Abstract
The photoperiodic response is critical for plants to adjust their reproductive phase to the most favorable season. Wheat heads earlier under long days (LD) than under short days (SD) and this difference is mainly regulated by the PHOTOPERIOD1 (PPD1) gene. Tetraploid wheat plants carrying the Ppd-A1a allele with a large deletion in the promoter head earlier under SD than plants carrying the wildtype Ppd-A1b allele with an intact promoter. Phytochromes PHYB and PHYC are necessary for the light activation of PPD1, and mutations in either of these genes result in the downregulation of PPD1 and very late heading time. We show here that both effects are reverted when the phyB mutant is combined with loss-of-function mutations in EARLY FLOWERING 3 (ELF3), a component of the Evening Complex (EC) in the circadian clock. We also show that the wheat ELF3 protein interacts with PHYB and PHYC, is rapidly modified by light, and binds to the PPD1 promoter in planta (likely as part of the EC). Deletion of the ELF3 binding region in the Ppd-A1a promoter results in PPD1 upregulation at dawn, similar to PPD1 alleles with intact promoters in the elf3 mutant background. The upregulation of PPD1 is correlated with the upregulation of the florigen gene FLOWERING LOCUS T1 (FT1) and early heading time. Loss-of-function mutations in PPD1 result in the downregulation of FT1 and delayed heading, even when combined with the elf3 mutation. Taken together, these results indicate that ELF3 operates downstream of PHYB as a direct transcriptional repressor of PPD1, and that this repression is relaxed both by light and by the deletion of the ELF3 binding region in the Ppd-A1a promoter. In summary, the regulation of the light mediated activation of PPD1 by ELF3 is critical for the photoperiodic regulation of wheat heading time.
Author summary
The coordination of reproductive development with the optimal season for seed production is critical to maximize grain yield in crop species. Plants can perceive the length of the day or night (photoperiod) and use this information to anticipate seasonal changes. In most eudicot plants, CONSTANS plays a central role in the perception of photoperiod, but in wheat the main photoperiod gene is PHOTOPERIOD1 (PPD1). In this study, we show that the clock gene EARLY FLOWERING 3 (ELF3) regulates the phytochrome-mediated light activation of PPD1. Loss-of-function mutations in ELF3 result in the upregulation of PPD1 at dawn, and in early heading under both long and short days, even in the absence of PHYB. A deletion in the PPD1 promoter including an ELF3 binding region also results in earlier heading under short days, indicating that ELF3 acts as a direct transcriptional repressor of PPD1. This study shows that ELF3 plays a critical role in the wheat photoperiod pathway by regulating the light signal between the phytochromes and PPD1. ELF3 provides an additional entry point to engineer heading time in wheat, an important trait for the development of better adapted varieties to a changing environment.
Introduction
Crop productivity depends on the precise alignment of flowering with the most favorable season for seed production and grain filling. For that to happen, plants need to anticipate the proximity of the favorable season to initiate the reproductive phase in a timely manner. The duration of days and nights (photoperiod) provides seasonal information that plants have evolved to sense and use to this end [1].
In Arabidopsis and other eudicot species CONSTANS (CO) is the main photoperiodic gene [2–4], but recent studies in wheat have shown that combined loss-of-function mutations in CO1 and its close paralog CO2 (co1 co2) have a limited effect on wheat photoperiodic response [5]. The earlier heading of wheat under long days (LD) than under short days (SD) is mainly regulated by the PHOTOPERIOD1 (PPD1) gene, also known as PSEUDO RESPONSE REGULATOR 37 (PRR37) [6,7]. Large deletions in the promoter of the A-genome (Ppd-A1a) [8] or the D-genome (Ppd-D1a) [6] homoeologs result in altered gene expression and earlier heading under SD than in genotypes carrying the wildtype alleles (Ppd1b). These deletions share a common ~900 bp region that has been hypothesized to include binding sites for one or more transcriptional repressors [8]. Wheat genotypes carrying the Ppd1b alleles head extremely late under SD and are referred to as photoperiod sensitive (PS). Genotypes carrying any dominant Ppd1a allele are referred to as photoperiod insensitive (PI), even though they still head significantly earlier under LD than under SD. Differences between PS and PI wheats have been shown to be associated with grain productivity in different environments [9,10].
The acceleration of heading time by PPD1 requires its transcriptional activation by light, which is mediated by phytochromes PHYB and PHYC [11–13]. Phytochromes are dimeric proteins that function both as red and far-red light sensors and as temperature sensors [14,15]. Phytochromes can transition from an inactive “Pr” form to an active “Pfr” form upon absorption of red light. In the darkness or upon absorption of far-red light, phytochromes revert to the inactive “Pr” state [16]. The rate of this dark reversion is temperature-dependent providing phytochromes the ability to perceive both light and temperature signals and integrate them to regulate photo- and thermo-morphogenetic processes [15].
The role of wheat PHYB and PHYC in the induction of PPD1 is particularly evident in night-break (NB) experiments, where a 15-min pulse of white light in the middle of the SD long nights (16h) is sufficient to induce PPD1 expression. However, at least two weeks of NBs (or LDs) are necessary to accelerate heading time [17]. Although 15 NBs accelerate heading time of PS wheat plants by more than three months, the same NBs in wheat lines carrying phyB or phyC loss-of-function mutations fail to induce PPD1 expression and plants head extremely late (~160–170 d) [17]. Similarly, wheat lines carrying ppd1 loss-of-function mutations are late heading under both LD (~100–130 d) [5,18] and NB (>150 d) conditions [17]. Taken together, these results indicate that PPD1 is central to the wheat NB and photoperiodic responses and that the duration of the night is critical for the perception of photoperiodic differences in this species.
The transcriptional activation of PPD1 by light results in the induction of FLOWERING LOCUS T1 (FT1) expression in the leaves [5,17]. FT1 encodes a protein (florigen) that is thought to migrate through the phloem to the shoot apical meristem in wheat as shown in Arabidopsis and rice [19,20]. FT1 interacts with 14-3-3 and FDL proteins to form a florigen activation complex that binds to the promoter of the MADS-box transcription factor VERNALIZATION1 (VRN1), which is critical to trigger the transition of the shoot apical meristem from the vegetative to the reproductive phase [21,22]. The upregulation of VRN1 results in the downregulation of the flowering repressor VERNALIZATION2 (VRN2), generating a positive feedback loop that accelerates heading time [23–25]
Significant genetic interactions have been detected between PPD1 and EARLY FLOWERING 3 (ELF3) in the regulation of heading time in both wheat and barley [26–28]. In Arabidopsis, it has been shown that ELF3 is part of the Evening Complex (EC), an important component of the circadian clock, that also includes LUX ARRHYTHMO (LUX) and EARLY FLOWERING 4 (ELF4) [29]. ELF3 functions as an adaptor between LUX and ELF4 and as a hub for multiple protein interactions [29]. In barley and wheat, loss-of-function mutations in ELF3 [26,27,30] or LUX [31–33] result in very early heading under both LD and SD conditions, suggesting that in these species the EC is critical for the photoperiodic response. Natural allelic variation at ELF3 has also been associated with differences in heading time in diploid Triticum monococcum and in hexaploid wheat [26,34,35].
In Arabidopsis, the expression of ELF3, ELF4, and LUX overlaps and peaks at dusk, maximizing their repressive effect early in the night [36]. Chromatin immunoprecipitation studies have shown that the EC acts as a direct transcriptional repressor of the circadian morning loop genes PSEUDORESPONSE REGULATOR 7 (PRR7) and PRR9 in Arabidopsis [37,38] and PRR37, PRR73, PRR95, GI, and GHD7 in rice [39] (GHD7 is the rice ortholog of the wheat LD flowering repressor VRN2 [40,41]). Since Arabidopsis PRR7/PRR3 are the closest homologs of wheat and barley PPD1, and elf3 mutants in barley and Brachypodium show altered PPD1 expression profiles [27,42], we hypothesized that a similar regulation of PPD1 by ELF3 may contribute to their observed epistatic interactions on heading time in the temperate cereals.
In this study, we show that the wheat ELF3 protein interacts with PHYB and PHYC, is modified by light, and binds to the promoter of PPD1. We also show that the repressed expression of PPD1 in the phyB mutant is restored in the elf3 phyB combined mutant, which flowers almost as early as the elf3 mutant. By contrast, the combined elf3 ppd1 mutant heads significantly later than elf3. Based on our results, we propose that ELF3 acts as a direct repressor of PPD1 and plays a critical role in the regulation of the light signals between the phytochromes and PPD1.
Results
The heading time delay of the phyB mutant mostly disappears in the absence of ELF3
Given the documented interactions between ELF3 and PHYB in Arabidopsis [29,43,44], we hypothesized that the large delay on wheat heading time observed in the phyB mutant in previous studies [12,13] could be mediated by ELF3. To test this, we generated loss-of-function mutants phyB, elf3 and elf3 phyB (Fig A in S1 Text) in two different tetraploid wheat cultivar Kronos backgrounds, one carrying the photoperiod insensitive (PI) Ppd-A1a allele with a promoter deletion (Fig A in S1 Text) and the other one carrying the photoperiod sensitive (PS) Ppd-A1b allele with an intact promoter. We evaluated these lines in growth chambers for heading time under SD (8 h light, 16 h dark) and LD (16 h light, 8 h dark).
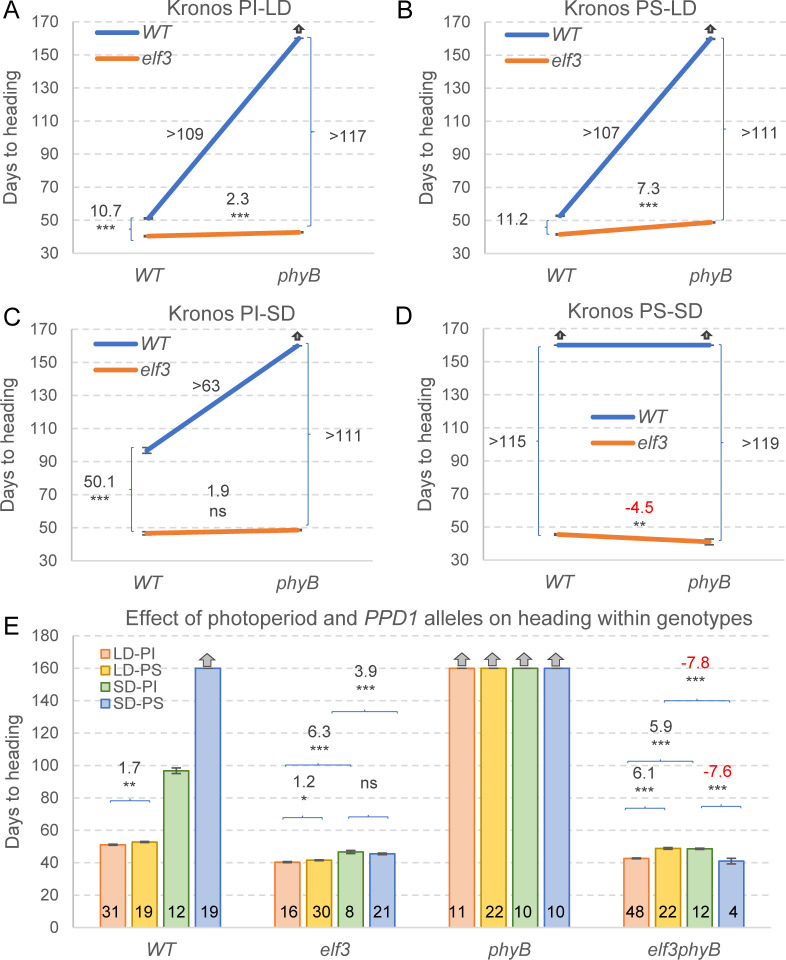
Under LD, strong genetic interactions between ELF3 and PHYB were detected in both PS and PI plants (Fig 1A and 1B). The phyB mutant failed to head before the experiment was terminated at 160 d, but the combined elf3 phyB mutant headed in 43 d in PI (Fig 1A) and in 49 d in PS (Fig 1B). In both genetic backgrounds, the elf3 mutation accelerated heading time by 11 d in the presence of the wildtype PhyB allele, but more than 110 days in the phyB mutant background. The early heading time of the elf3 mutant was associated with a reduced number of spikelets per spike as reported previously [26], but this number was restored to wildtype levels in elf3 phyB (Fig B in S1 Text and Data H in S1 Data).
