Abstract
A lipoprotein gene family first identified in Borrelia burgdorferi strain 297, designated 2.9 LP and recently renamed mlp, was found on circular and linear plasmids in the genome sequence of B. burgdorferi strain B31-M1. Sequence analyses of the B31 mlp genes and physically linked variant gene families indicated that mlp gene heterogeneity is unique and unrelated to location or linkage to divergent sequences. Evidence of recombination between B31 mlp alleles was also detected. Northern blot analysis of cultured strain B31 indicated that the mlp genes were not expressed at a temperature (23°C) characteristic of that of ticks in the environment. In striking contrast, expression of many mlp genes increased substantially when strain B31 was shifted to 35°C, a temperature change mimicking that occurring in the natural transmission cycle of the spirochete from tick to mammal. Primer extension analysis of the mlp mRNA transcripts suggested that sigma 70-like promoters are involved in mlp expression during temperature shift conditions. Antibodies were made against strain B31 Mlp proteins within the first 4 weeks after experimental mouse infection. Importantly, Lyme disease patients also had serum antibodies reactive with purified recombinant Mlp proteins from strain B31, a result indicating that humans are exposed to Mlp proteins during infection. Taken together, the data indicate that strain B31 mlp genes encode a diverse array of lipoproteins which may participate in early infection processes in the mammalian host.
Lyme disease caused by the bacterium Borrelia burgdorferi is the most prevalent arthropod-borne disease in the United States (47). Humans acquire the infection when the organism is transmitted by the bite of infected Ixodes ticks. Subsequent tissue invasion results in diverse clinical manifestations such as erythema migrans, flu-like symptoms, and neurologic, musculoskeletal, and cardiac problems (4, 21, 33, 36, 37, 46).
Most B. burgdorferi outer surface proteins are lipoproteins (29). B. burgdorferi strain B31-M1 has 21 extrachromosomal elements, which may carry up to 91 lipoprotein-encoding genes (9). The synthesis of several outer surface lipoproteins increases when B. burgdorferi cultures are shifted from 23 to 35°C (5, 34, 42, 45). Temperature-shifted cultures are presumed to mimic the warming that occurs when the tick attaches to the mammal and feeds. Several outer surface lipoproteins synthesized by B. burgdorferi grown at 35°C are recognized by sera from infected animals (34, 42), indicating that the mammal is exposed to these proteins during infection or transmission. Antigens expressed early in infection have potential serodiagnostic or vaccine utility.
A lipoprotein-encoding family of seven genes designated 2.9 LP located on 30- and 18-kb supercoiled plasmids was originally discovered and characterized in B. burgdorferi strain 297 (28). Recently, three new members of this gene family were characterized in strain 297 and renamed mlp (for “multicopy lipoprotein genes”) (49). Mlp homologues also are made by Borrelia hermsii (41) and Borrelia afzelii (44). The mlp genes in B. burgdorferi strain 297 can be assigned to categories on the basis of molecular size, protein sequence, and serologic reactivity (28). Two distinct categories of noncoding DNA sequences located immediately upstream of the ribosomal binding site of the mlp genes have been identified (28, 49).
B. burgdorferi strain B31-M1 may contain a combination of nine related 32-kb circular plasmids (designated cp32-1 through cp32-9) and a related linear plasmid (designated lp56) that contains an integrated cp32 plasmid (9, 10, 53, 54). Analysis of the seven cp32 plasmids and lp56 plasmid from the strain B31-M1 sequenced genome identified three families of loci named erp, orfC/orf3, and mlp. Two of these gene families (erp and mlp) encode lipoproteins, whereas the third (orfC/orf3) is believed to participate in plasmid partitioning (40).
Several lines of evidence suggest that members of the Mlp family participate in host-pathogen interactions. First, Akins et al. (2) demonstrated that one mlp operon in strain 297 was expressed only in vivo in dialysis chambers and not at 23, 34, or 37°C following a temperature shift. Second, Yang et al. (49) analyzed three other mlp genes in strain 297 and discovered that their expression increased when the cultures were shifted from 23 to 37°C and that they were antigenic in infected mice. Third, Mlps are lipoproteins, molecules that constitute a significant fraction of the spirochete outer surface and induce immunological responses in the host (16, 22, 24, 48). Fourth, a recent investigation has discovered that Mlp homologs in B. hermsii are antigenic in relapsing-fever patients (41).
Taken together, these observations suggest that Mlps are important molecules that may participate in the pathogenesis of human Lyme disease. The goal of the present study was to investigate molecular variation, expression, and antigenicity of nine mlps identified in the genome of B. burgdorferi strain B31.
MATERIALS AND METHODS
Bacterial strains.
B. burgdorferi strain B31 was originally isolated from an infected Ixodes scapularis tick collected on Shelter Island, N.Y. (7). This strain has been established in the laboratory by means of an infectious cycle between I. scapularis and mice (34). Clone B31-4A was derived from a single colony of infectious B31 plated on solid Barbour-Stoenner-Kelly (BSK) and retains mouse infectivity (10, 19). Clone B31-e1 was derived from a single colony of a high-passage, noninfectious culture of B31 (10).
B. burgdorferi was cultured in BSK-H broth (Sigma, St. Louis, Mo.) supplemented with 6% heat-inactivated rabbit serum (Sigma) at 23 or 35°C as previously described (34, 42). Briefly, 500-ml cultures were grown at 23°C to a density of approximately 107 bacteria per ml; this required approximately 3 weeks. For temperature shift conditions, a 100-ml volume of this culture was diluted into 500 ml of prewarmed fresh medium and grown at 35°C to a density not greater than 108 bacteria per ml (approximately 3 to 5 days). The bacteria were pelleted by centrifugation.
Cloning the B31 mlp genes.
Total genomic DNA from strains B31-e1 and B31-4A was purified from 500 ml of mid-log-phase BSK-H broth cultures with a DNA extraction kit (Stratagene, San Diego, Calif.). DNA was quantitated by UV spectroscopy, and 0.1 μg of DNA was used for PCRs. B31 mlp genes were given an alphabetical designation related to the cp32 plasmid (9, 10) on which they are found. The mlp gene names in relation to their respective plasmid location are designated as follows: mlpA, cp32-1; mlpB, cp32-2; mlpC, cp32-3; mlpD, cp32-4; mlpF, cp32-6; mlpG, cp32-7; mlpH, cp32-8; mlpI, cp32-9; and mlpJ, lp56. The sequences for the mlpB and mlpE genes were not available from The Institute for Genomic Research (TIGR) website (www.tigr.org) because the plasmids encoding these genes (cp32-2 and cp32-5) were not present in the sequenced B31-MI strain (9). For the mlpB gene, Expand Long Template PCR (Boehringer Mannheim, Indianapolis, Ind.) was performed, as recommended by the manufacturer, on B31-e1 genomic DNA with primers designated ErpC-prime and BB-2 (Table 1). A 12-kb fragment was amplified, and the region containing the mlpB gene was sequenced directly with primers ErpC-prime, Cp-2, ErpC-Lp2, and BB-2 (Table 1).
TABLE 1.
