Abstract
Autophagy is a dynamic process that regulates the selective and nonselective degradation of cytoplasmic components, such as damaged organelles and protein aggregates inside lysosomes to maintain tissue homeostasis. Different types of autophagy including macroautophagy, microautophagy, and chaperon-mediated autophagy (CMA) have been implicated in a variety of pathological conditions, such as cancer, aging, neurodegeneration, and developmental disorders. Furthermore, the molecular mechanism and biological functions of autophagy have been extensively studied in vertebrate hematopoiesis and human blood malignancies. In recent years, the hematopoietic lineage-specific roles of different autophagy-related (ATG) genes have gained more attention. The evolution of gene-editing technology and the easy access nature of hematopoietic stem cells (HSCs), hematopoietic progenitors, and precursor cells have facilitated the autophagy research to better understand how ATG genes function in the hematopoietic system. Taking advantage of the gene-editing platform, this review has summarized the roles of different ATGs at the hematopoietic cell level, their dysregulation, and pathological consequences throughout hematopoiesis.
1. Autophagy
The hallmark of autophagy is the cellular proteins and organelle clearance through lysosomal degradation and recycling of cytoplasmic materials, such as lipids, proteins, and organelles into smaller forms and release of amino acids, nucleotides, and fatty acids to maintain cellular homeostasis [1]. Various stress stimuli including oxidative stress, redox stress, endoplasmic reticulum (ER) stress, nutrient deprivation, mitochondrial damage, and hypoxia are responsible for inducing autophagy [2]. Autophagy machinery is orchestrated by the autophagy-related (ATG) genes, and till now, approximately 40 ATG genes have been identified in Saccharomyces cerevisiae yeast. In yeast, autophagy is initiated in response to various intracellular and extracellular stimulating factors, whereas ATG13 anchors unc-51 like autophagy-activating kinase 1 (ULK1) to form a cup-shaped preautophagosomal structure (PAS) [3], and subsequently, Atg proteins including Atg2 and Atg18 get localized to initiate autophagosome biogenesis [4, 5]. Most of the yeast ATG genes have mammalian counterparts which regulate various steps of the autophagy pathway [6]. Mammalian autophagy initiation is associated with the formation of a cup-like or omega-shaped (Ω) lipid bilayer membrane, also known as an “omegasome” near the endoplasmic reticulum (ER). Omegasomes further colocalize with phosphatidylinositol 3-phosphate (PI(3)P) and autophagosomal marker microtubule-associated protein light chain 3 (MAP-LC3) [7]. Therefore, at the initiation of PAS or omegasome, phagophores increase in size and elongate into cup-shaped structures before engulfing the cellular materials. Apart from yeast and mammals including Mus musculus, there are nematodes such as Caenorhabditis elegans and insects such as Drosophila melanogaster and Bombyx mori, where ATG genes have been identified [8–10]. Due to the diverse regulatory functions of ATG genes in metabolic homeostasis, autophagy plays an essential role in maintaining cellular development, and its dysregulations are associated with mammalian aging, developmental defects, neurodegeneration, and cancer [11–14].
To interpret autophagy more efficiently, measuring autophagy flux is critical, as it indicates whether autophagy cargos have been successfully degraded at the end of the autophagy process. The term “autophagic flux” refers to the entire autophagic process, which includes the formation, maturation, fusion, and disintegration of autophagosomes with lysosomes as well as the release of macromolecules recycled back into the cytosol. However, under nutrient-rich conditions, autophagy does not affect metabolic flux [15]. Flux can be quantitatively measured by LC3-II turnover using immunoblot analysis in the presence and absence of lysosomal inhibitors, such as chloroquine, bafilomycin A1, and NH4Cl [16]. Furthermore, an autophagic flux report system has been developed, which combines LC3 with the acid-resistant mCherry and the acid-sensitive GFP to measure autophagosome maturation and degradation. In this system, both LC3 with red fluorescent mCherry and green fluorescent GFP is fused to form yellow puncta and detected in autophagosomes [17]. Due to GFP degradation by acid lysosomal proteases, the green fluorescence of autophagosomes is lost when autophagosomes fuse with lysosomes, which results in LC3 emitting only red fluorescence. This dynamic switch indicates an active autophagic flux process when yellow fluorescence is switched to red. Additionally, p62 accumulation can be used as an autophagy flux indicator [18].
2. Hematopoiesis
Hematopoiesis is the process by which the multipotent hematopoietic stem cells (HSCs) give rise to all blood cell lineages such as myeloid cells, leukocytes, erythrocytes, and platelets through the generation of progenitor cells [19]. These progenitor cells further undergo gradual fate restriction and are eventually identified as mature blood cells. Physiologically, all these blood cells reside inside the bone marrow microenvironment known as the HSC niche [20]. In mammals, hematopoiesis occurs sequentially in four distinct areas including the yolk sac, aorta-gonad mesonephros (AGM), fetal liver, and bone marrow [19]. The first wave of blood cell production inside the yolk sac is called “primitive.” Primitive hematopoiesis generates red blood cells for tissue oxygenation as the embryo grows quickly. Hematopoiesis in the primitive state is transient and is quickly replaced by adult-type “definitive” hematopoiesis. Throughout embryogenesis, definitive hematopoiesis is associated with hematopoietic stem cells and multipotent progenitors (HSCs/MPPs), which later become mature blood cells and immune cells [21]. Normal hematopoiesis maintains a unique balance between cellular differentiation, quiescence, and self-renewal capacity of hematopoietic stem and progenitor cells (HSPCs).
3. Different Forms of Autophagy in Hematopoiesis
Macroautophagy, the most thoroughly researched form of autophagy, hereafter referred to as autophagy is the cellular proteins and organelle clearance mechanism through lysosomal degradation. Autophagy has been previously described as a nonselective or canonical nutrient recycling phenomenon that occurs ceaselessly at the basal level and delineates the random consumption as well as successive degradation of cytoplasmic organelles, pathogens, and aggregated proteins [1]. In nonselective macroautophagy, various external stimuli including pathogen infection, nutrient deprivation, and radiation therapy enhance autophagy by upregulating autophagy mediators such as ATG3, ATG5, ATG12, and ATG16L1 (Figure 1). During hypoxia-mediated nonselective macroautophagy, hypoxia-inducible factor-1α (HIF-1α) upregulates the expression of mitophagy marker BNIP3 (BCL2/adenovirus E1B 19 kDa-interacting protein 3) [22]. A detailed overview of the nonselective or canonical macroautophagy is narrated in Figure 2.
Figure 1.
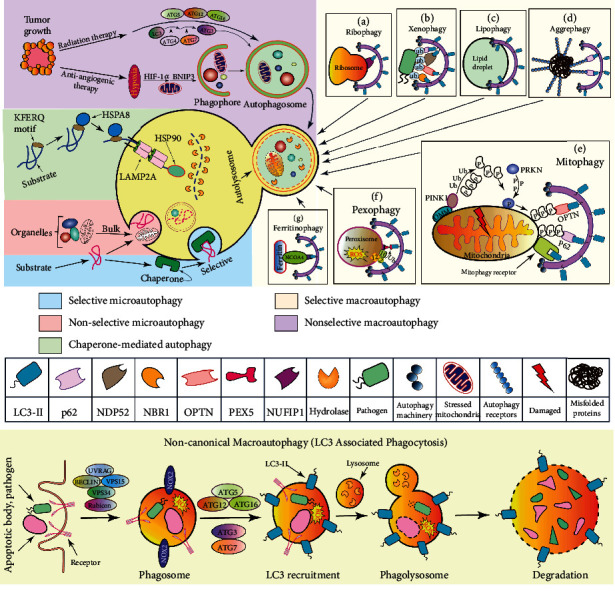
Schematic representation of the different types of autophagy. Selective macroautophagy is a strictly regulated cytosolic cargo degradation pathway for the removal of excess ribosomes—ribophagy (a), intracellular pathogens—xenophagy (b), lipid droplets—lipophagy (c), superfluous protein aggregates—aggrephagy (d), polluted mitochondria—mitophagy (e), dispensable peroxisomes—pexophagy (f), and ferritin iron—ferritinophagy (g). Under stress, tumorigenesis, anticancer therapy, and nonselective macroautophagy are initiated with the isolation of the phagophore membrane, autophagosome formation, maturation, and degradation in the lysosome. Likewise, macroautophagy and microautophagy can be selective, while most microautophagy is the nonselective and bulk degradation of cargo molecules. Chaperone-mediated autophagy (CMA) is the translocation of the motif-bearing substrate molecules to the lysosomal membrane, whereas the lysosomal hydrolases degraded the proteins and release amino acids. During LAP, a common form of noncanonical macroautophagy, ATGs from the PIK3 complex, ATG5-ATG12, and LC3 conjugation system incorporated in phagolysosome formation and its degradation without the ULK1 complex autophagy.
Figure 2.
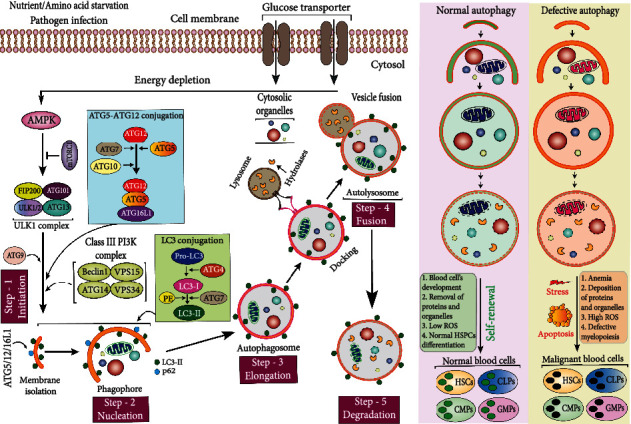
Macroautophagy and its role in hematopoiesis. During macroautophagy, AMPK activates the ULK1 complex and consequently induces the membrane isolation process where PI3Ks and ATG9 take part as positive regulators and enable autophagosome formation with the aid of ATG12-ATG5 and the LC3 conjugation systems. The ATG5–ATG12–ATG16L1 complex induces LC3 conjugation, whereas LC3 is cleaved by ATG4 protease to form LC3-I, and cytosolic LC3-1 is further conjugated with PE to form LC3-II. Afterward, autophagosomes come in contact with the lysosome which has the hydrolase enzymatic activity to fuse with autophagosome to form autolysosome for cytosolic cargo degradation. The normal state of autophagic activity is essential for the maintenance of blood cell homeostasis. Defective autophagy results in imbalanced hematopoiesis and impeded HSCs' self-renewal, leading to the generation of malignant blood cell types.
In selective autophagy, different autophagy receptors play critical roles in regulating cellular homeostasis by facilitating the turnover of proteins, aggregates, organelles, and pathogens via lysosomal recycling. As hematopoietic cells need to meet their specific intracellular turnover requirements, the evolutionary conserved selective degradation pathways are increasingly being recognized for their importance to orchestrate macromolecular complexes and organelles intracellularly. In ribophagy, nuclear FMR1-interacting protein 1 (NUFIP1) selectively targets ribosomes to the phagophore (Figure 1(a)) [23], while in xenophagy, pathogen-containing vacuole disruption leads to xenophagy, while pathogens are targeted by ubiquitin (Ub) ligases, following autophagy adaptor protein recruitment including nuclear dot protein 52 kDa (NDP52), a neighbor of BRCA1 gene 1 (NBR1), optineurin (OPTN), and p62 that binds LC3 on the phagophore membrane (Figure 1(b)) [24–27]. However, the biological functions and molecular mechanisms of both xenophagy and ribophagy in hematopoietic cells are yet to be elucidated. Another selective autophagy is lipophagy which includes phagophore formation and expansion on lipid droplets (Figure 1(c)) [28]. During starvation and nutrient stress, lipophagy releases fatty acids and other lipid molecules from lipid droplets and maintains HSCs and myeloid cell biology via lipoprotein lipase (LPL) mediated lipid catabolism [29, 30]. It has been indicated that LPL activity is essential to regulate definitive hematopoiesis. When LPL activity abrogated, HSC markers Runx1 and Cmyb, erythrocyte marker Betaglobin, and lymphocyte marker Rag1 expression have been diminished, which indicates the erythropoietic and lymphopoietic defects as well as hyperlipidemia [29]. In aggrephagy, aggregated proteins become ubiquitinated and selected for autophagic degradation by ubiquitin-binding autophagy receptors such as p62, NBR1, OPTN, NDP52, and Tollip (Figure 1(d)) [31]. However, the role of aggrephagy in hematopoiesis and leukemia remains poorly understood. There has been evidence implicating aggrephagy in autophagic degradation of aggregated oncoproteins in acute leukemia, such as promyelocytic leukemia (PML) retinoic acid receptor alpha (RARA) fusion protein degradation via p62-mediated aggrephagy [32]. A more common and widely studied form of selective autophagy is mitophagy. During mitophagy, mitochondria are selectively degraded by PTEN-induced kinase 1 (PINK1) dependent pathway and receptor-mediated mechanisms [33]. In PINK1-dependent mitophagy, PINK1 phosphorylates ubiquitin, and PRKN promotes outer mitochondrial membrane (OMM) protein ubiquitination. Ubiquitinated proteins are then linked to ubiquitin-binding autophagy receptors including optineurin (OPTN) and p62 that subsequently interact with LC3-II, triggering the degradation of mitochondria (Figure 1(e)) [34]. Most HSPCs are in a quiescent state at basal level in the bone marrow (BM) stem cell niche, and PINK1-dependent mitophagy plays important roles in maintaining hematopoietic niche, as well as quiescence, stress response, and self-renewal of HSPCs [35]. HSC quiescence and stemness are also maintained through mitophagy during aging, which actively removes healthy mitochondria to preserve HSC regenerative capacity [36]. Mice with conditional deletion of Atg12 have shown active mitochondrial mass, increased ROS, loss of quiescence and promyeloid differentiation, and hyperactive oxidative metabolism in HSCs. Further treatment with autophagy inhibitor bafilomycin A1 (BafA1) has exacerbated the condition in aged HSCs, indicating an Atg12 autophagy-dependent role of mitochondria in HSC homeostasis [36]. Furthermore, mitophagy is also essential for the self-renewal and expansion of HSCs [37]. Ito et al. have used Tie2-GFP+ HSCs as HSC markers, while their gene expression data have shown a higher expression of mitophagy-related genes, including Pink1, Parkin (Park2), Map1lc3a (Lc3), Optineurin, and p62/Sqstm1 in Tie2-GFP+ HSCs. Additionally, they found that silencing mitophagy via Park2 or Pink1 suppression not only eliminated Tie2-GFP+ cell growth but also inhibited the HSC cell maintenance, indicating that mitochondrial clearance is critical to maintaining HSC stemness by inducing mitophagosome formation [37]. Most of these genes were further upregulated by the activation of the PPAR-FAO pathway. However, mitophagy plays a paradoxical role in AML cells and normal hematopoietic cells. An increased level of mitophagy has been documented in AML cells, whereas macroautophagy inhibitors, like chloroquine (CQ), Lys05, and bafilomycin A1, are believed to decrease mitophagy in AML cells and increase antileukemic effects under hypoxia [38]. The results of this study open up a new therapeutic avenue for autophagy inhibitors in the treatment of AML. Selective degradation of peroxisomes refers to as pexophagy. A key component of mammalian pexophagy is the ubiquitination of peroxisomal proteins and the recognition of those proteins by SQSTM1 and NBR1 [39, 40]. Several studies identified the peroxisomal biogenesis factor 5 (PEX5) monoubiquitination as a cargo signal for peroxisome degradation [41, 42]. During pexophagy, PEX5 phosphorylation also induces PEX2-PEX10-PEX12 E3 ligase-mediated ubiquitination and Ub–PEX5 recognition (Figure 1(f)) [43]. Pexophagy can regulate mesenchymal stem cell aging in the bone marrow by inducing PEX10 and PEX14 activity through endothelial PAS domain-containing protein 1 (Epas1) regulation and suppressing ROS levels [44]. Pexophagy may also play a critical role in regulating monocyte biology [45]. As reported by Zhang et al., the PEX5 peroxisome import receptor binds serine/threonine kinase ataxia-telangiectasia mutated (ATM) and localizes it to peroxisomes. ATM kinase is activated by ROS which further phosphorylates PEX5 and subsequently monoubiquitinates it [46]. There is evidence that some widely used DNA-damaging agents in AML clinical treatment, such as doxorubicin, mitoxantrone, and etoposide, induced DNA damage response and ATM activation, which indicates that these agents could trigger pexophagy [47]. Further research is needed to determine whether pexophagy affects AML progression and therapeutic response. Ferritinophagy plays a critical role in the production and maintenance of erythrocytes and involves the iron-dependent ferritin (an iron-storing protein) breakdown. In ferritinophagy, ubiquitinated nuclear receptor coactivator 4 (NCOA4) selectively mediates the degradation of ferritin by targeting ferritin to LC3-II (Figure 1(g)) [48]. Using the zebrafish model and cultured cells, Mancias et al. have shown that ferritinophagy via selective cargo receptor NCOA4 regulates erythropoiesis [49]. Recently, a mice study has suggested that NCOA4-mediated ferritinophagy in macrophages plays essential roles to maintain erythropoiesis [50]. There is another form of selective autophagy termed crinophagy, whereas the secretory granules are directly fused with late endosomes or lysosomes for disposal [51]. As blood cell differentiation involves the secretion of granules or vesicles that may cause local tissue damage, crinophagy may play an important role to limit such damage through the uptake of granules by barrier cells and regulatory stromal cells while maintaining blood cell homeostasis.
