Abstract
Aims:
T cells play pathophysiologic roles in kidney ischemia-reperfusion injury (IRI), and the nuclear factor erythroid 2-related factor 2/kelch-like ECH-associated protein 1 (Nrf2/Keap1) pathway regulates T cell responses. We hypothesized that clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated Keap1-knockout (KO) augments Nrf2 antioxidant potential of CD4+ T cells, and that Keap1-KO CD4+ T cell immunotherapy protects from kidney IRI.
Results:
CD4+ T cell Keap1-KO resulted in significant increase of Nrf2 target genes NAD(P)H quinone dehydrogenase 1, heme oxygenase 1, glutamate-cysteine ligase catalytic subunit, and glutamate-cysteine ligase modifier subunit. Keap1-KO cells displayed no signs of exhaustion, and had significantly lower levels of interleukin 2 (IL2) and IL6 in normoxic conditions, but increased interferon gamma in hypoxic conditions in vitro. In vivo, adoptive transfer of Keap1-KO CD4+ T cells before IRI improved kidney function in T cell-deficient nu/nu mice compared with mice receiving unedited control CD4+ T cells. Keap1-KO CD4+ T cells isolated from recipient kidneys 24 h post IR were less activated compared with unedited CD4+ T cells, isolated from control kidneys.
Innovation:
Editing Nrf2/Keap1 pathway in murine T cells using CRISPR/Cas9 is an innovative and promising immunotherapy approach for kidney IRI and possibly other solid organ IRI.
Conclusion:
CRISPR/Cas9-mediated Keap1-KO increased Nrf2-regulated antioxidant gene expression in murine CD4+ T cells, modified responses to in vitro hypoxia and in vivo kidney IRI. Gene editing targeting the Nrf2/Keap1 pathway in T cells is a promising approach for immune-mediated kidney diseases.
Keywords: acute kidney injury, T cells, CRISPR/Cas9, Nrf2/Keap1 pathway, ischemia-reperfusion injury, cell-based therapy
Introduction
Acute kidney injury (AKI) is an important worldwide health care problem associated with increased mortality and morbidity (Ishani et al., 2009; James et al., 2011; Pechman et al., 2009). A major cause of AKI is ischemia-reperfusion injury (IRI) resulting from a temporary reduction of kidney blood flow. To date, there is no specific therapy available for AKI.
The role of immune cells in the pathogenesis of early injury during kidney IRI is well established and increasingly linked to repair processes in kidney (Anders et al., 2021; Battistone et al., 2020; D'Alessio et al., 2019; Dong et al., 2007; Jang and Rabb, 2015; Kinsey and Okusa, 2014). CD4+ T cells are important mediators as CD4+ T cell-deficient mice are protected during kidney IRI, and adoptive transfer of CD4+ T cells worsened postischemic renal injury in CD4+ T cell-deficient mice (Burne et al., 2001; Rabb et al., 2000).
Among CD4+ T cells, pro- and anti-inflammatory subgroups exist. AKI is promoted by T helper (Th)1 cells through tumor necrosis factor alpha (TNFα) and interferon gamma (IFNγ), as well as by Th17 through interleukin (IL)17A. The course of experimental kidney IRI can be improved by Th2 and regulatory T (Treg) cells (Dellepiane et al., 2020; Gandolfo et al., 2009; Gharaie Fathabad et al., 2020; Kinsey et al., 2009; Lee et al., 2019; Marques et al., 2006; Mehrotra et al., 2019; Mehrotra et al., 2017; Yokota et al., 2003). Thus, T cell-based therapies have potential for AKI.
The nuclear factor erythroid 2-related factor 2/kelch-like ECH-associated protein 1 (Nrf2/Keap1) pathway is a promising therapeutic target for kidney diseases (Stenvinkel et al., 2021). Nrf2 is an important transcription factor, which controls multiple antioxidant genes such as NAD(P)H quinone dehydrogenase 1 (Nqo1), heme oxygenase 1 (Hmox1), glutamate-cysteine ligase catalytic subunit (Gclc), and glutamate-cysteine ligase modifier subunit (Gclm).
In steady state, Keap1 regulates the cytoplasmic levels of Nrf2 by targeting it for proteasomal degradation. However, under oxidative stress such as during kidney IRI, the association between Keap1 and Nrf2 is lost, which leads to increased nuclear translocation of Nrf2 and upregulation of numerous antioxidant genes. The role of Nrf2 in kidney diseases is being actively investigated in various disease models, such as autosomal-dominant polycystic kidney disease, IgA nephropathy, and diabetic kidney disease.
There have been several clinical trials assessing pharmacological Nrf2 activators; for example, 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid methyl ester (CDDO-Me, bardoxolone methyl) (Kong et al., 2017; Lu et al., 2020; Stenvinkel et al., 2021; Yang et al., 2013). However, long-term treatment with CDDO-Me has been reported to be associated with cardiac adverse events and a development of albuminuria, which led to the termination of phase 3 clinical trial (de Zeeuw et al., 2013). Albuminuria has also been found in murine models after Nrf2 upregulation (Rush et al., 2021).
The Nrf2 pathway was initially shown to be activated in kidney IRI using microarray technology (Leonard et al., 2006). Subsequent studies showed that Nrf2-deficient mice had a worse course of ischemic and nephrotoxic AKI, and Nrf2 pharmacologic upregulation improved the course of kidney IRI (Liu et al., 2014; Liu et al., 2009). However, mice with deletion of Keap1 in renal tubular epithelial cells developed hydronephrosis due to reduced aquaporin 2 levels (Nezu et al., 2017a; Suzuki et al., 2017). Keap1 hypomorphic mice are protected from AKI in different models (Tan et al., 2016).
Considering the important role of T cells in kidney IRI, T cell-specific deletion of Keap1 is a potential therapeutic target. A previous study demonstrated that T cell-specific Keap1 deletion in mice (CD4-Keap1-knockout [KO]) led to structural and functional protection against kidney IRI. In addition, adoptive transfer of T cells from CD4-Keap1-KO mice with increased Nrf2 activity protected wild-type (WT) mice from kidney IRI (Noel et al., 2015).
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-based gene editing is a powerful novel tool that allows targeted modifications in genetic material to disrupt or enhance the activity of the target gene ex vivo (Doudna, 2020).
Recently, KEAP1-KO in human T cells using CRISPR/Cas9 led to successful upregulation of Nrf2 target genes; however, its therapeutic potential was not tested in kidney IRI or other kidney disease models (Noel et al., 2018). In this study, we demonstrate that ex vivo Keap1-KO using CRISPR/Cas9 in murine CD4+ T cells upregulated Nrf2-dependent antioxidant target genes, and adoptive transfer of Keap1-KO CD4+ T cells provided functional protection from kidney IRI.
Results
Keap1-KO using CRISPR/Cas9 augmented Nrf2 target gene expression
To test the efficiency of Keap1-KO on Nrf2 target gene expression in murine CD4+ T cells, three different single-guide RNAs (sgRNAs) were used. These sgRNAs targeted different sites of Keap1 gene, exon 2 (Table 1), and were tested individually (sgRNA1, sgRNA2, or sgRNA3) or in combination (sgRNA1 + 2 + 3). Primary mouse CD4+ T cells were cultured for 48 h before the ribonucleoprotein (RNP) complex, consisting of sgRNA and Cas9, was delivered. Cells were harvested 24 h after electroporation, and effect of Keap1-KO was assessed by quantifying Nrf2 target gene expression (Fig. 1).
Table 1.
Single-guide RNA Sequences Used to Target Keap1, Exon 2
| Keap1 target | Sequence |
|---|---|
| sgRNA1 | 5′-CGCGCGCTGCAAGGACTACC-3′ |
| sgRNA2 | 5′-TGACCCAGTCGATGCACGCG-3′ |
| sgRNA3 | 5′-ATCATCCCGGCTGATGAGCG-3′ |
Keap1, kelch-like ECH-associated protein 1; sgRNA, single-guide RNA.
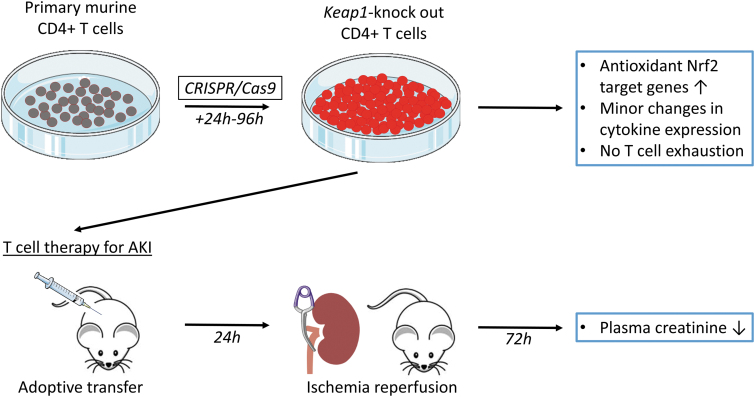
FIG. 1.
Graphic summary illustration of in vitro Nrf2/Keap1 editing use of CD4+ T cells and their impact on in vivo ischemia-reperfusion–induced AKI in a mouse model. Primary murine CD4+ T cells were isolated, and CRISPR/Cas9 technology was used for knocking out of Keap1 gene leading to upregulation of antioxidant Nrf2 target genes. After expansion of cells, adoptive transfer was performed in T cell-deficient nu/nu mice. Amelioration of kidney function after renal ischemia-reperfusion injury was auspicious, and Keap1-KO T cell therapy might be applicable for different kidney diseases. AKI, acute kidney injury; Cas9, CRISPR-associated protein 9; CRISPR, clustered regularly interspaced short palindromic repeats; Keap1, kelch-like ECH-associated protein 1; KO, knockout; Nrf2, nuclear factor erythroid 2-related factor 2.
