Abstract
The molecular and functional contributions of intratumoral nerves to disease remain largely unknown. We localized synaptic markers within tumors suggesting that these nerves form functional connections. Consistent with this, electrophysiological analysis shows that malignancies harbor significantly higher electrical activity than benign disease or normal tissues. We also demonstrate pharmacologic silencing of tumoral electrical activity. Tumors implanted in transgenic animals lacking nociceptor neurons show reduced electrical activity. These data suggest that intratumoral nerves remain functional at the tumor bed. Immunohistochemical staining demonstrates the presence of the neuropeptide, Substance P (SP), within the tumor space. We show that tumor cells express the SP receptor, NK1R, and that ligand/receptor engagement promotes cellular proliferation and migration. Our findings identify a mechanism whereby intratumoral nerves promote cancer progression.
Electro-physiologically active intratumoral nerves contribute to cancer progression through the release of substance P.
INTRODUCTION
A growing body of evidence supports the presence and importance of intratumoral nerves in cancer (1–4). For instance, genetic, chemical, and surgical ablations of tumor-infiltrating nerves in different cancer models demonstrate active roles for nerves in disease initiation and progression (5–7). Moreover, tumors actively recruit nerves by releasing small extracellular vesicles as well as soluble neurotrophic factors and neuropeptides (8–13). Together, these data suggest a requirement of neuronal infiltration of the tumor bed for disease progression. Consistent with this, neurotrophic factors and axonal guidance molecules may harbor protumorigenic activity (14–16), while neurotransmitter receptor blockade shows some antitumorigenic potential (17–20). One murine study even provides evidence for the recruitment of central nervous system neural progenitors to tumors further emphasizing the existence of intricate communication and interactions between cancers and the nervous system (21). Together, these data suggest that the nervous system is not a bystander but an active participant in cancer and indicate that extensive tumor innervation contributes to aggressive disease (1).
While the recruitment and presence of nerves in tumors is widely accepted, the neuronal functions and molecular signals released at the tumor bed remain largely undefined. However, a recent in vivo study has begun to shed light on this by documenting intratumoral electrical activity in a murine model of breast cancer (22). Here, mice implanted with breast cancer cells were maintained with indwelling microwire electrodes enabling intratumoral electrical recordings in vivo throughout the course of disease. Daily recordings reveal spikes in electrical activity that correlate with the emergence of metastasis. This electrophysiologic study suggests the intriguing possibility that intratumoral nerves remain functional at the tumor bed and actively promote disease progression.
To gain further molecular insights on the neural contributions to cancer, we elected to study two peripheral solid cancers: head and neck squamous cell carcinoma (HNSCC) and high-grade serous ovarian carcinoma (HGSOC). These cancers differ significantly in their disease sites (oral versus peritoneal cavities), prognosis, treatment responses, and survival. We reasoned that if tumor innervation is a common feature of peripheral solid tumors that promotes disease progression, then shared pathways will likely emerge from the study of these vastly different cancers.
HNSCCs are innervated by TRPV1 (transient receptor potential cation channel subfamily V member 1)-expressing sensory nerves (9). These cancers are generally induced by infection with high-risk human papillomavirus (HPV+) or associated with smoking cigarettes and drinking alcohol (HPV−) (23, 24). HGSOCs account for most ovarian cancer cases and deaths (25–27) and, like HNSCCs, are innervated with TRPV1-expressing nerves (28). While the literature demonstrates contributions of sympathetic and parasympathetic nerves to cancer (5, 20, 29), less is known about the influence of sensory nerves on malignancies. An understanding of sensory innervation in cancer is emerging (9, 11, 28, 30, 31). To expand this knowledge, we focused our efforts on TRPV1-expressing sensory nerves in these cancers. Using several different in vitro and in vivo approaches, we demonstrate that despite the inherent differences between HNSCC and HGSOC, the activity and molecular functions of their intratumoral nerves are remarkably similar.
RESULTS
Expression of neuron-associated genes correlates with poor survival
While the published electrophysiologic data support the presence of functional neural circuits in breast tumors (22), we wondered whether intratumoral nerves affect patient survival. Thus, we analyzed the expression of genes classically associated with neurons in ovarian cancer using the OncoLnc dataset assessing correlations between patient survival and gene expression. These patient datasets have been previously published and include tables summarizing the patient cohort characteristics (e.g., stage and grade) (32, 33). Of 171 neuron-associated genes, 56 were differentially expressed in ovarian cancer; in most genes, high expression correlated with poor survival (Fig. 1A). Survival plots of two of these genes, KCNT1 (a sodium-activated potassium channel) and RAMP3 (receptor activity modifying protein 3), are shown (fig. S1, A and B) as their elevated expression correlated highly with decreased patient survival. The ovarian cancer patient survival data used for these analyses is shown in fig. S1C, while a list of the differentially expressed genes and their correlation with survival are shown in table S1. To better understand the pathways affected by these neuron-associated genes, gene ontology (GO) enrichment analysis of these gene sets was used and showed that significant molecular and cellular functions include neuronal pathways (e.g., neuron projection and ion/gated channel activity) (Fig. 1, B and C). Given that HGSOCs account for most deaths from ovarian cancer, we specifically focused a second analysis on this disease subtype and the expression of neuron-associated genes with increasing disease grade. We found that a significant increase in PGP9.5 (neuronal marker) expression correlates with increasing grade (fig. S1D). These data suggest that increased expression of neuron-associated genes correlates with decreased HGSOC patient survival.
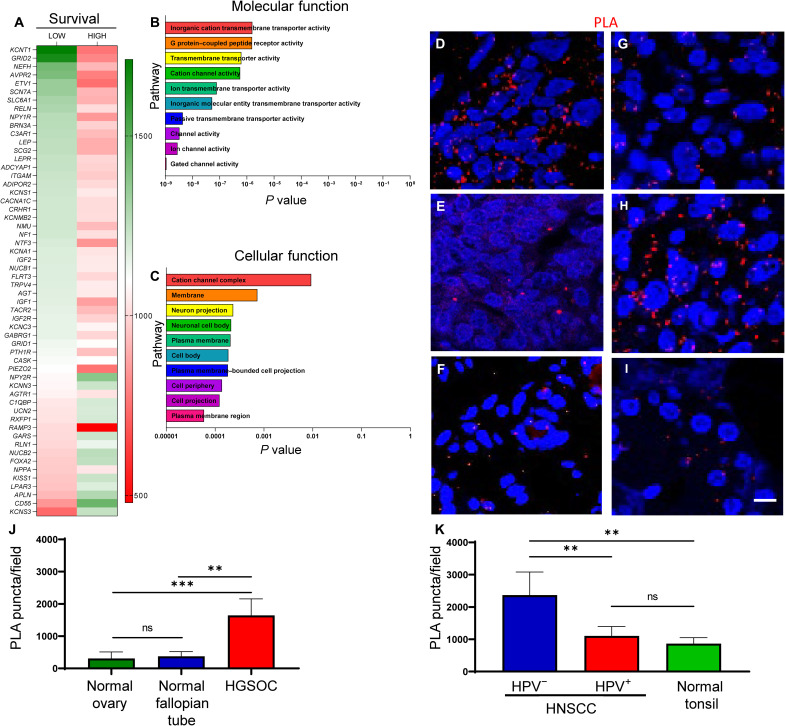
Fig. 1. Neuronal gene expression correlates with survival, neuronal pathways, and synapse-like structures in tumors.
Heatmap of neuronal genes expressed by ovarian cancers from OncoLnc, Gepia2, and Oncomine datasets (A). Correlations between gene expression and patient survival shown. GO enrichment analysis of neuronal genes demonstrating significant pathways important for molecular (B) and cellular (C) functions in ovarian cancers. Representative en face confocal images of HGSOC (n = 4) (D), normal ovary (n = 2) (E), normal fallopian tube (n = 2) (F), HPV+ HNSCC (n = 4) (G), HPV− HNSCC (n = 4) (H), and normal tonsil (n = 2) (I) stained by PLA for neurexin-3 and neuroligin-1. Red, positive PLA signal. Blue, 4′,6-diamidino-2-phenylindole nuclear stain. Scale bar, 100 μm. Quantification of PLA-positive signals from normal ovary, normal fallopian tube, and HGSOC (J) as well as HPV−, HPV+, and normal tonsil (K). Statistical analysis by unpaired Student’s t test. **P < 0.05; ***P < 0.01; ns, not significant.
A similar analysis focused on HNSCC was performed. Here, of 152 neuron-associated genes expressed, 79 were differentially expressed in HNSCC (33). The elevated expression of approximately half of these genes correlates with poor survival, while low expression of the remaining genes correlates with high survival (fig. S1E). This dichotomy in the contribution of neuron-associated genes to HNSCC patient survival likely reflects the mixed patient samples (HPV+ and HPV−) in the datasets.
Kaplan-Meier survival plots for two genes (GAL and MPZ) whose higher expression correlated most with decreased HNSCC patient survival are shown (fig. S1, F and G). The head and neck cancer patient survival data used for these analyses is shown in fig. S1H, while a list of the DEGs and their correlation with survival is shown on table S2. Similar to ovarian cancer, GO enrichment analysis of the HNSCC datasets underscore the significance of neuronal pathways (fig. S1, I and J) including axon/neuron projection and ion channel activity in cancer. Of the 56 (HGSOC) and 79 (HNSCC) differentially expressed neuron-associated genes, in silico analysis shows that 7 genes (AVPR2, KCNC3, KCNN3, KCNT1, RAMP3, TRPV4, and UCN2) overlap between the two datasets. Thus, despite the obvious differences between these cancers, they similarly harbor elevated expression of neuron-associated genes and pathways which correlate with poor patient outcomes. It is important to note that these genes should not be taken in isolation; but rather, when taken altogether, indicate that the overexpression of these gene classes indicates an increased activity of neurons which is coupled with a decreased patient survival.
Expression of pre- and postsynaptic markers in the tumor bed
Having previously established that HNSCCs and HGSOCs are innervated (9, 28) and that expression of neuron-associated genes correlate with poor survival, we wondered what tumor-infiltrating nerves do after they invade the tumor proper. To gain a more accurate understanding of the spatial relationships between intratumoral nerves and tumor cells, we immunohistochemically (IHC) stained HGSOCs (n = 4 cases) for PAX8 (lineage marker) (34, 35) and HNSCCs (n = 4 cases) for cytokeratin (tumor marker) and β-III tubulin (neuronal marker). In both cancers, many instances of neurites in close proximity to tumor cells were evident (fig. S2A). These data suggest the presence of intimate associations between tumor-infiltrating nerves and tumor cells in patient samples. Consistent with this, the presence of bona fide synapses in brain tumors has been demonstrated (36, 37), yet synapses within peripheral cancers have not been reported. We used proximity ligation assay (PLA) to probe this question since this method detects the close juxtaposition (less than 40 nm) of two proteins (38). Moreover, PLA has been used to identify synapses and their plasticity during learning (39). Thus, n = 4 cases each of HGSOC and HNSCC (n = 4 HPV+, n = 4 HPV−) were processed for PLA using pre- (neurexin-3) and post (neuroligin-1) synaptic markers. Tissue controls included normal ovary, normal fallopian tube, normal tonsil, as well as cases of glioblastoma that serve as positive and negative controls (n = 2; fig. S2, B to D). PLA positive signal was evident in all cancer cases as well as their normal tissue controls (Fig. 1, D to I). Quantification indicates a significant increase in PLA signal in HGSOC and HPV− HNSCC as compared to their normal tissue controls (Fig. 1, J and K). The PLA signal in cases of HPV+ HNSCC was not significantly different from the normal tonsil controls. HPV− cases (Fig. 1H) harbor a significantly increased number of PLA puntca per field as compared to HPV+ cases (Fig. 1G), suggesting that HPV− HNSCCs may harbor greater intratumoral nerves, increased formation of synapse-like connections, or both. We next combined PLA with immunofluorescence for β-III tubulin to assess whether synapse-like connections are formed between intratumoral nerves. While PLA puncta are found near β-III tubulin–positive nerves in HGSOC (fig. S2E) and HNSCC (fig. S2F) samples (n = 3 per tumor type), it is unclear at this level of resolution whether this indicates that synapses form between nerves in the tumor bed. Overall, these data suggest that tumor-infiltrating nerves likely establish functional interactions within the tumor microenvironment.
