Abstract
Surface-exposed proteins often play an important role in the interaction between pathogenic bacteria and their host. We isolated a pool of hydrophobic, surface-associated proteins of Streptococcus pneumoniae. The opsonophagocytic activity of hyperimmune serum raised against this protein fraction was high and species specific. Moreover, the opsonophagocytic activity was independent of the capsular type and chromosomal genotype of the pneumococcus. Since the opsonophagocytic activity is presumed to correlate with in vivo protection, these data indicate that the protein fraction has the potential to elicit species-specific immune protection with cross-protection against various pneumococcal strains. Individual proteins in the extract were purified by two-dimensional gel electrophoresis. Antibodies raised against three distinct proteins contributed to the opsonophagocytic activity of the serum. The proteins were identified by mass spectrometry and N-terminal amino acid sequencing. Two proteins were the previously characterized pneumococcal surface protein A and oligopeptide-binding lipoprotein AmiA. The third protein was the recently identified putative proteinase maturation protein A (PpmA), which showed homology to members of the family of peptidyl-prolyl cis/trans isomerases. Immunoelectron microscopy demonstrated that PpmA was associated with the pneumococcal surface. In addition, PpmA was shown to elicit species-specific opsonophagocytic antibodies that were cross-reactive with various pneumococcal strains. This antibody cross-reactivity was in line with the limited sequence variation of ppmA. The importance of PpmA in pneumococcal pathogenesis was demonstrated in a mouse pneumonia model. Pneumococcal ppmA-deficient mutants showed reduced virulence. The properties of PpmA reported here indicate its potential for inclusion in multicomponent protein vaccines.
Streptococcus pneumoniae is an important human pathogen which causes meningitis, otitis media, sepsis, and pneumonia. The precise molecular mechanisms by which the pneumococcus invades and damages host tissues are not fully understood. For many years, the polysaccharide capsule has been recognized as the major virulence factor and consequently was considered an important vaccine candidate (for reviews see references 5 and 34). The use of a 23-valent vaccine containing capsular polysaccharides from pneumococci commonly causing disease has had limited effect in reducing the morbidity and mortality associated with this organism (1, 16, 19, 41). The current pneumococcal vaccine strategy focuses on the use of conjugates, in which a limited number of capsular polysaccharides are linked to a carrier protein. The proteins in the conjugate vaccines cause a switch in the immune response to polysaccharides from T-cell independent to T-cell dependent. This results in an increase in the antibody response and the generation of memory T lymphocytes. Conjugate vaccines are more immunogenic in young children than polysaccharide vaccines (15, 18). Although the results of early trials look promising, the long-term efficacy is uncertain since large-scale vaccination may over time lead to a shift in serotype distribution towards capsular types that are poorly immunogenic or not included in the vaccine. Such a shift may be enhanced by the horizontal exchange of capsular genes, as described previously (8, 22, 23).
Over the past few years, much attention has been focused on the role of pneumococcal proteins in pathogenesis and protection. Proteins that are involved in the pathogenesis of infections by S. pneumoniae are considered interesting components for future conjugate or multicomponent protein vaccines. The immunological response against such proteins should provide protection against colonization and infection with S. pneumoniae strains of all capsular polysaccharide types. Immunization with pneumolysin (36), pneumococcal surface protein A (PspA) (33, 45, 53), pneumococcal surface adhesin A (PsaA) (44), and neuraminidase (28) clearly confers protection in animal models.
The purpose of this study was to identify additional pneumococcal proteins with abilities to elicit protective immune responses. We isolated a pool of hydrophobic, potentially surface-associated proteins of S. pneumoniae that were able to elicit cross-reactive, species-specific antibodies with opsonophagocytic activity. At least three distinct proteins contributed to the in vitro opsonophagocytic activity. Two proteins were the previously characterized surface proteins PspA and oligopeptide-binding protein AmiA. The third protein was identified as the putative proteinase maturation protein A (PpmA) (35a). The potential of PpmA to elicit protective immune responses and its role in the pathogenesis of pneumococcal infection are discussed.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and growth medium.
The pneumococcal strains used in this study are described in Table 1. Pneumococcal strain FT231 was used for protein purification. Bacteria were grown to logarithmic growth phase (optical density at 550 nm 0.3) in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% yeast extract (Difco Laboratories) (THY broth) at 37°C.
TABLE 1.
Bacterial strains used in this study
| Species and strain | Relevant characteristic(s) | Sourcea or reference |
|---|---|---|
| S. pneumoniae | ||
| FT231 | Serotype 19, clinical isolate | F. Tenover, CDC, Atlanta, Ga. |
| D39 | Serotype 2, clinical isolate | 7 |
| CDC205 | Serotype 3, clinical isolate | J. Butler, CDC, Atlanta, Ga. |
| EF3296 | Serotype 4, clinical isolate | C. Svanborg Eden, University of Göteborg, Sweden |
| ATCC6306 | Serotype 6A, clinical isolate | American Type Culture Collection |
| 911320 | Serotype 6A, clinical isolate | Our laboratory |
| 950357 | Serotype 9V, clinical isolate | Our laboratory |
| S1003 | Serotype 11, clinical isolate | Our laboratory |
| ATCC6314 | Serotype 14, clinical isolate | American Type Culture Collection |
| 950312 | Serotype 14, clinical isolate | Our laboratory |
| S1001 | Serotype 15, clinical isolate | Our laboratory |
| 800129 | Serotype 18C, clinical isolate | Our laboratory |
| 19F G | Serotype 19F, clinical isolate | Our laboratory |
| ATCC6323 | Serotype 23F, clinical isolate | American Type Culture Collection |
| 950110 | Serotype 23F, clinical isolate | Our laboratory |
| S3003 | Serotype 38, clinical isolate | Our laboratory |
| 19F K | Unencapsulated variant of 19F G | Our laboratory |
| Rx1 | Unencapsulated variant of D39 | 42 |
| P376 | Serotype 6A, opaque variant of clinical isolate p303 | 27 |
| P765 | Serotype 6B, transparent variant of clinical isolate p314 | 26 |
| P62 | Serotype 9V, opaque variant of clinical isolate p10 | 51 |
| R189 | ermAM-containing strain | 2 |
| MS9 | D39, ppmA::ermAM | This study |
| S. bovis 961008 | Clinical isolate | Our laboratory |
| E. faecalis ATCC29212 | Clinical isolate | American Type Culture Collection |
CDC, Centers for Disease Control.
Extraction of surface-associated hydrophobic proteins of S. pneumoniae.