In selective macroautophagy, the shape and size of the phagophore are regulated by the cargo itself and a variety of adaptor proteins including p62, optineurin (OPTN), NDP52, and NBR1 to link cargoes to the autophagy machinery [52]. Most of these adaptor proteins are associated with hematopoietic cell maintenance and blood malignancies such as acute myeloid leukemia (AML). AML is a heterogeneous disease and a common type of acute leukemia with poor survival and prognosis. It results from aberrant changes in the hematopoietic cells, which block myeloid differentiation and suppress hematopoiesis [53]. Selective autophagy receptor p62 is essential for hematopoietic cell growth and mitophagy to maintain mitochondrial integrity. Nguyen et al. have suggested that by depleting p62, mice develop myeloid leukemia and exhibit impaired mitophagy [54]. In hematological malignancies, OPTN has been identified as a prognostic biomarker for AML development and progression [55]. Moreover, simultaneous inhibition of autophagy receptors including p62, OPTN, and NDP52 sensitizes AML cells to the chemotherapy drug cytarabine [56]. In AML patients, quantitative real-time PCR analysis has suggested the downregulation of selective autophagy marker NBR1 expression along with other autophagy proteins such as LC3, Beclin1, UVRAG, and Rubicon [57].
Microautophagy can be either selective or nonselective. Nonselective microautophagy (generally refers to as microautophagy) is the random engulfment of intracellular organelles by tubular invagination or protrusions of the lysosomal membrane and the degradation of those bulk cytosolic cargos [58]. This kind of microautophagy is frequently spotted in mammalian cells. Conversely, selective microautophagy involves the degradation of substrates including lipid droplets (microlipophagy), peroxisomes (micropexophagy), and portions of the nucleus in a manner called piecemeal microautophagy of the nucleus (PMN) [59]. These forms of selective microautophagy are generally observed in yeasts. However, the molecular and functional details of microautophagy in hematopoietic cells and leukemia remain unknown.
Cellular proteins with a KFERQ-like motif are selectively degraded by lysosomes through CMA. During this process, KFERQ-like motif-bearing proteins are recognized and bound by a cytosolic chaperone named heat shock protein family A (Hsp70) member 8 (HSPA8) to form a translocation complex (Figure 1). Then, the complex is delivered to the lysosomal membrane where LAMP2A multimerization allows the channel formation to deliver proteins into the lysosomal lumen by lumenal chaperone heat shock protein 90 (HSP90). Lysosomal hydrolases break down the protein and release amino acids that are transported into the cytosol [60]. CMA is critical for HSC homeostasis. It has been suggested that quiescent HSCs derived from young mice have higher basal CMA activity than those derived from older mice [61]. In contrast, the deletion of Lamp2a blocks CMA, which indicates that CMA is vital for preserving HSC functionality and prevents the depletion of activated HSCs. Moreover, cells lacking CMA due to Lamp2a ablation produce less ATP, reduce glycolytic and oxidative phosphorylation activity for quiescent HSCs, generate more ROS, and accumulated proteins involve in metabolism. Myelodysplastic syndrome (MDS) AML patients exhibit resistance to azacytidine and poor survival as a result of a significant defect in CMA caused by a lack of LAMP2 expression, and lysosomal autophagy inhibitors such as CQ can be effective on AML cells lacking LAMP2 [62]. CMA activation in hematopoietic malignancies is also associated with the elimination of oncogenes such as mutated TP53 (tumor protein 53) and MLLT11 (myeloid/lymphoid or mixed-lineage leukemia translocated to 1q) [63, 64]. Similarly, AML patients with low LAMP2 expression levels have poor overall survival due to decreased CMA activity [65]. In chronic lymphocytic leukemia (CLL), AKT inhibitors and mTOR inhibitors (Torin-1) effectively restore CMA activity while removing prooncogenic proteins involved in leukemia initiation and progression [66]. Altogether, it appears that increasing CMA activity in the treatment of hematological malignancies will be an excellent therapeutic strategy, either alone or when combined with conventional chemotherapy.
While autophagy acts as a quality control process, the cargo selection process could be independent of autophagy. Noncanonical autophagy is a good example that utilizes some components of the macroautophagy machinery, while cargo identification and sequestration for autophagy occur upstream of autophagosome formation to process the extracellular cargo for lysosomal degradation [67, 68]. For instance, in LC3-associated phagocytosis (LAP), LC3 binds to phagosomes, modulating immune responses, antigen presentation, LC3-associated endocytosis, and tumor immune tolerance [69–72]. LAP functions in phagosome maturation and subsequent signaling events, while it is associated with the incorporation of the most upstream autophagic players such as Beclin1, VPS34, and VPS15 [73]. Importantly, LAP catalyzes the formation of a single membrane LAPosome other than double-membraned autophagosomes, whereas LAP exclusively utilizes the ultraviolet radiation resistance-associated gene (UVRAG) which contains the class III phosphatidylinositol 3-kinase (PI3K) complex (Figure 1, lower panel) [74]. Moreover, Rubicon (RUN domain protein as Beclin1-interacting and cysteine-rich-containing) has been identified as a key modulator of LAP that is noncanonical and independent of ULK kinases [75]. It is a negative regulatory protein of the class III PI3K complex in macroautophagy [76] and is critical for the localization of phosphatidylinositol 3-phosphate (PI(3)P) and stabilization of the active NOX2 (NADPH oxidase 2) complex to promote optimal ROS generation during successful LAP [74]. Several noncanonical autophagy has been previously identified, such as ammonia-induced ULK1/2-independent autophagy where it is suggested that ammonia-induced autophagy is independent of ULK1/2 kinases but requires ATG5 [77]. Recently, acetyltransferase p300-dependent class III phosphoinositide 3-kinase VPS34 activation and deacetylation have been demonstrated as noncanonical autophagy in which the upstream kinases of VPS34 such as ULK1 and AMPK can be bypassed [78]. This alternative autophagy underscores the involvement of canonical autophagosome biogenesis and the possibility of autophagy regulation in pathological consequences.
LC3-associated phagocytosis suppresses AML progression in bone marrow macrophages. The increased phagocytic activity of LAP led to the activation of the stimulator of the IFN genes (STING) pathway, which ultimately suppresses AML cell growth [79]. Furthermore, LAP promotes myeloid cell immunity to tumors, while single-cell RNA sequencing in tumor-associated macrophages (TAM) suggests that LAP defects trigger proinflammatory gene expression and type I interferon responses through STING modulation [72].
3.1. Macroautophagy Mechanism and Its Roles in Hematopoiesis
Autophagy induction is triggered through distinct signaling cascades under starved conditions and pathogen infection phage that results in the repression of the mammalian target of rapamycin (mTOR) [80, 81]. Opposingly, in nutrient-rich conditions, mTOR switches on and prevents ULK1 activation and disrupts the ULK1 association with adenosine monophosphate activated protein kinase (AMPK) [82]. During low glucose content, AMPK is activated and mTORC1 is inhibited, allowing ULK1 phosphorylation by AMPK interaction and eventually activating autophagy [82]. Next, ULK1 forms a tetrameric complex with FAK family kinase-interacting protein of 200 kDa (FIP200), ATG13, and ATG101 to recruit the VPS34 complex for phagophore isolation and autophagosome initiation [83, 84] (Figure 2). The class III PI3K catalytic subunit VPS34 then interacts with ATG14, VPS15, and Beclin1 to form a protein complex (PI3KC3) which is essential for the initiation and expansion of autophagosomes [85]. Furthermore, PI3KC3 synthesizes the lipid phosphatidylinositol 3-phosphate (PI(3)P), which recruited WD-repeat protein interacting with phosphoinositide (WIPI) proteins, and subsequently, WIPIs recruit Atg16L1 that conjugates with the autophagosome marker microtubule-associated protein 1A/1B-light chain 3 (LC3; mammalian ortholog of ATG8) through the Atg5/12/16L1 complex recruitment [86]. During autophagosome maturation, LC3 translocates from the cytosol to the isolation membrane where the cysteine protease ATG4 cleaves pro-LC3 to generate LC3-I. Then, LC3-I is subsequently transferred by ATG7 to the expanded phagophore membrane where LC3-I travels through Atg3, lipidated to LC3-II, and attached to the autophagosomal membrane [87]. In parallel, the ATG5/12/16L complex stimulates the conjugation of LC3-I to phosphatidylethanolamine (PE) to form lipidated LC3 (LC3-II) which binds to receptor molecule p62 inside the inner and outer membranes of the autophagosome [88] (Figure 2). Additionally, LC3-II is also involved in phagophore extension and closure during autophagosome formation. The next step is the fusion of the matured autophagosome with the hydrolase enzyme containing lysosome, referred to as autophagolysosome [89].
Hematopoietic cells are relatively low in basal autophagy, particularly in abundant nutrients and energy supply. The basal level of autophagy contributes to cellular homeostasis and protects cells against detrimental and dysfunctional organelles. Under nutrient-deprived conditions, normal blood cells undergo autophagy, which provides energy and building blocks for cellular homeostasis by breaking down misfolded proteins and damaged organelles [90]. During normal hematopoiesis, autophagy clears mitochondria, keeps a lower ROS level, and maintains the HSC stemness and genomic stability. Conversely, in malignant hematopoiesis, autophagy leads to excessive cell survival, low glucose consumption, and cell proliferation, which in turn induces malignant invasion by promoting the differentiation of leukemic cells. Besides, impaired autophagy during hematopoiesis may result in damaged organelles, protein aggregation, and excessive ROS and mitochondrial mass accumulation, leading to DNA damage and genomic mutation in the hematopoietic system (Figure 2) [91, 92]. Consequently, autophagy deficiency may cause HSC impairment, aberrant myeloproliferation, and severe anemia, while malignant blood cells, such as AML cells, exhibit a higher proliferation rate, apoptosis, and drug (chemotherapy and radiotherapy) resistance [93]. Interestingly, studies have also linked macroautophagy to AML development. In AML patients, high expression levels of key autophagic genes such as ATG7 and Beclin1 are associated with poor clinical outcomes and short remission duration [94, 95].
In contrast, genetic inhibition of murine Atg5 and Atg7 can prolong leukemia survival and delay the elimination of leukemia-initiating cells [96]. Altogether, these data indicated that both heightened and deficient autophagy activity may require for the malignant progression in AML. Currently, for most patients suffering from AML, primary and secondary drug resistance continues to be a major problem. Therefore, researchers are primarily exploring the mechanisms of drug resistance to develop next-generation AML therapies and designing combination regimens with the ultimate goal of eliminating all subclones of the disease and increasing the cure rate for AML. Overall, the canonical autophagy-dependent and canonical autophagy-independent (noncanonical) functions of the ATGs have been summarized in Table 1.
Table 1.
Canonical autophagy-dependent and canonical autophagy-independent function(s) of different autophagy marker proteins.
| Protein name | Autophagy step(s) | Canonical autophagy-dependent function(s) | Rf. | Canonical autophagy-independent function(s) | Rf. |
|---|---|---|---|---|---|
| ULK1 or ATG1 | Initiation | Mitochondrial respiration, ATP production, lipid metabolism, and mitochondrial and ribosomal clearance | [97, 98] | Cell death and apoptosis, endocytosis, immune signaling, antiproliferative and antineoplastic effects in MPNs, ER-to-Golgi trafficking, cellular homeostasis, ammonia-induced autophagy, and endosomal trafficking | [99–106] |
| ATG2A or ATG2B | Elongation | Regulates lipid homeostasis, promotes Atg9-Atg18 interaction, and programmed cell size reduction | [107–109] | iDISC-dependent caspase-8 activation, apoptosis, and lipid droplet localization | [110, 111] |
| ATG3 | Elongation | Induces HIV infection and cell death | [112] | LAP, endosomal trafficking, and apoptosis | [74, 113, 114] |
| ATG4B or ATG4D | Elongation | Sense balance and otoconial development induce HIV infection and cell death | [112, 115] | LAP, mitochondrial dysfunction, and apoptosis | [74, 116] |
| ATG5 | Elongation | Maintenance of innate lymphocytes, skeletal homeostasis, and antiviral immune responses | [117–119] | Immunity, intracellular pathogen killing, apoptosis, and adipogenesis | [120–123] |
| ATG6 or Beclin1 | Nucleation | Induces HIV infection and cell death | [112] | Apoptosis, cell death, cancer cell growth, embryogenesis, tumor suppression, STAT3 phosphorylation, DNA damage repair, receptor degradation, and cytokinesis and induces viral transmission and improves the life span | [124–132] |
| ATG7 | Elongation | Maintains cellular and behavioral responses and regulates potassium (K+) level in hypokalemia | [133, 134] | Cell shrinkage, cell cycle arrest, mitochondrial clearance, adipogenesis, and ISC integrity maintenance and promotes neuronal health and longevity | [109, 121, 135–137] |
| ATG8 or LC3 | Cargo selection | Maintains tissue homeostasis | [138] | LAP, apoptosis, virus replication, cancer cell survival, lysosome biogenesis, and exocytosis | [68, 120, 139–141] |
| ATG9 | Initiation | Pathogenesis of POI | [142] | Maintains lysosomal degradation, axonal degeneration, STING, and TBK1 assembly | [143, 144] |
| ATG10 | Elongation | Not known | - | Apoptosis, deficiency leads to ALS and FTD molecular defects, lysosomal degradation, and suppression of HCV replication | [145–147] |
| ATG12 | Elongation | Mitochondrial homeostasis, cell death, antiviral immune responses, osteoclast secretion, and pathogen control | [119, 148, 149] | Endosomal trafficking, mitochondrial apoptosis, and endosome to lysosome trafficking | [74, 150, 151] |
| ATG13 | Initiation | Cell cycle progression | [152] | Control virus replication and cardiac development |
[153, 154] |
| ATG14 | Nucleation | Autophagosome–endolysosome fusion | [155] | Autophagic cell death | [156] |
| ATG16l1 | Elongation | Urothelial vesicle trafficking | [157, 158] | Apoptosis | [120, 159] |
| ATG18 | Elongation | Programmed cell size reduction | [109] | Neural homeostasis | [160] |
| ATG101 | Initiation | Maintaining respiratory function | [161] | Not known | - |
| FIP200 | Initiation | Maintaining respiratory function | [161] | Control virus replication | [153] |
| VPS15 | Nucleation | Not known | - | Skeletal muscle function, endocytosis, and neuronal migration | [130, 162, 163] |
| VPS34 | Nucleation | T-cell homeostasis | [164] | Endocytosis, receptor degradation, and cytokinesis | [130] |
LAP: LC3-associated phagocytosis; iDISC: intracellular death-inducing signaling complex; Rf: reference(s); PAS: preautophagosomal structure; ISC: intestinal stem cell; STING: stimulator of IFN genes; TBK1: TANK-binding kinase 1; POI: primary ovarian insufficiency; FTD: frontotemporal dementia; ALS: amyotrophic lateral sclerosis; HCV: hepatitis C virus; VPS34: vacuolar protein sorting 34.