Electrical activity in tumors
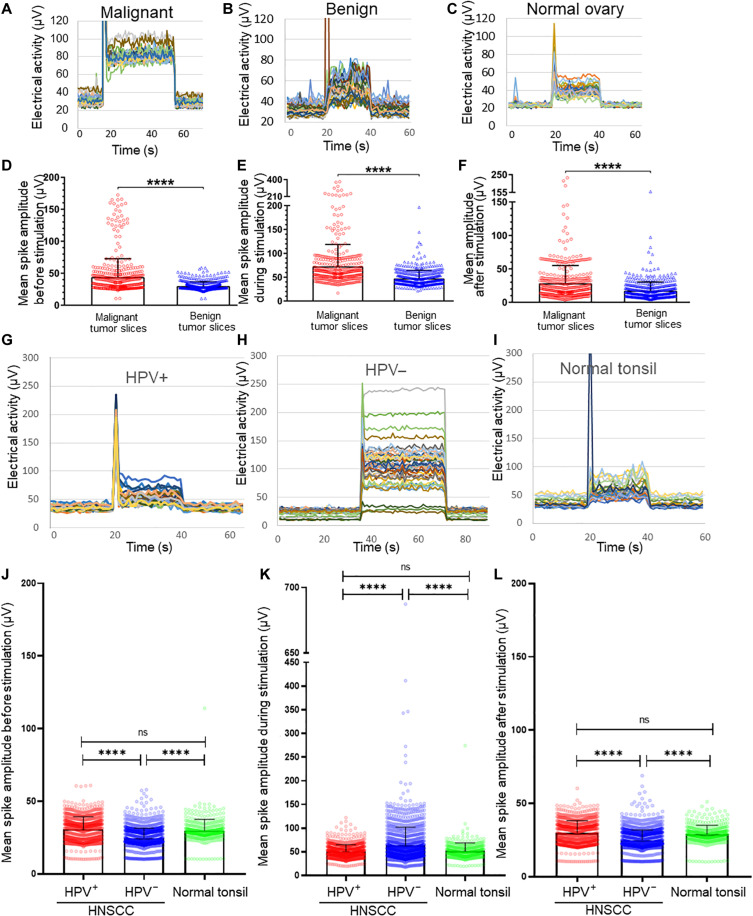
PLA-positive signal with pre- and postsynaptic markers in patient samples is consistent with the hypothesis that tumor-infiltrating nerves form functional connections within the tumor bed. Whether electrical or chemical in nature, these connections should elicit measurable electrical activity. To test this hypothesis, tumors were analyzed using microelectrode arrays (MEAs). Fresh HGSOC tumors (n = 13 cases) were sliced, maintained in artificial cerebrospinal fluid, and analyzed by MEA. HGSOC slices demonstrate a large evoked electrical activity (100 to 400 μV), while stimulus removal reverts this activity to baseline (Fig. 2A and fig. S2G). Each different colored line represents the electrical activity from each electrode within the MEA. We noted variability in electrical activity between patient samples and even between slices from the same patient tumor. Such variability likely reflects the level of neural circuit complexity captured in each slice. In addition, the stimulus peak differs between samples (e.g., Fig. 2, A to C). This stimulus peak is a well-recognized stimulus artifact that occurs with MEAs (40), and its peak relies upon the active cells captured within each slice. The stimulation itself was identical for all slices (see Material and Methods). The stimulus artifact generally consisted of 1 s of recorded data immediately following stimulation; all artifact data were removed before data analysis. For each patient sample, slices were histologically stained to confirm the presence of tumor (fig. S2, H to J). We wondered whether benign gynecological tumors harbor a similar ability to respond to electrical stimulation as malignant disease. MEA measurements show that while benign tumor slices (n = 5 cases) also respond when stimulated, the magnitude of these responses was lower than that from malignant slices (Fig. 2B). When slices of normal ovary (n = 2 cases; Fig. 2C) were analyzed, the magnitude of evoked electrical activity was similar to that of benign slices (fig. S2, K to M). Overall, malignant disease harbors significantly increased baseline, evoked, and poststimulation electrical activity as compared to benign disease (Fig. 2, D to F). Similar to HGSOC, cases of HNSCC (n = 6) harbor significantly increased evoked electrical activity. Specifically, HPV− HNSCCs (n = 4 cases) harbor significantly elevated electrical activity as compared to HPV+ tumors (n = 2 cases) and normal tonsil (n = 2 cases) (Fig. 2, G to I). This was in contrast to the baseline and poststimulus activity that were both significantly higher in HPV+ disease and normal tonsil. The electrical activity from HPV+ HNSCCs was not significantly different from that of normal tonsil (Fig. 2, J to L). Histological staining of samples verifies the presence of tumor (fig. S2, N to P).
Fig. 2. MEA analysis of tumors.
Snapshots of representative MEA recordings from malignant HGSOC (n = 13 cases) (A), benign gynecologic tumor (n = 5 cases) (B), and normal ovary (n = 2 cases) (C) patient samples before, during, and after stimulation. MEA quantification comparing malignant and benign disease at baseline (D), during evoked activation (E), and after removal of the artificial stimulus (F). Snapshots of representative MEA recordings from n = 2 HPV+ cases (G), n = 4 HPV− cases (H), and n = 2 normal tonsils (I) before, during, and after stimulation. Quantification of MEA data comparing baseline (J), during evoked activation (K), and after removal of the artificial stimulus (L). Signals from all electrodes and all tissue slices were pooled, averaged, and compared. Statistical analysis by one-way analysis of variance (ANOVA). SD, error bars. ****P < 0.0001. N ≥ 4 slices per sample were generated on the basis of tumor size.
Disease site may not influence tumor innervation
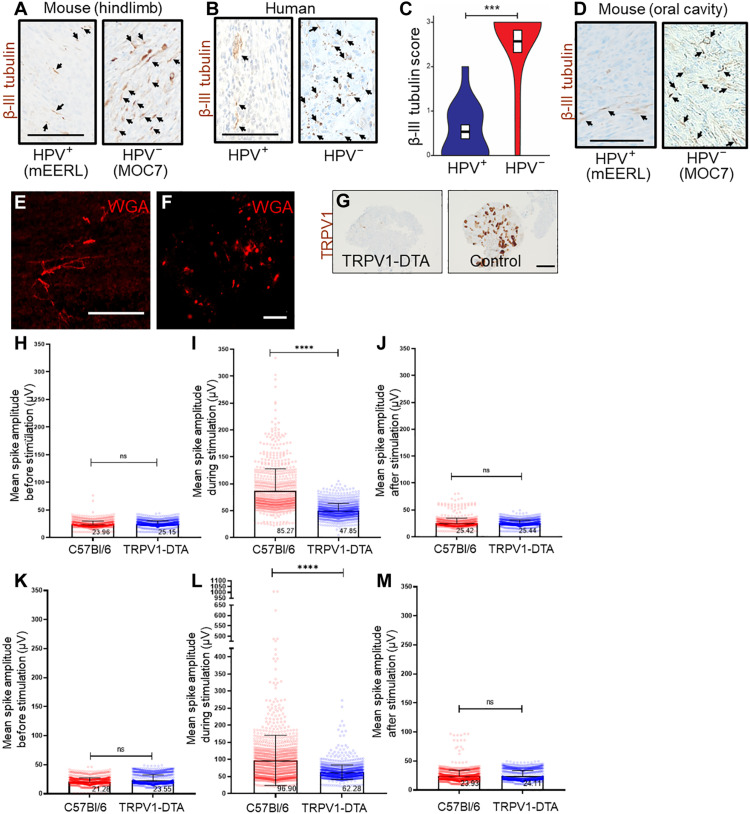
HPV+ and HPV− HNSCCs typically occur in different sites within the oral cavity. To test whether disease site affects tumor innervation, mice (n = 10 mice per group) were subcutaneously implanted in the hindlimb with mEERL (mouse HPV16 E6 and E7, Ras, Luciferase) (HPV+) (41–43) or MOC7 (mouse oral carcinoma) (HPV−) (44) tumors, thereby eliminating disease location as a variable. All animals were euthanized on day 30 after tumor implantation and tumors IHC stained for β-III tubulin. HPV− (MOC7) tumors were highly innervated, while HPV+ (mEERL) tumors were sparsely innervated (Fig. 3A). Because both tumors were implanted in the hindlimb, it suggests that disease site does not affect the extent of tumor innervation. These findings also suggest that the increased evoked electrical activity measured in HPV− patient samples (Fig. 2, H and J to L) reflects an increased innervation density. To test this hypothesis further, 15 HPV+ and 15 HPV− HNSCC patient tumors were IHC stained for β-III tubulin and blindly scored by a pathologist specializing in head and neck cancer. Again, HPV− tumors emerged as significantly more innervated (Fig. 3, B to C). These data suggest that the increased tumoral electrical activity of HPV− disease may stem from dense tumor innervation. In addition, mEERL (HPV+) and MOC7 (HPV−) cells were implanted into the oral cavity of C57Bl/6 mice to determine whether the tumors maintain this difference in innervation when orthotopically implanted. When tumors were removed and IHC stained for β-III tubulin, MOC7 (HPV−) tumors emerged as more densely innervated than mEERL (HPV+) tumors (Fig. 3D).
Fig. 3. Electrical activity from TRPV1-expressing neurons.
Photomicrographs of murine [(A), hindlimb] and human (B) HPV+ and HPV− HNSCCs immunohistochemically stained for β-III tubulin (brown). Arrows, positively stained intratumoral nerves. Scale bar, 100 μm. (C) Quantification of β-III tubulin score; score of 0, no positive staining; score of 1 indicates 10% positive staining; score of 2 indicates 30% positive staining; score of 3 indicates more than 30% positive staining. N = 15 cases each of HPV+ and HPV− HNSCCs. Statistical analysis by Student’s t test. ***P < 0.0001. Representative photomicrograph of orthotopic mEERL (HPV+) and MOC7 (HPV−) tumors IHC stained for β-III tubulin (arrowheads) (D). Scale bar, 100 μm. En face confocal image of orthotopic MOC7 (HPV−) tumor injected with wheat germ agglutinin (WGA) neural tracer. Tracer+ (red) nerves in the tumor bed (E) map to the ipsilateral TGM ganglion (F). Scale bar, 200 μm. (G) Photomicrographs of DRG from TRPV1cre::DTAfl/WT (TRPV1-DTA) and C57Bl/6 (control) mice IHC stained for TRPV1 (brown). Scale bar, 100 μm. MEA analysis of Trp53−/−;Pten−/− [HGSOC, (H) to (J)] and MOC7 [HPV− HNSCC, (K) to (M)] tumors subcutaneously implanted into either C57Bl/6 or TRPV1cre::DTAfl/WT (TRPV1-DTA) animals. Signals from all electrodes and all tissue slices were pooled, averaged, and compared. Statistical analysis by one-way ANOVA. SD, error bars. ****P < 0.0001.
Having already mapped intratumoral nerves in murine HGSOC (28), efforts focused on mapping them in murine HNSCC. Here, orthotopic implantation of tumor was chosen to closely mimic the human disease. Thus, C57Bl/6 mice were orally implanted with either mEERL (HPV+) or MOC7 (HPV−) cells. Intratumoral injection of fluorophore-conjugated wheat germ agglutinin (WGA), a neural tracer (45–47), was performed on palpable tumors [as in (28)] and tumors permitted to grow for an additional 3 to 7 days to allow for retrograde transport of the tracer. Following euthanasia, fluorescence microscopic examination of tumors (Fig. 3E) and regional ganglia shows that intratumoral nerves map to the ipsilateral trigeminal (TGM) ganglion (from MOC7 tumor, Fig. 3F; from mEERL tumor, fig. S3A).
Nociceptor neurons contribute to tumoral electrical activity
To determine whether nociceptor neurons contribute to tumoral electrical activity (9, 28), we used a double transgenic mouse, TRPV1cre::DTAfl/WT, in which all TRPV1-lineage neurons are ablated by diphtheria toxin A (48). The absence of nociceptor neurons was confirmed by IHC staining dorsal root ganglia (DRG) from control and nociceptor neuron ablated transgenic animals (Fig. 3G). TRPV1cre::DTAfl/WT and C57Bl/6 animals were implanted with MOC7 (HPV−) or mEERL (HPV+) cells. To similarly assess the contribution of nociceptor neurons to HGSOC electrical activity, mice were implanted with Trp53−/−;Pten−/− cells, a syngeneic model of HGSOC (28). Subcutaneous implantation into the hindlimb was chosen to eliminate the variable of disease site on tumor innervation. When sacrifice criteria were met (approximately 30 days after tumor implantation), animals were euthanized, and tumors harvested, sliced, and analyzed by MEA. Similar to our findings with patient samples, MOC7, mEERL, and Trp53−/−;Pten−/− tumors lacked spontaneous electrical activity. While head and neck as well as ovarian tumor slices from both C57Bl/6 and TRPV1cre::DTAfl/WT animals respond to stimulus with evoked activity, the magnitude of this response was significantly lower in tumors from TRPV1cre::DTAfl/WT animals (HGSOC, Fig. 3, H to J; MOC7, Fig. 3, K to M; and mEERL, fig. S3, B to D) suggesting that a significant component of tumoral evoked electrical responses originates from the TRPV1-lineage neurons.
