Abstract
The discovery of biomolecular condensates transformed our understanding of intracellular compartmentalization of molecules. To integrate interdisciplinary scientific knowledge about the function and composition of biomolecular condensates, we developed the crowdsourcing condensate database and encyclopedia (cd-code.org). CD-CODE is a community-editable platform, which includes a database of biomolecular condensates based on the literature, an encyclopedia of relevant scientific terms and a crowdsourcing web application. Our platform will accelerate the discovery and validation of biomolecular condensates, and facilitate efforts to understand their role in disease and as therapeutic targets.
Subject terms: Protein databases, Organelles
Crowdsourcing condensate database and encyclopedia is a community-editable platform for verified biomolecular condensates and their protein constituents. It also provides an encyclopedia for the scientific terms used in condensate biology and a crowdsourcing web application.
Main
Biomolecular condensates are membraneless organelles that selectively concentrate biomolecules (for example, proteins and nucleic acids) in the cell, with spatial and temporal precision1. In recent years, their role was implicated in several biochemical processes, in physiology and disease2. Consequently, biomolecular condensates are now leveraged as a new class of therapeutic targets3,4.
Basic science and drug discovery advances build upon published reports and the rate of new discoveries depends on timely accessibility to relevant data. However, as with every novel paradigm, new terms and concepts emerge and evolve as the field develops. Accordingly, currently available databases which catalog proteins involved in condensate formation use various definitions and criteria to define condensates and their constituent proteins and RNAs5–8. These are excellent databases curating proteins that phase separate. Specifically, LLPSDB7 and PhaSePro6 collect proteins that are thought to drive liquid–liquid phase separation, with the former curating exclusively in vitro data.
However, these databases do not answer the following questions regarding biomolecular condensates: What are the biomolecular condensates discovered and verified to date? What are their known protein components? Which condensates is a given protein known to belong to? What are the experimental evidences supporting the existence of a particular condensate? Our goal is to generate answers for these and other important questions, and to create a community-editable database to facilitate the dynamic data updates. Therefore, we designed a condensate-centric database, which is based on the scientific literature, and provides experimental evidences, scores and references for each condensate–protein relationship (Extended Data Figs. 1 and 2). This database is updated dynamically by contributors to keep up with the growing knowledge in the field. We call our platform CD-CODE, which consists of three main parts: (1) a database of biomolecular condensates and their protein constituents; (2) an encyclopedia for the scientific terms used in condensate biology; and (3) a crowdsourcing web application (Extended Data Fig. 3).
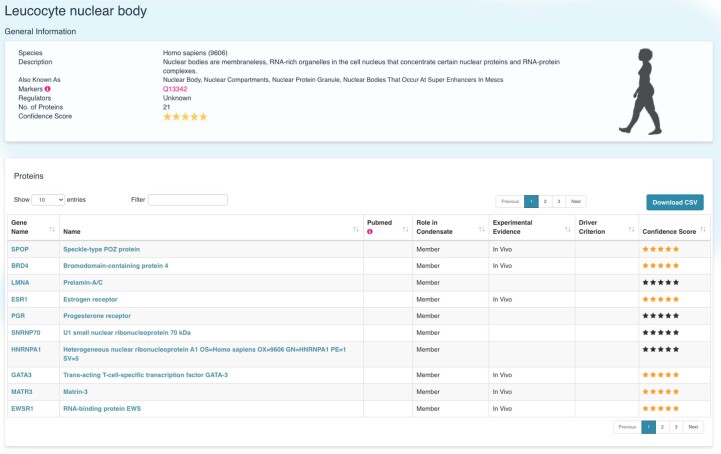
Extended Data Fig. 1. Detail page of the biomolecular condensate, Leucocyte nuclear body in humans.
The top section displays ‘General Information’ of the condensate whereas the section below shows ‘condensate-specific protein’ data in a tabular format.