Oligonucleotide primers and probes used in this study
| Primer designation | Sequence (5′-3′) | Purpose |
|---|---|---|
| ErpC-prime | CGAATGTATCAGAGTCTCCCTTCC | cp32-2 cloning and sequencing |
| BB-2 | TCCAGACTTATCAACACTAAAAGACGAACAC | cp32-2 cloning and sequencing |
| Cp-2 | AGGAATAACAATGAAAATTATCAACATATTATTTTG | cp32-2 sequencing |
| ErpC-Lp2 | AGGAGCTATTGATTGTAAAAAAACACTCCAC | cp32-2 sequencing |
| mlpA/F-5′ | AATTTTATTTTTACTCAACATATTGTACTGGAG | pCR2.1 cloning |
| mlpA-3′ | ATTAGGACCCATTGCCGCAGGTAG | pCR2.1 cloning |
| mlpB-5′ | ACGGTTCAAGCAATATACATGC | pCR2.1 cloning |
| mlpB-3′ | AATGTTTTTAGTTGTCCCAATCAC | pCR2.1 cloning |
| mlpC/D-5′ | GCTACTACCTCACTTAAAGAATATCAATTC | pCR2.1 cloning |
| mlpC-3′ | GGGCTGTTAGATTATTAGCCAC | pCR2.1 cloning |
| mlpD-3′ | TGAATTTTTGCACGTACTACTTGCAG | pCR2.1 cloning |
| mlpF-3′ | AGCTATTAGGAACCACCATTGTTG | pCR2.1 cloning |
| mlpG/H/I/J-5′ | CTACTACCTCACTTAAAACATATCAATTC | pCR2.1 cloning |
| mlpG-3′ | ATTGCAGGTAGCAGTTGCTTGATCTG | pCR2.1 cloning |
| mlpH-3′ | TATTAGGAACCGTTGCATGTAG | pCR2.1 cloning |
| mlpI-3′ | CGATATTATTGCTGAGCTTGGC | pCR2.1 cloning |
| mlpJ-3′ | TTGAGAAATGTTTTTAGTTTTGCC | pCR2.1 cloning |
| MlpA/C/F/H-His5′ | CCGCTCGAGAATTCTAATGATAATGACAC | Histine-tagged fusion primers |
| MlpB-His5′ | CCGCTCGAGAATGCTAATGATAATGATAC | Histine-tagged fusion primers |
| MlpD-His5′ | CCGCTCGAGAATTCTAATGATACTAATAATAGCC | Histine-tagged fusion primers |
| MlpG-His5′ | CCGCTCGAGAATTCTAATGATACAAATACCAAGC | Histine-tagged fusion primers |
| MlpI-His5′ | CCGCTCGAGAATTCTAATGATACTAATACTAGCC | Histine-tagged fusion primers |
| MlpJ-His5′ | CCGCTCGAGAATTCCAATGATAATGACAC | Histine-tagged fusion primers |
| MlpG-His3′ | CGGGATCCTTATTGCAGGTAGCAGTTGCTTG | Histine-tagged fusion primers |
| All(-G)His3′ | CGGGATCCTATGACCATGATTACGCCAAGC | Histine-tagged fusion primers |
| mlpA5′NP | TGTAATGGAAATGATGCAGACCAAC | Northern probe primers |
| mlpA3′NP | CATTGCCGCAGGTAGTAACTGC | Northern probe primers |
| mlpB5′NP | TGGAAAAGGTACGAACGAAAAGAG | Northern probe primers |
| mlpB3′NP | TTGTCCCAATCACTTGTAAGACCC | Northern probe primers |
| mlpC5′NP | TCAAGCAGAACAACAAAAAACCAC | Northern probe primers |
| mlpC3′NP | GCCACCATTATTGCAGTTACTAACC | Northern probe primers |
| mlpD5′NP | AATACCTTCAAGCAGGTCGTTCAG | Northern probe primers |
| mlpD3′NP | TTATGAATTTTTGCACGTACTACTTGC | Northern probe primers |
| mlpF5′NP | AGTTCAGGGTTTCTTTAGCGGC | Northern probe primers |
| mlpF3′NP | GCTATTAGGAACCACCATTGTTGC | Northern probe primers |
| mlpG5′NP | TGGAAATGATGAAGGAAAAAACACC | Northern probe primers |
| mlpG3′NP | TTAATTGCAGGTAGCAGTTGCTTG | Northern probe primers |
| mlpH5′NP | CAATGGAAAAGAAAATGGGGATG | Northern probe primers |
| mlpH3′NP | TTAGGAACCGTTGCATGTAGTAGTTG | Northern probe primers |
| mlpI5′NP | TTCAAAGAGGTGGTTAAGGGGG | Northern probe primers |
| mlpI3′NP | TTGCTGAGCTTGGCAGGTACTAC | Northern probe primers |
| mlpJ5′NP | GGGGATGGTGACAACTTAATAGAGC | Northern probe primers |
| mlpJ3′NP | TGCCAATTAGCTGTAAGACCAGC | Northern probe primers |
| mlpA/F-PE | GTGTCATTATCATTAGAATTGCAGCTATTTAGTAG | Primer extension primers |
| mlpC-PE | GGTTAAATCACGCTTTCCCCGT | Primer extension primers |
| mlpAprom-5′ | TGCCCTCTTCGAGGAACTTTATTACTTTG | Upstream region cloning |
| mlpAprom-3′ | ATTAGGACCCATTGCCGCAGGTAG | Upstream region cloning |
| mlpCprom-5′ | TGGAAACAGTGTCAACAAATATTGCAAGTG | Upstream region cloning |
| mlpCprom-3′ | GGGCTGTTAGATTATTAGCCAC | Upstream region cloning |
The primers used to amplify the mlpB gene from B31-e1 for cloning into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.) were designated mlpB-5′ and mlpB-3′ (Table 1). Primers for amplifying DNA fragments containing the mlpA, mlpC, mlpD, mlpF, mlpG, mlpH, mlpI, and mlpJ, for cloning in the pCR2.1 vector, were designed based on the B31 cp32 sequences available from the TIGR website. The same 5′ primer for several mlp genes (mlpA and mlpF, mlpC and mlpD, and mlpG, mlpH, mlpI, and mlpJ) was used in conjunction with a unique 3′ primer. The 5′ primers were designated mlpA/F-5′, mlpC/D-5′, and mlpG/H/I/J-5′, and the 3′ primers were designated mlpA-3′, mlpC-3′, mlpD-3′, mlpF-3′, mlpG-3′, mlpH-3′, mlpI-3′, and mlpJ-3′ (Table 1).
The mlpB-5′ and mlpB-3′ primers (Table 1) were used with B31-e1 genomic DNA. B31-e1 has been described as containing cp32-1, cp32-2, cp32-3, and cp32-4 but lacks the other cp32 plasmids (10). The mlpA/F-5′, mlpC/D-5′, mlpG/H/I/J-5′, mlpA-3′, mlpC-3′, mlpD-3′, mlpF-3′, mlpG-3′, mlpH-3′, mlpI-3′, and mlpJ 3′ primers were used with B31-4A DNA. B31-4A contains lp56 and all the cp32 plasmids except cp32-2 (10).
All PCRs were performed in a total volume of 100 μl under mineral oil with the following thermocycler conditions: heat denaturation at 94°C for 1 min, annealing at 50°C for 30 s, and extension at 72°C for 2 min. Twenty-five of these cycles were followed by 7 min of fill-in extension at 72°C. PCR fragments of 499, 676, 566, 525, 506, 524, 553, 538, and 729 bp containing the genes for mlpA, mlpB, mlpC, mlpD, mlpF, mlpG, mlpH, mlpI, and mlpJ, respectively, were produced.
The PCR fragments were excised from agarose gels, and the fragment was purified on (−)EtBr spin columns (Supelco, Bellefonte, Pa.) as specified by the manufacturer. Fragments were then quantitated by UV spectroscopy, ligated with the pCR2.1 vector, and transformed into INVαF′ cells as specified by the manufacturer (Invitrogen). Recombinants were selected, and the plasmids were purified and sequenced. The resulting plasmids were designated pCR-MlpA, pCR-MlpB, pCR-MlpC, pCR-MlpD, pCR-MlpF, pCR-MlpG, pCR-MlpH, pCR-MlpI, and pCR-MlpJ.
DNA sequencing.