Substance P promotes cellular proliferation in cancer cell lines
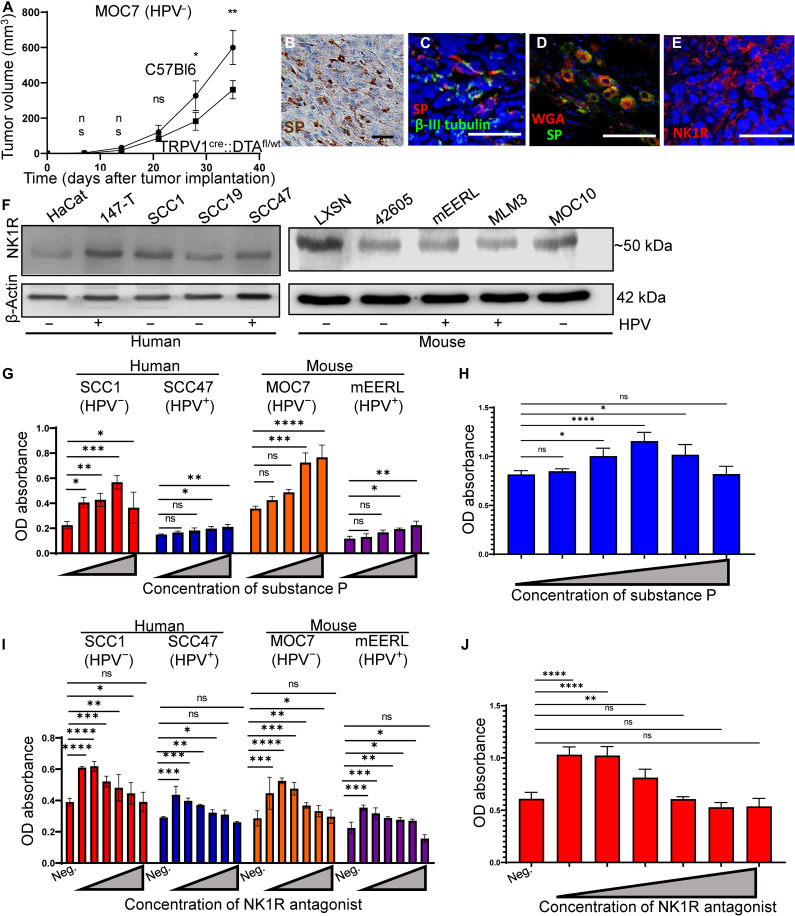
To test whether nociceptor intratumoral nerves contribute to tumor growth, we orthotopically implanted C57Bl/6 and nociceptor neuron ablated animals implanted with mEERL (HPV+) or MOC7 (HPV−) cells (n = 10 mice per group) and monitored tumor growth weekly. The absence of nociceptor neurons results in a modest, yet significant, slowing of tumor growth in both groups (MOC7, Fig. 4A; mEERL, fig. S3E). As an additional control, tumors were also implanted in TRPV1WT::DTAfl/WT or TRPV1cre::DTAWT/WT littermate control mice. This time, however, animals were implanted with a variant of mEERL (HPV+) cells that stably overexpress the axonal guidance cue, EphrinB1. These cells have been previously characterized and chosen because they generate densely innervated tumors (9).
Fig. 4. SP induces tumor cell proliferation.
(A) Tumor growth curve of MOC7 (HPV−) tumors orthotopically implanted into C57Bl/6 (control) or TRPV1cre::DTAfl/WT mice (n = 10 mice per group). Statistical analysis by multiple Student’s t test. **P < 0.01; *P < 0.05. (B) Photomicrograph of HNSCC (n = 4 cases) IHC stained for SP (brown). Scale bar, 50 μm. (C) En face confocal image of a HNSCC patient sample immunofluorescently stained for SP (red) and β-III tubulin (green). N = 4 tumors analyzed with similar results. Scale bar, 100 μm. (D) Ipsilateral TGM ganglion from tracer injected MOC7 (HPV−) tumor showing colocalization of tracer (red) and SP (green). Scale bar, 50 μm. (E) Representative en face confocal images of HNSCC patient sample immunofluorescently stained for NK1R (red) (n = 4 patient samples). Scale bar, 100 μm. (F) Western blot analysis of whole cell lysates from the indicated HNSCC cell lines for NK1R. β-Actin, loading control. Cellular proliferation of the indicated HNSCC (G) or HGSOC (H) cell lines in response to increasing concentrations of SP for 24 hours or with inclusion of an anti-NK1R antagonist that inhibits this effect [(I), HNSCC; (J), HGSOC]. Statistical test by one-way ANOVA with post hoc Tukey test. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001. OD, optical density.
Similar to our studies using C57Bl/6 J mice as controls, tumors implanted into nociceptor neuron ablated animals grew significantly slower than in the TRPV1WT::DTAfl/WT or TRPV1cre::DTAWT/WT littermate control mice (fig. S3F). Given these findings, we proceeded with C57Bl/6 J mice as controls. To define the potential neuronal basis for this effect, we focused on Substance P (SP), a major sensory neuropeptide with tumor supportive functions (49–51). IHC staining for SP in HNSCC and HGSOC patient tumors (n = 4 cases each) reveals neuropeptide expression in the tumor bed (Fig. 4B and fig. S3G, respectively). To define whether intratumoral nerves express SP and thus may be a source for this neuropeptide in these cancers, we immunofluorescently stained patient samples (n = 3 HNSCC) for coexpression of SP and β-III tubulin and found that they do indeed colocalize in the tumor bed (Fig. 4C). To verify this, tumor bearing animals were intratumorally injected with WGA (28), locoregional TGM ganglia harvested and immunofluorescently stained for SP. The presence of SP immunoreactivity within tracer-positive neurons in the ganglion confirms expression of SP by intratumoral nerves (Fig. 4D) and suggests its release into the tumor microenvironment. To determine whether SP is also secreted from tumor cells themselves, we cultured MOC7 (HPV−), mEERL (HPV+), and Trp53−/− Pten−/− cells in vitro, collected their conditioned media 48 hours later, and assessed it for the presence of SP by ELISA. We found that tumor cell conditioned media does not contain detectable levels of SP (fig. S3, H and I).
If SP is released into the tumoral space by intratumoral nerves, cells expressing its receptor, neurokinin 1 receptor (NK1R), could respond to it. Thus, HNSCCs (n = 3 HPV−, n = 2 HPV+) cases were immunofluorescently stained for NK1R. Immunofluorescent staining demonstrates NK1R expression by tumor cells (Fig. 4E). HNSCC and HGSOC cell lines (human and mouse) were also positive for NK1R by Western blot analysis (Fig. 4F and fig. S3J). To assess potential biological consequences of SP treatment, we incubated HNSCC and HGSOC cells with increasing concentrations of SP and cellular proliferation assessed at 24 (Fig. 4G, H) and 48 (fig. S4, A and B) hours. In all cases, SP treatment induces a modest, yet significant, increase in cellular proliferation. To determine whether this effect is mediated via NK1R, increasing concentrations of an NK1R antagonist (L-732,138) were tested in the presence of 100 nM SP and cellular proliferation was similarly assessed. In all cases, NK1R antagonism significantly blocks the ability of SP to promote cellular proliferation at 24 (Fig. 4I and J) and 48 hours (fig. S4, C and D), indicating that this effect is mediated by SP binding NK1R.
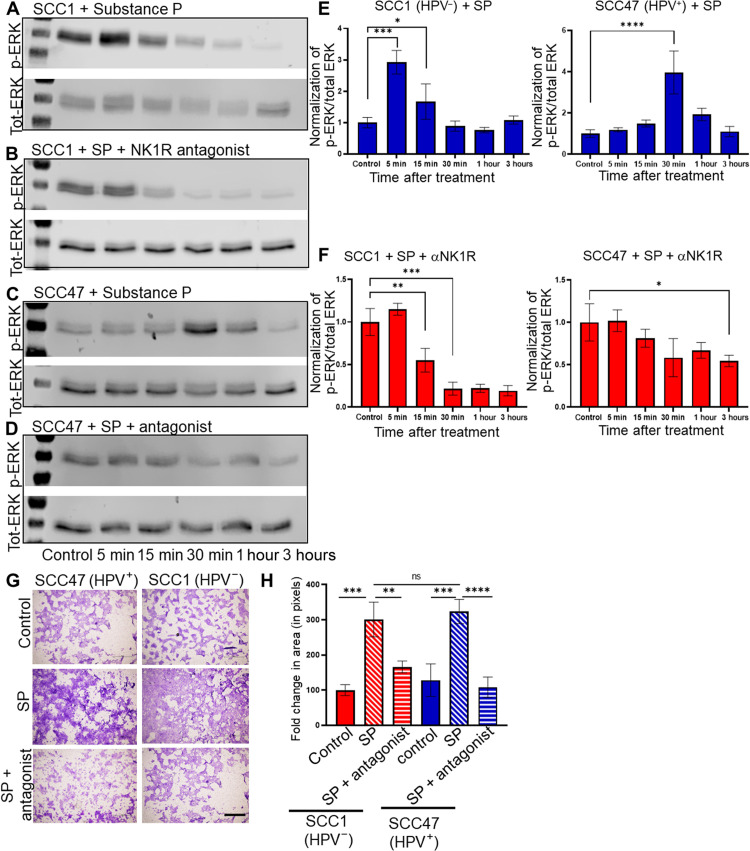
Ligation of SP and NK1R activates different downstream signals (52), including the mitogen-activated protein (MAP) kinase pathway that is highly active in many cancers. Using Western blot analysis of SP-treated cells, we found that SP treatment of HNSCC cells increases phosphorylation of ERK1/2 (Fig. 5, A, C, and E), which is inhibited by NK1R antagonism (Fig. 5B, D, and F). Whereas SP induces rapid ERK1/2 activation in SCC1 (HPV−) cells, the response is delayed in SCC47 (HPV+) cells. Similar results were found with the murine HNSCC cell lines (fig. S5, A to F) as well as the murine Trp53−/−;Pten−/− HGSOC cell line (fig. S5, G to J). Human NK1R exists in two isoforms, a full-length form, and a splice variant that has a truncated C terminus (which only extends seven amino acids into the cytosol after the seventh transmembrane domain) (52–54). Cells expressing full-length NK1R respond to SP stimulation with rapid extracellular signal–regulated kinase 1/2 (ERK1/2) phosphorylation. This is similar to the kinetics of ERK activation that we see in SP stimulated SCC1 (HPV−) cells (Fig. 5A). Cells expressing truncated NK1R respond with delayed kinetics to SP treatment, with a peak in phospho-ERK1/2 at 20 to 30 min (55, 56). This delayed kinetics of ERK1/2 activation is also found in SCC47 (HPV+) cells (Fig. 5C). To test whether this mechanism explains the difference in ERK phosphorylation kinetics that we see between SCC1 (HPV−) and SCC47 (HPV+) cells, quantitative reverse transcription polymerase chain reaction (RT-PCR) using NK1R primers that distinguish between full-length and truncated receptor was performed. We found that SCC1 (HPV−) cells predominantly express the full-length form of NK1R, while SCC47 (HPV+) cells express both truncated and full-length forms of the receptor (fig. S4E). These data suggest that the delayed MAP kinase response to SP stimulation evident in SCC47 (HPV+) cells likely occurs because of its expression of truncated NK1R.
Fig. 5. SP induces cellular migration.
Western blot analysis for p-ERK and total-ERK for SCC1 (HPV−) or SCC47 (HPV+) cells treated with SP alone (A and C, respectively) or in the presence of the NK1R antagonist (B and D, respectively) for indicated time points. Densitometric quantification (p-ERK/total ERK) for SCC1 and SCC47 cells treated with SP alone (E) or in the presence of NK1R antagonist (F). (G) Photomicrographs of Transwell migration assays of the indicated cells alone (control), treated with SP (SP), or treated with SP with the NK1R antagonist (SP + antagonist). Scale bar, 500 μm. (H) Quantification of migration assay [in (G)], n = 3 biological replicates, n = 4 technical replicates. Statistical test by one-way ANOVA with post hoc Tukey test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
SP promotes migration in cancer cell lines
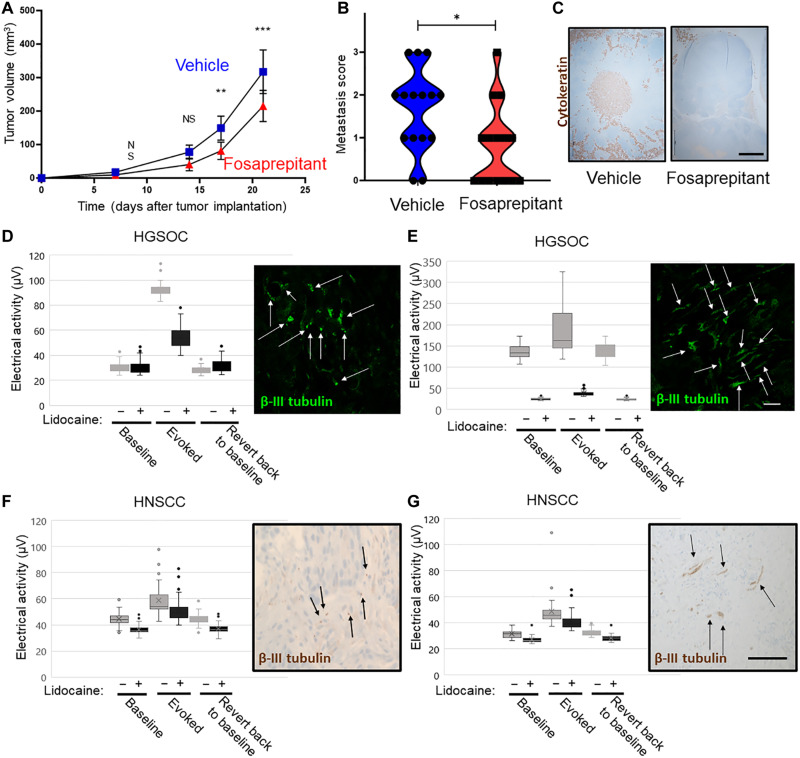
Because cancer progression involves the migration of tumor cells away from the primary site, we asked whether SP also affects migration of HNSCC and/or HGSOC cells. Using a Transwell assay, we found that SP treatment induces a nearly threefold increase in cell migration compared to vehicle treatment across all cell lines tested; pretreatment with the NK1R antagonist reduces cell migration to levels similar to that of vehicle (Fig. 5, G and H, and fig. S6, A to C). We further assessed the impact of SP on the ability of tumor cells to survive as a single cell and grow into a colony. Therefore, a clonogenic assay was performed on mEERL (HPV+), MOC7 (HPV−), and Trp53−/− Pten−/− cells. We found that treatment with SP does not alter the colony forming ability of any of the cells tested (fig. S6, D to F). The in vitro data suggest that blocking NK1R in vivo may significantly affect tumor growth and metastasis. To test this, C57Bl/6 mice were orthotopically implanted with MOC7 (HPV−) cells and randomized into two groups (n = 10 mice per group). One group received intraperitoneal treatment with fosaprepitant, a Food and Drug Administration (FDA)–approved NK1R antagonist, while the other group received vehicle. Tumor growth was monitored weekly. When euthanasia criteria were met, animals were euthanized. The draining lymph nodes were harvested, embedded in paraffin, cut, and IHC stained for cytokeratin to assess locoregional metastasis. Fosaprepitant treatment resulted in a modest, yet significant, decrease in tumor growth (Fig. 6A) as well as a significant decrease on metastasis to the draining lymph nodes (Fig. 6, B and C).