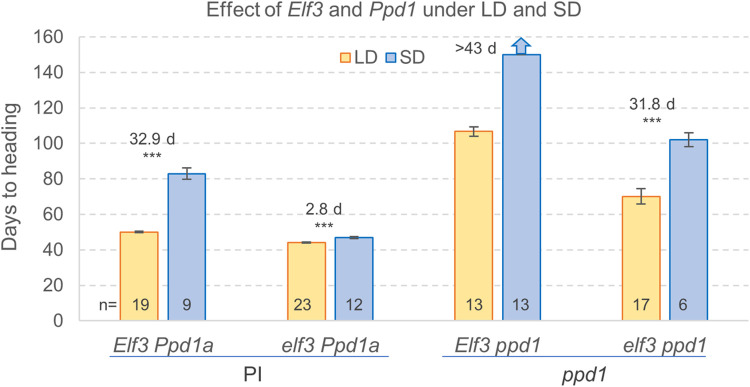
Fig 1. Effect of phyB and elf3 mutations on wheat heading time.
(A-B) Long days (LD). (C-D) Short days (SD). (A and C) Kronos photoperiod insensitive (PI) background and (B and D) photoperiod sensitive (PS) background. (E) Effect of the different photoperiods and genetic backgrounds on heading time within each mutant combination. Error bars are s.e.m. Numbers in the base of the bars indicate the number of plants analyzed in each genotype / photoperiod combination. * = P < 0.05, ** = P < 0.01, *** = P < 0.001 (Tukey tests). No statistical tests were performed for the plants that failed to head at 160 d when the experiment was terminated (indicated by gray arrows). Raw data and statistics are in Data A in S1 Data.
Under SD, PI plants carrying the Elf3 and PhyB wildtype alleles headed in 97 d (Fig 1C), whereas PS plants did not head (Fig 1D), as in previously published studies [5]. Similarly, both PI and PS phyB mutants failed to head before the end of the experiment. By contrast, plants carrying only the elf3 mutant allele headed very early in both PS and PI backgrounds (~46 d). Interestingly, when the elf3 mutation was present, the phyB loss-of-function mutation was associated with a non-significant 2 d delay in heading time in PI (Fig 1C), but with a 4.5 d significant acceleration in PS (Fig 1D) relative to the wildtype PhyB allele.
The same data used in Fig 1A–1D is reorganized in Fig 1E to facilitate comparisons among photoperiods and PPD1 alleles in the different mutant backgrounds. The wildtype plants headed significantly later under SD than under LD, but these differences were greatly reduced in the absence of ELF3. In the elf3 single mutant, SD was associated with a significant delay in heading time in both PI (6.3 d) and PS (3.9 d). However, in the combined elf3 phyB mutant, SD was associated with a significant delay in heading time in PI (5.9 d), but a significant acceleration in PS (7.8 d, P < 0.001, Fig 1E). Earlier heading under SD than under LD has been reported before both for the phyB and phyC mutants in Kronos [11,12], an interesting result considering that wheat is a LD-plant.
In summary, the heading time results indicate that i) the large delays in heading time caused by the phyB mutation are mediated in large part by ELF3, ii) the elf3 mutant is still able to respond to photoperiod, and iii) the elf3 phyB mutant heads earlier under LD than under SD in the PI background, but later in the PS background.
PPD1 and FT1 are downregulated in phyB but expression is restored in elf3 phyB
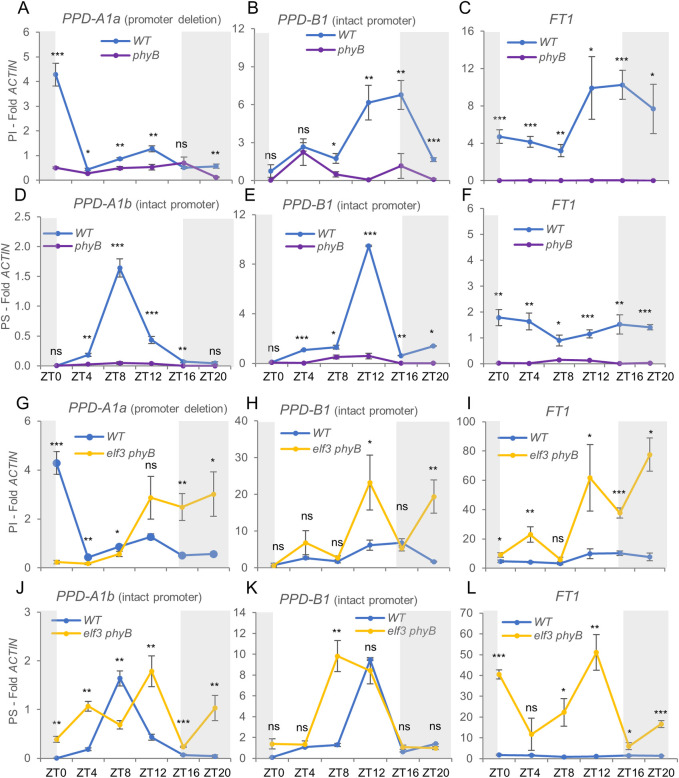
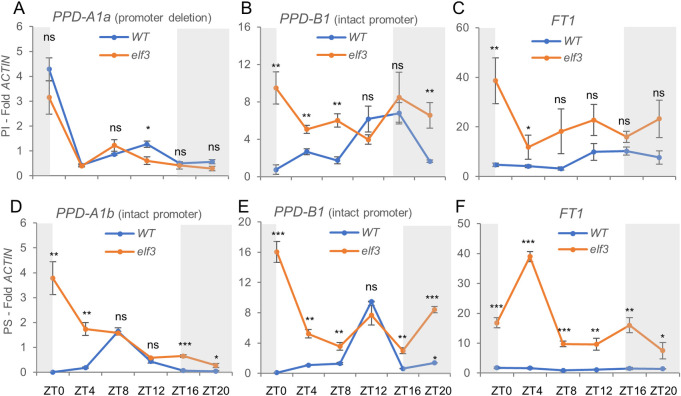
To better understand the strong genetic interaction observed between ELF3 and PHYB on heading time, we explored the expression of PPD-A1, PPD-B1, and FT1 in phyB and elf3 phyB mutants grown under LD (Fig 2). In PI, the PPD-A1a expression peak observed in the wildtype at dawn was no longer detected in the phyB mutant (Fig 2A), which also showed a significant downregulation of PPD-B1 from ZT8 to ZT20 (Fig 2B). The downregulation of both PPD1 homoeologs in phyB was reflected in reduced transcript levels of FT1 (Fig 2C) and VRN1 (Fig C in S1 Text) which, together with the upregulation of the VRN2 repressor (Fig C in S1 Text), resulted in late heading (Fig 1). The PI phyB mutant also showed upregulation of CO1 (ZT0-ZT8) and CO2 (ZT4-ZT8) (Fig C in S1 Text). In PS, the phyB mutant showed an even more pronounced downregulation of PPD-A1b and PPD-B1, which resulted in undetectable FT1 levels (Fig 2D–2F).
Fig 2. Transcript levels of PPD1 and FT1 in phyB and phyB elf3 mutants.
Samples were collected from leaves of 5-week-old Kronos photoperiod insensitive (PI) and sensitive (PS) plants grown under LD. (A-C) Wildtype vs. phyB in PI. (D-E) Wildtype vs. phyB in PS. (G-I) Wildtype vs. elf3 phyB in PI. (J-L) Wildtype vs. elf3 phyB in PS. (A & G) PPD-A1a allele with a deletion in the promoter. (D & J) PPD-A1b allele with intact promoter. (B, E, H, & K) PPD-B1 photoperiod sensitive allele. (C, F, I, & L) FT1 (both homoeologs). Expression in single elf3 mutants is discussed later in the ELF3 x PPD1 section. ACTIN was used as endogenous control. Error bars are s.e.m based on 5 biological replications. ns = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001 based on t-tests between mutants and wildtype at the different time points. Raw data and statistics are available in Data B in S1 Data.
The altered expression profile of PPD-A1a also resulted in an altered PPD-B1 expression profile (Fig 2B and 2E), a phenomenon that has been previously reported for different PPD1a alleles in hexaploid wheat [45,46]. These results suggest the existence of a feed-back regulatory loop where some of the genes differentially regulated by the PPD1a alleles affect the regulation of the other PPD1b homoeologs.
The transcriptional repression of PPD1 in the phyB mutant was greatly reduced in the elf3 phyB combined mutant (Fig 2G, 2H, 2J and 2K), resulting in higher FT1 (Fig 2I and 2L) and VRN1 (Fig C in S1 Text) transcript levels than in phyB. In elf3 phyB, VRN2 transcript levels remained elevated, whereas the upregulation observed in phyB for CO1 and CO2 at ZT4-ZT8 was no longer significant. In addition, CO1 showed a significant downregulation relative to the wildtype at dawn in elf3 phyB (Fig C in S1 Text). In summary, the increased expression of PPD1, FT1 and VRN1 in phyB elf3 correlates well with its early heading relative to phyB and the wildtype (Fig 1).
ELF3 interacts with PHYB and PHYC in yeast-two-hybrid assays
ELF3 is expressed at high levels in the leaves (4 to 15-fold ACTIN) and shows similar transcriptional profiles under SD and LD (Fig D in S1 Text). The phyC mutant showed no significant differences with the wildtype on ELF3 expression at any time point, while the phyB mutant showed a marginally significant downregulation (P < 0.05) between ZT12 and ZT20 relative to the wildtype (Fig D in S1 Text and Data J in S1 Data).
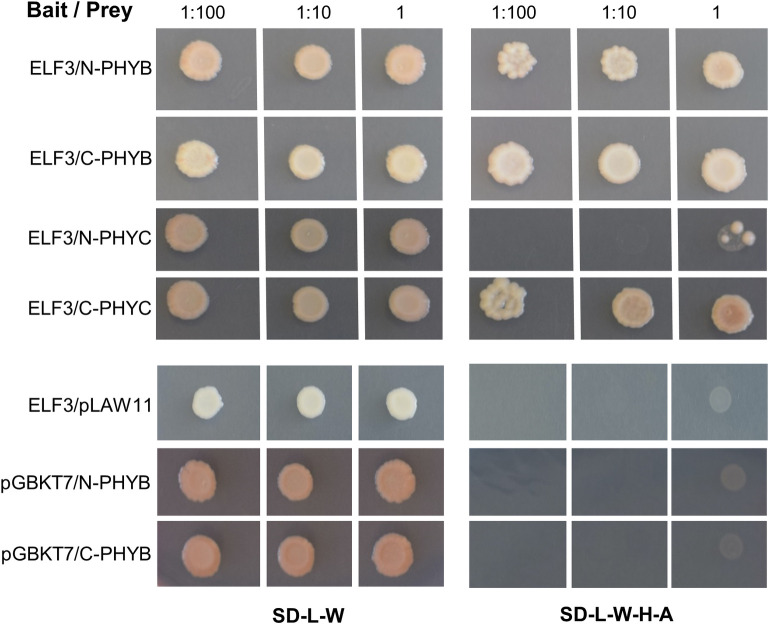
Since the transcriptional regulation of ELF3 by PHYB and PHYC was insufficient to explain the observed genetic interactions in heading time, we looked at possible interactions at the protein level using yeast-two-hybrid (Y2H) assays. The full-length coding region of the ELF3 gene was fused to the GAL4 DNA-binding domain and used as bait. The phytochrome genes PHYC and PHYB were each divided into two parts encoding the N-terminal photosensory and C-terminal regulatory regions, respectively. The N-terminal region included 625 amino acids in PHYB (N-PHYB) and 600 amino acids in PHYC (N-PHYC). The C-terminal region encoded amino acids 626–1166 in PHYB (C-PHYB) and amino acids 601 to 1139 in PHYC (C-PHYC).