Bacterial cells were harvested by centrifugation (1,500 × g, 15 min, room temperature) and washed three times with an equal volume of phosphate-buffered saline (PBS, pH 7.5). After the final wash, the bacteria were resuspended in 1/25 the volume of TE buffer (10 mM Tris-HCl, 1 mM EDTA). The cells were disrupted by ultrasonic treatment (Branson sonifier 250; Branson Ultrasonics, Danburry, Conn.). The method for extraction with sulfobetaine 14 (SB14) was adapted from that of Schouls and colleagues (40). In brief, cell walls, membranes, and other particulate material were collected by centrifugation at 48,400 × g for 20 min. The water-soluble cytoplasmic proteins were removed by washing the bacterial lysates five times with PBS. Pellets were resuspended in 150 mM NaCl and centrifuged for 20 min at 48,400 × g. The pellets were incubated for 2 h at room temperature with 0.25% N-tetradecyl-N,N-dimethylammonio-1-propanesulfonate (SB14; Serva, Heidelberg, Germany) in the presence of 150 mM NaCl–10 mM MgCl2–10 mM Tris-HCl (pH 8.0) with constant stirring. The hydrophobic, membrane-associated proteins were recovered as described by Wessel and Flügge (52). Protein concentrations were determined by the method of Bradford (13).
Protein gel electrophoresis and staining.
One-dimensional sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis was carried out in the Bio-Rad minigel system with 13% polyacrylamide gels. The samples were dissolved in sample buffer (10 mM Tris-HCl, 1 mM EDTA, 1% SDS, 10 mM dithiothreitol [DTT], 10% glycerol, 0.01% bromophenol blue indicator [Merck, Darmstadt, Germany]), boiled for 5 min, and subjected to electrophoresis (39).
Two-dimensional SDS-polyacrylamide gel electrophoresis includes separation of proteins by isoelectric point and by molecular weight, respectively. Isoelectric focusing (pI, 4 to 7) was performed with a Multiphor II electrophoresis unit and Immobiline DryStrips (Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's recommendations, but as modified by Rabilloud and colleagues (37). The proteins were separated in the second dimension by gradient (12 to 20%) polyacrylamide gel electrophoresis. Polyacrylamide gels were stained with Coomassie brilliant blue (CBB) (39).
Hyperimmune rabbit antiserum.
Hyperimmune antiserum was raised against the hydrophobic, surface-associated protein pool and distinct CBB-stained two-dimensional protein gel spots. Shortly after introduction into a one-dimensional SDS-polyacrylamide gel, the SB14-extracted protein pool of S. pneumoniae FT231 was stained with CBB and excised from the gel. The total protein fraction, as well as the individual protein spots cut from the two-dimensional polyacrylamide gel, were washed three times with 0.1 M sodium acetate–96% ethanol, ground into a fine suspension in 0.5 ml of PBS, and subsequently mixed with 0.5 ml of Freund's incomplete adjuvant (Pierce, Rockford, Ill.). New Zealand White rabbits were injected subcutaneously in four or five places. The primary injection was followed by three booster injections at 4-week intervals.
Indirect immunocytometric assay.
Pneumococci were grown to logarithmic phase in THY broth at 37°C and washed three times in ice-cold PBS. The bacterial pellet was dissolved in 5% rabbit serum in PBS (107 bacteria in a 20-μl final volume) and incubated for 15 min at 4°C with shaking. After being washed twice with ice-cold PBS, the bacteria were incubated for 15 min at 4°C with 20 μl (1:5 dilution) of fluorescein-conjugated goat anti-rabbit immunoglobulin G (IgG; Jackson ImmunoResearch Laboratories, West Grove, Pa.) with shaking. Finally, the bacteria were washed twice with ice-cold PBS and resuspended in 100 μl of ice-cold fresh paraformaldehyde (0.5%) in PBS. The samples were analyzed in a FACScan flow cytometer (Becton Dickinson, Mountain View, Calif.).
Phagocytosis assay.
Analysis of the opsonophagocytic activity of the sera was performed as described by Alonso DeVelasco et al. (4) and adapted by Jansen et al. (24). In our assay, S. pneumoniae was grown to logarithmic phase in THY broth without heat-inactivated human pooled serum, and the bacteria were not inactivated. Phagocytosis was defined as the uptake and binding of fluorescein isothiocyanate-labeled bacteria by human polymorphonuclear cells (PMNs) because of opsonization with antiserum. The opsonophagocytic activity is defined as the reciprocal of the serum concentration at which 25% (unless otherwise stated) of the human PMNs were fluorescent.
Western blot analysis.
The proteins separated by one-dimensional (0.5 μg) and two-dimensional (2.5 μg) SDS-polyacrylamide gel electrophoresis were transferred to Immobilon-P membranes (Millipore Corporation, Bedford, Mass.) as described by Sambrook et al. (39). The immunological detection of immobilized proteins was performed as described elsewhere (35a).
Amino acid sequence analysis.
For N-terminal amino acid sequence analysis, the proteins were separated by two-dimensional SDS-polyacrylamide gel electrophoresis as described above with a few modifications. Recrystallized SDS (Serva) was used for preparing the electrophoresis buffers, and sodium thioglycolate (100 mM) was added to the cathodal buffer compartment. The proteins in the gel were blotted to a Problot membrane (Applied Biosystems, San Jose, Calif.) with the Multiphor II system (Pharmacia Biotech) according to the manufacturer's instructions except that 0.02% β-mercaptoethanol was added to the blotting buffer. The blots were stained with amido black (Merck). Amino acid sequence analysis was performed with a model 473A protein sequenator (Applied Biosystems) as recommended by the manufacturer.
For mass spectrometric analysis, the proteins of interest were purified from the gel, digested with trypsin, and analyzed by mass spectrometry as described elsewhere (35a). With Peptide Search (29), the deduced (partial) amino acid sequences were analyzed for matching sequences in all possible translation products of the December 1998 version of the unfinished pneumococcal genome released by the Institute for Genomic Research (TIGR; http://www.tigr.org/data/s_pneumoniae/) to identify the proteins. With the BLAST algorithm (6), putative pneumococcal proteins were analyzed for similarity to sequences deposited in the November 1999 version of the nonredundant protein database at the National Center for Biotechnology Information (Washington, D.C.).
Immunoelectron microscopy.