Fig. 6. Pharmacologic blockade of neuronal function.
(A) Tumor growth curve of C57Bl/6 mice orally implanted with MOC7 (HPV−) tumors and treated with vehicle or fosaprepitant. N = 10 mice per group. Statistical analysis by Student’s t test. **P < 0.01; ***P < 0.001. (B) Quantification of metastases in the draining lymph nodes of mice from experiment in (A). (C) Representative photomicrographs of draining lymph nodes from the indicated conditions from mice in experiment A. Brown, cytokeratin IHC staining. Scale bar, 500 μm. Representative examples of HGSOC (D and E) and HNSCC (HPV−) (F and G) electrical activity of tumor slices before and after treatment with lidocaine. Gray bars, before lidocaine; black bars, after lidocaine. N = 7 cases of HGSOC and n = 2 cases of HNSCC were evaluated. Insets, immunofluorescent (D and E; green; scale bar, 10 μm) or immunohistochemical (F and G; brown; scale bar 20 μm) staining for β-III tubulin of each sample. Arrows highlight nerves.
Silencing tumoral electrical activity
While the above data suggest a mechanism whereby SP and NK1R promote disease progression, we tested another independent approach, that of silencing the electrical activity of intratumoral nerves. Here, we tested whether tumoral electrical activity could be pharmacologically blocked. Following recording of electrical activity, HGSOC (n = 4) and HNSCC (HPV−, n = 2), patient samples were incubated with lidocaine, a voltage-gated sodium channel blocker, and again analyzed the electrical activity. While lidocaine treatment decreased electrical activity (Fig. 6, D to G), those samples with high evoked activity (HGSOCs) were exquisitely sensitive to treatment (Fig. 6, D and E). Samples with low evoked responses (HNSCC) were modestly sensitive (Fig. 6, F and G). A slice of each sample was assessed by immunofluorescence or IHC staining for β-III tubulin (insets, Fig. 6, D to G) and suggests that differences in innervation density may account for this difference in response to lidocaine. These findings suggest that innervation density of the tumor needs to be considered when evaluating patients that may benefit from this potential therapeutic approach.
DISCUSSION
We focused on two vastly different cancers, HNSCC and HGSOC. Despite differences in disease site, therapy regimens, response to treatment and overall survival, the presence and activity of intratumoral nerves are remarkably similar. This notable similarity suggests that intratumoral neuronal contributions to disease may reflect their universal requirement in cancers. Our bioinformatic analyses of neuron-associated genes and patient survival support this concept. Consistent with this, PLA signal with pre- and postsynaptic markers suggests the establishment of functional connections within the tumor bed. PLA-positive signal is present within the normal tissues as well (albeit with diminished frequency) indicating that under normal conditions, resident nerves harbor tropic functions. The neurotrophin nerve growth factor is required for early follicular development in the ovary (57–59). The increased prevalence of PLA-positive puncta in HGSOC and HNSCC suggests that malignancies may appropriate, expand, and possibly alter the normal neuronal connections of the resident tissue. Our ability to measure electrical activity from patient tumors supports this model. Given that all tissues and organs are innervated, the ability to measure electrical activity from cancer slices may not be unexpected or unusual. However, our finding that malignant disease harbors significantly increased electrical activity as compared to benign or normal tissues suggests the presence of additional nerves or an enhanced complexity of their circuits within malignancies. In the context of HNSCC, we show that the increased electrical activity within HPV− samples likely stems from the increased presence of intratumoral nerves. Both HPV− HNSCCs and HGSOCs are aggressive diseases with poor survival rates. Patients with HPV+ HNSCC fair significantly better (60, 61). Although HPV+ tumors can induce immune-mediated tumor clearance, the sparsity of intratumoral nerves within this cancer type may also contribute to better outcomes, yet this remains to be defined.
Despite harboring significantly higher evoked electrical activity, HPV− HNSCC slices have significantly lower baseline and poststimulation activity compared to HPV+ cancers and normal tonsil tissues. This unexpected finding may speak to different types of neuronal connectivity that may be established within these cancers. Neuronal connections are structural, functional, or effective. Structural connectivity speaks to the anatomical connectome of neural circuits (how circuits are physically connected). Functional inferences about circuit connections are based on the timing of neuronal activities between groups of neurons. Last, effective connectivity refers to the ability of neurons to influence other neuronal populations that they are not necessarily directly connected to (62–64). We speculate that in the highly innervated HPV− HNSCC slices, effective connectivity results in the high evoked activity measured. However, in the absence of a stimulus, the effective influence becomes, in essence, an invisible MEA signal. The functional connectivity within HPV+ tumor and normal tonsil, on the other hand, are easily measured in the presence and absence of a stimulus. These findings emphasize the extent and complexity of neural circuits that may exist within peripheral cancers.
The differences in innervation between virus-induced (HPV+) and mutagen-induced (HPV−) HNSCCs were interesting. Moreover, the innervation of HGSOC is similar to that of HPV−HNSCC, although it is not mutagen-induced. On the basis of our previous publication (9) and the published literature, we hypothesize that the dense innervation shared between HPV− HNSCC (mutagen-induced) and HGSOC is mediated by shared nerve-recruiting signals that differ from those released by HPV+ HNSCC (viral-induced). For instance, a study focused on the microRNA (miRNA) profiles of sEVs (small extracellular vesicles) from HPV-positive and HPV-negative HNSCC cell lines identified two disease-unique miRNAs. mIR-205-5p was found exclusively in sEVs from HPV+ cells, while miR-1972 was exclusive to sEVs from HPV− cells (65). mIR-1972 functions as an oncogene in ovarian cancer (66) and is packaged in sEVs (67). Kaplan-Meier survival plots of patients with ovarian cancer and hsa-miR-1972 show that cases with high miR-1972 expression suffer a significantly worse prognosis than those with lower expression (66). TargetScan analysis of predicted biological processes and cellular components regulated by miR-1972 are neuron centric including: neuron differentiation, neuron development, cell morphogenesis involved in neuron differentiation, neuron projection development, neuron projection morphogenesis, synapse, synapse part, and synaptic vesicle (66). We speculate, therefore, that sEVs released from HPV− HNSCC and HGSOC carry miR-1972 as cargo that contributes to dense innervation of these tumors. sEVs from HPV+ HNSCC do not harbor miR-1972. Instead, these sEVs package miR-205-5p, which functions as a tumor suppressor (68), its packaging within sEVs inhibits progression of liver cancer (69), and it is also antitumorigenic in colorectal cancer (70). Consistent with this, miR-205-5p has been shown to regulate pathways important for cell adhesion, angiogenesis, and cell-cell communication (70–73). We speculate that additional innervation signals (miRNA, neurotrophins, and axonal guidance molecules) are likely shared between HPV− HNSCC and HGSOC and are distinct from those associated with HPV+ HNSCC and that, together, these factors dictate the innervation status of these cancers. Future studies will further clarify this mechanism.
While tumoral electrical activity suggests a neuronal basis, we tested this hypothesis by using nociceptor neuron ablated mice in which all TRPV1-lineage neurons were eliminated from embryogenesis. Tumors grown within these transgenic animal’s harbor significantly decreased electrical activity as compared to the same tumors grown in wild-type animals, strongly implicating nociceptor neurons as contributing to tumoral electrical activity. Despite the absence of nociceptor neurons, however, these tumors still harbor residual electrical activity. This activity may originate from other recruited nerves. The fact that neuronal activity is not lost altogether supports the hypothesis that nerve recruitment is necessary for persistence of disease.
Our finding that tumor growth is significantly attenuated in nociceptor neuron ablated animals implicates these nerves in disease progression. Consistent with this, tumor cells respond to SP treatment with proliferation in a dose-dependent manner. We noted that higher doses of SP result in either no additional increase in cellular proliferation or even a decrease in proliferation. This reflects the recycling kinetics of its receptor. When stimulated by low doses of SP, the NK1R receptor is rapidly endocytosed and recycled to the plasma membrane. However, at higher ligand concentrations, the receptor follows a different recycling pathway that is significantly slower. This results in a decreased number of receptors recycled to the plasma membrane and subsequently a decreased impact on proliferation (74). Nonetheless, the ability of NK1R antagonism to attenuate this effect implicates this ligand/receptor pair in mediating downstream signals contributing to cellular proliferation. In addition, despite a similar level of expression of NK1R, HPV− cells exhibit a greater proliferative response to the same dose of SP as HPV+ HNSCC cells, an effect that also likely contributes to tumor growth and disease outcomes in these patients. We also show that SP engagement of NK1R induces cellular migration, another key characteristic of aggressive disease. These data suggest that SP and NK1R (and the MAP kinase signaling they induce) may be potent therapeutic targets in oncology. The significant attenuation of locoregional metastasis by fosaprepitant supports the clinical utility of targeting this ligand/receptor system. Our findings are consistent with other studies (49, 50, 75–77). While this strategy requires testing, the ability to silence intratumoral electrical activity suggests a potentially unappreciated and potentially powerful therapeutic approach. However, not all tumors respond robustly to such treatment; those that are densely innervated do. Thus, identifying patients with highly innervated disease may be critical for the future testing of nerve-targeting therapies in oncology. MEA analysis may function not only to identify patients at risk of aggressive disease but also as a tool with which to identify robust nerve-targeting drugs for therapeutic intervention. A wealth of clinical information exists for FDA-approved drugs for neurological diseases; testing their ability to silence tumoral electrical activity may identify candidates for preclinical testing in oncology.
Despite the interesting findings, this study is not without limitations. One limitation centers on the use of nociceptor neuron ablated mice (TRPV1cre::DTAfl/WT). TRPV1-lineage ablation is likely broader than solely limited to adult TRPV1+ neurons. TRPV1 is broadly expressed during development (78), and the cre recombinase-mediated expression of diphtheria toxin during embryogenesis likely results in the loss of most NF200+ sensory neurons. Future work will aim to ascribe our findings to Tac1+ nociceptor neurons (the Tac1 gene encodes SP, neurokinin A, and neuropeptides K and γ) using targeted genetically engineered mice in combination with pharmacological blockade [QX-314; (79)] or chemoablation [RTX; (80)] of TRPV1+ neurons in adult mice. Another limitation focuses on the possible formation of synapses or synapse-like structures in the tumor bed. While the PLA data support the presence of these structures as do the MEA recordings (suggesting functional synapses), electron microscopy would be necessary to verify the presence of bona fide synapses within peripheral malignancies. In addition, while the in vivo fosaprepitant study demonstrates a statistically significant impact of the NK1R antagonist on tumor growth, the effect is modest. This may reflect a suboptimal dose or scheduling of the drug. A dose response study would clarify the clinical utility of fosaprepitant use in the context of oncology. Despite these limitations, this study emphasizes the critical contributions of intratumoral nerves to disease progression and identifies the potential clinical utility of nerve-specific interventions for cancer treatment.
MATERIALS AND METHODS
Oncomine gene expression analysis for ovarian cancer
The Oncomine (www.oncomine.com) contains data for normal and tumor tissues. Using Oncomine, we assessed transcript expression of PGP9.5 in patients with HGSOC (89 primary ovarian tumors and 36 ovarian metastases from breast cancer). Patient details can be found in the study by Meyniel et al. (81), and the dataset is publicly available on Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE20565.
OncoLnc gene expression analysis for ovarian and head and neck squamous cell carcinoma
OncoLnc (www.oncolnc.org) contains survival data for 8647 patients from 21 cancer studies performed by The Cancer Genome Atlas (TCGA). Using OncoLnc, we assessed the transcript expression of a user-defined list of 333 neuronal-enriched genes (neuronal membrane proteins, neural stem cell markers, transcription factors, ion channel receptors, and neuropeptides). Out of this list, 56 and 79 genes were detected in Ovarian and HNSCC, respectively. Heatmaps were generated to indicate the correlation between gene expression and patient survival. Details of patients can be found in TCGA, and computational analysis is defined by Anaya (82).
GO analysis
GO analysis was performed in PANTHER (Protein Analysis Through Evolutionary Relationships (http://pantherdb.org). All genes that showed a correlation between gene expression and patient survival from OncoLnc (56 genes in ovarian cancer and 79 genes in HNSCC) were further analyzed in Panther. The “Reference list” was PANTHER Generic Mapping File, as provided by PANTHER. For both lists, the organism selected was Mus musculus. Molecular function, biological process, and cellular component data were further analyzed to assess similarities between ovarian cancer and HNSCC gene expression pathways.