PCR fragments were sequenced by the following method. PCR products that were free of contaminating bands as assessed by agarose gel electrophoresis were purified and concentrated with a Centricon 100 concentrator (Millipore, Bedford, Mass.). PCR products with additional contaminating bands were purified in a different manner. The fragment of interest was excised from the gel and purified with a Supelco (−)EtBr spin column. DNA was quantitated by UV spectroscopy and diluted for automated DNA sequencing with an Applied Biosystems Inc. model 373 Stretch automated DNA sequencer and ABI PRISM dye terminator ready-reaction cycle-sequencing kit (PE Biosystems, Foster City, Calif.). DNA plasmids were purified with a Qiagen Midi kit (Qiagen, Valencia, Calif.).
DNA sequence analysis.
Nucleotide and deduced amino acid sequences were analyzed with MacVector version 6.5.1 (Oxford Molecular, Beaverton, Oreg.) and compared to reference sequences obtained from the TIGR website. Sequence alignments, phylogenetic tree construction, and calculation of protein sequence similarity values were performed with DNASTAR (Lasergene, Madison, Wis.). Protein sequence identity values were calculated by pairwise alignment of protein sequences in GAP (Genetics Computer Group, Madison, Wis.). Sequence and primer extension data were further analyzed with the ς70 consensus search program MACTARGSEARCH (25). Detection and analysis of recombination between mlp alleles were performed by previously described statistical methods (38). Bendability/curvature propensity plots were calculated with the bend.it server, with the DNase I-based trinucleotide bendability parameters described by Brukner et al. (6) and the consensus bendability scale (12).
Phylogenetic analyses also were performed with the PHYLIP phylogeny inference package, version 3.57c, written by Joseph Felsenstein, Department of Genetics, University of Washington, Seattle, Wash. The Jukes and Cantor method of nucleotide substitution (DNADIST) was used for comparative purposes, and distance matrices were analyzed by the neighbor-joining method (NEIGHBOR). Multiple bootstrapped data sets (SEQBOOT) were constructed with ClustalV alignments and then analyzed by using distance matrix construction (DNADIST) or parsimony analysis (DNAPARS). Trees were constructed from data sets by the neighbor-joining method NEIGHBOR or the Fitch Margoliash and Least-Squares method (KITCH). Majority-rule consensus trees were constructed with the program CONCENSE and RETREE. All phylogenetic trees were viewed with TreeView, version 1.5, written by Roderic D. M. Page, Division of Environmental and Evolutionary Biology, University of Glasgow, Glasgow, United Kingdom.
Northern blots.
Total RNA was extracted from B. burgdorferi B31-4A and B31-e1 cells grown at 23°C or after the temperature shift to 35°C, with Ultraspec isolation solution (Biotecx, Houston, Tex.). The RNA was denatured with glyoxal and dimethyl sulfoxide, separated by agarose gel electrophoresis in 10 mM sodium phosphate buffer (pH 7.0) (32), and transferred to nylon membranes (Micron Separations, Westborough, Mass.). Probes specific for each of the B31 mlp genes and the flaB (flagellin) gene were generated by PCR from pCR2.1 recombinant plasmids with the cognate gene. Probe specificities were first established by searching probe DNA sequences against the B31-M1 genome on the TIGR website. The oligonucleotides used to generate these PCR fragment probes are listed in Table 1. The flaB-specific probe was generated as previously described (39). DNA fragments generated by PCR were gel purified by using QIAEX II kit (QIAGEN) and radiolabeled with [α-32P]dATP (NEN Research Products, Du Pont, Boston, Mass.) by random priming. Unincorporated radiolabel was removed by column chromatography. To check for probe specificity, purified DNAs from the pCR2.1 recombinant clones were digested with EcoRI for 2 h at 37°C to linearize the DNA and 1 μg of the DNA was transferred to nitrocellulose with a vacuum dot-blot apparatus (Bio-Rad, Hercules, Calif.). Dot blot filters were hybridized overnight at 65°C in 2× Denhardt's solution–6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1 mM EDTA–100 μg of sonicated salmon sperm DNA per ml in a rotating hybridization oven (Robbin's Scientific Corp., Sunnyvale, Calif.). Following hybridization, the nitrocellulose filters were washed twice for 15 min at 65°C in 2× SSC containing 0.1% sodium dodecyl sulfate (SDS) and then four times for 15 min each at room temperature in 0.1× SSC containing 0.1% SDS. The filters then were subjected to autoradiography at −70°C for 4 to 48 h.
For Northern analysis, nylon membranes containing total RNA from B. burgdorferi B31-4A or B31-e1 were hybridized with 106 counts of each radiolabeled probe at 55°C in 1% (wt/vol) bovine serum albumin–7% (wt/vol) SDS–0.5 M sodium phosphate (pH 7.0)–1 mM EDTA. The membranes were washed at 55°C in 0.2× SSC–1% (wt/vol) SDS as previously described (5). The radioactivity was allowed to decay for more than 2 months until the signal diminished to a point no longer detectable by autoradiography and the membrane was reprobed with the fla probe as previously described (39) under similar hybridization conditions.
Primer extension analysis.
RNA for primer extension analysis was extracted from B. burgdorferi B31-4A or B31-e1 grown at 23°C or after the shift to 35°C by using Ultraspec isolation solution (Biotecx). The RNA was treated with DNase (Promega, Madison, Wis.), dissolved in sterile deionized water treated with 0.1% diethylpyrocarbonate, and quantitated by UV spectroscopy.
mlpA and mlpC were chosen for study on the basis of the uniqueness of their upstream and coding sequences for primer specificity. The DNA sequences of mlpA and mlpF are identical for the first 124 bases upstream and 275 bases downstream of the start codons. This high level of identity precluded the design of a primer that would differentiate between transcripts for these two genes. Therefore, a primer (designated mlpA/F-PE) that would bind equally well to mlpA or mlpF transcripts was made. Base differences within the regulatory and coding regions for mlpC, mlpD, mlpG, mlpH, mlpI, and mlpJ allowed the design of a primer specific for mlpC transcript expression, and this primer was designated mlpC-PE (Table 1). Alignment of the mlpA/F-PE and mlpC-PE primers against the B31-M1 total genome sequence established that these primers were specific for their targets.
Extension reactions were performed with the avian myeloblastosis virus reverse transcriptase primer extension system (Promega). The reaction products were loaded onto a 6% or 8% acrylamide sequencing gel. A 1,093-bp DNA fragment containing the start codon and upstream sequence for the mlpA gene was made with primers designated mlpAprom-5′ and mlpAprom-3′ (Table 1) and cloned into the pCR2.1 vector (Invitrogen). The plasmid DNA (designated pmlpA-prom) was purified and used as a sequencing marker. For primer extension analysis of the mlpC gene, a 1,174-bp DNA fragment containing the mlpC ATG start codon and upstream sequence was made with primers designated mlpCprom-5′ and mlpCprom-3′ (Table 1). This fragment was cloned into the pCR2.1 vector (designated pmlpC-prom) and used as a sequencing marker for mlpC primer extension analysis. All sequencing reactions were performed with the Amplicycle sequencing kit (PE Biosystems).
Recombinant Mlp proteins and Western blotting.
Genes encoding Mlp proteins were amplified with PCR primers that would result in a XhoI site and a BamHI site for restriction digestion and cloning in frame with an amino-terminal histidine tag in the pET-15b expression vector (Novagen, Madison, Wis.). The same 5′ primer (designated MlpA/C/F/H-His5′) was used for the mlpA, mlpC, mlpF, and mlpH genes (Table 1). For mlpB, mlpD, mlpG, mlpI, and mlpJ, unique 5′ primers were designated MlpB-His5′, MlpD-His5′, MlpG-His5′, MlpI-His5′, and MlpJ-His5′, respectively (Table 1). The same 3′ primer, designated All(-G)His3′ (Table 1), was used with all of the above-mentioned 5′ primers. Unique primers designated MlpG-His5′ and MlpG-His3′ were used for cloning and expression of the mlpG gene. The 5′ fusion primers begin with the codon for the first amino acid immediately downstream of the lipidated cysteine, and the 3′ primers correspond to the sequence in the pCR2.1 vector. The Mlp-His fusion proteins were terminated by the encoded Borrelia mlp termination sequence in each construct.