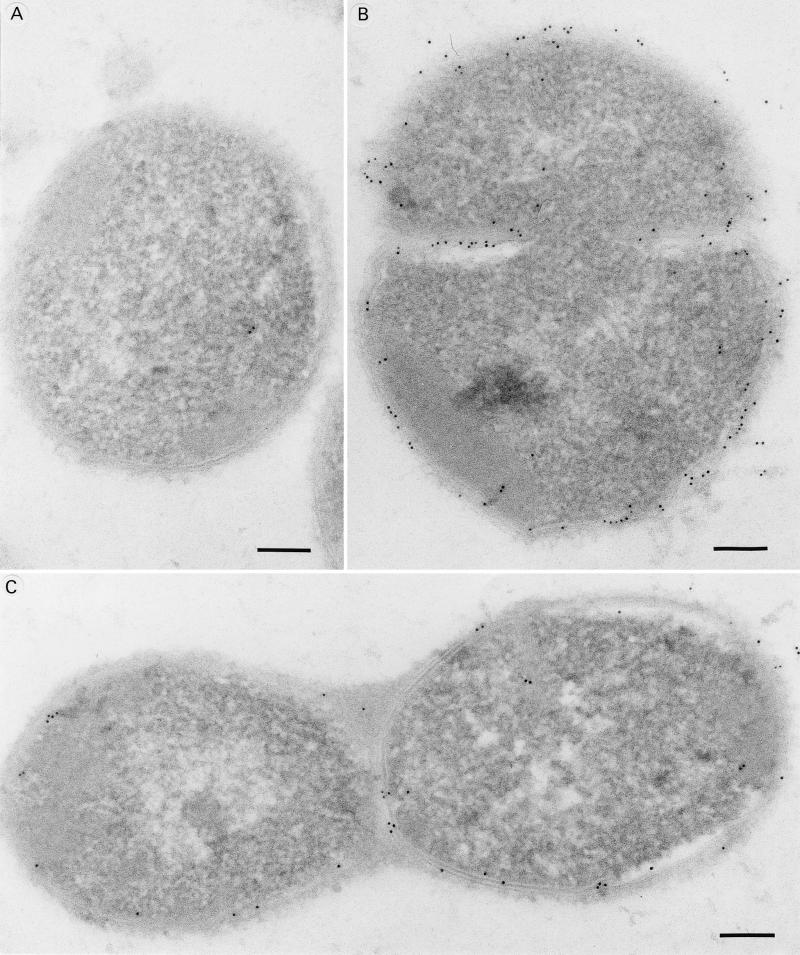
Colonies of S. pneumoniae strain FT231 were fixed in a mixture of 4% formaldehyde, freshly prepared from paraformaldehyde, and 0.1% glutaraldehyde in sodium cacodylate buffer, pH 7.4. The bacteria were embedded in 1.5% low-gelling-temperature agarose (Sigma), and fixation was continued for a total of 24 h. Small pieces of the agarose-embedded bacteria were dehydrated in ethanol in a CS Auto apparatus (Reichart, Vienna, Austria) with a progressive lowering of temperature and embedded in Lowicryl HM20 resin (TAAB, Reading, United Kingdom) at −40°C. Ultrathin sectioning was performed with an Ultracut E microtome (Reichart). Sections were collected on gold grids and immunolabeled by incubation with anti-PpmA rabbit serum and anti-PspA rabbit serum, respectively, diluted 1:200 with buffer A (1% normal goat serum, 1% bovine serum albumin [Biocell], 0.1% Tween 20 [Sigma] in Tris-buffered saline [pH 8.2]) for 2 h at room temperature. Control specimens were incubated with normal rabbit serum and in buffer A without primary antibody. Sections were washed in five 50-μl droplets of buffer A for 5 min each and incubated for 3 h at room temperature in goat anti-rabbit IgG antibody labeled with 5-nm-diameter colloidal gold (Biocell, Cardiff, United Kingdom) diluted 1:100 with buffer A. Final washing was done twice with buffer A and five times in distilled water. Sections were stained with uranyl acetate and examined with a Philips CM12 electron microscope (Philips, Eindhoven, The Netherlands) operating at 80 kV.
Nucleotide sequence analysis of ppmA.
Nucleotide sequencing was performed on ppmA from the pneumococcal strains D39, CDC205, EF3296, P376, P765, 950357, P62, S1003, 950312, S1001, 800129, FT231, 19F G, 950110, S3003, and Rx1. Two PCR products of 640 and 643 nucleotides that cover the whole gene were generated with an overlap of 140 nucleotides. The primers 5′-TCTCATGCTTCGTAAAAATG-3′ and 5′-AGCAAAATCAGCACCTTCTG-3′ were used to amplify the 5′ part of ppmA. The primers 5′-CTGAATTGACAGATGAAGCC-3′ and 5′-CCTTGTACTATGCGTTTTATTG-3′ were used to amplify the 3′ part of ppmA. PCR amplification of ppmA was performed in a 100-μl PCR mixture as described for penicillin-binding protein genotyping (35). The PCR products were purified by sodium acetate precipitation to remove the unincorporated nucleotides and primers (39). Purified templates were sequenced with the Thermo Sequenase dye terminator cycle sequencing premix kit (Pharmacia Amersham, Roosendaal, The Netherlands) and 50 pmol of each PCR primer. Sequencing was performed on the Applied Biosystems Prism 377 (PE Applied Biosystems, Nieuwerkerk, The Netherlands).
Cloning and insertional inactivation of ppmA.
ppmA was amplified from S. pneumoniae strain D39 genomic DNA by PCR with primers pmpp-FW1 (5′-GTTTGGAATTCGCAAGCAAATCACTCTCC-3′), positioned at nucleotides 369 to 340 upstream of the ATG start codon, and pmpp-REV1 (5′-CAGTAGGATCCTTGTACTATGCGTTTTATTG-3′), positioned at nucleotides 1073 to 1104 downstream of the ATG start codon. The forward and reverse primers contain EcoRI and BamHI recognition sequences, respectively. The amplified EcoRI-BamHI-digested ppmA DNA fragment was cloned into pBluescript KS+ (pMS1) and transformed into Escherichia coli DH5α (39). A ppmA mutant was constructed by insertion of an erythromycin resistance cassette (ermAM) in the gene. ermAM was amplified from S. pneumoniae strain R189 genomic DNA with primers ermAM-FW1 (5′-AAAGTTCGAAGCTTAAGTTCAAAACTACTTGCCC-3′) and ermAM-REV1 (5′-AAAGCTGCAGTTCGAATGTCTTCTCACCTTTAG-3′). The amplified ermAM DNA fragment was Csp45I-digested and cloned into the Csp45I site of ppmA (nucleotide position 64 downstream of the ATG start codon) of pMS1, forming pMS2. The EcoRI-BamHI-digested pMS2 DNA fragment was used to transform S. pneumoniae D39 as described previously (54). To confirm that ppmA was inactivated by ermAM, chromosomal DNA was analyzed by PCR with primers pmpp-FW1 and pmpp-REV1. Expression of PpmA was assessed by Western blot analysis with PpmA antibodies. The ppmA mutant MS9 was used in the described experiments.
Mouse pneumonia model.
Preparation of the challenge dose and intranasal challenge of mice were performed as described before (25). Significant differences in the survival time of mice challenged with the ppmA knockout mutants and parent strain D39 were determined by the nonparametric Mann-Whitney U test, with significance set at a value of P of ≤0.05.
RESULTS
Surface-associated protein fraction of S. pneumoniae is able to elicit opsonophagocytic activity.