Antibodies used for immunohistochemistry
Anti–β-III tubulin (Abcam, catalog no. ab78078, 1:250, RRID:AB_2256751) and anti-TRPV1 (Alomone Labs, catalog no. ACC-030, 1:400, RRID:AB_2313819) were used.
Antibodies used for immunofluorescence
Anti-SP (Thermo Fisher Scientific, catalog no. PA5-106934,1:750, RRID:AB_2854598) and anti-NK1R (Novus Biologicals, 1:500, catalog no. NB300-119) were used. For immunofluorescence combined with PLA, a chicken anti–β-III tubulin antibody was used (EMD Millipore, catalog no. AB9354, 1:100, RRID:AB_570918).
Antibodies used for PLA
The following antibodies were used: anti–neuroligin-1 (Invitrogen, 1:300, catalog no. MA5-27635, 1:400,RRID:AB_2735286), anti–neurexin-3 (Invitrogen, 1:300, catalog no. PA5-77521, 1:400, RRID:AB_2736259), and anti-TIM3 (T-cell immunoglobulin and mucin domain-containing protein 3) (Cell Signaling Technology, catalog no. 83882S, 1:400, RRID:AB_2800033).
Antibodies used for Western blot analysis
The following antibodies were used: anti–phospho-p44/42 MAP kinase (Erk1/2) (Thr202/Tyr204) (Cell Signaling Technology, catalog no. 9101, 1:1000, RRID:AB_331646), anti-p44/42 Erk1/2 (Cell Signaling Technology, catalog no. 9102, 1:1000, RRID:AB_330744), β-actin (Sigma Life Science, catalog no. A2228, 1:1000), and anti-NK1R (Novus Biologicals, 1:500, catalog no. NB300-119).
Proximity ligation assay
In situ PLAs were performed using the Duolink PLA kit (DUO92101-1KT). Formalin-fixed and paraffin-embedded sections were deparaffinized and rehydrated by using the following washes at room temperature (RT): 100% Histo-Clear (National Diagnostics) for 5 min, 100% ethanol for 1 min, 90% ethanol for 1 min, 70% ethanol for 1 min, and then in phosphate-buffered saline (PBS) for 1 min. A heat-induced antigen retrieval step was performed before staining as follows: Sections were incubated with 10 mM sodium citrate buffer [10 mM sodium citrate buffer and 0.05% Tween 20 (pH 6.0)] at 95°C for 1 hour. After cooling down at RT for 30 min, slides were washed with PBS and then blocked in Duolink blocking buffer (1× PBS, 10% goat serum, 0.5% TX-100, and 1% bovine serum albumin) for 1 hour at 37°C. Sections were incubated with primary antibodies overnight at +4°C. Slides were washed in PLA Wash Buffer A twice for 5 min at RT. Slides were incubated in Duolink PLA probe reagents from 1 hour at 37°C and then washed twice in PLA Wash Buffer A for 5 min at RT. Ligation was performed for 30 min at 37°C using Duolink ligation reagents and washed twice with PLA Wash Buffer A for 5 min at RT. Amplification was conducted using Duolink amplification reagents for 100 min at 37°C. Following amplification, slides were washed twice in PLA Wash Buffer B for 10 min and then in 0.01× Wash Buffer for 1 min. Last, slides were mounted with Duolink In Situ mounting media with 4′,6-diamidino-2-phenylindole and imaged. Negative controls included the omission of primary antibodies as well as the use of two antibodies to proteins that do not interact (we used neuroligin-1 and TIM3). All controls were performed on glioblastoma patient samples.
Proximity ligation assay with immunofluorescence
The same steps were followed as described above; however, following the 1-hour incubation (at 37°C) in blocking buffer, sections were incubated with primary antibodies overnight at +4°C in a humidity chamber. These primary antibodies were neuroligin-1, neurexin-3, and β-III tubulin. Slides were washed in PLA Wash Buffer A twice for 5 min at RT and then incubated in Duolink PLA probe reagents and immunofluorescent secondary antibody against β-III tubulin for 1 hour at 37°C, then washed twice in PLA Wash Buffer A for 5 min at RT. The remaining procedure remains the same as above.
IHC staining
Formalin-fixed, paraffin-embedded tissues were cut at 5 μm thickness. We used the BenchMark XT automated slide staining system (Ventana Medical Systems Inc.) to optimize antibody dilutions and staining. The antigen retrieval step (basic pH tris base buffer) was performed using the Ventana CC1 solution. Primary antibody incubation of sections was for 1 hour. The Ventana iView DAB detection kit was used as the chromogen, and the slides were counterstained with hematoxylin.
For double-IHC staining, the BenchMark XT automated slide staining system (Ventana Medical Systems Inc.) was used for deparaffinization and antigen retrieval. The antigen retrieval step was performed using the Ventana CC1 solution, which is a basic pH tris base buffer. Tissue was incubated with the antibody cocktail for 1 hour at 37°C. Tissue was then incubated with mouse AP (alkaline phosphatase) + rabbit horseradish peroxidase (HRP) polymer detection kit (Biocare Mach 2 Double stain 1) for 30 min at RT. Tissues were rinsed with tris-buffered saline and incubated with chromogens Betazoid DAB and Warp Red (both Biocare) for 5 min each, respectively. Slides were counterstained with hematoxylin, dehydrated, cleared, and coverslipped. The Aperio VERSA 8 slide scanning system from Leica Biosystems, equipped with a Point Grey Grasshopper3 color camera for bright-field scanning, was used to analyze stained sections.
Hematoxylin and eosin staining
The Sakura Tissue-Tek automated hematoxylin and eosin (H&E) stainer was used to stain sections as follows: deparaffinize and rehydrate tissue, stain in Gill’s III hematoxylin (2 min), differentiate running tap water, blue in ammonia water, counterstain in eosin (1 min), dehydrate, and clear.
Neural tracing of orthotopic HNSCC tumors
While the MEA findings speak to the functionality of intratumoral nerves, defining their site of origin could provide a greater understanding of their connectivity and impact on disease. Mice were anesthetized with isoflurane and oral tumors measured. When tumors reached approximately 5 mm by 5 mm in size, they were injected with neural tracer as follows. Mice were anesthetized with ketamine (87.5 mg/kg)/xylazine (10 mg/kg). A 10-μl Hamilton syringe with 30-G needle was loaded with WGA-Alexa 568 (1% in PBS). Bevel side up, the needle was slowly injected into the oral tumor from the inside of the oral cavity approximately midway through the tumor and then pulled back slightly to reduce pressure and leakage of tracer following injection. Tracer was injected slowly over the course of 10 min. Following injection, needle placement was maintained for two additional minutes and then slowly removed from the tumor and the oral cavity. Mice were returned to the appropriate cage on a heating pad until they recovered from anesthesia. Seven days following tracer injection, mice were deeply anesthetized and transcardially perfused with ice-cold PBS. The skin was removed from the back and top of the skull, and the top of the skull and brain were removed exposing the TGM ganglia (HNSCC). TGM ganglia were carefully removed from the base of the skull and placed in Hanks’ balanced salt solution in a 24-well plate kept on ice. Tumor tissue was also collected, fixed, sectioned by vibratome, and imaged for WGA-labeled nerve fibers under confocal microscopy.
Cell lines
mEERL (RRID:CVCL_B6J3), MOC7 (RRID:CVCL_ZD34), and Trp53−/−;Pten−/− (28) (RRID:CVCL_B6JA) cell lines have been previously described (41–44, 83). mEERL cells were maintained with E-medium: Dulbecco’s modifiedEagle's medium (DMEM) (Corning, catalog no. 10-017-CV)/Hams F12 (Corning, catalog no.10-080-CV), 10% fetal calf serum, 1% penicillin/streptomycin, hydrocortisone (0.5 μg/ml), transferrin (5 μg/ml), insulin (5 μg/ml), tri-iodo-thyonine (1.36 ng/ml), and epidermal growth factor (5 ng/ml). MOC7 cells were maintained in DMEM with 10% fetal calf serum. The Trp53−/−Pten−/− cells were grown in minimum essential medium alpha (500 ml, Gibco, 12571-048) with l-glutamine, supplemented with 10% fetal bovine serum, 500 ml of insulin/transferrin/sodium selenite (Roche no. 11074547001), 10 ml of β-estradiol (Sigma-Aldrich, no. E8875). All cell lines were maintained at 37°C and 5% CO2; culture medium was refreshed every 3 days.
Proliferation assay
mEERL, MOC7, SCC1, SCC47, and Trp53−/−Pten−/− cells were seeded in a 96-well plate at a density of 5000 cells per well in complete media overnight. Cells were then serum starved for 24 hours before being treated with various concentrations of SP (0, 5, 10, 50, and 100 nM) (Sigma-Aldrich, acetate salt hydrate, catalog no. S6883) for 24 or 48 hours. In studies including the NK1R antagonist, cells were pretreated with various concentrations (0, 20, 40, 40, 80, and 100 μM) of NK1R antagonist, L-732,138 (Tocris Bioscience), for 10 min before the addition of 100 nM of SP. Following incubation, wells were treated with Celltiter 96 MTT reagent (CellTiter 96Aqueous One Solution Cell Proliferation Assay, catalog no. G3582) for 4 hours. Optical densities (ODs) were measured using a Spectramax plate reader at 490 nm. Changes in proliferation was defined as OD (treatment)/OD (control).
Migration assay
mEERL, MOC7, SCC1, SCC47, and Trp53−/−Pten−/− cells were seeded in the upper compartment of a 24-well Transwell plate (8.0-μm polycarbonate membrane, Costar ref. no. 3422) with 1 ml of serum-free DMEM in the bottom compartment overnight. The following morning, media was removed from the bottom compartment and replated with serum-free DMEM containing 100 nM SP. In studies conducted with the NK1R antagonist, wells were pretreated with 100 μM NK1R antagonist for 10 min before the addition of 100 nM of SP. Twenty-four hours after treatment, inserts were removed, fixed with ethanol, and stained with crystal violet. Inserts were left to dry at RT for 15 min, and insert membranes removed and mounted on glass slides for imaging. The area of migration was analyzed by ImageJ software.
Clonogenic assay
Cells were plated in a six-well dish with 365 cells per well and were allowed to adhere overnight. The following day, cells were treated with 100 nM SP or PBS. Three days later, cells were fixed in 70% EtOH and stained with crystal violet so that the colonies could be counted. A Student’s t test was used to analyze for significance (P < 0.05).
MAP kinase signaling Western blot
To test the downstream signaling events initiated by binding of SP to NK1R, dishes of cells (approximately 80% confluent) were stimulated for 5 min, 15 min, 30 min, 1 hour, or 3 hours with 50 nM SP in the presence or absence of 80 μM NK1R antagonist. At the designated time points, cell lysates were generated and analyzed by Western blot as described below.
Western blot analysis
Cellular lysates were collected by removing media from cells and washing them twice with ice-cold PBS before the addition of 500 μl of lysis buffer (50 mM tris HCl, 150 mM NaCl, 5 mM EDTA, 2 mM Na3VO4, 100 mM NaF, 10 mM NaPPi, 10% glycerol, and 10% Tx-100) per well. Dishes were then placed on ice for 10 min before cell lysate was scraped, collected, sonicated, and centrifuged from 5 min at 10,000g at 4°C. Supernatant was collected and used for downstream Western blot analysis. Protein concentration was measured using BCA (Bicinchoninic acid) protein assay (Pierce, catalog no. 23225). For all Western blots, 30 ug of total protein was separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes for 1 hour at 350 mA. PDVF membranes were then blocked for 30 min at RT in 5% milk in PBS and incubated with primary antibody overnight at 4°C. Following washes in 1× TBST (Tris-buffered saline with 0.1% Tween-20 detergent), membranes were probed with appropriate secondary HRP-conjugated antibodies (1:10,000 dilution) for 1 hour at RT, washed, and imaged on a Li-COR Odyssey FC imaging system. Fold change in densitometry of Western blot bands was determined by ImageJ analysis.
Enzyme-linked immunosorbent assay
Conditioned media from mEERL (HPV+), MOC7 (HPV−) and Trp53−/−;Pten−/− cells was collected after 48 hours in culture (and a time when cells were approximately 90% confluent) and was analyzed for the presence of SP using an enzyme-linked immunosorbent assay (ELISA) (RayBio Substance P ELISA, catalog no. EIAM-SP) as per manufacturer’s instructions. Recombinant SP was used to establish a standard curve and serves as the positive control.