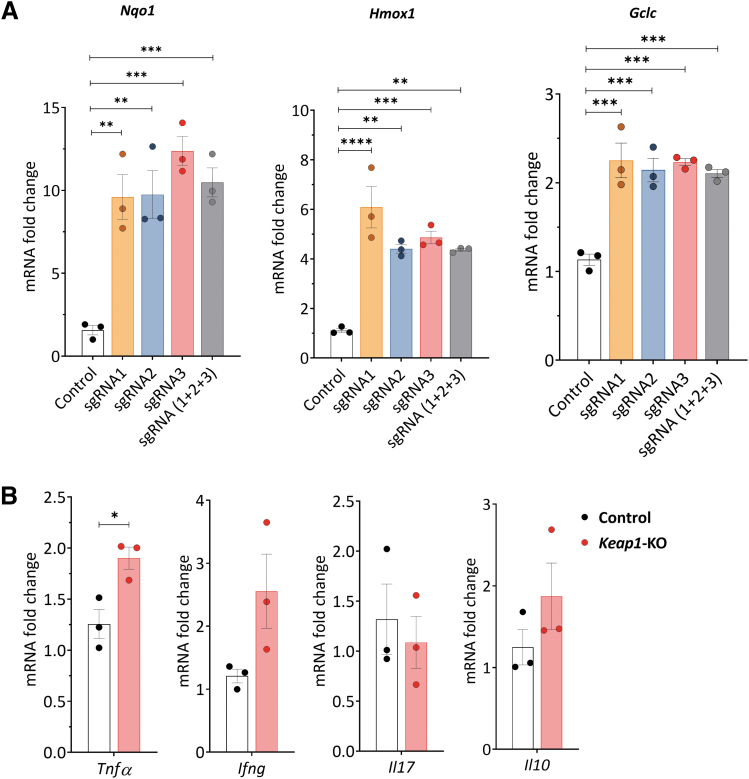
Quantitative real-time polymerase chain reaction (qRT-PCR) analyses revealed a significant increase in Nqo1 (>9.5-fold for all tested sgRNAs, p < 0.001), Hmox1 (>4.3-fold for all tested sgRNAs, p < 0.01), and Gclc (>2.1-fold for all tested sgRNAs, p < 0.0001) 24 h after electroporation for all tested sgRNAs both individually and in combination (Fig. 2A). We chose sgRNA3 for subsequent experiments, based on the strongest increase in Nqo1 mRNA expression and significant upregulation of Hmox1 and Gclc mRNA expression.
FIG. 2.
CRISPR/Cas9-induced Keap1-gene-KO increased Nrf2-regulated antioxidant and cytokine gene expression in cultured murine CD4+ T cells 24 h after electroporation. (A) Nrf2 target gene mRNA levels. We tested three different sgRNAs separately and in combination for Keap1-KO. All tested sgRNAs and their combination significantly increased Nrf2-regulated antioxidant mRNA expression in murine CD4+ T cells compared with control cells. (B) Tnfa showed significant changes using sgRNA3, whereas there were no changes in Ifng, Il17, and Il10 in Keap1-KO CD4+ T cells vs. control; n = 3 in each group. Statistical analyses were performed by one-way ANOVA with post hoc analysis using Tukey's honest significant difference test (A) and paired, two-tailed T-test (B). *p < 0.05, **p < 0.001, ***p < 0.001, ****p < 0.0001. Ifng, interferon gamma; Il10, interleukin 10; Il17, interleukin 17; sgRNA, single-guide RNA; Tnfa, tumor necrosis factor alpha.
We further assessed mRNA expression of pro- and anti-inflammatory cytokines, 24 h after RNP electroporation, to detect potential immunological changes caused by Keap1-KO. We observed a significant upregulation of Tnfa in Keap1-KO CD4+ T cells compared with control cells (1.9 ± 0.1-fold, p < 0.05). There was no difference in Ifng, Il17, and Il10 mRNA expression between Keap1-KO and control CD4+ T cells (Fig. 2B).
Keap1-KO resulted in sustained antioxidant gene expression
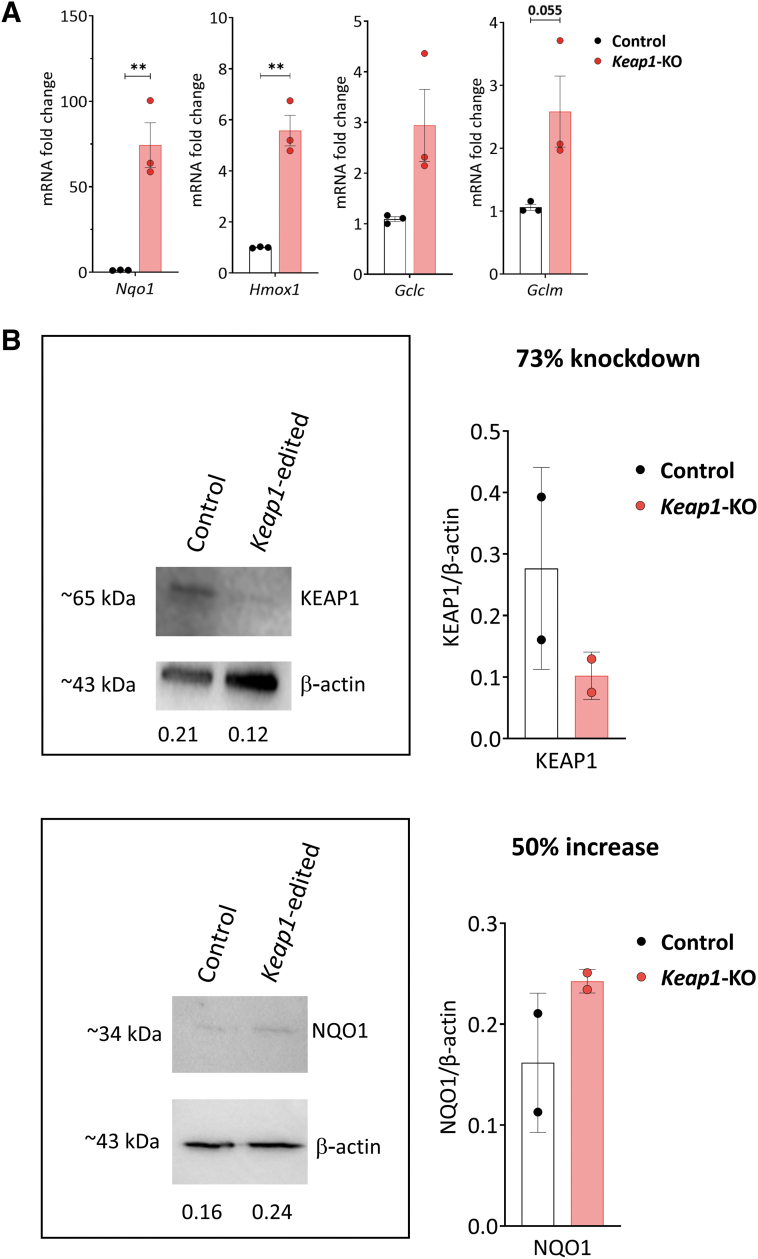
Since the development of immunotherapy requires extended culture time to produce sufficient number of cells for adoptive transfer, we assessed the effect of prolonged culture time on Nrf2 target gene expression in Keap1-KO CD4+ T cells. CD4+ T cells belonging to control and Keap1-KO groups were cultured up to 96 h after electroporation, and antioxidant gene expression assessed using qRT-PCR. We observed robust and sustained upregulation of Nrf2 target genes Nqo1 (74 ± 13.2-fold, p < 0.01), Hmox1 (5.5 ± 0.6-fold, p < 0.01), Gclc (2.9 ± 0.7-fold, p > 0.05), and Gclm (2.6 ± 0.6-fold, p = 0.055) in Keap1-KO T cells compared with control cells (Fig. 3A). This demonstrated that Keap1-KO using CRISPR/Cas9 resulted in sustained Nrf2 activation.