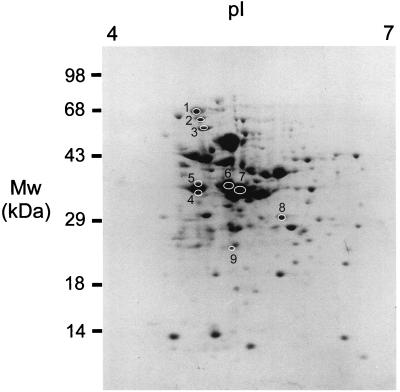
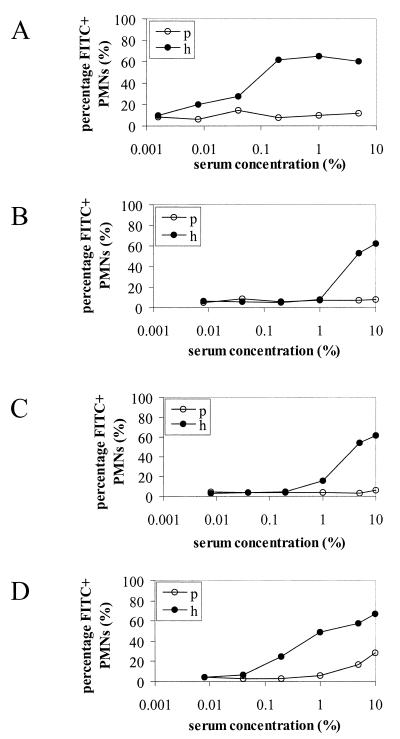
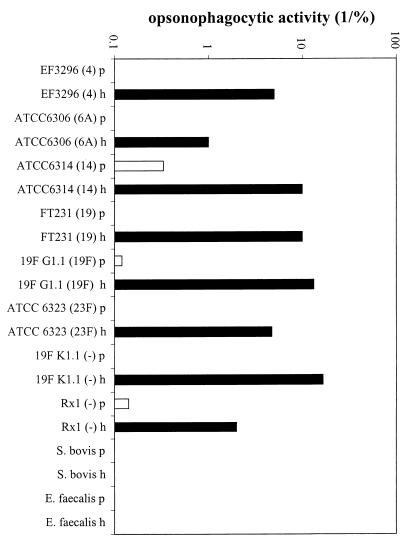
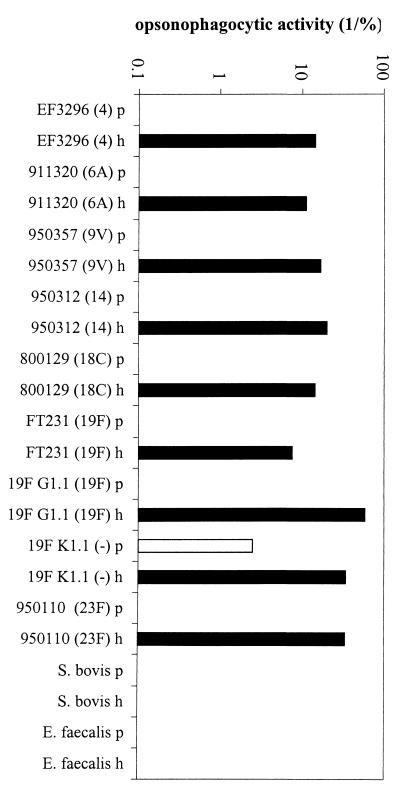
Approximately 30 polypeptides were isolated by the SB14 extraction procedure in relatively high concentrations, as shown by two-dimensional SDS-polyacrylamide gel electrophoresis (Fig. 1). Immunocytometric analysis demonstrated that serum raised against the extracted proteins recognized components at the surface of pneumococcal cells of the homologous strain FT231 (Fig. 2) and seven other pneumococcal strains (D39, EF3296, 911320, 950357, 800129, 19F, and 950110) that represent eight clinically important serotypes (types 2, 4, 6A, 9V, 14, 18C, 19F, and 23F, respectively) and display seven distinct genotypes (M. Sluijter, unpublished data). The in vitro serum opsonophagocytic activity was high (50/%) when determined with the homologous pneumococcal strain FT231 (Fig. 3A). In addition, the serum was invariably opsonophagocytically active against six genotypically distinct pneumococcal strains (EF3296, ATCC6306, ATCC6314, FT231, 19F G1.1, and ATCC6323) (M. Sluijter, unpublished data) representing serotypes 4, 6A, 14, 19, and 23F and two unencapsulated strains (19F K1.1. and Rx1). In contrast, a low serum opsonophagocytic activity was found against two strains of the genetically closely related species Streptococcus bovis and Enterococcus faecalis (Fig. 4).
FIG. 1.
Two-dimensional analysis of SB14-extracted surface-associated proteins from S. pneumoniae strain FT231. The proteins were separated by isoelectric focusing (pI, 4 to 7) and gradient SDS-polyacrylamide gel electrophoresis (10 to 100 kDa) and stained with CBB. Circles mark the proteins recognized by serum raised against the SB14-extracted proteins. The numbers 1 to 9 refer to the proteins discussed in the text.
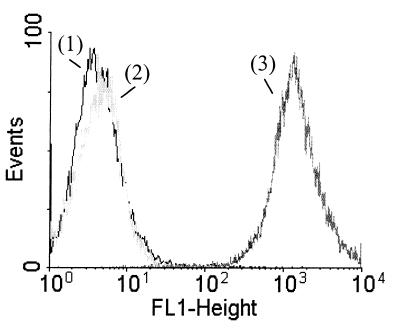
FIG. 2.
Indirect immunocytometric analysis demonstrating the presence of surface-exposed epitopes in the SB14 protein fraction of S. pneumoniae strain FT231. Y axis, number of pneumococci analyzed; x axis, degree of immunofluorescence. Numbers: 1, bacterial autofluorescence; 2, nonspecific binding of fluorescein-conjugated goat anti-rabbit IgG; 3, specific binding of serum to components at the surface of pneumococci.
FIG. 3.
Opsonophagocytic activity of hyperimmune rabbit sera raised against the surface-associated pneumococcal protein fraction (A) and sera raised against proteins 2 (B), 3 (C), and 4 (D) using the homologous strain FT231. The percentage of fluorescein isothiocyanate-positive (FITC+) human PMNs was determined at various serum concentrations. p, preimmune serum; h, hyperimmune serum.
FIG. 4.
Opsonophagocytic cross-reactivity of hyperimmune rabbit serum raised against the pneumococcal protein fraction using the heterologous pneumococcal strains EF3296, ATCC 6306, ATCC 6314, FT231, 19F G1.1, ATCC 6323, 19F K1.1, and Rx1, S. bovis strain 961008, and E. faecalis strain ATCC 29212. The serotype of the pneumococcal strains is indicated in parentheses. Y axis, opsonophagocytic activity, defined as the reciprocal of the serum concentration at which 25% of the human PMNs were fluorescent by phagocytosis of fluorescein isothiocyanate-labeled bacteria. p, preimmune serum; h, hyperimmune serum raised against the surface-associated protein fraction.
PspA, AmiA, and PpmA contribute to serum opsonophagocytic activity.