Animal studies
The Animal Resource Center (ARC) is a specific pathogen–free facility where mice are maintained in IVC Tecniplast Green line Seal Safe Plus cages, and cages are opened only under aseptic conditions in an animal transfer station. Aseptic technique is used to change animal cages every other week. All cages have individual HEPA filtered air and animal rooms are maintained at 75°F, 30 to 70% humidity, have a minimum of 15 air changes per hour, and have a 14:10 light/dark cycle. Corncob bedding and nesting materials, both autoclaved before use, are maintained in all cages. Animals are fed irradiated, sterile food (Envigo) and provided acidified water (pH 2.8 to 3.0) ad libitum. There is a maximum of five mice per cage and they are observed daily (technicians looking for abnormal behavior, signs of illness or distress, the availability of food and water and proper husbandry). Control (wild-type) animals injected with murine tumor cell lines were at least 6 weeks old. Control C57Bl/6 mice were each approximately 20 to 24 g in weight and purchased from The Jackson Laboratory. Double transgenic (TRPV1cre::DTAfl/WT) animals were generated by crossing TRPV1Cre (The Jackson Laboratories, #017769; RRID:IMSR_JAX017769) with Rosa26-DTA mice (The Jackson Laboratory, #009669; RRID: IMSR_JAX:009669). Progeny (TRPV1cre::DTAfl/WT) animals (including littermates) were used at 6 to 10 weeks of age and were approximately 20 to 23 g in weight. n = 2 C57Bl/6 and n = 2 TRPV1cre::DTAfl/WT mice were euthanized at 8 weeks of age and their DRG isolated, formalin-fixed, and IHC stained for TRPV1 to validate absence of these nerves in double transgenic animals. Both TRPV1Cre and Rosa26-DTA mice were back-crossed to C57Bl/6 by the depositing investigators for 10 generations and were again back-crossed by the Jackson Laboratory following deposit. Thus, the most appropriate control for the double transgenic (TRPV1cre::DTAfl/WT) animals are wild-type C57Bl/6 mice.
All animals were randomly assigned to a cage and group. When assessing animals (e.g., measuring tumors), investigators were blinded to the groups. Animals are numbered by ear punch and cage number only. No other identifiers are on the cages to maintain investigators blinded for the duration of the experiment. When measuring tumors, investigators do not have access to the identification key.
Tumors were initiated into age-matched C57Bl/6 or TRPV1cre::DTAfl/WT mice as follows: using a 23-gauge needle, cells (1 × 105) were implanted subcutaneously in the right hindlimb or orthotopically in the oral or peritoneal cavity. A total of n = 10 mice per group were used. Mice were euthanized when sacrifice criteria were met (e.g., tumor volume, emaciation, excessive edema, and ulceration). Following euthanasia, tumors were extracted and either fixed in neutral buffered formalin (for paraffin embedding and immunohistochemistry) or used fresh for microelectrode array analysis (electrophysiological measurement).
Human studies
Samples from the University of Pennsylvania were obtained through Ovarian Cancer Research Center Tumor BioTrust Collection (https://www.med.upenn.edu/OCRCBioTrust/) (RRID: SCR_02287). All samples were acquired from patients using informed consent under the University of Pennsylvania’s IRB Protocol #702679. Ovarian cancer cases used in this study consisted of HGSOC (n = 13 fresh tumors for MEA); fresh benign gynecologic tumors (n = 5) as well as normal ovary (n = 2) served as control tissues. The benign gynecologic tumors consisted of benign mucinous and serous cystadenomas. Head and neck squamous carcinoma cases were obtained from Sanford Health and consisted of n = 4 HPV− and n = 2 HPV+ cases for MEA and n = 15 each of HPV+ and HPV− for IHC staining. All samples were acquired from patients using informed consent under the Sanford Health IRB protocol no. 640. Normal tonsils were acquired from two separate donors using informed consent from the donor before death or from a legal next-of-kin post-mortem following the Uniform Anatomical Gift Act acknowledging use of the tissue for research purposes. These samples were collected by Dakota Lions Sight & Health in Sioux Falls, SD. Consented patients spanned 38 to 83 years of age.
Scoring of innervations
Formalin-fixed paraffin-embedded cases of HPV+ and HPV− HNSCC (n = 15 of each) were cut (5 μm) and IHC stained for β-III tubulin. Positively stained nerves were analyzed by bright-field microscopy and scored as follows: score of 0, no positive staining; score of 1 indicates 10% positive staining; score of 2 indicates 30% positive staining; score of 3 indicates more than 30% positive staining. Five fields per sample were analyzed. Statistical analysis by Student’s t test. The scorer was blinded to the HPV status of the cases.
Microelectrode array
Mouse tumors were quickly dissected and immediately sectioned using a scalpel. Fresh human samples were obtained from the Sanford Health Department of Pathology, Dakota Lions or shipped overnight on ice in Miltenyi Tissue Storage Solution (catalog no. 130-100-008) from the University of Pennsylvania OCRC BioTrust Collection (www.med.upenn.edu/OCRCBioTrust/) (SCR_02287). Tissues were sectioned using a scalpel; n ≥ 4 slices were analyzed at a minimum (larger tumors allowing for a larger number of slices). n = 1 slice was fixed in formalin, paraffin-embedded, and stained by H&E to assess the presence of tumor. To record electrical activity, an MEA1060-Inv-BC MEA system (Multichannel Systems) with a perforated MEA, 60pMEA100/30iR-Ti MEAs (catalog no. 890335, Harvard Apparatus) were used. These MEAs contain 60 electrodes, one of which is a reference electrode. Each electrode is 30 μm in diameter, and the electrodes are spaced 100 μm apart in a 6 × 10 grid (10 electrodes per row arranged in 6 columns generating a rectangle of electrodes onto which tissue slice is placed). The electrodes are made of Titanium nitride and are surrounded by a glass ring that enables maintenance of slices under oxygenated (95% O2, 5% CO2) artificial cerebrospinal fluid (119 mM NaCl, 2.5 mM KCl, 1 mM NaH2PO4, 26.2 mM NaHCO3, 11 mM glucose, 1.3 mM MgSO4, and 1.5 mM CaCl2), a buffer that preserves neuronal functions, at RT for the duration of recordings. MEAs are perforated to enable use of gentle suction, applied by a vacuum pump, to keep the tissue slice in place and in contact with all electrodes throughout the duration of recordings. Therefore, each tissue slice contacted all electrodes and activity from these electrodes was recorded every second for the duration of the recording. Given that one electrode is a reference electrode, recordings from 59 electrodes/s per slice were collected. Electrical activity was recorded at RT, with 25-kHz sampling frequency, using the Butterworth second-order digital filter set to high pass with a cutoff frequency of 10 Hz (to eliminate slow field potentials). For electrical stimulation, a STG4000 stimulus generator (Multichannel Systems) was used. Electrical stimulation (biphasic voltage, −0.5 and +0.5 V each for 100 μs and repeated after a 23-ms interval) was applied to selected electrodes and evoked spike activities recorded. Two types of stimulations were used, and each slice was separately stimulated in these two ways. The first stimulus consisted of a set of 14 electrodes and the second stimulation consisted of a set of 20 electrodes. The same sets of electrodes were stimulated for each slice so that the stimulation parameters were identical for all slices analyzed. When the electrical stimulus is shut off, recordings continued to demonstrate reversion of electrical activity back to baseline. For all slices, the electrical activity/s per electrode is continuously recorded (from 59 electrodes). Electrical activity was recorded and analyzed using the MC_Rack 4.6.2 software from Multichannel Systems.
MEA recordings from a total of 60 malignant HGSOC slices, 18 benign gynecologic slices, 8 slices of normal ovary, 21 slices of HPV+ HNSCC, 32 slices of HPV− HNSCC and 12 slices of normal tonsil were analyzed. For slices that were also exposed to lidocaine treatment, this consisted of first obtaining electrical activity as described. Then, the tumor slices were incubated in oxygenated lidocaine (20 mg/ml; Hospira, NDC 0409-4277-17) at RT for 30 to 40 min after which time they were again analyzed by MEA.
DRG isolation for IHC validation
N = 2 C57Bl/6 and n = 2 TRPV1cre::DTAfl/WT 8 weeks old, approximately 25-g male mice were euthanized by CO2 and cervical dislocation. The fur was sprayed with 70% ethanol and dorsal fur removed to expose the spinal column. Standard scissors were used to remove tissue and cut the ribs leaving only the spinal column intact (head and tail were cut and removed). Laying the ventral side of the spinal column face up, incisions were made on the left and right sides all along the length of the column. Once completed, the ventral half of the spine was lifted off, exposing the spinal marrow which was also removed. This exposed the DRG which were carefully removed from the surrounding tissue and placed into formalin. Following fixation, DRG were paraffin-embedded, cut, and IHC stained as described.
Quantitative PCR
Total RNA was extracted in Qiazol extraction reagent and further isolated through phenol-chloroform protocols. RNA quality and quantity were then determined (NanoDrop). cDNAs were generated by RT reaction using a High-Capacity cDNA Reverse Transcription kit (Applied Biosciences). Gene products were determined by real-time quantitative RT-PCR (CFX-96 BioRad) using listed primers. The mRNA levels were determined using a comparative threshold cycle method with the original expression level of each gene of interest normalized to the expression level of a housekeeping gene (β-actin). There were three biological replicates each with six technical replicates; all showing similar results.
The NK1R primers used were as follows: Full-length NK1R forward primer: TCTTCTTCCTCCTGCCCTACATC; full-length NK1R reverse primer: AGCACCGGAAGGCATGCTTGAAGCCCA; truncated NK1R forward primer: TCTTCTTCCTCCTGCCCTACATC and truncated NK1R reverse primer: TGGAGAGCTCATGGGGTTGGGATCCT.
Statistics
Clustering analysis of grade 1, 2, and 3 HGSOC samples
The clustering was first performed on reference samples (89 primary tumors and 36 ovarian metastases), then on 16 ovarian samples with ambiguous diagnosis were introduced in the dataset and the clustering was performed again. The following samples were also used: n = 3 for grade 1; n = 15 for grade 2; n = 51 for grade 3. Statistical analysis by one-way analysis if variance (ANOVA) with post hoc Dunnet.
GO enrichment analysis
GO analysis was performed in PANTHER (http://pantherdb.org). All genes that showed a link between gene expression and patient survival from OncoLnc (56 genes in ovarian cancer and 79 genes in HNSCC) were input in Panther. The “Reference list” was PANTHER Generic Mapping File, as provided by PANTHER. For both lists, the organism selected was M. musculus. Molecular function, Biological process, and cellular component data were further analyzed to assess similarities between ovarian cancer and HNSCC gene expression pathways.
MEA statistical analysis
The electrical activity for each electrode at each time point for all relevant tissue slices were pooled and averaged and then comparisons made with relevant groups/conditions. Statistical analysis was performed by one-way ANOVA.
Innervation scoring of patient samples
HNSCC patient samples (n = 15 HPV+, n = 15 HPV−) were blindly scored by a board-certified pathologist. Following unblinding, scores were statistically analyzed by Student’s t test.
ELISA for SP
The SP dose curve was used to interpolate the SP concentration in the conditioned media from mEERL (HPV+), MOC7 (HPV−) and Trp53−/−Pten−/− cells. One-way ANOVA with multiple comparisons was used to analyze the data.
Kaplan-Meier survival curves
The expression of neuronal genes and patient survival was assessed using log-rank test for significance.
Study approval
The cases for this study were obtained with patient consent and the study was approved by the Institutional Review Boards at Sanford Research, the University of Pennsylvania and Johns Hopkins University. Samples (normal tonsils) were also obtained by consent from donors through the Dakota Lions Sight and Health Eye and Tissue Donation Bank. Samples from Johns Hopkins were obtained through the Legacy Gift Rapid Autopsy Program (http://pathology.jhu.edu/RapidAutopsy/).
All animal experiments were performed under approved Sanford Research IACUC protocols, within institutional guidelines and comply with all relevant ethical regulations. All in vivo animal studies were performed within the ARC at Sanford Research whose Animal Welfare Assurance is on file with the Office of Laboratory Animal Welfare. The Assurance number is A-4568-01. Sanford Health is also a licensed research facility under the authority of the United States Department of Agriculture (USDA) with USDA certificate number 46-R-011. AAALAC, Intl has also accredited the Sanford Health Animal Research Program.
Acknowledgments
We thank the Histology and Imaging Core (supported by National Institutes of Health grants 5P20GM103548 and P20GM103620), specifically C. Evans, who provided their services and expertise toward this project. We also acknowledge O. C. Fink and S. Faber for help with initial experiments as well as during the revision process.
Funding: This study was supported by National Institutes of Health, National Institute of Dental and Craniofacial Research grant 1 R01 DE032712-01 (P.D.V.); Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health grant 5P20GM103548 (P.D.V.); DaCCoTA Scholar Award supported by the National Institute of General Medical Sciences of the National Institutes of Health grant U54 GM128729 (P.D.V.); University of Pennsylvania Basser Center for BRCA External Grant Program Innovation Award (P.D.V.); Sanford Research (P.D.V.); Canada Research Chair program grant 950-231859 (S.T.); Canadian Institutes of Health Research grants 162211, 461274, and 461275 (S.T.); New Frontiers in Research Fund grant NFRFE-2019-01326 (S.T.); Canadian Foundation for Innovation grant 37439 (S.T.); Natural Sciences and Engineering Research Council of Canada grant RGPIN-2019-06824 (S.T.); Fonds de Recherche du Québec Nature et technologies grant 253380 (S.T.); National Institutes of Health P50 grant CA228991 (R.D.); Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (R.D.); Penn ACC Translational Center of Excellence in Ovarian Cancer (R.D.); Honorable Tina Brozman Foundation for Ovarian Cancer Research (R.D.); Claneil Foundation (R.D.); Helene Ross Bogutz Fund for Ovarian Cancer Early Detection (R.D.); Marjorie S. Stanek and Lowell H. Dubrow Ovarian Cancer Research Center Endowed Fund (R.D.); and NIH SPORE in ovarian cancer P50CA228991 (R.D.).