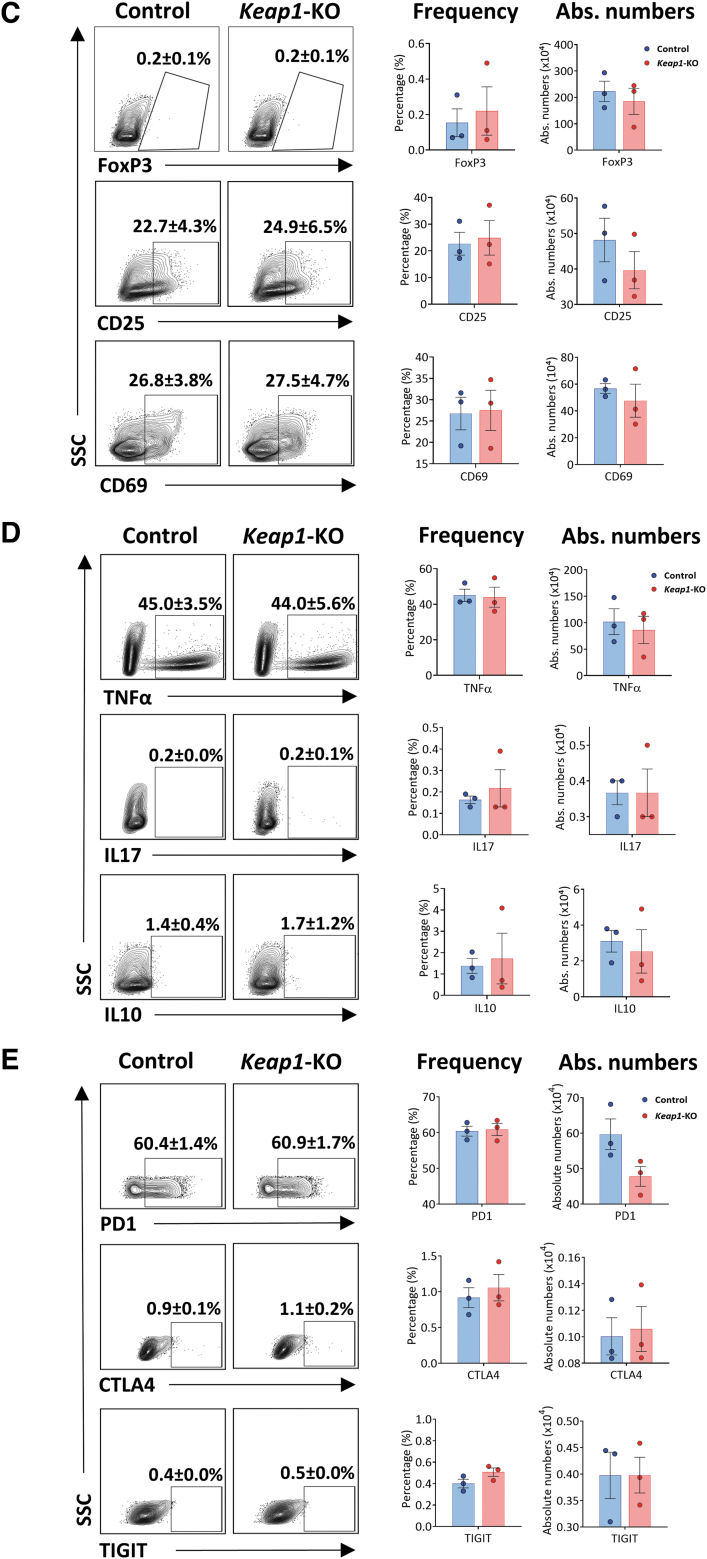
FIG. 3.
Analysis of control versus Keap1-KO CD4+ T cells. (A) Sustained increase of mRNA levels of Nrf2 target genes Nqo1, Hmox1, Gclc, and Gclm in Keap1-KO CD4+ T cells compared with control cells 96 h after electroporation. (B) Immunoblotting of control and Keap1-edited CD4+ T cells 96 h after electroporation and before adoptive transfer for KEAP1 with 73% knockdown and NQO1 with 50% increase of expression. (C) Comparable FoxP3 and activation marker expression and (D) comparable expression of TNFα, IL17, and IL10 72 h after electroporation. (E) Panel to test for T cell exhaustion markers showed no changes of inhibitory receptor expression 96 h after electroporation; n = 3 in each group in (A), (C), (D), and (E); n = 2 in each group in (B). Statistical analyses were conducted using paired, two-tailed T-test. **p < 0.001. Gclc, glutamate-cysteine ligase catalytic subunit; Gclm, glutamate-cysteine ligase modifier subunit; Hmox1, heme oxygenase 1; Nqo1, NAD(P)H quinone dehydrogenase 1.
Immunoblot analysis confirmed Keap1-KO
We further assessed the effect of CRISPR/Cas9-mediated Keap1 gene editing on KEAP1 protein expression, 96 h after RNP electroporation. Immunoblot analysis revealed significantly reduced KEAP1 protein expression (73% knockdown) in Keap1-edited CD4+ T cells compared with control CD4+ T cells. NQO1 protein expression showed 50% higher expression in Keap1-edited cells compared with control cells. These results confirmed that CRISPR/Cas9 successfully abolished KEAP1 protein expression in Keap1-KO CD4+ T cells with subsequent upregulation of Nrf2 target genes in vitro.
Sustained Nrf2 augmentation did not induce CD4+ T cell activation or exhaustion
We further assessed whether continuous Nrf2 activity induces CD4+ T cell activation or exhaustion. Flow cytometric analysis of Keap1-KO CD4+ T cells showed no difference in FoxP3 expression compared with control cells at 72 h time point. Activation markers CD25 and CD69 were comparable between the groups at 72 h time point (Fig. 3C). In addition, we observed no difference in cytokine expression of TNFα, IFNγ, IL17, and IL10 between Keap1-KO and control cells (Fig. 3D). Cell numbers between Keap1-KO and control CD4+ T cells were comparable 24 and 96 h after electroporation (Supplementary Fig. S1).
To further evaluate whether knocking out Keap1 using CRISPR/Cas9 technology results in CD4+ T cell exhaustion in ex vivo culture conditions, we assessed expression of coinhibitory molecules including programmed cell death protein 1 (PD1), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), and T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in Keap1-KO CD4+ T cells compared with control cells at 96 h time point after Keap1-KO. We did not observe any effect on above-mentioned coinhibitory molecules related to T cell exhaustion in Keap1-KO T cells compared with control cells (Fig. 3E).
Keap1-KO of CD4+ T cells changed their cytokines and Nrf2 target gene expression in normoxic and hypoxic conditions in vitro
To understand how enhanced Nrf2 antioxidant activity affected response of kidney-resident CD4+ T cells during kidney ischemic injury, cells were exposed to in vitro hypoxic conditions.
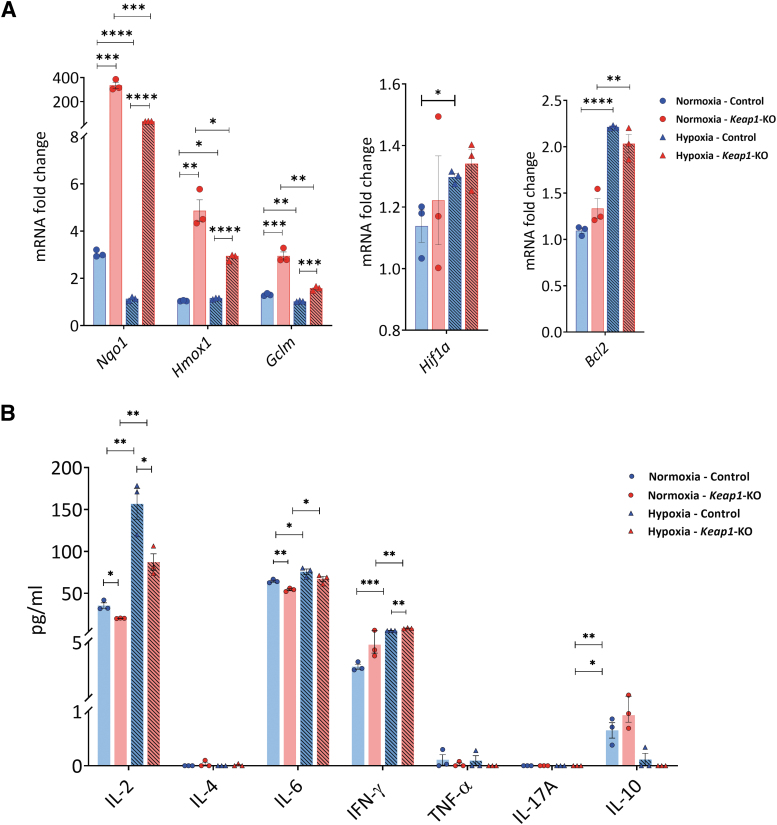
Keap1-KO CD4+ T cells showed an increase of mRNA levels of Nrf2 target genes in normoxia for Nqo1 (335.2 ± 25.7-fold, p < 0.001), Hmox1 (4.9 ± 0.5-fold, p < 0.01), and Gclm (3.0 ± 0.2-fold, p < 0.001) and in hypoxia for Nqo1 (36.7 ± 0.8-fold, p < 0.0001), Hmox1 (2.9 ± 0.1-fold, p < 0.0001), and Gclm (1.6 ± 0.1-fold, p < 0.001) compared with control cells (Fig. 4A). There were no significant changes in hypoxia-inducible factor 1α (Hif1a) and B-cell lymphoma 2 (Bcl2) expression between the groups in normoxic and hypoxic conditions (Fig. 4A).
FIG. 4.
Cytokine and chemokine expression in control and Keap1-KO CD4+ T cells under normoxic and hypoxic in vitro conditions. Control and Keap1-KO CD4+ T cells were cultured under normoxia or hypoxia for 72 h in vitro. (A) Significant Nrf2 target gene (Nqo1, Hmox1, Gclm) upregulation was confirmed under normoxia and hypoxia. There were no significant differences in Hif1a and Bcl2 between Keap1-KO and control cells under either condition. (B) Cytometric bead array data showed downregulation of IL2 in Keap1-KO CD4+ T cells compared with controls under normoxia and hypoxia. IL6 expression was significantly decreased in Keap1-KO CD4+ T cells under normoxia compared with controls. IFNγ upregulation was exacerbated under hypoxic conditions in Keap1-KO CD4+ T cells compared with control cells. For IL10, there were no significant changes between control and Keap1-KO cells in normoxic and hypoxic conditions. IL4, TNF-α, and IL17A expression was below detection level. n = 3 in each group. Statistical analyses were performed by paired, two-tailed T-test. *p < 0.05, **p < 0.001, ***p < 0.001, ****p < 0.0001. Bcl2, B-cell lymphoma 2 receptor; Hif1a, hypoxia-inducible factor 1 alpha.
