Abstract
A fundamental feature of cell signaling is the conversion of extracellular signals into adaptive transcriptional responses. The role of RNA modifications in this process is poorly understood. The small nuclear RNA 7SK prevents transcriptional elongation by sequestering the cyclin dependent kinase 9/cyclin T1 (CDK9/CCNT1) positive transcription elongation factor (P-TEFb) complex. We found that epidermal growth factor signaling induces phosphorylation of the enzyme methyltransferase 3 (METTL3), leading to METTL3-mediated methylation of 7SK. 7SK methylation enhanced its binding to heterogeneous nuclear ribonucleoproteins, causing the release of the HEXIM1 P-TEFb complex subunit1 (HEXIM1)/P-TEFb complex and inducing transcriptional elongation. Our findings establish the mechanism underlying 7SK activation and uncover a previously unknown function for the m6A modification in converting growth factor signaling events into a regulatory transcriptional response via an RNA methylation–dependent switch.
m6A methylation of the non-coding RNA 7SK promotes transcriptional activity upon growth factor stimulation.
INTRODUCTION
Extracellular signals regulate intracellular pathways that control adaptive responses such as growth, quiescence, differentiation, and cell migration. Ultimately, these signaling pathways converge in the nucleus to elicit specific transcriptional programs. 7SK (RN7SK) is a 332-nucleotide small nuclear RNA (snRNA) that interacts with multiple RNA binding proteins to regulate transcriptional elongation. 7SK sequesters the positive transcription elongation factor (P-TEFb) complex CDK9/CCNT1 by interacting with the protein HEXIM1 (1, 2). Under conditions that demand an increase in transcriptional elongation, such as cell proliferation, 7SK dissociates from HEXIM1/P-TEFb and binds to heterogeneous nuclear ribonucleoprotein (HNRNP) proteins, including HNRNPA1, HNRNPA2B1, HNRNPR, and HNRNPQ (SYNCRIP) (3–5). Upon release from 7SK, P-TEFb phosphorylates the C-terminal domain (CTD) of the RNA polymerase (Pol) II, allowing transcription elongation to proceed (6, 7).
RNA modifications are a key feature of posttranscriptional gene expression control. Methylation of RNA on the N6 nitrogen of adenosine (m6A) is the most abundant posttranscriptional modification in mRNA (8). We and others have implicated m6A, as well as the enzymatic complex responsible for RNA methylation METTL3/METTL14 (9–11), in regulation of RNA stability (12), microRNA (miRNA) processing (13, 14), RNA splicing (15, 16), and translation (17, 18). These processes affect cellular functions such as meiosis (19, 20), cell proliferation (17, 21, 22), and embryonic stem cell differentiation (22, 23), as well as pathophysiological states such as cancer (24, 25). Despite the known general roles for m6A in cellular homeostasis, it remains unclear how signaling pathways affect METTL3 function and regulate downstream transcriptional activity.
RESULTS
METTL3 methylates the 7SK small noncoding RNA
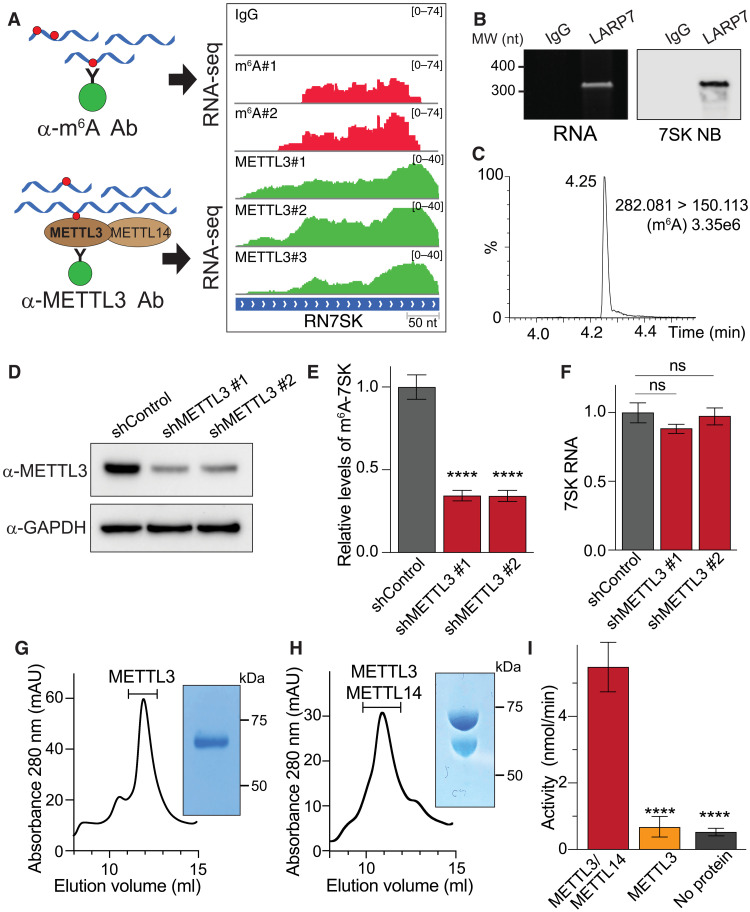
Nuclear m6A sequencing (m6A-seq) experiments revealed the presence of m6A modifications on 7SK (Fig. 1A) (13). Furthermore, high-throughput sequencing of RNA isolated in cross-linking immunoprecipitation (HITS-CLIP) of METTL3 revealed direct binding of the m6A-methylating enzyme METTL3 to the 7SK RNA (Fig. 1A), suggesting that METTL3 may recognize and methylate 7SK (13). To further validate these results with an independent method, we pulled down endogenous 7SK and used RNA mass spectrometry (MS) to assess its m6A content. To isolate endogenous 7SK, we took advantage of its known interaction with the protein LARP7 (26, 27). LARP7 binds 7SK constitutively, thus protecting it from degradation (28), which contrasts with the binding of HNRNPs or HEXIM1/P-TEFb to 7SK, which depends on a proliferative or nonproliferative cell state, respectively. We therefore generated a HeLa cell line stably expressing Flag-tagged LARP7 protein at physiological levels (fig. S1A). To facilitate the isolation of native LARP7-7SK complexes, we performed mechanical disruption of cells at cryogenic temperature (cryomilling) using liquid nitrogen, thus preserving posttranslational states and high-order complexes (29). As expected, immunoprecipitation of Flag-tagged LARP7 (Flag-IP) efficiently pulled down endogenous 7SK as the main RNA species, as revealed by acrylamide gel electrophoresis and Northern blotting (Fig. 1B). We extracted 7SK from the gel and, after purification, digested it with a mixture of enzymes to generate single nucleotides. Levels of m6A were then measured by ultra-high-performance liquid chromatography coupled with tandem MS (UHPLC-MS/MS). MS detected the presence of m6A in endogenous 7SK, thus validating its methylation status and diminishing the possibility of cross-recognition of the m6A antibody with other potential RNA modifications (Fig. 1C and fig. S1B).
Fig. 1. 7SK is m6A methylated by METTL3.
(A) Tracks of m6A-seq and METTL3 HITS-CLIP on 7SK. The top shows an immunoglobulin G (IgG)–negative control. On the left, a schematic representation of the experiments. Ab, antibody. (B) Isolation of endogenous 7SK by IP of Flag-tagged LARP7. Purified endogenous 7SK as shown by a urea-acrylamide gel (left) and Northern blot (right). IgG was used as controls. MW, molecular weight; nt, nucleotide; NB, Northern blot. (C) Detection of m6A in 7SK through UHPLC-MS/MS. The figure shows a representative spectrum from three biological replicates. (D) Knockdown of METTL3 using two independent shRNAs. Total cell extracts were analyzed by Western blot using the indicated antibodies. (E) m6A-methylated nuclear RNA was immunoprecipitated and the levels of 7SK were quantified by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Total endogenous 7SK levels were used for normalization. The graph shows means ± SD from three biological replicates. One-way analysis of variance (ANOVA) with Dunnett’s posttest; ****P < 1 × 10−4. (F) Normalized levels of 7SK were quantified by qRT-PCR. The bar graph depicts the effect of METTL3 depletion using two independent shRNAs. The graph shows means ± SD from three biological replicates. One-way ANOVA with Dunnett’s posttest. (G and H) METTL3 (G) and METTL3/METTL14 protein complexes (H) were expressed in insect cells, affinity-purified using Ni beads, and separated using size exclusion chromatography. The inset shows a representative image of a Coomassie staining of SDS–polyacrylamide gel electrophoresis (SDS-PAGE) from one of the fractions corresponding to the peak (indicated in the chromatogram, absorbance 280 nm was measured in milli-absorbance units (mAU). (I) Methylation of in vitro–transcribed (IVT) 7SK using proteins obtained from (G) and (H). The graph shows means ± SD from a representative experiment out of three biological replicates. One-way ANOVA with Dunnett’s posttest; ****P < 1 × 10−4.
To confirm that this methylation is mediated by METTL3, we depleted METTL3 by RNA interference and quantified the effect on abundance of methylated 7SK. To do this, we generated stable cell lines expressing two independent short hairpin RNAs (shRNAs) against METTL3 and performed immunoprecipitation of m6A-methylated RNA (m6A-IP) of nuclear RNA followed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) of 7SK. Depleting METTL3 reduced methylated 7SK levels by almost threefold, without affecting total 7SK levels (Fig. 1, D to F). We next tested the ability of the purified METTL3/METTL14 complex or METTL3 (expressed in insect cells) to methylate in vitro–transcribed (IVT) 7SK. As shown in Fig. 1 (G to I), METTL3/METTL14 complex methylates 7SK in vitro. 7SK methylation required both the catalytic METTL3 and noncatalytic METTL14 proteins of the methylation complex, as METTL3 alone was insufficient to drive methylation (Fig. 1I). Together, these results demonstrate that the METTL3/METTL14 complex methylates 7SK.
m6A favors binding of HNRNP proteins to 7SK
Although the structure of 7SK is not fully elucidated and could be altered by binding of different protein complexes, secondary structure predictions and previous Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) experiments identify several distinct stem-loops in 7SK (30–32). HEXIM1 binds a stem-loop close to the 5′ end of the RNA, and the crystal structure of a partial HEXIM1 protein associated with a double-stranded RNA fragment corresponding to 7SK nucleotides 24 to 87 has been determined (33). HNRNPA1, HNRNPA2B1, HNRNPQ, and HNRNPR mainly bind to a hairpin corresponding to 7SK nucleotides 196 to 277 (31, 34). LARP7 protects 7SK from degradation by binding to the junction of the 5′ and 3′ ends (Fig. 2A) (28, 35–37).