Author contributions: Conceptualization: P.D.V. Methodology: P.D.V., A.C.R., A.K., and J.B. Investigation: A.C.R., C.T.L., H.R., S.J.V., R.R.F., D.W.V., and J.B. Visualization: P.D.V., A.C.R., A.W., and J.B. Supervision: P.D.V. and S.T. Writing—original draft: P.D.V. Writing—review and editing: P.D.V., A.C.R., H.R., S.T., R.D., R.R.F., D.W.V., and J.B., W.C.S. Formal analysis: A.C.R., P.D.V., R.R.F., T.E., S.T., and J.B. Funding acquisition: P.D.V., R.D., and S.T. Project administration: P.D.V. Resources: R.D., E.J., L.E.S., M.B., W.C.S., W.C.S., D.K.O., D.M.M., J.E.H., and C.S.W. Validation: A.C.R. and R.R.F.
Competing interests: S.T. is a minority stake holder in Nocion Therapeutics and received funding from Nocion Therapeutics. R.D. serves on the advisory board of Repare Therapeutics and VOC Health. D.W.V. has a licensing agreement with Nant for an HPV vaccine. All other authors declare that they have no competing interests.
Data and materials availability: The cell lines generated and used in this study are freely available at Applied Biological Materials (mEERL cells), Kerafest (MOC cells) or by contacting P.D.V. or R.D. (Trp53−/−Pten−/−) directly. All data needed to evaluate the conclusions are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
REFERENCES AND NOTES
- 1.P. D. Vermeer, Exosomal induction of tumor innervation. Cancer Res. 79, 3529–3535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.H. D. Reavis, H. I. Chen, R. Drapkin, Tumor innervation: Cancer has some nerve. Trends Cancer 6, 1059–1067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. M. Gysler, R. Drapkin, Tumor innervation: Peripheral nerves take control of the tumor microenvironment. J. Clin. Invest. 131, e147276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. C. Restaino, P. D. Vermeer, Neural regulations of the tumor microenvironment. FASEB Bioadv. 4, 29–42 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C. Magnon, S. J. Hall, J. Lin, X. Xue, L. Gerber, S. J. Freedland, P. S. Frenette, Autonomic nerve development contributes to prostate cancer progression. Science 341, 1236361 (2013). [DOI] [PubMed] [Google Scholar]
- 6.D. Albo, C. L. Akay, C. L. Marshall, J. A. Wilks, G. Verstovsek, H. Liu, N. Agarwal, D. H. Berger, G. E. Ayala, Neurogenesis in colorectal cancer is a marker of aggressive tumor behavior and poor outcomes. Cancer 117, 4834–4845 (2011). [DOI] [PubMed] [Google Scholar]
- 7.L. Zhang, L. L. Wu, H. B. Huan, X. J. Chen, X. D. Wen, D. P. Yang, F. Xia, Sympathetic and parasympathetic innervation in hepatocellular carcinoma. Neoplasma 64, 840–846 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Y. Hayakawa, K. Sakitani, M. Konishi, S. Asfaha, R. Niikura, H. Tomita, B. W. Renz, Y. Tailor, M. Macchini, M. Middelhoff, Z. Jiang, T. Tanaka, Z. A. Dubeykovskaya, W. Kim, X. Chen, A. M. Urbanska, K. Nagar, C. B. Westphalen, M. Quante, C. S. Lin, M. D. Gershon, A. Hara, C. M. Zhao, D. Chen, D. L. Worthley, K. Koike, T. C. Wang, Nerve growth factor promotes gastric tumorigenesis through aberrant cholinergic signaling. Cancer Cell 31, 21–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Madeo, P. L. Colbert, D. W. Vermeer, C. T. Lucido, J. T. Cain, E. G. Vichaya, A. J. Grossberg, D. R. Muirhead, A. P. Rickel, Z. Hong, J. Zhao, J. M. Weimer, W. C. Spanos, J. H. Lee, R. Dantzer, P. D. Vermeer, Cancer exosomes induce tumor innervation. Nat. Commun. 9, 4284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. T. Lucido, E. Wynja, M. Madeo, C. S. Williamson, L. E. Schwartz, B. A. Imblum, R. Drapkin, P. D. Vermeer, Innervation of cervical carcinoma is mediated by cancer-derived exosomes. Gynecol. Oncol. 154, 228–235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.M. Amit, H. Takahashi, M. P. Dragomir, A. Lindemann, F. O. Gleber-Netto, C. R. Pickering, S. Anfossi, A. A. Osman, Y. Cai, R. Wang, E. Knutsen, M. Shimizu, C. Ivan, X. Rao, J. Wang, D. A. Silverman, S. Tam, M. Zhao, C. Caulin, A. Zinger, E. Tasciotti, P. M. Dougherty, A. el-Naggar, G. A. Calin, J. N. Myers, Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 578, 449–454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Y. Ye, D. Dang, J. Zhang, C. T. Viet, D. K. Lam, J. C. Dolan, J. L. Gibbs, B. L. Schmidt, Nerve growth factor links oral cancer progression, pain, and cachexia. Mol. Cancer Ther. 10, 1667–1676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.E. Vanhecke, E. Adriaenssens, S. Verbeke, S. Meignan, E. Germain, N. Berteaux, V. Nurcombe, X. le Bourhis, H. Hondermarck, Brain-derived neurotrophic factor and neurotrophin-4/5 are expressed in breast cancer and can be targeted to inhibit tumor cell survival. Clin. Cancer Res. 17, 1741–1752 (2011). [DOI] [PubMed] [Google Scholar]
- 14.A. A. Geldof, E. P. Van Haarst, D. W. Newling, Neurotrophic factors in prostate and prostatic cancer. Prostate Cancer Prostatic Dis. 1, 236–241 (1998). [DOI] [PubMed] [Google Scholar]
- 15.A. Chedotal, G. Kerjan, C. Moreau-Fauvarque, The brain within the tumor: New roles for axon guidance molecules in cancers. Cell Death Differ. 12, 1044–1056 (2005). [DOI] [PubMed] [Google Scholar]
- 16.J. K. Allen, G. N. Armaiz-Pena, A. S. Nagaraja, N. C. Sadaoui, T. Ortiz, R. Dood, M. Ozcan, D. M. Herder, M. Haemmerle, K. M. Gharpure, R. Rupaimoole, R. A. Previs, S. Y. Wu, S. Pradeep, X. Xu, H. D. Han, B. Zand, H. J. Dalton, M. Taylor, W. Hu, J. Bottsford-Miller, M. Moreno-Smith, Y. Kang, L. S. Mangala, C. Rodriguez-Aguayo, V. Sehgal, E. L. Spaeth, P. T. Ram, S. T. C. Wong, F. C. Marini, G. Lopez-Berestein, S. W. Cole, S. K. Lutgendorf, M. de Biasi, A. K. Sood, Sustained adrenergic signaling promotes intratumoral innervation through BDNF induction. Cancer Res. 78, 3233–3242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Ortega, A new role for GABA: Inhibition of tumor cell migration. Trends Pharmacol. Sci. 24, 151–154 (2003). [DOI] [PubMed] [Google Scholar]
- 18.K. Masur, B. Niggemann, K. S. Zanker, F. Entschladen, Norepinephrine-induced migration of SW 480 colon carcinoma cells is inhibited by beta-blockers. Cancer Res. 61, 2866–2869 (2001). [PubMed] [Google Scholar]
- 19.J. Mu, W. Huang, Z. Tan, M. Li, L. Zhang, Q. Ding, X. Wu, J. Lu, Y. Liu, Q. Dong, H. Xu, Dopamine receptor D2 is correlated with gastric cancer prognosis. Oncol. Lett. 13, 1223–1227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Kamiya, Y. Hayama, S. Kato, A. Shimomura, T. Shimomura, K. Irie, R. Kaneko, Y. Yanagawa, K. Kobayashi, T. Ochiya, Genetic manipulation of autonomic nerve fiber innervation and activity and its effect on breast cancer progression. Nat. Neurosci. 22, 1289–1305 (2019). [DOI] [PubMed] [Google Scholar]
- 21.P. Mauffrey, N. Tchitchek, V. Barroca, A. P. Bemelmans, V. Firlej, Y. Allory, P. H. Roméo, C. Magnon, Progenitors from the central nervous system drive neurogenesis in cancer. Nature 569, 672–678 (2019). [DOI] [PubMed] [Google Scholar]
- 22.G. A. McCallum, J. Shiralkar, D. Suciu, G. Covarrubias, J. S. Yu, E. Karathanasis, D. M. Durand, Chronic neural activity recorded within breast tumors. Sci. Rep. 10, 14824 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L. Dal Maso, N. Torelli, E. Biancotto, M. di Maso, A. Gini, G. Franchin, F. Levi, C. la Vecchia, D. Serraino, J. Polesel, Combined effect of tobacco smoking and alcohol drinking in the risk of head and neck cancers: A re-analysis of case-control studies using bi-dimensional spline models. Eur. J. Epidemiol. 31, 385–393 (2016). [DOI] [PubMed] [Google Scholar]
- 24.N. J. Farsi, M. C. Rousseau, N. Schlecht, G. Castonguay, P. Allison, P. F. Nguyen-Tan, D. Souliéres, F. Coutlée, M. Hier, S. Madathil, E. L. Franco, B. Nicolau, Aetiological heterogeneity of head and neck squamous cell carcinomas: The role of human papillomavirus infections, smoking and alcohol. Carcinogenesis 38, 1188–1195 (2017). [DOI] [PubMed] [Google Scholar]
- 25.M. A. Lisio, L. Fu, A. Goyeneche, Z. H. Gao, C. Telleria, High-grade serous ovarian cancer: Basic sciences, clinical and therapeutic standpoints. Int. J. Mol. Sci. 20, 952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.P. T. Kroeger Jr., R. Drapkin, Pathogenesis and heterogeneity of ovarian cancer. Curr. Opin. Obstet. Gynecol. 29, 26–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.U. A. Matulonis, A. K. Sood, L. Fallowfield, B. E. Howitt, J. Sehouli, B. Y. Karlan, Ovarian cancer. Nat. Rev. Dis. Primers 2, 16061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. L. Barr, A. Kruse, A. C. Restaino, N. Tulina, S. Stuckelberger, S. J. Vermeer, C. S. Williamson, D. W. Vermeer, M. Madeo, J. Stamp, M. Bell, M. Morgan, J.-Y. Yoon, M. A. Mitchell, A. Budina, D. K. Omran, L. E. Schwartz, R. Drapkin, P. D. Vermeer, Intra-tumoral nerve-tracing in a novel syngeneic model of high-grade serous ovarian carcinoma. Cell 10, 3491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.B. W., β2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell 33, 75–90.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Balood, M. Ahmadi, T. Eichwald, A. Ahmadi, A. Majdoubi, K. Roversi, K. Roversi, C. T. Lucido, A. C. Restaino, S. Huang, L. Ji, K. C. Huang, E. Semerena, S. C. Thomas, A. E. Trevino, H. Merrison, A. Parrin, B. Doyle, D. W. Vermeer, W. C. Spanos, C. S. Williamson, C. R. Seehus, S. L. Foster, H. Dai, C. J. Shu, M. Rangachari, J. Thibodeau, S. V. del Rincon, R. Drapkin, M. Rafei, N. Ghasemlou, P. D. Vermeer, C. J. Woolf, S. Talbot, Nociceptor neurons affect cancer immunosurveillance. Nature 611, 405–412 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.L. A. McIlvried, M. A. Atherton, N. L. Horan, T. N. Goch, N. N. Scheff, Sensory neurotransmitter calcitonin gene-related peptide modulates tumor growth and lymphocyte infiltration in oral squamous cell carcinoma. Adv. Biol. 6, 2200019 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.N., Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.N. Cancer Genome Atlas , Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. R. Laury, R. Perets, H. Piao, J. F. Krane, J. A. Barletta, C. French, L. R. Chirieac, R. Lis, M. Loda, J. L. Hornick, R. Drapkin, M. S. Hirsch, A comprehensive analysis of PAX8 expression in human epithelial tumors. Am. J. Surg. Pathol. 35, 816–826 (2011). [DOI] [PubMed] [Google Scholar]
- 35.H. W. Cheung, G. S. Cowley, B. A. Weir, J. S. Boehm, S. Rusin, J. A. Scott, A. East, L. D. Ali, P. H. Lizotte, T. C. Wong, G. Jiang, J. Hsiao, C. H. Mermel, G. Getz, J. Barretina, S. Gopal, P. Tamayo, J. Gould, A. Tsherniak, N. Stransky, B. Luo, Y. Ren, R. Drapkin, S. N. Bhatia, J. P. Mesirov, L. A. Garraway, M. Meyerson, E. S. Lander, D. E. Root, W. C. Hahn, Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 12372–12377 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.V. Venkataramani, D. I. Tanev, C. Strahle, A. Studier-Fischer, L. Fankhauser, T. Kessler, C. Körber, M. Kardorff, M. Ratliff, R. Xie, H. Horstmann, M. Messer, S. P. Paik, J. Knabbe, F. Sahm, F. T. Kurz, A. A. Acikgöz, F. Herrmannsdörfer, A. Agarwal, D. E. Bergles, A. Chalmers, H. Miletic, S. Turcan, C. Mawrin, D. Hänggi, H. K. Liu, W. Wick, F. Winkler, T. Kuner, Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573, 532–538 (2019). [DOI] [PubMed] [Google Scholar]
