Abstract
Significance:
Cells depend on well-functioning mitochondria for essential processes such as energy production, redox signaling, coordination of metabolic pathways, and cofactor biosynthesis. Mitochondrial dysfunction, metabolic decline, and protein stress have been implicated in the etiology of multiple late-onset diseases, including various ataxias, diabetes, sarcopenia, neuromuscular disorders, and neurodegenerative diseases such as parkinsonism, amyotrophic lateral sclerosis, and glaucoma.
Recent Advances:
New evidence supports that increased energy metabolism protects neuron function during aging. Key energy metabolic enzymes, however, are susceptible to oxidative damage making it imperative that the mitochondrial proteome is protected. More than 40 different enzymes have been identified as important factors for guarding mitochondrial health and maintaining a dynamic pool of mitochondria.
Critical Issues:
Understanding shared mechanisms of age-related disorders of neurodegenerative diseases such as glaucoma, Alzheimer's disease, and Parkinson's disease is important for developing new therapies. Functional mitochondrial shape and dynamics rely on complex interactions between mitochondrial proteases and membrane proteins. Identifying the sequence of molecular events that lead to mitochondrial dysfunction and metabolic stress is a major challenge.
Future Directions:
A critical need exists for new strategies that reduce mitochondrial protein stress and promote mitochondrial dynamics in age-related neurological disorders. Discovering how mitochondria-associated degradation is related to proteostatic mechanisms in mitochondrial compartments may reveal new opportunities for therapeutic interventions. Also, little is known about how protein and membrane contacts in the inner and outer mitochondrial membrane are regulated, even though they are pivotal for mitochondrial architecture. Future work will need to delineate the molecular details of these processes.
Keywords: mitochondria, quality control, protein homeostasis, mitochondrial dynamics, UPRmt, ISR
Introduction
Mitochondria are complex and essential organelles that now have emerged as key metabolic and signaling hubs in a cell. In addition to their well-known energy conversion functions, mitochondria are recognized as critical sites for heme and iron–sulfur cofactor synthesis, calcium buffering, metabolic and redox signaling, and innate immune responses. Moreover, a plethora of cellular metabolic pathways is known to either converge on or emerge from the mitochondria. The metabolic requirement of oxygen for these pathways puts tremendous burden on mitochondria to maintain enzyme complexes of the electron transport chain (ETC) and adenosine triphosphate (ATP) synthase. Aberrant reactions of oxygen with redox complexes in the ETC can lead to reactive oxygen species (ROS) initiating a cascade of damage to the organelle. It is therefore not surprising that the functional integrity of mitochondria is critical for cellular physiology. This is especially true for postmitotic cells with high bioenergetic demand such as neurons, myotubes, and cardiomyocytes.
Mitochondria are highly dynamic organelles and, in many cell types, are organized into interconnected networks, whose shape and morphology support and modulate cellular metabolic responses to homeostatic challenges including nutrient deprivation, mitochondrial uncoupling, or hypoxic insults (Giacomello et al, 2020; Hackenbrock, 1966; Sprenger and Langer, 2019). Changes in mitochondrial network behavior, commonly referred to as fusion (network expansion) and fission (network fragmentation), lead to changes in metabolic and bioenergetic outputs of mitochondria. Such alterations are now recognized as important physiological contributors to cellular differentiation, “stemness” and senescent states, and cell death.
Mitochondria comprise 1000–1300 proteins of dual genetic origin, in which the vast majority of mitochondrial proteome is encoded by nuclear genome, synthesized in cytosol, and imported in the organelle (Morgenstern et al, 2021; Pagliarini et al, 2008; Rath et al, 2021; Schmidt et al, 2010). Many of these polypeptides are then stoichiometrically paired with a handful of mitochondria-encoded proteins to form the ETC complexes in the inner mitochondrial membrane (IM). The IM and mitochondrial matrix harbor the majority of mitochondrial proteome, and are the main sites for vital mitochondrial activities. A highly protein-rich and metastable environment in these mitochondrial subcompartments necessitates tight homeostatic control. Failure to fold, assemble, or rearrange protein complexes in these mitochondrial locales is associated with perturbations to protein homeostasis (proteostasis), increased oxidative damage, ion imbalance, and bioenergetic deficit.
Consequently, these impediments impinge on the fusion and fission balance of mitochondrial network, thereby leading to downstream physiological alterations in the cell (Higuchi-Sanabria et al, 2018).
The functional integrity of mitochondria is established and maintained through several interrelated mechanisms, many facets of which are highly conserved throughout the eukaryotes. Operating at both the molecular and organellar levels, these multilayered and overlapping mechanisms protect the organelle from the aforementioned homeostatic challenges to safeguard mitochondrial and cellular health. At the molecular level, mitochondrial welfare is surveyed and corrected by molecular machines localized outside and within the organelle. The former group includes several mechanisms converging on the ubiquitin–proteasomal system (UPS), and the latter group is represented by molecular chaperones and proteases localized to mitochondrial subcompartments, with the major fraction of these factors being integral to or associated with the IM. At the organellar level, mitochondrial health is protected by mechanisms encompassing mitochondrial fusion and fragmentation, selective autophagy of mitochondria (known as mitophagy), and interorganellar retrograde signaling.
In this study, we briefly discuss these mechanisms and their molecular links to mitochondrial form, proteostasis, function, and metabolism and attempt to review these molecular pathways in the context of glaucoma.
Mitochondrial Metabolism and Oxidative Stress
Energy pathways
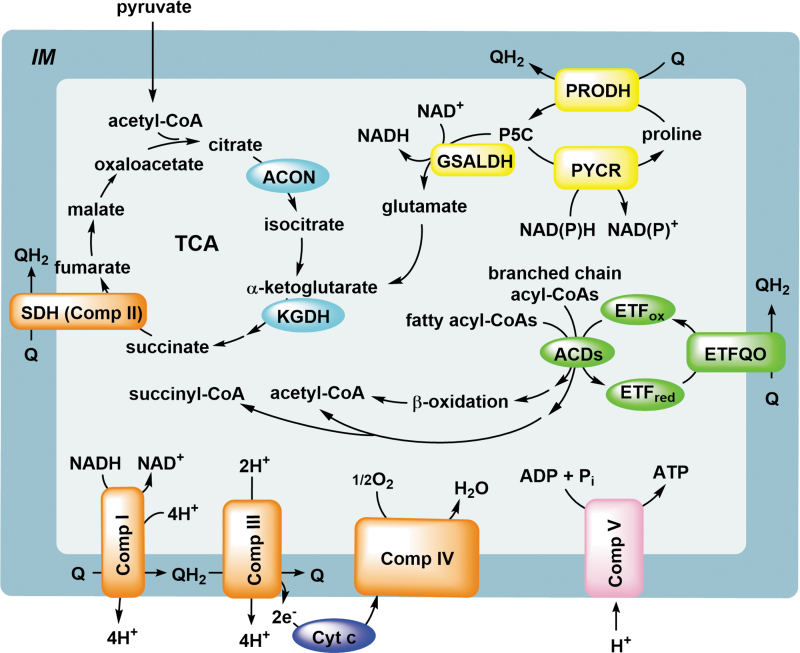
The mitochondrion is home to several metabolic pathways that are interconnected and integrate into the respiratory chain. The tricarboxylic acid (TCA) cycle, located in the matrix of mitochondria, serves as the central hub for connecting different metabolic pathways and provides the majority of ATP in the cell via reduced nicotinamide adenine dinucleotide (NADH). Acetyl-CoA generated via pyruvate and pyruvate dehydrogenase (PDH) and also fatty oxidation is the key feeder molecule into the TCA cycle with other metabolites also supplied to maintain cycle flux. Metabolites of the TCA cycle are kept properly balanced by linkages to glycolysis and gluconeogenesis, fatty acid oxidation and biosynthesis, amino acid metabolism, and nucleic acid metabolism.
NADH produced by the TCA cycle is oxidized by complex I (NADH dehydrogenase) of the mitochondrial ETC. Complex I catalyzes a hydride transfer from NADH to the flavin mononucleotide (FMN) and then funnels electrons to the ubiquinone (Q) reduction site via multiple Fe-S clusters (eight in mammals) thereby adding reduced ubiquinone (QH2) to the coenzyme Q (CoQ) pool of the IM (Baradaran et al, 2013; Fiedorczuk et al, 2016; Parey et al, 2020). QH2 serves as an electron carrier in the membrane by delivering electrons to complex III via the QH2 oxidation sites (Iwata et al, 1998).
Through a series of one-electron transfer steps involving cytochrome c, electrons eventually are taken up by cytochrome c oxidase (complex IV), which catalyzes the reduction of oxygen to water (Hartley et al, 2019; Tsukihara et al, 1996). Proton transfer (10 protons total) that is coupled to the different electron transport steps in complexes I, III, and IV generates a proton gradient between the IM space and the mitochondrial matrix. This gradient drives complex V (ATP synthetase) production of ATP, thus providing the needed energy to the cell.
Molecules that are able to support TCA cycle activity and/or provide reducing equivalents directly to the respiratory chain are critical for cellular metabolism and energy production. For instance, fatty acid oxidation and branched chain amino acids (leucine, isoleucine, and valine) impact TCA flux and directly provide reducing power to the respiratory chain. Fatty acid oxidation involves four successive enzymatic steps in each oxidative cycle that provide one molecule of acetyl-CoA and 2e− directly to the ubiquinone pool via the acyl-CoA dehydrogenase/electron transfer flavoprotein (ETF)/ETF ubiquinone oxidoreductase (ETFQO) complex (Zhang et al, 2006). Catabolism of branched-chain amino acids involves different CoA derivatives that are metabolized by their respective acyl-CoA dehydrogenases, which are coupled to ETF/ETFQO. The downstream products are acetyl-CoA and succinyl-CoA (Mann et al, 2021).
Similar to fatty acid oxidation and branched-chain amino acids, proline oxidation provides substrates for the TCA cycle and directly funnels electrons to the ubiquinone pool. One intermediate in particular, α-ketoglutarate, is replenished from glutamate, which is provided by the catabolism of proline, histidine, arginine, and glutamine. Figure 1 summarizes these pathways.
FIG. 1.
Metabolic pathways of the mitochondrion. The TCA cycle is shown with pyruvate, acetyl-CoA, and glutamate feeding into the cycle. The roles of complexes I, II, III, IV, and V in mitochondrial energy production are highlighted. Complexes I and II reduce ubiquinone (Q), which provides QH2 for the reduction of Cyt c by Complex III. The electron transfer events in the IM are coupled with proton transport out of the matrix to ultimately drive ATP production by complex V. Catabolism of fatty acids (and branched chain amino acids) and proline also leads to QH2 via the ETFQO and PRODH, respectively. ACD, acyl-CoA dehydrogenase; ACON, aconitase; ATP, adenosine triphosphate; Cyt c, cytochrome c; ETF, electron transfer flavoprotein; ETFQO, ETF ubiquinone oxidoreductase; GSALDH, l-glutamate-γ-semialdehyde dehydrogenase; IM, inner mitochondrial membrane; KGDH, α-ketoglutarate dehydrogenase; PRODH, proline dehydrogenase; PYCR, Δ1-pyrroline-5-carboxylate reductase; Q, ubiquinone; QH2, reduced ubiquinone; SDH, succinate dehydrogenase; TCA, tricarboxylic acid.
Reactive oxygen species
ROS are by-products of mitochondrial metabolism, a phenomenon that was shown 50 years ago using isolated mitochondria (Loschen et al, 1971). Complex I is a major contributor to superoxide (O2•−) formation, with the amount of ROS generated dependent on NADH/NAD+ (nicotinamide adenine dinucleotide) and the abundance of QH2 (Murphy, 2009). A reducing pool of quinone has been proposed to drive reverse electron transfer via complex I resulting in a burst of ROS during ischemia–reperfusion injury (Chouchani et al, 2016; Kim et al, 2018; Niatsetskaya et al, 2012). In addition, complex II has been shown to be a significant contributor to ROS both in the forward and reverse directions of electron flow, with again high levels of QH2 helping drive reactions in the reverse direction (Quinlan et al, 2012). Thus, enzymes that directly feed electrons into the quinone pool also contribute to ROS production by providing reducing equivalents that support reverse electron transfer.
These enzymes include those shown in Figure 1 (e.g., ETFQO and proline dehydrogenase), and others such as α-glycerophosphate dehydrogenase, dihydroorotate dehydrogenase, and hydrogen sulfide CoQ oxidoreductase.
Oxidative and proteostatic stress of mitochondrial enzymes
In considering the circumstance of proteostatic stress, it seems that some proteins may be more susceptible to oxidative modification than others. The different sensitivity can be due to the nature of the enzyme reaction and the cofactors and active site residues required to catalyze the reaction. A well-known example is aconitase, which does not catalyze an electron transfer reaction but contains an essential Fe-S cluster (Humphries et al, 2006). The cluster functions as a Lewis-acid in the rearrangement reaction of citrate to isocitrate. One of the Fe atoms in the cluster is reactive to hydrogen peroxide, the result of which is destruction of the Fe-S cluster and loss of aconitase activity (Humphries et al, 2006).
Thus, measurement of aconitase activity is used as a readout of oxidative stress levels. Another TCA cycle enzyme that is also sensitive to oxidative insults is succinate dehydrogenase (complex II), which contains a flavin and an Fe-S cluster and catalyzes the oxidation of succinate to fumarate (Wallace, 2005). Again, the Fe-S cluster is the target of oxidative damage, which leads to loss of enzyme activity.
The respiratory chain of course contains a number of Fe-S clusters in complex I, III, and IV. These complexes can also be damaged via loss of Fe-S clusters, but their reactivity toward oxidative molecules is somewhat protected by their location in membrane-bound proteins. Thus, these clusters are not necessarily considered to be the “first hit” under oxidative or proteostatic stress. By the time these complexes are being hit with oxidative damage, the integrity of the mitochondria is likely far gone and headed toward a path of mitophagy.
In addition to the destruction of metal cofactors, oxidative stress can lead to the modification of proteins that reduce the overall stability of the protein. Oxidative damage in proteins that is associated with aging and different pathologies can occur through different types of mechanisms. Oxidative carbonylation of protein occurs, namely, by direct oxidation of amino acid side chains of Arg, Lys, Pro, His, and Thr, but also can occur at the protein backbone (Mannaa and Hanisch, 2020). Oxidative carbonylation is a significant mechanism of irreversible protein damage with the total amount of protein carbonylation in a cell or tissue used as a marker of oxidative damage (Mannaa and Hanisch, 2020). Indeed, it is well established that the carbonyl content of proteins generally increases with age in different species resulting in lower protein stability in old age (de Graff et al, 2016).
Oxidative modifications of proteins occur randomly, but proteins with a high surface charge may be destabilized more easily (de Graff et al, 2016). In addition to carbonylation, proteins can also be modified by lipid peroxidation products (e.g., 4-hydroxyl-2-nonenal [HNE] and malondialdehyde) and by glycoxidation resulting in advanced glycation end-products (Hamon et al, 2020).
An interesting study of senescent cells identified enzymes that were oxidized in energy metabolism relative to matching young cells. In senescent fibroblasts, several mitochondrial enzymes were identified (Ahmed et al, 2010), including l-glutamate-γ-semialdehyde dehydrogenase (GSALDH or ALDH4A1), which is responsible for catalyzing the second step in proline catabolism of the NAD+-dependent oxidation of Δ1-pyrroline-5-carboxylate (P5C) to glutamate (Hamon et al, 2020). Lack of GSALDH causes accumulation of P5C and proline, a condition known as type II hyperprolinemia. P5C is a highly reactive metabolic intermediate prone to damage proteins (Lerma-Ortiz et al, 2016). Thus, oxidative damage of GSALDH would not only limit glutamate production, but also potentially lead to increased levels of reactive P5C contributing to a cascade effect of damage. Other mitochondrial enzymes found to be modified in senescent fibroblasts included malate dehydrogenase, α-ketoglutarate dehydrogenase (KGDH), succinate dehydrogenase, and ornithine aminotransferase (Ahmed et al, 2010; Hamon et al, 2020).