3.2. Autophagy in the Maintenance and Survival of HSCs
Autophagy is crucial to the proper development, survival, and maintenance of HSCs during acute metabolic stress. HSCs lacking autophagy exhibit impaired self-renewal activity as well as other characteristics similar to those of aging HSCs, which indicates the essential role of autophagy in regulating HSC pluripotency and preventing HSC aging (Table 2). Mice with HSC-specific conditional deletion of Atg12 result in defective HSC self-renewal and metabolism, myeloid cell expansion, and premature blood aging [36]. Furthermore, MitoTracker Green (MTG) labeling, immunofluorescence staining for TOM20, and elevated levels of p62 have suggested an increased mitochondrial mass and depleted bulk autophagy in Atg12-deficient HSCs. This suggests that PINK1-dependent mitophagy is activated in HSCs. Conditional deletion of Atg7 in adult mice HSCs leads to impaired HSC activity, enhanced mitochondrial metabolism, and severe myeloproliferation, which indicates that Atg7-deficient bone marrow cells are only capable of sustaining short-term hematopoiesis [165, 166]. However, Atg7-deficient neonatal HSCs have minimal effect on hematopoiesis or metabolic state, as they display long-term regenerative capabilities similar to wild-type neonatal HSCs [166]. Similarly, mice with HSC-specific ablation of Fip200 and Atg5 have shown significant reduction in HSCs, increased ROS and HSC proliferation, decreased HSC reconstitution ability in the bone marrow, and survival defects including severe anemia and lymphopenia [167, 168]. Mechanistically, it appears that impaired autophagy-mediated mitochondrial clearance underlies the functional defects in HSCs. In addition to maintaining normal hematopoiesis, macroautophagy plays an important role in regulating leukemic cell survival and the progression of AML [169]. It has been demonstrated that the inactivation of ATG7 or ATG5 in mice increases survival and suppresses functional leukemic HSCs as these are the principal cell types in AML development [96]. Similar to the murine AML model, AML patients have also shown increased Notch signaling and lower levels of autophagy in their HSCs [170]. Since inhibiting the Akt-mTOR pathway can trigger autophagy activation and control HSC homeostasis, thus this network is essential for self-renewal, survival, differentiation, and preventing HSCs from becoming leukemia stem cells (LSCs) [171]. Collectively, these findings have demonstrated the indispensable role of autophagy in HSC biology.
Table 2.
Autophagy defects during hematopoietic differentiation in mice.
| Model system | Target cells | Hematopoietic outcomes | Rf. |
|---|---|---|---|
| Fip200flox/flox × Tie2-Cre | HSCs | Myeloproliferation, HSC apoptosis, and severe anemia. | [167] |
| Atg5flox/flox × Vav-Cre | Lymphopenia, anemia, accumulation of monocytes, macrophages, and neutrophils. | [200] | |
| Atg12flox/flox × Mx1-cre | Increased apoptosis in HSCs and loss of HSCs. | [201] | |
| Atg7flox/flox × Vav-Cre | Loss of HSC function, severe myeloproliferation, defective megakaryocyte homeostasis, platelet aggregation, apoptosis in bone marrow, defective HSPC quiescence, and cell cycle arrest. | [165, 202, 203] | |
|
| |||
| Atg5flox/flox × CD19-Cre | B cells | Defective antibody responses in B cells, increased cell death in BM, depletion in B-1 B cells, defective B cell homeostasis, EMH, and anemia. | [183, 184, 187] |
| Atg5flox/− × CD21-Cre | Decreased T1 B cells and follicular B-cell numbers, reduced B-1a and B-2 B-cell proportion. | [185] | |
|
| |||
| Atg5flox/flox × Gzmb-Cre Atg7flox/flox × Gzmb-Cre |
T cells | Pathogen infection, loss of memory T cell function. | [197] |
| Atg7flox/flox × CD4-Cre | Loss of iNKT in lymphoid organs, lymphopenia, severely compromised CD8+ memory T cells. | [192] | |
| Atg3flox/flox × Lck-Cre | Decreased T cell numbers. | [195] | |
| Atg5flox/flox × Cre-ERT2 | Loss of CD8+ T cells, a severe reduction in lymphoid-specific memory T cells. | [198] | |
| Atg5flox/flox × CD4-Cre, Atg7flox/flox × Lck-Cre | Significant reduction in iNKT, CD4, and CD8 T cell numbers. | [193, 194] | |
| Vps34flox/flox;CD4-Cre | Impaired T cell homeostasis and anemia. | [189] | |
| Atg7flox/flox;CD4-Cre | Progressive anemia perturbed iNKT cell development. | [191] | |
| Atg16l1flox/flox;Cd11c-Cre | GVHD with increased T cell proliferation. | [199] | |
| Atg16l1flox/flox;CD4-Cre | Expanded T cell proliferation, impaired innate NK T lymphocyte development. | [190] | |
|
| |||
| Atg3flox/flox Ubc;cre-ERT2 | NK cells | Loss of memory NK cells. | [204] |
| Atg5flox/flox;NKp46-Cre | Impairment in NK cell development, reduction in iNKs and mNKs in the spleen and BM. | [205] | |
|
| |||
| ATG5flox/flox-Lyz-Cre | Macrophages | Perturbed lymphoid and myeloid cell homeostasis, altered macrophage differentiation. | [206] |
|
| |||
| Atg7Flox/Flox; Mx-Cre | Mast cells | Impairment of mast cell degranulation. | [207] |
|
| |||
| Vav-Cre × Atg7flox/flox Cebpa-cre × Atg7flox/flox Mx1-cre × Atg5flox/flox |
Neutrophils and Eosinophils | Impaired neutrophil differentiation. | [208] |
| Atg7flox/flox;Lyz2-Cr | Eosinophilic inflammation. | [209] | |
| Atg7flox/flox;LysM-cre | Reduced neutrophil degranulation, increased circulating neutrophil numbers, decreased inflammatory potential of neutrophils. | [210] | |
|
| |||
| Atg5flox/flox;CD11c-Cre | Dendritic cells | Reduced migration of DCs. | [211] |
|
| |||
| Atg7flox/flox;PF4-Cre | Megakaryocytes and Platelets | Impaired thrombosis, robust bleeding, and platelet aggregation. | [212] |
| VPS34flox/flox;PF4-Cre | Impeded thrombus formation. | [213] | |
| Atg5flox/flox;PF4-Cre | Delayed thrombus formation, pulmonary thrombosis, and significantly reduced platelet aggregation. | [214] | |
mNKs: mature natural killers; BM: bone marrow; iNKT: invariant natural killer T: EMH: extramedullary hematopoiesis; CD: cluster of differentiation; DCs: dendritic cells.
3.3. Autophagy in the Maintenance of Hematopoietic Progenitor Cells
Autophagy plays an important role in the maintenance of hematopoietic progenitors in the bone marrow (BM) (Figure 3). Mice with Atg7 deficiency in the HSPC compartment result in significantly reduced common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs), increased mitochondrial ROS, DNA damage, apoptosis, and consequently induced severe myeloid proliferation leading to AML [172]. Further, histological analyses have suggested that Atg7-deficient mice develop myeloid leukemia and die as a result of BM failure. Together, this study has demonstrated that autophagy plays a crucial role in maintaining adult HSPCs and protection against leukemia development. Using flow cytometry-based autophagic vesicle quantification, as well as measuring LC3-II and p62 levels, it has been reported that human HSPCs have a higher basal autophagic flux than more differentiated myeloid or erythroid progenitor cells [173]. Since autophagy suppression via ATG5 or ATG7 knockdown results in decreased HSPC frequencies, higher autophagy flux is essential for the maintenance of myeloid and erythroid progenitor cell function [173]. However, such a decrease in HSPCs occurs not as a result of impaired differentiation but rather due to reduced cell cycle progression and increased apoptosis. Another study has suggested that the loss of Atg5 increases the mitochondrial oxidative stress in neonatal HSPCs [92]. Even though p62 accumulates in immature bone marrow cells of Atg5-deficient mice, p62 deletion does not restore defective HSC functions, which indicates that p62 is not necessary for Atg5-dependent hematopoietic regulation [92]. Autophagy inhibition by mTOR signaling modulates myeloid progenitor-derived macrophage differentiation [174]. Together, these data have shown the importance of autophagy in reducing cellular stress, promoting survival, and regulating HSPCs.
Figure 3.
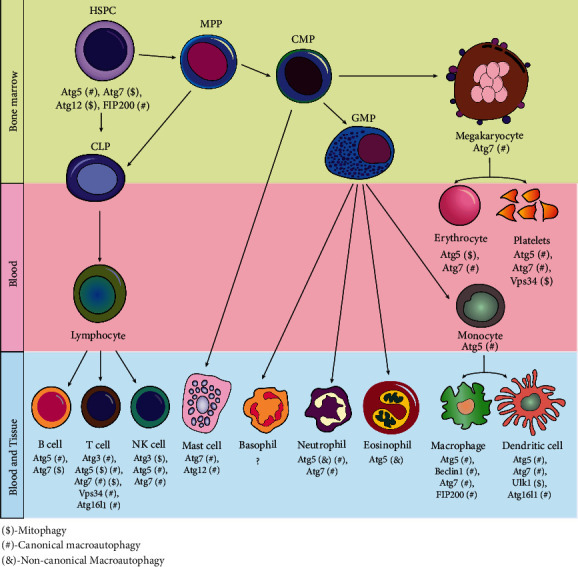
Autophagy-related genes involved in hematopoietic differentiation. Schematic representation indicated that different autophagy and marker proteins play vital roles during hematopoiesis. Specific stages of hematopoietic cell differentiation require the putative mechanistic involvement of different autophagy genes as well as multiple autophagy factors. HSPC: hematopoietic stem and progenitor cell; CMP: common myeloid progenitor; CLP: common lymphoid progenitor; GMP: granulocyte-macrophage progenitor; NK: natural killer cell; PP: multipotent progenitor.
3.4. Autophagy in Lymphopoiesis
Autophagy renders a pivotal role in lymphoid differentiation and maturation. Mechanistic insights using experimental mouse models suggest that the deletion of Atg5 or Atg7 inside the T-cell and B-cell compartments has perturbed the fundamental autophagy process and dysregulated the cellular renewal, differentiation, and immune cell functions during lymphoid maturation (Figure 3).
3.4.1. Autophagy in B Lymphocyte Development
During early embryogenesis, the development of B lymphocytes initiates in a stepwise manner such as pro-B cells, pre-B cells, and immature B cells from HSCs inside the bone marrow [175–178]. Afterward, immature B cells migrate and secrete antibodies to the secondary lymphoid organs where they get fully matured. B lymphocytes are categorized into B-1 and B-2 lymphocytes depending on their cell surface marker expression properties [179]. Matured B cells further differentiate into either quiescent memory B cells or long-lived antibody-secreting plasma cells [180]. As a result of apoptosis, autophagy-defective plasma cells and memory B cells are unable to synthesize proteins continuously and eventually come across misfolded protein aggregation [181, 182]. The first evidence concerning the necessity of autophagy during B-cell development comes from the conditional knockout of the Atg5 mice model, whereas B lymphocyte-specific Atg5 ablation results in defective B-cell development during pro-B-cell to pre-B-cell transition stages with a substantial decrease in B-1 lymphocytes inside the bone marrow [183]. B-1 lymphocytes are more likely to undergo cell death, which affects their numbers more than unaffected peripheral B-2 cells. This indicates that autophagy may be a critical process in peripheral B-1 cells. Conditional deletion of Atg5 in B cells caused endoplasmic reticulum stress and impaired antibody responses inside the plasma cells, impairing plasma cell homeostasis and long-term humoral immunity [184–186]. Furthermore, Atg5 autophagy-deficient mice develop anemia and extramedullary hematopoiesis (EMH), most likely due to the absence of proinflammatory cytokines [187].
3.4.2. Autophagy in the Quiescence and Development of T Lymphocytes
Autophagy plays an important role in T lymphocyte homeostasis. Deficiency of Atg5 in mice embryonic stem cells shows full T lymphocyte maturation, but the peripheral T and B lymphocytes and total thymocytes are reduced [188]. Targeted deletion of Vps34 and Atg16l1 in the T-cell compartment of aged mice models impairs the normal development of innate natural killer (NK) T lymphocytes [189, 190]. Under normal circumstances, invariant natural killer T (iNKT) cells display upregulated mitophagy during thymus development. However, Atg7-ablated mice within the T-cell compartment perturb cellular differentiation of iNKT cells including the enhanced susceptibility of iNKT cells to apoptosis and the accumulation of mitochondrial superoxide species [191], impaired peripheral survival of memory CD8+ T cells, and subsequently block mature iNKT cell formation in the peripheral lymphoid organs [192]. Such autophagy deficits result in the accumulation of intracellular mitochondrial ROS and apoptosis. Similarly, mice with a T lymphocyte-specific deletion of Atg5 or Atg7 indicate a cell-autonomous profound decrease in the iNKT cell population, increased ROS and mitochondrial content, and increased apoptosis and survival defects in T cells [193, 194]. Together, these findings highlight the unique roles of Atg5 and Atg7 for the quiescence and development of T lymphocytes. Similar scenarios are also observed in mice with Atg3-deficient T lymphocytes [195]. Furthermore, T-cell-specific deletion Atg5 or Atg7 results in autophagy deficiency and defective T lymphocyte production [196]. Moreover, conditional ablation of Atg5 or Atg7 in peripheral blood-derived CD8+ T cells (also known as cytotoxic T lymphocytes) induces defective autophagy flux, pathogen infection, and survival defects in memory T cells [197]. Inducible knockout of Atg5 in mouse CD8+ T cells results in concomitant p53 activation, higher ROS, apoptosis, hypoxia in lymphoid tissues, and increased susceptibility to viral infection [198]. Autophagy is also essential for cell transplantation studies, as Atg16l1 deficiency leads to an exacerbated graft-versus-host disease (GVHD) in a mouse model of allogeneic HSC transplantation (allo-HSCT), whereas Atg16l1-deficient allo-HSCT recipients with GVHD displayed increased T-cell proliferation due to increased dendritic cell (DC) numbers [199]. Collectively, these results demonstrate that T lymphocytes require autophagy to suppress cell death and maintain survival in response to virus and pathogen infection.