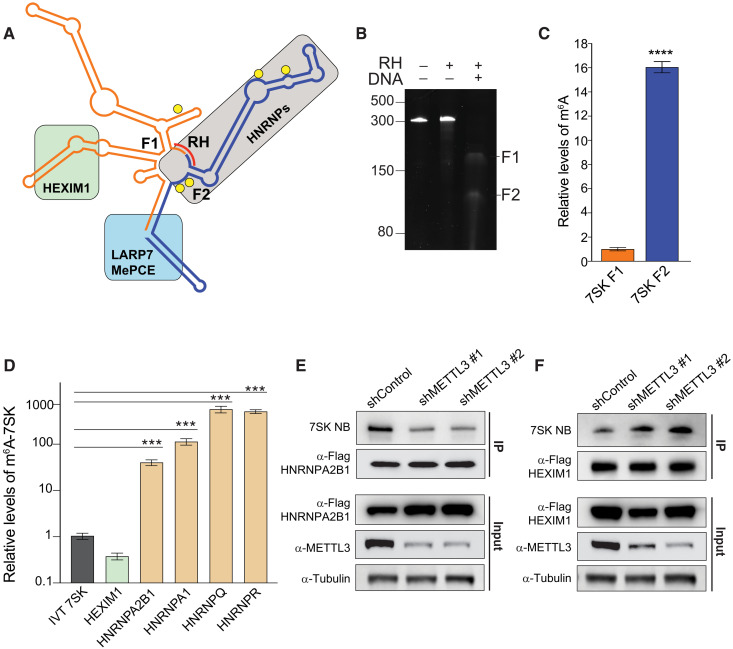
Fig. 2. HEXIM1 preferentially binds unmethylated 7SK.
(A) 7SK secondary structure representation. Proteins known to interact with 7SK are depicted as a green (HEXIM1/P-TEFb), gray (HNRNPs), or light blue (LARP7 and MePCE) box. Predicted m6A motifs are shown as yellow circles, and the complementary DNA oligo used to fragment 7SK in the RNase H (RH) digestion is shown as a red line. (B) RNase H digestion of endogenous 7SK. Tris-borate EDTA (TBE)–urea acrylamide RNA gel showing untreated 7SK and treated with RNase H, with or without the addition of the complementary DNA oligo (DNA). (C) Relative m6A levels in the two fragments generated by RNase H treatment of endogenous 7SK pulled down using Flag-tagged LARP7 as shown in (B). After purification, 7SK was digested by RNase H using the DNA complementary oligo depicted in (A). After fragmentation, the purified fragments were subjected to m6A-IP followed by qRT-PCR quantification. The m6A-immunoprecipitated 7SK fragments were normalized to each fragment before the m6A-IP. The graph shows means ± SD from three biological replicates. Student’s two-tailed t test; ****P < 1 × 10−4 (D) Stably expressing Flag-tagged proteins were used to pull down endogenous 7SK. After purification of the pulled-down 7SK RNA, a second IP using an m6A antibody was performed, and the levels of 7SK were quantified by qRT-PCR. Nonmethylated IVT 7SK RNA was used as a negative control for the m6A-IP. The graph shows means ± SD from three biological replicates. One-way ANOVA with Dunnett’s posttest; ***P < 1 × 10−3. (E and F) 7SK-protein interaction with HNRNPA2B1 (E) and HEXIM1 (F) upon METTL3 depletion using two independent shRNAs. The top two panels represent the pull-down of endogenous 7SK measured by Northern blot. The immunoprecipitated proteins are detected by Western blot using the indicated antibodies. The bottom three panels show the input protein levels before the IP. Blots are representatives of at least two biological repeats.
Sequence analysis reveals that 7SK contains five canonical METTL3 recognition sequences consisting of the ribonucleotide sequence DRACH located between nucleotides 170 and 290. Four of these motifs are located within the known hairpin that is recognized by HNRNP proteins (yellow circles in Fig. 2A and fig. S1C). To better understand the location of m6A modifications experimentally, we used ribonuclease H (RNAse H) to fragment 7SK and analyzed the m6A levels in each fragment. We first pulled down endogenous 7SK using Flag-LARP7 and extracted 7SK from the complex. We then used a DNA oligonucleotide complementary to the sequence between positions 190 and 198. The hybridization of this DNA oligo will guide RNase H to cleave the resulting RNA-DNA hybrids and split 7SK into two fragments, F1 and F2 (Fig. 2, A and B, and fig. S1, C and D). After RNase H cleavage, the fragments were isolated and subjected to m6A-IP. After this IP, the fragments were further purified and quantified by qRT-PCR. We found that the 3′ fragment (F2), which contains four of the five predicted m6A motifs, exhibited the highest levels of m6A (Fig. 2C).
To understand the effects of m6A modifications on 7SK, we tested the ability of the known 7SK-interacting proteins to interact with methylated 7SK. To do this, we generated independent cell lines stably expressing the main 7SK direct interactors (HEXIM1, HNRNPA2B1, HNRNPA1, HNRNPQ, and HNRNPR) as Flag-tagged proteins in HeLa cells (fig. S2A). The generation of these multiple cell lines facilitates the normalization of the IP protocol, making it less dependent on the intrinsic differences among antibodies against the various proteins. After Flag-IPs of each independent protein, 7SK was extracted and purified, and a second IP using an m6A antibody or control beads was performed. The methylated immunoprecipitated 7SK was then analyzed by qRT-PCR and normalized by the total amount of 7SK immunoprecipitated by the Flag-IP. As a negative control for m6A-IP, we used an IVT nonmethylated 7SK. As shown in Fig. 2D, all the HNRNP proteins pulled down m6A-methylated 7SK between 50- and 500-fold more efficiently than the background IVT unmethylated control. In contrast, HEXIM1 pulled down m6A-methylated 7SK to a similar degree as the negative control (Fig. 2D), indicating that HNRNP proteins but not HEXIM1 interact with m6A-methylated 7SK. We then tested the impact of METTL3 depletion on the ability of these proteins to interact with endogenous 7SK. We depleted METTL3 from cells expressing Flag-HNRNPA2B1 as a representative of the HNRNP proteins and from cells expressing Flag-HEXIM1. As shown in Fig. 2E, METTL3 depletion reduced the levels of 7SK that co-immunoprecipitated with HNRNPA2B1, while not affecting the association of 7SK with LARP7 (fig. S2B). However, knock down of METTL3 increased the levels of 7SK that immunoprecipitated with HEXIM1 (Fig. 2F) as well as the levels of HEXIM1 and P-TEFb associated with the 7SK complex (fig. S2C). Together, these results demonstrate that the region of 7SK recognized by HNRNP proteins is the most m6A methylated and that this methylation facilitates the interaction of 7SK with HNRNPs, thus decreasing its binding to HEXIM1.
Epidermal growth factor stimulates 7SK methylation and transcriptional activity
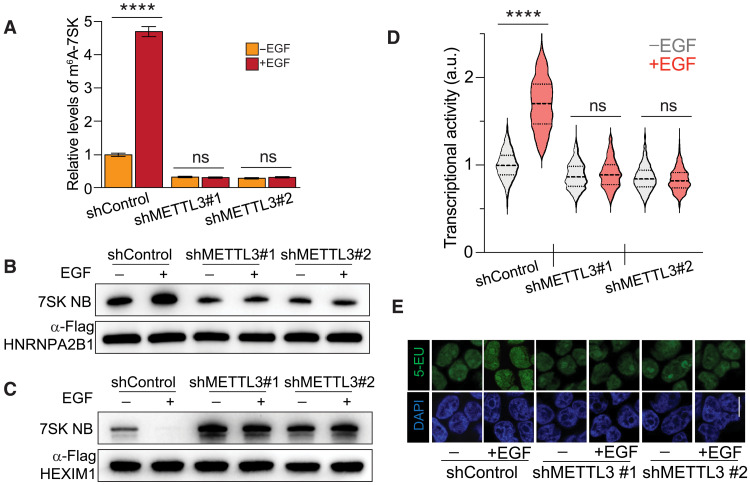
The effect of 7SK methylation on the formation of the 7SK/HNRNP complex and the release of HEXIM1/P-TEFb to promote transcriptional activity is consistent with the known positive effect of m6A on cell proliferation. On the basis of this relationship, we hypothesized that growth factors might regulate the methylation of 7SK. We focused on the epidermal growth factor (EGF) because of its established role in cell proliferation. To determine the impact of EGF on 7SK methylation, we immunoprecipitated m6A-methylated nuclear RNA from unstimulated and EGF-treated cells using an m6A antibody, followed by a qRT-PCR for 7SK. As shown in Fig. 3A, EGF stimulation increased the levels of m6A-methylated 7SK several-fold. This effect was prevented by METTL3 depletion, indicating that METTL3 is required for EGF-stimulated methylation of 7SK (Fig. 3A). Because EGF stimulation induces 7SK methylation, and METTL3 depletion reduces HNRNPA2B1 binding to 7SK, we predicted that EGF stimulation should increase the interaction between HNRNPA2B1 and 7SK. As expected, EGF stimulation enhanced HNRNPA2B1 binding to 7SK, as shown by Northern blotting for 7SK after IP of HNRNPA2B1. This effect was lost in cells depleted of METTL3, indicating that the ability of EGF to induce the interaction between HNRNPA2B1 and 7SK depends on METTL3 (Fig. 3B). In contrast, EGF stimulation reduced the interaction between HEXIM1 and 7SK, an outcome that was eliminated by depletion of METTL3 (Fig. 3C). Consistent with the results shown in Fig. 2 (E and F), basal levels of 7SK bound to HNRNPA2B1 decreased upon METTL3 depletion, with the opposite observed for HEXIM1 (Fig. 3, B and C).