Two-dimensional Western blot analysis was performed to identify the proteins that were serologically recognized at the surface of pneumococci and responsible for the in vitro opsonophagocytic activity. The hyperimmune rabbit serum recognized nine proteins, designated 1 to 9 (Fig. 1). Monospecific rabbit sera were raised against the individual proteins. The monospecific sera raised against proteins 2, 3, and 4 were able to facilitate phagocytosis of pneumococci with an opsonophagocytic activity of 0.5/, 0.7/, and 5/%, respectively (Fig. 3B to D). Protein 2 was analyzed by N-terminal amino acid sequencing, and proteins 3 and 4 were analyzed by mass spectrometry. Eighteen of the first 21 amino acids of protein 2 were successfully identified (Table 2). The amino acid sequence was identical to that of PspA as published by McDaniel and colleagues (GenBank accession number AAC62252) (31). The molecular size of PspA deduced from the two-dimensional protein gel was approximately 65 kDa and correlates with the size range of PspA (60 to 200 kDa) (17, 50). Mass spectrometric analysis of protein 3 resulted in three peptides that were identical to AmiA (GenBank accession number P18791) (Table 2) (30). Mass spectrometric analysis of protein 4 resulted in 12 peptides (Table 2) that were part of a hypothetical translation product present in the TIGR pneumococcal genome encoding a protein of 322 amino acids (7,659 to 8,597, contig 33). The calculated size of this hypothetical protein (35.4 kDa) correlated with the size of protein 4 (approximately 35 kDa) deduced from the protein gel. This protein has been described before and designated putative proteinase maturation protein A (PpmA) due to its homology to proteinase maturation proteins (PrtM) of lactic acid bacteria (35a).
TABLE 2.
Partial amino acid sequences of surface proteins 2, 3, and 4 identified by amino acid sequence analysisa
| Protein | Name | Start-end positions | Sequence |
|---|---|---|---|
| 2 | PspA | 1–21 | EEAPVAXQXKAEKDYDAAVXK |
| 3 | AmiA | 40–49 | 1. VYTADPETLD |
| 148–159 | 2. DYLSGTSTDFST | ||
| 387–394 | 3. VAAQLPAY | ||
| 4 | PpmA | 51–59 | 1. PSAQQVLLN |
| 64–66 | 2. QVF | ||
| 74–83 | 3. LDDQEVDDTL | ||
| 120–123 | 4. LAVQ | ||
| 125–137 | 5. VAEAELTDEAYQQ | ||
| 142–146 | 6. YTPDV | ||
| 161–165 | 7. EVLEQ | ||
| 195–198 | 8. FDSA | ||
| 221–235 | 9. VLTATGTQAYSSQYY | ||
| 248–251 | 10. NLDD | ||
| 260–264 | 11. LLTQQ | ||
| 268–277 | 12. STFVQSLLGK |
It is not possible to distinguish between Ile and Leu, Gln and Lys, or Phe and oxidized Met by mass spectrometry.
PpmA is located at the surface of S. pneumoniae.
Since the three proteins PspA, AmiA, and PpmA are able to elicit opsonophagocytically active antibodies, they are presumed to be surface associated. The monospecificity of PpmA rabbit serum was confirmed by Western blot analysis. PpmA serum recognized a single protein band with the correct molecular size (35 kDa) in whole-cell lysates (Fig. 5). We performed indirect immunoelectron microscopy to identify the subcellular location of PpmA. For this purpose, anti-PpmA serum was used. Rabbit serum raised against PspA and normal rabbit serum were used as positive and negative controls, respectively. Immunoelectron microscopy demonstrated that both PpmA and PspA antibodies bound to the surface of pneumococci (Fig. 6).
FIG. 5.
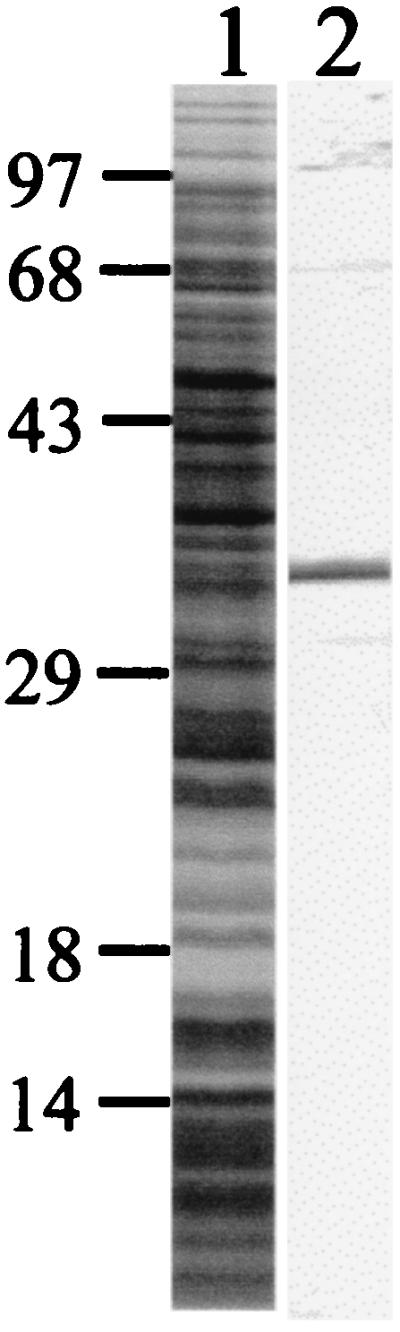
Specificity of PpmA rabbit serum. Whole-cell lysates of S. pneumoniae strain FT231 were separated by one-dimensional protein gel electrophoresis, stained with CBB (lane 1), and analyzed by Western blotting with PpmA serum (lane 2). The positions of the size markers are indicated (in kilodaltons).
FIG. 6.
Cellular localization of PpmA (B) and PspA (C) demonstrated by immunoelectron microscopy. Normal rabbit serum was used as negative control (A). Electron-dense immunogold particles are located mainly on the bacterial surface. Magnification, ×1,125,000. Bar, 100 nm.
PpmA is able to elicit species-specific opsonophagocytic antibodies that are cross-reactive against various pneumococcal strains.
The PpmA antibodies were shown to facilitate phagocytosis of eight genetically distinct pneumococcal strains representing serotypes 4, 6A, 9V, 14, 18C, 19F, and 23F. Preimmune rabbit serum was only opsonophagocytically active when the unencapsulated variant of strain 19F was used. The opsonophagocytic activity of the PpmA antibodies was very low when S. bovis and E. faecalis were used (Fig. 7).
FIG. 7.
Opsonophagocytic cross-reactivity of the monospecific hyperimmune sera raised against PpmA using the heterologous pneumococcal strains EF3296, 911320, 950312, FT231, 19F G1.1, 950110, 19F K1.1, and Rx1, S. bovis strain 961008, and E. faecalis strain ATCC 29212. The serotype of the pneumococcal strains is indicated in parentheses. Y axis, opsonophagocytic activity, defined as the reciprocal of the serum concentration at which 10% of the human PMNs were positive by phagocytosis of fluorescein isothiocyanate-labeled bacteria. p, preimmune serum; h, hyperimmune serum raised against PpmA.
ppmA is conserved among pneumococcal strains.
DNA sequence analysis of ppmA from 16 pneumococcal strains representing 15 distinct genotypes and 13 serotypes, and including the seven serotypes that cause most of the infections in young children, revealed limited genetic variation. The variation in ppmA was randomly distributed, and most of the point mutations were synonymous. Compared to the TIGR genome sequence, we found variability in six nucleotides: A33G (n = 1), T81A (n = 10), T81G (n = 2), C87T (n = 1), T114C (n = 13), G146A (n = 15), T339C (n = 3) and G818A (n = 1). Except for G146A and G818A, none of the point mutations resulted in an amino acid substitution. Mutation G146A results in a Ser49Asp substitution, and mutation G818A results in a Ser272Asp substitution.