3.4.3. Autophagy in Natural Killer Cell Development, Maturation, and Function
Natural killer (NK) cells are specialized large lymphocytes that play critical roles in recognizing and clearing virally infected and targeted tumor cells while ensuring innate and adaptive immune responses against viral infections and pathogen attacks [215]. NK cells are derived from CLPs, while their development takes place in the bone marrow, and differentiated into immature or innate NK cells (iNKs) and eventually into mature NKs (mNKs) [215–218]. In addition to their well-established roles in antitumor and antiviral immune response, NK cells are also responsible for the production of proinflammatory cytokines and chemokines, such as interferon gamma (IFN-γ), tumor necrosis factor (TNF), chemokine (C-C motif) ligand 3 (CCL3), CCL4, and CCL5, regulating tissue and immune homeostasis [219].
Autophagy plays a crucial role in regulating NK cell survival, development, and responses against infections. Mouse bone marrow-derived iNK cells show basal autophagy activity including LC3 lipidation and p62 degradation that may be attributed to the highly proliferative murine mNK cells. It has been reported that canonical autophagy component Atg5 is essential for the proper development of mNK cells, while mice with Atg5 deficiency displayed high viral titers due to a drastically reduced number of peripheral mNK cells [220]. Wang et al. suggested that mice with NK cell-specific ablation of Atg5 showed severe reduction in iNKs and mNKs within the bone marrow and spleen, leading to the accumulation of ROS and damaged mitochondria [205]. They also indicated that Atg7 silencing impairs iNK cell autophagy and functional NK cell development in mice. Therefore, autophagy may have a role to play in different stages of NK cell maturation and proliferation during homeostasis.
Autophagy may not be required during activated NK cell proliferation, but it is critical during the transition from effector to long-lived memory cells, particularly when the autophagosome machinery component Atg3 is absent [204]. Moreover, autophagy modulator mTOR (mechanistic target of rapamycin), a serine/threonine kinase, also plays an essential role in NK cell development. Studies also suggested that mTOR and its related complexes including RPTOR (regulatory associated protein of mTOR complex 1) and RICTOR (RPTOR independent companion of mTOR complex 2) participate in NK cell development. When RPTOR or RICTOR is depleted, two distinct mTOR complexes (TORC1 and MTORC2) are destabilized, while rapamycin (mTOR inhibitor) administration impairs NKP differentiation without affecting autophagy [205]. This implies that mTOR participates in NK cell development but may be independent of autophagy. Nevertheless, these findings indicated that autophagic responses may influence NK cell biology.
3.5. Autophagy in Myelopoiesis and Granulopoiesis
Myelopoiesis is a stepwise differentiation and maturation of HSCs to CMPs by terminal differentiation which led to the production of monocytes and granulocytes including neutrophils, basophils, and eosinophils. This process is subdivided into monocytopoiesis and granulopoiesis [221]. However, it remains elusive how autophagy mediates monocyte and granulocyte differentiation.
3.5.1. Autophagy in Neutrophil Differentiation and Degranulation
Granulopoiesis is the sequential differentiation of GMPs to become eosinophils, neutrophils, basophils, and macrophages inside the bone marrow [221]. Granulocytes, also known as polymorphonuclear leukocytes (PMNL), are white blood cells, while neutrophil granulocytes are the short-lived and widely abundant cells of the host immune system, and their functional impairments lead to serious immunodeficiency syndromes [222]. There are at least four types of granules in neutrophils. Endoplasmic reticulum (ER) Golgi networks produce primary, secondary, and tertiary granules, while secretory vesicles are derived from endocytic origin. Neutrophil degranulation is an important process by which neutrophils kill the pathogen and modulate the immune response to an infection, while its inhibition can promote bacterial survival and pathogen infection. It has been shown that not only phagocytosis kills microbes but extracellular traps (ET) named neutrophil extracellular traps (NETs) for neutrophils also kill microbes in the extracellular space. NETs are made up of extracellular fibers, granular proteins, and deoxyribonucleic acid (DNA), while NET formation serves as a defense against infection by the innate immune system [223]. Therefore, neutrophil plays important roles in the body's innate immune defense via migrating to the inflammation site and controlling microbial infection by phagocytosis, degranulation, forming NETs, and secreting antimicrobial compounds. However, there is no conclusive evidence that autophagy is involved in the NET formation. Germic et al. indicated that the formation of extracellular traps by eosinophils and neutrophils is not dependent on Atg5-regulated autophagy, as both murine Atg5 knockout eosinophils and neutrophils exhibit normal extracellular trap (ET) formation [224]. Using human and mouse neutrophils, their further investigation has demonstrated that NET formation was significantly blocked after pretreatment with class III PI3K inhibitors such as 3-methyladenine (3-MA) and wortmannin. Conversely, bafilomycin A1 and chloroquine (CQ), so-called late autophagy inhibitors, have no effect on NET formation, which suggest the autophagy-independent role of Atg5 in NET formation [224]. Taken together, the picture remains unclear whether autophagy is indeed involved in the NET formation.
During neutrophil granulopoiesis and degranulation, autophagy plays an important role in neutrophil granule secretion and its degranulation (Figure 3) [225]. For instance, Atg5-deficient mice have shown minimal degranulation from primary and secondary granules. To test whether Atg5 plays an autophagy-independent role in neutrophil degranulation, Bhattacharya et al. generated the Atg5-deficient mice model from myeloid cells that resulted in reduced degranulation of primary, secondary, and tertiary granules from bone marrow neutrophils (BMNs) [225]. Although myeloid-specific autophagy deficiency leads to increased circulating neutrophil numbers and subsequent recruitment to inflammation sites, these neutrophils were unable to effectively mediate inflammation due to their reduced effector functions. As with many biological phenomena, there is still much to learn about the consequences of suppressed autophagy on neutrophil degranulation. Atg5 deletion from mice myeloid cells accelerated the neutrophil differentiation process and the proliferation of neutrophil precursor cells inside the bone marrow, which eventually leads to an accumulation of neutrophils in the bone marrow, blood, spleen, and lymph nodes [226]. Furthermore, Atg7-deficient mice exhibit impaired mitochondrial respiration, excessive glycolysis, reduced ATP production, and defective lipophagy during neutrophil differentiation. Nevertheless, the exact pathway by which lipophagy is directed into the autophagic machinery requires further investigation.
3.5.2. Autophagy in Eosinophil Biogenesis
Similar to neutrophils, eosinophils and basophils are also granulocytes that arise from the bone marrow but occur in relatively low numbers in human peripheral blood. A response to inflammation triggers the maturation of eosinophils in the bone marrow and then leads to its migration and activation in inflamed tissues. In contrast to neutrophils, it remains largely unknown how autophagy regulates eosinophil hematopoiesis and functions because eosinophils are rare and poorly studied. During an innate immune response, no defects in the formation of eosinophil extracellular traps (EETs) were observed in Atg5 knockout mouse eosinophils or autophagy-deficient human eosinophils (Figure 3) [224]. Rapamycin, however, effectively inhibits eosinophil differentiation by blocking mTORC1 and significantly reduces eosinophil counts in the peripheral blood and bone marrow [227]. Surprisingly, an increased number of eosinophil lineage-committed progenitors were observed in the bone marrow after rapamycin inhibited eosinophil differentiation. The same group also found an increase in eosinophil lineage-committed progenitors upon the murine myeloid cell-specific deletion of mTOR [228].
However, their LysM-cre mice strain is questionable because this group used that mice strain to suppress mTOR expression in eosinophils, which is normally used to determine neutrophilic, macrophagic, and dendritic cell populations, but not eosinophils. Collectively, these two studies indicated that mTOR modulates eosinophil differentiation in different ways, possibly because mTORC1 and mTORC2 may have distinct functions. So far, autophagy has not been reported to be involved in basophil hematopoiesis.
3.5.3. Autophagy in Mast Cell Function and Degranulation
Mast cells (MCs) are granule-containing immune cells that are found in nearly all vascularized tissues and contribute to the generation of protective innate immunity against infection. Furthermore, activated MCs regulate adaptive immune responses by influencing lymph node composition [229]. Despite this, very little is known about how autophagy contributes to MC function. It has been reported that under adequate nutrient supply, bone marrow-derived mast cells (BMMCs) exhibit autophagic flux, and LC3-II is found in their secretory granules [230]. When Atg7 is deleted from BMMCs, degranulation has been severely impaired, but cytokine production is not affected, suggesting that autophagy may not be essential for mast cell differentiation but critical for their degranulation.
3.5.4. Autophagy in Monocyte and Macrophage Differentiation
Monocytes are bone marrow-derived white blood cells or leukocytes, circulating inside the blood, and upon migrating from the bloodstream into tissues, monocytes are terminally differentiated into macrophages and dendritic cells [231]. Such immune cells are involved in various cytokine induction and antigen presentation while conferring innate immunity and tissue homeostasis [232], while macrophages are multifaceted innate immune phagocytes that serve as a first-line host defense against intracellular pathogen invasion by mounting proinflammatory responses via phagocytosis, releasing cytokines and renovating damaged tissues [233]. A study reported that colony-stimulating factor-1 (CSF-1) or granulocyte/macrophage colony-stimulating factor (GM-CSF) induces autophagy and is essential for proper macrophage differentiation in human and murine monocytes [234]. Moreover, the differentiation signal prevents ATG5 cleavage and activates c-Jun N-terminal kinase (JNK) to release Beclin1 from BCL-2, both of which are essential for the initiation of autophagy. Since the Beclin1 knockdown and autophagy inhibitors including 3-MA and CQ cause apoptosis via caspase activation and prevent differentiation and cytokine production, therefore autophagy is critical in transitioning from monocyte apoptosis to differentiation (Figure 3). In 2020, Wang et al. reported that mice with myeloid cell-specific deletion of beclin1 and Fip200 induce defective lymphoid and myeloid cell homeostasis and impaired macrophage differentiation [206]. Conversely, excision of other core autophagy genes including Atg5, Atg7, Atg14, and Atg16l1 from the mice macrophages showed moderate to severe level vulnerability to pathogen infection and bacterial burden. To determine the autophagic activity of all these genes, degradation of the p62 protein and lipidation of LC3 are measured. Collectively, it appears that certain autophagy genes, but not all, have a key role in maintaining immunity quiescence within tissue-resident macrophages. Circulating monocytes derived from CMPs can generate macrophages and dendritic cells inside tissues through cellular differentiation [235]. However, detailed mechanistic insights and molecular events have not been elucidated. In contrast, mice monocytes and macrophages lacking Atg5 are more likely to be infected by intracellular pathogens, suggesting that Atg5 expression in such phagocytic cells supports intracellular pathogen resistance [236]. However, in this study, classical autophagosomes were missing during lysosomal fusion with phagosomes and Atg5-dependent intracellular killing of pathogens, which suggests a classical autophagy-independent role of Atg5 in intracellular membrane dynamics. Similarly, myeloid lineage-specific Atg7-deficient mice's macrophages showed increased bacterial uptake with accumulating p62 level when infected with Mycobacterium tuberculosis [237]. It is presumable that autophagy plays similar roles in monocytes/macrophages and granulocytes of hematopoietic origin, although the direct mechanistic insights of autophagy in blood monocytes and macrophages remain poorly understood.
3.5.5. Autophagy in Dendritic Cell Immune Response
Dendritic cells (DCs) are a special kind of antigen-presenting cells, derived from HSCs, which sequentially differentiate into CMPs and CLPs. DCs from the CMPs capture antigens in peripheral tissue and then migrate to the lymphoid organs for immunity. On the other hand, DCs arise from the CLP origin and are found in the lymph node T cells and thymic medulla to initiate immune tolerance and T-cell immunity [238]. As previously mentioned, autophagy machinery is utilized to deliver the pathogenic organelles into autophagosomes and its subsequent fusion with lysosomes, it is believed that cargos from lysosomal degradation are delivered to T helper cells by major histocompatibility complex (MHC) class II molecule to mediate the adaptive immune response [239]. Lee et al. have suggested that mice with Atg5 deficiency in DCs exhibited impaired CD4+ T-cell priming and rapid viral infection [240]. Atg5-deficient DCs also display a prominent defect in phagocytized antigen processing for MHC class-II molecule and phagosome-to-lysosome fusion. Previously, it has been stated that cytosolic receptors may be activated by pathogens, which then trigger autophagy. Therefore, Atg16l1 autophagy is induced by the activation of bacterial sensors called nucleotide-binding oligomerization domain 1 (NOD1) and NOD2 [241]. Cooney et al. demonstrated that DCs from individuals with Crohn's disease expressing NOD2 or Atg16l1 risk variants show abnormalities in autophagy induction, antigen presentation, and bacterial trafficking [242]. Autophagy activating kinase Ulk1 plays a critical role in mitochondrial quality control in bone marrow-derived DCs (BMDCs). Mice lacking Ulk1 in BMDCs have increased accumulation of damaged mitochondrial mass and cell death via caspase-1 activation [243]. Moreover, BMDC cytokine response is dependent on Beclin1 autophagy. BMDCs infected with H1N1 influenza and lacking Beclin1 are detected with lower levels of inflammatory cytokine response [244]. Thus, the ability to boost DC immunogenicity by inducing autophagy is a novel strategy for vaccination.
3.5.6. Autophagy in the Maintenance of Megakaryopoiesis and Thrombopoiesis
Several human diseases are associated with a lack of autophagy, while their role in platelet function has only recently been explored. Thrombocytes or platelets are colorless, small, and flowing cell fragments in the bloodstream, generated from the megakaryocyte progenitors (MPs), that immediately respond to blood vessel injury, form blood clotting, prevent bleeding, and assist in hemostasis [245]. It has been reported that resting human and mice platelets exhibit a wide range of autophagy proteins including ULK1, FIP200, ATG3, ATG5, Beclin1, ATG7, LC3-II, ATG12, ATG14, VPS15, VPS34, and UVRAG [246]. Autophagy is constitutively active in resting platelets, while platelet activation triggers autophagy and autophagy flux, as evidenced by agonist-induced loss of autophagy marker LC3-II [247]. Megakaryocyte and thrombocyte-specific ablation of Atg7 indicated a markedly reduced LC3-II level, defective platelet aggregation, abrogated granule cargo packaging, impaired hemostasis, and thrombus formation. Similarly, hematopoietic cells lacking Atg7 indicate an absence of LC3-II formation due to disrupted LC3 lipidation, which indicates the absence of autophagy. This leads to mitochondrial mass accumulation as detected by increased mitochondrial superoxide production and cell cycle dysfunction in Atg7−/− bone marrow Lin− cells, which eventually disrupts megakaryopoiesis, megakaryocyte differentiation, and thrombopoiesis and results in aberrant platelet production, prolonged bleeding, and impeded hemostasis [248]. In another study, mice with megakaryocyte/platelet-lineage-specific ablation of Atg5 contributed to impaired mitochondrial quality control and expanded mitochondrial mass inside the thrombocytes and blocked mitophagy [249]. Using VPS34- (vacuolar protein sorting 34-) deficient mice, Liu et al. further indicated that VPS34−/− mice exhibited impaired mTOR signaling and arterial thrombosis, significantly reduced thrombus formation, and altered basal level of autophagy in resting platelets [213]. Collectively, these data suggested that platelet autophagy, especially platelet mitophagy, is crucial for hemostasis and thrombus formation.
Autophagy has also been studied in invertebrate hematopoiesis to a lesser extent. Drosophila's hematopoietic system is an excellent genetic model for studying invertebrate hematopoiesis. It has been reported that homozygous Atg6 loss causes melanotic blood cell masses in Drosophila, whereas ubiquitous expression of Atg6 completely rescues the phenotype of blood cell tumors [250]. Melanotic blood cell masses are also observed in Atg7 and Atg13 mutated Drosophila. Immunohistochemical analyses using the GFP antibody labeling have suggested that homozygous Atg6 mutant larvae have blood-like melanotic masses that confirm that hematopoietic cells are the source of these masses.