Fig. 3. METTL3 depletion favors HEXIM1 binding to 7SK and decreases EGF-induced transcriptional activation.
(A) m6A methylation levels on 7SK upon EGF stimulation in wild-type cells and cells depleted of METTL3. Nuclear m6A-IP followed by qRT-PCR quantification of 7SK upon EGF stimulation. A control shRNA and two independent shRNAs targeting METTL3 were stably expressed in HeLa cells. The m6A-immunoprecipitated 7SK RNA was normalized to total 7SK. The graph shows means ± SD from three biological replicates. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4. ns, not significant. (B and C) 7SK-protein interaction with HNRNPA2B1 (B) and HEXIM1 (C) upon EGF stimulation in control cells and cells depleted of METTL3 using two independent shRNAs. The top panels show the pull-down of endogenous 7SK measured by Northern blot. The immunoprecipitated proteins are detected by Western blot using the indicated antibodies. Blots are a representation of two biological replicates. (D) Cellular transcriptional activity upon EGF stimulation in control cells and cells depleted of METTL3 using two independent shRNAs, as measured by RNA Click-iT. Violin plots of fluorescent measurement of 600 cells per condition. (E) Micrographs of representative cell nuclei of (D). Scale bar, 10 μm. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4. DAPI, 4′,6-diamidino-2-phenylindole; 5-EU, 5-ethynyl uridine.
Because it is stablished that the release of HEXIM1 from 7SK results in transcriptional elongation via the activity of free P-TEFb, we tested the functional role of m6A on EGF-stimulated transcription. We found that depletion of METTL3 resulted in the inability of P-TEFb to phosphorylate CTD of RNA Pol II at position S2 upon EGF stimulation (fig. S3). We then used the quantitative imaging technique called RNA Click-iT, which enables the detection of global RNA transcription in cells (38). As shown in Fig. 3 (D and E), EGF stimulated transcription in control cells. However, the depletion of METTL3 eliminated the ability of EGF to stimulate transcription, indicating that m6A is required for this EGF-induced response (Fig. 3, D and E). These results indicate that m6A is required for the EGF-induced exchange in 7SK binding of HEXIM1 for HNRNPs, as well as subsequent transcriptional activity.
EGF modulates the interaction between METTL3 and HEXIM1
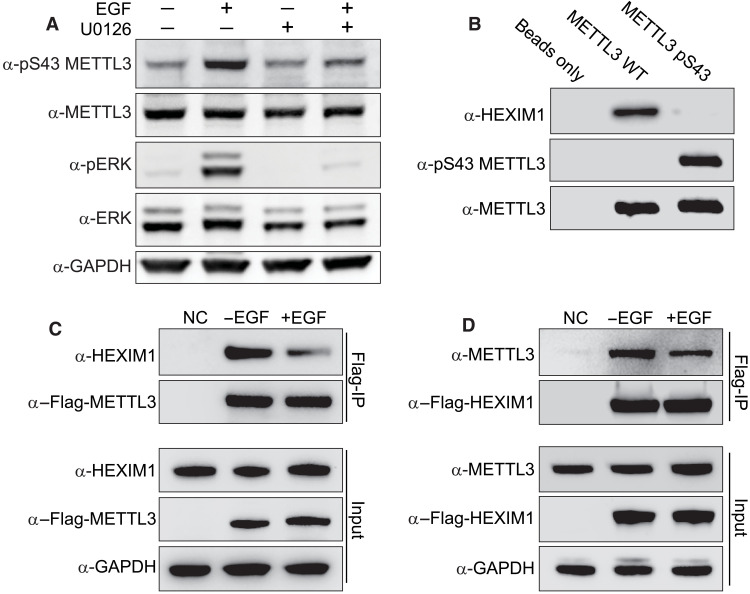
On the basis of the requirement of METTL3 for EGF-induced 7SK methylation, we hypothesized that growth factors might regulate METTL3 activity. Because protein phosphorylation is a primary mechanism for regulating protein function in response to extra- and intracellular signals, we used IP coupled to phospho-MS to investigate whether growth factor stimulation alters METTL3 phosphorylation. EGF stimulation induced METTL3 phosphorylation at serine 43 (pS43) (fig. S4A). On the basis of these results, we generated polyclonal antibodies against pS43-METTL3 and validated the EGF-induced phosphorylation of METTL3 (Fig. 4A). Because pS43 is a predicted extracellular signal–regulated kinase 1 (ERK1) phosphorylation site (Ser/Thr-Pro), we next tested the requirement of ERK for the EGF-induced S43 METTL3 phosphorylation. We used the general mitogen-activated protein kinase kinase (MEK) inhibitor U0126 to block ERK activation. As shown in Fig. 4A, U0126 completely abolished EGF-induced ERK activation in HeLa cells and prevented the EGF-dependent increase in S43-METTL3 phosphorylation (Fig. 4A). To determine whether ERK can phosphorylate METTL3 directly, we performed in vitro kinase assays using recombinant ERK1. We expressed and isolated human METTL3 as a glutathione S-transferase (GST) fusion protein in bacteria to eliminate endogenous phosphorylation events that may occur in eukaryotic cells (fig. S4B). Incubation of ERK with METTL3 in vitro resulted in robust phosphorylation of METTL3 on S43 as measured by phospho-MS (fig. S4C). These results demonstrate that ERK phosphorylates METTL3 at S43 downstream of growth factor stimulation.
Fig. 4. METTL3 phosphorylation prevents its interaction with HEXIM1.
(A) Effect of U0126 on S43-METTL3 phosphorylation induced by EGF in HeLa cells. Western blot analysis of total cell extracts using the indicated antibodies. Blots in (A) to (D) are representatives of three biological repeats. (B) Pull-down of endogenous HEXIM1 from HeLa cell lysates using beads containing GST-tagged unphosphorylated or pS43 METTL3, produced in bacteria. Beads alone were used as a negative control. Western blot analysis using the indicated antibodies. Blots are a representation of three biological repeats. (C) IP of stably expressed Flag-METTL3. The top two panels represent the IP and the bottom three the input. Western blot analysis using the indicated antibodies. Wild-type HeLa cells were used as a negative control (NC) for the IP. (D) IP of stably expressed Flag-HEXIM1.
We hypothesized that METTL3 phosphorylation on S43 could affect the interaction between METTL3 and 7SK-interacting proteins. To test this, we generated recombinant GST-METTL3 phosphorylated at position S43 using the orthogonal translation system SepOTS (39). This system is based on an Escherichia coli strain engineered to fully incorporate phosphoserine (Sep) genetically into recombinant proteins (39). We validated the presence of pS43 by Western blot techniques (Fig. 4B). We then used glutathione beads loaded with GST-pS43-METTL3 and the nonphosphorylated GST-METTL3 counterpart protein fusions as “baits” to pull down interacting proteins from HeLa cell lysates obtained by cryomilling. We found that 7SK-interacting protein HEXIM1 specifically interacts with nonphosphorylated METTL3 (Fig. 4B). Because EGF stimulates the phosphorylation of METTL3 on S43, we next tested the effect of EGF stimulation on METTL3/HEXIM1 interactions in a cellular context. Using cryomilled lysates from serum-starved cells stimulated or not with EGF, we immunoprecipitated Flag-METTL3 and performed Western blots for endogenous HEXIM1 (Fig. 4C). Consistent with the GST fusion pull-down experiments, we found that EGF stimulation reduced the interaction between METTL3 and HEXIM1. These findings were confirmed with reciprocal co-IP using Flag-HEXIM1 (Fig. 4D). The interaction between METTL3 and HEXIM1 seems to be the only interaction of METTL3 with any other component of the 7SK complex, suggesting that HEXIM1 and METTL3 interaction is independent of the 7SK-interacting proteins and the 7SK RNA (fig. S4, D and E). These results strongly suggest that S43 phosphorylation disrupts the interaction between METTL3 and HEXIM1.
On the basis of these results, we hypothesize that HEXIM1 might keep METTL3 in an inhibited state in close proximity to the 7SK complexes and ready to be activated by growth factors. To test this hypothesis, we knock down HEXIM1 using two independent shRNAs. We then extracted nuclear RNA from those two cell lines and a control line, and we used an antibody against m6A to isolate methylated RNA followed by qRT-PCR of the methylated 7SK. As shown in fig. S4, F and G, depletion of HEXIM1 increased the levels of methylated 7SK, particularly under unstimulated conditions, suggesting that HEXIM1 has a negative effect on the activity of METTL3 and that HEXIM1 depletion could diminish the need of EGF-induced phosphorylation of METTL3.
7SK interactome switch depends on METTL3 phosphorylation
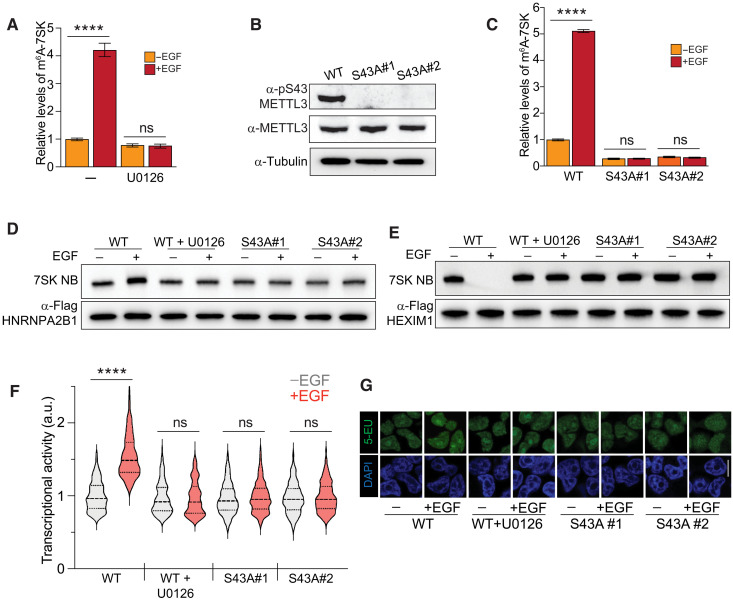
We decided to test whether the effect of EGF on the methylation levels of 7SK was mediated by ERK. We found that the MEK inhibitor U0126 blocked the EGF-induced increase in 7SK methylation, indicating that ERK activity is required for this process (Fig. 5A). To determine whether METTL3 phosphorylation downstream of ERK is required for 7SK methylation, we performed a loss-of-function experiment using CRISPR-Cas9 to eliminate the S43 phosphorylation site of endogenous METTL3. We obtained positive clones carrying the S43A homozygous mutation (METTL3S43A; fig. S5A). These mutated cell lines, as expected, were defective in EGF-induced S43 phosphorylation (Fig. 5B). We then used these clones to assess the impact of pS43 on the ability of METTL3 to methylate 7SK. As shown in Fig. 5C, wild-type cells efficiently increased the 7SK methylation upon EGF stimulation. However, two independent METTL3S43A clones lost the EGF-induced capacity for the m6A methylation of 7SK (Fig. 5C). These results indicate that the ERK-mediated phosphorylation of METTL3 is required for the ability of the methyltransferase to methylate 7SK upon EGF stimulation.