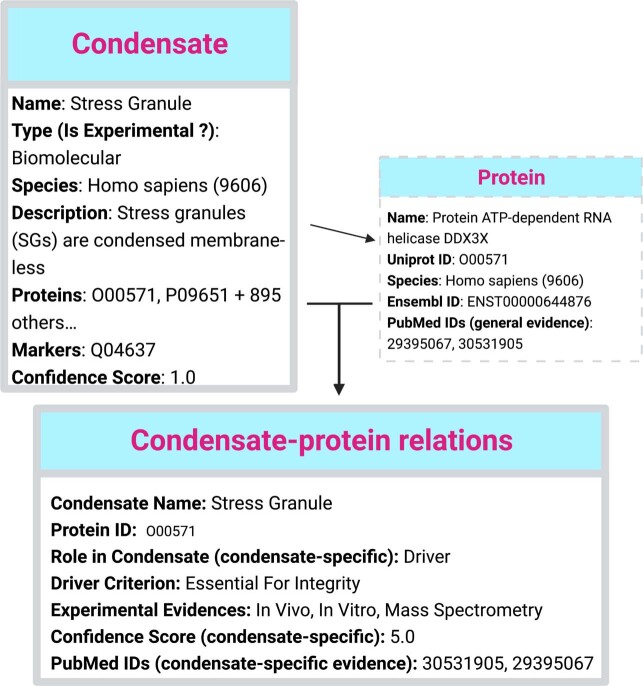
Extended Data Fig. 2. CD-CODE is a condensate centric database.
Example data structure of a condensate and one of its member proteins. We defined two distinct entity collections – one for condensates and one for proteins, the latter comprising of general details regarding the members of the former. We also store mappings collection of condensate-protein relations, comprising of data points that relate to specific condensate-protein pairs.
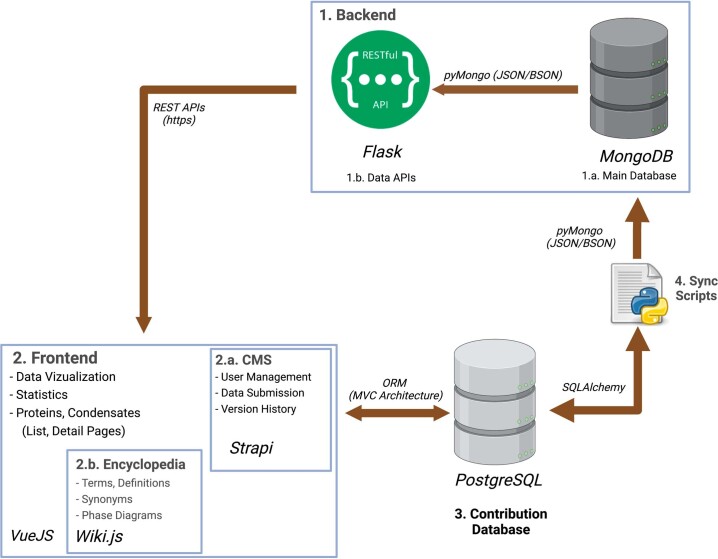
Extended Data Fig. 3. Schematics of the technical architecture of CD-CODE web application.
The web application is internally divided into four distinct components. 1. The main database contains the condensates, proteins, and other related datasets. The API layer exposes consumable data for the frontend to visualize data. This can also be used for programmatic access to the data by statisticians and bioinformaticians. It also facilitates filter, search, sort, and some basic aggregate functionalities. 2. The frontend allows visualization of data, for example detail pages for condensates and proteins (Extended Data Figs. 1 and 7). A sub-component embedded within the frontend is the Content Management System (CMS) which facilitates user management, and workflow of each update item submitted by users. The CMS also helps maintain a history of contribution for each data record. 3. The contribution database stores the history of data from the crowdsourcing effort and helps versioning the database. All data edits submitted by contributors and review actions performed by maintainers are stored here to aid transparency. 4. The Sync script is a scheduled process that runs at regular time intervals (daily/weekly) to copy the update items safely to the main database. This python program interacts with both PostgreSQL from CMS and MongoDB from the main database in a producer-consumer paradigm.