4. Expression Profile of ATG Proteins in Hematopoietic Cells and AMLs
The catabolic nature of dynamic autophagy enables cells to maintain their self-renewal capacity, cellular homeostasis, and survival under stress conditions. In humans, ATG proteins are expressed differentially during normal hematopoiesis (Figure 4). Autophagy-activating kinase ULK1 and other core ATG proteins such as ATG6, ATG7, ATG13, autophagy factor FIP200, and autophagy receptor protein NBR1 are highly expressed in polymorphonuclear leukocytes and rarely expressed in MPPs, CMPs, MEPs, and GMPs. Conversely, core autophagy protein ATG5 is predominantly expressed in CMPs, MEPs, and monocytes. Autophagy receptor protein SQMST1 and tumor suppressor as well as an important macroautophagy regulator UVRAG are significantly expressed in PMNs, monocytes, GMPs, and CMPs. However, the basal levels of all autophagy proteins in the HSCs are relatively low.
Figure 4.

Expression of different autophagy markers in hematopoietic cells and AMLs. Schematic representation indicated the expression level of different ATG genes, autophagy-related factors, and different ATG receptors in both hematopoietic cells and various types of acute myeloid leukemia. PMN: polymorphonuclear cells; MPP: multipotential progenitors; MLL: mixed-lineage leukemia; AML t(15;17): AML with t(15;17); AML inv(16)/t(16;16): AML with inv(16)/t(16;16); AML t(11q23)/MLL: AML with t(11q23)/MLL; AML complex: AML with complex aberrant karyotype. Data were generated from normal human samples and samples from the human AML cells using microarray-based gene expression profiling. The “BloodSpot” (https://servers.binf.ku.dk/bloodspot/) database was used to generate the hierarchical differentiation tree.
The role of autophagy in various types of cancers has been extensively studied. However, the relationship between autophagy and hematopoietic malignancies remains controversial. Autophagy-related proteins such as ULK1, ATG3, ATG5, ATG13, and ATG14 were markedly expressed in various acute myeloid leukemias when comparing the microarray-based expression profiling of distinct ATG proteins from human AML samples with the human normal hematopoiesis. Nevertheless, the expression profiling of ATG proteins in AML samples is paradoxical. For instance, Jin et al. reported a lower expression of ULK1 in human AML cell lines [251], while Hwang et al. indicated a significantly higher level of ULK1 in FLT3-ITD-mutated AML cells compared to FLT3-wild-type AML cells, which ultimately suggested that inhibiting ULK1 in FLT3-ITD AML cells might be a promising therapeutic approach against FLT3-ITD-mutated AML [252]. Similar to the later study, it has been demonstrated that Atg7 knockdown from patients' AML samples prolonged the overall survival after receiving chemotherapy [95]. As a result, increasing chemosensitivity by decreasing Atg7 may be a potential strategy to improve treatment results for AML. Jin et al. also found significantly decreased ATG5 expression levels in AML samples. In contrast, a recent study found that an Egyptian cohort with newly diagnosed acute lymphoblastic leukemia (ALL) had considerably greater levels of ATG5 expression [253]. Moreover, Liu et al. suggested that Atg5-dependent autophagy may contribute to the development of murine AML induced by the MLL-AF9 fusion oncogene, resulting from the t(9;11)(p22;q23) translocation [254].
However, researchers suggested a Beclin1 autophagy-dependent but ATG5 autophagy-independent role in murine chronic myeloid leukemia (CML) model, whereas Beclin1 knockdown, but not Atg5 ablation, leads to a reduced leukemic burden and a significantly higher survival rate of targeted mice [255]. Collectively, autophagy plays dual roles depending on the kind of hematological malignancies. Nevertheless, more research is needed to clarify the relationship between autophagy and blood malignancies.
5. Current Challenges and Future Perspectives
Autophagy plays a critical role in the hematopoietic system by regulating the self-renewal of HSCs, differentiation, and development of lymphoid and myeloid progenitors and their precursor cells in response to cytokine signaling [256–258]. Several studies indicated that the deletion of ATGs such as FIP200, ATG5, and ATG7 decreases the number of HSCs and diminishes the reconstituting capacity of normal HSCs [167, 259]. Moreover, ATG7 or FIP200 deficiency in HSC results in abnormal myeloid expansion, accompanied by ROS accumulation and genomic instability, which contribute to the development of aggressive phenotypes in the hematopoietic system similar to anemia, lymphopenia, and splenomegaly. Besides, ubiquitous deletion of the core autophagy gene(s) from in vivo mice models leads to embryonically and neonatally lethal phenotypes [260–263]. To avoid such embryonic and neonatal lethality at the germline and to overcome the secondary effects associated with altered gene function in other tissues upon ubiquitous gene knockout, tissue-specific gene disruption strategies have been developed to study particular gene functions. Studying autophagy within a particular hematopoietic cell type or lineage-specific manner will allow us to determine the actual function of different ATGs in the hematopoietic system. However, such strategies might be difficult to apply in humans due to the molecular complexity of autophagy regulatory genes and ethical issues. Therefore, previous studies have been done using cell lines and animal model platforms. As an alternative approach, bone marrow transplantation strategies have been utilized to generate hematopoietic cell-specific knockout animals. To date, Atg5, Atg7, and Beclin1 are the most studied autophagy genes concerning hematological physiology and pathobiology, while the other core autophagy genes such as Ulk1, Atg3, Atg9, Atg10, Atg12, Atg13, Atg18, and Atg101 need further investigations. Moreover, direct mechanistic insights between impaired autophagy, ROS, and mitochondrial mass accumulation were partly lacking, possibly due to the nontransparent nature of the widely used in vivo mice model. To overcome the nontransparent nature of the mice models, the introduction of the in vivo zebrafish model having the optical transparency, high fertility, commercially available subcellular fluorescent dye, and ubiquitous and tissue-specific GFP : Lc3-expressing transgenic lines with time-lapse in vivo live-imaging technology has immensely alleviated the technical downfall and geared up the experimental outcomes to monitor dynamic autophagy in hematopoietic development, differentiation, and blood malignancies.
Autophagy is involved in the complex molecular signaling pathway; while numerous studies have demonstrated that autophagy promotes both cell survival and cell death, these studies have yet been unable to answer the basic question about autophagy's two functions. Moreover, autophagy is poorly understood at the molecular level and how tumor genetics, tissue type, and disease state affect its specific functions. Further research should be directed at understanding autophagy's role and status in cellular survival and death and clarify whether autophagy directly causes cell death and cell survival or whether such changes in the cell microenvironments are secondary to autophagy. Thus, it is also necessary to determine whether autophagy results from or causes changes in cellular metabolic processes. It is more likely that drug resistance may occur due to autophagy activation in leukemia cells during drug-induced apoptosis if autophagy acts as a secondary change.
Immune cells' activation and differentiation are also regulated by autophagy, while those immune cells' functions are regulated by their metabolism. Therefore, it would be beneficial to study tumor and host autophagy in immune cell metabolism to develop novel anti-AML therapies. We know that autophagy recycling is essential for the survival, metabolism, and proper functioning of AML cells, but does the same apply to other leukemias? Is autophagy in the host responsible for promoting leukemia cell growth, and if so, how does it work? How can the autophagy flux in human tissue be measured? Which steps should be targeted in the autophagy pathway for the development of inhibitors? Nevertheless, drugs that regulate autophagy are now being researched for the treatment of leukemia. A key feature of targeted therapy will be the selective induction of autophagy in leukemic cells. In such a scenario, a better starting point would be to investigate autophagy under hypoxic conditions. It is also challenging to determine which stages of AML blast formation require autophagy to protect the tumor cells and to clarify how autophagy functions during different stages of AMLs. Therefore, it is necessary to investigate specific pathways that activate autophagy in AMLs and other associated cellular processes including apoptosis and cell differentiation to identify targeted treatments and reverse AML cell growth. Recent advances have led to the development of more potent and specific lysosome inhibitors than chloroquine to develop a wide combination of different therapies, such as concanamycin A, which prevents the acidification of lysosomes and endosomes by inhibiting V-ATPase; pepstatin A, which inhibits cathepsins D and E; and E64d, which inhibits cathepsins B, H, and L [264]. However, autophagosome formation and cargo sequestration are not affected by the prevention of autophagosome degradation by lysosomal inhibitors. Mitophagy, for example, removes damaged mitochondria, preventing oxidative stress and the activation of apoptosis. Lysosomal inhibitors, nonetheless, may not be effective in reducing the rate at which mitochondria are sequestered by autophagosomes, and this could limit the drug's efficacy against AML effects that are reliant on mitophagy. Therefore, what level of the autophagy pathway would be optimal for inhibition remains an unanswered question. If the later steps, such as the lysosome, are inhibited, other metabolic scavenging pathways may also be blocked, which has also been demonstrated to be vital to tumor cell metabolism and growth. It is also possible for tumor cells to become toxic due to the aggregation of undelivered autophagosomes if the later stages of the autophagy pathway are targeted. Alternatively, inhibiting autophagy at the earlier stages, such as those phages involved in autophagosome biogenesis, would result in a buildup of damaged mitochondria and toxic protein aggregates, which would not be contained by the autophagosome anymore, resulting in constant exposure to these toxic insults to tumor cells. Nevertheless, a better outcome may be achieved in this context with drugs that block the initial phases of autophagy, such as ULK1 inhibitor (MRT68921) and VPS34 inhibitor (PIK-III or SAR405) [265–267]. Furthermore, it is important to determine whether specific autophagy pathways can be targeted or if targeting the general macroautophagy pathway will be sufficient. It may be possible to enhance therapeutic efficacy and minimize toxicity by targeting specific cargo adaptors in selective autophagy. Thus, developing more selective and potent autophagy/lysosome pathway inhibitors at different levels will facilitate preclinical validation studies. Collectively, a concrete knowledge and complete understanding of the complex autophagy network will help us to better develop more precise autophagy-modulating therapies and to evaluate which patients will be benefited from such treatments.
Basal autophagy could be monitored by isolating HSCs from the blood of AML patients. However, it is extremely challenging to study autophagy in living organisms, particularly in patients, despite methods for studying the autophagic flux in vitro. Therefore, further research is also needed to develop reliable and new methods to quantify autophagy flux in patient samples to adjust therapy and better target the autophagy pathway. Transcriptomic and proteomic analysis of patient cells could provide valuable insight into how treatment affects autophagy gene expression and how their expression might predict patient survival. Using RNA sequencing, an innovative approach to transcriptome analysis, it could be possible to predict whether or not these cells are undergoing autophagy activation. It is still possible to develop new therapeutic approaches in the future by investigating cargo receptors and selective autophagy. A growing body of evidence suggests that autophagy inhibition in normal tissues without driver oncogene mutations does not cause leukemia progression to fully invasive stages such as AML. As a consequence of these previous concerns, autophagy inhibition studies have been focused exclusively on AML; however, the new findings indicate that autophagy inhibition may be of critical importance for early-stage blood cancers, either as a prevention of metastases or as a prevention of leukemia from developing initially. It is believed that targeting either tumor or host autophagy is an important treatment strategy for AML because both are involved in the growth of the disease. While autophagy plays an important role in AML, there is still a need to fully understand its mechanisms across AML subtypes and treatment options.
6. Concluding Remarks
Autophagy has become an intriguing subject in hematopoiesis and related malignancies such as AML. Throughout this review, we have summarized the different forms of autophagy and discussed the hematopoietic-lineage-specific roles of different autophagy regulators. Since the blood progenitors and precursors derive from HSCs, targeting HSCs may be a better approach for treating AML. In addition, autophagy inhibitors might also be successfully used in clinical settings to treat AML patients by using newly developed drug delivery systems. However, introducing autophagy modulators in AML therapy should be considered in light of the potential toxicity they may cause in HSCs, even during the progression of malignant phenotypes. To overcome this potential toxicity, autophagy modulators need to be studied further.