Fig. 5. EGF transcriptional activation depends on METTL3 phosphorylation.
(A) Effect of U0126 on the EGF-induced m6A methylation of 7SK. The experimental design is similar to Fig. 4A. m6A-methylated RNA was immunoprecipitated from cells under the treatments shown in the figure, and 7SK was quantified by qRT-PCR. The m6A-immunoprecipitated 7SK RNA was normalized to the total 7SK. The graph shows means ± SD from three biological replicates. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4. (B) CRISPR-Cas9 mutagenesis of S43A of METTL3. Western blot of METTL3 pS43 upon EGF stimulation in wild-type (WT) cells and two METTL3S43A homozygous clones. (C) Similar to (A), but in this case, wild-type cells and two METTL3S43A homozygous clones were used. The graph shows means ± SD from three biological replicates. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4. (D and E) 7SK-protein interaction with HNRNPA2B1 (D) and HEXIM1 (E), upon EGF stimulation in wild-type cells and two independent METTL3S43A homozygous clones. Blots are representatives of two biological replicates. (F and G) Effect of U0126 and mutagenesis of S43A-METTL3 on transcriptional activity induced by EGF stimulation, as measured by RNA Click-iT. Violin plots of fluorescent measurement of 600 cells per condition. (G) Micrographs of representative cell nuclei of (F). Scale bar, 10 μm. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10−4.
To assess the impact of S43 METTL3 phosphorylation on the 7SK interactome switch upon EGF stimulation, we immunoprecipitated HNRNPA2B1 and assessed the levels of associated 7SK by Northern blot. As shown in Fig. 5D, U0126 blocked the ability of EGF to induce the HNRNPA2B1/7SK interaction. This effect was mimicked by the METTL3S43A clones (Fig. 5D). As expected, MEK inhibition and S43A mutagenesis blocked the suppressive effect of EGF on the HEXIM1/7SK interaction (Fig. 5E). Consistent with the data shown so far, inhibiting ERK activation prevented the association of HNRNPA2B1 with and the release of HEXIM1 from 7SK upon EGF stimulation, an effect that was phenocopied by eliminating the S43 phosphorylation site in the METTL3S43A clones (Fig. 5, D and E). Endogenous METTL3S43A increased its affinity for HEXIM1, consistent with the inhibitory role of S43 phosphorylation on the interaction between METTL3 and HEXIM1 (fig. S5B).
We further tested whether the EGF requirement of m6A for transcriptional activity relies on the ability of EGF to induce ERK-mediated phosphorylation of METTL3. As anticipated, the effect of EGF on transcriptional activity was blocked by both the MEK inhibitor U0126 and by the elimination of the S43 METTL3 phosphorylation site (Fig. 5, F and G). Together, these results demonstrate that METTL3 and its ERK-mediated phosphorylation of S43 are required for the EGF-induced 7SK-m6A methylation and the subsequent transition from 7SK-bound HEXIM1 to HNRNP complexes, resulting in enhanced transcriptional activity. Our findings, thus far, reveal a model whereby EGF induces phosphorylation of METTL3, this, in turn, causes 7SK methylation, leading to transcription activation.
7SK methylation is required for interactome switch and transcriptional activity
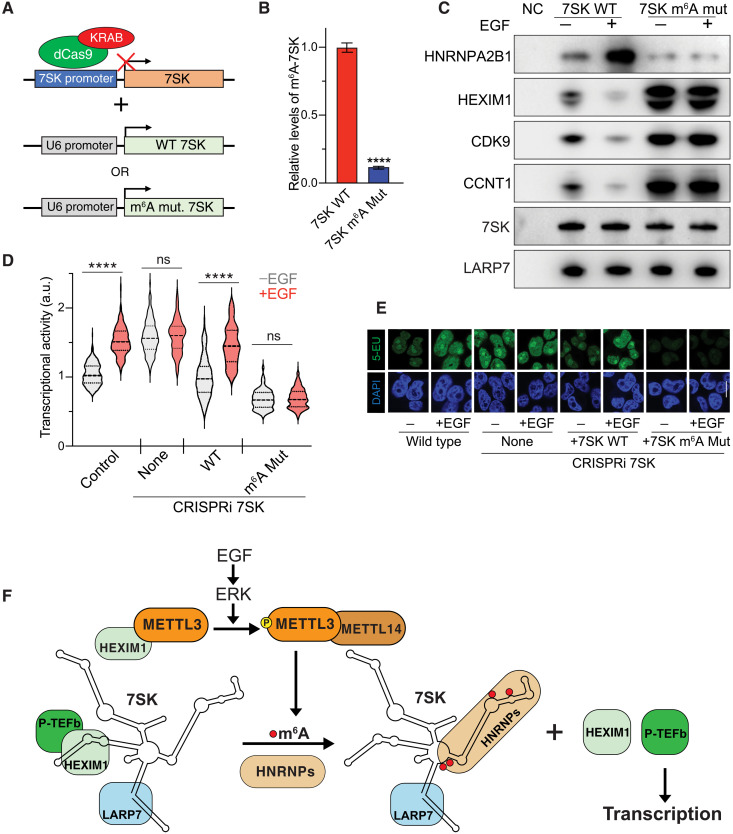
To test whether 7SK methylation is responsible for the interactome switch and transcriptional activity, we mutated the adenosines corresponding to the predicted methylation sites described above. To achieve this, we used CRISPR interference (CRISPRi) (40) to repress the expression of endogenous 7SK. The use of a guide RNA complementary to the 7SK promoter region in conjunction with dCas9-KRAB led to a 90% reduction of endogenous 7SK (Fig. 6A and fig. S5A). We then used a lentiviral expression system to express exogenous 7SK, either wild-type or m6A-mutant, under the RNA Pol III U6 promoter (Fig. 6A and fig. S6A). The mutations introduced were A172G, A220T, A228G, A281G, and A288G. As shown in Fig. 6B, elimination of the predicted m6A sites markedly reduces the levels of m6A on 7SK.
Fig. 6. 7SK m6A methylation is required for HEXIM1 dissociation and transcriptional activation downstream EGF signaling.
(A) CRISPRi was used to down-regulate the expression of 7SK. Once cells were depleted of endogenous 7SK, lentiviral vectors were used to stably express exogenous wild-type or m6A-mutant 7SK under the U6 promoter. (B) IP of m6A-methylated RNA followed by qRT-PCR of 7SK from cells depleted of 7SK and transduced with wild-type 7SK (red bar) or 7SK m6A mutant (blue bar). The m6A-immunoprecipitated 7SK RNA was normalized to total 7SK. The graph shows means ± SD from three biological replicates. Student’s two-tailed t test; ****P < 1 × 10−4 (C) Flag-LARP7 IP of CRISPRi 7SK plus wild-type 7SK or 7SK mutant for m6A sites upon EGF stimulation. Western blot for the interacting proteins using the indicated antibodies. Northern blot using a probe against 7SK was performed. (D and E) Effect of mutagenesis of m6A sites of 7SK on transcriptional activity induced by EGF stimulation, as measured by RNA Click-iT. Violin plots of fluorescent measurement of 600 cells per condition. (E) Micrographs of representative cell nuclei of (D). Scale bar, 10 μm. Two-way ANOVA with Tukey’s posttest; ****P < 1 × 10–4. (F) Model. The work presented here is summarized in this model. Upon EGF stimulation, the downstream effector ERK kinase is activated and phosphorylates METTL3 at position S43. This phosphorylation results in the release of METTL3 from its association with HEXIM1, thus allowing the methylation of 7SK. Subsequently, m6A methylation of 7SK results in its association with HNRNP proteins and release of the HEXIM1/P-TEFb complex. Free P-TEFb is then available to induce transcription activation.
On the basis of the experiments presented thus far, we predicted that elimination of the m6A sites would result in the inability of 7SK to interact with HNRNPs, would not release HEXIM1 and P-TEFb, and therefore would impair the stimulation of transcription upon EGF stimulation. As expected, wild-type 7SK efficiently increased the binding of HNRNPA2B1 and released HEXIM1 and CDK9/CCNT upon EGF stimulation, while the m6A-mutant version of 7SK was inert to the interactome switch by the growth factor (Fig. 6C and fig. S6B). Elimination of the m6A sites decreased the basal interaction with HNRNPA2B1 and enhanced the basal interaction between 7SK and the HEXIM1/P-TEFb complex (Fig. 6C and fig. S6B). 7SK depletion by CRISPRi, consistent with its role in sequestering the transcription elongation factors, showed an enhanced basal transcriptional activity compared to control wild-type cells. Moreover, EGF failed to induce transcription in the absence of 7SK, demonstrating the requirement of 7SK for the downstream transcriptional functions of EGF (Fig. 6, D and E). Reintroduction of wild-type 7SK rescued the basal transcriptional levels and the regulation by EGF stimulation, while the m6A-mutant 7SK was unresponsive to EGF stimulation and failed to induce transcriptional activity (Fig. 6, D and E). Together, these results demonstrate that the methylation of 7SK is required for the release of HEXIM1/P-TEFb and induction of transcription upon EGF stimulation.