CD-CODE is a ‘living database’ designed for dynamic and rapid addition and review of information about condensates and proteins by users (Fig. 1) and is open to any expert researcher who wishes to contribute. Our user management system supports three types of users: viewers, contributors and maintainers. Viewers can read and download the curated information. Contributors can suggest edits and propose new condensate and protein entries (Extended Data Figs. 4 and 5). Maintainers are part of the development team, who curate the changes and accept or reject suggestions by contributors, who are then notified about the status of their suggestions and can engage in further discussion. To keep up with the rapidly evolving definitions, nomenclature and growing scientific evidence, the crowdsourcing platform allows the community to aggregate scientific findings in condensate biology.
Fig. 1. CD-CODE information flow.
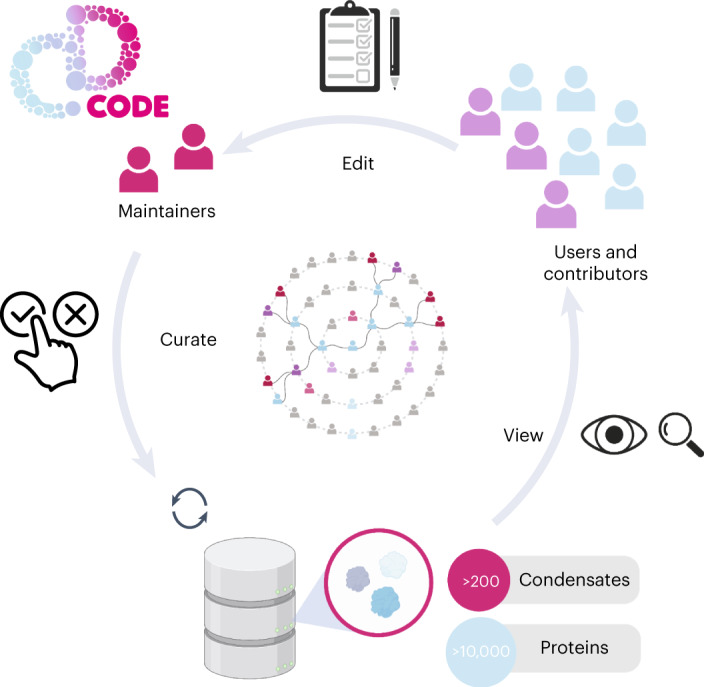
Users can view and search the data, or become contributors after registration and edit the content of the database via the community-editable web application. The maintainers assure quality control and only approved edits will be part of the dynamically updated database. Figure created with BioRender.com.
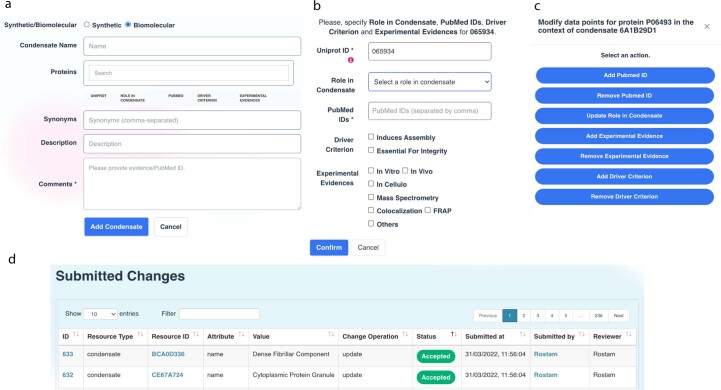
Extended Data Fig. 4. Data curation from the user interface.
a) Add new condensate form. b) Add new protein form. c) Edit data actions. d) History of entries is recorded.