All PCRs were performed in a total volume of 100 μl under mineral oil with the appropriate primer set and the appropriate pCR2.1 recombinant DNA as the template. The PCR conditions used were 25 cycles of 94°C for 1 min, 50°C for 30 s, and 72°C for 2 min followed by one cycle of 7 min at 72°C. The pET15b vector (Novagen) was digested with XhoI and BamHI enzymes, purified with (−)EtBr spin columns, and ligated to the fusion PCR fragments. Ligation reaction mixtures were transformed into BLR(DE3) competent cells (Novagen). Recombinants were induced with isopropyl-β-d-thiogalactopyranoside (IPTG) and the proteins encoded by mlpA, mlpC, mlpD, mlpF, mlpG, mlpH, mlpI, and mlpJ were identified in supernatant fractions by SDS-polyacrylamide gel electrophoresis analysis. These proteins were purified as recommended by the manufacturer (Novagen). The MlpB fusion protein was present in the cell pellet fraction, and attempts to purify the protein under denaturing conditions were unsuccessful.
Purified proteins were quantitated with the Bio-Rad protein assay kit, and 2 μg of each protein was loaded onto 15% polyacrylamide gels and subjected to SDS-polyacrylamide gel electrophoresis. The gels were stained with Coomassie blue or transferred to nitrocellulose. Mouse serum collected 4 weeks (two mice) or 8 weeks (three mice) after infection of the mouse, via a tick bite, with a low-passage, nonclonal population of B31, was provided by Tom Schwan, Rocky Mountain Laboratories (39). The individual 4- and 8-week mouse sera were pooled separately prior to immunoblotting of nitrocellulose membranes. Control sera were collected from two mice not infected with B. burgdorferi. Eleven human sera were collected from Lyme disease patients and provided by Tom Schwan. The diagnosis of Lyme disease was based on p39 immunoreactivity and clinical presentation consistent with Lyme disease (35). In addition, sera collected from five noninfected humans, with no previous exposure to B. burgdorferi, were used in immunoblotting experiments.
Western blotting was performed as previously described with slight modifications (39). Briefly, primary incubations were performed for 1 h with mouse or human sera at a 1:100 dilution. A rabbit anti-mouse or anti-human horseradish peroxidase-conjugated secondary antibody at 1:20,000 dilution was incubated with the membrane for 1 h in phosphate-buffered saline–Tween. Membranes were developed with the ECL kit as specified by the manufacturer (Amersham, Piscataway, N.J.).
Nucleotide sequence accession number.
The sequence for the B31 mlpB gene has been deposited in GenBank under accession number AF245449.
RESULTS
mlp genes in strain B31.
B. burgdorferi strain B31 can contain nine different cp32 plasmids and a related linear plasmid designated lp56. The B. burgdorferi strain recently sequenced (B31-M1) contains seven cp32 mlp genes (cp32-2 and cp32-5 are absent) and one lp56 mlp gene. We used DNA purified from a B31 passage variant known to contain cp32-2 to amplify a large fragment that extended from the erpC gene to the intergenic region located 3′ of the mlp gene on cp32-2. Sequence analysis of this PCR fragment indicated that a mlp gene homologue (designated mlpB) was present. Our attempt to clone the mlp gene located on the cp32-5 plasmid with a similar strategy was unsuccessful.
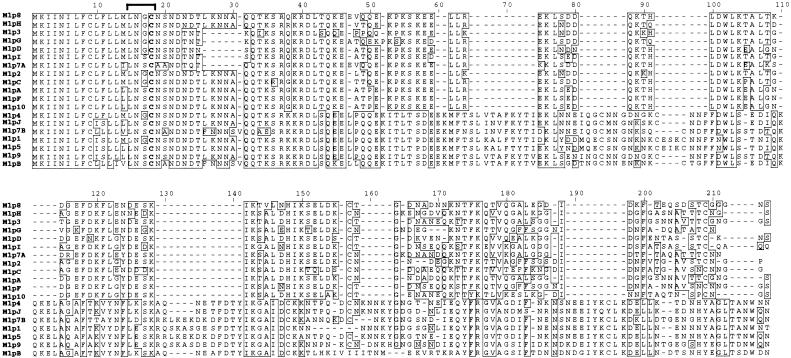
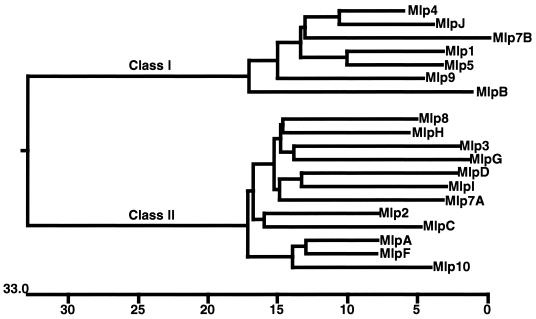
Alignment of the Mlp protein sequences encoded by mlp genes in strains 297 and B31 identified a relatively conserved consensus signal peptidase II cleavage site for all Mlps (Fig. 1). Mlp proteins have been previously assigned to two categories or classes on the basis of sequence homology, reactivity with polyclonal antisera, and molecular weight (28, 49). Phylogenetic tree construction with the amino acid sequences of 9 strain B31 Mlps and 10 strain 297 Mlps confirmed that two distinct sequence families exist and that mlpB encodes a class I protein (Fig. 2). Identity values ranged from 22.1 to 43.1% for comparison of Mlp class I proteins with class II proteins within or between the two strains. Within the two classes, the identity values were 68.7 to 86.4% for class I proteins and 65.8 to 88.6% for class II proteins. The phylogenetic analysis also found that divergent or unique Mlp proteins exist between the two strains (Fig. 2). For example, only four of nine strain B31 Mlp proteins were related to strain 297 Mlp proteins (Mlp-2 and MlpC, 81.1% identity; Mlp-4 and MlpJ, 88.6% identity; Mlp-8 and MlpH, 81.4% identity; and Mlp-3 and MlpG, 74.4% identity). Hence, there are no closely related Mlp homologs (>95%) in strains B31 and 297.
FIG. 1.
Alignment of Mlp proteins from strain B31 and 297. The protein alignment was constructed with ClustalV (15). Mlp proteins from strain 297 were numerically designated (Mlp1, Mlp2, etc.), and Mlp proteins from strain B31 have alphabetical designations (MlpA, MlpB, etc.). The bracket identifies the consensus signal peptidase II sequence. The lipidated cysteine is shown in bold type. Boxed residues indicate perfect matches with consensus sequence. Dashes indicate gaps created during the alignment process.
FIG. 2.
Phylogenetic tree of Mlp proteins from strains B31 and 297. The phylogenetic tree was constructed by using ClustalV (15) and the nearest-neighbor joining method (31). Class I and class II groupings are shown. Mlp proteins from strain 297 are numerically designated (Mlp1, Mlp2, etc.), and Mlp proteins from strain B31 have alphabetical designations (MlpA, MlpB, etc.). The scale represents the number of amino acid replacements, expressed as a percentage of the number of residue positions compared.