ELF3 showed strong interactions with both C-PHYB and C-PHYC. However, its interaction with the N-PHYB was much stronger than with N-PHYC (Fig 3). Autoactivation tests showed no interaction with empty vectors for ELF3 bait and PHYB preys (Fig 3). N-PHYC and C-PHYC preys have been tested previously and showed no autoactivation [11].
Fig 3. Yeast-two-hybrid interactions between ELF3 and phytochromes PHYB and PHYC.
SD medium lacking Leucine and Tryptophan (-L-W) was used to select for yeast transformants containing both bait and prey vectors. Interactions were determined on SD media lacking Leucine, Tryptophan, Histidine and Adenine (-L-W-H-A). Autoactivation was tested for ELF3 bait using the empty prey vector pLAW11, and for N-PHYB and C-PHYB preys using the empty bait vector pGBKT7. We did not add a chromophore, so we are likely seeing the interaction with the inactive Pr form.
The ELF3 protein is modified by light
Given that the ELF3 protein directly interacts with phytochromes, we hypothesized that light may affect its stability. To monitor the ELF3 protein, we generated transgenic Kronos PS lines constitutively expressing a C-terminal HA-tagged ELF3 under the maize UBIQUITIN promoter (UBI::ELF3-HA). The transgene was expressed at high levels in the leaves of both Kronos PS and the elf3 mutant, and was associated with the downregulation of PPD1 and FT1 and delayed heading, including in plants carrying the elf3 mutation (Fig E in S1 Text). These results confirmed that the UBI::ELF3-HA transgene is functional and can complement the elf3 mutant phenotype. The ELF3-HA protein detected by immunoblotting using an anti-HA antibody was approximately 110 kDa, which is slightly higher than the predicted 88.5 kDa both in protein extracted from UBI::ELF3-HA transgenic plants and from transformed protoplasts (Fig F in S1 Text). This was the only band detected in the blots (except for rubisco) and it disappeared in the non-transgenic controls confirming that it is ELF3-HA. A higher than expected size for the ELF3 protein was also observed in western blots in rice [39].
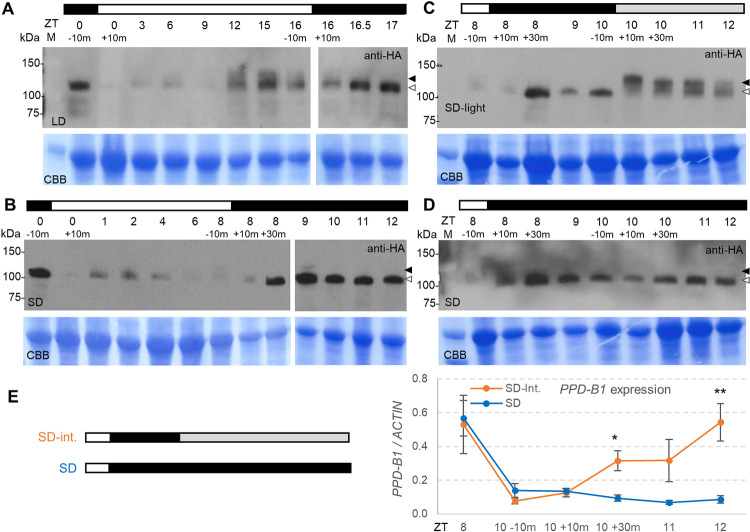
We then looked at the stability of the ELF3 protein in UBI::ELF3-HA transgenic plants grown under LD (Fig 4A) and SD (Fig 4B) by immunoblotting using an anti-HA antibody. Under LD, two bands were detected during the day. The intensity of these bands varied during the day but were stronger later in the afternoon. The higher and more diffuse band disappeared in the dark, whereas the lower band became more intense after the lights were turned off (Fig 4A). Under SD, we also observed a strong lower ELF3 band 30 min after the lights were turned off, which faded away when the lights were turned on. A higher, fainter, and more diffused band fluctuated during the day (Fig 4B).
Fig 4. ELF3-HA protein in UBI::ELF3-HA transgenic plants grown under LD, SD and SD interrupted night (SD-int.).
ELF3 protein levels were analyzed by immunoblotting using an anti-HA antibody (samples were harvested at the indicated ZT times). (A) Plants grown under LD. (B) Plants grown under SD. (C) Plants were grown under SD, but on the day when samples were collected, lights were turned on at ZT10, 2 h after the start of the night (SD-int.). (D) Plants under SD grown simultaneously with those in B but without turning on the light before sampling (collected at the same time points as in C). The black arrowhead indicates the higher and more diffuse band and the white arrowhead the sharper lower band detected in the dark. The bottom panel is a Coomassie Brilliant Blue (CBB) stained membrane used as a loading control. The white bar indicates lights on and the black bar lights off. The gray bar in C indicates that the lights were turned on during the subjective night. (E) Quantitative reverse transcription PCR (qRT-PCR) analysis of PPD-B1 expression in leaves collected at the same time points as in C. Raw data and statistics are available in Data C in S1 Data.
The difference between the two ELF3 bands is easier to see in the experiment in Fig 4C, where we grew plants under SD and, on the night of the experiment, we turned on the lights 2 h after the start of the night and kept them on during the subjective night (gray bar, Fig 4C). A large increase in intensity of a higher and more diffused band and a decrease in intensity of the lower band were observed 10 min after the lights were turned on at ZT10 (Fig 4C), confirming that the transition between the lower and higher bands was mediated by light, rather than by the circadian clock alone. However, the intensity of the upper band 10 min after the lights were turned on was very different at ZT0 than at ZT10 (Fig 4C), suggesting that not only the light but also the time of the day is important in the regulation of the ELF3 protein. In the SD control where lights were not turned on during the night, the higher band was not detected in the dark (Fig 4D).
In RNA samples collected from leaves at the same time points as in Fig 4C and 4D, PPD1 was significantly upregulated 30 min after the lights were turned on (Fig 4E, SD-int.), but remained low in the SD control where the night was not interrupted by light. These results suggest that the lower ELF3 band is the critical active form for the repression of PPD1 during the night.
Loss-of-function mutations in PPD1 delay heading time in the absence of ELF3
The upregulation of PPD1 in elf3 phyB relative to phyB (Fig 2) and its downregulation in the UBI::ELF3-HA transgenic plants (Fig E in S1 Text) suggested that ELF3 acts as a transcriptional repressor of PPD1, so we combined loss of function mutations on both genes in the PI background to study their epistatic interactions on heading time. As controls, we included Kronos PI and the elf3 mutant in the PI background.
For all genotypes, heading time was significantly later in SD than in LD, but the differences were reduced when the elf3 mutant allele was present (Fig 5). Under LD, the elf3 mutants headed 6 d earlier than the wildtype, while the elf3 ppd1 combined mutants headed 20 d later than the wildtype, but still 36 d earlier than the ppd1 lines. Under SD, the relative order of heading time of the genotypes was similar to the LD, but the differences were larger. The elf3 mutant headed 36 d earlier than the wildtype, the ppd1 lines failed to head by the time the experiment was terminated at 150 d, and elf3 ppd1 combined mutants headed >48 d earlier than the single ppd1 mutant (Fig 5).
Fig 5. Effect of elf3 and ppd1 mutations on heading time under LD and SD conditions.
Bars are means and error bars are s.e.m. The blue arrow on top of the Elf3 ppd1 SD treatment indicates that plants did not head by the time the experiment was terminated at 150 d. Differences in heading times between SD and LD are indicated for each genotype above the bars with the corresponding t-tests (*** = P < 0.001). No t-test is provided for Elf3 ppd1 because plants failed to head under SD. Numbers at the base of the bars indicate the number of plants analyzed in each genotype / photoperiod combination. Raw data and statistics are available in Data D in S1 Data.
A factorial ANOVA including ELF3 (wildtype and elf3), PPD1 (wildtype and ppd1) and photoperiod (SD and LD) revealed highly significant effects for these three factors and also for all two-way and three-way interactions (P < 0.001, Data D in S1 Data). These results indicate that i) there are strong genetic interactions between ELF3, PPD1, and photoperiod, ii) the Ppd-A1a allele can accelerate heading time in the absence of ELF3 under both LD and SD, iii) ELF3 can delay heading time in a PPD1-independent manner, and iv) there is a residual photoperiodic response that is independent of PPD1 and ELF3.
Effect of elf3 mutations on PPD1 and FT1 expression
To understand the effect of the elf3 mutations on heading time at the molecular level, we analyzed the transcript levels of PPD1 and FT1 in the youngest leaves of five-week-old Kronos PI and PS plants grown under LD (Fig 6). In the PI lines (Fig 6A–6C), the presence of two PPD1 homoeologs with contrasting photoperiodic regulation provided interesting insights on the regulation of this gene. The PPD-A1a homoeolog showed a similar expression profile between the wildtype and the elf3 mutant, with a strong peak at dawn (ZT0) (Fig 6A). By contrast, in the PPD-B1 homoeolog this peak was present in the elf3 mutant but disappeared in the wildtype. The elf3 mutant also showed a significant upregulation of PPD-B1 relative to wildtype at ZT20-ZT8 (Fig 6B), which was paralleled by a significant upregulation of FT1 at ZT0-ZT4 (Fig 6C).
Fig 6. qRT-PCR analysis of transcript levels of PPD1 and FT1 in wildtype and elf3 mutants.
Leaf samples collected from 5-week-old Kronos photoperiod insensitive (PI) and sensitive (PS) plants grown under LD. (A-C) Wildtype vs. elf3 in PI. (D-F) Wildtype vs. elf3 in PS. (A) PPD-A1a allele with a deletion in the promoter associated with earlier heading under SD. (D) PPD-A1b allele with the intact promoter and late heading under SD. (B & E) PPD-B1 with intact promoter in PI and PS. (C & F) FT1 (both homoeologs combined). ACTIN was used as endogenous control. Error bars are s.e.m based on 5 biological replications. ns = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001 based on t-tests between mutants and wildtype at the different time points. Raw data and statistics are available in Data E in S1 Data.
We hypothesized that the difference in the expression profiles between the two PPD1 homoeologs at dawn was associated with the presence of the 1,027 bp deletion in the promoter of Ppd-A1a [8]. To test this hypothesis, we repeated the experiment in the PS lines, which carry the Ppd-A1b allele with an intact promoter. Indeed, we found that the ZT0 peak was absent in PPD-A1b (Fig 6D). Consistent with the earlier heading of PI relative to PS, the wildtype transcript levels of FT1 were 2.6- to 8.5-fold higher in PI than in PS throughout the day (P = 0.028, Fig 6C and 6F). In the elf3 mutant, FT1 transcript levels were significantly higher than in the wildtype in both PI and PS, which agrees with their very early heading time (Fig 1).
Effect of ppd1 and elf3 mutations on the expression of VRN1, VRN2, and FT1
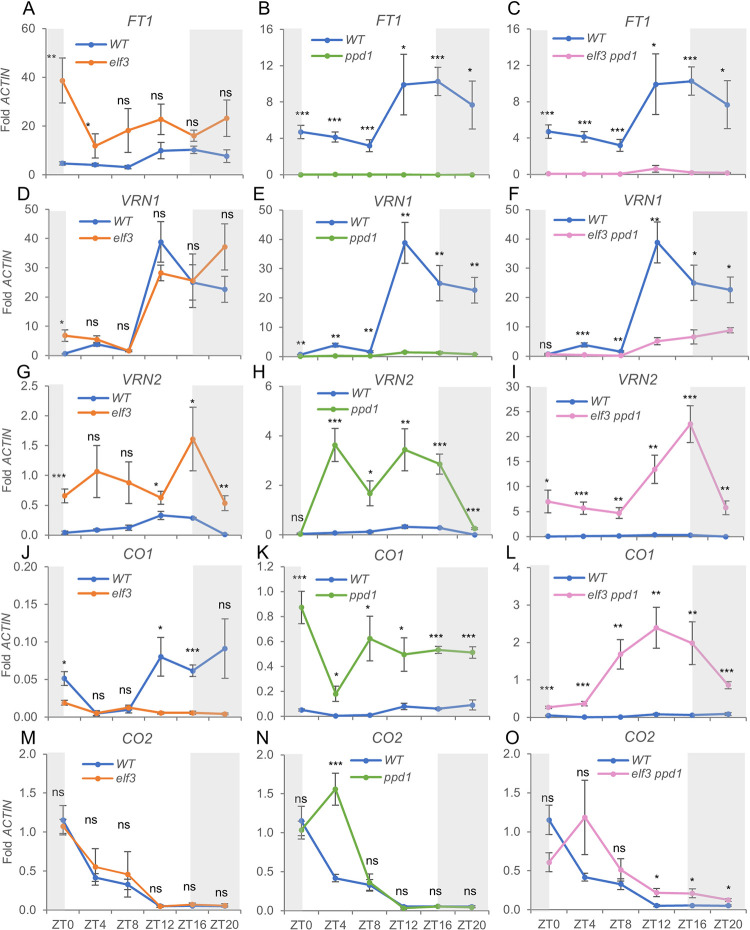
To further characterize the interaction between ELF3 and PPD1 on heading time, we evaluated the expression levels of the main wheat flowering genes in five-week-old PI wildtype, elf3, ppd1, and elf3 ppd1 plants grown under LD. The significant upregulation of FT1 (Fig 7A–7C) at dawn in the elf3 mutant was correlated with a significant upregulation of VRN1 (Fig 7D–7F) at the same time point. Consistent with previous reports [28,39,42], the lack of a functional ELF3 also resulted in the upregulation of the flowering repressor VRN2 (Fig 7G–7I).