Acknowledgments
The basic and experimental research of this study was funded by the Hong Kong Polytechnic University, Hong Kong. The manuscript contains some parts that have been published in Kazi Md Mahmudul Hasan's doctoral dissertation titled “Role of Ulk1b-Regulated Autophagy in Zebrafish Definitive Hematopoiesis” and presented in international conferences such as the “11th European Zebrafish Meeting 2020”, “Keystone eSymposia on Autophagy: Mechanisms and Disease 2020”, and the “Cell Bio 2020 Virtual-A Virtual ASCB/EMBO Meeting”.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nature Reviews Molecular Cell Biology . 2018;19(6):349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G., Mariño G., Levine B. Autophagy and the integrated stress response. Molecular Cell . 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto H., Fujioka Y., Suzuki S. W., et al. The intrinsically disordered protein Atg13 mediates supramolecular assembly of autophagy initiation complexes. Developmental Cell . 2016;38(1):86–99. doi: 10.1016/j.devcel.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. The EMBO Journal . 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotani T., Kirisako H., Koizumi M., Ohsumi Y., Nakatogawa H. The Atg 2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proceedings of the National Academy of Sciences . 2018;115(41):10363–10368. doi: 10.1073/pnas.1806727115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohsumi Y. Historical landmarks of autophagy research. Cell Research . 2014;24(1):9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axe E. L., Walker S. A., Manifava M., et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of Cell Biology . 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y., Li Z., Hu W., et al. _C. elegans_ Screen Identifies Autophagy Genes Specific to Multicellular Organisms. Cell . 2010;141(6):1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 9.Guo T., Nan Z., Miao C., et al. The autophagy-related gene _Atg101_ in _Drosophila_ regulates both neuron and midgut homeostasis. The Journal of Biological Chemistry . 2019;294(14):5666–5676. doi: 10.1074/jbc.RA118.006069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian L., Ma L., Guo E., et al. 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy . 2013;9(8):1172–1187. doi: 10.4161/auto.24731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cecconi F., Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Developmental Cell . 2008;15(3):344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinsztein D. C., Mariño G., Kroemer G. Autophagy and aging. Cell . 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 13.Menzies F. M., Fleming A., Caricasole A., et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron . 2017;93(5):1015–1034. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Mathew R., Karantza-Wadsworth V., White E. Role of autophagy in cancer. Nature Reviews. Cancer . 2007;7(12):961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J. Y., Teng X., Laddha S. V., et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes & Development . 2016;30(15):1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju J. S., Varadhachary A. S., Miller S. E., Weihl C. C. Quantitation of "autophagic flux" in mature skeletal muscle. Autophagy . 2010;6(7):929–935. doi: 10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy . 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy . 2007;3(6):542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 19.Orkin S. H., Zon L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell . 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison S. J., Scadden D. T. The bone marrow niche for haematopoietic stem cells. Nature . 2014;505(7483):327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y., Wang T., Gu J., et al. Characterization and generation of human definitive multipotent hematopoietic stem/progenitor cells. Cell Discovery . 2020;6(1):p. 89. doi: 10.1038/s41421-020-00213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S., Zhang C., Ni L., et al. Stabilization of HIF-1α alleviates osteoarthritis via enhancing mitophagy. Cell Death & Disease . 2020;11(6):p. 481. doi: 10.1038/s41419-020-2680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin M., Klionsky D. J. Finding a ribophagy receptor. Autophagy . 2018;14(9):1479–1480. doi: 10.1080/15548627.2018.1483672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wild P., Farhan H., McEwan D. G., et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science . 2011;333(6039):228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y. T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J. H. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. Journal of Immunology . 2009;183(9):5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 26.Thurston T. L. M., Wandel M. P., von Muhlinen N., Foeglein Á., Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature . 2012;482(7385):414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reggio A., Buonomo V., Grumati P. Eating the unknown: xenophagy and ER-phagy are cytoprotective defenses against pathogens. Experimental Cell Research . 2020;396(1, article ???) doi: 10.1016/j.yexcr.2020.112276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zechner R., Madeo F., Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nature Reviews Molecular Cell Biology . 2017;18(11):671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 29.Liu C., Han T., Stachura D. L., et al. Lipoprotein lipase regulates hematopoietic stem progenitor cell maintenance through DHA supply. Nature Communications . 2018;9(1):p. 1310. doi: 10.1038/s41467-018-03775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang C. L., Garcia-Arcos I., Nyrén R., et al. Lipoprotein lipase deficiency impairs bone marrow myelopoiesis and reduces circulating monocyte levels. Arteriosclerosis, Thrombosis, and Vascular Biology . 2018;38(3):509–519. doi: 10.1161/ATVBAHA.117.310607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lystad A. H., Simonsen A. Assays to monitor aggrephagy. Methods . 2015;75:112–119. doi: 10.1016/j.ymeth.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Cao L., Kang R., et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy . 2011;7(4):401–411. doi: 10.4161/auto.7.4.14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nature Cell Biology . 2018;20(9):1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 34.Tang C., Han H., Yan M., et al. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy . 2018;14(5):880–897. doi: 10.1080/15548627.2017.1405880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami K., Kurotaki D., Kawase W., et al. OGT regulates hematopoietic stem cell maintenance via PINK1-dependent mitophagy. Cell Reports . 2021;34(1, article 108579) doi: 10.1016/j.celrep.2020.108579. [DOI] [PubMed] [Google Scholar]
- 36.Ho T. T., Warr M. R., Adelman E. R., et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature . 2017;543(7644):205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito K., Turcotte R., Cui J., et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science . 2016;354(6316):1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fay H. R. S., Dykstra K. M., Johnson M., et al. Mitophagy plays a key role in the anti-leukemic activity of autophagy inhibitors under hypoxia in acute myeloid leukemia. Blood . 2019;134(Supplement_1):1278–1278. doi: 10.1182/blood-2019-127024. [DOI] [Google Scholar]
- 39.Kim P. K., Hailey D. W., Mullen R. T., Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proceedings of the National Academy of Sciences of the United States of America . 2008;105(52):20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deosaran E., Larsen K. B., Hua R., et al. NBR1 acts as an autophagy receptor for peroxisomes. Journal of Cell Science . 2013;126(Part 4):939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 41.Nordgren M., Francisco T., Lismont C., et al. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy . 2015;11(8):1326–1340. doi: 10.1080/15548627.2015.1061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demers N. D., Riccio V., Jo D. S., et al. PEX13 prevents pexophagy by regulating ubiquitinated PEX5 and peroxisomal ROS. Autophagy . 2023:1–22. doi: 10.1080/15548627.2022.2160566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Germain K., Kim P. K. Pexophagy: a model for selective autophagy. International Journal of Molecular Sciences . 2020;21(2):p. 578. doi: 10.3390/ijms21020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houri K., Mori T., Onodera Y., et al. miR-142 induces accumulation of reactive oxygen species (ROS) by inhibiting pexophagy in aged bone marrow mesenchymal stem cells. Scientific Reports . 2020;10(1):p. 3735. doi: 10.1038/s41598-020-60346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geng S., Zhang Y., Yi Z., Lu R., Li L. Resolving monocytes generated through TRAM deletion attenuate atherosclerosis. JCI Insight . 2021;6(20) doi: 10.1172/jci.insight.149651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J., Tripathi D. N., Jing J., et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nature Cell Biology . 2015;17(10):1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinz M., Stilmann M., Arslan S., Khanna K. K., Dittmar G., Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation. Molecular Cell . 2010;40(1):63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Ajoolabady A., Aslkhodapasandhokmabad H., Libby P., et al. Ferritinophagy and ferroptosis in the management of metabolic diseases. Trends in Endocrinology and Metabolism . 2021;32(7):444–462. doi: 10.1016/j.tem.2021.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Mancias J. D., Pontano Vaites L., Nissim S., et al. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife . 2015;4 doi: 10.7554/eLife.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nai A., Lidonnici M. R., Federico G., et al. NCOA4-mediated ferritinophagy in macrophages is crucial to sustain erythropoiesis in mice. Haematologica . 2021;106(3):795–805. doi: 10.3324/haematol.2019.241232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weckman A., Di Ieva A., Rotondo F., et al. Autophagy in the endocrine glands. Journal of Molecular Endocrinology . 2014;52(2):R151–R163. doi: 10.1530/JME-13-0241. [DOI] [PubMed] [Google Scholar]
- 52.Zaffagnini G., Martens S. Mechanisms of selective autophagy. Journal of Molecular Biology . 2016;428(9):1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Short N. J., Konopleva M., Kadia T. M., et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discovery . 2020;10(4):506–525. doi: 10.1158/2159-8290.CD-19-1011. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen T. D., Shaid S., Vakhrusheva O., et al. Loss of the selective autophagy receptor p62 impairs murine myeloid leukemia progression and mitophagy. Blood . 2019;133(2):168–179. doi: 10.1182/blood-2018-02-833475. [DOI] [PubMed] [Google Scholar]
- 55.Chen X. X., Li Z. P., Zhu J. H., Xia H. T., Zhou H. Systematic analysis of autophagy-related signature uncovers prognostic predictor for acute myeloid leukemia. DNA and Cell Biology . 2020;39(9):1595–1605. doi: 10.1089/dna.2020.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Putyrski M., Vakhrusheva O., Bonn F., et al. Disrupting the LC3 interaction region (LIR) binding of selective autophagy receptors sensitizes AML cell lines to cytarabine. Frontiers in Cell and Development Biology . 2020;8:p. 208. doi: 10.3389/fcell.2020.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang P. Q., Miao M., Liu Z. G., et al. Expression of autophagy genes in acute myeloid leukemia: associations with clinical characteristics and prognosis. Neoplasma . 2018;65(5):807–814. doi: 10.4149/neo_2018_171028N691. [DOI] [PubMed] [Google Scholar]
- 58.Mijaljica D., Prescott M., Devenish R. J. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy . 2011;7(7):673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 59.Li W. W., Li J., Bao J. K. Microautophagy: lesser-known self-eating. Cellular and Molecular Life Sciences . 2012;69(7):1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dokladny K., Myers O. B., Moseley P. L. Heat shock response and autophagy--cooperation and control. Autophagy . 2015;11(2):200–213. doi: 10.1080/15548627.2015.1009776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong S., Wang Q., Kao Y. R., et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature . 2021;591(7848):117–123. doi: 10.1038/s41586-020-03129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dubois A., Furstoss N., Calleja A., et al. LAMP2 expression dictates azacytidine response and prognosis in MDS/AML. Leukemia . 2019;33(6):1501–1513. doi: 10.1038/s41375-018-0336-1. [DOI] [PubMed] [Google Scholar]
- 63.Vakifahmetoglu-Norberg H., Kim M., Xia H. G., et al. Chaperone-mediated autophagy degrades mutant p53. Genes & Development . 2013;27(15):1718–1730. doi: 10.1101/gad.220897.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li P., Ji M., Lu F., et al. Degradation of AF1Q by chaperone-mediated autophagy. Experimental Cell Research . 2014;327(1):48–56. doi: 10.1016/j.yexcr.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 65.Robert G., Auberger P. Azacitidine resistance caused by LAMP2 deficiency: a therapeutic window for the use of autophagy inhibitors in MDS/AML patients. Autophagy . 2019;15(5):927–929. doi: 10.1080/15548627.2019.1586259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arias E., Koga H., Diaz A., Mocholi E., Patel B., Cuervo A. M. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Molecular Cell . 2015;59(2):270–284. doi: 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Codogno P., Mehrpour M., Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self- eating? Nature Reviews Molecular Cell Biology . 2012;13(1):7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 68.Heckmann B. L., Green D. R. LC3-associated phagocytosis at a glance. Journal of Cell Science . 2019;132(5) doi: 10.1242/jcs.222984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heckmann B. L., Teubner B. J. W., Tummers B., et al. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. Cell . 2019;178(3):536–551.e14. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henault J., Martinez J., Riggs J. M., et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity . 2012;37(6):986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez J., Cunha L. D., Park S., et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature . 2016;533(7601):115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Cunha L. D., Yang M., Carter R., et al. LC3-associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell . 2018;175(2):429–441.e16. doi: 10.1016/j.cell.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanjuan M. A., Milasta S., Green D. R. Toll-like receptor signaling in the lysosomal pathways. Immunological Reviews . 2009;227(1):203–220. doi: 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- 74.Martinez J., Malireddi R. K. S., Lu Q., et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature Cell Biology . 2015;17(7):893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Boyle K. B., Randow F. Rubicon swaps autophagy for LAP. Nature Cell Biology . 2015;17(7):843–845. doi: 10.1038/ncb3197. [DOI] [PubMed] [Google Scholar]
- 76.Matsunaga K., Saitoh T., Tabata K., et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature Cell Biology . 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 77.Cheong H., Lindsten T., Wu J., Lu C., Thompson C. B. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proceedings of the National Academy of Sciences . 2011;108(27):11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Su H., Yang F., Wang Q., et al. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Molecular Cell . 2017;67(6):907–921.e7. doi: 10.1016/j.molcel.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 79.Moore J. A., Mistry J. J., Hellmich C., et al. LC3-associated phagocytosis in bone marrow macrophages suppresses acute myeloid leukemia progression through STING activation. The Journal of Clinical Investigation . 2022;132(5) doi: 10.1172/JCI153157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.González A., Hall M. N. Nutrient sensing and TOR signaling in yeast and mammals. The EMBO Journal . 2017;36(4):397–408. doi: 10.15252/embj.201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saxton R. A., Sabatini D. M. mTOR signaling in growth, metabolism, and disease. Cell . 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J., Kundu M., Viollet B., Guan K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology . 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ganley I. G., Lam D. H., Wang J., Ding X., Chen S., Jiang X. ULK1·ATG13·FIP200 Complex Mediates mTOR Signaling and Is Essential for Autophagy ∗. Journal of Biological Chemistry . 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mercer C. A., Kaliappan A., Dennis P. B. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy . 2009;5(5):649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 85.Bilanges B., Posor Y., Vanhaesebroeck B. PI3K isoforms in cell signalling and vesicle trafficking. Nature Reviews Molecular Cell Biology . 2019;20(9):515–534. doi: 10.1038/s41580-019-0129-z. [DOI] [PubMed] [Google Scholar]
- 86.Dooley Hannah C., Razi M., Polson Hannah E. J., Girardin Stephen E., Wilson Michael I., Tooze Sharon A. WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12-5-16L1. Molecular Cell . 2014;55(2):238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsapras P., Nezis I. P. Caspase involvement in autophagy. Cell Death and Differentiation . 2017;24(8):1369–1379. doi: 10.1038/cdd.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martens S., Fracchiolla D. Activation and targeting of ATG8 protein lipidation. Cell Discovery . 2020;6(1):p. 23. doi: 10.1038/s41421-020-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu L., Chen Y., Tooze S. A. Autophagy pathway: cellular and molecular mechanisms. Autophagy . 2018;14(2):207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lahiri V., Hawkins W. D., Klionsky D. J. Watch what you (self-) eat: autophagic mechanisms that modulate metabolism. Cell Metabolism . 2019;29(4):803–826. doi: 10.1016/j.cmet.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Czarny P., Pawlowska E., Bialkowska-Warzecha J., Kaarniranta K., Blasiak J. Autophagy in DNA damage response. International Journal of Molecular Sciences . 2015;16(2):2641–2662. doi: 10.3390/ijms16022641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nomura N., Ito C., Ooshio T., et al. Essential role of autophagy in protecting neonatal haematopoietic stem cells from oxidative stress in a p62-independent manner. Scientific Reports . 2021;11(1):p. 1666. doi: 10.1038/s41598-021-81076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shahrabi S., Paridar M., Zeinvand-Lorestani M., et al. Autophagy regulation and its role in normal and malignant hematopoiesis. Journal of Cellular Physiology . 2019;234(12):21746–21757. doi: 10.1002/jcp.28903. [DOI] [PubMed] [Google Scholar]
- 94.Hu X., Mei S., Meng W., et al. CXCR4-mediated signaling regulates autophagy and influences acute myeloid leukemia cell survival and drug resistance. Cancer Letters . 2018;425:1–12. doi: 10.1016/j.canlet.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 95.Piya S., Kornblau S. M., Ruvolo V. R., et al. Atg 7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood . 2016;128(9):1260–1269. doi: 10.1182/blood-2016-01-692244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sumitomo Y., Koya J., Nakazaki K., et al. Cytoprotective autophagy maintains leukemia-initiating cells in murine myeloid leukemia. Blood . 2016;128(12):1614–1624. doi: 10.1182/blood-2015-12-684696. [DOI] [PubMed] [Google Scholar]