Oxidative stress and disease
The importance of various pathways in energy metabolism of course is dependent on the context of the cellular environment and metabolic regulation. Some cells are content with glycolytic survival under conditions of limiting oxygen, whereas other cells are highly dependent on mitochondrion oxidative metabolism and oxygen levels. In addition, the fuel that is used to drive mitochondrial metabolism will be dependent on the nutritional sources available to the cell. For example, it is interesting to compare the metabolic preferences of skeletal muscle and the brain. The main fuel for skeletal muscle is glucose, with robust anaerobic and aerobic glycolytic metabolism. Under poor nutrient conditions, muscle cells are able to mobilize amino acids via proteolysis to help with energy demands. In the brain, demand for acetyl-CoA by the TCA cycle is met by glucose and ketones.
In age-related neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, Huntington disease, frontotemporal dementia, and amyotrophic lateral sclerosis, it is thought that disruption of brain energy metabolism is a key contributor to disease onset (Cunnane et al, 2020). Various redox proteomic studies have identified critical energy metabolic enzymes to be susceptible to damage leading to inactivation and destabilization. For example, malondialdehyde was shown to decrease the activities of PDH, KGDH, and respiratory complex I and II, and complex V in rat brain mitochondria (Long et al, 2009). PDH and KGDH are also inactivated by HNE in rat heart mitochondria (Humphries and Szweda, 1998; Humphries et al, 2006). Another enzyme found to be modified by oxidative stress is malate dehydrogenase.
In a large proteomic study of adult brain, a higher abundance of mitochondrial enzymes, such as PDH complex (PDHA1), aconitase (ACO2), nicotinamide nucleotide transhydrogenase (NNT), several subunits of complex I, and a subunit of complex III (UQCRFS1) (Wingo et al, 2019), were associated with higher cognitive stability in older adult brains. Thus, indicating that maintenance of energy metabolism is a key factor in protecting against the declining function of the brain during aging. Another target of oxidative inactivation in the brain is glutamine synthetase, which leads to glutamate accumulation and potential neurotoxicity (Oliver et al, 1990).
Proteostatic Mechanisms in Mitochondria
Outer mitochondrial membrane
Mechanisms that protect mitochondrial enzymes and proteins are critical for maintaining mitochondrial integrity. As indicated above, the vast majority of mitochondrial proteins are encoded by the nuclear genome and produced in the cytosol as precursor proteins that are subsequently imported in the mitochondria. Virtually all mitochondria-destined polypeptides are imported in an unfolded state through the protein translocase of the outer mitochondrial membrane (TOM) complex in the outer mitochondrial membrane (OM); a large fraction of proteins subsequently undergo translocation via the two translocases of the inner mitochondrial membrane (TIM) complexes (Neupert and Herrmann, 2007; Pfanner et al, 2019). To avoid undesired accumulation of unfolded mitochondrial precursor proteins in the cytosol, the synthesis and import thereof are tightly coordinated (Bykov et al, 2020).
In many cases, mitochondrial precursor proteins are synthesized by cytosolic ribosomes that are localized on the OM, thereby allowing for nearly simultaneous import of nascent polypeptides in the mitochondria (Gold et al, 2017; Williams et al, 2014). Furthermore, the synthesis of cytosolic and mitochondrial proteins appears to occur in a concerted manner (Couvillion et al, 2016), likely to facilitate stoichiometric pairing of imported and mitochondria-borne polypeptides. In addition to tightly coordinated synthesis and import, cytosol-borne mitochondrial precursor proteins are chaperoned by the cytosolic molecular chaperones of the Hsp70- and Hsp90-family to ensure their import-competent state (Bykov et al, 2020; Fan et al, 2006; Opalinski et al, 2018; Young et al, 2003; Zara et al, 2009).
Mitochondrial protein import, however, represents one of the challenges to mitochondrial proteostasis, as translocation stalling and accumulation of unimported polypeptide on the mitochondrial surface—for example, due to reduced mitochondrial membrane potential required for vectorial transfer of newly synthesized polypeptide through the protein translocation pore of the TOM complex. A result is depleted pools of mitochondrial enzymes and transporters with short half-lives (Bomba-Warczak et al, 2021), thereby impinging on the organelle's bioenergetic and metabolic capacity. In addition, accumulated polypeptides can prematurely fold or misfold and further impact the functional integrity of mitochondria and other organelles in the cell (Liu et al, 2019). In extreme cases, mitochondrial precursor accumulation can result in proteostatic stress known as mitochondrial precursor overaccumulation stress, mPOS, which can trigger an unfolded protein response activated by mistargeting of proteins (UPRam) subsequently leading to cellular damage, and—in most extreme cases—death (Wang and Chen, 2015; Wrobel et al, 2015).
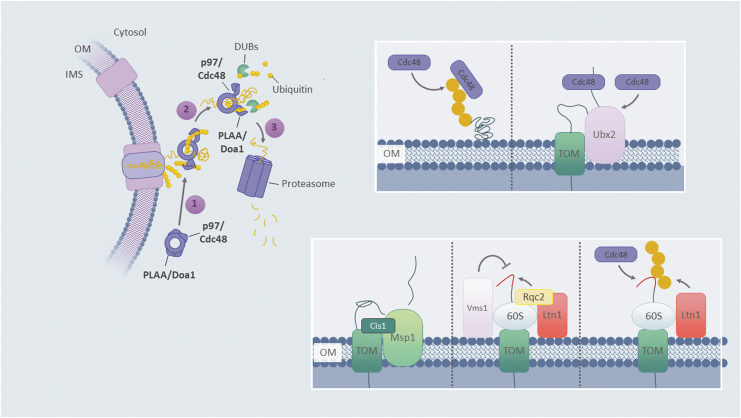
To avoid such unfavorable scenarios, a series of molecular machines cooperate with the UPS machinery to facilitate proteolytic turnover of the OM-associated precursor proteins (Fig. 2). In addition to ubiquitylation and UPS-mediated removal of proteins en route to mitochondria, a more specialized conserved mechanism, termed mitochondria-associated degradation (MAD; also known as OMMAD), is in place to ensure proteostasis at the OM. MAD comprises AAA+ ATPase Cdc48/p97/VCP and its substrate adapters such as dominant optic atrophy mitofusins (Doa) 1/PLAA—the components shared with other UPS-interceded degradation pathways such as ERAD. These two factors act in concert to facilitate the removal of stalled polypeptides by UPS (Heo et al, 2010; Wu et al, 2016; Xu et al, 2011).
FIG. 2.
Quality control factors external to mitochondria. A cartoon summarizing ubiquitin–proteasome system-mediated quality control events at the OM, known as MAD. General steps include protein ubiquitylation and binding of a complex containing VCP/p97/Cdc48 AAA-ATPase and its adaptor protein PLAA/Doa1 to mitochondrial substate (1), followed by protein extraction (2), substrate deubiquitylation by DUBs (3), and degradation by the proteasome. Insets show specific subtypes of MAD that thus far have been mainly characterized in yeast but are likely to be conserved in eukaryotes. These degradation scenarios include direct ubiquitylation of OM-associated substrates, followed by Cdc48-assisted degradation (upper left inset); extraction of substrates crossing the OM import pore (TOM complex) with the help of bridging factors such as Ubx2 (upper right inset) or Cis1 and OM-anchored AAA-ATPase Msp1 (lower left inset); or specific modification and protection/degradation of nascent mitochondria-destined polypeptides translated on mitochondria-associated ribosomes in the vicinity of the TOM machinery, via the components of ribosomal quality control pathway: protein Rqc2, peptidyl-tRNA hydrolase Vms1, and E3 ubiquitin ligase Ltn1 (lower middle and right insets). See Outer Mitochondrial Membrane section for additional details. AAA, ATPases associated with diverse cellular activities; Doa, dominant optic atrophy mitofusins; DUBs, deubiquitylases; MAD, mitochondria-associated degradation; OM, outer mitochondrial membrane; TOM, translocase of the outer mitochondrial membrane.
Interestingly, the results of a recent study by Liao et al (2020) suggest that MAD could extend beyond the OM and may also be responsible for the degradation of oxidatively damaged mitochondrial intermembrane space (IMS) and matrix proteins. In addition, variations of MAD include the ribosomal quality control of mitochondrial polypeptides (mitoRQC) (Izawa et al, 2017; Verma et al, 2013; Zurita Rendon et al, 2018), and p97-mediated extraction of polypeptides halted within the TOM translocase, a process referred to as TAD (Martensson et al, 2019). In addition to p97, the mitoRQC mechanism involves peptidyl-tRNA hydrolase Vms1/ANKZF1 and E3 ubiquitin ligases Rkr1/LTN1 and Rqc2/NEMF. These factors interact with the OM-associated cytosolic ribosomes to facilitate import and expulsion of the stalled nascent polypeptides (Izawa et al, 2017; Zurita Rendon et al, 2018).
The mitoRQC pathway appears to work on polypeptides that are imported into the mitochondria in a cotranslational manner. Stalling of these polypeptides results in so-called CAT-tailing—the Rqc2-mediated attachment of alanyl/threonyl sequences on the C-termini of stalled polypeptides on 60S ribosomes; such modification promotes aggregation of said polypeptides rendering them inaccessible to UPS (Shen et al, 2015). To counteract this process, Vms1 is recruited to the 60S ribosome to displace Rqc2 and curtail CAT-tailing, thereby either stimulating their translocation into mitochondria or allowing for Rkr1-mediated ubiquitylation and p97-mediated disaggregation of the stalled polypeptides and their subsequent removal by UPS (Izawa et al, 2017; Verma et al, 2013; Zurita Rendon et al, 2018).
Finally, another AAA+ ATPase, Msp1/ATAD1, has been shown to represent yet another facet of MAD (Matsumoto et al, 2019). Distributed in the OM as well as on peroxisomes, Msp1 functions as an ATP-fueled extraction factor facilitating degradation of mistargeted polypeptides such as tail-anchored proteins, thereby ensuring their proper subcellular localization (Chen et al, 2014; Matsumoto et al, 2019; Okreglak and Walter, 2014). In addition, Msp1 has been postulated to participate in the clearance mechanism for mitochondrial precursor proteins with bipartite targeting sequences to prevent their overaccumulation on the OM (Weidberg and Amon, 2018). Such a mechanism termed mitoCPR (for the mitochondrial compromised protein response) includes a protein Cis1, whose expression is increased in response to precursor overaccumulation at the translocation pore; Cis1 then binds to accumulated precursor polypeptides and helps to recruit Msp1 for their retrotranslocation, and the subsequent removal by UPS (Weidberg and Amon, 2018).
At present, however, it is unclear if such a mechanism is fully conserved as no Cis1 orthologs have been identified outside of fungi. Several other IMS-localized proteins have been reported to be retrotranslocated to the cytosol via the TOM40 import pore (Bragoszewski et al, 2015); however, whether this process involves Msp1 and/or Cis1 has not been investigated. Going forward, it will be interesting to determine if and how these various MAD-related mechanisms act in concert to mediate the OM proteostasis, and to examine their conservation in higher eukaryotes.
Inner mitochondrial compartments
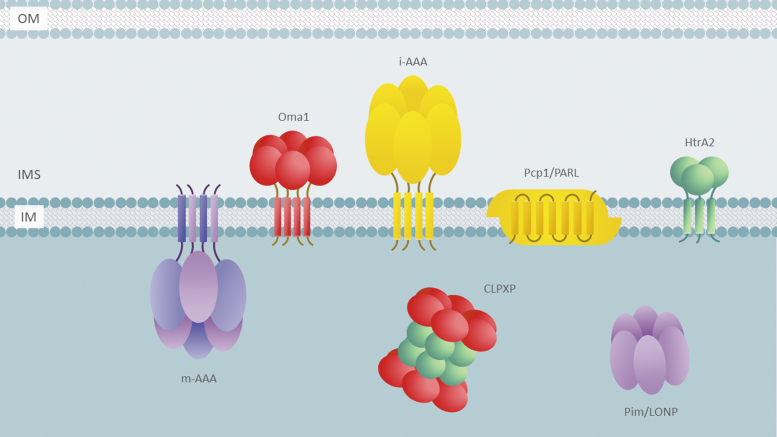
Mitochondrial proteins are not uniformly distributed across the organelle's compartments. The IM and mitochondrial matrix harbor the majority of the mitochondrial proteome, with the IM being among the most protein-rich compartments in a cell. Such a metastable proteome creates a challenging environment with a high probability of protein misfolding, malfunction, and aggregation. Failure to insert, fold, assemble, or rearrange protein complexes in these mitochondrial compartments—which happen to be the site of many vital mitochondrial activities—is associated with altered proteostasis, increased oxidative damage, ion dyshomeostasis, and bioenergetic deficit. Naturally, such homeostatic challenges necessitate a tight control of protein quality in mitochondria. Although some reports on ubiquitylation of mitochondrial proteins exist (Lavie et al, 2018; Liao et al, 2020; Sulkshane et al, 2020), it is generally believed that once within the organelle, mitochondrial proteins become largely inaccessible to UPS. Therefore, another molecular surveillance mechanism is required to ensure the quality of mitochondrial proteins within the organelle. Indeed, mitochondria are equipped with a variety of conserved molecular chaperones and proteases that mediate folding, degradation, or controlled proteolysis of mitochondrial polypeptides (Bohovych et al, 2015a; Deshwal et al, 2020) (Fig. 3). This diverse group of quality control factors encompasses more than 40 different enzymes, with nearly half of these being bona fide proteolytic enzymes (Bohovych et al, 2015a; Deshwal et al, 2020; Quiros et al, 2015). Table 1 provides a list of known mitochondrial proteases.
FIG. 3.
Mitochondrial quality control proteases. Schematic depiction of key intrinsic mitochondrial quality control proteases in the IM and the matrix compartments. Some of the factors are not depicted for simplicity. IMS, mitochondrial intermembrane space.
Table 1.
Intrinsic Human Mitochondrial Proteases
| Protease | Yeast ortholog | Category | Class | Mitochondrial localization | Function |
|---|---|---|---|---|---|
| CLPP | — | ATP-dependent | Serine | Matrix | PQC, mitochondrial biogenesis |
| LONP1 | Pim1 | ATP-dependent | Serine | Matrix | PQC, mitochondrial biogenesis |
| SPG7 (m-AAA) | Yta10 | ATP-dependent | Metallo | IM/Matrix | PQC, mitochondrial biogenesis |
| AFG3L2 (m-AAA) | Yta12 | ATP-dependent | Metallo | IM/Matrix | PQC, mitochondrial biogenesis |
| YME1L (i-AAA) | Yme1 | ATP-dependent | Metallo | IM/IMS | PQC, mitochondrial biogenesis |
| PARL | Pcp1 | ATP-independent processing | Serine | IM | Mitochondrial biogenesis, mitophagy, apoptosis |
| OMA1 | Oma1 | ATP-independent processing | Metallo | IM/IMS | PQC, mitochondrial biogenesis, dynamics, mitophagy, apoptosis |
| ATP23 | Atp23 | ATP-independent processing | Metallo | IMS | PQC, mitochondrial biogenesis |
| IMMP1L | Imp1 | ATP-independent processing | Serine | IM/IMS | Mitochondrial biogenesis |
| IMMP2L | Imp2 | ATP-independent processing | Serine | IM/IMS | Mitochondrial biogenesis |
| PMPCB | Mas1 | ATP-independent processing | Metallo | Matrix | Mitochondrial biogenesis, protein import |
| METAP1D | Map1 | ATP-independent processing | Metallo | Matrix | Mitochondrial biogenesis, protein import/activation |
| OSGEPL1 | Qri7 | ATP-independent processing | Metallo | Matrix | Mitochondrial biogenesis |
| XPNPEP3 | Icp55 | ATP-independent processing | Metallo | Matrix | Mitochondrial biogenesis, protein import/activation |
| MIP | Oct1 | ATP-independent processing | Metallo | Matrix | Mitochondrial biogenesis, protein import/activation |
| MEP | Prd1 | ATP-independent oligopeptidase | Metallo | IMS | PQC |
| PITRIM1 | Mop112 | ATP-independent oligopeptidase | Metallo | Matrix | PQC |
| HTRA2 | — | ATP-independent | Serine | IMS | PQC, mitophagy, apoptosis |
| LACTB | — | ATP-independent | Serine | IMS | Mitochondrial biogenesis |
| USP30 | Ubp16 | ATP-independent | Cysteine | OM | Mitochondrial dynamics, mitophagy |
ATP, adenosine triphosphate; i-AAA, intermembrane space-facing AAA+ protease; IM, inner mitochondrial membrane; IMS, mitochondrial intermembrane space; LONP1 Lon peptidase 1; m-AAA, matrix-facing AAA+ protease; Metallo, metallopeptidase; OM, outer mitochondrial membrane; PQC, protein quality control.