Fig 7. Transcript levels of flowering genes FT1, VRN1, VRN2, CO1, and CO2 in Kronos PI, elf3, ppd1 and elf3 ppd1 mutants.
(A-C) Flowering promoter gene FT1. (D-F) Flowering promoter gene VRN1. (G-I) Flowering repressor gene VRN2. (J-L) CO1. (M-O) CO2. (A, D, G, J, & M) Wildtype vs. elf3. (B, E, H, K, & N) Wildtype vs. ppd1. (C, F, I, L, & O) Wildtype vs. elf3 ppd1. Primers used for all genes amplify both homoeologs. The WT data is the same within each row but at different scales. Fig 7A is the same as Fig 6C, and is included again to facilitate comparisons. Error bars are s.e.m based on 5 biological replications. ns = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001 based on t-tests between mutants and wildtype at the different time points. Raw data and statistics are available in Data F in S1 Data.
In the absence of functional copies of PPD1, the transcript levels of FT1 and VRN1 were almost undetectable, whereas VRN2 was highly upregulated (Fig 7B, 7E and 7H). The transcription profiles of these three genes are consistent with the delayed heading time of the ppd1 mutant (Fig 5). When the ppd1 mutant was combined with elf3, the FT1 transcripts remained severely downregulated (Fig 7C), but VRN1 showed a gradual upregulation from ZT12 to ZT20 (Fig 7F), which is consistent with the earlier heading of elf3 ppd1 relative to ppd1 (Fig 5). The VRN2 flowering repressor was upregulated to higher levels in elf3 ppd1 than in the elf3 or ppd1 single mutants (Fig 7I). In summary, the upregulation of VRN1 likely contributes to the accelerated heading time of the elf3 ppd1 relative to ppd1 (Fig 5) although this contribution may be partially offset by the increase in VRN2 transcript levels.
Finally, we showed that in elf3, CO1 transcript levels were significantly downregulated relative to the wildtype (Fig 7J), whereas those of CO2 were not affected. CO1 was upregulated in ppd1 at all time points, and that upregulation was higher in the elf3 ppd1 combined mutant (Fig 7K and 7L). The effect of ppd1 on CO2 was significant only at ZT4, whereas that of elf3 ppd1 only at ZT12-ZT20.
It is interesting to point out that at dawn (ZT0), PPD1, VRN1, VRN2, CO1, and CO2 are expressed at lower levels in elf3 phyB (Figs 2 and C in S1 Text) than in elf3 (Figs 6 and 7), suggesting that at this time point PHYB is required for the upregulation of these genes in the elf3 mutant background. However, given the complex feedback regulatory loops that exist among these genes, we do not know which of these effects are direct or indirect.
ELF3 binds directly to the PPD1 promoter in vivo
The elevated expression levels of PPD1 in the elf3 mutant suggest that ELF3 acts as a transcriptional repressor of this gene. To test the binding of ELF3 to the PPD1 promoter in vivo, we performed replicated chromatin immunoprecipitation (ChIP) experiments using PS plants carrying the UBI::ELF3-HA transgene combined with the elf3 mutation and non-transgenic elf3 plants as a control. Samples were collected from the aerial part of plants grown under SD at ZT10 in the dark, when ELF3 levels are elevated (Fig 4).
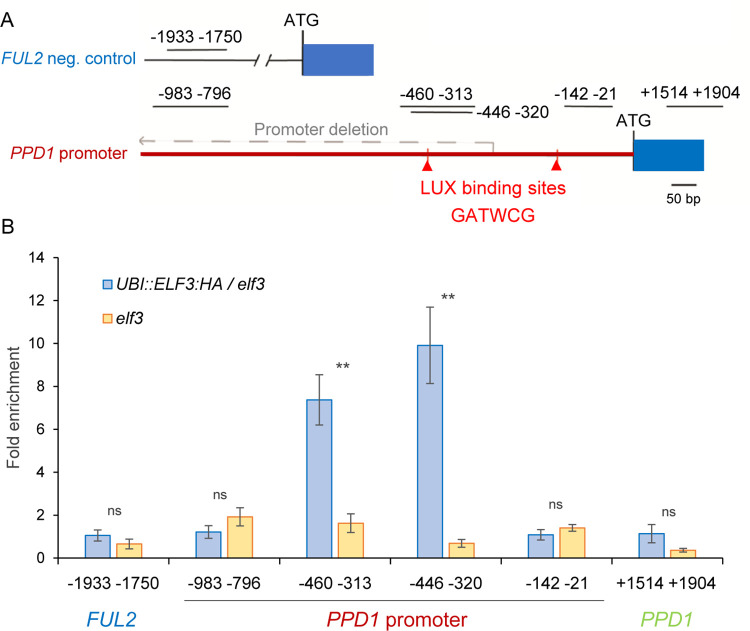
We observed significant enrichment of ELF3 in two PCR amplified fragments covering PPD1 promoter regions -460 to -313 and -446 to -320 bp from the start codon (Fig 8 and Table D in S1 Text), which include a LUX binding motif (GATWCG) [36,37]. This suggests that the binding of ELF3 to this promoter region is likely mediated by LUX, which is part of the EC with ELF3 and ELF4. This LUX binding motif (in the minus DNA strand) is deleted in the Ppd-A1a allele (Fig 8), likely affecting the binding of the EC.
Fig 8. Chromatin immunoprecipitation (ChIP) analysis of the PPD1 promoter.
(A) Gene diagram of the promoter of PPD-B1 showing the regions -983 to -796, -460 to -313, -446 to -320, -142 to -21, and a control coding region at +1514 to +1904 analyzed by ChIP, followed by qRT-PCR. The grey dashed arrow demarks the location deleted within the Ppd-A1a promoter (PI). The red triangles mark the locations of predicted LUX binding sites (GATWCG). The PPD1 promoter is indicated by a horizontal red line and a rectangular box represent the first exon (ATG indicates the start codon). (B) Fold enrichment of ELF3 at the PPD1 promoter in the PS-elf3 mutant and transgenic UBI::ELF3-HA in a mutant PS-elf3 background. A FUL -promoter region between -1933 and -1750 that contained no LUX binding sites was used as a negative control. Bars represent the mean ± s.e.m. from four biological replicate experiments. ** = P < 0.01 and ns = not significant. Primer sequences are provided in Table D in S1 Text. Raw data and statistics are in Data G in S1 Data.
We did not observe enrichment of ELF3 for a region between -142 to -21 bp (close to another putative LUX binding site), a region in the coding region of PPD1 between +1514 and +1904 bp, and a region further upstream in the PPD1 promoter between -983 and -796 bp from the start codon (Fig 8). Lastly, no enrichment of ELF3 was observed at the FRUITFUL 2 (FUL2) promoter (-1933 to -1750), which does not have any putative LUX binding site and was included as a negative control. Taken together, these experiments confirm that ELF3 acts as a direct transcriptional repressor of PPD1 in planta (Fig 8), likely as part of the EC.
Discussion
ELF3 regulates wheat heading time and spike development
The role of ELF3 on the regulation of flowering time has been extensively studied given the importance of this trait in the adaptation of crop species to different latitudes [27,30,47–51]. The early heading time of the elf3 Kronos mutants in LD and SD and the delayed heading time of the UBI::ELF3-HA transgenic plants indicate that ELF3 functions as a heading time repressor in wheat. Early flowering of elf3 mutants has also been reported in barley [27,30] and Brachypodium [42], suggesting a conserved function of ELF3 as a flowering repressor in the temperate grasses. A similar function has been observed in Arabidopsis, where elf3 mutants flower early under both SD and LD [44,52].
The opposite effect has been observed in rice, where the individual elf3-1 and elf3-2 mutants flower later than the wildtype under both SD and LD conditions [53,54] and the double elf3-1 elf3-2 fails to flower in both photoperiods [39]. In spite of their opposite effects on heading time, the elf3 mutants show some similarities between wheat and rice. In both species, elf3 shows increased mRNA levels of the LD flowering repressor GHD7/VRN2 [53]. In addition, although the phyC and phyB mutants have opposite effects on heading time in wheat and rice, those are mostly cancelled when combined with the elf3 mutant in both species [55]. This indicates that the effect of the phytochromes on heading time is mainly mediated by ELF3 in both wheat and rice [55]. The reverse roles of ELF3 and the phytochromes in wheat and rice flowering time are likely associated with the reverse role of their downstream target PPD1/PRR37, which acts as a LD flowering promoter in wheat and as a LD flowering repressor in rice [56].
The elf3 ppd1 combined mutant headed earlier than the ppd1 mutant both in LD (37 d) and SD (>48 d) indicating that ELF3 can delay wheat heading time independently of PPD1. The same result was reported for Brachypodium plants grown under 20 h light / 4 h darkness, where elf3 ppd1 headed 16 d earlier than ppd1 [57]. However, when the same Brachypodium genotypes were grown under SD (8 h light) or LD (16 h light), the elf3 ppd1 plants headed significantly later (24.4 and 13.7 d) than the ppd1 plants [57]. These results suggest differences between Brachypodium and wheat in the PPD1-independent effect of ELF3.
In addition to its effects on heading time, the Kronos elf3 mutants showed a significant reduction in spikelet number per spike (Fig B in S1 Text). Effects of ELF3 on heading time and spikelet number per spike have been previously reported in diploid wheat T. monococcum¸ where the ELF3 locus was originally described as Eps-Am1 [26,58,59]. Interestingly, the differences in heading time and spikelet number per spike associated with Eps-Am1 were both modulated by temperature, an effect that was also reported for the differences in heading time associated with the EPS-D1 locus in hexaploid wheat [60]. These observations are likely related to the known role of ELF3 in the temperature entrainment of the circadian clock in Arabidopsis [14,15,36,37,61–63] and barely [64].
Interactions between ELF3 and phytochromes PHYB and PHYC
Previous studies have shown that the wheat photoperiodic response is strongly affected by phytochromes PHYB and PHYC [11–13] and by ELF3 [26], which prompted our study of the genetic interactions among these genes and their encoded proteins. The characterization of the different elf3 and phyB combinations revealed that the strong heading time delay observed in the phyB mutant almost disappeared in the early heading elf3 phyB. The combined mutant also restored the expression of PPD1, which was repressed in phyB. Similar genetic interactions between elf3 and phyC are reported in the companion study in Brachypodium [57], suggesting that these interactions are conserved in the temperate grasses. Taken together, these results indicate that the critical role of PHYB or PHYC in the light activation of PPD1 is mediated by ELF3 in both wheat and Brachypodium.