PpmA plays a role in the pathogenesis of pneumococcal infections in vivo.
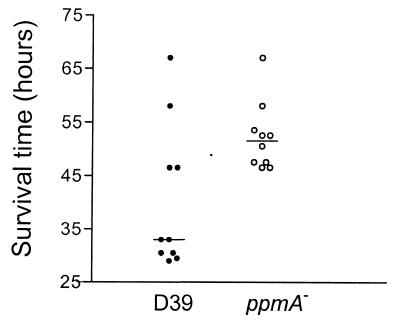
A ppmA knockout mutant of strain D39 was generated by insertion mutagenesis. Interruption of ppmA in erythromycin-resistant transformants was confirmed by PCR analysis (data not shown). In addition, absence of PpmA expression was demonstrated by Western blot analysis (data not shown). To confirm that the ppmA mutation did not affect the in vitro growth rate, both the mutant and the parent strain were grown overnight on blood agar, inoculated into THY broth, and incubated at 37°C for 8 h. During this period, there was no significant difference in growth rate between the ppmA-deficient strain and wild-type D39, as judged from the optical density of the culture (data not shown). To determine the effect of inactivation of ppmA on virulence, mice were challenged via the intranasal route with strain D39 and the ppmA mutant, respectively. Mice challenged with the ppmA mutant survived significantly longer than mice challenged with the parent strain D39 (P = 0.023); median survival times were 51.5 and 33.0 h, respectively (Fig. 8). These data demonstrate that pneumococcal ppmA deficiency results in an extended survival time for mice during infection.
FIG. 8.
Intranasal challenge of mice with ppmA-deficient S. pneumoniae. Groups of 10 mice were challenged with 106 CFU of S. pneumoniae strain D39 (solid circles) and its ppmA-deficient derivative (open circles). The survival time of each mouse is presented. Bars represent the median survival time for each group.
DISCUSSION
Various pneumococcal proteins are displayed on the cell surface. These proteins have a wide range of functions, including adherence to host tissues, binding to specific immune system components, protein processing, nutrient acquisition, and uptake of DNA from the environment. Immunization with several pneumococcal surface proteins has been shown to confer protection against pneumococcal infection in animal models. These include PspA (45), PsaA (44), autolysin (9), and neuraminidase (28). To identify novel protection-eliciting pneumococcal proteins, we isolated a pool of SB14-soluble, potentially surface-associated proteins of S. pneumoniae strain FT231. This protein fraction was able to induce antibodies that facilitated phagocytosis in vitro. Phagocytosis is a major defense mechanism against pneumococci, and the induction of opsonophagocytic antibodies is presumed to correlate with in vivo protection against S. pneumoniae infection (4). Among the proteins, at least three surface-associated proteins contributed to the in vitro opsonophagocytic activity.
The pneumococcal proteins were selected for hydrophobicity, immunogenic characteristics, and the ability to induce opsonophagocytic antibodies. One of the proteins was the previously characterized protein PspA. PspA inhibits complement activation and is proposed to exert a virulence function by recruitment of the alternative complement pathway, thereby reducing the effectiveness of complement receptor-mediated pathways of clearance (48). In addition, PspA functions as a lactoferrin-binding protein and is suggested to be involved in iron uptake and thus to contribute to pneumococcal growth under iron-limited conditions, i.e., in the human host (21). Surface exposure of PspA has been demonstrated previously (32, 46), and this characteristic was confirmed in this study by immunoelectron microscopy. The immunogenic nature of PspA observed in this study has also been demonstrated in previous studies (32). In line with the ability of PspA-specific antibodies to induce opsonophagocytic activity against strains expressing distinct capsular types, PspA has shown to possess immune protective potential (14, 32) with cross-protection (17, 47).
The second protein that induced opsonophagocytic antibodies was AmiA, which is a membrane-bound lipoprotein in S. pneumoniae (2) and part of the AmiA-AliAB oligopeptide permease that mediates the uptake of oligopeptides (2, 3). Since S. pneumoniae is auxotrophic for valine, leucine, arginine, asparagine, histidine, and glutamine, uptake of oligopeptides is important from a nutritional point of view (43). So far, no data are available on the possible protective abilities of this protein. Our data derived from phagocytosis experiments with AmiA antibodies are the first indications that AmiA may be protective against pneumococcal infections. The contribution of the AmiA-AliAB oligopeptide permease system to pneumococcal virulence is currently under investigation.
The third surface-associated protein which possessed the ability to induce opsonophagocytic antibodies was PpmA. PpmA is a recently identified pneumococcal protein with significant sequence homology to the proteinase maturation protein (PrtM) of lactic acid bacteria (Overweg et al., submitted for publication). Like PrtM from lactic acid bacteria (20), PpmA contains an N-terminal signal sequence, which serves as a label for translocation and cell membrane anchoring. In this study, the surface location of PpmA was confirmed by immunoelectron microscopy. The protein was able to induce antibodies in rabbits with opsonophagocytic activity. Although the affinity of the antibodies is unknown, the higher opsonophagocytic activity of PpmA antibodies compared to PspA and AmiA antibodies indicates the presence of relatively more PpmA molecules at the surface of the pneumococcus. Importantly, the opsonophagocytic activity of the PpmA antibodies was species specific and cross-reactive among heterologous pneumococcal strains. The observed opsonophagocytic activity of preimmune serum when the unencapsulated pneumococcal strain 19G K was used might be due to the absence of the capsule leading to the exposure of epitopes that are recognized by the preimmune serum. The cross-reactivity of the PpmA antibodies was in line with the limited sequence variation of ppmA. Like PrtM, PpmA is suggested to function as a membrane-bound isomerase (35a). PrtM is a trans-acting protein involved in the processing of precursors of serine protease PrtP into active enzymes (49) and belongs to the family of peptidyl-prolyl cis/trans isomerases. These enzymes are thought to assist in protein folding by catalyzing the cis/trans isomerization of the peptidyl-prolyl bonds in peptides and proteins (38). However, the pneumococcal protein(s) that is activated by PpmA is currently unknown. The differential expression of PpmA in phenotypic variants of S. pneumoniae indicates that PpmA may play a role in the pathogenesis of pneumococcal infections (35a). In the transparent phenotype that is selected for during nasopharyngeal colonization, PpmA is more prevalent, and therefore the protein may be involved in adherence through maturation of surface components or by the activation of proteases or other secreted proteins. In this study, PpmA was demonstrated to be involved in virulence. Inactivation of ppmA significantly reduced the virulence of strain D39 for mice as judged by survival time after intranasal challenge. However, the ppmA mutant was not completely avirulent. Like D39 mutants deficient in the production of pneumolysin (12), PspA (10), NanA (10), and LytA (11) that were also reduced in virulence, the ppmA mutant was still capable of killing mice in our animal model. The proposed role of PpmA in speeding up the folding reactions (cis/trans isomerization of the peptidyl-prolyl bonds) is consistent with the significant but limited reduction in virulence of the mutant strain. The rate of maturation of target proteins is presumed to slow in the absence of PpmA. This will subsequently result in a reduction but not elimination of target proteins that are modified in their biologically active configuration. Another explanation for the limited reduction in virulence of PpmA-deficient mutants might be the presence of other as yet unknown cis/trans isomerases that partially substitute for the PpmA activity. We conclude that PpmA contributes to pneumococcal virulence. Based on the surface location of PpmA and its ability to elicit protective species-specific antibodies, we also conclude that PpmA may be an interesting candidate for inclusion in future multicomponent protein vaccines.