DISCUSSION
Here, we uncovered 7SK as a novel substrate of METTL3/m6A methylation and uncovered a new mechanism for transcriptional control downstream of this important extracellular signaling pathway (Fig. 6F). On the basis of our results, we propose a model whereby, in unstimulated conditions, METTL3 associates with HEXIM1. Growth-promoting signals such as EGF elicit ERK-mediated phosphorylation of METTL3, causing its dissociation from HEXIM1 and enabling methylation of 7SK. Methylated 7SK then associates with HNRNP proteins, thereby releasing HEXIM1 and the P-TEFb complex. Last, the release of P-TEFb drives enhanced transcriptional activity (Fig. 6F).
7SK is an abundant noncoding RNA that is evolutionarily conserved in vertebrates, with orthologs in insects and other animals (41). The secondary and tertiary structures of 7SK are not entirely elucidated; however, 7SK is known to fold into several hairpins recognized by distinct binding proteins. 7SK associates with a subset of HNRNPs when it is not associated with HEXIM1. Several posttranslational modifications on the 7SK-associated proteins have been proposed to increase or decrease their affinity for 7SK. However, the mechanism for exchanging between the two protein complexes is not well understood. Our identification of m6A methylation of 7SK provides a novel regulatory mechanism for the 7SK transition from an anti- to a pro-proliferative state. In this novel mechanism, the RNA itself is modified upon growth factor stimulation (Fig. 6F). This novel growth factor–induced methylation by METTL3 of the mature 7SK, an RNA Pol III transcript, contrasts with the known co-transcriptional activity of METTL3 on RNA Pol II transcripts such as mRNA and primary miRNAs (14), thus increasing the type of RNA target molecules of METTL3.
Our results show that the m6A sites on 7SK are localized in the hairpin recognized by HNRNPs. A recent individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) experiment of HNRNPR shows a substantial overlap between its binding regions and the m6A motifs (34). The 7SK “HNRNP hairpin” is opposite to the one recognized by HEXIM1. Thus, when the HEXIM1/P-TEFb complex is bound to 7SK, it is likely that the HNRNP hairpin is free and available for METTL3 recognition and methylation. It has been shown that HNRNPC, HNRNPG, and HNRNPA2B1 bind methylated RNA indirectly, through a structural switching mechanism (42–44). Thus, it is plausible that all the 7SK-interacting HNRNP proteins, none of which have specialized m6A binding domains seen in YTHD readers, use a structural switching mechanism to bind methylated 7SK. Whether the various HNRNPs that recognize methylated 7SK represent redundancy, respond to different stimuli, or are regulated in a cell type–specific manner remains to be elucidated.
METTL3 phosphorylation has been associated with changes in its subcellular localization and probable activity (45), exposing this as one regulatory mechanism of m6A methylation. Here, we show that a major signaling pathway induces functional METTL3 phosphorylation to regulate transcriptional activity. We found that ERK phosphorylates METTL3 at S43 upon EGF stimulation. This phosphorylation releases METTL3 from HEXIM1 binding, thereby allowing methylation of 7SK. Others have recently shown the importance of S43 phosphorylation in different contexts (45, 46), supporting the relevance of this phosphorylation site for METTL3 activity. As EGF induces the activation of METTL3, resulting in transcription activity through a 7SK-mediated mechanism, note that, once activated, METTL3 could have additional downstream consequences for the newly transcribed mRNAs either in regulating translation, splicing, or degradation. As EGF and other signaling pathways are altered in diseases such as cancer, it will be important to assess the role of m6A as a downstream effector of such dysregulations.
MATERIALS AND METHODS
Tissue culture
HeLa cells were cultured in vitro in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin-streptomycin, 2 mM l-glutamine, and 1 mM sodium pyruvate (Gibco). For growth factor stimulation, cells were previously serum-starved for 16 hours. Cells were incubated with recombinant human EGF protein [100 ng/ml; R&D Systems 236-EG-200 in 0.1% bovine serum albumin (BSA) and phosphate-buffered saline (PBS)] for 30 min or the indicated time. Pretreatment of 1 hour with 10 μM U0126 (EMD Millipore Corp., #662005) was performed when indicated.
Generation of stable cell lines was performed as described previously (47). Plasmids for the expression of shRNA or Flag-tagged proteins were cotransfected with lentivirus packaging plasmids in human embryonic kidney 293T cells to generate lentivirus. Two shRNAs targeting METTL3 (TRCN0000034715 and TRCN0000034717) and a control shRNA (SHC002) were used for experiments. To express Flag-tagged proteins, they were independently cloned into pLenti-C-Myc-DDK-IRES-Puro (OriGene, PS100069).
Mutant METTL3 S43A cell lines were generated using the CRISPR-Cas9 system (48). Four nucleotides were modified in the donor oligonucleotide. Those changes include the ones required for the codon change (Ser to Ala), one to produce a silent mutation to achieve resistance to CRISPR-Cas9, and one to create a novel restriction site (Nar I) for screening clones using digestion of genomic DNA.
Cryomilling
We followed the protocol as previously described (49). Briefly, 2 × 108 to 8 × 108 cells were pelleted and dripped from a syringe directly into liquid nitrogen to generate frozen pellets. Frozen pellets were cryomilled in a Retsch Planetary Ball Mill PM100. We performed three cycles of milling with the following program: 400 rpm with direction reversal for 3 min, with intervals of 1 min.
Immunoprecipitations
Flag-IP from 1 to 2 g of cryomilled HeLa Flag-LARP7 cells was performed to isolate endogenous 7SK RNA for RNA MS and to assess the localization of m6A sites. Lysis, incubation with anti-Flag beads, and washes were performed in IP buffer A (20 mM Hepes Na, 150 mM NaCl, 0.3% CHAPS, and protease and RNase inhibitors). Beads were resuspended in Tris-EDTA (TE) buffer, and RNA was extracted using phenol:chloroform, followed by ethanol precipitation and resuspension in RNase-free water.
For the RNase experiment, cell lysates were treated or not with 400 μg of RNase A (EN0531) for 60 min at 37°C before the Flag-IP. For endogenous protein IP, we performed the same protocol as in Flag-IPs, but 10 g of rabbit α-HEXIM1 (ab25388, Abcam) or rabbit α-LARP7 (ab134746, Abcam) antibodies was incubated with protein A bead (Dynabeads 1002D) in buffer A at room temperature for 30 min with agitation. For IP performed from cross-linked cells, 6 × 107 to 20 × 107 cells per sample were washed with PBS, ultraviolet (UV) cross-linked at 254 nm (400 mJ/cm2), and lysed using lysis buffer [50 mM Hepes-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Triton X-100, and protease and RNase inhibitors]. For the METTL3 and HEXIM1 interaction analysis, 1 g of cryomilled cells was resuspended in IP buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, and protease and phosphatase inhibitors.
For the cross-linked experiments, beads were resuspended in 500 mM NaCl in PBS.
After taking an input aliquot, lysates were incubated with anti-Flag M2 magnetic beads (Sigma-Aldrich) for 4 hours at 4°C. Beads were washed twice with 150 mM NaCl and once with PBS. After the washes, the sample was treated with proteinase K (Ambion), and RNA was extracted using phenol:chloroform followed by ethanol precipitation. A fraction of each sample was treated with RNase A (Thermo Fisher Scientific), and the reaction was stopped by adding one volume of 4× sample buffer (Bio-Rad), 10% β-mercaptoethanol. In all cases, lysates were sonicated until complete lysis was observed, spun for 15 min at 25,000g, and filtered using 0.22-μm syringe filters.
m6A-IP qRT-PCR
A total of 1.4 × 108 cells per sample were lysed using lysis buffer 1 [50 mM Hepes-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.01% Triton X-100, and protease inhibitors]. Nuclei were pelleted for 15 min at 3500 rpm. RNA was extracted with the Total RNA Purification Maxi Kit (Norgen Biotek).
m6A-IP was performed as previously published (15). Fifty micrograms of nuclear RNA or 1 μg of Flag-immunoprecipitated RNA was immunoprecipitated in triplicates, and a negative control without antibody was included. Rabbit anti-m6A antibody (Synaptic Systems) and protein A Dynabeads (Invitrogen) were used for the IPs. The immunoprecipitated RNA was eluted with N6-methyladenosine (Sigma-Aldrich), ethanol precipitated, and resuspended in water. RNA was retrotranscribed with the SuperScript III Reverse Transcriptase (Invitrogen), using random hexamers and incubating samples at 55°C for 1 hour. qPCR was performed with the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) in CFX Opus 384 (Bio-Rad) following the manufacturer’s instructions. The following primers were used: 7SK, TAAGAGCTCGGATGTGAGGGCGATCTG (forward) and CGAATTCGGAGCGGTGAGGGAGGAAG (reverse); GAPDH, AGCCTCAAGATCATCAGCAATG (forward) and ATGGACTGTGGTCATGAGTCCTT (reverse); 18S rRNA, CGGCGACGACCCATTCGAAC (forward) and GAATCGAACCCTGATTCCCCGTC (reverse); 7SK Frag2, CCTGCTAGAACCTCCAAACAA (forward) and CATGCAGCGCCTCATTTG (reverse). For quantification, 7SK measurements in the m6A-IPs were normalized to their respective control and to the input levels.
Northern blot
Fifty to 200 ng of endogenous 7SK was run in 6% tris-borate EDTA (TBE)–urea gels (Invitrogen). We used TBE buffer (American Bio) and Low Range ssRNA Ladder [New England Biolabs (NEB)]. Samples were prepared using 2× gel loading buffer (Invitrogen) and denatured for 2 min at 80°C. Gels were run at 200 V in 1× TBE buffer, visualized using Sybr Gold (Invitrogen), and then transferred to a nylon membrane (Cytiva) at 0.06 A for 45 min in 0.5×TBE buffer. Membranes were UV cross-linked at 254 nm (240 mJ/cm2). Membranes were pretreated with hybridization buffer (DIG Easy Hyb Granules, Roche) at 37°C for 30 min in a hybridization oven. The probe was denatured at 95°C for 5 min and added to the hybridization buffer at 37°C overnight. Membranes were washed twice with low stringent buffer for 5 min, twice with high stringent buffer for 10 min, and once with washing buffer at 37°C for 10 min. Then, membranes were incubated in blocking buffer (DIG wash and block buffer set, Roche) for 1 hour at room temperature, and the anti–digoxigenin-AP antibody (Roche) was added (1:15,000 dilution) and incubated at room temperature for an extra 30 min. The membrane was washed in a DIG washing buffer four times for 15 min each at room temperature. Last, membranes were incubated in a detection buffer for 5 min and developed with CDP-Star (Roche). Images were acquired in Bio-Rad Chemidoc, using the probe 5′/5DigN/rGrGrArArGrCrUrUrGrArCrUrArCrCrC (Integrated DNA Technologies).