The genetic interactions between ELF3 and the phytochromes were also reflected in a physical interaction between the ELF3 protein and both PHYB and PHYC proteins in Y2H assays (Fig 3). Although these interactions still need to be validated in planta for wheat, in planta interactions between phytochromes and all three members of the EC have been reported before in Arabidopsis, where they have been proposed to be important in the connection between the circadian clock and the light signaling pathways [65,66]. We hypothesize that these physical interactions might be associated with the rapid reduction of ELF3 when the lights were turned on at ZT0, and with the rapid replacement of the ELF3 lower band in the Western blots by a higher and more diffuse ELF3 band when the lights were turned on during the night (Fig 4C). The light modification of the lower ELF3 band was followed by the upregulation of PPD1 30 min after the lights were turned on, suggesting that the lower ELF3 band represents the active repressor of PPD1. A similar upregulation of PPD1 was previously reported in wheat after 15 min pulse of light in the middle of a 16 h night [17]. The same study showed that 15 consecutive NBs were sufficient to accelerate wheat heading under SD, but that this acceleration disappeared in the phyB and phyC mutants [17]. These results suggest that PHYB and PHYC are required for the light modification of ELF3 and the resulting induction of PPD1 after NBs.
In rice, Western blots using an ELF3 antibody revealed two forms of the ELF3-1 protein, a post-translationally modified higher band detected during the light and dark periods, and a relatively lower band detected only during the night [39]. This lower band was also detected during the day in the rice phyB mutant, suggesting that PHYB is necessary for the efficient modification of the lower band by light [39]. The rice ELF3 protein is known to be a direct substrate of the E3 ubiquitin ligase HAF1, which plays a role in the determination of rice heading time under LD [67]. In Arabidopsis, the ubiquitin-ligases CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and XBAT31 can also mediate the ubiquitination and proteasomal degradation of ELF3 [68,69]. These studies suggest that ubiquitination can be responsible for the observed changes in the wheat and rice ELF3 protein when exposed to light during the night, but we cannot completely rule out phosphorylation or other posttranscriptional modifications.
The interactions between the phytochromes and ELF3 in wheat differ from those reported in Arabidopsis, where PHYB contributes to ELF3 accumulation in the light, likely by competing for COP1 and limiting the degradation of ELF3 [70]. The contrasting effects of the phytochromes on ELF3 protein stability may contribute to their different effect in wheat and Arabidopsis flowering time. In the temperate cereals, both the phyB and phyC mutants flower extremely late under LD [11,13], whereas in Arabidopsis the phyB mutants flower earlier under both SD and LD and phyC flowers earlier under SD [71]. However, the different photoperiod pathways affected in these species may also contribute to the differences in flowering time caused by the phytochrome mutations. In Arabidopsis, PHYB is involved in the morning degradation of the CO protein, so the phyB mutation results in the accumulation of CO and early flowering [2,72]. By contrast, in the temperate cereals PHYB and PHYC operate as flowering promoters in the PPD1-dependent photoperiod pathway, and the mutants are extremely late [11,13,73,74].
Interactions between ELF3 and PPD1
The ppd1 mutant delays heading time even in an elf3 background, indicating that it operates downstream of ELF3. This is also evident in the changes of PPD1 transcription profiles observed in the elf3 mutants (Fig 6A, 6B, 6D and 6E). The upregulation of Ppd-A1b and Ppd-B1b (intact promoter) at ZT0 in the absence of ELF3 (Fig 6D, PS) is mimicked in the wild type by the Ppd-A1a allele carrying the promoter deletion in the presence of ELF3 (Fig 6A, PI). A similar upregulation at dawn has been observed in the Ppd-D1a allele [6] and in a different Ppd-A1a allele [8], all carrying overlapping deletions in the PPD1 promoter (Fig A in S1 Text). Our ChIP-PCR experiments support the hypothesis that this is the result of the direct transcriptional regulation of PPD1 by ELF3 (Fig 8). A similar interaction has been reported in Brachypodium [75], rice [39] and maize [51] suggesting that is conserved in the grass species.
The ELF3 repression of PPD1 transcription is likely the result of the binding of the EC to the LUX binding site present in the region deleted in the promoter of the Ppd-A1a allele. This is supported by the dawn upregulation of PPD1 in lux mutants in barley [31], diploid wheat T. monococcum [33,76], and hexaploid wheat with combined mutations in all three LUX homoeologs [32]. LUX is a member of the EC that has a specific DNA-binding domain targeting the GATWCG motif and related promoter elements in Arabidopsis [37,38,77,78]. Therefore, the similar effects of elf3 and lux on the transcriptional regulation of PPD1 suggests that the EC plays an important role in the repression of PPD1 during the night and at dawn in the temperate grasses. In Arabidopsis, it has been shown that the EC represses the expression of PRR7 (a close homoeolog of wheat PPD1), PRR9, GIGANTEA, and LUX itself [37,38,77,78].
The PRR3 –PRR7 paralogs in Arabidopsis and the PRR37 (PPD1)–PRR73 paralogs in the grass lineage belong to the same clade, suggesting a common history [79,80]. However, since the duplication that originated PRR3 and PRR7 in Arabidopsis is independent of the duplication that originated PPD1—PRR73 in the grasses, the potential sub-functionalization of these two pairs of paralogs is also independent [79]. In Arabidopsis, PRR7 is an important component of the circadian clock with no major effects on the photoperiodic regulation of flowering, whereas natural mutations in PPD1 selected during wheat [6,8,18,81] and barley [7,82] domestication have a strong effect on the photoperiodic regulation of heading time but limited effects on the transcription profiles of the central oscillator [46,79]. However, transformation of the Arabidopsis prr7-11 mutant with the barley photoperiod sensitive (Ppd-H1) and insensitive (ppd-H1) alleles driven by the Arabidopsis PRR7 promoter complements circadian leaf movements in Arabidopsis [83], suggesting a conserved clock function. We hypothesize that selection during wheat and barley domestication may have favored PPD1 mutations with limited effects on the clock that minimize negative pleiotropic effects. Taken together, these results suggest that grasses may have repurposed an ancient interaction between clock genes (EC and PRR7) to develop a photoperiod pathway that can operate independently of CO.
Interactions between ELF3 and CO1 / CO2
In a previous study, we showed that combined loss-of-function mutants co1 co2 headed 3 d earlier than the wildtype under LD and 13.5 d under SD, suggesting that CO1 and CO2 function as weak heading time repressors in the presence of functional PPD1 alleles [5]. However, under LD the ppd1 mutant headed more than 60 d earlier than the ppd1 co1 plants, which failed to head before the experiment was terminated at 180 d [5]. A similar effect was reported in barley, where a strong QTL for heading time under LD was found at the CO1 locus, but only among accessions carrying the ppd-H1 photoperiod insensitive allele [84]. These results demonstrate that in wheat and barley CO1 can accelerate heading time under LD in the absence of PPD1, and may explain the residual photoperiodic response observed in the elf3 ppd1 mutant, which headed earlier under LD than under SD (Fig 5).
A comparison of the CO1 expression results in wheat and Brachypodium revealed differences between these two species. In wheat, CO1 is upregulated in phyB (Fig C in S1 Text) and ppd1 (Fig 7K), whereas in Brachypodium CO1 is downregulated in phyC and ppd1 [57]. These differences may explain the earlier heading of the co1 mutant relative to the wildtype in wheat [5], and the delayed heading of CO1 RNA interference knock-down plants in Brachypodium under LD [85]. Differences in CO1 regulation may also contribute to the contrasting effect of ELF3 in the ppd1 background under 16h LD, where elf3 ppd1 headed 36 d earlier than ppd1 in wheat but 2.5 d later in Brachypodium. In summary, even though Brachypodium and wheat have similar photoperiodic genes, changes in the interactions among these widely interconnected genes can result in different flowering outcomes.
A working model for the wheat photoperiod pathway
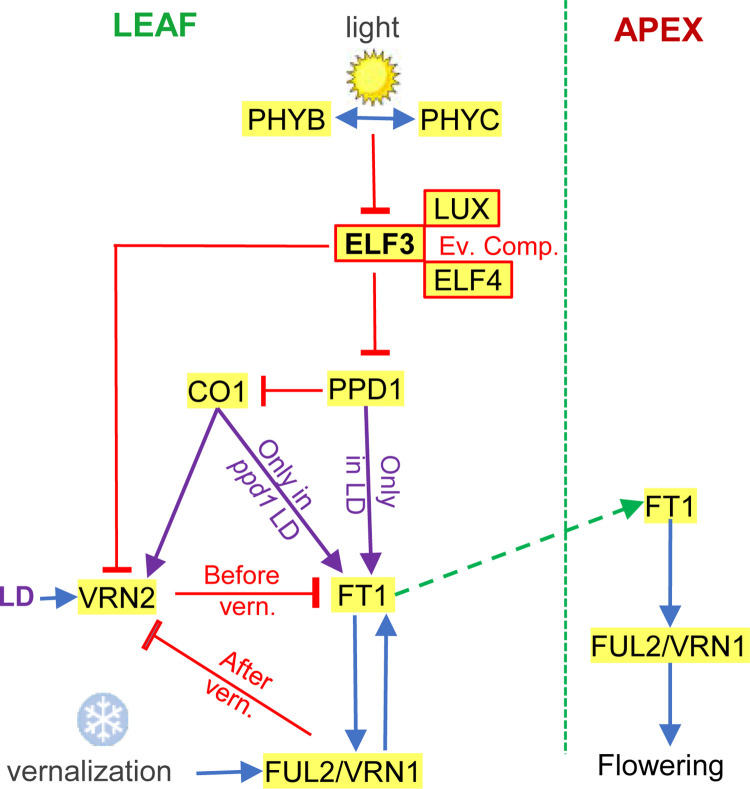
Based on the interactions among flowering genes described in this and previous studies, we proposed a working model of the wheat photoperiodic pathways that is presented in Fig 9. In this model, we place ELF3 between the phytochromes and PPD1, as a critical component of the transcriptional activation of PPD1 by light. We hypothesize that a PHYB-PHYC heterodimer is required to repress the activity of ELF3 since mutations in both phytochromes result in very late flowering and reduced PPD1 expression [11].
Fig 9. Working model for the regulation of heading time in wheat.
The model integrates photoperiod and vernalization signals into the regulation of FT1 expression in wheat leaves. FT1 is then transported to the shoot apical meristem (dotted green arrow) where it induces the transition from the vegetative to the reproductive phase. Blue arrows indicate promotion of gene expression or activity and red lines ending in a crossed-bar indicate repression. ELF3, LUX and ELF4 proteins form the evening complex, which binds to the PPD1 promoter and inhibits its transcription. The complex interactions between VRN2 and CO1, CO2 and PPD1 [5] are not included in the figure for clarity.
In this model, ELF3 acts as a transcriptional repressor of PPD1 by direct binding of the EC to a LUX-binding site in the PPD1 promoter, with the promoter deletions in Ppd-A1a and Ppd-D1a limiting the ability of the EC to repress their expression. This model explains the upregulation of PPD1 in the elf3 (Fig 6) and lux mutants at dawn [31–33,76], and its repression in the UBI::ELF3-HA transgenic plants (Fig E in S1 Text).
According to this model, PPD1 is required for the upregulation of FT1 under LD, but this induction requires at least two weeks of NBs or LD to induce the expression of FT1 and accelerate heading time [17]. The NB experiments also suggest that the connection between PPD1 and FT1 may be gated by circadian clock-regulated genes. Although the maximum acceleration of heading time occurs when the NBs are applied in the middle of the night, the maximum induction of PPD1 occurs when the NBs are applied at the end of the night [17]. The proposed gating mechanism of the PPD1 activity is also supported by the induction of FT1 transcription under LD but not under SD in spite of the induction of PPD1 transcription during the light phase both under SD and LD conditions [5,17].
PPD1 interferes with CO1 function, which induces FT1 and accelerates heading time under LD only in a ppd1 mutant background [5]. CO1 also acts as a transcriptional promoter of VRN2 [5], which may explain the upregulation of VRN2 transcript levels observed in the ppd1 mutant (Fig 7H). The interactions between VRN2 and both CO1 and PPD1 are not shown in the model for simplicity. VRN2 is also negatively regulated by ELF3 (Fig 7G and [28]), an interaction that has been also reported in rice. In this species, OsELF3 promotes flowering under LD mainly by direct repression of GHD7, the rice ortholog of VRN2 [39,54,86]. In wheat, VRN2 acts as a LD flowering repressor of FT1 [24], so the low transcript levels of FT1 in elf3 ppd1 (Fig 7C) can be caused by a combination of high levels of VRN2 and absence of PPD1.