While the function and role of some of these enzymes are only beginning to emerge, others have been relatively well-characterized. Mitochondrial proteases can be broadly divided into ATP-dependent and ATP-independent enzymes. For example, several conserved protein quality control modules survey and correct the IM and matrix subproteomes, including ATP-fueled enzymatic complexes: the i-AAA (for the intermembrane space-facing AAA+ protease), the m-AAA (for the matrix-facing AAA+ protease), Lon peptidase 1 (LONP1), and chaperone subcomplex attachment (CLPXP) (Deshwal et al, 2020; Glynn, 2017; Quiros et al, 2015; Steele and Glynn, 2019). Additional key factors include ATP-independent proteases OMA1 and PARL (Deshwal et al, 2020; Levytskyy et al, 2017).
Growing evidence indicates functional versatility and well-orchestrated actions of these proteolytic machineries that appear to be functionally interconnected and function beyond simple quality control (Alavi, 2021; Deshwal et al, 2020; Levytskyy et al, 2017). Such an integration and partial functional redundancy of these factors likely exist to ensure the flexibility and resilience of mitochondrial protein quality control mechanisms.
The matrix-residing LONP and CLPXP are serine proteases equipped with the protein unfolding/translocating AAA+ module—which is either integrated with the same polypeptide (LONP1) or present as a CLPXP. LONP1, a homo-hexameric complex, in which the protease units are organized into a trimer of dimers (Shin et al, 2021; Shin et al, 2020), has been shown to act as a versatile regulator of mitochondrial proteostasis (Venkatesh et al, 2012). For example, mammalian LONP1 plays a role in mitochondrial genome maintenance and expression, achieved through selective degradation of the mitochondrial DNA (mtDNA)-associated factor TFAM (Lu et al, 2013; Matsushima et al, 2010), and proteolytic processing of mitochondrial gene expression-related factors SLIRP, MTERFD3, and FASTKD2 (Bota and Davies, 2016; Lagouge et al, 2015; Zurita Rendon and Shoubridge, 2018).
In addition, this protease is known to regulate mitochondrial bioenergetics and metabolism through proteolytic removal of normoxic form of the COX4 subunit of the respiratory complex IV under low oxygen conditions, thereby mediating hypoxic retuning of mitochondrial ETC, and by degrading oxidatively damaged TCA cycle enzyme aconitase, respectively (Bota and Davies, 2002; Fukuda et al, 2007; Sepuri et al, 2017).
The highly conserved proteolytic complex CLPXP comprised the 14 copies of serine caseinolytic protease CLPP assembled into a double-ringed barrel-like structure capped at each end by an AAA-ATPase chaperone CLPX (Gatsogiannis et al, 2019; Kang et al, 2005; Sauer et al, 2022). This large, proteasome-like molecule resides in the mitochondrial matrix. This enzymatic complex is responsible for degrading various excess proteins, including ribosome-stalled translation polypeptides (Hofsetz et al, 2020; Szczepanowska and Trifunovic, 2021). While the exact role of mitochondrial CLPXP is not yet fully understood, recent studies implicated it as a critical regulator of protein homeostasis in the organelle, especially complex I and complex II (Nguyen et al, 2022; Seo et al, 2016; Szczepanowska et al, 2020), underscoring its importance for mitochondrial metabolism.
Recent studies have also demonstrated the importance of the CLPXP machinery in mitochondrial protein synthesis through its role in the proteolytic control of mitochondrial ribosome assembly regulator ERAL1 (Szczepanowska et al, 2016). In addition, the nonproteolytic CLPX chaperone moiety of CLPXP has been shown to facilitate mitochondrial heme metabolism and erythropoiesis via activation of 5-aminolevulinic acid synthase (ALAS), a key enzyme in heme biosynthetic pathway catalyzing the condensation of glycine and succinyl CoA to form 5-aminolevulinic acid (Kardon et al, 2020; Kardon et al, 2015).
The IM-anchored m-AAA and i-AAA proteases are paralogous zinc metalloproteases equipped with an integrated AAA+ module facing the mitochondrial matrix and the IMS, respectively (Deshwal et al, 2020; Leonhard et al, 1996; Levytskyy et al, 2017). Both m-AAA and i-AAA proteases combine unfoldase/protein extraction function with subsequent proteolytic processing, although in certain instances, these enzymes have also been shown to act as bona fide molecular chaperones (Arlt et al, 1996; Leonhard et al, 1999; Schreiner et al, 2012). In mammalian mitochondria, m-AAA protease—a conserved hexameric complex exists either as a homo-hexamer composed of six identical AFG3L2 subunits or a hetero-hexametric complex encompassing several copies of AFG3L2 and its paralogous subunit SPG7/paraplegin (Atorino et al, 2003; Koppen et al, 2007; Puchades et al, 2019).
Of note, the latter molecular architecture appears to be exclusive for the yeast m-AAA, in which the enzyme comprised the Yta10 and Yta12 subunits—the orthologs of SPG7 and AFG3L2, respectively (Arlt et al, 1996; Leonhard et al, 1996). In human mitochondria, the SPG7 does not appear to form homo-oligomeric assemblies, most likely because its maturation is dependent on the proteolytic activity of AFG3L2 (Koppen et al, 2009). Interestingly, murine mitochondria contain yet another variant of the m-AAA complex harboring a close paralog of AFG3L2, Afg3L1, that morphed into a pseudogene in humans (Koppen et al, 2007; Kremmidiotis et al, 2001). The m-AAA protease is a multifaceted enzyme responsible for proteolytic and chaperoning functions pertinent to the maturation and quality control of membrane and peripheral proteins on the matrix side of the IM; these functions are critical to mitochondrial physiology (Arlt et al, 1996; Glynn, 2017).
For example, in addition to its role in the degradation of OXPHOS subunits, the m-AAA is crucial for mtDNA stability and protein synthesis through its role in the biogenesis of the bL32m subunit of the mitochondrial ribosome (Bonn et al, 2011; Nolden et al, 2005). Likewise, the enzyme plays a major role in the maturation of the regulatory subunit EMRE of the mitochondrial calcium uniporter complex—a key regulator of mitochondrial calcium homeostasis, which is also linked to the organelle's metabolic activity via calcium-dependent activity of PDH (Hurst et al, 2019; Konig et al, 2016; Patron et al, 2018). Not surprisingly, this function is vital for the well-being and survival of postmitotic cells.
The i-AAA complex exists exclusively as a homo-oligomer that comprised six copies of the Yme1/YME1L protease (Puchades et al, 2017; Shi et al, 2016). The i-AAA is believed to be the main factor mediating degradation of the IMS-facing IM polypeptides such as OXPHOS subunits, and other IMS-resident proteins (Baker et al, 2012; Schreiner et al, 2012; Stiburek et al, 2012).
The i-AAA protease is implicated in a variety of critical processes, including the regulation of the mitochondrial network through controlled proteolysis of the membrane-shaping GTPase OPA1, which is discussed later, and maintenance of IM proteins and lipids via the proteolytic turnover of the IM translocase TIM23-related proteins TIMM17 and ROMO1, and phosphatidic acid-shuttling protein Ups1/PRELID (Anand et al, 2014; Consolato et al, 2018; Ehses et al, 2009; MacVicar et al, 2019; Ohba et al, 2020; Potting et al, 2013; Potting et al, 2010; Rainbolt et al, 2013; Richter et al, 2019; Richter et al, 2015; Wai et al, 2015).
The ATP-independent proteases remain less characterized. The best-studied enzymes in this diverse group are the rhomboid protease Pcp1/PARL, and metallopeptidase Oma1. PARL is a serine intramembrane protease with its serine-histidine catalytic dyad site buried within the IM (Ha, 2009; Quiros et al, 2015). The substrate repertoire of PARL remains somewhat elusive, although several of its substrates have been identified. These include serine protease HTRA2 (Chao et al, 2008), lipid transport-related protein Stard7 (Yang et al, 2017), apoptotic factor SMAC/Diablo (Saita et al, 2017), and—perhaps most notably—the serine/threonine ubiquitin kinase PINK1 (Deas et al, 2011; Jin et al, 2010), and serine/threonine phosphatase PGAM5 (Sekine et al, 2012; Wai et al, 2016).
Of note, PARL is also a remarkable example of an evolutionarily divergence, in which the enzyme's function in higher eukaryotes is shifted away from processing of the IM GTPase Mgm1 (yeast ortholog of OPA1); instead, this process is mediated through a well-tuned action of the YME1L and OMA1 proteases (Anand et al, 2014; MacVicar and Langer, 2016; Wang et al, 2021).
OMA1 is an ATP-independent zinc metallopeptidase, originally identified as a backup protease for the m-AAA proteolytic complex (Kaser et al, 2003). However, subsequent studies on the yeast and mammalian enzyme established OMA1 as a key stress-activated protease in the IMM (Bohovych et al, 2016; Bohovych et al, 2014; Jiang et al, 2014; MacVicar and Langer, 2016; Rainbolt et al, 2016; Richter et al, 2015). Oma1 exists as a homo-oligomeric complex and is largely dormant under normal physiological conditions. However, the enzyme becomes rapidly activated when mitochondrial homeostasis is challenged; such a perturbation can be caused by a variety of homeostatic insults (Baker et al, 2014; Bohovych et al, 2014; Khalimonchuk et al, 2012; Murata et al, 2020; Zhang et al, 2014). Stress activation of Oma1 appears to involve conformational changes within its oligomeric complex (Bohovych et al, 2014), but the exact mechanisms through which Oma1 senses stress and is activated remain elusive.
Studies in mammalian mitochondria established that OMA1 activation in mammalian cells is associated with autocatalytic processing of the enzyme (Baker et al, 2014; Rainbolt et al, 2015; Zhang et al, 2014), which has been proposed to be a measure for the spatiotemporal control of OMA1 activity (Baker et al, 2014); this, however, is not the case for the yeast enzyme (Bohovych et al, 2014). In addition, the mammalian OMA1 appears to act in concert with the i-AAA protease, and the two enzymes were shown to exhibit condition-dependent reciprocity in neuronal cells (Rainbolt et al, 2016). Known substrates of Oma1 include mutant variants of IM translocase Oxa1, phosphatidyl serine decarboxylase Psd1 (Ogunbona et al, 2017), and cytochrome c oxidase subunit Cox1 in yeast (Bohovych et al, 2014; Khalimonchuk et al, 2012).
In mammals, OMA1 is involved in the processing of a different cohort of IMM proteins including respiratory complex III assembly factor UQCC3 (Desmurs et al, 2015), integrated stress response (ISR) activator DELE1 (Fessler et al, 2020; Guo et al, 2020), and GTPase OPA1 (Anand et al, 2014; Ehses et al, 2009; Head et al, 2009), which is discussed below. Recent studies shown that mammalian OMA1 can also coordinate its activity with PARL, thereby contributing to the proteolytic processing of PGAM5 phosphatase (Wai et al, 2016), and a mutant variant of ubiquitin kinase PINK1 (Sekine et al, 2019).
Mechanisms of Mitochondrial Form and Shape
Several mitochondrial stress-mitigating mechanisms—both at the molecular and organellar quality control levels—are coupled to one another through the action of mitochondrial proteases. Indeed, protein quality control enzymes have emerged as important regulators of many mitochondrial processes. In this study, we mainly focus on mitochondrial network dynamics, one of the central and perhaps most understood of such processes.
Mitochondrial network dynamics
Mitochondrial morphology and organization vary across cell types, but one common theme appears to be their dynamic organization into tubular networks, which can be further elongated through the process known as mitochondrial fusion or partitioned via the mechanism commonly referred to as mitochondrial division (also known as fission) (Giacomello et al, 2020; Labbe et al, 2014; Pernas and Scorrano, 2016). These opposing effects are mediated by conserved IM- and OM-associated dynamin-like GTP hydrolases (GTPases), with a membrane-tethering activity, that function in concert. Of note, however, the fusion and fission processes at the IM and OM are believed to be highly coordinated but physically separate membrane remodeling events (Malka et al, 2005), although this view has been recently challenged (Kondadi et al, 2020a).
The molecular mechanisms behind mitochondrial fusion and fission have been extensively studied for the past two and a half decades (Labbe et al, 2014; Pernas and Scorrano, 2016) and are beyond the scope of this review. Briefly, and relevant to the present review, the main fusion factors for the OM are mitofusins—the semiredundant mitofusin 1 (MFN1) and mitofusin 2 (MFN2) in mammals (Chen et al, 2003; Giacomello et al, 2020), and the single yeast ortholog thereof, Fzo1 (Westermann, 2010). Of note, the MFN2 has also been shown to contribute to the formation of endoplasmic reticulum (ER)-mitochondria contacts, although the protein's exact role in this process is debated (de Brito and Scorrano, 2008; Filadi et al, 2017; Naon et al, 2017). The profusion activity of these proteins is countered by the dynamin GTPase Dnm1/dynamin-related protein 1 (DRP1) (Giacomello et al, 2020; Labbe et al, 2014; Labrousse et al, 1999).
The OM-anchored mitofusins have their active domains exposed to the cytosol and therefore are easily accessible to the UPS, which can modulate these molecules in either a degradative or nondegradative way (Dietz et al, 2019; Kraus et al, 2021). For example, MFN1 and MFN2 are ubiquitylated by the PINK1-dependent E3 ubiquitin ligase Parkin in depolarized mitochondria (Chen and Dorn, 2013; Gegg et al, 2010) to be unfolded and removed by the proteasome, thus resulting in detachment of defective mitochondria from the ER and their further segregation (McLelland et al, 2018). On the contrary, MFN2 oligomerization has been shown to be dependent on the GTPase's ubiquitylation by another ubiquitin ligase, MARCH5 (also known as MITOL) (Nagashima et al, 2014; Sugiura et al, 2013).
Of note, such a modification has been shown to promote ubiquitylation of other OMM proteins such as VDAC and Miro, thereby further facilitating the removal of depolarized mitochondria via mitophagy (McLelland et al, 2018).
Unlike the mitofusins, DRP1 is not anchored in the OM, but is recruited to the mitochondrial surface with the help of OM-associated adapter proteins: mitochondrial fission factor MFF, and MiD49 and MiD51 (Gandre-Babbe and van der Bliek, 2008; Loson et al, 2013; Palmer et al, 2011). In yeast, Dnm1 recruitment to the mitochondrial surface is mediated by the adapter proteins Caf4 and Mdv1 (Griffin et al, 2005; Tieu et al, 2002). Another conserved factor, FIS1, has been postulated to facilitate DRP1 association with the OM although its exact role in mitochondrial fission remains to be clarified (Koirala et al, 2013; Loson et al, 2013; Osellame et al, 2016; Yu et al, 2019). Post-translational modifications of these factors are vast (Elgass et al, 2013; Kraus et al, 2021), and provide a powerful regulatory handle to control mitochondrial fission.
Of relevance to the present review, ubiquitylation of DRP1 and FIS1 by MARCH5 has been shown to play a role in the regulation of mitochondrial fission (Dietz et al, 2019; Nagashima et al, 2014; Sugiura et al, 2013). In line with this notion, loss of MARCH5 impairs mitochondrial dynamics, leading to hyperfusion of mitochondrial network, which can be mitigated by DRP1 overexpression (Karbowski et al, 2007; Park et al, 2010).
In the IM, the GTPase OPA1 (Mgm1 in yeast) is one of the central elements in the regulation of mitochondrial fusion and fragmentation (Faelber et al, 2019; Yan et al, 2020). This process has been extensively studied in mammalian cell models. Under normal physiological conditions, OPA1 exists as a roughly balanced mix of long, IMM-anchored form (L-OPA1) and short, soluble variant (S-OPA1). The L-OPA1 variant can exist in several splice variants harboring specific cleavage sites—a well-defined S1 and S2 (Dietz et al, 2019; MacVicar and Langer, 2016) as well as recently described S3 site (Wang et al, 2021). Processing at the S2 and S3 sites occurs under basal conditions and is mediated by the i-AAA protease, resulting in acertain amount of S-OPA1 that is in equilibrium with S2/S3-less L-OPA1 (Dietz et al, 2019; MacVicar and Langer, 2016; Wang et al, 2021).