The cp32 orfC/orf3 genes encode proteins similar to ParA and SopA (3, 54), products required for efficient partitioning of low-copy-number plasmids in other bacteria (1, 27). Sequence analysis of orfC/orf3 loci that have physically linked erp loci was used to assess the possibility of past recombination events within the erp genes on the cp32 plasmids (40). Comparison of strain B31 mlp phylogenetic trees with previously published trees for the B31 orfC/orf3 loci (40) indicated that the sequence differences observed among the mlp genes were distinct and did not correlate with differences observed in cp32 plasmids or orfC/orf3 loci. For example, mlpB (cp32-2) and mlpJ (lp56), both encoding members of the class I proteins, are most homologous to each other, whereas the cp32-2 orfC/orf3 locus is most similar to the cp32-7 orfC/orf3 locus and the lp56 orfC/orf3 locus is most similar to the cp32-8 orfC/orf3 locus (40). Similar sequence analysis of the strain B31 erp genes did not identify a correlation between linkage and sequence heterogeneity for the erp and mlp loci (data not shown). These data suggest that strain B31 mlp sequence diversity is distinct and unrelated to the diversity observed in two other linked, heterogeneous loci on strain B31 cp32 plasmids.
To determine if intragenic recombination between the mlp genes had occurred and therefore may have contributed to the distinct sequence heterogeneity observed for these genes in strain B31, seven members of the B31 class II Mlp protein-encoding genes were analyzed. The strain B31 class I protein genes were not included because they demonstrated bias due to their small sample size and significant sequence divergence from class II protein-encoding genes. Recombination identification was performed by a statistical phylogenetic partitioning method that comparatively analyzes polymorphic sites in each allele (38). The results indicated that the seven strain B31 class II protein-encoding alleles contain significant clustered sites that partition into distinct, conflicting groups (data not shown), thereby demonstrating that strong evidence of recombination exists among these alleles.
Northern analysis of mlp gene expression at tick (23°C) and mammalian (35°C) temperatures.
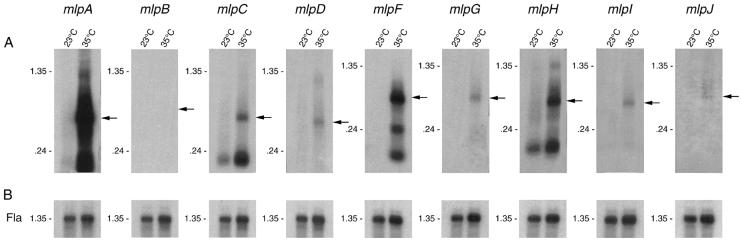
The expression of several B. burgdorferi outer surface lipoproteins increases when cultures are shifted from 23 to 35°C (34, 42). We investigated if the mlp genes of B31 were differentially expressed at temperatures normally associated with the tick (23°C) or mammal (35°C) (42). Oligonucleotide primers were designed that amplify unique regions of the B31 mlp genes. The resulting PCR products varied in size from 84 to 160 bp for mlpA through mlpJ. Each probe was confirmed to be specific by being tested against the full-length cloned mlp genes digested with EcoRI.
When the PCR products were probed against B. burgdorferi B31-4A RNA under stringent hybridization conditions, it was found that full-length mlp gene transcripts were not present at 23°C (Fig. 3). In striking contrast, predicted full-length transcripts were identified when the cultures were shifted from 23 to 35°C (Fig. 3). The RNA transcripts smaller than the predicted full-length transcript observed for mlpA, mlpC, mlpF, and mlpH (Fig. 3) may be caused by functional inactivation, a process involving endonucleolytic cleavage or degradation of the 5′ terminus of the transcript (18). The intensities of RNA transcripts for mlpA, mlpC, mlpF, and mlpH were considerably greater than those observed for mlpD, mlpG, mlpI, and mlpJ. Variance in transcript intensity could be due, in part, to differences in mlp promoter sequences. For example, mlpA and mlpF, which contain nearly identical upstream sequences, had a high level of band intensity, whereas mlpC, mlpD, mlpG, mlpH, mlpI, and mlpJ, with a different upstream sequence, had comparatively lower band intensity.
FIG. 3.
Northern blot analysis of mlp mRNA transcripts. B. burgdorferi B31-4A was grown in culture medium at 23°C (lanes 23°C) or shifted from 23°C to 35°C (lanes 35°C). RNA in gels was transferred to filters, which were individually incubated with radiolabeled probes specific for the B31 gene indicated above each panel. Arrows indicate the predicted sizes of mRNA transcripts for each gene. Cp32-2 does not exist in B31-4A, and therefore, as anticipated, no reactivity was observed in the mlpB-probed lane. (B) After 2 months of decay and loss of signal, each filter was rehybridized with a probe specific for the constitutively expressed flaB gene. RNA molecular size markers (in kilobases) are indicated to the left of each panel.
Primer extension analysis of mlpA/F and mlpC.
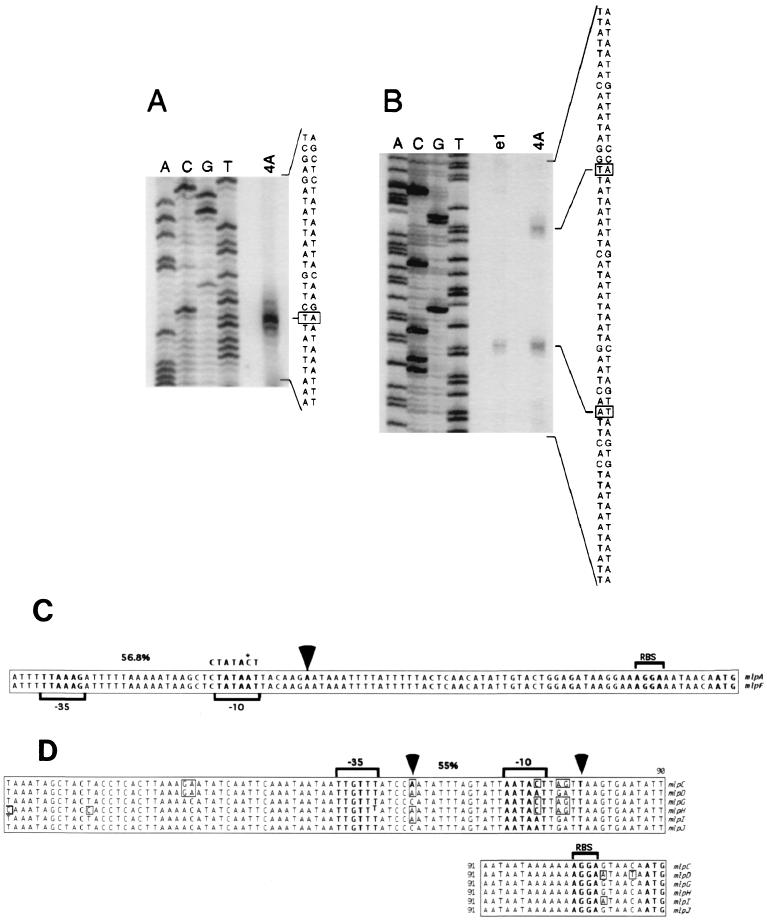
Primer extension analysis was performed to identify the mRNA initiation sites and promoter regions responsible for transcript initiation. The DNA sequence of mlpC, mlpD, mlpG, mlpH, mlpI, and mlpJ in strain B31 differs completely from that of the analogous region of mlpA and mlpF beginning 6 bases upstream of the ATG start codon. The high homology between the coding and noncoding regions of mlpA and mlpF precluded the design of a primer which would bind specifically to one gene but not the other. Therefore, the primer used (designated mlpA/F-PE) had the potential to bind to both mlpA or mlpF transcripts. Primer extension experiments were performed with RNA isolated from temperature-shifted strain B31-e1, a passage variant known to contain cp32-1 (mlpA present) but not cp32-6 (mlpF absent). No product was detected in primer extension experiments (data not shown). Northern blot analysis with the mlpA-specific probe also failed to detect an mRNA transcript for the mlpA gene in RNA from temperature-shifted strain B31-e1 (data not shown). PCR amplification and sequence analysis with the mlpA/F-5′ and mlpA-3′ primers confirmed the presence of the mlpA gene in DNA from strain B31-e1.