- 97.Ro S. H., Jung C. H., Hahn W. S., et al. Distinct functions of Ulk 1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy . 2013;9(12):2103–2114. doi: 10.4161/auto.26563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Honda S., Arakawa S., Nishida Y., Yamaguchi H., Ishii E., Shimizu S. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nature Communications . 2014;5(1):p. 4004. doi: 10.1038/ncomms5004. [DOI] [PubMed] [Google Scholar]
- 99.Joo J. H., Wang B., Frankel E., et al. The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Molecular Cell . 2016;62(4):491–506. doi: 10.1016/j.molcel.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheong H., Lindsten T., Thompson C. B. Autophagy and ammonia. Autophagy . 2012;8(1):122–123. doi: 10.4161/auto.8.1.18078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Longatti A., Lamb C. A., Razi M., Yoshimura S., Barr F. A., Tooze S. A. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. The Journal of Cell Biology . 2012;197(5):659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scott R. C., Juhász G., Neufeld T. P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Current Biology . 2007;17(1):1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Joshi A., Iyengar R., Joo J. H., et al. Nuclear ULK1 promotes cell death in response to oxidative stress through PARP1. Cell Death and Differentiation . 2016;23(2):216–230. doi: 10.1038/cdd.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou X., Babu J. R., da Silva S., et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proceedings of the National Academy of Sciences . 2007;104(14):5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Konno H., Konno K., Barber G. N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell . 2013;155(3):688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saleiro D., Mehrotra S., Kroczynska B., et al. Central role of ULK1 in type I interferon signaling. Cell Reports . 2015;11(4):605–617. doi: 10.1016/j.celrep.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Valverde D. P., Yu S., Boggavarapu V., et al. ATG2 transports lipids to promote autophagosome biogenesis. The Journal of Cell Biology . 2019;218(6):1787–1798. doi: 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gómez-Sánchez R., Rose J., Guimarães R., et al. Atg 9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. The Journal of Cell Biology . 2018;217(8):2743–2763. doi: 10.1083/jcb.201710116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chang T.-K., Shravage B. V., Hayes S. D., et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nature Cell Biology . 2013;15(9):1067–1078. doi: 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tang Z., Takahashi Y., Chen C., et al. Atg2A/B deficiency switches cytoprotective autophagy to non-canonical caspase-8 activation and apoptosis. Cell Death and Differentiation . 2017;24(12):2127–2138. doi: 10.1038/cdd.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Velikkakath A. K., Nishimura T., Oita E., Ishihara N., Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Molecular Biology of the Cell . 2012;23(5):896–909. doi: 10.1091/mbc.e11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang X., Gao Y., Tan J., et al. HIV-1 and HIV-2 infections induce autophagy in Jurkat and CD4+ T cells. Cellular Signalling . 2012;24(7):1414–1419. doi: 10.1016/j.cellsig.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 113.Yoo B. H., Zagryazhskaya A., Li Y., et al. Upregulation of ATG3 contributes to autophagy induced by the detachment of intestinal epithelial cells from the extracellular matrix, but promotes autophagy-independent apoptosis of the attached cells. Autophagy . 2015;11(8):1230–1246. doi: 10.1080/15548627.2015.1056968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu K., Zhao Q., Liu P., et al. ATG3-dependent autophagy mediates mitochondrial homeostasis in pluripotency acquirement and maintenance. Autophagy . 2016;12(11):2000–2008. doi: 10.1080/15548627.2016.1212786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mariño G., Fernández A. F., Cabrera S., et al. Autophagy is essential for mouse sense of balance. The Journal of Clinical Investigation . 2010;120(7):2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Betin V. M., Lane J. D. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. Journal of Cell Science . 2009;122(14):2554–2566. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yin X., Zhou C., Li J., et al. Autophagy in bone homeostasis and the onset of osteoporosis. Bone Research . 2019;7(1):p. 28. doi: 10.1038/s41413-019-0058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arbogast F., Arnold J., Hammann P., et al. ATG5 is required for B cell polarization and presentation of particulate antigens. Autophagy . 2019;15(2):280–294. doi: 10.1080/15548627.2018.1516327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jounai N., Takeshita F., Kobiyama K., et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proceedings of the National Academy of Sciences of the United States of America . 2007;104(35):14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Young M. M., Takahashi Y., Khan O., et al. Autophagosomal Membrane Serves as Platform for Intracellular Death-inducing Signaling Complex (iDISC)-mediated Caspase-8 Activation and Apoptosis ∗. The Journal of Biological Chemistry . 2012;287(15):12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nishida Y., Arakawa S., Fujitani K., et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature . 2009;461(7264):654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 122.Galluzzi L., Green D. R. Autophagy-Independent Functions of the Autophagy Machinery. Cell . 2019;177(7):1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baerga R., Zhang Y., Chen P. H., Goldman S., Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy . 2009;5(8):1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu Y., Zhao L., Liu L., et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein & Cell . 2010;1(5):468–477. doi: 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hu F., Li G., Huang C., et al. The autophagy-independent role of BECN1 in colorectal cancer metastasis through regulating STAT3 signaling pathway activation. Cell Death & Disease . 2020;11(5):p. 304. doi: 10.1038/s41419-020-2467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scarlatti F., Maffei R., Beau I., Codogno P., Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death and Differentiation . 2008;15(8):1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 127.Cao Y., Klionsky D. J. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Research . 2007;17(10):839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 128.Xu F., Fang Y., Yan L., et al. Nuclear localization of Beclin 1 promotes radiation-induced DNA damage repair independent of autophagy. Scientific Reports . 2017;7(1):p. 45385. doi: 10.1038/srep45385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Elgendy M., Ciro M., Abdel-Aziz A. K., et al. Beclin 1 restrains tumorigenesis through Mcl-1 destabilization in an autophagy-independent reciprocal manner. Nature Communications . 2014;5(1):p. 5637. doi: 10.1038/ncomms6637. [DOI] [PubMed] [Google Scholar]
- 130.Thoresen S. B., Pedersen N. M., Liestøl K., Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Experimental Cell Research . 2010;316(20):3368–3378. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 131.Sun P., Nie K., Zhu Y., et al. A mosquito salivary protein promotes flavivirus transmission by activation of autophagy. Nature Communications . 2020;11(1):p. 260. doi: 10.1038/s41467-019-14115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fernández Á. F., Sebti S., Wei Y., et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature . 2018;558(7708):136–140. doi: 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Su L. Y., Luo R., Liu Q., et al. Atg5- and Atg7-dependent autophagy in dopaminergic neurons regulates cellular and behavioral responses to morphine. Autophagy . 2017;13(9):1496–1511. doi: 10.1080/15548627.2017.1332549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim W.-Y., Nam S. A., Choi A., et al. Atg7-dependent canonical autophagy regulates the degradation of aquaporin 2 in prolonged hypokalemia. Scientific Reports . 2019;9(1):p. 3021. doi: 10.1038/s41598-019-39702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee I. H., Kawai Y., Fergusson M. M., et al. Atg7 modulates p 53 activity to regulate cell cycle and survival during metabolic stress. Science (New York, N.Y.) . 2012;336(6078):225–228. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Trentesaux C., Fraudeau M., Pitasi C. L., et al. Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity. Proceedings of the National Academy of Sciences of the United States of America . 2020;117(20):11136–11146. doi: 10.1073/pnas.1917174117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Juhász G., Erdi B., Sass M., Neufeld T. P. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes & Development . 2007;21(23):3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Salah F. S., Ebbinghaus M., Muley V. Y., et al. Tumor suppression in mice lacking GABARAP, an Atg8/LC3 family member implicated in autophagy, is associated with alterations in cytokine secretion and cell death. Cell Death & Disease . 2016;7(4):e2205–e2205. doi: 10.1038/cddis.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reggiori F., Monastyrska I., Verheije M. H., et al. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host & Microbe . 2010;7(6):500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Scherz-Shouval R., Weidberg H., Gonen C., Wilder S., Elazar Z., Oren M. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proceedings of the National Academy of Sciences . 2010;107(43):18511–18516. doi: 10.1073/pnas.1006124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gu Y., Princely Abudu Y., Kumar S., et al. Mammalian Atg8 proteins regulate lysosome and autolysosome biogenesis through SNAREs. The EMBO Journal . 2019;38(22, article e101994) doi: 10.15252/embj.2019101994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Delcour C., Amazit L., Patino L. C., et al. ATG7 and ATG9A loss-of-function variants trigger autophagy impairment and ovarian failure. Genetics in medicine: official journal of the American College of Medical Genetics . 2019;21(4):930–938. doi: 10.1038/s41436-018-0287-y. [DOI] [PubMed] [Google Scholar]
- 143.Jia S., Wang Y., You Z., Liu B., Gao J., Liu W. Mammalian Atg9 contributes to the post-Golgi transport of lysosomal hydrolases by interacting with adaptor protein-1. FEBS Letters . 2017;591(24):4027–4038. doi: 10.1002/1873-3468.12916. [DOI] [PubMed] [Google Scholar]
- 144.Yamaguchi J., Suzuki C., Nanao T., et al. Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy . 2018;14(5):764–777. doi: 10.1080/15548627.2017.1314897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shen H., Lin Z., Shi H., et al. MiR-221/222 promote migration and invasion, and inhibit autophagy and apoptosis by modulating ATG10 in aggressive papillary thyroid carcinoma. 3 Biotech . 2020;10(8):p. 339. doi: 10.1007/s13205-020-02326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Catanese A., Olde Heuvel F., Mulaw M., et al. Retinoic acid worsens ATG10-dependent autophagy impairment in TBK1-mutant hiPSC-derived motoneurons through SQSTM1/p62 accumulation. Autophagy . 2019;15(10):1719–1737. doi: 10.1080/15548627.2019.1589257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao Q., Hu Z. Y., Zhang J. P., et al. Dual Roles of Two Isoforms of Autophagy-related Gene _ATG10_ in HCV- Subgenomic replicon Mediated Autophagy Flux and Innate Immunity. Scientific Reports . 2017;7(1):p. 11250. doi: 10.1038/s41598-017-11105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.DeSelm C. J., Miller Brian C., Zou W., et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Developmental Cell . 2011;21(5):966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hwang S., Maloney N. S., Bruinsma M. W., et al. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host & Microbe . 2012;11(4):397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rubinstein Assaf D., Eisenstein M., Ber Y., Bialik S., Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Molecular Cell . 2011;44(5):698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 151.Murrow L., Malhotra R., Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nature Cell Biology . 2015;17(3):300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li Z., Tian X., Ji X., et al. ULK1-ATG13 and their mitotic phospho-regulation by CDK1 connect autophagy to cell cycle. PLoS Biology . 2020;18(6, article e3000288) doi: 10.1371/journal.pbio.3000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mauthe M., Langereis M., Jung J., et al. An siRNA screen for ATG protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. The Journal of Cell Biology . 2016;214(5):619–635. doi: 10.1083/jcb.201602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kaizuka T., Mizushima N. Atg13 is essential for autophagy and cardiac development in mice. Molecular and Cellular Biology . 2016;36(4):585–595. doi: 10.1128/MCB.01005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Diao J., Liu R., Rong Y., et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature . 2015;520(7548):563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu Y., Shoji-Kawata S., Sumpter R. M., et al. Autosis is a Na+,K+-ATPase–regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia–ischemia. Proceedings of the National Academy of Sciences . 2013;110(51):20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cadwell K., Liu J. Y., Brown S. L., et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature . 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang C., Bauckman K. A., Ross A. S. B., et al. A non-canonical autophagy-dependent role of the ATG16L1(T300A) variant in urothelial vesicular trafficking and uropathogenic Escherichia coli persistence. Autophagy . 2019;15(3):527–542. doi: 10.1080/15548627.2018.1535290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Heckmann B. L., Teubner B. J. W., Boada-Romero E., et al. Noncanonical function of an autophagy protein prevents spontaneous Alzheimer’s disease. Science Advances . 2020;6(33, article eabb9036) doi: 10.1126/sciadv.abb9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ji C., Zhao H., Li D., et al. Role of Wdr45b in maintaining neural autophagy and cognitive function. Autophagy . 2020;16(4):615–625. doi: 10.1080/15548627.2019.1632621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Morishita H., Kanda Y., Kaizuka T., et al. Autophagy is required for maturation of surfactant-containing lamellar bodies in the lung and swim bladder. Cell Reports . 2020;33(10, article 108477) doi: 10.1016/j.celrep.2020.108477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nemazanyy I., Blaauw B., Paolini C., et al. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Molecular Medicine . 2013;5(6):870–890. doi: 10.1002/emmm.201202057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gstrein T., Edwards A., Přistoupilová A., et al. Mutations in Vps15 perturb neuronal migration in mice and are associated with neurodevelopmental disease in humans. Nature Neuroscience . 2018;21(2):207–217. doi: 10.1038/s41593-017-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Willinger T., Flavell R. A. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proceedings of the National Academy of Sciences of the United States of America . 2012;109(22):8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mortensen M., Soilleux E. J., Djordjevic G., et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. The Journal of Experimental Medicine . 2011;208(3):455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hashimoto M., Umemoto T., Nakamura-Ishizu A., et al. Autophagy is dispensable for the maintenance of hematopoietic stem cells in neonates. Blood Advances . 2021;5(6):1594–1604. doi: 10.1182/bloodadvances.2020002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu F., Lee J. Y., Wei H., et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood . 2010;116(23):4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jung H. E., Shim Y. R., Oh J. E., Oh D. S., Lee H. K. The autophagy protein Atg5 plays a crucial role in the maintenance and reconstitution ability of hematopoietic stem cells. Immune Network . 2019;19(2, article e12) doi: 10.4110/in.2019.19.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Du W., Xu A., Huang Y., et al. The role of autophagy in targeted therapy for acute myeloid leukemia. Autophagy . 2021;17(10):2665–2679. doi: 10.1080/15548627.2020.1822628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cao Y., Cai J., Zhang S., et al. Autophagy sustains hematopoiesis through targeting Notch. Stem Cells and Development . 2015;24(22):2660–2673. doi: 10.1089/scd.2015.0176. [DOI] [PubMed] [Google Scholar]
- 171.Wu F., Chen Z., Liu J., Hou Y. The Akt-mTOR network at the interface of hematopoietic stem cell homeostasis. Experimental Hematology . 2021;103:15–23. doi: 10.1016/j.exphem.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 172.Mortensen M., Watson A. S., Simon A. K. Lack of autophagy in the hematopoietic system leads to loss of hematopoietic stem cell function and dysregulated myeloid proliferation. Autophagy . 2011;7(9):1069–1070. doi: 10.4161/auto.7.9.15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Phadwal K., Watson A. S., Simon A. K. Tightrope act: autophagy in stem cell renewal, differentiation, proliferation, and aging. Cellular and Molecular Life Sciences . 2013;70(1):89–103. doi: 10.1007/s00018-012-1032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang M., Liu F., Zhou P., et al. The MTOR signaling pathway regulates macrophage differentiation from mouse myeloid progenitors by inhibiting autophagy. Autophagy . 2019;15(7):1150–1162. doi: 10.1080/15548627.2019.1578040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Carsetti R. The development of B cells in the bone marrow is controlled by the balance between cell-autonomous mechanisms and signals from the microenvironment. The Journal of Experimental Medicine . 2000;191(1):5–8. doi: 10.1084/jem.191.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lim V. Y., Zehentmeier S., Fistonich C., Pereira J. P. A chemoattractant-guided walk through lymphopoiesis: from hematopoietic stem cells to mature B lymphocytes. Advances in Immunology . 2017;134:47–88. doi: 10.1016/bs.ai.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Arsov I., Li X., Matthews G., et al. BAC-mediated transgenic expression of fluorescent autophagic protein Beclin 1 reveals a role for Beclin 1 in lymphocyte development. Cell Death and Differentiation . 2008;15(9):1385–1395. doi: 10.1038/cdd.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Clarke A. J., Ellinghaus U., Cortini A., et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Annals of the Rheumatic Diseases . 2015;74(5):912–920. doi: 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Martin F., Kearney J. F. B1 cells: similarities and differences with other B cell subsets. Current Opinion in Immunology . 2001;13(2):195–201. doi: 10.1016/S0952-7915(00)00204-1. [DOI] [PubMed] [Google Scholar]