The OMA1 protease has emerged as a key controller of mitochondrial morphology and metabolic activity via specific processing of L-OPA1 at the S1 site in response to changes in metabolic demands or homeostatic insults (Ehses et al, 2009; Head et al, 2009; MacVicar and Langer, 2016). As all the L-OPA1 variants contain the S1 site, rapid proteolytic conversion of this variant by OMA1 results in accumulation of S-OPA1, which promotes IM remodeling in coordination with the OMM constriction, and subsequently, fragmentation of the mitochondrial network—a critical event required for multiple downstream mechanisms such as apoptosis or mitophagy (Anand et al, 2014; Frezza et al, 2006; Lee et al, 2017; Patten et al, 2014; Rambold et al, 2011; Yamaguchi et al, 2008).
Consistently, preservation of the unprocessed OPA1 variant through its overexpression or OMA1 depletion can stabilize the mitochondrial network, maintain normal cristae structure, and exert antiapoptotic effects (Acin-Perez et al, 2018; Anand et al, 2014; Quiros et al, 2012; Varanita et al, 2015).
Why do mitochondria fuse and divide? Elongation and partitioning of the mitochondrial network are believed to be important for the organellar and cellular physiology. Mitochondrial fusion appears to be crucial for the organelle's metabolic activity, ATP synthesis, and mtDNA maintenance (Chen et al, 2010; Elachouri et al, 2011). Furthermore, mitochondrial network elongation is believed to have a protective effect and aid cells in coping with various homeostatic challenges such as nutrient deprivation and hypoxia (Gomes et al, 2011; Khacho et al, 2014; Rambold et al, 2011; Tondera et al, 2009). Indeed, extensive mitochondrial elongation response—termed stress-induced mitochondrial hyperfusion (SIMH)—has been observed under the aforementioned conditions, although the physiological relevance of this response is not entirely clear and warrants additional investigations.
One model posits that extended mitochondrial networks formed during starvation-induced SIMH may be the way to spare mitochondria from degradation via mitophagy, which requires mitochondrial network fragmentation (Abeliovich et al, 2013; Gomes et al, 2011; Kageyama et al, 2014; MacVicar and Lane, 2014; Mao et al, 2013; Rambold et al, 2011; Tanaka et al, 2010). Of note—in addition to the aforementioned fusion factors—SIMH is dependent on the IM-anchored protein SLP2 that appears to function as a platform to organize the YME1L and PARL proteases within the inner membrane (Tondera et al, 2009; Wai et al, 2016), further linking function of the mitochondrial proteases to the regulation of mitochondrial dynamics and mitophagy. It is important to note, however, that the significance of mitochondrial fragmentation as an absolute prerequisite for mitophagy remains debated (Le Guerroue et al, 2017; Mendl et al, 2011; Yang and Yang, 2013).
More detailed reviews on this account can be found elsewhere (see e.g., a recent review by Ng et al, 2021). In addition to being important for mitophagy, mitochondrial fragmentation is required for mtDNA inheritance (Hanekamp et al, 2002) and regulation of cell death (Jenner et al, 2022; Jiang et al, 2014; Prudent et al, 2015; Varanita et al, 2015; Yamaguchi et al, 2008). Indeed, inhibition of mitochondrial fragmentation—for example, via overexpression of L-OPA1 or OMA1 depletion—precludes BAX-mediated release of cytochrome c, effectively blocking early steps of apoptosis (Acin-Perez et al, 2018; Jiang et al, 2014; Viana et al, 2021; Wai et al, 2015).
Another piecemeal mitophagy-resembling, but presumably distinct cytoprotective mechanism is mitochondria-derived vesicles (MDVs). It involves formation of small (up to 100 nm in diameter) single- or double-membrane-bound vesicles encompassing the OM or both the OM and IM and containing various mitochondrial proteins as cargo (Konig et al, 2021; Neuspiel et al, 2008; Sugiura et al, 2014). MDVs were shown to harbor oxidized mitochondrial proteins and are selectively delivered to various subcellular locales such as multivesicular bodies and lysosomes (Konig et al, 2021; Soubannier et al, 2012). They are formed under both basal and oxidative stress conditions and their formation appears to be stimulated in the latter case (Cadete et al, 2016; Neuspiel et al, 2008).
Interestingly, MDV formation occurs independently of DRP1 and core autophagy components such as ATG5; however, at least in some cases, appears to require mitophagy-related factors PINK1 and Parkin (McLelland et al, 2014; Neuspiel et al, 2008; Ryan et al, 2020).
A similar phenomenon has been described recently in yeast, in which larger, MDV-like structures termed mitochondria-derived compartments (MDCs) are formed in aged or nutrient-stressed cells, or cells with depolarized mitochondria (Hughes et al, 2016; Schuler et al, 2021). Currently, there is no complete agreement as to whether MDCs and MDVs are analogous or distinct structures. However, the fact that formation of MDCs requires Dnm1, Gem1, and core autophagy proteins such as Atg5 (English et al, 2020; Hughes et al, 2016) supports the latter notion. Similarly, it is presently unknown whether and how mitochondrial quality control mechanisms and factors could influence MDV- and MDC-mediated processes. However, considering that some of the key proteins behind MDV and MDC formation are known to be directly or indirectly influenced by mitochondrial contact site factors, it is tempting to speculate that such regulation and/or functional cross talk between said mechanisms could indeed exist.
Mitochondrial architecture
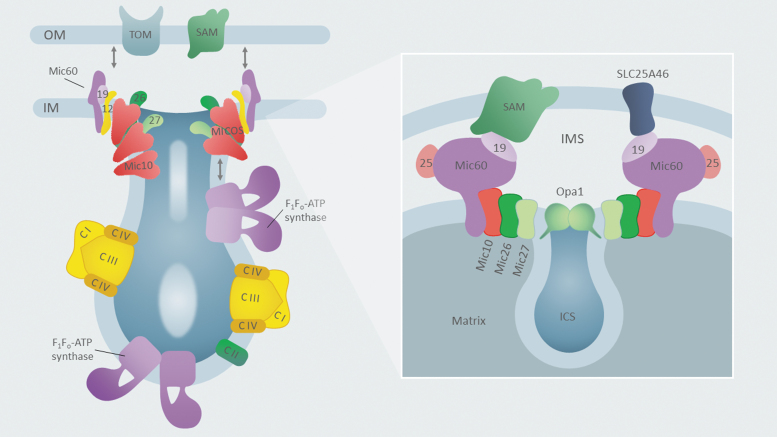
Ultrastructurally, the IM comprises two dynamic domains: the inner boundary membrane (IBM) and the extended and folded IM invaginations, called cristae (Cogliati et al, 2016; Klecker and Westermann, 2021; Kondadi et al, 2020b). The cristae membranes are specifically enriched in respiratory chain supercomplexes (RCS) and are important for RCS stability and retention of cytochrome c pools, thus influencing mitochondrial bioenergetics and susceptibility to apoptosis (Cogliati et al, 2016; Cogliati et al, 2013; Varanita et al, 2015; Wolf et al, 2019). The IBM is physically connected to the OM through the mitochondrial contact site and cristae organizing system (MICOS) machinery (Fig. 4). MICOS is a large evolutionarily conserved complex that resides on the cusp of the IBM and cristae IM domains. It establishes IM-OM contacts between said membranes and contributes to the stabilization of cristae junctions (Van der Laan et al, 2016).
FIG. 4.
Factors mediating mitochondrial architecture. A diagram summarizing factors involved in the shaping of mitochondrial cristae and intermembrane contacts between the inner and OMs. Cristae harbor respiratory chain complexes (Complex I, II, III, and IV) along with the F1Fo-ATPase (Complex V), which plays a role in shaping these membrane structures, whereas the MICOS machinery supports cristae formation at the so-called inner boundary membrane sites, and mediates contacts between the inner and OMs. The inset provides a more detailed look at the MICOS machinery and its key components. See Mitochondrial Architecture section for additional details. MICOS, mitochondrial contact site and cristae organizing system.
In mammalian mitochondria, additional factors important for the IM ultrastructure include F1FO-ATP synthase (Daum et al, 2013; Davies et al, 2012; Quintana-Cabrera et al, 2018) and OPA1 (Glytsou et al, 2016; Lee et al, 2017). Moreover, the OPA1 and MICOS oligomers are in physical contact and were proposed to reciprocally regulate cristae shape in a semicooperative manner (Barrera et al, 2016; Darshi et al, 2011; Glytsou et al, 2016; Lee et al, 2017).
Studies in yeast and mammalian cells established that MICOS and OM-IM contact sites are pivotal for mitochondrial architecture, lipid transport, and IM homeostasis (Schorr and van der Laan, 2018; Van der Laan et al, 2016). Consistent with the critical roles of MICOS machinery in mitochondrial and cell physiology, a number of neuromuscular disorders associated with defects in MICOS or its interacting partners have been reported in recent years (Khosravi and Harner, 2020).
While it is clear that OM-IM contacts are highly dynamic structures, little is known about the regulation thereof and factors involved in this process. Proteolytic processing of OPA1 by the i-AAA and OMA1 proteases discussed above is most likely to be one such mechanism. However, the role of OMA1 in MICOS regulation appears to extend beyond L-OPA1 processing as recent studies reported that OMA1 is associated with the MICOS machinery and may modulate its stability independently of OPA1 (Tang et al, 2020; Viana et al, 2021). The OMA1-MICOS-mediated intermembrane connectivity appears to be important for reorganization and/or stabilization of respiratory complexes, and consequently, bioenergetic plasticity in response to various physiological or stress stimuli (Viana et al, 2021).
In line with this notion, Oma1-deficient MEFs exhibit destabilized respiratory supercomplexes and are bioenergetically compromised under conditions that demand maximal respiratory output (Bohovych et al, 2015b; Korwitz et al, 2016; Quiros et al, 2012). A similar physiological effect has been reported in yeast (Bohovych et al, 2016), underscoring the conserved nature of OMA1-MICOS association.
Similarly, the OMA1-MICOS-mediated intermembrane connectivity is important for regulation of cell death. Earlier studies have established that preservation of L-OPA1 oligomers through L-OPA1 overexpression or OMA1 depletion has an antiapoptotic effect, likely due to the preservation of tight cristae junctions and the consequent hindrance of cytochrome c release from the intercristae space (Frezza et al, 2006; Jiang et al, 2014; Korwitz et al, 2016; Quiros et al, 2012; Varanita et al, 2015; Wai et al, 2015; Yamaguchi et al, 2008). In addition, disruption of the OMA1-MICOS functional axis was shown to promote the formation of OM-disconnected but “locked” cristae able to encapsulate and retain the majority of available cytochrome c pools, thereby leading to an apoptotic resistance (Viana et al, 2021). While we have some understanding of these critical events, the molecular details thereof remain to be clarified.
Mitochondrial Retrograde Signaling Responses
Accumulation of aberrant mitochondrial proteins and their aggregation can be a major threat to organellar and cellular homeostasis. To prevent such scenarios, perturbations to mitochondrial homeostasis can initiate several retrograde signaling responses to regulate the expression of certain mitochondrial proteases and protein chaperones. Detailed reviews on this account can be found elsewhere (Higuchi-Sanabria et al, 2018; Ng et al, 2021). Briefly, these target proteins include the i-AAA, and m-AAA proteases described above, and the matrix-localized AAA+ protease LON. Additional targets include a suite of specialized molecular chaperones/disaggregases that act in concert to counter proteostatic challenges within the organelle (Fig. 5). These enzymes can power protein disaggregation, extraction of proteins from the lipid bilayer, and protein refolding/reactivation in various mitochondrial subcompartments (Bohovych et al, 2015a; Ng et al, 2021).
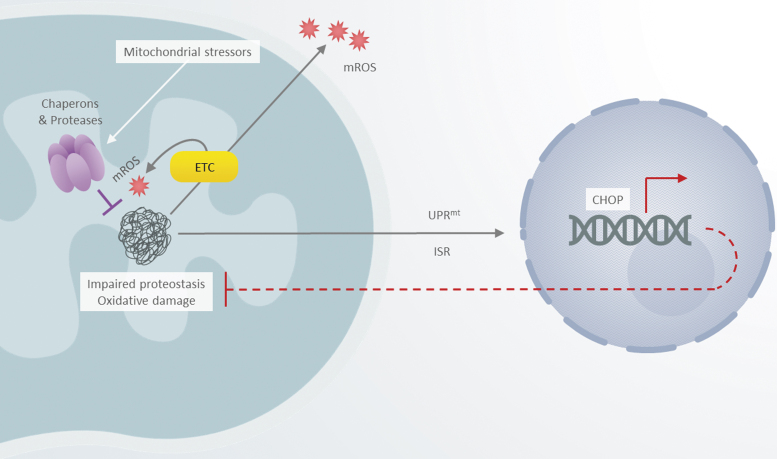
FIG. 5.
Mitochondria-to-nucleus communication. A schematic depicting a minimal model of mitochondrial dyshomeostasis and relevant retrograde signaling responses in mitochondria with perturbed proteostasis. CHOP, C/EBP homologous protein transcription factor; ETC, electron transport chain; ISR, integrated stress response; mROS, mitochondria-borne reactive oxygen species; UPRmt, mitochondrial unfolded protein response.
In higher eukaryotes, the abundance of the aforementioned factors is regulated through several retrograde signaling mechanisms, most notably the mitochondrial unfolded protein response (UPRmt). Originally described in mammalian cells and extensively characterized in the roundworm Caenorhabditis elegans model, the UPRmt pathway comprises two main molecular facets—one involving generation of signaling peptides through proteolytic processing of unassembled or misfolded polypeptides by the CLPXP protease and their subsequent export via the HAF-1 peptide transporter (Haynes et al, 2010; Haynes et al, 2007). Another likely more prominent facet of UPRmt includes bZip transcription factor ATFS-1 equipped with both a mitochondrial targeting sequence and a nuclear localization signal (Nargund et al, 2012). Under basal conditions, ATFS-1 is targeted to mitochondria and inactivated through degradation by the Lon protease. However, upon homeostatic insults, the import of ATFS-1 is impeded, which permits its localization to the nucleus; this process also requires the Ub-like protein 5 (UBL-5).
Another transcriptional factor working in concert with ATFS-1 is DVE-1/SATB2. UPRmt-associated chromatin remodeling allows for binding of nuclear-localized ATFS-1 and DVE-1 to targeted sequences (Haynes et al, 2007; Shao et al, 2020). This promotes the expression of some 400 gene encoding proteins involved in mitochondrial proteostasis, metabolism, and innate immunity (Nargund et al, 2015; Pellegrino et al, 2014). Of note, challenging a paradigm that UPRmt is a transcriptional response specific to higher eukaryotes, a UPRmt-like response—the so-called early UPRmt—has been recently reported in yeast (Poveda-Huertes et al, 2020). The early UPRmt is triggered by aggergating unprocessed precursor proteins and involves relocalization of transcription factor Rox1 to mitochondria, in which it binds to mtDNA to preserve mitochondrial genome expression and promote cell survival (Poveda-Huertes et al, 2020).
The role of UPRmt in mitochondrial proteostasis in mammalian cells is less understood. Evidently, it is more complex and stochastic, and appears to be part of a multifaceted cellular response mechanism known as ISR (Anderson and Haynes, 2020; Costa-Mattioli and Walter, 2020). This complex signaling cascade requires sequential activation of the c-Jun N-terminal kinase, a component of AP-1 transcription factor c-Jun, and the transcriptional factor C/EBP homologous protein transcription factor (CHOP) (Aldridge et al, 2007; Anderson and Haynes, 2020; Horibe and Hoogenraad, 2007). Ultimately, ISR drives significant changes in cellular transcriptome and attenuates cytosolic translation (Klann et al, 2020; Quiros et al, 2017).
Concomitantly it promotes the expression of a gene set including the canonical UPRmt genes encoding mitochondrial proteostasis-related factors. A recent quantitative proteomic profiling of mitochondrial protein uptake in cells under the ISR-inducing mitochondrial stress conditions determined that protein uptake is drastically modulated by concerted action of the import and translation machineries to accommodate such changes (Schafer et al, 2022).
Metabolic Output and Mitochondrial Sculpturing in Neurological Disease
Incidence of neurological disease significantly increases with age, and since aging is accompanied by proteostatic stress and diminished metabolic function, therapies that protect the proteasome system and mitochondrial function are highly sought. For example, a significant decline in NAD+ is observed with aging and is now a major focus of therapeutic approaches for mitochondrial health in age-related diseases (Pirinen et al, 2020; Rajman et al, 2018). Supplements containing NAD+ precursors such as nicotinamide riboside are available with the intention of helping maintain cellular energy metabolism and function by boosting NAD+ levels in the aging population (Rajman et al, 2018). Below we highlight advances in understanding and treating pathological mechanisms of mitochondrial decline in glaucoma, which is a neurodegenerative disease of high global importance and is predicted to impact ∼112 million people worldwide by 2040 (Tham et al, 2014).