Cytometric bead array (CBA) analysis of culture supernatants from Keap1-KO CD4+ T cells revealed significantly reduced IL2 in Keap1-KO compared with nonedited control cells in both normoxia (Keap1-KO vs. control: 20.2 ± 0.2 ng/dL vs. 35.9 ± 3.3 ng/dL, p < 0.05) and hypoxia (Keap1-KO vs. control: 87.2 ± 9.8 ng/dL vs. 156.5 ± 18.4 ng/dL, p < 0.05).
In addition, reduced levels of IL6 in Keap1-KO compared with controls were found under normoxic conditions (Keap1-KO vs. control: 54.3 ± 1.2 ng/mL vs. 64.5 ± 1.2 ng/mL, p < 0.01). IFNγ levels were significantly increased in Keap1-KO cells in hypoxia compared with control cells (Keap1-KO vs. control: 8.3 ± 0.4 ng/mL vs. 5.5 ± 0.2 ng/mL, p < 0.01). Keap1-KO and control cells showed comparable IL10 expression in normoxic and hypoxic conditions. IL4, TNF-α, and IL17A expression was below detection level (Fig. 4B).
Keap1-KO CD4+ T cell immunotherapy improved kidney function after IRI
We next assessed the therapeutic potential of Keap1-KO CD4+ T cell immunotherapy in an established mouse model of kidney IRI. We adoptively transferred 10 × 106 Keap1-KO or control CD4+ T cells into nu/nu mice, ∼96 h after electroporation. Nu/nu mice lack CD4+ T cells and are therefore ideal to track adoptively transferred CD4+ T cells. C57BL/6J background was chosen to avoid rejection of infused cells.
To test for adoptive transfer, an additional group of mice received PBS alone. Detection of CD4+ T cells in recipient kidneys 24 h after adoptive transfer confirmed successful transfer of cells (Supplementary Fig. S2). Mice belonging to both groups were subjected to IR surgery 24 h after adoptive transfer of control or Keap1-KO CD4+ T cells.
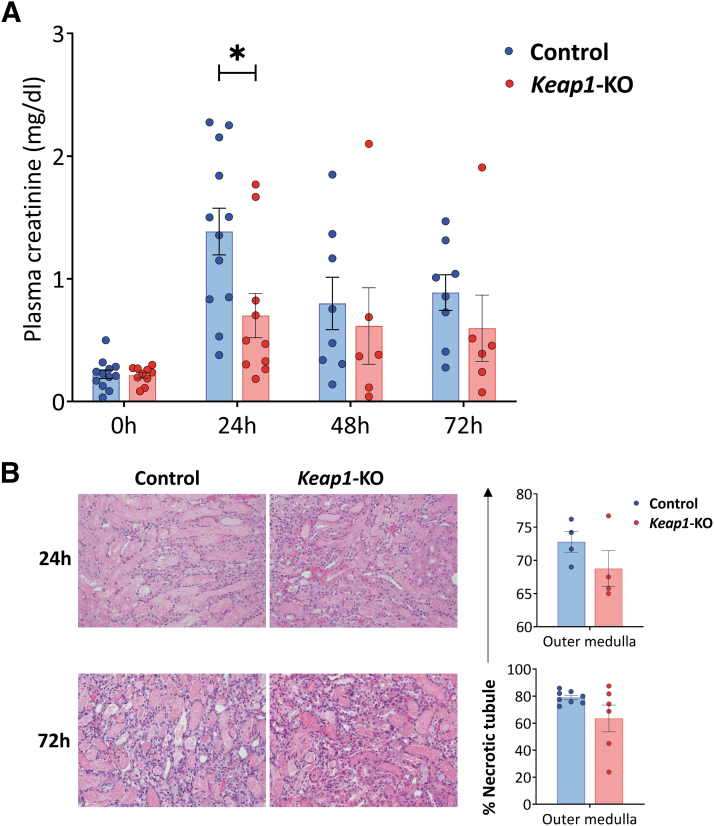
Keap1-KO CD4+ T cell recipient mice showed significantly (p < 0.05) reduced plasma creatinine levels (0.7 ± 0.2 mg/dL) compared with mice that received control CD4+ T cells at 24 h time point (1.4 ± 0.2 mg/dL) (Fig. 5A).
FIG. 5.
Adoptive transfer of Keap1-KO CD4+ T cells resulted in functional protection from IRI. (A) Plasma creatinine concentrations were significantly lower in recipient mice of Keap1-KO CD4+ T cells compared with recipients of control CD4+ T cells at 24 h. (B) Proportions of necrotic tubules and representative images of H&E-stained kidney sections of each experimental group. Original magnification × 200; n in experimental groups at 24 h: Control CD4+ T cells: 12; Keap1-KO CD4+ T cells: 10; n in experimental groups at 72 h: control CD4+ T cells: 8; Keap1-KO CD4+ T cells: 6 (three independent experiments). Statistical analysis was performed using paired, two-tailed T-test; *p < 0.05. H&E, hematoxylin and eosin; IRI, ischemia-reperfusion injury.
Histological assessment of recipient kidneys showed no significant differences of necrotic tubules in the outer medulla in mice that received Keap1-KO CD4+ T cells compared with recipients of control cells at 24 h (Keap1-KO vs. control CD4+ T cells: 68.8 ± 2.7% vs. 72.7 ± 1.6%, p = 0.244) and 72 h after IRI (Keap1-KO vs. control CD4+ T cells: 63.5 ± 10.0% vs. 79.0 ± 1.6%, p = 0.100) (Fig. 5B).
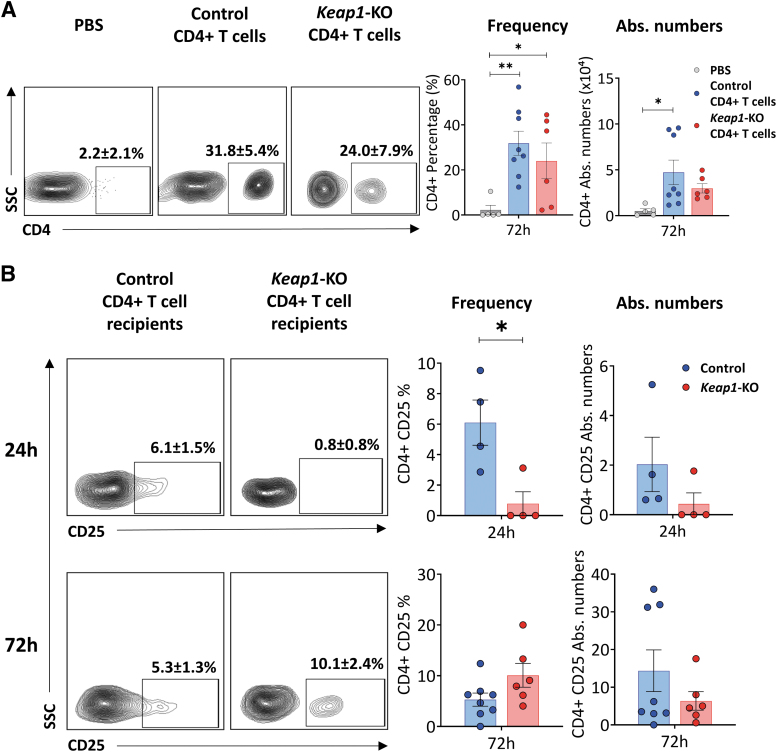
Immunophenotyping of nu/nu mice kidneys 72 h after surgery showed significant numbers of CD4+ T cells, demonstrating that adoptively transferred cells trafficked to the kidneys of recipient mice (Fig. 6A). Furthermore, there was no difference in the percentage or absolute number of CD4+ T cells between the control or Keap1-KO group.
FIG. 6.
Confirmation of adoptive transfer and immunophenotyping of recipient kidneys. (A) Detection of control or Keap1-KO CD4+ T cells in recipient kidneys confirmed successful adoptive transfer into T cell-deficient nu/nu mice. Immunophenotyping of recipient mice kidneys 72 h after IRI surgery presented significant increase of CD4+ T cell frequencies and absolute numbers in both experimental groups vs. PBS group; n in experimental groups at 72 h: PBS: 5; control CD4+ T cells: 8; Keap1-KO CD4+ T cells: 6. One-way ANOVA with unprotected Fisher LSD was performed. (B) Immunophenotyping of recipient kidney CD4+ T cells after IRI revealed significantly lower CD25 expression in Keap1-KO CD4+ T cells compared with control CD4+ T cells at 24 h, whereas expression after 72 h was comparable; n in experimental groups at 24 h: control CD4+ T cells: 4; Keap1-KO CD4+ T cells: 4; n in experimental groups at 72 h: control CD4+ T cells: 8; Keap1-KO CD4+ T cells: 6. Statistical analyses were performed using paired, two-tailed T-test. *p < 0.05, **p < 0.001. LSD, least significant difference.
Examination of CD4+ T cells from recipient kidneys 24 h after IRI showed significantly (p < 0.05) reduced CD25 expression in Keap1-KO (0.8 ± 0.8%) CD4+ T cells compared with controls (6.1 ± 1.5%) (Fig. 6B).
Furthermore, there was a trend for lower TNFα in Keap1-KO CD4+ T cells compared with control CD4+ T cells (p = 0.056) (Supplementary Fig. S3B). There was no difference in CD69 expression or intracellular levels of IFNγ between Keap1-KO cells isolated from postischemic kidneys compared with controls (Supplementary Fig. S3A, S3C). Analysis of CD4+ T cells from kidneys of recipient mice showed an extremely week signal for FoxP3 likely due to small cell numbers.