Primer extension analysis of RNA obtained from temperature-shifted strain B31-4A with the mlpA/F-PE primer produced a single, dominant extension product (Fig. 4A). This product is most probably the result of extension from mlpA and mlpF transcripts known to be present in this preparation (Fig. 3). The calculated point of initiation is 95 bases from the 3′ end of the primer sequence and 56 bases upstream of the ATG start codon for mlpA and mlpF. No additional extension products were detected during brief or prolonged runs of the gel or subsequent repeated experiments (data not shown). The faint bands observed immediately above and below the primary initiation site (Fig. 4A) may represent initiation just before and after the adenine 56 bases upstream of the ATG start codon for mlpA and mlpF during transcription initiation, a phenomenon reported in studies of ospC transcription initiation (23).
FIG. 4.
Primer extension analysis of the transcriptional initiation site for the mlpA, mlpF, and mlpC genes and alignment of the upstream sequences for the mlp genes, with primer extension initiation sites and potential promoters identified. Alignments were performed with ClustalV (15). (A) cDNA synthesized from B. burgdorferi B31-4A (4A) mRNA using the mlpA/F-PE primer was loaded adjacent to lanes containing a DNA-sequencing reaction with the same primer and pmlpAprom recombinant DNA. Lanes A, C, G, and T represent the mlpA upstream noncoding sequence. (B) cDNA synthesized from B. burgdorferi B31-e1 (e1) or B31-4A (4A) mRNA using the mlpC-PE primer was loaded adjacent to lanes containing a DNA-sequencing reaction performed using the same primer and pmlpCprom recombinant DNA. Lanes A, C, G, and T represent the mlpC upstream noncoding sequence. Minus-strand sequence and coding-strand sequences are shown, with lines and boxes denoting the cDNA products and the initiating nucleotides, respectively. Numbers 1 and 2 designate the two initiation sites relative to distance from the ATG start codon for mlpC. (C) mlpA and mlpF upstream alignment. The ATG start codon is shown in bold type on the right, and the potential ribosomal binding site (RBS) is noted. The transcript initiation site is marked with an arrow, and the potential promoter element with −10 (−10) and −35 (−35) regions (bracketed) is shown. The consensus ςS sequence CTATACT is shown above the −10 region, along with the one base difference (∗). The percent similarity score for the ς70 consensus is listed above the spacer region. (D) mlpC, mlpD, mlpG, mlpH, mlpI, and mlpJ upstream alignment. The ATG start codon is shown in bold type on the right, and the potential ribosomal binding site (RBS) is noted. The initiation sites are indicated by arrows and designated 1 and 2, and the potential promoter element for site 1 with −10 (−10) and −35 (−35) regions (bracketed) is shown. The percent similarity score for ς70 consensus is listed above the sequences. Boxed residues signify bases different from the consensus.
Primer extension analysis with the mlpC-PE primer, specific for mlpC, and RNA from temperature-shifted B31-4A identified two products (Fig. 4B). The first initiation site was located 34 bases upstream from the ATG start codon of the mlpC gene, and the second site was 57 bases upstream. No other extension products were detected in primer extension reactions during short or prolonged runs of the gel or in subsequent repeated experiments (data not shown). Interestingly, for strain B31-e1 temperature-shifted RNA, only the lower-molecular-weight product (initiation at the first site) was observed (Fig. 4B). Primer extension experiments with the mlpC-PE and mlpA/F-PE primers were repeated 11 times with different acrylamide gels, cultures, and RNA preparations of strain B31-4A and strain B31-e1. Results identical to those shown in Fig. 4 were always obtained (data not shown).
Computer analyses of the region 7 bases upstream of the initiation site for mlpA and mlpF identified a potential ς70 promoter with a similarity score of 56.8% (Fig. 4C). Moreover, further analysis of this promoter indicated the presence of an Escherichia coli ςS (RpoS) consensus sequence (11) in the −10 region (Fig. 4C). Consistent with this observation, a curvature-propensity plot calculated with DNase I-based trinucleotide parameters (6, 12) identified an upstream sequence capable of adopting a curved conformation. We note that E. coli ςS promoters usually are located immediately downstream of DNA capable of adopting a curved conformation (11).
A ς70 consensus promoter sequence with a homology score of 55% was found immediately upstream of the first initiation site for mlpC (Fig. 4D). Alignment of the regulatory regions of mlpC, mlpD, mlpG, mlpH, mlpI, and mlpJ and the first and second initiation sites is shown in Fig. 4D. Surprisingly, no consensus sequence with a homology score greater than 45%, the cutoff value for functional ς70 promoter sequences in E. coli, was identified upstream of the second initiation site (25). In addition, no ςS consensus sequence or curved DNA was identified upstream of the first or second initiation site.
Immunoblot analysis of the B31 Mlp proteins with sera from infected mice and Lyme disease patients.
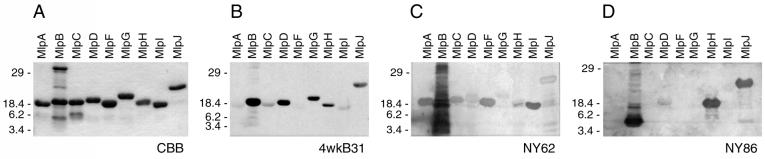
Identification of temperature-induced mRNA transcripts corresponding to the B31 mlp genes led us to test if the protein products were antigenic in mammalian hosts. Each Mlp protein was expressed as a His-tagged fusion protein and purified to homogeneity (Fig. 5A). MlpB was present in insoluble form in the E. coli pellet fraction and was intractable to purification (Fig. 5A).
FIG. 5.
Immunoblot analysis of recombinant B31 Mlp proteins. (A) Coomassie blue-stained gel of the purified Mlp fusion proteins (CBB). (B) Nitrocellulose filter incubated with a 1:100 dilution of serum collected from mice 4 weeks after infection with strain B31 by tick feeding (4wkB31). Sera from uninfected mice did not react with any of the Mlp proteins (data not shown). MlpB, a 20-kDa protein band, could not be purified to homogeneity from E. coli proteins (higher- and lower-molecular-weight bands). (C and D) Two immunoblots probed with patient sera (NY62 and NY86, respectively). Sera from uninfected humans did not react with any of the Mlp proteins (data not shown). Molecular masses (in kilodaltons) are indicated to the left.
Western blot analysis was conducted with pooled mouse sera obtained 4 weeks after tick inoculation with low-passage, noncloned strain B31, which contains cp-1 through cp-9 and lp56 (10) (Fig. 5B). The mouse sera contained antibodies that reacted with the Mlp proteins (Fig. 5B). Identical results were obtained with pooled mouse sera collected 8 weeks post inoculation. No increased reactivity was identified to the proteins observed to be weakly reactive with the sera collected at 4 weeks (data not shown). Differences in mouse antibody production or reactivity to the Mlp proteins were apparent (Fig. 5B), a result suggesting that Mlps may differ in antigenicity or levels of expression during this early stage of infection.
We next determined if antibodies were produced against Mlp proteins during natural human infections. Sera obtained from Lyme disease patients were used. Sera obtained from two patients (designated NY62 and NY86) had differential antibody reactivities to the panel of B31 Mlp proteins (Fig. 5C and D). For example, serum from patient NY62 reacted strongly with MlpA, MlpF, and MlpI, reacted weakly with MlpC, MlpD, and MlpJ, and did not react with MlpB, MlpG and MlpH (Fig. 5C). In contrast, serum from patient NY86 reacted strongly with MlpH and MlpJ, reacted weakly with MlpD, and did not react with MlpA, MlpB, MlpC, MlpF, MlpG, and MlpI (Fig. 5D). Normal human serum did not contain anti-Mlp antibodies (data not shown).