A limitation of the model presented in Fig 9 is the absence of GIGANTEA (GI), which has not yet been functionally characterized in wheat. Loss-of-function mutants in GI have a reduced photoperiodic response in both rice [87] and Arabidopsis [88]. In Arabidopsis, ELF3 is known to interact with COP1 in vivo to promote GI protein degradation [68]. In addition, ChIP studies have shown that the EC binds to the GI promoter and regulates its transcription [37]. Therefore, interactions between ELF3 and GI can also contribute to the photoperiodic response in the temperate cereals and are an important target for future studies in our laboratory.
Although much remains to be learned on the wheat PPD1-mediated photoperiod pathway, this study clearly shows that the ELF3 protein operates downstream of PHYB and PHYC, is regulated by light and acts as a direct repressor of PPD1. This information provides additional entry points to engineer heading time in wheat, an important trait for the development of better adapted varieties to a changing environment.
Materials and methods
Plant materials
Plant materials used in this study were derived from a sequenced EMS-mutagenized TILLING population of the tetraploid wheat variety Kronos (Triticum turgidum ssp. durum, 2n = 28, genomes AABB) [89,90]. Kronos is a spring PI wheat that carries a functional Vrn-A1 allele for spring growth habit (Vrn-A1c allele) [23,91], a functional Vrn-B2 long day repressor locus (ZCCT-B2a and ZCCT-B2b) [92], and the Ppd-A1a allele for reduced photoperiodic response [8]. Kronos lines carrying loss-of function mutations in the A- and B- genome copies of genes ELF3 [26], PHYB [13], and PPD1 [17] were described before and are summarized in Fig A in S1 Text.
Mutants for the individual homoeologs are indicated with the capital letter of the genome (e.g. ppd-A1 and ppd-B1) whereas the double mutants without any functional copy of the gene are indicated without any genome letter (e.g. ppd1). We intercrossed these different mutants and selected different mutant combinations using marker-assisted selection. All lines had at least two backcrosses to parental line Kronos to reduce the number of background mutations. The elf3 phyB combined mutant was also crossed with a Kronos near-isogenic line carrying the photoperiod sensitive Ppd-A1b allele [93] to test the mutant combinations in both Kronos-PI (PI) and Kronos-PS (PS) backgrounds.
Genotyping of mutant lines
Kompetitive Allele Specific PCR (KASP) markers (LGC-Genomics, UK) [94] were developed to detect the EMS-induced nucleotide changes that resulted in premature stop codons in genes ELF3, PHYB, and PPD-A1. The mutants’ identification numbers, the type and position of the loss-of-function mutations, and the primers used in the diagnostic KASP assays are listed in Table A in S1 Text. To detect the PPD-B1 deletion, we used a TaqMan assay described previously [81].
KASP reactions were carried out using 5 μl of genomic DNA diluted to a concentration of 5–50 ng μl− 1, 5 μl of 2x KASP Master Mix, and 0.14 μl of KASP primer mix. For every 100 μl, the primer mix included 12 μl of the VIC primer (100 μM), 12 μl of the FAM primer (100 μM), 30 μl of the genome-specific common primer (100 μM), and 46 μl of distilled water. A two-step touchdown PCR program was used, with an initial denaturalization step of 94°C for 15 min, followed by ten cycles of touchdown of 94°C for 20 s and annealing/extension at 61–55°C for 1 min (dropping 0.6°C per cycle), followed by 36 cycles of 94°C for 20 s and annealing/extension at 55°C for 1 min. KASP results were analyzed with a FLUOstar Omega F plate reader (BMG Labtech, Ortenberg, Germany) using the software KlusterCaller (LGC Genomics, Teddington, UK).
Phenotypic characterization of mutant lines
After the two backcrosses to Kronos, BC2F3 plants homozygous for the different allelic combinations were stratified for 3 days at 4°C in the dark and then planted in PGR15 growth chambers (Conviron, Manitoba, Canada). Lights were set to 350 μmol m−2 s−1 at canopy level and were kept on for 16 h in the LD experiments and for 8 h in the SD experiments. Temperature was set to 22°C during light periods and 17°C during dark periods. Heading time was recorded as the number of calendar days from planting in the soil to full emergence of the spike from the sheath.
Quantitative reverse transcription-PCR (qRT-PCR) analyses of flowering time genes
The transcriptional profiles of genes regulating wheat flowering time were analyzed during a 24-hour period in PI and PS plants with different mutant combinations. Plants were grown in a Conviron growth chamber under LD conditions and tissue samples from the last fully-expanded leaf were collected simultaneously from five-week-old plants every 4 h, starting at Zeitgeber time 0 (ZT 0 = 6:00 am, lights on). Because of variation in leaf developmental rates of the different mutants, the last fully-expanded leaf at week five was leaf four in ppd1 and ppd1 elf3; leaf five in wildtype and phyB, and leaf seven in elf3 and elf3 phyB. Five plants per genotype were analyzed at each time point. Different plants were sampled for each time point.
Harvested tissue was grounded to a fine powder in liquid nitrogen and RNA was extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA) following the manufacturer’s recommendations. First-strand cDNAs were synthesized from 500 ng of total RNA using the High Capacity Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). The qRT-PCR experiments were performed using 2X USB VeriQuest Fast SYBR Green qPCR Master Mix (Affymetrix, Inc., Santa Clara, CA, USA) in a 7500 Fast Real-Time PCR system. Primers used for quantitative qRT-PCR are listed in Table B in S1 Text. ACTIN was used as an endogenous control. Transcript levels for all genes are expressed as linearized fold-ACTIN levels calculated by the formula 2(ACTIN CT–TARGET CT) ± standard error (SE) of the mean (s.e.m.).
Yeast-two-hybrid (Y2H) assays
The ELF3 full-length coding region was first cloned into the pDONR-Zeo vector by BP reaction (Thermo Fisher, http://www.thermofisher) using primers ELF3-GW-F and ELF3-GW-R (Table C in S1 Text). It was then transferred to the pLAW10 vector (with the GAL4 DNA-binding domain) with the LR cloning Gateway strategy (Thermo Fisher, http://www.thermofisher) and used as bait. The cloning of the N-PHYC (1–600 AA) and C-PHYC (601-1139AA) was described previously [11]. The N-PHYB (1-625AA) was cloned into the pGADT7 vector, which contains the GAL4 activation domain, between EcoRI and XmaI restriction sites using primers N-PHYB-F and N-PHYB-R, and the C-PHYB (626-1166AA) was cloned into pGADT7 between EcoRI and ClaI sites using primers C-PHYB- F and C-PHYB-R (Table C in S1 Text). The PHYB and PHYC proteins were used as preys. SD medium lacking Leucine and Tryptophan (-L-W) was used to select for yeast transformants containing both bait and prey vectors. Interactions were determined on SD media lacking Leucine, Tryptophan, Histidine, and Adenine (-L-W-H-A).
Autoactivation tests for the ELF3-bait was tested against empty vector pLAW11 as prey. N-PHYC and C-PHYC preys have been previously shown to result in no autoactivation with the empty bait vector pGBKT7 [11] and the N-PHYB and C-PHYB preys were confirmed to show no autoactivation in this study.
Generation of transgenic wheat plants constitutively expressing ELF3
The full-length coding region of ELF3 from T. monococcum accession G3116 was fused to a C-terminal 3xHA tag and cloned downstream of the maize UBIQUITIN promoter in the binary vector pLC41, which has the kanamycin resistance for selection in E. coli and Agrobacterium, and hygromycin resistance for selection in planta. Agrobacterium strain EHA105 was used to infect Kronos immature embryos using the Japan Tobacco transformation technology licensed to UC Davis. The following primers were used to confirm the presence of the transgene: Ubi-F2 CAGAGATGCTTTTTGTTCGC and ELF3-genotyping-R3 AAAGCCTCCCAGATGTAGCA.
Immunoblot analysis
Tissue from newly fully-expanded leaves was harvested from UBI::ELF3-HA transgenic plants. Protein extraction was performed as described previously [39]. Whole leaf tissue was ground to powder in liquid nitrogen. For each timepoint, 50 mg of powder was transferred into a 1.5 mL microcentrifuge tube with 150 ul of 2 x Laemmli buffer (Bio-Rad, Hercules, CA, USA). The sample was immediately vortexed for 1 min followed by incubation at 70°C for 10 min. The supernatants were harvested after centrifugation at 20,000 x g for 10 min. The protein extract was subjected to immunoblot analysis with mono-HA antibody. Briefly, proteins were separated on a 7.5% Mini-PROTEAN TGX stain-free precast gel (Bio-Rad, Hercules, CA, USA). Trans-blot TURBO system was used to transfer proteins from the gel to a PVMF membrane. The membrane was blocked with TBST buffer (20 mM Tris, 137 mM sodium chloride, 1% Tween20 (v/v)) with 5% milk powder for 1 h. The blot was incubated in TBST solution containing 1:2000 Anti-HA-Peroxidase (Roche) for 1 h. The blot was washed 3 times, then developed with chemiluminescent detection reagent SuperSignal West Femto (Thermo Fisher, http://www.thermofisher).
Chromatin immunoprecipitation analysis
Kronos PS plants carrying the elf3 mutation or a combination of this mutation and the UBI::ELF3-HA transgene were grown to the fourth-leaf stage under SD (8h light). Eight grams of above-ground seedling tissue (four replicates, 2 g / replicate) were harvested at ZT10 and fixed in formaldehyde. Plants were harvested in the dark prior to fixation to avoid degradation of the ELF3 protein in the light. To aid in fixation penetration, harvested tissue was cut into 2–4 mm pieces and placed into metal tea infusers. Tissue was immediately cross linked under vacuum for 10 minutes in buffer containing 400 mM sucrose, 10 mM Tris (pH 8), 1% formaldehyde, then the vacuum was turned off and the tissue sat for an additional 5 minutes. To quench the cross-linking reaction 0.25 M glycine was used and the vacuum re-applied for an additional 5 minutes. Tissue was rinsed with milli-q water three times. After fixation, tissue was frozen in liquid nitrogen and stored at −80°C until performing the chromatin immunoprecipitation as described before [95]. Anti-HA magnetic beads from Thermo Fisher (http://www.thermofisher) were used for the immunoprecipitation.