- 37.H. S. Venkatesh, W. Morishita, A. C. Geraghty, D. Silverbush, S. M. Gillespie, M. Arzt, L. T. Tam, C. Espenel, A. Ponnuswami, L. Ni, P. J. Woo, K. R. Taylor, A. Agarwal, A. Regev, D. Brang, H. Vogel, S. Hervey-Jumper, D. E. Bergles, M. L. Suvà, R. C. Malenka, M. Monje, Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O. Soderberg, M. Gullberg, M. Jarvius, K. Ridderstråle, K.-J. Leuchowius, J. Jarvius, K. Wester, P. Hydbring, F. Bahram, L.-G. Larsson, U. Landegren, Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 39.K. Dore, Y. Pao, J. Soria Lopez, S. Aronson, H. Zhan, S. Ghosh, S. Merrill, A. M. Zador, R. Malinow, J. M. Kebschull, SYNPLA, a method to identify synapses displaying plasticity after learning. Proc. Natl. Acad. Sci. U.S.A. 117, 3214–3219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. A. Brown, J. D. Ross, R. A. Blum, Y. Nam, B. C. Wheeler, S. P. DeWeerth, Stimulus-artifact elimination in a multi-electrode system. IEEE Trans. Biomed. Circuits Syst. 2, 10–21 (2008). [DOI] [PubMed] [Google Scholar]
- 41.A. C. Hoover, W. C. Spanos, G. F. Harris, M. E. Anderson, A. J. Klingelhutz, J. H. Lee, The role of human papillomavirus 16 E6 in anchorage-independent and invasive growth of mouse tonsil epithelium. Arch. Otolaryngol. Head Neck Surg. 133, 495–502 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D. W. Vermeer, W. C. Spanos, P. D. Vermeer, A. M. Bruns, K. M. Lee, J. H. Lee, Radiation-induced loss of cell surface CD47 enhances immune-mediated clearance of human papillomavirus-positive cancer. Int. J. Cancer 133, 120–129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.W. C. Spanos, P. Nowicki, D. W. Lee, A. Hoover, B. Hostager, A. Gupta, M. E. Anderson, J. H. Lee, Immune response during therapy with cisplatin or radiation for human papillomavirus-related head and neck cancer. Arch. Otolaryngol. Head Neck Surg. 135, 1137–1146 (2009). [DOI] [PubMed] [Google Scholar]
- 44.H. Cash, S. Shah, E. Moore, A. Caruso, R. Uppaluri, C. van Waes, C. Allen, mTOR and MEK1/2 inhibition differentially modulate tumor growth and the immune microenvironment in syngeneic models of oral cavity cancer. Oncotarget 6, 36400–36417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.L. F. Borges, R. L. Sidman, Axonal transport of lectins in the peripheral nervous system. J. Neurosci. 2, 647–653 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Dumas, M. E. Schwab, H. Thoenen, Retrograde axonal transport of specific macromolecules as a tool for characterizing nerve terminal membranes. J. Neurobiol. 10, 179–197 (1979). [DOI] [PubMed] [Google Scholar]
- 47.R. H. Fabian, J. D. Coulter, Transneuronal transport of lectins. Brain Res. 344, 41–48 (1985). [DOI] [PubMed] [Google Scholar]
- 48.S. K. Mishra, S. M. Tisel, P. Orestes, S. K. Bhangoo, M. A. Hoon, TRPV1-lineage neurons are required for thermal sensation. EMBO J. 30, 582–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Rosso, M. J. Robles-Frias, R. Covenas, M. V. Salinas-Martin, M. Munoz, The NK-1 receptor is expressed in human primary gastric and colon adenocarcinomas and is involved in the antitumor action of L-733,060 and the mitogenic action of substance P on human gastrointestinal cancer cell lines. Tumour Biol. 29, 245–254 (2008). [DOI] [PubMed] [Google Scholar]
- 50.X. Li, G. Ma, Q. Ma, W. Li, J. Liu, L. Han, W. Duan, Q. Xu, H. Liu, Z. Wang, Q. Sun, F. Wang, E. Wu, Neurotransmitter substance P mediates pancreatic cancer perineural invasion via NK-1R in cancer cells. Mol. Cancer Res. 11, 294–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.T. Goto, H. Iwai, E. Kuramoto, A. Yamanaka, Neuropeptides and ATP signaling in the trigeminal ganglion. Jpn Dent. Sci. Rev. 53, 117–124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. Garcia-Recio, P. Gascon, Biological and pharmacological aspects of the NK1-receptor. Biomed. Res. Int. 2015, 495704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.T. M. Fong, S. A. Anderson, H. Yu, R. R. Huang, C. D. Strader, Differential activation of intracellular effector by two isoforms of human neurokinin-1 receptor. Mol. Pharmacol. 41, 24–30 (1992). [PubMed] [Google Scholar]
- 54.J. P. Lai, A. Cnaan, H. Zhao, S. D. Douglas, Detection of full-length and truncated neurokinin-1 receptor mRNA expression in human brain regions. J. Neurosci. Methods 168, 127–133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. P. Lai, S. Lai, F. Tuluc, M. F. Tansky, L. E. Kilpatrick, S. E. Leeman, S. D. Douglas, Differences in the length of the carboxyl terminus mediate functional properties of neurokinin-1 receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 12605–12610 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Spitsin, V. Pappa, S. D. Douglas, Truncation of neurokinin-1 receptor-negative regulation of substance P signaling. J. Leukoc. Biol. 103, 1043–1051 (2018). [DOI] [PubMed] [Google Scholar]
- 57.G. A. Dissen, C. Romero, A. N. Hirshfield, S. R. Ojeda, Nerve growth factor is required for early follicular development in the mammalian ovary. Endocrinology 142, 2078–2086 (2001). [DOI] [PubMed] [Google Scholar]
- 58.S. R. Ojeda, C. Romero, V. Tapia, G. A. Dissen, Neurotrophic and cell-cell dependent control of early follicular development. Mol. Cell. Endocrinol. 163, 67–71 (2000). [DOI] [PubMed] [Google Scholar]
- 59.G. A. Dissen, J. A. Parrott, M. K. Skinner, D. F. Hill, M. E. Costa, S. R. Ojeda, Direct effects of nerve growth factor on thecal cells from antral ovarian follicles. Endocrinology 141, 4736–4750 (2000). [DOI] [PubMed] [Google Scholar]
- 60.V. B. Wookey, A. K. Appiah, A. Kallam, V. Ernani, L. M. Smith, A. K. Ganti, HPV status and survival in non-oropharyngeal squamous cell carcinoma of the head and neck. Anticancer Res 39, 1907–1914 (2019). [DOI] [PubMed] [Google Scholar]
- 61.S. Sethi, R. Ali-Fehmi, S. Franceschi, L. Struijk, L. J. van Doorn, W. Quint, B. Albashiti, M. Ibrahim, I. Kato, Characteristics and survival of head and neck cancer by HPV status: a cancer registry-based study. Int. J. Cancer 131, 1179–1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.K. J. Friston, Functional and effective connectivity: A review. Brain Connect. 1, 13–36 (2011). [DOI] [PubMed] [Google Scholar]
- 63.D. Poli, V. P. Pastore, P. Massobrio, Functional connectivity in in vitro neuronal assemblies. Front Neural Circuits 9, 57 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.P. Bonifazi, P. Massobrio, Reconstruction of functional connectivity from multielectrode recordings and calcium imaging. Adv. Neurobiol. 22, 207–231 (2019). [DOI] [PubMed] [Google Scholar]
- 65.S. Ludwig, P. Sharma, P. Wise, R. Sposto, D. Hollingshead, J. Lamb, S. Lang, M. Fabbri, T. L. Whiteside, mRNA and miRNA profiles of exosomes from cultured tumor cells reveal biomarkers specific for HPV16-positive and HPV16-negative head and neck cancer. Int. J. Mol. Sci. 21, 8570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Guo, H. Pan, Long noncoding RNA LINC01125 enhances cisplatin sensitivity of ovarian cancer via miR-1972. Med. Sci. Monit. 25, 9844–9854 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Y. Feng, W. Hang, Z. Sang, S. Li, W. Xu, Y. Miao, X. Xi, Q. Huang, Identification of exosomal and non-exosomal microRNAs associated with the drug resistance of ovarian cancer. Mol. Med. Rep. 19, 3376–3392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.X. Chen, L. Zhang, J. Geng, Z. Chen, X. Cui, MiR-205-5p functions as a tumor suppressor in gastric cancer cells through downregulating FAM84B. J. Oncol. 2022, 8267891 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Q. Sun, X. Zhang, Z. Tan, H. Gu, S. Ding, Y. Ji, Bone marrow mesenchymal stem cells-secreted exosomal microRNA-205-5p exerts inhibitory effect on the progression of liver cancer through regulating CDKL3. Pathol. Res. Pract. 225, 153549 (2021). [DOI] [PubMed] [Google Scholar]
- 70.S. Chen, Y. Wang, Y. Su, L. Zhang, M. Zhang, X. Li, J. Wang, X. Zhang, miR-205-5p/PTK7 axis is involved in the proliferation, migration and invasion of colorectal cancer cells. Mol. Med. Rep. 17, 6253–6260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.J. Zhang, J. Zhang, X. Pang, Z. Chen, Z. Zhang, L. Lei, H. Xu, L. Wen, J. Zhu, Y. Jiang, Y. Cui, G. Chen, X. Wang, MiR-205-5p suppresses angiogenesis in gastric cancer by downregulating the expression of VEGFA and FGF1. Exp. Cell Res. 404, 112579 (2021). [DOI] [PubMed] [Google Scholar]
- 72.D. Gulei, L. Magdo, A. Jurj, L. Raduly, R. Cojocneanu-Petric, A. Moldovan, C. Moldovan, A. Florea, S. Pasca, L. A. Pop, V. Moisoiu, L. Budisan, C. Pop-Bica, C. Ciocan, R. Buiga, M. S. Muresan, R. Stiufiuc, C. Ionescu, I. Berindan-Neagoe, The silent healer: miR-205-5p up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell Death Dis. 9, 66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.H. N. Li, H. M. Zhang, X. R. Li, J. Wang, T. Xu, S. Y. Li, M. L. Dong, G. Wang, X. Q. Cui, X. Yang, Y. L. Wu, X. H. Liao, Y. Y. Du, MiR-205-5p/GGCT attenuates growth and metastasis of papillary thyroid cancer by regulating CD44. Endocrinology 163, bqac022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D. Roosterman, G. S. Cottrell, F. Schmidlin, M. Steinhoff, N. W. Bunnett, Recycling and resensitization of the neurokinin 1 receptor. Influence of agonist concentration and Rab GTPases. J. Biol. Chem. 279, 30670–30679 (2004). [DOI] [PubMed] [Google Scholar]
- 75.J. R. Schank, M. Heilig, Substance P and the neurokinin-1 receptor: The new CRF. Int. Rev. Neurobiol. 136, 151–175 (2017). [DOI] [PubMed] [Google Scholar]
- 76.M. A. Gonzalez-Moles, P. Ramos-Garcia, F. Esteban, Significance of the overexpression of substance P and its receptor NK-1R in head and neck carcinogenesis: A systematic review and meta-analysis. Cancer 13, 1349 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.S. Ebrahimi, F. Mirzavi, S. H. Aghaee-Bakhtiari, S. I. Hashemy, SP/NK1R system regulates carcinogenesis in prostate cancer: Shedding light on the antitumoral function of aprepitant. Biochim. Biophys. Acta. Mol. Cell. Res. 1869, 119221 (2022). [DOI] [PubMed] [Google Scholar]
- 78.S. C. Goswami, S. K. Mishra, D. Maric, K. Kaszas, G. L. Gonnella, S. J. Clokie, H. D. Kominsky, J. R. Gross, J. M. Keller, A. J. Mannes, M. A. Hoon, M. J. Iadarola, Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J. Pain 15, 1338–1359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.A. M. Binshtok, B. P. Bean, C. J. Woolf, Inhibition of nociceptors by TRPV1-mediated entry of impermeant sodium channel blockers. Nature 449, 607–610 (2007). [DOI] [PubMed] [Google Scholar]
- 80.F. A. Pinho-Ribeiro, B. Baddal, R. Haarsma, M. O’Seaghdha, N. J. Yang, K. J. Blake, M. Portley, W. A. Verri, J. B. Dale, M. R. Wessels, I. M. Chiu, Blocking neuronal signaling to immune cells treats streptococcal invasive infection. Cell 173, 1083–1097.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.J. P. Meyniel, P. H. Cottu, C. Decraene, M. H. Stern, J. Couturier, I. Lebigot, A. Nicolas, N. Weber, V. Fourchotte, S. Alran, A. Rapinat, D. Gentien, S. Roman-Roman, L. Mignot, X. Sastre-Garau, A genomic and transcriptomic approach for a differential diagnosis between primary and secondary ovarian carcinomas in patients with a previous history of breast cancer. BMC Cancer 10, 222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.J. Anaya, OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comp. Sci. 2, e67 (2016). [Google Scholar]
- 83.V. Chalivendra, K. L. Kanchi, M. D. Onken, A. E. Winkler, E. Mardis, R. Uppaluri, Genomic analysis to define molecular basis of aggressiveness in a mouse model of oral cancer. Genom. Data 3, 61–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2