Discussion
The Nrf2/Keap1 pathway is an important mediator and promising target for the treatment of many kidney diseases, including IR-induced AKI (Nezu et al., 2017a; Stenvinkel et al., 2021). It was previously demonstrated that Nrf2-regulated antioxidant response is protective during AKI, and that CD4+ T cell-specific Nrf2 augmentation protects mice from IRI (Noel et al., 2015). In this study, we increased Nrf2 activity of CD4+ T cells via Keap1-KO using CRISPR/Cas9 and tested its therapeutic effect on kidney IRI outcome.
Our optimized CRISPR/Cas9 protocol resulted in significant induction of the Nrf2-regulated genes Nqo1, Hmox1, Gclc, and Gclm in murine CD4+ T cells. Immunotherapy using Keap1-KO murine CD4+ T cells improved kidney function in an in vivo model of kidney IRI. This study demonstrates the feasibility of T cell gene editing with potential for human translation to treat kidney diseases.
Our approach to edit Keap1 gene using CRISPR/Cas9 technology led to sustained augmentation of Nrf2-regulated antioxidant gene expression in CD4+ T cells. The results were confirmed on protein level verifying a sufficient KEAP1-knockdown and NQO1 upregulation. This observation is in accordance with previous results using Keap1-KO in human T cells and Treg cells as well as Cre/lox-based Keap1 deletion in murine T cells (Noel et al., 2018; Noel et al., 2015).
Immunophenotyping of Keap1-KO CD4+ T cells revealed a reduced frequency of CD25+ CD4+ T cells compared with control recipient mice 24 h after adoptive transfer. Interestingly, after 72 h the trend was reversed. We speculate that the cells' activation status after anti-CD3/anti-CD28 antibody stimulation in the beginning vanished. We found a significant upregulation of Tnfa mRNA levels 24 h after electroporation. However, there was no difference in protein levels at both 24 and 72 h time points.
Furthermore, other measured cytokines revealed no major differences in Keap1-KO cells compared with control cells, specifically at later time points. However, a previous study using genetic Keap1 deletion in murine CD4+ T cells demonstrated significant reduction in TNFα, IFNγ, and IL17 expression (Noel et al., 2015). The discrepancy between these two studies could be due to heterogeneous characteristics and partial KO status of CRISPR/Cas9 gene-edited cells, or the use of CD4+ T cells in this study compared with pan T cells by prior study. In fact, it has been described that different T cell subsets can develop toward an either pro- or anti-inflammatory phenotype after Nrf2 upregulation (Ohl and Tenbrock, 2021).
In vitro studies examined responses of Keap1-KO and unedited control cells in hypoxic and normoxic conditions after 72 h. Antioxidant Nrf2 target gene upregulation was maintained under hypoxia, however, in an overall lower fold change than in normoxia, which has previously been shown for Hmox1 and Gclm (Roman et al., 2010). Hif1a expression was comparable between the groups in our model contrary to the results shown in renal epithelial cells, indicating possible differences in Keap1 effects on Hif1a-regulated responses in renal epithelial cells versus the CD4+ T cells, and could also be due to different time points studied.
Analysis of culture supernatants revealed a decrease of IL2 in Keap1-KO cells compared with control cells in both conditions. Hypoxia-induced upregulation of IL2 in WT CD4+ T cells has previously been found (Roman et al., 2010). However, the same study demonstrated an upregulation of IL2 under in vitro hypoxic conditions in Nrf2 KO CD4+ T cells, which is different from our findings in Keap1-KO CD4+ T cells (Roman et al., 2010).
In addition, IL6 levels were significantly lower in Keap1-KO CD4+ T cells compared with control cells in normoxia. This is consistent with the finding that CDDO-imidazolide treatment of WT mice before IR-induced AKI resulted in lower IL6 levels compared with controls (Liu et al., 2014). We also demonstrated that Keap1-KO cells produced significantly more IFNγ than control cells, which was confirmed on mRNA level. Splenic CD4+ cells from Nrf2 KO mice had reduced IFNγ production in hypoxic in vitro conditions (Roman et al., 2010).
Although IFNγ is known as proinflammatory cytokine, recent evidence suggests that it also plays an important role in T cell maintenance, which might explain its upregulation (Borges da Silva et al., 2015). Furthermore, there was no difference in inhibitory receptor expression and IL2 and IFNγ production following Keap1-KO. These results suggest that neither Nrf2 upregulation nor electroporation leads to T cell exhaustion in CD4+ T cells and aligns with previously published studies (Nüssing et al., 2020).
Successful adoptive transfer of Keap1-KO CD4+ T cells into nu/nu mice resulted in significantly improved renal function 24 h after IR compared with mice that had received control CD4+ T cells. Histological assessment revealed no significant differences of necrotic tubules in mice that received Keap1-KO T cells compared with controls. These findings are in concordance but more modest than previous observations using T cell-specific Keap1-KO mice that were structurally more protected from kidney IRI (Noel et al., 2015).
It is important to note that improvement in kidney function is the most important factor for patient outcomes, and changes in function do not always equate to structural changes. Future experiments with different dosing or reagents may also demonstrate changes in structure.
Renal fibrotic changes occur at least one week after induction of AKI, and were therefore not expected in our model with a follow-up time of 72 h only (Black et al., 2018; Chung et al., 2018; Guan et al., 2019). The assessment of the development of kidney fibrosis and/or recovery using, for example, Masson's trichrome staining would be an interesting aspect for future investigations in an AKI-to-chronic kidney disease model with longer follow-up time (Farris and Alpers, 2014).
Analysis of Keap1-KO CD4+ T cells isolated from nu/nu mice that were adoptively transferred and exposed to IR showed significantly reduced CD25, 24 h after IRI. We did not assess the suppression function of Keap1-KO cells in this study, but an abrupt increase in antioxidant status and/or electroporation may have affected CD25 expression. Future studies are warranted to understand this effect. It is also to be investigated if additional mechanisms, such as M2 action, contribute to this effect on top of the antioxidant effect of Keap1-KO CD4+ T cells preventing a burst of reactive oxygen species during AKI.
Nu/nu mice have been used in the past in many AKI models (Ascon et al., 2006). However, they are a leaky model with residual T cell populations (Burne et al., 2001). Immunophenotyping of CD8+ and double negative T cells from recipient kidneys did not show significant differences, confirming the impact of adoptively transferred CD4+ T cells on kidney IRI outcomes.
Keap1 KOs, pharmacological Nrf2 activators, and Keap1-KO by CRISPR/Cas9 might show different effects due to potentially different molecular changes. We aimed for an immunotherapeutic approach to bypass potential side effects and poor elimination of systemically applied drugs targeting the cells' redox system such as pharmacological Nrf2 activators or malonate ester prodrugs, which protect from murine AKI (Beach et al., 2020). CDDO derivates led to cardiovascular side effects and proteinuria in clinical studies (Nezu et al., 2017b).
Furthermore, administration of malonate ester as prodrugs shows reduced toxicity (Beach et al., 2020). A functional crosstalk between Nrf2/Keap1 with other pathways such as nuclear factor kappa B (NF-κB) and HIF1α, all involved in pathogenesis of ischemic AKI and interconnected, has been described (Agarwal et al., 2016; Willam, 2014).
In physiologic conditions, Keap1 regulates IκB kinase-β, which phosphorylates the NF-κB inhibitor IκB by autophagic degradation and therefore upregulates NF-κB signaling (Kim et al., 2010). We did not assess these potential differences between a physiologic and artificially induced Nrf2 upregulation (Pedruzzi et al., 2012; Wardyn et al., 2015). It will be important for future studies to investigate Keap1-KO effects on related cellular pathways and to assess potential side effects.
Although our study provided important initial data on feasibility of gene editing CD4+ T cells, expansion in vitro, adoptive transfer, and preliminary results in an ischemic AKI model, this study had various limitations. CRISPR/Cas9-based gene editing can have undesirable off-target effects. Despite the fact that we did not observe any gross abnormalities in CD4+ T cells in this study, we cannot reject the possibility of off-target effects of our Nrf2/Keap1 gene editing.
Second, we were not able to enrich Keap1-KO cells before adoptive transfer. Since CRISPR/Cas9-based gene editing results in ∼60%–70% of electroporated T cells in mouse and in human, enriched edited cells before adoptive transfer could augment protective effect in the immunotherapy. However, enrichment of intracellular targets such as Keap1 is technically difficult.
Nonetheless, ongoing effort to improve CRISPR/Cas9 specificity will substantially reduce off-target effects and heterogeneity of the infused cells (Michlits et al., 2017). In vivo gene editing using CRISPR/Cas9 technology has already successfully been performed in clinical trials for other diseases, and would allow for bypassing obstacles such as electroporation, cell death, and adequate T cell expansion (Gillmore et al., 2021).
CRISPR/Cas9 technology is a promising approach for upregulating antioxidant capacity of T cells, which can be further developed as an effective therapy for kidney diseases (Miyagi et al., 2016). Possible applications for a future T cell therapy for humans could be preoperative treatment to prevent ischemic AKI due to cardiac surgery or deceased donor kidney transplantation.
Recently, novel T cell-based therapies such as immune checkpoint inhibitors and chimeric antigen receptor T cells for cancer treatment have been associated with an increased incidence of AKI highlighting the pathogenic role of T cells (Gutgarts et al., 2020; Herrmann and Perazella, 2020). In addition, gene editing of the Keap1 gene or other target genes may help ameliorate AKI, and therefore prevent this adverse side effect. Furthermore, other immune-mediated kidney disease models can be studied, including allograft rejection and glomerulonephritis.
