L, Halwe S, Rohde C, Schmidt J, Morty RE, Danov O, Braun A, et al. Cyclosporin A reveals potent antiviral effects in preclinical models of SARS-CoV-2 infection. Am J Respir Crit Care Med. 2022;205:964–968. doi: 10.1164/rccm.202108-1830LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Campione E, Lanna C, Cosio T, Rosa L, Conte MP, Iacovelli F, Romeo A, Falconi M, Vecchio CD, Franchin E, et al. Lactoferrin as antiviral treatment in COVID-19 management: preliminary evidence. Int J Environ Res Public Health. 2021;18:10985. doi: 10.3390/ijerph182010985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takahashi W, Yoneda T, Koba H, Ueda T, Tsuji N, Ogawa H, Asakura H. Potential mechanisms of nafamostat therapy for severe COVID-19 pneumonia with disseminated intravascular coagulation. Int J Infect Dis. 2021;102:529–531. doi: 10.1016/j.ijid.2020.10.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hempel T, Elez K, Krüger N, Raich L, Shrimp JH, Danov O, Jonigk D, Braun A, Shen M, Hall MD, et al. Synergistic inhibition of SARS-CoV-2 cell entry by otamixaban and covalent protease inhibitors: pre-clinical assessment of pharmacological and molecular properties. Chem Sci. 2021;12:12600–12609. doi: 10.1039/d1sc01494c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.The RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021; 384:693–704. doi: 10.1056/NEJMoa2021436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Selvaraj V, Finn A, Lal A, Khan MS, Dapaah-Afriyie K, Carino GP. Baricitinib in hospitalised patients with COVID-19: a meta-analysis of randomized controlled trials. EClinicalMedicine. 2022;49:101489. doi: 10.1016/j.eclinm.2022.101489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Skayem C, Ayoub N. Carvedilol and COVID-19: a potential role in reducing infectivity and infection severity of SARS-CoV-2. Am J Med Sci. 2020;360:300. doi: 10.1016/j.amjms.2020.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aronson KI, Podolanczuk AJ. Lungs after COVID-19: evolving knowledge of post-COVID-19 interstitial lung disease. Ann Am Thorac Soc. 2021;18:773–774. doi: 10.1513/AnnalsATS.202102-223ED [DOI] [PMC free article] [PubMed] [Google Scholar]