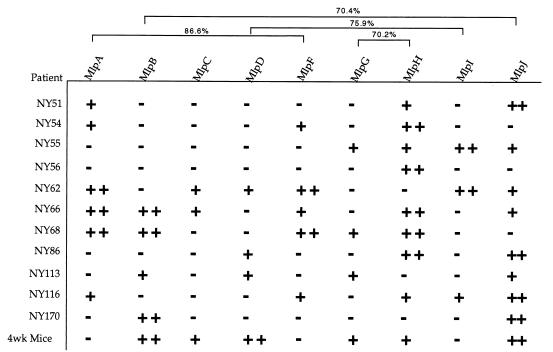
We next tested sera obtained from nine additional patients diagnosed with Lyme disease. On average, these nine Lyme disease patients had antibodies against relatively few of the strain B31 Mlp proteins (Fig. 6). Similarly, some proteins had more reactivity with the patient sera than others did. For example, MlpA and MlpF (86.6% identical) had similar patterns of reactivity (Fig. 6), whereas proteins with 75.9% identity or less did not have similar patterns of reactivity. MlpD and MlpI belong to the same class of proteins (class II) and have 75.9% sequence identity, but they had very different reactivity with the mouse sera (Fig. 5B).
FIG. 6.
Reactivity of Lyme disease patient antisera (diluted 1:100) with recombinant B31 Mlp proteins. Sera were scored for strong reactivity (++), weak reactivity (+), or no reactivity (−). The numbers above the brackets represent percent identity values for MlpA and MlpF (86.6%), MlpB and MlpJ (70.4%), MlpD and MlpI (75.9%), and MlpG and MlpH (70.2%).
DISCUSSION
Sequence analysis.
Phylogenetic analysis of the B31 cp32 mlp genes compared with two other physically linked heterogeneous gene families established that the sequence diversity present in the mlp genes is unique and is unrelated to location or linkage to variant sequences. Caimano et al. (8) concluded that the importance of recombination as a mechanism for generating sequence diversity at the B31 mlp loci was less apparent than their results obtained for the strain 297 mlp genes, presumably because of the lower level of sequence diversity seen among the strain B31 mlp alleles. However, our focused analysis of polymorphic sites within each of the seven class II mlp alleles demonstrated that these genes, and presumably the class I mlp genes, have been diversified by recombination events. Comparative analysis of the mlp genes present in strains 297 and B31 (including the newly discovered mlpB gene) demonstrated that only four mlp genes were relatively homologous. Taken together, the data suggest that recombination events contribute to the diversity of the mlp genes and suggest that the rate of generating new allelic variants may be relatively high. Although the host processes that select new Mlp variants are unknown, immune avoidance or functional selection processes may participate.
mlp Northern blot analysis.
We discovered that upregulation of the mlp genes in B. burgdorferi strain B31 occurs when cultures are shifted from 23 to 35°C, a result in agreement with data obtained from analysis of three newly identified mlp genes in B. burgdorferi strain 297 (49). We note that unlike the Northern blot results reported for the erp genes (40), no mlp-specific transcripts were detected in bacteria grown at 23°C. Hence, our results add the strain B31 mlp gene family to other Borrelia lipoprotein genes that are differentially upregulated by growth at 35°C, a temperature characteristic of many mammalian hosts, including humans (2, 34, 42).
Primer extension analysis of mlp expression.
Primer extension analysis identified the transcriptional initiation sites of the mlp transcripts produced during temperature shift conditions. These experiments identified ς70 promoter elements that may participate in the expression of mlpA, mlpF, and mlpC. Promoters with ς70-like homology in B. burgdorferi have been described previously for the genes encoding variable, lipidated proteins such as OspA, OspC, OspD, and OspE (20, 23, 26) and flagellin (13). We note that primer extension studies with the mlp genes of strain 297 were not performed due to high sequence identity between genes and the lack of defined cp32 passage variants for strain 297 (28).
Promoters under the control of ς70 can be recognized in vitro by ςS, a sigma factor involved in stationary-growth gene expression (43). Our discovery of a consensus ςS sequence (CTATCT) in the −10 region of the promoter region of mlpA and mlpF, together with identification of a region of potential curved DNA upstream, suggests that these genes may be regulated by ςS. In E. coli, the presence of a −35 region in a ςS promoter generally favors recognition by ς70 (11). However, direct evidence of molecular interaction between the mlpA and mlpF promoters and ςS and/or ς70 was not sought in the present study.
The promoter region identified for the second transcription initiation site located upstream of the mlpC gene was not homologous to ς70 or ςS sequences, suggesting a distinct molecular mechanism of mRNA initiation. We discovered that this cryptic promoter is no longer active in strain B31-e1 (which lacks cp32 plasmids and some other plasmids and is noninfectious [10]), a result mimicking that seen for the promoter element of the mlpA gene. It is probable that cis and trans factors responsible for expression of mlpA and mlpC are not present, are present at greatly reduced levels, or are not active in B31-e1. Altered gene expression has been described in high-passage B31 variants (17, 30).
Among the B31 mlpC, mlpD, mlpG, mlpH, mlpI, and mlpJ genes, mlpC and mlpH had the highest level of expression by Northern analysis. These two genes also had identical sequences in the spacer region and the −10 region of the promoter identified for the first upstream initiation site. In contrast, mlpJ had the lowest level of expression and the greatest divergence in sequence of this promoter region. It is likely that the number and location of minor base changes in this promoter region affect the level of expression of these genes in vitro. Minor base changes in promoter sequences that lead to altered promoter activity are well described in many organisms.
Antigenicity of purified Mlp recombinant proteins.
We discovered that mice made anti-Mlp antibodies within the first 4 weeks of experimental infection. This antibody reactivity was not Mlp class specific, because Mlps from both classes reacted similarly with the mouse sera. In addition, there was no simple relationship between the Mlp proteins that were immunoreactive and the upstream regulatory region of their structural genes. Importantly, similar results were obtained with sera from the Lyme disease patients. The mlp complement of the organisms that infected these Lyme disease patients is not known. It is possible that some of the structural features of the Mlp proteins from B31 may be conserved among different B. burgdorferi strains or isolates. In addition, we do not know if these Mlp-reactive antibodies were produced early or late in these human infections, although the mouse data favor early production. We note that our data are consistent with results obtained from recent work on B. hermsii Mlp homologs, also shown to be antigenic in infected humans (41).
Potential pathogenesis roles have been attributed to protein families such as Vls, BlyAB, and the Opps on the basis of functional assays or sequence homologies (5, 14, 50–52). Putative functions have yet to be assigned to the Erp, 39-kDa, and Mlp proteins and numerous other paralog proteins of B. burgdorferi. The identification of multiple mlp alleles in strain B31, together with evidence of intragenic recombination, suggests that more alleles will arise in a strain by recombination. In addition, other B. burgdorferi strains will undoubtedly have diversity in mlp sequence. The extensive diversity of mlp genes within and between strains could constrain the serodiagnostic and vaccine utility of these proteins unless broadly conserved epitopes are identified.
Conclusions.
Our data and those contributed by others (2, 8, 28, 44, 49) suggest that Mlp proteins may play an important, albeit undefined role in B. burgdorferi transmission, establishment of infection, or immune evasion. Analysis of early expression of the mlp genes in the tick during the transmission cycle and their expression within different host tissues may provide important new insights into Lyme disease pathogenesis.
ACKNOWLEDGMENTS
We thank Thomas S. Whittam for help in analysis of the B31 mlp alleles, Tom Schwan for providing sera from Lyme disease patients and infected mice, and Gary Hettrick for help with the graphics. J. M. Musser edited the manuscript.