Innovation
AKI is a major health care problem without specific therapy. T cells are important pathophysiologic players, and parallel studies showed that the Nrf2/Keap1 pathway regulating antioxidant responses is protective during IR-induced AKI and other kidney diseases such as Alport syndrome.
This study demonstrates that ex vivo Keap1-KO of murine T cells using CRISPR/Cas9 technology is a promising approach for developing immunotherapy for AKI. CRISPR/Cas9-induced Keap1 gene KO in CD4+ T cells resulted in upregulation of antioxidant Nrf2 target genes. Adoptively transferred Keap1-KO T cells into nu/nu mice improved kidney function. Gene editing of T cells targeting Nrf2/Keap1 pathway is a promising technology for ischemic AKI and other immune-mediated kidney diseases (see graphic summary illustration, Fig. 1).
Methods
Animals
Eight- to eleven-week-old male C57BL/6J WT and homozygous B6.Cg-Foxn1nu/J (nu/nu) on a C57BL/BJ background mice (strain number: 000819) were purchased from Jackson Laboratory (Bar Harbor, ME). For cryopreserved colony management, mouse embryos were frozen after 51 backcrosses of nu/nu male to C57BL/6J female mice in alternating generations and used for breeding up to 10 generations. Mice were housed at the central animal facility of the Johns Hopkins University under specific pathogen-free conditions. Animal protocols used for this study were approved by the Johns Hopkins University Institutional Animal Care and Use Committee.
CD4+ T cell isolation and culture
Spleens and inguinal lymph nodes were harvested from WT mice, passed through a 70-μm strainer (Fisherbrand, Pittsburgh, PA; 22363548), and washed with PBS containing 2% fetal bovine serum (FBS) and 1 mM EDTA. Pan CD4+ T cells were enriched by negative selection using the EasySep Mouse CD4+ T cell Isolation Kit (STEMCELL Technologies, Cambridge, MA; 19852).
Primary pan mouse CD4+ T cells were isolated and cultured in six-well plates with anti-CD3e (Clone: 145-2C11; Tonbo Bioscience, San Diego, CA; 70-0031) and anti-CD28 antibodies (Clone: 37.51; Tonbo Bioscience; 70-0281) (10 μg/mL) for 48 h in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 mM nonessential amino acid, 1% penicillin and streptomycin, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 55 mM β-mercaptoethanol, and 2 mM L-glutamine.
Optimization of CRISPR/Cas9-based Keap1-KO and RNP complex preparation for Keap1-gene-KO
Adjusted conditions with a sgRNA:Cas9 of 1.8:1 ratio and electroporation settings of 1600 V, 3 pulses, 10 ms were used for Keap1-gene-KO per published protocol with minor modifications (Seki and Rutz, 2018).
CRISPR/Cas9-mediated Keap1-KO was achieved by delivering RNP complexes. RNP complexes were prepared by assembling different sgRNAs (Synthego) separately or in combination (Table 1; 66 pmol/500,000 cells) with Cas9 protein (36 pmol/500,000 cells) (IDT, Newark, NJ). To improve electroporation efficiency, Alt-R Cas9 electroporation enhancer oligo (1.8 μM; IDT) was added before transferring into CD4+ T cells using electroporation by Neon Transfection kit and device (Invitrogen, Waltham, MA; kit: MPK10025; device: MPK5000) set at 1600 V, 3 pulses, and 10 ms. Control CD4+ T cells were electroporated without sgRNA and Cas9.
After electroporation, the cells were cultured in antibiotic-free complete DMEM supplemented with 10% FBS, 100 mM nonessential amino acid, 10 mM HEPES, 55 mM β-mercaptoethanol, 2 mM L-glutamine, and IL2 (100 U/mL) for 24 h in 12-well plates. After 24 h, culture medium was changed to complete DMEM containing 1% penicillin and streptomycin, and the cells were harvested at various time points depending on the experimental design. Medium was changed every 48 h. Downstream analyses included qRT-PCR, immunophenotyping, and cytokine/chemokine of culture supernatants assessment via Immunology Multiplex Assay (ThermoFisher Scientific, Waltham, MA; EXP110-20820-901).
Analysis of Nrf2 targets, cytokine, Bcl2 and Hif1a gene expression
To analyze Nrf2-dependent antioxidant gene expression, total RNA was isolated with RNeasy Mini and Micro Kit (Qiagen, Hilden, Germany; Mini Kit: 74104; Micro Kit: 74004) from CD4+ T cells. cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Waltham, MA; 4368814).
Ready-made TaqMan primer and probe sets (ThermoFisher Scientific) for Nqo1 (Mm00500821_m1), Hmox1 (Mm00516005_m1), Gclc (Mm00802655_m1), Gclm (Mm01324400_m1), Hif1a (Mm00468869_m1), Bcl2 (Mm00477631_m1), Tnfa (Mm00443258_m1), Ifng (Mm01168134_m1), Il10 (Mm00439614_m1), and Il17 (Mm00439618_m1) were used to assess the effect of Keap1-gene-KO in CFX96 qRT-PCR (Bio-Rad, Hercules, CA). The expression of each gene was normalized to Rpl13a expression, and relative fold expressions were calculated using a ΔΔ cycle threshold method (Livak and Schmittgen, 2001).
Immunoblotting
Cell lysis was performed using RIPA buffer (Sigma-Aldrich, St. Louis, MO; R0278) together with protease cocktail inhibitor (Sigma-Aldrich; P8340). Cell lysates were loaded on protein gel for separation and blotted on a polyvinylidene difluoride membrane. The membrane was incubated with antibodies specific for KEAP1 (E-20; Santa Cruz Biotechnology, Dallas, TX; sc-15246), NQO1 (ProteinTech, Roselmont, IL; 11451-1-AP), or β-actin (Santa Cruz Biotechnology; sc-58673).
For NQO1, membrane was probed with NQO1 first, and membrane stripped and probed with β-actin in the second step. For detection, HyGlo ECL kit (Denville Scientific, Inc., South Plainfield, NJ; E2400) and Bio-Rad Gel Doc system were applied.
In vitro hypoxia experiments
Cells were exposed to either hypoxic (1% O2) or normoxic (19% O2) conditions 48 h after gene editing in complete DMEM for 72 h. For hypoxic conditions, culture plates were placed in a modular incubator chamber. After flushing the chamber with a 1% O2/5% CO2/94% N2 gas mixture for 2 min, it was sealed. Normoxic conditions included incubation of cells at 37°C in a humidified atmosphere containing 19% O2/5% CO2/70% N2 (Wenger et al., 2015). After 72 h, cells were harvested, and the supernatant was assessed for cytokine production. Cells were lysed in RNA lysis buffer, and gene expression measured by qRT-PCR as described above.
CBA mouse Th1/Th2/Th17 cytokine assay
Cytokines (IL2, IL4, IL6, TNFα, IFNγ, IL17, IL10) were measured in culture supernatants using CBA kit (BD Biosciences; 560485) (Morgan et al., 2004).
In brief, 50 μL of capture beads were mixed with 50 μL of samples, followed by adding 50 μL Th1/Th2/Th17 Phycoerythrin detection reagent. The mixture was incubated at room temperature while protected from light for 2 h. Samples were washed and acquired using LSRII flow cytometer (BD Biosciences). Data were analyzed using FCAP Array software (BD Biosciences; 641488 [PC]). Protein levels below detection level were considered 0 pg/mg protein.
Adoptive transfer of Keap1-KO CD4+ T cells
Keap1-KO CD4+ T cells were harvested ∼96 h after electroporation and intravenously transferred (10 × 106 cells per animal) into 9- to 11-week-old male nu/nu mice 24 h pre-IRI. The control group received the same number of CD4+ T cells, which were electroporated without RNP complex. A third group received only PBS serving as a negative control for adoptive transfer confirmation. Mice were subjected to an established bilateral IRI model to induce AKI (Fig. 5A).
Kidney IRI
An established experimental IRI model in mice was used after anesthetizing mice by intraperitoneal sodium pentobarbital injection (75 mg/kg body weight) (Rabb et al., 1996).
The renal pedicels were approached by a medial abdominal incision. After dissection, a microvascular clamp (Roboz Surgical Instrument, Gaithersburg, MD) was placed on each pedicle for 30 min. Hydration of the animals was assured by application of 1 mL warm saline, and body temperature was kept at 37°C. Clamps were removed, and reperfusion was assessed. After suturing of the abdominal incision, animals were kept on surgery table for 30 min during reperfusion. Afterward, animals were provided food and water for appropriate recovery.
Assessment of renal function
To assess the effect of Keap1-KO CD4+ T cell immunotherapy on kidney function after IRI, blood was collected at baseline (0 h) and 24, 48, and 72 h after IRI surgery. Plasma creatinine levels were analyzed with the Cobas Mira Plus automated analyzer system (Roche, Basel, Switzerland) using creatinine measurement reagents (Pointe Scientific, Canton, MI). This technique has been validated with inulin clearances (O'Donnell et al., 2002).
Tissue histological analysis
Mice were sacrificed 24 or 72 h after IRI surgery, and a transverse piece from right kidneys was fixed in 10% phosphate buffered formalin and embedded with paraffin. Hematoxylin and eosin staining was performed, and a renal pathologist assessed the kidney injury under blinded conditions. The percentage of necrotic tubules was assessed in cortex and outer medulla.
Isolation of kidney mononuclear cells
Kidney mononuclear cells (KMNCs) were isolated using an established protocol based on percoll density gradient (Ascon et al., 2006).