Supporting information
Fig A. Schematic representations of PHYB, ELF3, and PPD1 mutants. Mutations introducing premature stop codons are indicated with red triangles and deletions with dotted red lines. Exons are shown as gray rectangles and introns as black lines. (A) The phyB line combines a premature stop codon on the A-genome homoeolog that truncates the last 641 amino acids of the protein, spanning the entire regulatory module, and a premature stop codon in the B-genome copy that eliminates the distal 140 amino acids, including the histidine kinase domain [13]. (B) The elf3 line carries premature stop codons that eliminate the last 241 (A-genome) and 244 (B-genome) amino acids of the C-terminal region, including the third and fourth conserved blocks of the ELF3 protein [26]. (C) The ppd1 line carries a premature stop codon in ppd-A1 that eliminates 514 amino acids of the PPD-A1 protein, including the highly conserved CCT domain, and ppd-B1 is a gamma ray-induced deletion the eliminates the complete gene [5,17]. (D) Natural deletions in the promoter of the Ppd-A1a [8] and Ppd-D1a [6] alleles in tetraploid and hexaploid wheat, respectively. Fig B. Effect of elf3 and elf3 phyB mutations on spikelet number per spike (SNS). (A) Kronos photoperiod insensitive (PI). (B) Kronos photoperiod sensitive (PS). In both experiment plants were grown under LD (16h light). Different letters above the bars indicate significant differences in pair-wise non-parametric Kruskal-Wallis tests (P < 0.05). The non-parametric test was used because no transformation was able to restore normality of residuals and homogeneity of variances simultaneously. The reduced SNS in the elf3 mutant was restored in the phyB elf3 combined mutant. Raw data is available in Data H in S1 Data. Fig C. Transcript levels of flowering genes VRN1, VRN2, CO1, and CO2 in Kronos PI, phyB and elf3 phyB. (A-B) VRN1, (C-D) VRN2, (E-F) CO1, and (G-H) CO2. (A, C, E, and G) Wildtype vs. phyB. (B, D, F, and H) Wildtype vs. elf3 phyB. Primers used for qRT-PCR amplify both homoeologs of each gene. The WT data is the same within each row but can be at different scales. Error bars are s.e.m based on 5 biological replications. ns = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001 based on t-tests between mutants and wildtype at the different time points. Raw data and statistics are available in Data I in S1 Data. Fig D. ELF3 transcript levels in leaves in the presence of phyB and phyC mutants under different photoperiods. (A) Transcript levels of Elf3 during the day in PS under SD. (B) Elf-A3 and Elf-B3 transcripts per million (TPM) in PI WT, phyB and phyC in leaves collected at ZT4 from 4-w old plants grown under LD and 8-w old plants grown under SD. Data are from previously published RNA-seq [12]. (C) Transcript levels during the day in PI and phyC under LD (D) Transcript levels during the day in PI and phyB under LD. Transcript levels were determined by qRT-PCR using ACTIN as endogenous control. NS = P > 0.05. Raw data and statistics are in Data J in S1 Data Fig E. Effect of the constitutive expression of ELF3 under the maize UBIQUITIN promoter (UBI::ELF3-HA). (A-C) Transcription profiles in the 7th leaf collected at ZT0, ZT4, ZT12 and ZT20 from five-week-old Kronos PS plants grown under LD. ACTIN was used as endogenous control. (A) ELF3, (B) PPD1 (conserved primers that amplify both Ppd-A1b and Ppd-B1b) and (C) FT1. Different letters indicate significant differences in Tukey tests (P < 0.05). The colors of the letters match the color of the respective treatment. OE = UBI::ELF3-HA and NT = non-transgenic sister line. (D) Complementation of the elf3 mutant by UBI::ELF3-HA. A factorial ANOVA showed highly significant effects on days to heading (DTH) for both the mutant and transgenic genotypes and for their interaction. Simple effects were tested by contrasts: ns = not significant, ** = P < 0.01, *** = P < 0.001. Raw data and statistics are available in Data K in S1 Data. Fig F. ELF3-HA protein size in western blots. (A-B) Samples extracted from leaves of Kronos-PS transgenic plants over-expressing ELF3 and a C-terminal 3xHA tag under the maize UBIQUITIN promoter (UBI::ELF3-HA). The ELF3-HA protein was detected by immunoblotting using an anti-HA antibody. Leaf samples were collected from plants grown under short day either 10 minutes before the lights were turned off (ZT8 -10m) or four hours after the lights were turned off (ZT12). (A) The expected size of the ELF3-HA protein is 88.5 kDa but the estimated size of the lowest band detected with the HA antibody was ~110 kDa based on both pre-stained and unstained protein markers. No bands were detected in the negative untransformed controls confirming the identity of the ELF3-HA protein. An additional and more diffuse band was detected at ZT8 -10m between 110 and 130 kDa. (B) Replicated experiment showing the complete blot. The lower strong band is rubisco (Rub). Top: Chemiluminescence, middle: CBB stained color-matrix, and bottom: merged images obtained by ChemiDoc imaging system (BioRad). (C) Wheat protoplasts transformed with UBI::ELF3:HA grown under both light and dark conditions. The unstained molecular marker was used to estimate band size. Table A. EMS-induced nucleotide changes that resulted in premature stop codon mutations in ELF3, PHYB, and PPD-A1. The ID of each of the mutant lines, the position of the loss-of-function mutations in the CDS and protein, and the primers used in the diagnostic KASP assays are listed. Capital letters in the primer sequences indicate the VIC and FAM tails. The 3′ allele-specific nucleotides are underlined. Table B. Primers used for qRT-PCR analysis. Table C. Primers used for cloning ELF3 and PHYB in yeast-two-hybrid assays. Table D. Primers used for chromatin immunoprecipitation.
(DOCX)
Data A. Supporting data for Fig 1. Data B. Supporting data for Fig 2. Data C. Supporting data for Fig 4. Data D. Supporting data for Fig 5. Data E. Supporting data for Fig 6. Data F. Supporting data for Fig 7. Data G. Supporting data for Fig 8. Data H. Supporting data for Fig B in S1 Text. Data I. Supporting data for Fig C in S1 Text. Data J. Supporting data for Fig D in S1 Text. Data K. Supporting data for Fig E in S1 Text.
(XLSX)
Data Availability
Mutant lines were deposited in the National Small Grains Collection under ID numbers PI 701905 (Kronos-PS, introgression of photoperiod sensitive allele Ppd-A1b), PI 701906 (Kronos elf3 phyB combined knock-outs), and PI 701907 (Kronos elf3 ppd1). Additional information about these accessions and/or seed requests can be done at GRIN-Global https://npgsweb.ars-grin.gov/gringlobal/search. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
J.D. acknowledges support from the Howard Hughes Medical Institute (https://www.hhmi.org/) and by competitive Grants 2016-67013-24617 and 2022-68013-36439 (WheatCAP) from the United States Department of Agriculture, National Institute of Food and Agriculture (https://nifa.usda.gov/). D.P.W. was a Howard Hughes Medical Institute Post-Doctoral Fellow of the Life Sciences Research Foundation. The funders played no role in this research beyond providing the funding.
References
- 1.Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res. 1919;18:0553–606. WOS:000188347200038. [Google Scholar]
- 2.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80(6):847–57. doi: 10.1016/0092-8674(95)90288-0 . [DOI] [PubMed] [Google Scholar]
- 3.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410(6832):1116–20. doi: 10.1038/35074138 . [DOI] [PubMed] [Google Scholar]
- 4.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303(5660):1003–6. doi: 10.1126/science.1091761 . [DOI] [PubMed] [Google Scholar]
- 5.Shaw LM, Li CX, Woods DP, Alvarez MA, Lin HQ, Lau MY, et al. Epistatic interactions between PHOTOPERIOD1, CONSTANS1 and CONSTANS2 modulate the photoperiodic response in wheat. PLoS Genet. 2020;16(7):e1008812. doi: 10.1371/journal.pgen.1008812 WOS:000552626900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet. 2007;115(5):721–33. doi: 10.1007/s00122-007-0603-4 . [DOI] [PubMed] [Google Scholar]
- 7.Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310(5750):1031–4. doi: 10.1126/science.1117619 . [DOI] [PubMed] [Google Scholar]
- 8.Wilhelm EP, Turner AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.). Theor Appl Genet. 2009;118(2):285–94. doi: 10.1007/s00122-008-0898-9 . [DOI] [PubMed] [Google Scholar]
- 9.Worland T, Snape JW. Genetic basis of worldwide wheat varietal improvement. In: Bonjean AP, Angus WJ, editors. The world wheat book: a history of wheat breeding. Paris, France: Lavoisier Publishing; 2001. p. 59–100. [Google Scholar]
- 10.Worland AJ. The influence of flowering time genes on environmental adaptability in European wheats. Euphytica. 1996;89(1):49–57. doi: 10.1038/sj.gt.3300937 [DOI] [Google Scholar]
- 11.Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, et al. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc Natl Acad Sci U S A. 2014;111(28):10037–44. doi: 10.1073/pnas.1409795111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kippes N, VanGessel C, Hamilton J, Akpinar A, Budak H, Dubcovsky J, et al. Effect of phyB and phyC loss-of-function mutations on the wheat transcriptome under short and long day photoperiods. BMC Plant Biol. 2020;20(1):297. doi: 10.1186/s12870-020-02506-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearce S, Kippes N, Chen A, Debernardi JM, Dubcovsky J. RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol. 2016;16(1):141. doi: 10.1186/s12870-016-0831-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao MJ, et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354(6314):886–9. doi: 10.1126/science.aaf6005 WOS:000388531900040. [DOI] [PubMed] [Google Scholar]
- 15.Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016;354(6314):897–900. doi: 10.1126/science.aaf5656 . [DOI] [PubMed] [Google Scholar]
- 16.Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Bio. 2002;3(2):85–93. doi: 10.1038/nrm728 WOS:000173821700018. [DOI] [PubMed] [Google Scholar]
- 17.Pearce S, Shaw LM, Lin H, Cotter JD, Li C, Dubcovsky J. Night-break experiments shed light on the Photoperiod1-mediated flowering. Plant Physiol. 2017;174(2):1139–50. doi: 10.1104/pp.17.00361 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw LM, Turner AS, Herry L, Griffiths S, Laurie DA. Mutant alleles of Photoperiod-1 in wheat (Triticum aestivum L.) that confer a late flowering phenotype in long days. PLoS One. 2013;8(11):e79459. doi: 10.1371/journal.pone.0079459 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–3. ISI:000246554000047. doi: 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- 20.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316(5827):1033–6. ISI:000246554000048. doi: 10.1126/science.1141753 [DOI] [PubMed] [Google Scholar]
- 21.Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55:543–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Lin H, Dubcovsky J. Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 2015;84:70–82. doi: 10.1111/tpj.12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8(12):e1003134. doi: 10.1371/journal.pgen.1003134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009;12(2):178–84. doi: 10.1016/j.pbi.2008.12.010 . [DOI] [PubMed] [Google Scholar]
- 25.Shaw LM, Lyu B, Turner R, Li C, Chen F, Han X, et al. FLOWERING LOCUS T2 regulates spike development and fertility in temperate cereals. J Exp Bot. 2019;70:193–204. doi: 10.1093/jxb/ery350 WOS:000459348500016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez MA, Tranquilli G, Lewis S, Kippes N, Dubcovsky J. Genetic and physical mapping of the earliness per se locus Eps-Am1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct Integr Genomic. 2016;16(4):365–82. doi: 10.1007/s10142-016-0490-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, et al. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci U S A. 2012;109(21):8328–33. doi: 10.1073/pnas.1120496109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner AS, Faure S, Zhang Y, Laurie DA. The effect of day-neutral mutations in barley and wheat on the interaction between photoperiod and vernalization. Theor Appl Genet. 2013;126(9):2267–77. doi: 10.1007/s00122-013-2133-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Nusinow DA. Into the evening: complex interactions in the Arabidopsis circadian clock. Trends Genet. 2016;32(10):674–86. doi: 10.1016/j.tig.2016.08.002 WOS:000384856300008. [DOI] [PubMed] [Google Scholar]
- 30.Zakhrabekova S, Gough SP, Braumann I, Muller AH, Lundqvist J, Ahmann K, et al. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci USA. 2012;109(11):4326–31. doi: 10.1073/pnas.1113009109 WOS:000301426700061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M. HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 2013;199(4):1045–59. doi: 10.1111/nph.12346 WOS:000322598700019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno N, Kinoshita M, Kinoshita S, Nishida H, Fujita M, Kato K, et al. Loss-of-function mutations in three homoeologous PHYTOCLOCK 1 genes in common wheat are associated with the extra-early flowering phenotype. PLoS One. 2016;11(10):e0165618. doi: 10.1371/journal.pone.0165618 WOS:000389604900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno N, Nitta M, Sato K, Nasuda S. A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotypeto einkorn wheat. Genes Genet Syst. 2012;87:357–67. [DOI] [PubMed] [Google Scholar]