Western blot
A total of 7 × 106 cells per sample were lysed using radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) with protease (Sigma-Aldrich) and phosphatase inhibitors (Roche). Chromatin fractions were purified as previously described (50). Lysates were sonicated or passed through QIAshredder and ran in NuPAGE 4 to 12% bis-tris gel (Invitrogen). After transfer to nitrocellulose membrane (Bio-Rad) and block with milk, samples were incubated with the respective antibody overnight at 4°C in 5% BSA tris-buffered saline with Tween-20 (TBS-T). Membranes were washed three times for 10 min with TBS-T, incubated for 1 hour with secondary antibody in 1% milk (chemiluminescence) or 3% BSA (fluorescence), washed three times more, and developed with ECL prime (Amersham). Images were acquired in Bio-Rad Chemidoc. The following antibodies were used: mouse α-Flag (A8592, Sigma-Aldrich); rabbit α-METTL3 (ab195352, Abcam); mouse α-METTL3 (H00056339-B01P, Abnova); rabbit α–alpha-tubulin [2144, Cell Signaling Technology (CST)]; mouse α-HNRNPA2B1 (Sc-32316, Santa Cruz Biotechnology); rabbit α-HEXIM1 (ab25388, Abcam); rabbit α-LARP7 (ab134746, Abcam); rabbit α-HNRNPR (ab30930, Abcam); rabbit α-HNRNPQ (ab184946, Abcam); mouse α-HNRNPA1 (ab5832, Abcam); rabbit α-pERK1/2 (9101, CST); mouse α-ERK1/2 (4696, CST); rabbit α-GAPDH horseradish peroxidase (HRP) conjugate (8884, CST); rabbit α-CDK9 (ab76320, Abcam); rabbit α–cyclin T1 (A303-496A, Bethyl Laboratories Inc.); rabbit α-MEPCE (14917-1-AP, Proteintech); rabbit α-RNA Pol II CTD repeat YSPTSPS (phospho S2) (ab5095, Abcam); mouse α-RNA Pol II CTD repeat YSPTSPS antibody [8WG16] (ab817); donkey α-rabbit immunoglobulin G (IgG), HRP linked (NA934V, GE Healthcare); sheep α-mouse IgG, HRP linked (NA931V, GE Healthcare); donkey α-rabbit IgG H&L (IRDye 680RD) preadsorbed (ab216779, Abcam); and donkey α-mouse IgG H&L (IRDye 800CW) preadsorbed (ab216774, Abcam). Polyclonal antibody against pS43 METTL3 was generated by Thermo Fisher Scientific.
RNase H
To fragment 7SK, we used endogenous 7SK, obtained from pull-down of Flag-LARP7 in growing cells, and purified the 332-nucleotide band from the 6% TBE-urea RNA gel. A total of 250 ng of RNA was incubated with 500 ng of the complementary oligo (CTCGTATAC) in RNase H buffer in a final volume of 14 μl. For the annealing, samples were boiled at 95°C for 5 min, cooled down to 37°C at 0.1°C/s, and then let sit at 37°C for 5 min. One microliter of RNase H (NEB, #M0297) was added, and the reaction was performed at 37°C for 30 min. Fragments were extracted using phenol:chloroform followed by ethanol precipitation and resuspension in water. m6A-IP qRT-PCR was performed as described above, using at least 100 ng of purified bands and normalizing the m6A-immunoprecipitated RNA to the IP control and the pre-IP input fragment.
RNA Click-iT
Transcriptional activity was measured using the Click-iT RNA Alexa Fluor 488 Imaging Kit (Invitrogen, C10329), following the manufacturer’s instructions. Reactions were scaled down for 24-well plates. Cells were grown in poly-l-lysine Cellware 12-mm round coverslips (Corning, #354085). After 16 hours of serum starvation, cells were treated for 1 hour with 1 mM 5-ethynyl uridine (5-EU). Treatment with EGF (100 ng/ml) was performed for 1 hour, adding EGF at the same time as 5-EU. Pretreatment with U0126 was performed as described above. Cells were fixed with 3.7% formaldehyde in PBS and mounted using Fluoromount-G (Invitrogen, #00-4958-02). Images were acquired with a confocal microscope Leica SP8. Images were analyzed using ImageJ, generating masks of the nuclei and measuring the mean intensity of the particles.
Recombinant protein expression and purification
For proteins expressed in insect cells, full-length human METTL3 and METTL14 were subcloned into MultiBac vector with a His6 affinity tag fused to the N terminus. pFL was a gift from T. Richmond (Addgene, plasmid no. 110739) (51). Bacmids were generated in DH10Bac (Geneva Biotech) cells, and baculoviruses were generated and amplified in Sf-9 insect cells. High Five (Trichoplusia ni) cells grown in Ex-Cell 405 medium (Sigma-Aldrich) were infected by baculoviruses at a density of 2.0 × 106 cells/ml. The METTL3/METTL14 complex was coexpressed in High Five cells at 27°C for 48 hours. METTL3 alone was infected and expressed in High Five cells at 27°C for 48 hours as well. Cells were harvested by centrifugation at 4000 rpm for 30 min at 4°C and homogenized in ice-cold lysis buffer containing 25 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM tris-(2-carboxyethyl)-phosphine (TCEP) and supplemented with protease inhibitor cocktail tablets (Sigma-Aldrich). The cell debris was separated by ultracentrifugation (20,000g) for 1 hour at 4°C. The supernatant was first incubated with the Ni-Penta Agarose-Base Resin (Marvelgent Biosciences) and washed with lysis buffer supplemented with 20 mM imidazole. Proteins were eluted with buffer containing 25 mM Hepes (pH 7.5), 150 mM NaCl, 400 mM imidazole, and 1 mM TCEP. Eluted proteins were concentrated and further purified by size exclusion chromatography with a Superdex 200 Increase 100/300 GL size exclusion column (GE Healthcare) equilibrated in 20 mM Hepes (pH 7.5) with 150 mM NaCl and 1 mM TCEP. Fractions were evaluated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), then concentrated in a Centricon 10-kDa spin concentrator (Millipore), and used for further study.
GST fusion METTL3 was expressed and purified from bacteria cells. For recombinant protein GST-METTL3 phosphorylated at position S43, an E. coli strain engineered to incorporate phosphoserine (Sep) genetically in recombinant proteins using the orthogonal translation system SepOTS (enhanced phosphoserine insertion during E. coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion) was used.
Human METTL3 was expressed using the pGEX-6P-1 vector (Amersham). SepOTSλ was a gift from J. Rinehart (Addgene, plasmid no. 68292). E. coli strain C321.ΔA was a gift from J. Rinehart (Addgene, bacterial strain no. 68306) (39). Overnight starter cultures for METTL3 WT and METTL3-phospho-S43 were inoculated into LB medium supplemented with Carb (100 μg/ml), Kan (25 μg/ml), bleomycin (1:4000 from stock), and O-phospho-serine (2 mM at a final concentration) and grew at 30°C, 220 rpm until OD600 (optical density at 600 nm) reached 0.8 and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 20 hours. Cells were harvested by centrifugation at 4000 rpm for 30 min at 4°C and homogenized in ice-cold lysis buffer containing 50 mM tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM TCEP and supplemented with protease and phosphatase inhibitors. After ultracentrifugation (20,000g) for 1 hour at 4°C to remove cell debris, the supernatant was incubated with glutathione-agarose beads (Thermo Fisher Scientific) at 4°C for 1 hour, and beads were washed thoroughly with wash buffer [50 mM tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM TCEP] after incubation. Proteins were eluted with wash buffer containing 10 mM reduced glutathione. Proteins were concentrated and further purified by size exclusion chromatography with a Superdex 200 Increase 100/300 GL size exclusion column (GE Healthcare) equilibrated in 20 mM Hepes (pH 7.5) with 150 mM NaCl and 1 mM TCEP. Fractions were evaluated by SDS-PAGE and used for pull-down assay.
In vitro transcription
Full-length 7SK was amplified from cDNA using the Herculase II Fusion Enzyme (Agilent Technologies). In vitro transcription was performed using the MEGAshortscript T7 Transcription Kit (Invitrogen AM1354), incubating the gel-purified PCR product with the reagents for 4 hours at 37°C. The product was incubated with TURBO deoxyribonuclease for 15 min at 37°C and extracted using phenol:chloroform, followed by ethanol precipitation and resuspension in water.
In vitro methyltransferase activity assay
In vitro methyltransferase activity assay was performed using SAM-fluoro: SAM Methyltransferase Assay (GBiosciences, catalog no. 786-431). A total of 0.7 μM purified protein and 10 μM transcribed RNA were incubated for 1 hour at 37°C, and fluorescence was measured every 2 min in Biotek Synergy H1, with excitation wavelength: 535 nm and emission wavelength: 590 nm. The data were analyzed as indicated in the product protocol.
In vitro kinase activity assay
In vitro kinase activity assay was performed incubating 0.4 μg of ERK1, active, untagged, human PRECISIO kinase recombinant (Sigma-Aldrich, #E7407) with 4 μM GST-tagged METTL3 (bound to glutathione agarose beads) following the manufacturer’s instructions. Proteins were incubated for 30 min at 30°C with 1500 rpm of agitation.