Mice were anesthetized with ketamine hydrochloride (130 mg/kg) and xylazine (7 mg/kg), and exsanguinated to remove circulating immune cells. Kidneys were harvested, decapsulated, cut into small pieces, and incubated in collagenase D solution (2 mg/mL; Sigma-Aldrich; 11088882001) for 30 min at 37°C. The digested tissue was passed through a 70 μm strainer (BD Biosciences), and KMNCs were isolated using percoll density gradient (GE Healthcare, Chicago, IL; 17-0891-01). KMNCs were washed and resuspended in RPMI media with 5% FBS.
Immunophenotyping of kidney CD4+ T cells or KMNCs
To assess the immunological effect of Keap1-KO during in vitro experiments, CD4+ T cells were analyzed by flow cytometry at 72 or ∼96 h of culture after gene editing. For in vivo experiments, KMNCs were isolated from mouse kidneys 24 or 72 h after IRI surgery.
Cells were stimulated with cell activation cocktail (BD Biosciences; 423304) containing phorbol 12-myristate 13-acetate, ionomycin, and brefeldin A for 4 h before incubation with anti-CD16/CD32 Fc block (Clone: 2.4G2; BD Biosciences; 156604), and stained for the following surface markers and intracellular cytokines:
CD45-BUV395 (Clone: 30-F11; BD Biosciences; 564279), TCR-BV421 (Clone: H57-597; BioLegend, San Diego, CA; 109230), TCR-AF488 (Clone: H57-595; BioLegend; 109230), CD4-PerCP-Cy5.5 (Clone: GK1.5; BioLegend; 100434), CD8a-BV605 (Clone: 53-6.7; BioLegend; 100744), PD1-APC Fire 750 (Clone: 29F.1412; BioLegend; 135240), CTLA4-BV421 (Clone: UC10-4B9; BioLegend; 106302), TIGIT-APC (Clone: 1G9; BioLegend; 142105), CD25-BV605 (Clone: PC61; BioLegend; 102036), CD25-APCCy7 (Clone: PC61; BioLegend; 102026), FoxP3-Alexa 488 (Clone: 150D; BioLegend; 320012), CD69-APC-Cy7 (Clone: H1.2F3; BioLegend; 104526), TNFα-PE (BD Biosciences; 561063), IFNγ-APC (Clone: XMG1.2; BioLegend; 505810), IFNγ-BV605 (Clone XMG 1.2; BioLegend; 505839), IL10-APC (Clone: JES5-16E3; BD Biosciences; 505010) and IL17-PE (Clone: eBio1787; Invitrogen; 12-7177-81), IL2-PE (Clone: JES6-5H4; BD Biosciences; 554428).
Cells were analyzed with LSRII flow cytometer (BD Biosciences). Unstained and unstimulated samples as well as fluorescence minus one controls were used for correct gating analyses using FlowJo software (BD Biosciences). The gating strategy included identification of KMNCs based on forward scatter and side scatter.
In the next step, all nonsingle cells were excluded followed by the gating based on specific antibody staining. For calculation of absolute numbers, the total numbers of KMNCs or cultured T cells were counted manually using a hemocytometer. The number of each population acquired by flow cytometry was multiplied with the total number of KMNCs or cultured T cells counted, followed by adjustment using the total number of cells acquired using flow cytometry.
Multiplex cytokine assay
To examine cytokines and chemokines in culture supernatants as well as lysates of recipient kidneys, protein levels of IL2, IL6, IFNγ, and IL10 were analyzed using Milliplex Mouse Cytokine/Chemokine Magnetic Bead Panel–Immunology Multiplex Assay (Millipore Sigma). In brief, lysates were incubated at −80°C overnight, sonicated and centrifuged at 4500 g for 5 min. The Bioplex 200 platform (Bio-Rad) was used to determine the concentration of cytokines in the extracted specimens.
Luminex bead-based immunoassays (Millipore, Billerica, NY) were performed after Immune Monitoring Core standard operating procedures, and concentrations were determined using five-parameter log curve fits (using Bioplex Manager 6.0) with vendor-provided standards and quality controls. The concentration of cytokines and chemokines was normalized using raw protein concentration (measured by Pierce BCA Protein Assay Kit; ThermoFisher Scientific).
Data collection and statistical analysis
Electronic laboratory notebook was not used. Data were expressed as mean ± standard error of mean. Comparison between two groups was performed with paired, two-tailed T-test, and multiple groups with one-way ANOVA using Microsoft Excel 2016 or Graph Pad Prism 9 for Windows. p Value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgment
The assistance for in vitro hypoxia experiments provided by Mr. Nick Dordai from Prof. Gregg Semenza's laboratory was greatly appreciated.
Abbreviations Used
- AKI
acute kidney injury
- Bcl2
B-cell lymphoma 2
- Cas9
CRISPR-associated protein 9
- CBA
cytometric bead array
- CDDO-Me
2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid methyl ester
- CRISPR
clustered regularly interspaced short palindromic repeats
- CTLA4
cytotoxic T-lymphocyte-associated protein 4
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- Gclc
glutamate-cysteine ligase catalytic subunit
- Gclm
glutamate-cysteine ligase modifier subunit
- H&E
hematoxylin and eosin
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- Hif1a
hypoxia-inducible factor 1α
- Hmox1
heme oxygenase 1
- IFNγ
interferon gamma
- IL
interleukin
- IRI
ischemia reperfusion injury
- Keap1
kelch-like ECH-associated protein 1
- KMNCs
kidney mononuclear cells
- KO
knockout
- NF-κB
nuclear factor kappa B
- Nqo1
NAD(P)H quinone dehydrogenase 1
- Nrf2
nuclear factor erythroid 2-related factor 2
- PD1
programmed cell death protein 1
- PE
Phycoerythrin
- qRT-PCR
quantitative real-time polymerase chain reaction
- RNP
ribonucleoprotein
- sgRNA
single-guide RNA
- Th
T helper
- TIGIT
T-cell immunoreceptor with immunoglobulin and ITIM domains
- TNFα
tumor necrosis factor alpha
- Treg
regulatory T cells
- WT
wild type
Authors' Contributions
J.T.K., S.N., M.S., A.R.A.H., S.P.R., and H.R. designed the study. J.T.K., S.N., K.L., S.G., A.A., A.N.-R., and J.G. conducted experiments. L.J.A. analyzed and scored histology slides and took representative pictures. J.T.K., S.N., M.S., K.L., and A.A. analyzed data. J.T.K. prepared the figures, and J.T.K. and S.N. drafted the article. S.N., K.L., M.S., S.G., S.A.L., A.R.A.H., and H.R. helped with writing the article. All authors approved the final version of the article.
Author Disclosure Statement
The authors have declared that no conflict of interest exists.
Funding Information
J.T.K. was supported by a Dr. Werner Jackstädt-Foundation grant (project number S 134–10.117). S.N. was supported by Carl W. Gottschalk research scholar grant from American Society of Nephrology (grant number: 134535) and Ed Kraus award from Johns Hopkins Nephrology. K.L. was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) (grant number: HI19C1337), and the National Research Foundation of Republic of Korea (NRF) funded by the Ministry of Education (grant number: 2021R1A6A3A03039863). S.P.R. received an NIH grant (R01HL136946). H.R. was supported by the NIDDK (R01DK111209) and philanthropic gifts from Rogelio Miro of Panama.