Metabolic decline in glaucoma
Pathologies of the optic nerve such as glaucoma are age-related disorders that share hallmarks of brain neurodegenerative diseases. Similar to the brain, the retina has a high demand for oxygen and generates ATP via oxidative phosphorylation (Casson et al, 2021). In addition, a neurovascular structure forms a blood–retinal barrier and supplies blood to the optic nerve and retina. Retinal ganglion cells (RGCs) are neurons with a three-part structure that comprised a soma, dendrites, and axons from which the optic nerve is formed (Casson et al, 2021). Glaucoma is associated with dysfunctional RGCs with the primary pathology found in the axons at the optic nerve head (Ito and Di Polo, 2017).
Metabolism is not the same across the three structural regions of RGCs as nutrient conditions and energy needs can fluctuate (Casson et al, 2021). Glucose, pyruvate, and lactate have been shown to be substrates for energy metabolism in RGCs (Casson et al, 2021). Glycolysis and oxidative phosphorylation are critical to meet the energy demands in RGCs, with a higher rate of oxidative phosphorylation occurring in dendrites and axons as indicated by increased oxygen consumption and mitochondrial content (Casson et al, 2021).
Primary open-angle glaucoma (POAG) is the most common form of glaucoma and is associated with elevated intraocular pressure (IOP). Increased IOP that occurs with aging is a significant risk factor for glaucoma that eventually damages RGCs (Ito and Di Polo, 2017). Mechanisms by which RGCs are impaired are complex and involve a multitude of factors such as oxidative stress, ischemia–hypoxia, glutamate excitotoxicity, inflammation, protein accumulation, and diminished mitochondrial function (Ito and Di Polo, 2017; Leruez et al, 2018; Weinreb and Khaw, 2004). Deficiencies in mitochondrial metabolism and mitochondrial quality control are associated with Leber's hereditary optic neuropathy (LHON) and autosomal dominant optic atrophy (ADOA), respectively.
The importance of mitochondrial metabolism in RGCs is evident in LHON (Brown et al, 2002; Ito and Di Polo, 2017). LHON is caused by maternal inheritance of missense mutations in mitochondria DNA that result in partial deficiency of complex I function (Brown et al, 2002; Ito and Di Polo, 2017). The disease is characterized by RGC dysfunction and optic nerve atrophy with symptoms of blurred vision by adolescence or early adulthood, and eventual blindness (Brown et al, 2002).
In POAG, evidence for lower complex I-dependent ATP synthesis has been reported (Lee et al, 2012; Van Bergen et al, 2015). In patients with clinical POAG (average age of 80 ± 7 years), decreased complex I activity and mitochondrial ATP synthesis were observed in patient lymphocytes relative to age-matched controls (Van Bergen et al, 2015). A mitochondrial genome sequencing study of POAG patients found that one-third of the patients had mtDNA mutations in complex I (Sundaresan et al, 2015). These studies provide evidence that complex I dysfunction contributes to the pathology of glaucoma paralleling what is found frequently in neuromuscular disorders (Sundaresan et al, 2015). The predominant role of mitochondrial dysfunction in glaucoma is the basis for discovery of mtDNA biomarkers of POAG disease (Singh et al, 2018).
Deficits in mitochondrial energy metabolism have also been evidenced by finding lower NAD+ levels in RGCs of aging mice (Williams et al, 2017). The ability of nicotinamide (vitamin B3) supplementation to protect against glaucoma in mice further suggests that available NAD+ is severely lacking in RGCs (Williams et al, 2017). In glaucoma rodent models of RGC degeneration, nicotinamide was also shown to increase oxidative phosphorylation (Tribble et al, 2021). A recent phase 2 clinical trial examining nicotinamide and pyruvate supplementation was observed to improve vision in patients consistent with lower NAD+ limiting energy metabolism in RGCs (De Moraes et al, 2022). Metabolic profiles of plasma from glaucoma patients have also uncovered defective mitochondrial oxidative metabolism, and interestingly, lower levels of spermidine and spermine, which are known neuroprotective molecules (Leruez et al, 2018).
Administration of CoQ10 and vitamin E drops has shown improved retinal function in a study of glaucoma patients (Parisi et al, 2014). CoQ10 also delays apoptosis in RGCs exposed to ocular pressure (Nebbioso et al, 2013; Nucci et al, 2007).
In an effort to gain new insights into the pathology of glaucoma, an interesting large-scale proteomic study was performed on the eye from glaucoma and nonglaucoma human postmortem samples. Thirty-two retinal proteins related to mitochondrial function and oxidative phosphorylation were identified in their data set, of which 24 were significantly downregulated in the glaucoma group (Mirzaei et al, 2017). The proteins found to be lower in abundance are part of respiratory complexes I–IV and ATP synthase, indicating that glaucoma subjects had a lower capacity for mitochondrial energy metabolism in the retina. Glutathione-S-transferases (GSTs) were also observed to be less abundant in the retina of glaucoma patients suggesting that antioxidant defense systems have a lower capacity as well. GSTs have a critical role in helping eliminate reactive free radical species, and electrophilic and xenobiotic compounds.
Disruption of GST and glutathione metabolism in general would weaken cellular defenses against oxidative stress and possibly contribute to the disease pathology of glaucoma.
Mitochondrial dynamics and proteostatic stress in glaucoma
ADOA or Kjer's optic neuropathy is caused by mutations in the OPA1 (Optic Atrophy Gene 1) gene, the loss of which impairs mitochondrial dynamics (Van Bergen et al, 2011). As noted already, OPA1 has a critical role in regulating mitochondrial fusion and networks, and promotes oxidative phosphorylation and ATP production. IOP induces oxidative stress in RGCs and disrupts the mitochondrial dynamics of fusion/fission leading to the eventual loss of mitochondrial integrity and function. In mouse models of glaucoma, IOP triggers mitochondrial fission via expression changes in OPA1 and complex IV subunit 1 (cytochrome c oxidase [COX]), and also causes the release of cytochrome c leading to apoptosis (Ju et al, 2008). A protective role for OPA1 was demonstrated in mice by showing that overexpression of OPA1 increased RGC survival (Ju et al, 2010).
Thus, loss of functional OPA1 in ADOA significantly impairs mitochondria function and energy metabolism in RGCs (Ito and Di Polo, 2017).
Genetic evidence for mitochondrial fission and degradation contributing to glaucoma is from a mutation in the autophagy adaptor protein optineurin (OPTN), which has roles in regulating nuclear factor kappa B (NF-κB) signaling and autophagy (Rezaie et al, 2002). The OPTN variant E50K found in normal-tension glaucoma patients was shown to increase oxidative stress and mitochondrial fission when overexpressed in RGCs. Consistent with these observations, inhibition of DRP1, which drives mitochondrial fission, has been shown to promote RGC survival in mouse models of glaucoma (Kim et al, 2015). As a result, DRP1 has been proposed to be a therapeutic target for protecting RGCs against unwanted mitochondrial fission in optic neuropathies, including glaucoma (Kim et al, 2015). Inefficient clearing of damaged mitochondria is also thought to be an important factor in RGC degeneration.
A recent study showed that RGCs rely on the endolysosomal pathway rather than the proteasome pathway for clearing damaged mitochondria and preventing apoptosis (Das et al, 2020). Another study reported that knockout of the uncoupling protein 2 (Ucp2) in the retina diminished RGC death in a mouse glaucoma model by increasing mitophagy and thereby enhancing overall mitochondrial quality control (Hass and Barnstable, 2019).
Besides mitochondrial and metabolic issues, proteostatic stress in general also contributes to disease progression in glaucoma. Elevated levels of the heat shock protein 27 (Hsp27) was observed in retinal sections from glaucoma patients relative to the age-matched control group (Tezel et al, 2000). Cytidine-5′-diphosphocholine (CDP-choline), also known as citicoline, is known for its several neuroprotective properties, which include enhancing 20S proteasome activity and supporting protein homeostasis (Sbardella et al, 2020). Treatment of glaucoma patients with citicoline has resulted in improved retinal function and vision (Parisi et al, 2008), presumably by diminishing proteostatic stress. Chemical chaperones such as 4-phenylbutyric acid (4-PBA) have been explored as potential therapies for decreasing protein aggregation in retinal cells (Athanasiou et al, 2013; Zode et al, 2015).
Conclusion
Mitochondrial dysfunction, metabolic decline, and protein stress have been implicated in the etiology of multiple late-onset diseases, including parkinsonism, various ataxias, glaucoma, muscle function decline, and diabetes. Mechanisms of neuron and muscle cell death involve apoptosis, necrosis, and autophagy with the importance of the various forms depending on the pathophysiologic condition (e.g., aging, denervation, inflammation, and cancer) (Higuchi-Sanabria et al, 2018). Although abnormal mitochondrial function is well documented in the death of neurons and muscle cells, it is not clear whether mitochondrial dysfunction precedes the accumulation of misfolded proteins, or if unfolded protein stress leads to impaired mitochondrial function. In addition, the inability of a cell to remove damaged mitochondria accentuates oxidative stress conditions.
Therefore, the impact of oxidative stress in aging postmitotic cells is likely twofold: by disrupting mitochondrial respiratory function and by impeding clearance of damaged cell material by the unfolded protein response (UPR) and autophagy. Interestingly, mitochondria and the ER have distinct UPR mechanisms, suggesting that the ability to handle proteotoxic stress is vital for the cell. Understanding how redox homeostasis can be maintained to increase the longevity of neuronal and muscle cells is a strategic area of research that needs to continue, being high priority.
The type of mitochondrial dysfunction that occurs with age and the degree to which it impacts protein clearance mechanisms, however, vary greatly, resulting in wide ranging effects on cellular fidelity. A problem in the field is the lack of a system-wide understanding of how communication between mitochondria and the proteostasis network breaks down during aging and in age-related diseases. The mitochondria and the proteostasis network involve multiple dynamic processes and pathways that adapt to environmental stress and change over time. Current knowledge is mainly from approaches that artificially impair mitochondrial function leading to acute and quite severe consequences that are not necessarily relevant to a neurological disease. In addition, because of the complexity of the proteostasis network, conclusions from studies are often limited to select pathways. Thus, new approaches for exploring the entirety of the proteostatis network need to be developed.
Understanding the role of mitochondrial dysfunction in postmitotic cells during disease progression and other age-related problems is of central importance. Besides oxidative phosphorylation and energy metabolism, mitochondria can provide multiple benefits to other organelles and serve as a communication hub for preserving cellular health during aging. Therapeutic advances for glaucoma that target mitochondrial include a recent phase 2 clinical trial that involves supplementing with nicotinamide and pyruvate. Results thus far show promising improvement of vision in patients (De Moraes et al, 2022) and provide motivation to continue these trials.
Acknowledgments
We apologize to those authors whose work we were unable to cite due to space constraints. We wish to thank Carey Goddard for her help with illustrations, and the members of Khalimonchuk and Becker laboratories for critical reading of the article.
Abbreviations Used
- AAA
ATPases associated with diverse cellular activities
- ACD
acyl-CoA dehydrogenase
- ACON
aconitase
- ADOA
autosomal dominant optic atrophy
- ATP
adenosine triphosphate
- CHOP
C/EBP homologous protein transcription factor
- CLPXP
chaperone subcomplex attachment
- CoQ
coenzyme Q
- COX
cytochrome c oxidase
- Cyt c
cytochrome c
- Doa
dominant optic atrophy mitofusins
- DRP1
dynamin-related protein 1
- DUBs
deubiquitylases
- ER
endoplasmic reticulum
- ETC
electron transport chain
- ETF
electron transfer flavoprotein
- ETFQO
ETF ubiquinone oxidoreductase
- GSALDH
l-glutamate-γ-semialdehyde dehydrogenase
- GST
glutathione-S-transferase
- HNE
4-hydroxyl-2-nonenal
- i-AAA
intermembrane space-facing AAA+ protease
- IBM
inner boundary membrane
- IM
inner mitochondrial membrane
- IMS
mitochondrial intermembrane space
- IOP
intraocular pressure
- ISR
integrated stress response
- KGDH
α-ketoglutarate dehydrogenase
- LHON
Leber's hereditary optic neuropathy
- LONP1
Lon peptidase 1
- L-OPA1
long, IMM-anchored form of OPA1
- m-AAA
matrix-facing AAA+ protease
- MAD
mitochondria-associated degradation
- MDCs
mitochondria-derived compartments
- MDVs
mitochondria-derived vesicles
- Metallo
metallopeptidase
- MFN1
mitofusin 1
- MFN2
itofusin 2
- MICOS
mitochondrial contact site and cristae organizing system
- mitoRQC
ribosomal quality control of mitochondrial polypeptides
- mROS
mitochondria-borne reactive oxygen species
- mtDNA
mitochondrial DNA
- NAD+
nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
- OM
outer mitochondrial membrane
- OPA1
optic atrophy gene 1
- OPTN
optineurin
- P5C
Δ1-pyrroline-5-carboxylate
- PDH
pyruvate dehydrogenase
- POAG
primary open-angle glaucoma
- PQC
protein quality control
- PRODH
proline dehydrogenase
- PYCR
Δ1-pyrroline-5-carboxylate reductase
- Q
ubiquinone
- QH2
reduced ubiquinone
- RCS
respiratory chain supercomplexes
- RGC
retinal ganglion cell
- ROS
reactive oxygen species
- SDH
succinate dehydrogenase
- SIMH
stress-induced mitochondrial hyperfusion
- S-OPA1
short, soluble variant of OPA1
- TCA
tricarboxylic acid
- TIM
translocase of the inner mitochondrial membrane
- TOM
translocase of the outer mitochondrial membrane
- UPR
unfolded protein response
- UPRmt
mitochondrial unfolded protein response
- UPS
ubiquitin–proteasomal system
Authors' Contributions
O.K. and D.F.B. contributed equally to the content, and writing and preparation of figures.
Author Disclosure Statement
The authors declare no conflicts of interest.
Funding Information
This publication was supported, in part, by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM132640 (D.F.B.) and R35GM GM131701 (O.K.).