RNA MS
Measurements of the level of m6A were performed by UHPLC-MS/MS using a method similar to a previously described method (52–54). Briefly, 200 ng of purified endogenous 7SK was digested with the Nucleoside Digestion Mix (NEB) according to the manufacturer’s instructions. The digested samples were then reconstituted in 100 μl of RNAse-free water with 0.01% formic acid before UHPLC-MS/MS analysis. The UHPLC-MS/MS analysis was accomplished on a Waters XEVO TQ-S (Waters Corporation, USA) triple quadruple tandem mass spectrometer equipped with an electrospray source (ESI) maintained at 150°C and a capillary voltage of 1 kV. Nitrogen was used as the nebulizer gas, maintained at 7-bar pressure, with a flow rate of 1,000 liter/hour and a temperature of 500°C. UHPLC-MS/MS analysis was performed in ESI positive-ion mode using multiple reaction monitoring from ion transitions previously determined for m6A [mass/charge ratio (m/z) of 282.08 > 150.11] and adenosine (m/z of 268.1 > 136.1). The transition for internal standard [guanosine (13C15N)] was m/z of 299 > 162. A Waters ACQUITY UPLC HSS T3 guard column (2.1 × 5 mm, 1.8 μm), attached to an HSS T3 column (2.1 × 50 mm, 1.7 μm), was used for the separation. Mobile phases included RNAse-free water (18 megohm cm−1) containing 0.01% formic acid (buffer A) and 50% acetonitrile (ACN) (v/v) in buffer A (buffer B). The digested nucleotides were eluted at a flow rate of 0.4 ml/min with a gradient as follows: 0 to 2 min, 0 to 10% B; 2 to 3 min, 10 to 15% B; 3 to 4 min, 15 to 100% B; and 4 to 4.5 min, 100% B. The total run time was 7 min. The column oven temperature was kept at 35°C, and the sample injection volume was 10 μl. Data acquisition and analysis were performed using MassLynx V4.1 and TargetLynx. Calibration curves were plotted using linear regression with a weight factor of 1/x.
Protein extraction and digestion for MS
The cell pellets with 10 M urea lysis buffer, complete protease inhibitor cocktail, and phosphatase inhibitor were ultrasonically lysed by sonication at 4°C for 2 min using the Vial Tweeter device (Hielscher-Ultrasound Technology). After lysis, the lysates were cleared up by centrifugation at 20,000g for 1 hour to remove the insoluble material. The protein mixtures were reduced by 10 mM TCEP for 1 hour at 37°C and alkylated by 20 mM iodoacetamide (IAA) in the dark for 45 min at room temperature. The protein was first precipitated by precooled precipitation buffer (50% acetone, 50% ethanol, and 0.1% acetic acid) overnight at −20°C and washed by precooled 50% acetone and 70% ethanol with centrifugation at 20,000g for 40 min at 4°C. The pellet was dry on SpeedVac for 5 min to remove any chemical residues. After directly adding 300 𝜇l of 100 mM NH4HCO3 into the pellet, the samples were digested at a protease/protein ratio of 1:20 overnight at 37°C. The peptide mixture was acidified with formic acid and then desalted with a C18 column (MarocoSpin Columns, NEST Group Inc.).
For IP samples, the eluate was reduced by 10 mM dithiothreitol for 1 hour at 56°C and 20 mM IAA in the dark for 45 min at room temperature. After reduction and alkylation, the sample was digested with sequencing grade porcine trypsin (Promega) at 1:20 overnight at 37°C. The peptides were purified with the C18 column. The amount of the final peptides was determined by Nanodrop (Thermo Fisher Scientific). About 1 𝜇g of the total peptide digest was injected into MS.
Sample preparation for phosphoproteomics
The phosphopeptide enrichment was performed using the High-Select Fe-NTA kit (Thermo Fisher Scientific, A32992) according to the manufacturer’s instructions. Briefly, the resins of the spin column were aliquoted, incubated with 200 μg of total peptides for 30 min at room temperature, and transferred into the filter tip (TF-20-L-R-S, Axygen). The supernatant was then removed by centrifugation. Then, the resins adsorbed with phosphopeptides were washed three times with 200 μl of washing buffer (80% ACN and 0.1% trifluoroacetic acid) and twice 200 μl of water to remove nonspecifically adsorbed peptides. The phosphopeptides were then eluted off the resins twice with 100 μl of elution buffer (50% ACN and 5% NH3•H2O). The centrifugation steps above were all kept at 500g for 30 s. The eluates were dried by SpeedVac and stored at −80°C before MS measurements.
DIA-MS measurement
The samples were measured using the Data-independent acquisition mass spectrometry (DIA-MS) method described previously (55–57). LC separation was performed on EASY-nLC 1200 systems (Thermo Fisher Scientific, San Jose, CA) using a self-packed analytical PicoFrit column (New Objective, Woburn, MA, USA) (75 μm by 50 cm in length) using C18 material of ReproSil-Pur 120A C18-Q 1.9 μm (Dr. Maisch GmbH, Ammerbuch, Germany). A 120-min measurement with buffer B (80% ACN containing 0.1% formic acid) from 5 to 37% and corresponding buffer A (0.1% formic acid) during the gradient was used to elute peptides from the LC. The flow rate was kept at 300 nl/min, temperature-controlled at 60°C using a column oven (PRSO-V1, Sonation GmbH, Biberach, Germany).
The Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) instrument coupled to a nanoelectrospray ion source (NanoFlex, Thermo Scientific) was calibrated using Tune (version 3.0) instrument control software. The spray voltage was set to 2000 V and the heating capillary temperature at 275°C. The DIA-MS methods consisted of 1 MS1 scan and 33 MS2 scans of variable isolated windows. The MS1 scan range is 350 to 1650 m/z, and the MS1 resolution is 120,000 at m/z of 200. The MS1 full scan AGC target value was 2 × 106, and the maximum injection time was 50 ms. The MS2 resolution was set to 30,000 at m/z of 200 with the MS2 scan range 200 to 1800 m/z, and the normalized higher-energy collisional dissociation (HCD) collision energy was 28%. The MS2 AGC was set to be 1.5 × 106, and the maximum injection time was 50 ms. The default peptide charge state was set to 2. Both MS1 and MS2 spectra were recorded in profile mode.
DIA-MS data analyses were performed using Spectronaut v15 with a direct DIA search against the Uniprot-downloaded human fasta file (58). The oxidation at methionine was set as a variable modification, whereas carbamidomethylation at cysteine was set as a fixed modification. Both peptide and protein false discovery rate (FDR) cutoffs (Q value) were controlled at 1%, and the data matrix was strictly filtered by Q value. Overall, both peptide and protein FDRs (based on Q value) were controlled at 1%, and the data matrix was filtered by Q value. For the phosphoproteomics, the serine/threonine/tyrosine (S/T/Y) was enabled as a variable modification, and the posttranslational modification (PTM) score > 0.75 for all the samples. All the other settings in Spectronaut are kept as default.
Statistics
Student’s two-tailed t test and one-way or two-way analysis of variance (ANOVA) were used according to the experimental design. Multiple comparisons were performed with Tukey’s or Dunnett’s posttest. Analyses were performed in GraphPad Prism 9. All experiments were performed at least twice. Bar graphs show the mean ± SD.
Acknowledgments
We thank M. Lemmon and S. F. Tavazoie for reading earlier versions of this manuscript. We acknowledge support from our colleagues at the Yale Cancer Biology Institute. We thank the Yale West Campus Imaging Core for the support and assistance in this work. We thank C. Osuji and P. Rahmani for technical support. Funding: This work was supported by National Institutes of Health grant R35GM138185 (to C.R.A., A.W.D., and H.L.), National Institutes of Health grant R01GM137031 (to Y.L.), National Institutes of Health grant R01CA248532 (to D.E.K.), and National Institutes of Health grant R01CA244634 (to H.G.). C.R.A. is a Louis Goodman and Alfred Gilman Yale Scholar.