- 34.Zikhali M, Wingen LU, Griffiths S. Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum). J Exp Bot. 2016;67(1):287–99. doi: 10.1093/jxb/erv458 WOS:000367816300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittern L, Steed G, Taylor LJ, Ramirez DC, Pingarron-Cardenas G, Gardner K, et al. Wheat EARLY FLOWERING 3 affects heading date without disrupting circadian oscillations. Plant Physiol. 2023;191(2):1383–403. doi: 10.1093/plphys/kiac544 PMID: WOS:000913128500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva CS, Nayak A, Lai XL, Hutin S, Hugouvieux V, Jung JH, et al. Molecular mechanisms of Evening Complex activity in Arabidopsis. Proc Natl Acad Sci USA. 2020;117(12):6901–9. doi: 10.1073/pnas.1920972117 WOS:000521821800079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezer D, Jung JH, Lan H, Biswas S, Gregoire L, Box MS, et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants. 2017;3(7):17087. doi: 10.1038/nplants.2017.87 WOS:000406037200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, et al. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014;55(5):958–76. doi: 10.1093/pcp/pcu030 WOS:000336491400008. [DOI] [PubMed] [Google Scholar]
- 39.Andrade L, Lub Y, Cordeiro A, Costa JMF, Wigge PA, Saibo NJM, et al. The evening complex integrates photoperiod signals to control flowering in rice. Proc Natl Acad Sci USA. 2022;119:e2122582119. doi: 10.1073/pnas.2122582119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods DP, McKeown MA, Dong Y, Preston JC, Amasino RM. Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 2016;170(4):2124–35. doi: 10.1104/pp.15.01279 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303(5664):1640–4. doi: 10.1126/science.1094305 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouche F, Woods DP, Linden J, Li WY, Mayer KS, Amasino RM, et al. EARLY FLOWERING 3 and photoperiod sensing in Brachypodium distachyon. Front Plant Sci. 2022;12:769194. doi: 10.3389/fpls.2021.769194 WOS:000745196300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang H, Alvarez S, Nusinow DA. Data on the identification of protein interactors with the Evening Complex and PCH1 in Arabidopsis using tandem affinity purification and mass spectrometry (TAP-MS). Data Brief. 2016;8:56–60. doi: 10.1016/j.dib.2016.05.014 WOS:000453168700011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu XL, Covington MF, Fankhauser C, Chory J, Wanger DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13(6):1293–304. doi: 10.1105/tpc.13.6.1293 WOS:000169519600005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gauley A, Boden SA. Stepwise increases in FT1 expression regulate seasonal progression of flowering in wheat (Triticum aestivum). New Phytol. 2021;229(2):1163–76. doi: 10.1111/nph.16910 WOS:000583516200001. [DOI] [PubMed] [Google Scholar]
- 46.Shaw LM, Turner AS, Laurie DA. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J. 2012;71(1):71–84. doi: 10.1111/j.1365-313X.2012.04971.x . [DOI] [PubMed] [Google Scholar]
- 47.Weller JL, Liew LC, Hecht VFG, Rajandran V, Laurie RE, Ridge S, et al. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Natl Acad Sci USA. 2012;109(51):21158–63. doi: 10.1073/pnas.1207943110 WOS:000313123700081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu SJ, Zhao XH, Hu YL, Liu SL, Nan HY, Li XM, et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet. 2017;49(5):773–9. doi: 10.1038/ng.3819 WOS:000400051400017. [DOI] [PubMed] [Google Scholar]
- 49.Rubenach AJS, Hecht V, Vander Schoor JK, Liew LC, Aubert G, Burstin J, et al. EARLY FLOWERING3 redundancy fine-tunes photoperiod sensitivity. Plant Physiol. 2017;173(4):2253–64. doi: 10.1104/pp.16.01738 WOS:000402054300023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bu TT, Lu SJ, Wang K, Dong LD, Li SL, Xie QG, et al. A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc Natl Acad Sci USA. 2021;118(8):e2010241118. doi: 10.1073/pnas.2010241118 WOS:000621797000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y, Zhao B, Xie Y, Jia H, Li Y, Xu M, et al. The evening complex promotes maize flowering and adaptation to temperate regions. Plant Cell. 2022. doi: 10.1093/plcell/koac296 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13(6):1305–15. doi: 10.1105/tpc.13.6.1305 WOS:000169519600006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, et al. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 2012;53(4):717–28. doi: 10.1093/pcp/pcs029 WOS:000302809300012. [DOI] [PubMed] [Google Scholar]
- 54.Zhao JM, Huang X, Ouyang XH, Chen WL, Du AP, Zhu L, et al. OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One. 2012;7(8):e43705. doi: 10.1371/journal.pone.0043705 WOS:000308063700128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itoh H, Tanaka Y, Izawa T. Genetic relationship between phytochromes and OsELF3-1 reveals the mode of regulation for the suppression of phytochrome signaling in rice. Plant Cell Physiol. 2019;60(3):549–61. doi: 10.1093/pcp/pcy225 WOS:000467887100007. [DOI] [PubMed] [Google Scholar]
- 56.Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, et al. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant. 2013;6(6):1877–88. doi: 10.1093/mp/sst088 . [DOI] [PubMed] [Google Scholar]
- 57.Woods DP, Li W, Sibout R, Laudencia-Chingcuanco D, Shao M, Vogel JP, et al. PHYTOCHROME C regulation of photoperiodic flowering via PHOTOPERIOD1 is mediated by EARLY FLOWERING 3 in Brachypodium distachyon. PLoS Genet. 2023; 19(5):e1010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Appendino ML, Slafer GA. Earliness per se and its dependence upon temperature in diploid wheat lines differing in the major gene Eps-Am1 alleles. Journal of Agricultural Science, Cambridge. 2003;141:149–54. [Google Scholar]
- 59.Lewis S, Faricelli ME, Appendino ML, Valarik M, Dubcovsky J. The chromosome region including the earliness per se locus Eps-Am1 affects the duration of early developmental phases and spikelet number in diploid wheat. J Exp Bot. 2008;59:3595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ochagavia H, Prieto P, Zikhali M, Griffiths S, Slafer GA. Earliness per se by temperature interaction on wheat development. Sci Rep-Uk. 2019;9:2584. doi: 10.1038/s41598-019-39201-6 WOS:000459399400053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ronald J, Wilkinson AJ, Davis SJ. EARLY FLOWERING3 sub-nuclear localization responds to changes in ambient temperature. Plant Physiol. 2021;187(4):2352–5. doi: 10.1093/plphys/kiab423 WOS:000733403700037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salome PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the arabidopsis circadian clock. Plant Cell. 2005;17(3):791–803. doi: 10.1105/tpc.104.029504 WOS:000227685600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raschke A, Ibanez C, Ullrich KK, Anwer MU, Becker S, Glockner A, et al. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 2015;15:197. doi: 10.1186/s12870-015-0566-6 WOS:000359400100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ford B, Deng WW, Clausen J, Oliver S, Boden S, Hemming M, et al. Barley (Hordeum vulgare) (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an EARLY FLOWERING 3-dependent manner. J Exp Bot. 2016;67(18):5517–28. doi: 10.1093/jxb/erw317 WOS:000386067000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang H, Alvarez S, Bindbeutel R, Shen ZX, Naldrett MJ, Evans BS, et al. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics. 2016;15(1):201–17. doi: 10.1074/mcp.M115.054064 WOS:000367461000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeom M, Kim H, Lim J, Shin AY, Hong S, Kim JI, et al. How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis? Mol Plant. 2014;7(11):1701–4. doi: 10.1093/mp/ssu086 WOS:000345832200010. [DOI] [PubMed] [Google Scholar]
- 67.Zhu C, Peng Q, Fu D, Zhuang D, Yu Y, Duan M, et al. The E3 ubiquitin ligase HAF1 modulates circadian accumulation of EARLY FLOWERING3 to control heading date in rice under long-day conditions. Plant Cell. 2018;30(10):2352–67. doi: 10.1105/tpc.18.00653 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu JW, Rubio V, Lee NY, Bai SL, Lee SY, Kim SS, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008;32(5):617–30. doi: 10.1016/j.molcel.2008.09.026 WOS:000261539700004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang LL, Shao YJ, Ding L, Wang MJ, Davis SJ, Liu JX. XBAT31 regulates thermoresponsive hypocotyl growth through mediating degradation of the thermosensor ELF3 in Arabidopsis. Sci Adv. 2021;7(19):eabf4427. doi: 10.1126/sciadv.abf4427 WOS:000648332700027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nieto C, Lopez-Salmeron V, Daviere JM, Prat S. ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol. 2015;25(2):187–93. doi: 10.1016/j.cub.2014.10.070 WOS:000348129100019. [DOI] [PubMed] [Google Scholar]
- 71.Monte E, Alonso JM, Ecker JR, Zhang YL, Li X, Young J, et al. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell. 2003;15(9):1962–80. doi: 10.1105/Tpc.012971 ISI:000185357900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–39. doi: 10.1038/nrg3291 . [DOI] [PubMed] [Google Scholar]
- 73.Woods DP, Ream TS, Minevich G, Hobert O, Amasino RM. PHYTOCHROME C is an essential light receptor for photoperiodic flowering in the temperate grass, Brachypodium distachyon. Genetics. 2014;198(1):397–408. doi: 10.1534/genetics.114.166785 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishida H, Ishihara D, Ishii M, Kaneko T, Kawahigashi H, Akashi Y, et al. Phytochrome C is a key factor controlling long-day flowering in barley (Hordeum vulgare L.). Plant Physiol. 2013;163:804–14. doi: 10.1104/pp.113.222570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao M, Geng F, Klose C, Staudt A, Huang H, Nguyen D, et al. Phytochromes measure photoperiod in Brachypodium. bioRxiv. 2019;697169. doi: doi.org/10.1101/697169 [Google Scholar]
- 76.Gawronski P, Ariyadasa R, Himmelbach A, Poursarebani N, Kilian B, Stein N, et al. A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics. 2014;196(4):1253–+. doi: 10.1534/genetics.113.158444 WOS:000334179300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475(7356):398–U161. doi: 10.1038/nature10182 WOS:000292911200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21(2):126–33. doi: 10.1016/j.cub.2010.12.021 WOS:000286680800022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campoli C, Shtaya M, Davis SJ, von Korff M. Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 2012;12:97. doi: 10.1186/1471-2229-12-97 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Farre EM, Liu T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr Opin Plant Biol. 2013;16(5):621–9. doi: 10.1016/j.pbi.2013.06.015 WOS:000326432800012. [DOI] [PubMed] [Google Scholar]
- 81.Diaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One. 2012;7(3):e33234. doi: 10.1371/journal.pone.0033234 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alqudah AM, Youssef HM, Graner A, Schnurbusch T. Natural variation and genetic make-up of leaf blade area in spring barley. Theor Appl Genet. 2018;131(4):873–86. doi: 10.1007/s00122-018-3053-2 WOS:000427584400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kusakina J, Rutterford Z, Cotter S, Marti MC, Laurie DA, Greenland AJ, et al. Barley Hv CIRCADIAN CLOCK ASSOCIATED 1 and Hv PHOTOPERIOD H1 are circadian regulators that can affect circadian rhythms in Arabidopsis. PLoS One. 2015;10(6):e0127449. doi: 10.1371/journal.pone.0127449 WOS:000356329900013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alqudah AM, Sharma R, Pasam RK, Graner A, Kilian B, Schnurbusch T. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS One. 2014;9(11):e113120. doi: 10.1371/journal.pone.0113120 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qin Z, Bai Y, Muhammad S, Wu X, Deng P, Wu J, et al. Divergent roles of FT-like 9 in flowering transition under different day lengths in Brachypodium distachyon. Nat Commun. 2019;10(1):812. doi: 10.1038/s41467-019-08785-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y, Peng Q, Chen GX, Li XH, Wu CY. OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol Plant. 2013;6(1):202–15. doi: 10.1093/mp/sss062 WOS:000314117100021. [DOI] [PubMed] [Google Scholar]
- 87.Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422(6933):719–22. doi: 10.1038/nature01549 . [DOI] [PubMed] [Google Scholar]
- 88.Mishra P, Panigrahi KC. GIGANTEA—an emerging story. Front Plant Sci. 2015;6:8. doi: 10.3389/fpls.2015.00008 WOS:000348446200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krasileva KV, Vasquez-Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, et al. Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci U S A. 2017;114(6):E913–E21. doi: 10.1073/pnas.1619268114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fu D, Szűcs P, Yan L, Helguera M, Skinner J, Hayes P, et al. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4 [DOI] [PubMed] [Google Scholar]
- 92.Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 2009;149(1):245–57. doi: 10.1104/pp.108.129353 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013;163(3):1433–45. doi: 10.1104/pp.113.225854 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed. 2014;33(1):1–14. doi: 10.1007/s11032-013-9917-x WOS:000329670900001. [DOI] [Google Scholar]
- 95.Woods DP, Ream TS, Bouche F, Lee J, Thrower N, Wilkerson C, et al. Establishment of a vernalization requirement in Brachypodium distachyon requires REPRESSOR OF VERNALIZATION1. Proc Natl Acad Sci USA. 2017;114(25):6623–8. doi: 10.1073/pnas.1700536114 WOS:000403687300052. [DOI] [PMC free article] [PubMed] [Google Scholar]