References
- Abeliovich H, Zarei M, Rigbolt KT, et al. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat Commun 2013;4:2789; doi: 10.1038/ncomms3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acin-Perez R, Lechuga-Vieco AV, Del Mar Munoz M, et al. Ablation of the stress protease OMA1 protects against heart failure in mice. Sci Transl Med 2018;10(434):eaan4935; doi: 10.1126/scitranslmed.aan4935 [DOI] [PubMed] [Google Scholar]
- Ahmed EK, Rogowska-Wrzesinska A, Roepstorff P, et al. Protein modification and replicative senescence of WI-38 human embryonic fibroblasts. Aging Cell 2010;9(2):252–272; doi: 10.1111/j.1474-9726.2010.00555.x [DOI] [PubMed] [Google Scholar]
- Alavi MV. OMA1—An integral membrane protease? Biochim Biophys Acta Proteins Proteom 2021;1869(2):140558; doi: 10.1016/j.bbapap.2020.140558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One 2007;2(9):e874; doi: 10.1371/journal.pone.0000874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Wai T, Baker MJ, et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol 2014;204(6):919–929; doi: 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NS, Haynes CM. Folding the mitochondrial UPR into the integrated stress response. Trends Cell Biol 2020;30(6):428–439; doi: 10.1016/j.tcb.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H, Tauer R, Feldmann H, et al. The YTA10–YTA12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 1996;85(6):875–885; doi: 10.1016/s0092-8674(00)81271-4 [DOI] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Bevilacqua D, et al. The cell stress machinery and retinal degeneration. FEBS Lett 2013;587(13):2008–2017; doi: 10.1016/j.febslet.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atorino L, Silvestri L, Koppen M, et al. Loss of m-AAA protease in mitochondria causes complex i deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J Cell Biol 2003;163(4):777–787; doi: 10.1083/jcb.200304112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Lampe PA, Stojanovski D, et al. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J 2014;33(6):578–593; doi: 10.1002/embj.201386474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Mooga VP, Guiard B, et al. Impaired folding of the mitochondrial small TIM chaperones induces clearance by the i-AAA protease. J Mol Biol 2012;424(5):227–239; doi: 10.1016/j.jmb.2012.09.019 [DOI] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS, et al. Crystal structure of the entire respiratory complex I. Nature 2013;494(7438):443–448; doi: 10.1038/nature11871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera M, Koob S, Dikov D, et al. OPA1 functionally interacts with MIC60 but is dispensable for crista junction formation. FEBS Lett 2016;590(19):3309–3322; doi: 10.1002/1873-3468.12384 [DOI] [PubMed] [Google Scholar]
- Bohovych I, Chan SS, Khalimonchuk O. Mitochondrial protein quality control: The mechanisms guarding mitochondrial health. Antioxid Redox Signal 2015a;22(12):977–994; doi: 10.1089/ars.2014.6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I, Donaldson G, Christianson S, et al. Stress-triggered activation of the metalloprotease OMA1 involves its C-terminal region and is important for mitochondrial stress protection in yeast. J Biol Chem 2014;289(19):13259–13272; doi: 10.1074/jbc.M113.542910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I, Fernandez MR, Rahn JJ, et al. Metalloprotease OMA1 fine-tunes mitochondrial bioenergetic function and respiratory supercomplex stability. Sci Rep 2015b;5:13989; doi: 10.1038/srep13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I, Kastora S, Christianson S, et al. OMA1 links mitochondrial protein quality control and TOR signaling to modulate physiological plasticity and cellular stress responses. Mol Cell Biol 2016;36(17):2300–2312; doi: 10.1128/MCB.00156-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomba-Warczak E, Edassery SL, Hark TJ, et al. Long-lived mitochondrial cristae proteins in mouse heart and brain. J Cell Biol 2021;220(9):e202005193; doi: 10.1083/jcb.202005193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn F, Tatsuta T, Petrungaro C, et al. Presequence-dependent folding ensures MrpL32 processing by the m-AAA protease in mitochondria. EMBO J 2011;30(13):2545–2556; doi: 10.1038/emboj.2011.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol 2002;4(9):674–680; doi: 10.1038/ncb836 [DOI] [PubMed] [Google Scholar]
- Bota DA, Davies KJ. Mitochondrial Lon protease in human disease and aging: Including an etiologic classification of Lon-related diseases and disorders. Free Radic Biol Med 2016;100:188–198; doi: 10.1016/j.freeradbiomed.2016.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragoszewski P, Wasilewski M, Sakowska P, et al. Retro-translocation of mitochondrial intermembrane space proteins. Proc Natl Acad Sci U S A 2015;112(25):7713–7718; doi: 10.1073/pnas.1504615112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Starikovskaya E, Derbeneva O, et al. The role of mtDNA background in disease expression: A new primary LHON mutation associated with Western Eurasian haplogroup J. Hum Genet 2002;110(2):130–138; doi: 10.1007/s00439-001-0660-8 [DOI] [PubMed] [Google Scholar]
- Bykov YS, Rapaport D, Herrmann JM, et al. Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem Sci 2020;45(8):650–667; doi: 10.1016/j.tibs.2020.04.001 [DOI] [PubMed] [Google Scholar]
- Cadete VJ, Deschenes S, Cuillerier A, et al. Formation of mitochondrial-derived vesicles is an active and physiologically relevant mitochondrial quality control process in the cardiac system. J Physiol 2016;594(18):5343–5362; doi: 10.1113/JP272703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson RJ, Chidlow G, Crowston JG, et al. Retinal energy metabolism in health and glaucoma. Prog Retin Eye Res 2021;81:100881; doi: 10.1016/j.preteyeres.2020.100881 [DOI] [PubMed] [Google Scholar]
- Chao JR, Parganas E, Boyd K, et al. Hax1-mediated processing of HtrA2 by PARL allows survival of lymphocytes and neurons. Nature 2008;452(7183):98–102; doi: 10.1038/nature06604 [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 2003;160(2):189–200; doi: 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 2010;141(2):280–289; doi: 10.1016/j.cell.2010.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dorn GW, 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 2013;340(6131):471–475; doi: 10.1126/science.1231031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Umanah GK, Dephoure N, et al. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J 2014;33(14):1548–1564; doi: 10.15252/embj.201487943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, James AM, et al. A unifying mechanism for mitochondrial superoxide production during ischemia-reperfusion injury. Cell Metab 2016;23(2):254–263; doi: 10.1016/j.cmet.2015.12.009 [DOI] [PubMed] [Google Scholar]
- Cogliati S, Enriquez JA, Scorrano L. Mitochondrial cristae: Where beauty meets functionality. Trends Biochem Sci 2016;41(3):261–273; doi: 10.1016/j.tibs.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 2013;155(1):160–171; doi: 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consolato F, Maltecca F, Tulli S, et al. m-AAA and i-AAA complexes coordinate to regulate OMA1, the stress-activated supervisor of mitochondrial dynamics. J Cell Sci 2018;131(7):jcs213546; doi: 10.1242/jcs.213546 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P. The integrated stress response: From mechanism to disease. Science 2020;368(6489):eaat5314; doi: 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, et al. Synchronized mitochondrial and cytosolic translation programs. Nature 2016;533(7604):499–503; doi: 10.1038/nature18015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnane SC, Trushina E, Morland C, et al. Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat Rev Drug Discov 2020;19(9):609–633; doi: 10.1038/s41573-020-0072-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi M, Mendiola VL, Mackey MR, et al. Chchd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem 2011;286(4):2918–2932; doi: 10.1074/jbc.M110.171975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Bell CM, Berlinicke CA, et al. Programmed switch in the mitochondrial degradation pathways during human retinal ganglion cell differentiation from stem cells is critical for RGC survival. Redox Biol 2020;34:101465; doi: 10.1016/j.redox.2020.101465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum B, Walter A, Horst A, et al. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci U S A 2013;110(38):15301–15306; doi: 10.1073/pnas.1305462110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Anselmi C, Wittig I, et al. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A 2012;109(34):13602–13607; doi: 10.1073/pnas.1204593109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008;456(7222):605–610; doi: 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- de Graff AM, Hazoglou MJ, Dill KA. Highly charged proteins: The Achilles' heel of aging proteomes. Structure 2016;24(2):329–336; doi: 10.1016/j.str.2015.11.006 [DOI] [PubMed] [Google Scholar]
- De Moraes CG, John SWM, Williams PA, et al. Nicotinamide and pyruvate for neuroenhancement in open-angle glaucoma: A phase 2 randomized clinical trial. JAMA Ophthalmol 2022;140(1):11–18; doi: 10.1001/jamaophthalmol.2021.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 2011;20(5):867–879; doi: 10.1093/hmg/ddq526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshwal S, Fiedler KU, Langer T. Mitochondrial proteases: Multifaceted regulators of mitochondrial plasticity. Annu Rev Biochem 2020;89:501–528; doi: 10.1146/annurev-biochem-062917-012739 [DOI] [PubMed] [Google Scholar]
- Desmurs M, Foti M, Raemy E, et al. C11orf83, a mitochondrial cardiolipin-binding protein involved in bc1 complex assembly and supercomplex stabilization. Mol Cell Biol 2015;35(7):1139–1156; doi: 10.1128/MCB.01047-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz JV, Bohovych I, Viana MP, et al. Proteolytic regulation of mitochondrial dynamics. Mitochondrion 2019;49:289–304; doi: 10.1016/j.mito.2019.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 2009;187(7):1023–1036; doi: 10.1083/jcb.200906084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elachouri G, Vidoni S, Zanna C, et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res 2011;21(1):12–20; doi: 10.1101/gr.108696.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgass K, Pakay J, Ryan MT, et al. Recent advances into the understanding of mitochondrial fission. Biochim Biophys Acta 2013;1833(1):150–161; doi: 10.1016/j.bbamcr.2012.05.002 [DOI] [PubMed] [Google Scholar]
- English AM, Schuler MH, Xiao T, et al. ER-mitochondria contacts promote mitochondrial-derived compartment biogenesis. J Cell Biol 2020;219(12):e202002144; doi: 10.1083/jcb.202002144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelber K, Dietrich L, Noel JK, et al. Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature 2019;571(7765):429–433; doi: 10.1038/s41586-019-1372-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan AC, Bhangoo MK, Young JC. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J Biol Chem 2006;281(44):33313–33324; doi: 10.1074/jbc.M605250200 [DOI] [PubMed] [Google Scholar]
- Fessler E, Eckl EM, Schmitt S, et al. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 2020;579(7799):433–437; doi: 10.1038/s41586-020-2076-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorczuk K, Letts JA, Degliesposti G, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature 2016;538(7625):406–410; doi: 10.1038/nature19794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filadi R, Greotti E, Turacchio G, et al. On the role of mitofusin 2 in endoplasmic reticulum-mitochondria tethering. Proc Natl Acad Sci U S A 2017;114(12):E2266–E2267; doi: 10.1073/pnas.1616040114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 2006;126(1):177–189; doi: 10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007;129(1):111–122; doi: 10.1016/j.cell.2007.01.047 [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 2008;19(6):2402–2412; doi: 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatsogiannis C, Balogh D, Merino F, et al. Cryo-EM structure of the CLPXP protein degradation machinery. Nat Struct Mol Biol 2019;26(10):946–954; doi: 10.1038/s41594-019-0304-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/Parkin-dependent manner upon induction of mitophagy. Hum Mol Genet 2010;19(24):4861–4870; doi: 10.1093/hmg/ddq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello M, Pyakurel A, Glytsou C, et al. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol 2020;21(4):204–224; doi: 10.1038/s41580-020-0210-7 [DOI] [PubMed] [Google Scholar]
- Glynn SE. Multifunctional mitochondrial AAA proteases. Front Mol Biosci 2017;4:34; doi: 10.3389/fmolb.2017.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glytsou C, Calvo E, Cogliati S, et al. Optic atrophy 1 is epistatic to the core MICOS component MIC60 in mitochondrial cristae shape control. Cell Rep 2016;17(11):3024–3034; doi: 10.1016/j.celrep.2016.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold VA, Chroscicki P, Bragoszewski P, et al. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep 2017;18(10):1786–1800; doi: 10.15252/embr.201744261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 2011;13(5):589–598; doi: 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol 2005;170(2):237–248; doi: 10.1083/jcb.200503148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Aviles G, Liu Y, et al. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 2020;579(7799):427–432; doi: 10.1038/s41586-020-2078-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y. Structure and mechanism of intramembrane protease. Semin Cell Dev Biol 2009;20(2):240–250; doi: 10.1016/j.semcdb.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenbrock CR. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J Cell Biol 1966;30(2):269–297; doi: 10.1083/jcb.30.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon MP, Ahmed EK, Baraibar MA, et al. Proteome oxidative modifications and impairment of specific metabolic pathways during cellular senescence and aging. Proteomics 2020;20(5–6):e1800421; doi: 10.1002/pmic.201800421 [DOI] [PubMed] [Google Scholar]
- Hanekamp T, Thorsness MK, Rebbapragada I, et al. Maintenance of mitochondrial morphology is linked to maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Genetics 2002;162(3):1147–1156; doi: 10.1093/genetics/162.3.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley AM, Lukoyanova N, Zhang Y, et al. Structure of yeast cytochrome C oxidase in a supercomplex with cytochrome bc1. Nat Struct Mol Biol 2019;26(1):78–83; doi: 10.1038/s41594-018-0172-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass DT, Barnstable CJ. Mitochondrial uncoupling protein 2 knock-out promotes mitophagy to decrease retinal ganglion cell death in a mouse model of glaucoma. J Neurosci 2019;39(18):3582–3596; doi: 10.1523/JNEUROSCI.2702-2718.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, et al. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell 2007;13(4):467–480; doi: 10.1016/j.devcel.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, et al. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 2010;37(4):529–540; doi: 10.1016/j.molcel.2010.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, et al. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 2009;187(7):959–966; doi: 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Livnat-Levanon N, Taylor EB, et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell 2010;40(3):465–480; doi: 10.1016/j.molcel.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Frankino PA, Paul JW, 3rd, et al. A futile battle? Protein quality control and the stress of aging. Dev Cell 2018;44(2):139–163; doi: 10.1016/j.devcel.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsetz E, Demir F, Szczepanowska K, et al. The mouse heart mitochondria N terminome provides insights into CLPXP-mediated proteolysis. Mol Cell Proteomics 2020;19(8):1330–1345; doi: 10.1074/mcp.RA120.002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horibe T, Hoogenraad NJ. The Chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One 2007;2(9):e835; doi: 10.1371/journal.pone.0000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Hughes CE, Henderson KA, et al. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. Elife 2016;5:e13943; doi: 10.7554/eLife.13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries KM, Szweda LI. Selective inactivation of alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry 1998;37(45):15835–15841; doi: 10.1021/bi981512h [DOI] [PubMed] [Google Scholar]
- Humphries KM, Szweda PA, Szweda LI. Aging: A shift from redox regulation to oxidative damage. Free Radic Res 2006;40(12):1239–1243; doi: 10.1080/10715760600913184 [DOI] [PubMed] [Google Scholar]
- Hurst S, Baggett A, Csordas G, et al. SPG7 targets the m-AAA protease complex to process MCU for uniporter assembly, Ca(2+) influx, and regulation of mitochondrial permeability transition pore opening. J Biol Chem 2019;294(28):10807–10818; doi: 10.1074/jbc.RA118.006443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito YA, Di Polo A. Mitochondrial dynamics, transport, and quality control: A bottleneck for retinal ganglion cell viability in optic neuropathies. Mitochondrion 2017;36:186–192; doi: 10.1016/j.mito.2017.08.014 [DOI] [PubMed] [Google Scholar]
- Iwata S, Lee JW, Okada K, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 1998;281(5373):64–71; doi: 10.1126/science.281.5373.64 [DOI] [PubMed] [Google Scholar]
- Izawa T, Park SH, Zhao L, et al. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell 2017;171(4):890..e18–903.e18; doi: 10.1016/j.cell.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Jenner A, Pena-Blanco A, Salvador-Gallego R, et al. DRP1 interacts directly with BAX to induce its activation and apoptosis. EMBO J 2022;41(8):e108587; doi: 10.15252/embj.2021108587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Jiang H, Shen Z, et al. Activation of mitochondrial protease OMA1 by Bax and Bak promotes cytochrome C release during apoptosis. Proc Natl Acad Sci U S A 2014;111(41):14782–14787; doi: 10.1073/pnas.1417253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 2010;191(5):933–942; doi: 10.1083/jcb.201008084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju WK, Kim KY, Duong-Polk KX, et al. Increased optic atrophy type 1 expression protects retinal ganglion cells in a mouse model of glaucoma. Mol Vis 2010;16:1331–1342. [PMC free article] [PubMed] [Google Scholar]