Author contributions: On the basis of https://credit.niso.org/. Conceptualization: C.R.A. Formal analysis: M.P.-P. Methodology: M.P.-P. and C.R.A. Investigation: M.P.-P., A.W.D., H.L., W.L., B.H., Q.L., and H.G. Visualization: M.P.-P. and C.R.A. Supervision: D.E.K., Y.L., and C.R.A. Writing—original draft: M.P.-P. and C.R.A.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Constructs will be through Addgene, plasmid IDs: 200724 to 200734.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
REFERENCES AND NOTES
- 1.S. A. Dames, A. Schönichen, A. Schulte, M. Barboric, B. M. Peterlin, S. Grzesiek, M. Geyer, Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl. Acad. Sci. U.S.A. 104, 14312–14317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. H. N. Yik, R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, Q. Zhou, Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12, 971–982 (2003). [DOI] [PubMed] [Google Scholar]
- 3.C. Barrandon, F. Bonnet, V. T. Nguyen, V. Labas, O. Bensaude, The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 27, 6996–7006 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Y. K. Kim, U. Mbonye, J. Hokello, J. Karn, T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 410, 896–916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.X. Contreras, M. Barboric, T. Lenasi, B. M. Peterlin, HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLOS Pathog. 3, 1459–1469 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.V. T. Nguyen, T. Kiss, A. A. Michels, O. Bensaude, 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414, 322–325 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Z. Yang, Q. Zhu, K. Luo, Q. Zhou, The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414, 317–322 (2001). [DOI] [PubMed] [Google Scholar]
- 8.K. Chen, B. S. Zhao, C. He, Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 23, 74–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.P. Sledz, M. Jinek, Structural insights into the molecular mechanism of the m6A writer complex. eLife 5, e18434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.P. Wang, K. A. Doxtader, Y. Nam, Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.X. Wang, J. Feng, Y. Xue, Z. Guan, D. Zhang, Z. Liu, Z. Gong, Q. Wang, J. Huang, C. Tang, T. Zou, P. Yin, Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 (2016). [DOI] [PubMed] [Google Scholar]
- 12.X. Wang, Z. Lu, A. Gomez, G. C. Hon, Y. Yue, D. Han, Y. Fu, M. Parisien, Q. Dai, G. Jia, B. Ren, T. Pan, C. He, N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.C. R. Alarcón, H. Lee, H. Goodarzi, N. Halberg, S. F. Tavazoie, N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.P. Knuckles, S. H. Carl, M. Musheev, C. Niehrs, A. Wenger, M. Bühler, RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat. Struct. Mol. Biol. 24, 561–569 (2017). [DOI] [PubMed] [Google Scholar]
- 15.C. R. Alarcón, H. Goodarzi, H. Lee, X. Liu, S. Tavazoie, S. F. Tavazoie, HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.W. Xiao, S. Adhikari, U. Dahal, Y.-S. Chen, Y.-J. Hao, B.-F. Sun, H.-Y. Sun, A. Li, X.-L. Ping, W.-Y. Lai, X. Wang, H.-L. Ma, C.-M. Huang, Y. Yang, N. Huang, G.-B. Jiang, H.-L. Wang, Q. Zhou, X.-J. Wang, Y.-L. Zhao, Y.-G. Yang, Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016). [DOI] [PubMed] [Google Scholar]
- 17.S. Lin, J. Choe, P. Du, R. Triboulet, R. I. Gregory, The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.K. D. Meyer, D. P. Patil, J. Zhou, A. Zinoviev, M. A. Skabkin, O. Elemento, T. V. Pestova, S.-B. Qian, S. R. Jaffrey, 5' UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. J. Clancy, M. E. Shambaugh, C. S. Timpte, J. A. Bokar, Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30, 4509–4518 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. F. Hongay, T. L. Orr-Weaver, Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 14855–14860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Q. Fei, Z. Zou, I. A. Roundtree, H.-L. Sun, C. He, YTHDF2 promotes mitotic entry and is regulated by cell cycle mediators. PLOS Biol. 18, e3000664 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Y. Wang, Y. Li, J. I. Toth, M. D. Petroski, Z. Zhang, J. C. Zhao, N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.P. J. Batista, B. Molinie, J. Wang, K. Qu, J. Zhang, L. Li, D. M. Bouley, E. Lujan, B. Haddad, K. Daneshvar, A. C. Carter, R. A. Flynn, C. Zhou, K.-S. Lim, P. Dedon, M. Wernig, A. C. Mullen, Y. Xing, C. C. Giallourakis, H. Y. Chang, m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Q. Cui, H. Shi, P. Ye, L. Li, Q. Qu, G. Sun, G. Sun, Z. Lu, Y. Huang, C. G. Yang, A. D. Riggs, C. He, Y. Shi, m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Y. Xiang, B. Laurent, C.-H. Hsu, S. Nachtergaele, Z. Lu, W. Sheng, C. Xu, H. Chen, J. Ouyang, S. Wang, D. Ling, P.-H. Hsu, L. Zou, A. Jambhekar, C. He, Y. Shi, RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.N. He, N. S. Jahchan, E. Hong, Q. Li, M. A. Bayfield, R. J. Maraia, K. Luo, Q. Zhou, A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 29, 588–599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.B. J. Krueger, C. Jeronimo, B. B. Roy, A. Bouchard, C. Barrandon, S. A. Byers, C. E. Searcey, J. J. Cooper, O. Bensaude, E. A. Cohen, B. Coulombe, D. H. Price, LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36, 2219–2229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L. Muniz, S. Egloff, T. Kiss, RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res. 41, 4686–4698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. LaCava, K. R. Molloy, M. S. Taylor, M. Domanski, B. T. Chait, M. P. Rout, Affinity proteomics to study endogenous protein complexes: Pointers, pitfalls, preferences and perspectives. Biotechniques 58, 103–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. E. Brogie, D. H. Price, Reconstitution of a functional 7SK snRNP. Nucleic Acids Res. 45, 6864–6880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.E. Van Herreweghe, S. Egloff, I. Goiffon, B. E. Jády, C. Froment, B. Monsarrat, T. Kiss, Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 26, 3570–3580 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D. A. Wassarman, J. A. Steitz, Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 11, 3432–3445 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D. Martinez-Zapien, P. Legrand, A. McEwen, F. Proux, T. Cragnolini, S. Pasquali, A.-C. Dock-Bregeon, The crystal structure of the 5 functional domain of the transcription riboregulator 7SK. Nucleic Acids Res. 45, 3568–3579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Briese, L. Saal-Bauernschubert, C. Ji, M. Moradi, H. Ghanawi, M. Uhl, S. Appenzeller, R. Backofen, M. Sendtner, hnRNP R and its main interactor, the noncoding RNA 7SK, coregulate the axonal transcriptome of motoneurons. Proc. Natl. Acad. Sci. U.S.A. 115, E2859–E2868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. D. Eichhorn, Y. Yang, L. Repeta, J. Feigon, Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc. Natl. Acad. Sci. U.S.A. 115, E6457–E6466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Y. Xue, Z. Yang, R. Chen, Q. Zhou, A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 38, 360–369 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Y. Yang, C. D. Eichhorn, Y. Wang, D. Cascio, J. Feigon, Structural basis of 7SK RNA 5′-γ-phosphate methylation and retention by MePCE. Nat. Chem. Biol. 15, 132–140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.C. Y. Jao, A. Salic, Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 105, 15779–15784 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. L. Pirman, K. W. Barber, H. R. Aerni, N. J. Ma, A. D. Haimovich, S. Rogulina, F. J. Isaacs, J. Rinehart, A flexible codon in genomically recoded Escherichia coli permits programmable protein phosphorylation. Nat. Commun. 6, 8130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.P. I. Thakore, A. M. D'Ippolito, L. Song, A. Safi, N. K. Shivakumar, A. M. Kabadi, T. E. Reddy, G. E. Crawford, C. A. Gersbach, Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. Marz, A. Donath, N. Verstraete, V. T. Nguyen, P. F. Stadler, O. Bensaude, Evolution of 7SK RNA and its protein partners in metazoa. Mol. Biol. Evol. 26, 2821–2830 (2009). [DOI] [PubMed] [Google Scholar]
- 42.N. Liu, Q. Dai, G. Zheng, C. He, M. Parisien, T. Pan, N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.N. Liu, K. I. Zhou, M. Parisien, Q. Dai, L. Diatchenko, T. Pan, N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. Wu, S. Su, D. P. Patil, H. Liu, J. Gan, S. R. Jaffrey, J. Ma, Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9, 420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.C. Zhang, L. Chen, D. Peng, A. Jiang, Y. He, Y. Zeng, C. Xie, H. Zhou, X. Luo, H. Liu, L. Chen, J. Ren, W. Wang, Y. Zhao, METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol. Cell 79, 425–442.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 46.H. L. Sun, A. C. Zhu, Y. Gao, H. Terajima, Q. Fei, S. Liu, L. Zhang, Z. Zhang, B. T. Harada, Y.-Y. He, M. B. Bissonnette, M.-C. Hung, C. He, Stabilization of ERK-Phosphorylated METTL3 by USP5 Increases m6A Methylation. Mol. Cell 80, 633–647.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.S. F. Tavazoie, C. Alarcón, T. Oskarsson, D. Padua, Q. Wang, P. D. Bos, W. L. Gerald, J. Massagué, Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J.-B. Renaud, C. Boix, M. Charpentier, A. de Cian, J. Cochennec, E. Duvernois-Berthet, L. Perrouault, L. Tesson, J. Edouard, R. Thinard, Y. Cherifi, S. Menoret, S. Fontanière, N. de Crozé, A. Fraichard, F. Sohm, I. Anegon, J.-P. Concordet, C. Giovannangeli, Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep. 14, 2263–2272 (2016). [DOI] [PubMed] [Google Scholar]
- 49.J. LaCava, H. Jiang, M. P. Rout, Protein complex affinity capture from cryomilled mammalian cells. J. Vis. Exp., 54518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. Mayer, L. S. Churchman, Genome-wide profiling of RNA polymerase transcription at nucleotide resolution in human cells with native elongating transcript sequencing. Nat. Protoc. 11, 813–833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. J. Fitzgerald, P. Berger, C. Schaffitzel, K. Yamada, T. J. Richmond, I. Berger, Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3, 1021–1032 (2006). [DOI] [PubMed] [Google Scholar]
- 52.J. Lorent, E. P. Kusnadi, V. van Hoef, R. J. Rebello, M. Leibovitch, J. Ristau, S. Chen, M. G. Lawrence, K. J. Szkop, B. Samreen, P. Balanathan, F. Rapino, P. Close, P. Bukczynska, K. Scharmann, I. Takizawa, G. P. Risbridger, L. A. Selth, S. A. Leidel, Q. Lin, I. Topisirovic, O. Larsson, L. Furic, Translational offsetting as a mode of estrogen receptor alpha-dependent regulation of gene expression. EMBO J. 38, e101323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. Sas-Chen, J. M. Thomas, D. Matzov, M. Taoka, K. D. Nance, R. Nir, K. M. Bryson, R. Shachar, G. L. S. Liman, B. W. Burkhart, S. T. Gamage, Y. Nobe, C. A. Briney, M. J. Levy, R. T. Fuchs, G. B. Robb, J. Hartmann, S. Sharma, Q. Lin, L. Florens, M. P. Washburn, T. Isobe, T. J. Santangelo, M. Shalev-Benami, J. L. Meier, S. Schwartz, Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Tardu, J. D. Jones, R. T. Kennedy, Q. Lin, K. S. Koutmou, Identification and quantification of modified nucleosides in Saccharomyces cerevisiae mRNAs. ACS Chem. Biol. 14, 1403–1409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Mehnert, W. Li, C. Wu, B. Salovska, Y. Liu, Combining rapid data independent acquisition and CRISPR gene deletion for studying potential protein functions: A case of HMGN1. Proteomics 19, e1800438 (2019). [DOI] [PubMed] [Google Scholar]
- 56.W. Li, H. Chi, B. Salovska, C. Wu, L. Sun, G. Rosenberger, Y. Liu, Assessing the relationship between mass window width and retention time scheduling on protein coverage for data-independent acquisition. J. Am. Soc. Mass Spectrom. 30, 1396–1405 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Y. Liu, Y. Mi, T. Mueller, S. Kreibich, E. G. Williams, A. van Drogen, C. Borel, M. Frank, P.-L. Germain, I. Bludau, M. Mehnert, M. Seifert, M. Emmenlauer, I. Sorg, F. Bezrukov, F. S. Bena, H. Zhou, C. Dehio, G. Testa, J. Saez-Rodriguez, S. E. Antonarakis, W.-D. Hardt, R. Aebersold, Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol. 37, 314–322 (2019). [DOI] [PubMed] [Google Scholar]
- 58.The UnitProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6