ACKNOWLEDGMENTS
We thank J. Timmermans, G. Roemen, A. Jorna, D. van Nispen, N. Overbeeke, W. Jansen, A. Verheul, H. Meiring, J. ten Hove, C. L. Whitley, and D. Hockley for excellent technical assistance; J.-P. Claverys for fruitful discussions about AmiA; D. Morrison for kindly providing CSP; P. V. Adrian for critically reading the manuscript; and A. van Belkum for his interest in and advice during the project.
This work was financially supported by the Sophia Foundation for Medical Research, Rotterdam, The Netherlands (grant 183).
REFERENCES
- 1.Aaberge I S, Hvalbye B, Lovik M. Enhancement of Streptococcus pneumoniae serotype 6B infection in mice after passive immunization with human serum. Microb Pathog. 1996;21:125–137. doi: 10.1006/mpat.1996.0048. [DOI] [PubMed] [Google Scholar]
- 2.Alloing G, de Philip P, Claverys J-P. Three highly homologous membrane-bound lipoproteins participate in oligopeptide transport by the Ami system of the gram-positive Streptococcus pneumoniae. J Mol Biol. 1994;241:44–58. doi: 10.1006/jmbi.1994.1472. [DOI] [PubMed] [Google Scholar]
- 3.Alloing G, Trombe M-C, Claverys J-P. The ami locus of the gram-positive bacterium Streptococcus pneumoniae is similar to binding protein-dependent transport operons of gram-negative bacteria. Mol Microbiol. 1990;4:633–644. doi: 10.1111/j.1365-2958.1990.tb00632.x. [DOI] [PubMed] [Google Scholar]
- 4.Alonso DeVelasco E, Dekker B A T, Verheul A F, Feldman R G, Verhoef J, Snippe H. Anti-polysaccharide immunoglobin isotype levels and opsonic activity of antisera: relationship with protection against Streptococcus pneumoniae infection in mice. J Infect Dis. 1995;172:562–565. doi: 10.1093/infdis/172.2.562. [DOI] [PubMed] [Google Scholar]
- 5.Alonso DeVelasco E, Verheul A F, Verhoef J, Snippe H. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev. 1995;59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul S F, Miller G W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;251:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Avery O T, MacLeod C M, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 9.Berry A M, Lock R A, Hansman D, Paton J C. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989;57:2324–2330. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry A M, Paton J C. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence factors. Infect Immun. 2000;68:133–140. doi: 10.1128/iai.68.1.133-140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry A M, Paton J C, Hansman D. Effect of insertional inactivation of the genes encoding pneumolysin and autolysin on the virulence of Streptococcus pneumoniae type 3. Microb Pathog. 1992;12:87–93. doi: 10.1016/0882-4010(92)90111-z. [DOI] [PubMed] [Google Scholar]
- 12.Berry A M, Yother J, Briles D E, Hansman D, Paton J C. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect Immun. 1989;57:2037–2042. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 14.Briles D E, King J D, Gray M A, McDaniel L S, Swiatlo E, Benton K A. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine. 1996;14:858–867. doi: 10.1016/0264-410x(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 15.Butler J C. Epidemiology of pneumococcal serotypes and conjugate vaccine formulations. Microb Drug Resist. 1997;3:125–129. doi: 10.1089/mdr.1997.3.125. [DOI] [PubMed] [Google Scholar]
- 16.Butler J C, Dowell S F, Breiman R F. Epidemiology of emerging pneumococcal drug resistance: implications for treatment and prevention. Vaccine. 1998;16:1693–1697. doi: 10.1016/s0264-410x(98)00132-7. [DOI] [PubMed] [Google Scholar]
- 17.Crain M J, Waltman W D, Turner J S, Yother J, Talkington D F, McDaniel L S, Gray B M, Briles D E. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect Immun. 1990;58:3293–3299. doi: 10.1128/iai.58.10.3293-3299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagan R, Melamed R, Muallem M, Piglansky L, Yagupsky P. Nasopharyngeal colonization in southern Israel with antibiotic-resistant pneumococci during the first 2 years of life: relation to serotypes likely to be included in pneumococcal conjugate vaccines. J Infect Dis. 1996;174:1352–1355. doi: 10.1093/infdis/174.6.1352. [DOI] [PubMed] [Google Scholar]
- 19.Douglas R M, Paton J C, Duncan S J, Hansman D J. Antibody response to pneumococcal vaccination in children younger than five years of age. J Infect Dis. 1983;148:131–137. doi: 10.1093/infdis/148.1.131. [DOI] [PubMed] [Google Scholar]
- 20.Haandrikman A J, Kok J, Venema G. Lactococcal proteinase maturation protein PrtM is a lipoprotein. J Bacteriol. 1991;173:4517–4525. doi: 10.1128/jb.173.14.4517-4525.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt S, Bethe G, Remane P H, Chhatwal G S. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect Immun. 1999;67:1683–1687. doi: 10.1128/iai.67.4.1683-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermans P W, Sluijter M, Dejsirilert S, Lemmens N, Elzenaar K, van Veen A, Goessens W H, de Groot R. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb Drug Resist. 1997;3:243–251. doi: 10.1089/mdr.1997.3.243. [DOI] [PubMed] [Google Scholar]
- 23.Hermans P W, Sluijter M, Elzenaar K, van Veen A, Schonkeren J J, Nooren F M, van Leeuwen W J, de Neeling A J, van Klingeren B, Verbrugh H A, de Groot R. Penicillin-resistant Streptococcus pneumoniae in the Netherlands: results of a 1-year molecular epidemiologic survey. J Infect Dis. 1997;175:1413–1422. doi: 10.1086/516474. [DOI] [PubMed] [Google Scholar]
- 24.Jansen W T M, Gootjes J, Zelle M, Madore D V, Verhoef J, Snippe H, Verheul A F M. Use of highly encapsulated Streptococcus pneumoniae strains in a flow-cytometric assay for assessment of the phagocytic capacity of serotype-specific antibodies. Clin Diagn Lab Immunol. 1998;5:703–710. doi: 10.1128/cdli.5.5.703-710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadioglu A, Gingles N A, Grattan K, Kerr A, Mitchell T J, Andrew P W. Host cellular immune response to pneumococcal lung infection in mice. Infect Immun. 2000;68:492–501. doi: 10.1128/iai.68.2.492-501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J O, Romero-Steiner S, Sorensen U B S, Blom J, Carvalho M, Barnard S, Carlone G, Weiser J N. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect Immun. 1999;67:2327–2333. doi: 10.1128/iai.67.5.2327-2333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J O, Weiser J N. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis. 1998;177:368–377. doi: 10.1086/514205. [DOI] [PubMed] [Google Scholar]