Supplementary Material
References
- Agarwal A, Dong Z, Harris R, et al. Cellular and molecular mechanisms of AKI. J Am Soc Nephrol 2016;27:1288–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders HJ, Wilkens L, Schraml B, et al. One concept does not fit all: The immune system in different forms of acute kidney injury. Nephrol Dial Transplant 2021;36:29–38. [DOI] [PubMed] [Google Scholar]
- Ascon DB, Lopez-Briones S, Liu M, et al. Phenotypic and functional characterization of kidney-infiltrating lymphocytes in renal ischemia reperfusion injury. J Immunol 2006;177:3380–3387. [DOI] [PubMed] [Google Scholar]
- Battistone MA, Mendelsohn AC, Spallanzani RG, et al. Proinflammatory P2Y14 receptor inhibition protects against ischemic acute kidney injury in mice. J Clin Invest 2020;130:3734–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach TE, Prag HA, Pala L, et al. Targeting succinate dehydrogenase with malonate ester prodrugs decreases renal ischemia reperfusion injury. Redox Biol 2020;36:101640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LM, Lever JM, Traylor AM, et al. Divergent effects of AKI to CKD models on inflammation and fibrosis. Am J Physiol Renal Physiol 2018;315:F1107–F1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges da Silva H, Fonseca R, Alvarez JM, et al. IFN-γ priming effects on the maintenance of effector memory CD4(+) T cells and on phagocyte function: Evidences from infectious diseases. J Immunol Res 2015;2015:202816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne MJ, Daniels F, El Ghandour A, et al. Identification of the CD4(+) T cell as a major pathogenic factor in ischemic acute renal failure. J Clin Invest 2001;108:1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Overstreet JM, Li Y, et al. TGF-β promotes fibrosis after severe acute kidney injury by enhancing renal macrophage infiltration. JCI Insight 2018;3:e123563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio FR, Kurzhagen JT, Rabb H. Reparative T lymphocytes in organ injury. J Clin Invest 2019;129:2608–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zeeuw D, Akizawa T, Audhya P, et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N Engl J Med 2013;369:2492–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellepiane S, Leventhal JS, Cravedi P. T Cells and acute kidney injury: A two-way relationship. Front Immunol 2020;11:1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Swaminathan S, Bachman LA, et al. Resident dendritic cells are the predominant TNF-secreting cell in early renal ischemia-reperfusion injury. Kidney Int 2007;71:619–628. [DOI] [PubMed] [Google Scholar]
- Doudna JA. The promise and challenge of therapeutic genome editing. Nature 2020;578:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris AB, Alpers CE. What is the best way to measure renal fibrosis? A pathologist's perspective. Kidney Int Suppl (2011) 2014;4:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandolfo MT, Jang HR, Bagnasco SM, et al. Foxp3+ regulatory T cells participate in repair of ischemic acute kidney injury. Kidney Int 2009;76:717–729. [DOI] [PubMed] [Google Scholar]
- Gharaie Fathabad S, Kurzhagen JT, Sadasivam M, et al. T lymphocytes in acute kidney injury and repair. Semin Nephrol 2020;40:114–125. [DOI] [PubMed] [Google Scholar]
- Gillmore JD, Gane E, Taubel J, et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med 2021;385:493–502. [DOI] [PubMed] [Google Scholar]
- Guan Y, Nakano D, Zhang Y, et al. A mouse model of renal fibrosis to overcome the technical variability in ischaemia/reperfusion injury among operators. Sci Rep 2019;9:10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutgarts V, Sathick IJ, Zheng J, et al. Incidence and risk factors for acute and chronic kidney injury after adult cord blood transplantation. Biol Blood Marrow Transplant 2020;26:758–763. [DOI] [PubMed] [Google Scholar]
- Herrmann SM, Perazella MA. Immune checkpoint inhibitors and immune-related adverse renal events. Kidney Int Rep 2020;5:1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishani A, Xue JL, Himmelfarb J, et al. Acute kidney injury increases risk of ESRD among elderly. J Am Soc Nephrol 2009;20:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MT, Ghali WA, Knudtson ML, et al. Associations between acute kidney injury and cardiovascular and renal outcomes after coronary angiography. Circulation 2011;123:409–416. [DOI] [PubMed] [Google Scholar]
- Jang HR, Rabb H. Immune cells in experimental acute kidney injury. Nat Rev Nephrol 2015;11:88–101. [DOI] [PubMed] [Google Scholar]
- Kim JE, You DJ, Lee C, et al. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal 2010;22:1645–1654. [DOI] [PubMed] [Google Scholar]
- Kinsey GR, Okusa MD. Expanding role of T cells in acute kidney injury. Curr Opin Nephrol Hypertens 2014;23:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey GR, Sharma R, Huang L, et al. Regulatory T cells suppress innate immunity in kidney ischemia-reperfusion injury. J Am Soc Nephrol 2009;20:1744–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Wang Y, Luo M, et al. Prevention of streptozotocin-induced diabetic nephropathy by MG132: Possible roles of Nrf2 and IκB. Oxid Med Cell Longev 2017;2017:3671751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Bae E, Kwon SH, et al. Transcriptional modulation of the T helper 17/interleukin 17 axis ameliorates renal ischemia-reperfusion injury. Nephrol Dial Transplant 2019;34:1481–1498. [DOI] [PubMed] [Google Scholar]
- Leonard MO, Kieran NE, Howell K, et al. Reoxygenation-specific activation of the antioxidant transcription factor Nrf2 mediates cytoprotective gene expression in ischemia-reperfusion injury. FASEB J 2006;20:2624–2626. [DOI] [PubMed] [Google Scholar]
- Liu M, Grigoryev DN, Crow MT, et al. Transcription factor Nrf2 is protective during ischemic and nephrotoxic acute kidney injury in mice. Kidney Int 2009;76:277–285. [DOI] [PubMed] [Google Scholar]
- Liu M, Reddy NM, Higbee EM, et al. The Nrf2 triterpenoid activator, CDDO-imidazolide, protects kidneys from ischemia-reperfusion injury in mice. Kidney Int 2014;85:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- Lu Y, Sun Y, Liu Z, et al. Activation of NRF2 ameliorates oxidative stress and cystogenesis in autosomal dominant polycystic kidney disease. Sci Transl Med 2020;12. [DOI] [PubMed] [Google Scholar]
- Marques VP, Goncalves GM, Feitoza CQ, et al. Influence of TH1/TH2 switched immune response on renal ischemia-reperfusion injury. Nephron Exp Nephrol 2006;104:e48–e56. [DOI] [PubMed] [Google Scholar]
- Mehrotra P, Collett JA, McKinney SD, et al. IL-17 mediates neutrophil infiltration and renal fibrosis following recovery from ischemia reperfusion: Compensatory role of natural killer cells in athymic rats. Am J Physiol Renal Physiol 2017;312:F385–F397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra P, Sturek M, Neyra JA, et al. Calcium channel Orai1 promotes lymphocyte IL17 expression and progressive kidney injury. J Clin Invest 2019;129:4951–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlits G, Hubmann M, Wu SH, et al. CRISPR-UMI: Single-cell lineage tracing of pooled CRISPR-Cas9 screens. Nat Methods 2017;14:1191–1197. [DOI] [PubMed] [Google Scholar]
- Miyagi A, Lu A, Humphreys BD. Gene editing: Powerful new tools for nephrology research and therapy. J Am Soc Nephrol 2016;27:2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E, Varro R, Sepulveda H, et al. Cytometric bead array: A multiplexed assay platform with applications in various areas of biology. Clin Immunol 2004;110:252–266. [DOI] [PubMed] [Google Scholar]
- Nezu M, Souma T, Yu L, et al. Transcription factor Nrf2 hyperactivation in early-phase renal ischemia-reperfusion injury prevents tubular damage progression. Kidney Int 2017a;91:387–401. [DOI] [PubMed] [Google Scholar]
- Nezu M, Suzuki N, Yamamoto M. Targeting the KEAP1-NRF2 system to prevent kidney disease progression. Am J Nephrol 2017b;45:473–483. [DOI] [PubMed] [Google Scholar]
- Noel S, Lee SA, Sadasivam M, et al. KEAP1 editing using CRISPR/Cas9 for therapeutic NRF2 activation in primary human T lymphocytes. J Immunol 2018;200:1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel S, Martina MN, Bandapalle S, et al. T lymphocyte-specific activation of Nrf2 protects from AKI. J Am Soc Nephrol 2015;26:2989–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüssing S, House IG, Kearney CJ, et al. Efficient CRISPR/Cas9 gene editing in uncultured naive mouse T cells for in vivo studies. J Immunol 2020;204:2308–2315. [DOI] [PubMed] [Google Scholar]
- O'Donnell MP, Burne M, Daniels F, et al. Utility and limitations of serum creatinine as a measure of renal function in experimental renal ischemia-reperfusion injury. Transplantation 2002;73:1841–1844. [DOI] [PubMed] [Google Scholar]
- Ohl K, Tenbrock K. Oxidative stress in SLE T cells, is NRF2 really the target to treat? Front Immunol 2021;12:633845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechman KR, De Miguel C, Lund H, et al. Recovery from renal ischemia-reperfusion injury is associated with altered renal hemodynamics, blunted pressure natriuresis, and sodium-sensitive hypertension. Am J Physiol Regul Integr Comp Physiol 2009;297:R1358–R1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi LM, Stockler-Pinto MB, Leite M Jr, et al. Nrf2-keap1 system versus NF-κB: The good and the evil in chronic kidney disease? Biochimie 2012;94:2461–2466. [DOI] [PubMed] [Google Scholar]
- Rabb H, Daniels F, O'Donnell M, et al. Pathophysiological role of T lymphocytes in renal ischemia-reperfusion injury in mice. Am J Physiol Renal Physiol 2000;279:F525–F531. [DOI] [PubMed] [Google Scholar]
- Rabb H, Ramirez G, Saba SR, et al. Renal ischemic-reperfusion injury in L-selectin-deficient mice. Am J Physiol 1996;271:F408–F413. [DOI] [PubMed] [Google Scholar]
- Roman J, Rangasamy T, Guo J, et al. T-cell activation under hypoxic conditions enhances IFN-gamma secretion. Am J Respir Cell Mol Biol 2010;42:123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush BM, Bondi CD, Stocker SD, et al. Genetic or pharmacologic Nrf2 activation increases proteinuria in chronic kidney disease in mice. Kidney Int 2021;99:102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med 2018;215:985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvinkel P, Chertow GM, Devarajan P, et al. Chronic inflammation in chronic kidney disease progression: Role of Nrf2. Kidney Int Rep 2021;6:1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Seki S, Hiramoto K, et al. Hyperactivation of Nrf2 in early tubular development induces nephrogenic diabetes insipidus. Nat Commun 2017;8:14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RJ, Chartoumpekis DV, Rush BM, et al. Keap1 hypomorphism protects against ischemic and obstructive kidney disease. Sci Rep 2016;6:36185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardyn JD, Ponsford AH, Sanderson CM. Dissecting molecular cross-talk between Nrf2 and NF-kappaB response pathways. Biochem Soc Trans 2015;43:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger RH, Kurtcuoglu V, Scholz CC, et al. Frequently asked questions in hypoxia research. Hypoxia (Auckl) 2015;3:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willam C. HIF meets NF-κB signaling. Kidney Int 2014;85:232–234. [DOI] [PubMed] [Google Scholar]
- Yang SM, Ka SM, Hua KF, et al. Antroquinonol mitigates an accelerated and progressive IgA nephropathy model in mice by activating the Nrf2 pathway and inhibiting T cells and NLRP3 inflammasome. Free Radic Biol Med 2013;61:285–297. [DOI] [PubMed] [Google Scholar]
- Yokota N, Burne-Taney M, Racusen L, et al. Contrasting roles for STAT4 and STAT6 signal transduction pathways in murine renal ischemia-reperfusion injury. Am J Physiol Renal Physiol 2003;285:F319–F325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.