- Ju WK, Kim KY, Lindsey JD, et al. Intraocular pressure elevation induces mitochondrial fission and triggers OPA1 release in glaucomatous optic nerve. Invest Ophthalmol Vis Sci 2008;49(11):4903–4911; doi: 10.1167/iovs.07-1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, et al. Parkin-independent mitophagy requires DRP1 and maintains the integrity of mammalian heart and brain. EMBO J 2014;33(23):2798–2813; doi: 10.15252/embj.201488658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Dimitrova MN, Ortega J, et al. Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J Biol Chem 2005;280(42):35424–35432; doi: 10.1074/jbc.M507240200 [DOI] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for DRP1 dependent mitochondrial division. J Cell Biol 2007;178(1):71–84; doi: 10.1083/jcb.200611064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Moroco JA, Engen JR, et al. Mitochondrial ClpX activates an essential biosynthetic enzyme through partial unfolding. Elife 2020;9:e54387; doi: 10.7554/eLife.54387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Yien YY, Huston NC, et al. Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell 2015;161(4):858–867; doi: 10.1016/j.cell.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser M, Kambacheld M, Kisters-Woike B, et al. OMA1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J Biol Chem 2003;278(47):46414–46423; doi: 10.1074/jbc.M305584200 [DOI] [PubMed] [Google Scholar]
- Khacho M, Tarabay M, Patten D, et al. Acidosis overrides oxygen deprivation to maintain mitochondrial function and cell survival. Nat Commun 2014;5:3550; doi: 10.1038/ncomms4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Jeong MY, Watts T, et al. Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants. J Biol Chem 2012;287(10):7289–7300; doi: 10.1074/jbc.M111.313148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi S, Harner ME. The MICOS complex, a structural element of mitochondria with versatile functions. Biol Chem 2020;401(6–7):765–778; doi: 10.1515/hsz-2020-0103 [DOI] [PubMed] [Google Scholar]
- Kim KY, Perkins GA, Shim MS, et al. DRP1 inhibition rescues retinal ganglion cells and their axons by preserving mitochondrial integrity in a mouse model of glaucoma. Cell Death Dis 2015;6:e1839; doi: 10.1038/cddis.2015.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Stepanova A, Niatsetskaya Z, et al. Attenuation of oxidative damage by targeting mitochondrial complex I in neonatal hypoxic-ischemic brain injury. Free Radic Biol Med 2018;124:517–524; doi: 10.1016/j.freeradbiomed.2018.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann K, Tascher G, Munch C. Functional translatome proteomics reveal converging and dose-dependent regulation by mTORC1 and eIF2alpha. Mol Cell 2020;77(4):913..e4–925.e4; doi: 10.1016/j.molcel.2019.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker T, Westermann B. Pathways shaping the mitochondrial inner membrane. Open Biol 2021;11(12):210238; doi: 10.1098/rsob.210238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S, Guo Q, Kalia R, et al. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci U S A 2013;110(15):E1342–E1351; doi: 10.1073/pnas.1300855110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondadi AK, Anand R, Hansch S, et al. Cristae undergo continuous cycles of membrane remodelling in a MICOS-dependent manner. EMBO Rep 2020a;21(3):e49776; doi: 10.15252/embr.201949776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondadi AK, Anand R, Reichert AS. Cristae membrane dynamics—A paradigm change. Trends Cell Biol 2020b;30(12):923–936; doi: 10.1016/j.tcb.2020.08.008 [DOI] [PubMed] [Google Scholar]
- Konig T, Nolte H, Aaltonen MJ, et al. MIROs and DRP1 drive mitochondrial-derived vesicle biogenesis and promote quality control. Nat Cell Biol 2021;23(12):1271–1286; doi: 10.1038/s41556-021-00798-4 [DOI] [PubMed] [Google Scholar]
- Konig T, Troder SE, Bakka K, et al. The m-AAA protease associated with neurodegeneration limits MCU activity in mitochondria. Mol Cell 2016;64(1):148–162; doi: 10.1016/j.molcel.2016.08.020 [DOI] [PubMed] [Google Scholar]
- Koppen M, Bonn F, Ehses S, et al. Autocatalytic processing of m-AAA protease subunits in mitochondria. Mol Biol Cell 2009;20(19):4216–4224; doi: 10.1091/mbc.E09-03-0218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Metodiev MD, Casari G, et al. Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia. Mol Cell Biol 2007;27(2):758–767; doi: 10.1128/MCB.01470-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korwitz A, Merkwirth C, Richter-Dennerlein R, et al. Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J Cell Biol 2016;212(2):157–166; doi: 10.1083/jcb.201507022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus F, Roy K, Pucadyil TJ, et al. Function and regulation of the divisome for mitochondrial fission. Nature 2021;590(7844):57–66; doi: 10.1038/s41586-021-03214-x [DOI] [PubMed] [Google Scholar]
- Kremmidiotis G, Gardner AE, Settasatian C, et al. Molecular and functional analyses of the human and mouse genes encoding AFG3L1, a mitochondrial metalloprotease homologous to the human spastic paraplegia protein. Genomics 2001;76(1–3):58–65; doi: 10.1006/geno.2001.6560 [DOI] [PubMed] [Google Scholar]
- Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol 2014;30:357–391; doi: 10.1146/annurev-cellbio-101011-155756 [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, et al. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell 1999;4(5):815–826; doi: 10.1016/s1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Lagouge M, Mourier A, Lee HJ, et al. SLIRP regulates the rate of mitochondrial protein synthesis and protects LRPPRC from degradation. PLoS Genet 2015;11(8):e1005423; doi: 10.1371/journal.pgen.1005423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie J, De Belvalet H, Sonon S, et al. Ubiquitin-dependent degradation of mitochondrial proteins regulates energy metabolism. Cell Rep 2018;23(10):2852–2863; doi: 10.1016/j.celrep.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Le Guerroue F, Eck F, Jung J, et al. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol Cell 2017;68(4):786..e6–796.e6; doi: 10.1016/j.molcel.2017.10.029 [DOI] [PubMed] [Google Scholar]
- Lee H, Smith SB, Yoon Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J Biol Chem 2017;292(17):7115–7130; doi: 10.1074/jbc.M116.762567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sheck L, Crowston JG, et al. Impaired complex-I-linked respiration and ATP synthesis in primary open-angle glaucoma patient lymphoblasts. Invest Ophthalmol Vis Sci 2012;53(4):2431–2437; doi: 10.1167/iovs.12-9596 [DOI] [PubMed] [Google Scholar]
- Leonhard K, Herrmann JM, Stuart RA, et al. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J 1996;15(16):4218–4229. [PMC free article] [PubMed] [Google Scholar]
- Leonhard K, Stiegler A, Neupert W, et al. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature 1999;398(6725):348–351; doi: 10.1038/18704 [DOI] [PubMed] [Google Scholar]
- Lerma-Ortiz C, Jeffryes JG, Cooper AJ, et al. ‘Nothing of chemistry disappears in biology’: The top 30 damage-prone endogenous metabolites. Biochem Soc Trans 2016;44(3):961–971; doi: 10.1042/BST20160073 [DOI] [PubMed] [Google Scholar]
- Leruez S, Marill A, Bresson T, et al. A Metabolomics profiling of glaucoma points to mitochondrial dysfunction, senescence, and polyamines deficiency. Invest Ophthalmol Vis Sci 2018;59(11):4355–4361; doi: 10.1167/iovs.18-24938 [DOI] [PubMed] [Google Scholar]
- Levytskyy RM, Bohovych I, Khalimonchuk O. Metalloproteases of the inner mitochondrial membrane. Biochemistry 2017;56(36):4737–4746; doi: 10.1021/acs.biochem.7b00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PC, Wolken DMA, Serrano E, et al. Mitochondria-associated degradation pathway (MAD) function beyond the outer membrane. Cell Rep 2020;32(2):107902; doi: 10.1016/j.celrep.2020.107902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang X, Coyne LP, et al. Mitochondrial carrier protein overloading and misfolding induce aggresomes and proteostatic adaptations in the cytosol. Mol Biol Cell 2019;30(11):1272–1284; doi: 10.1091/mbc.E19-01-0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Liu C, Sun L, et al. Neuronal mitochondrial toxicity of malondialdehyde: Inhibitory effects on respiratory function and enzyme activities in rat brain mitochondria. Neurochem Res 2009;34(4):786–794; doi: 10.1007/s11064-008-9882-7 [DOI] [PubMed] [Google Scholar]
- Loschen G, Flohe L, Chance B. Respiratory chain linked H(2)O(2) production in pigeon heart mitochondria. FEBS Lett 1971;18(2):261–264; doi: 10.1016/0014-5793(71)80459-3 [DOI] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, et al. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 2013;24(5):659–667; doi: 10.1091/mbc.E12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Lee J, Nie X, et al. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 2013;49(1):121–132; doi: 10.1016/j.molcel.2012.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar T, Langer T. OPA1 processing in cell death and disease—The long and short of it. J Cell Sci 2016;129(12):2297–2306; doi: 10.1242/jcs.159186 [DOI] [PubMed] [Google Scholar]
- MacVicar T, Ohba Y, Nolte H, et al. Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature 2019;575(7782):361–365; doi: 10.1038/s41586-019-1738-6 [DOI] [PubMed] [Google Scholar]
- MacVicar TD, Lane JD. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. J Cell Sci 2014;127(Pt 10):2313–2325; doi: 10.1242/jcs.144337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malka F, Guillery O, Cifuentes-Diaz C, et al. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep 2005;6(9):853–859; doi: 10.1038/sj.embor.7400488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann G, Mora S, Madu G, et al. Branched-chain amino acids: Catabolism in skeletal muscle and implications for muscle and whole-body metabolism. Front Physiol 2021;12:702826; doi: 10.3389/fphys.2021.702826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannaa A, Hanisch FG. Redox proteomes in human physiology and disease mechanisms. J Proteome Res 2020;19(1):1–17; doi: 10.1021/acs.jproteome.9b00586 [DOI] [PubMed] [Google Scholar]
- Mao K, Wang K, Liu X, et al. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell 2013;26(1):9–18; doi: 10.1016/j.devcel.2013.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martensson CU, Priesnitz C, Song J, et al. Mitochondrial protein translocation-associated degradation. Nature 2019;569(7758):679–683; doi: 10.1038/s41586-019-1227-y [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Nakatsukasa K, Kakuta C, et al. Msp1 clears mistargeted proteins by facilitating their transfer from mitochondria to the ER. Mol Cell 2019;76(1):191..e10–205.e10; doi: 10.1016/j.molcel.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Goto Y, Kaguni LS. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor a (TFAM). Proc Natl Acad Sci U S A 2010;107(43):18410–18415; doi: 10.1073/pnas.1008924107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Goiran T, Yi W, et al. Mfn2 ubiquitination by PINK1/Parkin gates the P97-dependent release of ER from mitochondria to drive mitophagy. Elife 2018;7:e32866; doi: 10.7554/eLife.32866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Soubannier V, Chen CX, et al. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J 2014;33(4):282–295; doi: 10.1002/embj.201385902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendl N, Occhipinti A, Muller M, et al. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci 2011;124(Pt 8):1339–1350; doi: 10.1242/jcs.076406 [DOI] [PubMed] [Google Scholar]
- Mirzaei M, Gupta VB, Chick JM, et al. Age-related neurodegenerative disease associated pathways identified in retinal and vitreous proteome from human glaucoma eyes. Sci Rep 2017;7(1):12685; doi: 10.1038/s41598-017-12858-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern M, Peikert CD, Lubbert P, et al. Quantitative high-confidence human mitochondrial proteome and its dynamics in cellular context. Cell Metab 2021;33(12):2464..e18–2483.e18; doi: 10.1016/j.cmet.2021.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata D, Yamada T, Tokuyama T, et al. Mitochondrial safeguard: A stress response that offsets extreme fusion and protects respiratory function via flickering-induced OMA1 activation. EMBO J 2020;39(24):e105074; doi: 10.15252/embj.2020105074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 2009;417(1):1–13; doi: 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima S, Tokuyama T, Yonashiro R, et al. Roles of mitochondrial ubiquitin ligase MITOL/MARCH5 in mitochondrial dynamics and diseases. J Biochem 2014;155(5):273–279; doi: 10.1093/jb/mvu016 [DOI] [PubMed] [Google Scholar]
- Naon D, Zaninello M, Giacomello M, et al. Reply to Filadi et al.: Does mitofusin 2 tether or separate endoplasmic reticulum and mitochondria? Proc Natl Acad Sci U S A 2017;114(12):E2268–E2269; doi: 10.1073/pnas.1618610114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, et al. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes oxphos recovery during the UPR(mt). Mol Cell 2015;58(1):123–133; doi: 10.1016/j.molcel.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, et al. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 2012;337(6094):587–590; doi: 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso M, Scarsella G, Tafani M, et al. Mechanisms of ocular neuroprotection by antioxidant molecules in animal models. J Biol Regul Homeost Agents 2013;27(1):197–209. [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem 2007;76:723–749; doi: 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- Neuspiel M, Schauss AC, Braschi E, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol 2008;18(2):102–108; doi: 10.1016/j.cub.2007.12.038 [DOI] [PubMed] [Google Scholar]
- Ng MYW, Wai T, Simonsen A. Quality control of the mitochondrion. Dev Cell 2021;56(7):881–905; doi: 10.1016/j.devcel.2021.02.009 [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Gronauer TF, Nast-Kolb T, et al. Substrate profiling of mitochondrial caseinolytic protease P via a site-specific photocrosslinking approach. Angew Chem Int Ed Engl 2022;61(10):e202111085; doi: 10.1002/anie.202111085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niatsetskaya ZV, Sosunov SA, Matsiukevich D, et al. The oxygen free radicals originating from mitochondrial complex I contribute to oxidative brain injury following hypoxia-ischemia in neonatal mice. J Neurosci 2012;32(9):3235–3244; doi: 10.1523/JNEUROSCI.6303-6311.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, et al. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 2005;123(2):277–289; doi: 10.1016/j.cell.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Nucci C, Tartaglione R, Cerulli A, et al. Retinal damage caused by high intraocular pressure-induced transient ischemia is prevented by coenzyme Q10 in rat. Int Rev Neurobiol 2007;82:397–406; doi: 10.1016/S0074-7742(07)82022-8 [DOI] [PubMed] [Google Scholar]
- Ogunbona OB, Onguka O, Calzada E, et al. Multitiered and cooperative surveillance of mitochondrial phosphatidylserine decarboxylase 1. Mol Cell Biol 2017;37(17):e00049-17; doi: 10.1128/MCB.00049-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba Y, MacVicar T, Langer T. Regulation of mitochondrial plasticity by the i-AAA protease YME1L. Biol Chem 2020;401(6–7):877–890; doi: 10.1515/hsz-2020-0120 [DOI] [PubMed] [Google Scholar]
- Okreglak V, Walter P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci U S A 2014;111(22):8019–8024; doi: 10.1073/pnas.1405755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver CN, Starke-Reed PE, Stadtman ER, et al. Oxidative damage to brain proteins, loss of glutamine synthetase activity, and production of free radicals during ischemia/reperfusion-induced injury to gerbil brain. Proc Natl Acad Sci U S A 1990;87(13):5144–5147; doi: 10.1073/pnas.87.13.5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinski L, Song J, Priesnitz C, et al. Recruitment of cytosolic J-proteins by TOM receptors promotes mitochondrial protein biogenesis. Cell Rep 2018;25(8):2036..e5–2043.e5; doi: 10.1016/j.celrep.2018.10.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame LD, Singh AP, Stroud DA, et al. Cooperative and independent roles of the DRP1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J Cell Sci 2016;129(11):2170–2181; doi: 10.1242/jcs.185165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008;134(1):112–123; doi: 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 2011;12(6):565–573; doi: 10.1038/embor.2011.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parey K, Wirth C, Vonck J, et al. Respiratory complex I—Structure, mechanism and evolution. Curr Opin Struct Biol 2020;63:1–9; doi: 10.1016/j.sbi.2020.01.004 [DOI] [PubMed] [Google Scholar]
- Parisi V, Centofanti M, Gandolfi S, et al. Effects of coenzyme Q10 in conjunction with vitamin E on retinal-evoked and cortical-evoked responses in patients with open-angle glaucoma. J Glaucoma 2014;23(6):391–404; doi: 10.1097/IJG.0b013e318279b836 [DOI] [PubMed] [Google Scholar]
- Parisi V, Coppola G, Centofanti M, et al. Evidence of the neuroprotective role of citicoline in glaucoma patients. Prog Brain Res 2008;173:541–554; doi: 10.1016/S0079-6123(08)01137-0 [DOI] [PubMed] [Google Scholar]
- Park YY, Lee S, Karbowski M, et al. Loss of MARCH5 mitochondrial E3 ubiquitin ligase induces cellular senescence through dynamin-related protein 1 and mitofusin 1. J Cell Sci 2010;123(Pt 4):619–626; doi: 10.1242/jcs.061481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron M, Sprenger HG, Langer T. m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration. Cell Res 2018;28(3):296–306; doi: 10.1038/cr.2018.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten DA, Wong J, Khacho M, et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J 2014;33(22):2676–2691; doi: 10.15252/embj.201488349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, et al. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 2014;516(7531):414–417; doi: 10.1038/nature13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L, Scorrano L. Mito-morphosis: Mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol 2016;78:505–531; doi: 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Warscheid B, Wiedemann N. Mitochondrial proteins: From biogenesis to functional networks. Nat Rev Mol Cell Biol 2019;20(5):267–284; doi: 10.1038/s41580-018-0092-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen E, Auranen M, Khan NA, et al. Niacin cures systemic NAD(+) deficiency and improves muscle performance in adult-onset mitochondrial myopathy. Cell Metab 2020;31(6):1078..e5–1090.e5; doi: 10.1016/j.cmet.2020.04.008 [DOI] [PubMed] [Google Scholar]