- 180.O’Connor B. P., Gleeson M. W., Noelle R. J., Erickson L. D. The rise and fall of long-lived humoral immunity: terminal differentiation of plasma cells in health and disease. Immunological Reviews . 2003;194(1):61–76. doi: 10.1034/j.1600-065X.2003.00055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Oliva L., Cenci S. Autophagy in plasma cell pathophysiology. Frontiers in Immunology . 2014;5:103–103. doi: 10.3389/fimmu.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen M., Hong M. J., Sun H., et al. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nature Medicine . 2014;20(5):503–510. doi: 10.1038/nm.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Miller B. C., Zhao Z., Stephenson L. M., et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy . 2008;4(3):309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 184.Pengo N., Scolari M., Oliva L., et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nature Immunology . 2013;14(3):298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 185.Arnold J., Murera D., Arbogast F., Fauny J. D., Muller S., Gros F. Autophagy is dispensable for B-cell development but essential for humoral autoimmune responses. Cell Death and Differentiation . 2016;23(5):853–864. doi: 10.1038/cdd.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Arbogast F., Gros F. Lymphocyte autophagy in homeostasis, activation, and inflammatory diseases. Frontiers in Immunology . 2018;9:p. 1801. doi: 10.3389/fimmu.2018.01801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Weindel C. G., Richey L. J., Bolland S., Mehta A. J., Kearney J. F., Huber B. T. B cell autophagy mediates TLR7-dependent autoimmunity and inflammation. Autophagy . 2015;11(7):1010–1024. doi: 10.1080/15548627.2015.1052206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.McLeod I. X., He Y. Roles of autophagy in lymphocytes: reflections and directions. Cellular & Molecular Immunology . 2010;7(2):104–107. doi: 10.1038/cmi.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Parekh V. V., Wu L., Boyd K. L., et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. Journal of Immunology (Baltimore, Md : 1950) . 2013;190(10):5086–5101. doi: 10.4049/jimmunol.1202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kabat A. M., Harrison O. J., Riffelmacher T., et al. The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. eLife . 2016;5, article e12444 doi: 10.7554/eLife.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang G., Driver J. P., Van Kaer L. The Role of Autophagy in iNKT Cell Development. Frontiers in Immunology . 2018;9:p. 2653. doi: 10.3389/fimmu.2018.02653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Salio M., Puleston D. J., Mathan T. S., et al. Essential role for autophagy during invariant NKT cell development. Proceedings of the National Academy of Sciences of the United States of America . 2014;111(52):E5678–E5687. doi: 10.1073/pnas.1413935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pei B., Zhao M., Miller B. C., et al. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. Journal of Immunology (Baltimore, Md : 1950) . 2015;194(12):5872–5884. doi: 10.4049/jimmunol.1402154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pua H. H., Guo J., Komatsu M., He Y. W. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. Journal of Immunology (Baltimore, Md : 1950) . 2009;182(7):4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 195.Jia W., He Y. W. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. Journal of Immunology (Baltimore, Md : 1950) . 2011;186(9):5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 196.Puleston D. J., Zhang H., Powell T. J., et al. Autophagy is a critical regulator of memory CD8+ T cell formation. Elife . 2014;3 doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Botbol Y., Guerrero-Ros I., Macian F. Key roles of autophagy in regulating T-cell function. European Journal of Immunology . 2016;46(6):1326–1334. doi: 10.1002/eji.201545955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schlie K., Westerback A., DeVorkin L., et al. Survival of effector CD8+ T cells during influenza infection is dependent on autophagy. Journal of Immunology (Baltimore, Md : 1950) . 2015;194(9):4277–4286. doi: 10.4049/jimmunol.1402571. [DOI] [PubMed] [Google Scholar]
- 199.Hubbard-Lucey V. M., Shono Y., Maurer K., et al. Autophagy Gene Atg16l1 Prevents Lethal T Cell Alloreactivity Mediated by Dendritic Cells. Immunity . 2014;41(4):579–591. doi: 10.1016/j.immuni.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Watson A. S., Riffelmacher T., Stranks A., et al. Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Discovery . 2015;1(1):1–10. doi: 10.1038/cddiscovery.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Warr M. R., Binnewies M., Flach J., et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature . 2013;494(7437):323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cao Y., Zhang A., Cai J., et al. Autophagy regulates the cell cycle of murine HSPCs in a nutrient-dependent manner. Experimental Hematology . 2015;43(3):229–242. doi: 10.1016/j.exphem.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 203.Riffelmacher T., Simon A. K. Mechanistic roles of autophagy in hematopoietic differentiation. The FEBS Journal . 2017;284(7):1008–1020. doi: 10.1111/febs.13962. [DOI] [PubMed] [Google Scholar]
- 204.O'Sullivan T. E., Johnson L. R., Kang H. H., Sun J. C. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity . 2015;43(2):331–342. doi: 10.1016/j.immuni.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang S., Xia P., Huang G., et al. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nature Communications . 2016;7(1):p. 11023. doi: 10.1038/ncomms11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang Y.-T., Zaitsev K., Lu Q., et al. Select autophagy genes maintain quiescence of tissue-resident macrophages and increase susceptibility to _Listeria monocytogenes_. Nature Microbiology . 2020;5(2):272–281. doi: 10.1038/s41564-019-0633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nakano H., Ushio H. An unexpected role for autophagy in degranulation of mast cells. Autophagy . 2011;7(6):657–659. doi: 10.4161/auto.7.6.15384. [DOI] [PubMed] [Google Scholar]
- 208.Riffelmacher T., Clarke A., Richter F. C., et al. Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity . 2017;47(3):466–480.e5. doi: 10.1016/j.immuni.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Choi G. E., Yoon S. Y., Kim J. Y., Kang D. Y., Jang Y. J., Kim H. S. Autophagy deficiency in myeloid cells exacerbates eosinophilic inflammation in chronic rhinosinusitis. The Journal of Allergy and Clinical Immunology . 2018;141(3):938–950.e12. doi: 10.1016/j.jaci.2017.10.038. [DOI] [PubMed] [Google Scholar]
- 210.Skendros P., Mitroulis I., Ritis K. Autophagy in Neutrophils: From Granulopoiesis to Neutrophil Extracellular Traps. Frontiers in Cell and Developmental Biology . 2018;6:1–12. doi: 10.3389/fcell.2018.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wildenberg M. E., Koelink P. J., Diederen K., et al. The ATG16L1 risk allele associated with Crohn's disease results in a Rac1-dependent defect in dendritic cell migration that is corrected by thiopurines. Mucosal Immunology . 2017;10(2):352–360. doi: 10.1038/mi.2016.65. [DOI] [PubMed] [Google Scholar]
- 212.Luo X. L., Jiang J. Y., Huang Z., Chen L. X. Autophagic regulation of platelet biology. Journal of Cellular Physiology . 2019;234(9):14483–14488. doi: 10.1002/jcp.28243. [DOI] [PubMed] [Google Scholar]
- 213.Liu Y., Hu M., Luo D., et al. Class III PI3K positively regulates platelet activation and thrombosis via PI(3)P-directed function of NADPH oxidase. Arteriosclerosis, Thrombosis, and Vascular Biology . 2017;37(11):2075–2086. doi: 10.1161/ATVBAHA.117.309751. [DOI] [PubMed] [Google Scholar]
- 214.Lee S. H., Du J., Stitham J., et al. Inducing mitophagy in diabetic platelets protects against severe oxidative stress. EMBO Molecular Medicine . 2016;8(7):779–795. doi: 10.15252/emmm.201506046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Freud A. G., Mundy-Bosse B. L., Yu J., Caligiuri M. A. The broad spectrum of human natural killer cell diversity. Immunity . 2017;47(5):820–833. doi: 10.1016/j.immuni.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Vivier E., Raulet D. H., Moretta A., et al. Innate or adaptive immunity? The example of natural killer cells. Science . 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Fathman J. W., Bhattacharya D., Inlay M. A., Seita J., Karsunky H., Weissman I. L. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood . 2011;118(20):5439–5447. doi: 10.1182/blood-2011-04-348912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Artis D., Spits H. The biology of innate lymphoid cells. Nature . 2015;517(7534):293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 219.Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. Functions of natural killer cells. Nature Immunology . 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 220.O’Sullivan Timothy E., Geary Clair D., Weizman O.-E., et al. Atg5 Is Essential for the Development and Survival of Innate Lymphocytes. Cell Reports . 2016;15(9):1910–1919. doi: 10.1016/j.celrep.2016.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ward J. M., Cherian S., Linden M. A. 19 - Hematopoietic and lymphoid tissues. In: Treuting P. M., Dintzis S. M., Montine K. S., editors. Comparative Anatomy and Histology (Second Edition) San Diego: Academic Press; 2018. pp. 365–401. [Google Scholar]
- 222.Athens J. W., Haab O. P., Raab S. O., et al. Leukokinetic studies. IV. The total blood, circulating and marginal granulocyte pools and the granulocyte turnover rate in normal subjects. The Journal of Clinical Investigation . 1961;40(6):989–995. doi: 10.1172/JCI104338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yousefi S., Mihalache C., Kozlowski E., Schmid I., Simon H. U. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death and Differentiation . 2009;16(11):1438–1444. doi: 10.1038/cdd.2009.96. [DOI] [PubMed] [Google Scholar]
- 224.Germic N., Stojkov D., Oberson K., Yousefi S., Simon H. U. Neither eosinophils nor neutrophils require ATG5-dependent autophagy for extracellular DNA trap formation. Immunology . 2017;152(3):517–525. doi: 10.1111/imm.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bhattacharya A., Wei Q., Shin Jin N., et al. Autophagy is required for neutrophil-mediated inflammation. Cell Reports . 2015;12(11):1731–1739. doi: 10.1016/j.celrep.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 226.Rožman S., Yousefi S., Oberson K., Kaufmann T., Benarafa C., Simon H. U. The generation of neutrophils in the bone marrow is controlled by autophagy. Cell Death and Differentiation . 2015;22(3):445–456. doi: 10.1038/cdd.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hua W., Liu H., Xia L. X., et al. Rapamycin inhibition of eosinophil differentiation attenuates allergic airway inflammation in mice. Respirology . 2015;20(7):1055–1065. doi: 10.1111/resp.12554. [DOI] [PubMed] [Google Scholar]
- 228.Zhu C., Xia L., Li F., et al. mTOR complexes differentially orchestrates eosinophil development in allergy. Scientific Reports . 2018;8(1):p. 6883. doi: 10.1038/s41598-018-25358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Johnson-Weaver B., Choi H. W., Abraham S. N., Staats H. F. Mast cell activators as novel immune regulators. Current Opinion in Pharmacology . 2018;41:89–95. doi: 10.1016/j.coph.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ushio H., Ueno T., Kojima Y., et al. Crucial role for autophagy in degranulation of mast cells. The Journal of Allergy and Clinical Immunology . 2011;127(5):1267–1276.e6. doi: 10.1016/j.jaci.2010.12.1078. [DOI] [PubMed] [Google Scholar]
- 231.Guilliams M., Mildner A., Yona S. Developmental and functional heterogeneity of monocytes. Immunity . 2018;49(4):595–613. doi: 10.1016/j.immuni.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 232.Ginhoux F., Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nature Reviews Immunology . 2014;14(6):392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 233.Pathria P., Louis T. L., Varner J. A. Targeting tumor-associated macrophages in cancer. Trends in Immunology . 2019;40(4):310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 234.Zhang Y., Morgan M. J., Chen K., Choksi S., Liu Z. G. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood . 2012;119(12):2895–2905. doi: 10.1182/blood-2011-08-372383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ricardo S. D., van Goor H., Eddy A. A. Macrophage diversity in renal injury and repair. The Journal of Clinical Investigation . 2008;118(11):3522–3530. doi: 10.1172/JCI36150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhao Z., Fux B., Goodwin M., et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host & Microbe . 2008;4(5):458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bonilla D. L., Bhattacharya A., Sha Y., et al. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity . 2013;39(3):537–547. doi: 10.1016/j.immuni.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Banchereau J., Steinman R. M. Dendritic cells and the control of immunity. Nature . 1998;392(6673):245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 239.Tallóczy Z., Jiang W., Virgin H. W., et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proceedings of the National Academy of Sciences of the United States of America . 2002;99(1):190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lee H. K., Mattei L. M., Steinberg B. E., et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity . 2010;32(2):227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Travassos L. H., Carneiro L. A. M., Ramjeet M., et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunology . 2010;11(1):55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 242.Cooney R., Baker J., Brain O., et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Medicine . 2010;16(1):90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 243.Lupfer C., Thomas P. G., Anand P. K., et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nature Immunology . 2013;14(5):480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zang F., Chen Y., Lin Z., et al. Autophagy is involved in regulating the immune response of dendritic cells to influenza A (H1N1) pdm09 infection. Immunology . 2016;148(1):56–69. doi: 10.1111/imm.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Machlus K. R., Italiano J. E., Jr. The incredible journey: from megakaryocyte development to platelet formation. The Journal of Cell Biology . 2013;201(6):785–796. doi: 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Banerjee M., Huang Y., Ouseph M. M., et al. Autophagy in platelets. Methods in Molecular Biology . 2019;1880:511–528. doi: 10.1007/978-1-4939-8873-0_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ouseph M. M., Huang Y., Banerjee M., et al. Autophagy is induced upon platelet activation and is essential for hemostasis and thrombosis. Blood . 2015;126(10):1224–1233. doi: 10.1182/blood-2014-09-598722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Cao Y., Cai J., Zhang S., et al. Loss of autophagy leads to failure in megakaryopoiesis, megakaryocyte differentiation, and thrombopoiesis in mice. Experimental Hematology . 2015;43(6):488–494. doi: 10.1016/j.exphem.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 249.Zhang W., Ren H., Xu C., et al. Hypoxic mitophagy regulates mitochondrial quality and platelet activation and determines severity of I/R heart injury. eLife . 2016;5, article e21407 doi: 10.7554/eLife.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shravage B. V., Hill J. H., Powers C. M., Wu L., Baehrecke E. H. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development . 2013;140(6):1321–1329. doi: 10.1242/dev.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Jin J., Britschgi A., Schläfli A. M., et al. Low autophagy (ATG) gene expression is associated with an immature AML blast cell phenotype and can be restored during AML differentiation therapy. Oxidative Medicine and Cellular Longevity . 2018;2018:16. doi: 10.1155/2018/1482795.1482795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hwang D. Y., Eom J. I., Jang J. E., et al. ULK1 inhibition as a targeted therapeutic strategy for FLT3-ITD-mutated acute myeloid leukemia. Journal of Experimental & Clinical Cancer Research . 2020;39(1):p. 85. doi: 10.1186/s13046-020-01580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Soliman A. A., Ibrahim R. I., Saeed A. M., Mohamed H. S., Nageeb M. G., Mahmoud H. M. Study of expression of autophagy related gene 5 (ATG5) in Egyptian patients newly diagnosed with acute lymphoblastic leukemia (ALL) QJM: An International Journal of Medicine . 2021;114(Supplement_1) doi: 10.1093/qjmed/hcab100.066. [DOI] [Google Scholar]
- 254.Liu Q., Chen L., Atkinson J. M., Claxton D. F., Wang H.-G. Atg5-dependent autophagy contributes to the development of acute myeloid leukemia in an MLL-AF9-driven mouse model. Cell Death & Disease . 2016;7(9):e2361–e2361. doi: 10.1038/cddis.2016.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Chuanjiang Y., Sivahari P. G., Alina M.-R., et al. Phosphorylation of Beclin-1 by BCR-ABL suppresses autophagy in chronic myeloid leukemia. Haematologica . 2020;105(5):1285–1293. doi: 10.3324/haematol.2018.212027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mortensen M., Ferguson D. J. P., Edelmann M., et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proceedings of the National Academy of Sciences . 2010;107(2):832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Xu X., Araki K., Li S., et al. Autophagy is essential for effector CD8+ T cell survival and memory formation. Nature Immunology . 2014;15(12):1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mouttie L. L.-E., Vu T., Lineburg K. E., et al. Autophagy is required for stem cell mobilization by G-CSF. Blood . 2015;125(19):2933–2936. doi: 10.1182/blood-2014-03-562660. [DOI] [PubMed] [Google Scholar]
- 259.Gomez-Puerto M. C., Folkerts H., Wierenga A. T., et al. Autophagy proteins ATG5 and ATG7 are essential for the maintenance of human CD34+ hematopoietic stem-progenitor cells. Stem Cells (Dayton, Ohio) . 2016;34(6):1651–1663. doi: 10.1002/stem.2347. [DOI] [PubMed] [Google Scholar]
- 260.Kuma A., Hatano M., Matsui M., et al. The role of autophagy during the early neonatal starvation period. Nature . 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 261.Komatsu M., Waguri S., Ueno T., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of Cell Biology . 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Saitoh T., Fujita N., Jang M. H., et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature . 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 263.Sou Y. S., Waguri S., Iwata J., et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Molecular Biology of the Cell . 2008;19(11):4762–4775. doi: 10.1091/mbc.e08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yang Y. P., Hu L. F., Zheng H. F., et al. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacologica Sinica . 2013;34(5):625–635. doi: 10.1038/aps.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pasquier B. SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy . 2015;11(4):725–726. doi: 10.1080/15548627.2015.1033601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Dowdle W. E., Nyfeler B., Nagel J., et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis _in vivo_. Nature Cell Biology . 2014;16(11):1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 267.Petherick K. J., Conway O. J., Mpamhanga C., et al. Pharmacological Inhibition of ULK1 Kinase Blocks Mammalian Target of Rapamycin (mTOR)-dependent Autophagy. The Journal of Biological Chemistry . 2015;290(18):11376–11383. doi: 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]