REFERENCES
- 1.Abeles A L, Snyder K M, Chattoraj D K. P1 plasmid replication: replicon structure. J Mol Biol. 1984;173:307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- 2.Akins D R, Bourell K W, Caimano M J, Norgard M V, Radolf J D. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Investig. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour A G, Carter C J, Bundoc V, Hinnebusch J. The nucleotide sequence of a linear plasmid of Borrelia burgdorferi reveals similarities to those of circular plasmids of other prokaryotes. J Bacteriol. 1996;178:6635–6639. doi: 10.1128/jb.178.22.6635-6639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthold S W, Persing D H, Armstrong A L, Peeples R A. Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am J Pathol. 1991;139:263–273. [PMC free article] [PubMed] [Google Scholar]
- 5.Bono J L, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 1998;144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 6.Brukner I, Sanchez R, Suck D, Pongor S. Trinucleotide models for DNA bending propensity: comparison of models based on DNaseI digestion and nucleosome packaging data. J Biomol Struct Dyn. 1995;13:309–317. doi: 10.1080/07391102.1995.10508842. [DOI] [PubMed] [Google Scholar]
- 7.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 8.Caimano M J, Yang X, Popova T G, Clawson M L, Akins D R, Norgard M V, Radolf J D. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infect Immun. 2000;68:1574–1586. doi: 10.1128/iai.68.3.1574-1586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 10.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espinosa-Urgel M, Chamizo C, Tormo A. A consensus sequence for ςS-dependent promoters. Mol Microbiol. 1996;21:657–659. doi: 10.1111/j.1365-2958.1996.tb02573.x. [DOI] [PubMed] [Google Scholar]
- 12.Gabrielian A, Pongor S. Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Lett. 1996;393:65–68. doi: 10.1016/0014-5793(96)00855-1. [DOI] [PubMed] [Google Scholar]
- 13.Ge Y, Old I G, Saint Girons I, Charon N W. The flgK motility operon of Borrelia burgdorferi is initiated by a ς70-like promoter. Microbiology. 1997;143:1681–1690. doi: 10.1099/00221287-143-5-1681. [DOI] [PubMed] [Google Scholar]
- 14.Guina T, Oliver D B. Cloning and analysis of a Borrelia burgdorferi membrane-interactive protein exhibiting haemolytic activity. Mol Microbiol. 1997;24:1201–1213. doi: 10.1046/j.1365-2958.1997.4291786.x. [DOI] [PubMed] [Google Scholar]
- 15.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. CABIOS. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 16.Hirschfeld M, Kirschning C J, Schwandner R, Wesche H, Weis J H, Wooten R M, Weis J J. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J Immunol. 1999;163:2382–2386. [PubMed] [Google Scholar]
- 17.Jonsson M, Bergstrom S. Transcriptional and translational regulation of the expression of the major outer surface proteins in Lyme disease Borrelia strains. Microbiology. 1995;141:1321–1329. doi: 10.1099/13500872-141-6-1321. [DOI] [PubMed] [Google Scholar]
- 18.King T C, Schlessinger D. Processing of mRNA transcripts. In: Neidhardt F C, Ingram J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1990. pp. 703–718. [Google Scholar]
- 19.Kurtti T J, Munderloh U G, Johnson R C, Ahlstrand G G. Colony formation and morphology in Borrelia burgdorferi. J Clin Microbiol. 1987;25:2054–2058. doi: 10.1128/jcm.25.11.2054-2058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam T T, Nguyen T-P K, Montgomery R R, Kantor F S, Fikrig E, Flavell R A. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logigian E L, Kaplan R F, Steere A C. Chronic neurologic manifestations of Lyme disease. N Engl J Med. 1990;323:1438–1444. doi: 10.1056/NEJM199011223232102. [DOI] [PubMed] [Google Scholar]
- 22.Ma Y, Seiler K P, Tai K F, Yang L, Woods M, Weis J J. Outer surface lipoproteins of Borrelia burgdorferi stimulate nitric oxide production by the cytokine-inducible pathway. Infect Immun. 1994;62:3663–3671. doi: 10.1128/iai.62.9.3663-3671.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marconi R T, Samuels D S, Garon C F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marietta E V, Weis J J, Weis J H. CD28 expression by mouse mast cells is modulated by lipopolysaccharide and outer surface protein A lipoprotein from Borrelia burgdorferi. J Immunol. 1997;159:2840–2848. [PubMed] [Google Scholar]
- 25.Mulligan M E, Hawley D K, Entriken R, McClure W R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura T, Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 28.Porcella S F, Popova T G, Akins D R, Li M, Radolf J D, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and a lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosa P A. Microbiology of Borrelia burgdorferi. Semin Neurol. 1997;17:5–10. doi: 10.1055/s-2008-1040906. [DOI] [PubMed] [Google Scholar]
- 30.Sadziene A, Wilske B, Ferdows M S, Barbour A G. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schutzer S E. Lyme disease: molecular and immunological approaches. Vol. 6. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 34.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson W J, Schrumpf M E, Schwan T G. Reactivity of human Lyme borreliosis sera with a 39-kilodalton antigen specific to Borrelia burgdorferi. J Clin Microbiol. 1990;28:1329–1337. doi: 10.1128/jcm.28.6.1329-1337.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steere A C. Lyme disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 37.Steere A C, Batsford W P, Weinberg M, Alexander J, Berger H J, Wolfson S, Malawista S E. Lyme carditis: cardiac abnormalities of Lyme disease. Ann Intern Med. 1980;93:8–16. doi: 10.7326/0003-4819-93-1-8. [DOI] [PubMed] [Google Scholar]
- 38.Stephens J C. Statistical methods of DNA sequence a analysis: detection of intragenic recombination or gene conversion. Mol Biol Evol. 1985;2:539–556. doi: 10.1093/oxfordjournals.molbev.a040371. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson B, Bono J L, Schwan T G, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick-bite and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson B, Casjens S, Rosa P. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology. 1998;144:1869–1879. doi: 10.1099/00221287-144-7-1869. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson B, Porcella S F, Oie K L, Fitzpatrick C A, Raffel S J, Lubke L, Schrumpf M E, Schwan T G. The relapsing fever spirochete Borrelia hermsii contains multiple antigen-encoding circular plasmids Homologous to the cp32 plasmids of Lyme disease spirochetes. Infect Immun. 2000;7:3900–3908. doi: 10.1128/iai.68.7.3900-3908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci USA. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theisen M. Molecular cloning and characterization of nlpH, encoding a novel, surface-exposed, polymorphic, plasmid-encoded 33-kilodalton lipoprotein of Borrelia afzelii. J Bacteriol. 1996;178:6435–6442. doi: 10.1128/jb.178.22.6435-6442.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tilly K, Casjens S, Stevenson B, Bono J L, Samuels D S, Hogan D, Rosa P. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 46.Van der Linde M R. Lyme carditis: clinical characteristics of 105 cases. Scand J Infect Dis Suppl. 1991;77:81–84. [PubMed] [Google Scholar]
- 47.Walker D H. Tick-transmitted infectious diseases in the United States. Annu Rev Public Health. 1998;19:237–269. doi: 10.1146/annurev.publhealth.19.1.237. [DOI] [PubMed] [Google Scholar]
- 48.Wooten R M, Morrison T B, Weis J H, Wright S D, Thieringer R, Weis J J. The role of CD14 in signaling mediated by outer membrane lipoproteins of Borrelia burgdorferi. J Immunol. 1998;160:5485–5492. [PubMed] [Google Scholar]
- 49.Yang X, Popova T G, Hagman K E, Wikel S K, Schoeler G B, Caimano M J, Radolf J D, Norgard M V. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J-R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J-R, Norris S J. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J-R, Norris S J. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect Immun. 1998;66:3689–3697. doi: 10.1128/iai.66.8.3689-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zückert W R, Filipuzzi-Jenny E, Meister-Turner J, Stålhammar-Carlemalm M, Meyer J. Repeated DNA sequences on circular and linear plasmids of Borrelia burgdorferi sensu lato. In: Axford J S, Rees D H E, editors. Lyme borreliosis. New York, N.Y: Plenum Press; 1994. pp. 253–260. [Google Scholar]
- 54.Zückert W R, Meyer J. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]