- Potting C, Tatsuta T, Konig T, et al. TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab 2013;18(2):287–295; doi: 10.1016/j.cmet.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Potting C, Wilmes C, Engmann T, et al. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J 2010;29(17):2888–2898; doi: 10.1038/emboj.2010.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda-Huertes D, Matic S, Marada A, et al. An early mtUPR: Redistribution of the nuclear transcription factor Rox1 to mitochondria protects against intramitochondrial proteotoxic aggregates. Mol Cell 2020;77(1):180..e9–188.e9; doi: 10.1016/j.molcel.2019.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent J, Zunino R, Sugiura A, et al. MAPL SUMOylation of Drp1 stabilizes an ER/mitochondrial platform required for cell death. Mol Cell 2015;59(6):941–955; doi: 10.1016/j.molcel.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Puchades C, Ding B, Song A, et al. Unique structural features of the mitochondrial AAA+ protease AFG3L2 reveal the molecular basis for activity in health and disease. Mol Cell 2019;75(5):1073..e6–1085.e6; doi: 10.1016/j.molcel.2019.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchades C, Rampello AJ, Shin M, et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 2017;358(6363):eaao0464; doi: 10.1126/science.aao0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan CL, Orr AL, Perevoshchikova IV, et al. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem 2012;287(32):27255–27264; doi: 10.1074/jbc.M112.374629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Cabrera R, Quirin C, Glytsou C, et al. The cristae modulator optic atrophy 1 requires mitochondrial ATP synthase oligomers to safeguard mitochondrial function. Nat Commun 2018;9(1):3399; doi: 10.1038/s41467-018-05655-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol 2015;16(6):345–359; doi: 10.1038/nrm3984 [DOI] [PubMed] [Google Scholar]
- Quiros PM, Prado MA, Zamboni N, et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 2017;216(7):2027–2045; doi: 10.1083/jcb.201702058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Ramsay AJ, Sala D, et al. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J 2012;31(9):2117–2133; doi: 10.1038/emboj.2012.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt TK, Atanassova N, Genereux JC, et al. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab 2013;18(6):908–919; doi: 10.1016/j.cmet.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt TK, Lebeau J, Puchades C, et al. Reciprocal degradation of YME1L and OMA1 adapts mitochondrial proteolytic activity during stress. Cell Rep 2016;14(9):2041–2049; doi: 10.1016/j.celrep.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt TK, Saunders JM, Wiseman RL. YME1L degradation reduces mitochondrial proteolytic capacity during oxidative stress. EMBO Rep 2015;16(1):97–106; doi: 10.15252/embr.201438976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman L, Chwalek K, Sinclair DA. Therapeutic potential of NAD-boosting molecules: The in vivo evidence. Cell Metab 2018;27(3):529–547; doi: 10.1016/j.cmet.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, et al. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A 2011;108(25):10190–10195; doi: 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S, Sharma R, Gupta R, et al. Mitocarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res 2021;49(D1):D1541–D1547; doi: 10.1093/nar/gkaa1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaie T, Child A, Hitchings R, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 2002;295(5557):1077–1079; doi: 10.1126/science.1066901 [DOI] [PubMed] [Google Scholar]
- Richter F, Dennerlein S, Nikolov M, et al. ROMO1 is a constituent of the human presequence translocase required for YME1L protease import. J Cell Biol 2019;218(2):598–614; doi: 10.1083/jcb.201806093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, et al. Quality control of mitochondrial protein synthesis is required for membrane integrity and cell fitness. J Cell Biol 2015;211(2):373–389; doi: 10.1083/jcb.201504062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan TA, Phillips EO, Collier CL, et al. Tollip coordinates Parkin-dependent trafficking of mitochondrial-derived vesicles. EMBO J 2020;39(11):e102539; doi: 10.15252/embj.2019102539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saita S, Nolte H, Fiedler KU, et al. PARL mediates Smac proteolytic maturation in mitochondria to promote apoptosis. Nat Cell Biol 2017;19(4):318–328; doi: 10.1038/ncb3488 [DOI] [PubMed] [Google Scholar]
- Sauer RT, Fei X, Bell TA, et al. Structure and function of CLPXP, a AAA+ proteolytic machine powered by probabilistic ATP hydrolysis. Crit Rev Biochem Mol Biol 2022;57(2):188–204; doi: 10.1080/10409238.2021.1979461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbardella D, Coletta A, Tundo GR, et al. Structural and functional evidence for citicoline binding and modulation of 20S proteasome activity: Novel insights into its pro-proteostatic effect. Biochem Pharmacol 2020;177:113977; doi: 10.1016/j.bcp.2020.113977 [DOI] [PubMed] [Google Scholar]
- Schafer JA, Bozkurt S, Michaelis JB, et al. Global mitochondrial protein import proteomics reveal distinct regulation by translation and translocation machinery. Mol Cell 2022;82(2):435..e7–446.e7; doi: 10.1016/j.molcel.2021.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat Rev Mol Cell Biol 2010;11(9):655–667; doi: 10.1038/nrm2959 [DOI] [PubMed] [Google Scholar]
- Schorr S, van der Laan M. Integrative functions of the mitochondrial contact site and cristae organizing system. Semin Cell Dev Biol 2018;76:191–200; doi: 10.1016/j.semcdb.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Schreiner B, Westerburg H, Forne I, et al. Role of the AAA protease YME1 in folding of proteins in the intermembrane space of mitochondria. Mol Biol Cell 2012;23(22):4335–4346; doi: 10.1091/mbc.E12-05-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MH, English AM, Xiao T, et al. Mitochondrial-derived compartments facilitate cellular adaptation to amino acid stress. Mol Cell 2021;81(18):3786..e13–3802.e13; doi: 10.1016/j.molcel.2021.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Kanamaru Y, Koike M, et al. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J Biol Chem 2012;287(41):34635–34645; doi: 10.1074/jbc.M112.357509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Wang C, Sideris DP, et al. Reciprocal roles of Tom7 and OMA1 during mitochondrial import and activation of PINK1. Mol Cell 2019;73(5):1028..e5–1043.e5; doi: 10.1016/j.molcel.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Seo JH, Rivadeneira DB, Caino MC, et al. The mitochondrial unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. PLoS Biol 2016;14(7):e1002507; doi: 10.1371/journal.pbio.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepuri NBV, Angireddy R, Srinivasan S, et al. Mitochondrial LON protease-dependent degradation of cytochrome C oxidase subunits under hypoxia and myocardial ischemia. Biochim Biophys Acta Bioenerg 2017;1858(7):519–528; doi: 10.1016/j.bbabio.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao LW, Peng Q, Dong M, et al. Histone deacetylase HDA-1 modulates mitochondrial stress response and longevity. Nat Commun 2020;11(1):4639; doi: 10.1038/s41467-020-18501-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 2015;347(6217):75–78; doi: 10.1126/science.1259724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Rampello AJ, Glynn SE. Engineered AAA+ proteases reveal principles of proteolysis at the mitochondrial inner membrane. Nat Commun 2016;7:13301; doi: 10.1038/ncomms13301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Puchades C, Asmita A, et al. Structural basis for distinct operational modes and protease activation in AAA+ protease Lon. Sci Adv 2020;6(21):eaba8404; doi: 10.1126/sciadv.aba8404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Watson ER, Song AS, et al. Structures of the human LONP1 protease reveal regulatory steps involved in protease activation. Nat Commun 2021;12(1):3239; doi: 10.1038/s41467-021-23495-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LN, Crowston JG, Lopez Sanchez MIG, et al. Mitochondrial DNA variation and disease susceptibility in primary open-angle glaucoma. Invest Ophthalmol Vis Sci 2018;59(11):4598–4602; doi: 10.1167/iovs.18-25085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McLelland GL, Zunino R, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol 2012;22(2):135–141; doi: 10.1016/j.cub.2011.11.057 [DOI] [PubMed] [Google Scholar]
- Sprenger HG, Langer T. The good and the bad of mitochondrial breakups. Trends Cell Biol 2019;29(11):888–900; doi: 10.1016/j.tcb.2019.08.003 [DOI] [PubMed] [Google Scholar]
- Steele TE, Glynn SE. Mitochondrial AAA proteases: A stairway to degradation. Mitochondrion 2019;49:121–127; doi: 10.1016/j.mito.2019.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiburek L, Cesnekova J, Kostkova O, et al. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol Biol Cell 2012;23(6):1010–1023; doi: 10.1091/mbc.E11-08-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, McLelland GL, Fon EA, et al. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J 2014;33(19):2142–2156; doi: 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A, Nagashima S, Tokuyama T, et al. MITOL regulates endoplasmic reticulum-mitochondria contacts via mitofusin2. Mol Cell 2013;51(1):20–34; doi: 10.1016/j.molcel.2013.04.023 [DOI] [PubMed] [Google Scholar]
- Sulkshane P, Duek I, Ram J, et al. Inhibition of proteasome reveals basal mitochondrial ubiquitination. J Proteomics 2020;229:103949; doi: 10.1016/j.jprot.2020.103949 [DOI] [PubMed] [Google Scholar]
- Sundaresan P, Simpson DA, Sambare C, et al. Whole-mitochondrial genome sequencing in primary open-angle glaucoma using massively parallel sequencing identifies novel and known pathogenic variants. Genet Med 2015;17(4):279–284; doi: 10.1038/gim.2014.121 [DOI] [PubMed] [Google Scholar]
- Szczepanowska K, Maiti P, Kukat A, et al. ClpP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J 2016;35(23):2566–2583; doi: 10.15252/embj.201694253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowska K, Senft K, Heidler J, et al. A salvage pathway maintains highly functional respiratory complex I. Nat Commun 2020;11(1):1643; doi: 10.1038/s41467-020-15467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowska K, Trifunovic A. Tune instead of destroy: How proteolysis keeps oxphos in shape. Biochim Biophys Acta Bioenerg 2021;1862(4):148365; doi: 10.1016/j.bbabio.2020.148365 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, et al. Proteasome and P97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 2010;191(7):1367–1380; doi: 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Zhang K, Dong J, et al. Sam50-Mic19-Mic60 axis determines mitochondrial cristae architecture by mediating mitochondrial outer and inner membrane contact. Cell Death Differ 2020;27(1):146–160; doi: 10.1038/s41418-019-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel G, Hernandez R, Wax MB. Immunostaining of heat shock proteins in the retina and optic nerve head of normal and glaucomatous eyes. Arch Ophthalmol 2000;118(4):511–518; doi: 10.1001/archopht.118.4.511 [DOI] [PubMed] [Google Scholar]
- Tham YC, Li X, Wong TY, et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014;121(11):2081–2090; doi: 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- Tieu Q, Okreglak V, Naylor K, et al. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol 2002;158(3):445–452; doi: 10.1083/jcb.200205031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 2009;28(11):1589–1600; doi: 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble JR, Otmani A, Sun S, et al. Nicotinamide provides neuroprotection in glaucoma by protecting against mitochondrial and metabolic dysfunction. Redox Biol 2021;43:101988; doi: 10.1016/j.redox.2021.101988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, et al. The whole structure of the 13-subunit oxidized cytochrome C oxidase at 2.8 A. Science 1996;272(5265):1136–1144; doi: 10.1126/science.272.5265.1136 [DOI] [PubMed] [Google Scholar]
- Van Bergen NJ, Crowston JG, Craig JE, et al. Measurement of systemic mitochondrial function in advanced primary open-angle glaucoma and leber hereditary optic neuropathy. PLoS One 2015;10(10):e0140919; doi: 10.1371/journal.pone.0140919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bergen NJ, Crowston JG, Kearns LS, et al. Mitochondrial oxidative phosphorylation compensation may preserve vision in patients with OPA1-linked autosomal dominant optic atrophy. PLoS One 2011;6(6):e21347; doi: 10.1371/journal.pone.0021347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Laan M, Horvath SE, Pfanner N. Mitochondrial contact site and cristae organizing system. Curr Opin Cell Biol 2016;41:33–42; doi: 10.1016/j.ceb.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Varanita T, Soriano ME, Romanello V, et al. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab 2015;21(6):834–844; doi: 10.1016/j.cmet.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Lee J, Singh K, et al. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim Biophys Acta 2012;1823(1):56–66; doi: 10.1016/j.bbamcr.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania RS, Kolawa NJ, et al. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife 2013;2:e00308; doi: 10.7554/eLife.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana MP, Levytskyy RM, Anand R, et al. Protease OMA1 modulates mitochondrial bioenergetics and ultrastructure through dynamic association with MICOS complex. iScience 2021;24(2):102119; doi: 10.1016/j.isci.2021.102119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Garcia-Prieto J, Baker MJ, et al. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science 2015;350(6265):aad0116; doi: 10.1126/science.aad0116 [DOI] [PubMed] [Google Scholar]
- Wai T, Saita S, Nolte H, et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep 2016;17(12):1844–1856; doi: 10.15252/embr.201642698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet 2005;39:359–407; doi: 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Mishra P, Garbis SD, et al. Identification of new OPA1 cleavage site reveals that short isoforms regulate mitochondrial fusion. Mol Biol Cell 2021;32(2):157–168; doi: 10.1091/mbc.E20-09-0605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015;524(7566):481–484; doi: 10.1038/nature14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Amon A. MitoCPR—A surveillance pathway that protects mitochondria in response to protein import stress. Science 2018;360(6385):eaan4146; doi: 10.1126/science.aan4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb RN, Khaw PT. Primary open-angle glaucoma. Lancet 2004;363(9422):1711–1720; doi: 10.1016/S0140-6736(04)16257-0 [DOI] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial dynamics in model organisms: What yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin Cell Dev Biol 2010;21(6):542–549; doi: 10.1016/j.semcdb.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 2014;346(6210):748–751; doi: 10.1126/science.1257522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PA, Harder JM, Foxworth NE, et al. Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science 2017;355(6326):756–760; doi: 10.1126/science.aal0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingo AP, Dammer EB, Breen MS, et al. Large-scale proteomic analysis of human brain identifies proteins associated with cognitive trajectory in advanced age. Nat Commun 2019;10(1):1619; doi: 10.1038/s41467-019-09613-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DM, Segawa M, Kondadi AK, et al. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J 2019;38(22):e101056; doi: 10.15252/embj.2018101056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 2015;524(7566):485–488; doi: 10.1038/nature14951 [DOI] [PubMed] [Google Scholar]
- Wu X, Li L, Jiang H. Doa1 targets ubiquitinated substrates for mitochondria-associated degradation. J Cell Biol 2016;213(1):49–63; doi: 10.1083/jcb.201510098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Peng G, Wang Y, et al. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell 2011;22(3):291–300; doi: 10.1091/mbc.E10-09-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Lartigue L, Perkins G, et al. OPA1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol Cell 2008;31(4):557–569; doi: 10.1016/j.molcel.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Qi Y, Ricketson D, et al. Structural analysis of a trimeric assembly of the mitochondrial dynamin-like GTPase Mgm1. Proc Natl Acad Sci U S A 2020;117(8):4061–4070; doi: 10.1073/pnas.1919116117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Yang WY. Bit-by-bit autophagic removal of Parkin-labelled mitochondria. Nat Commun 2013;4:2428; doi: 10.1038/ncomms3428 [DOI] [PubMed] [Google Scholar]
- Yang L, Na CL, Luo S, et al. The phosphatidylcholine transfer protein Stard7 is required for mitochondrial and epithelial cell homeostasis. Sci Rep 2017;7:46416; doi: 10.1038/srep46416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 2003;112(1):41–50; doi: 10.1016/s0092-8674(02)01250-3 [DOI] [PubMed] [Google Scholar]
- Yu R, Jin SB, Lendahl U, et al. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J 2019;38(8):e99748; doi: 10.15252/embj.201899748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zara V, Ferramosca A, Robitaille-Foucher P, et al. Mitochondrial carrier protein biogenesis: Role of the chaperones Hsc70 and Hsp90. Biochem J 2009;419(2):369–375; doi: 10.1042/BJ20082270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Frerman FE, Kim JJ. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc Natl Acad Sci U S A 2006;103(44):16212–16217; doi: 10.1073/pnas.0604567103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Li H, Song Z. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep 2014;15(5):576–585; doi: 10.1002/embr.201338240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zode GS, Kuehn MH, Nishimura DY, et al. Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma. J Clin Invest 2015;125(8):3303; doi: 10.1172/JCI82799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita Rendon O, Fredrickson EK, Howard CJ, et al. Vms1p is a release factor for the ribosome-associated quality control complex. Nat Commun 2018;9(1):2197; doi: 10.1038/s41467-018-04564-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita Rendon O, Shoubridge EA. LONP1 is required for maturation of a subset of mitochondrial proteins, and its loss elicits an integrated stress response. Mol Cell Biol 2018;38(20):e00412-17; doi: 10.1128/MCB.00412-17 [DOI] [PMC free article] [PubMed] [Google Scholar]