- 28.Lock R A, Paton J C, Hansman D. Comparative efficacy of pneumococcal neuraminidase and pneumolysin as immunogens protective against Streptococcus pneumoniae. Microb Pathog. 1988;5:461–467. doi: 10.1016/0882-4010(88)90007-1. [DOI] [PubMed] [Google Scholar]
- 29.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 30.Martin B, Alloing G, Boucraut C, Claverys J P. The difficulty of cloning Streptococcus pneumoniae mal and ami loci in Escherichia coli: toxicity of malX and amiA gene products. Gene. 1989;80:227–238. doi: 10.1016/0378-1119(89)90287-4. [DOI] [PubMed] [Google Scholar]
- 31.McDaniel L S, McDaniel D O, Hollingshead S K, Briles D E. Comparison of the PspA sequence from Streptococcus pneumoniae EF5668 to the previously identified PspA sequence from strain Rx1 and the ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect Immun. 1998;66:4748–4754. doi: 10.1128/iai.66.10.4748-4754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel L S, Scott G, Kearney J F, Briles D E. Monoclonal antibodies against protease-sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J Exp Med. 1984;160:386–397. doi: 10.1084/jem.160.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDaniel L S, Sheffield J S, Delucchi P, Briles D E. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–228. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell T J, Alexander J E, Morgan P J, Andrew P W. Molecular analysis of virulence factors of Streptococcus pneumoniae. Soc Appl Bacteriol Symp Ser. 1997;26:62S–71S. [PubMed] [Google Scholar]
- 35.Overweg K, Hermans P W M, Trzcinski K, Sluijter M, de Groot R, Hryniewicz W. Multidrug-resistant Streptococcus pneumoniae in Poland: identification of emerging clones. J Clin Microbiol. 1999;37:1739–1745. doi: 10.1128/jcm.37.6.1739-1745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Overweg, K., C. D. Pericone, G. G. C. Verhoef, J. N. Weiser, H. D. Meiring, A. P. J. M. de Jong, R. de Groot, and P. W. M. Hermans. Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 36.Paton J C, Lock R A, Hansman D J. Effect of immunization with pneumolysin on survival time of mice challenged with Streptococcus pneumoniae. Infect Immun. 1983;40:548–552. doi: 10.1128/iai.40.2.548-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabilloud T, Valette C, Lawrence J J. Sample application by in-gel rehydration improves the resolution of two-dimensional electrophoresis with immobilized pH gradients in the first dimension. Electrophoresis. 1994;15:1552–1558. doi: 10.1002/elps.11501501223. [DOI] [PubMed] [Google Scholar]
- 38.Rudd K E, Sofia H J, Koonin E V, Plunkett II G, Lazar S, Rouveire P E. A new family of peptidyl-prolyl isomerases. Trends Biochem Sci. 1995;20:12–14. doi: 10.1016/s0968-0004(00)88940-9. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schouls L M, Ijsselmuiden O E, Weel J, van Embden J D. Overproduction and purification of Treponema pallidum recombinant-DNA-derived proteins TmpA and TmpB and their potential use in serodiagnosis of syphilis. Infect Immun. 1989;57:2612–2623. doi: 10.1128/iai.57.9.2612-2623.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro E D, Berg A T, Austrian R, Schroeder D, Parcells V, Margolis A, Adair R K, Clemens J D. The protective efficacy of polyvalent pneumococcal polysaccharide vaccine. N Engl J Med. 1991;325:1453–1460. doi: 10.1056/NEJM199111213252101. [DOI] [PubMed] [Google Scholar]
- 42.Shoemaker N B, Guild W R. Destruction of low efficiency markers is a slow process occurring at a heteroduplex stage of transformation. Mol Gen Genet. 1974;128:283–290. doi: 10.1007/BF00268516. [DOI] [PubMed] [Google Scholar]
- 43.Sicard A M. A new synthetic medium for Diplococcus pneumoniae and its use for the study of reciprocal transformation at the amiA locus. Genetics. 1964;50:31–44. doi: 10.1093/genetics/50.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talkington D F, Brown B G, Tharpe J A, Koenig A, Russell H. Protection of mice against fatal pneumococcal challenge by immunization with pneumococcal surface adhesin A (PsaA) Microb Pathog. 1996;21:17–22. doi: 10.1006/mpat.1996.0038. [DOI] [PubMed] [Google Scholar]
- 45.Talkington D F, Crimmins D L, Voellinger D C, Yother J, Briles D E. A 43-kilodalton pneumococcal surface protein, PspA: isolation, protective abilities, and structural analysis of the amino-terminal sequence. Infect Immun. 1991;59:1285–1289. doi: 10.1128/iai.59.4.1285-1289.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talkington D F, Voellinger D C, McDaniel L S, Briles D E. Analysis of pneumococcal PspA microheterogeneity in SDS polyacrylamide gels and the association of PspA with the cell membrane. Microb Pathog. 1992;13:343–355. doi: 10.1016/0882-4010(92)90078-3. [DOI] [PubMed] [Google Scholar]
- 47.Tart R C, McDaniel L S, Ralph B A, Briles D E. Truncated Streptococcus pneumoniae PspA molecules elicit cross-protective immunity against pneumococcal challenge in mice. J Infect Dis. 1996;173:380–386. doi: 10.1093/infdis/173.2.380. [DOI] [PubMed] [Google Scholar]
- 48.Tu A T, Fulgham R L, McCrory M A, Briles D E, Szalai A J. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun. 1999;67:4720–4724. doi: 10.1128/iai.67.9.4720-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vos P, van Asseldonk M, van Jeveren F, Siezen R, Simons G, de Vos W M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine protease located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waltman W D, McDaniel L S, Gray B M, Briles D E. Variation in the molecular weight of PspA (pneumococcal surface protein A) among Streptococcus pneumoniae. Microb Pathog. 1990;8:61–69. doi: 10.1016/0882-4010(90)90008-e. [DOI] [PubMed] [Google Scholar]
- 51.Weiser J N, Austrian R, Sreenivasan P K, Masure H R. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun. 1994;62:2582–2589. doi: 10.1128/iai.62.6.2582-2589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wessel D, Flugge U I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 53.Wu M H N, Guo Y, Russel M W, Briles D E. Intranasal immunization of mice with pspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with Streptococcus pneumoniae. J Infect Dis. 1997;175:839–846. doi: 10.1086/513980. [DOI] [PubMed] [Google Scholar]
- 54.Yother J, McDaniel L S, Briles D E. Transformation of encapsulated Streptococcus pneumoniae. J Bacteriol. 1986;168:1463–1465. doi: 10.1128/jb.168.3.1463-1465.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]