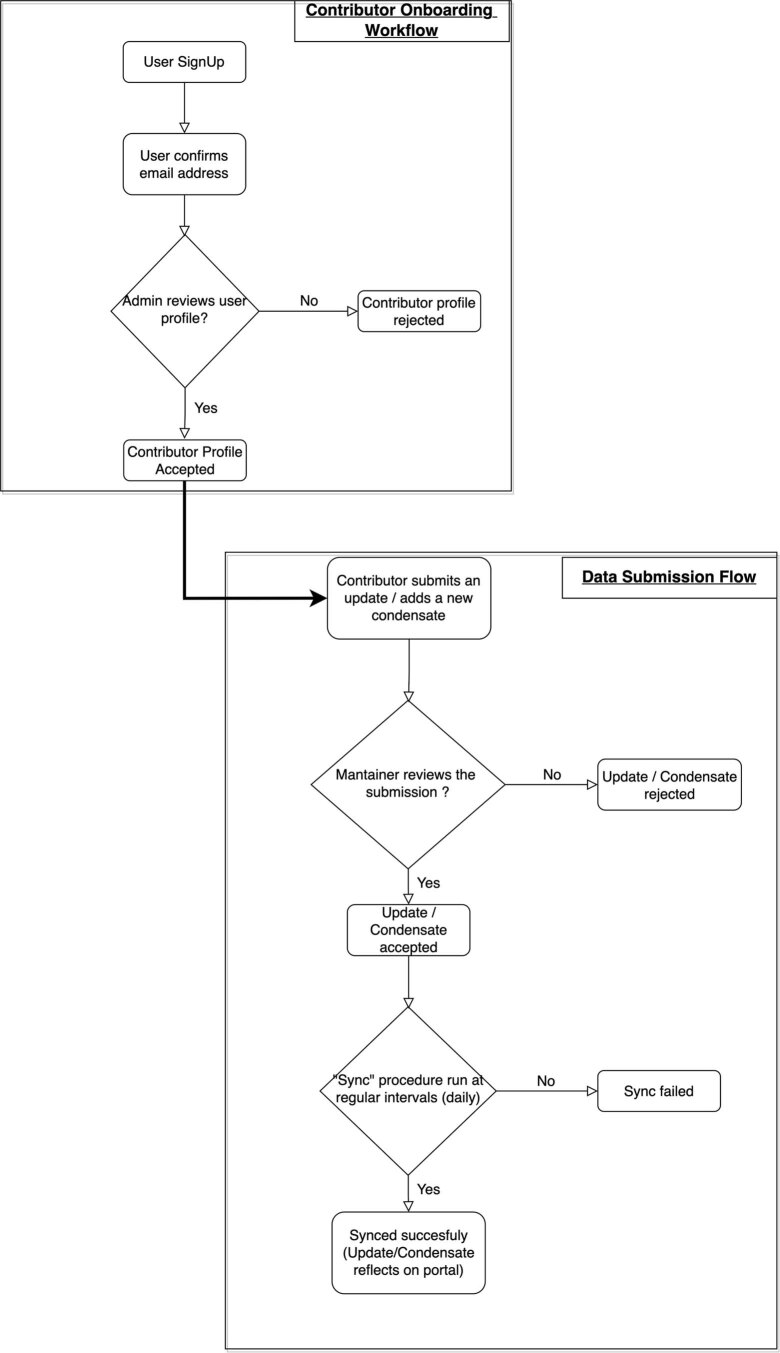
Extended Data Fig. 5. Flowchart of contributions.
After submitting the registration form, the user receives an email to confirm the given address by clicking a link. After the user’s email address is confirmed, an administrator checks the pending contributor signup requests and approves them. A signed-in contributor can see editable attributes besides selected data points on detail pages, in addition to a new webpage form for submission of novel condensates. The new submissions are then moderated by maintainers first and later by the Sync script.
At the time of this report, CD-CODE (cd-code.org) contains 9,861 proteins linked to 244 unique biomolecular condensates (and 375 in vitro synthetic condensates) across 49 different organisms. Notably, these numbers are continuously changing as contributors add and review more data. CD-CODE, as a semi-manually curated and annotated resource, aggregates information from the primary literature (to date, PubMed references published until 1 June 2022 were manually curated) and other databases5–8 (Extended Data Fig. 6 and Extended Data Tables 1 and 2). To promote easy integration with other resources, protein entries are cross-referenced with UniProt9, Ensembl10 and the Human Protein Atlas (proteinatlas.org)11. Common sequence properties of condensate proteins are also displayed graphically, such as disorder score12 and amino acid composition (Extended Data Fig. 7), facilitating the identification of regions that may drive condensate partitioning.
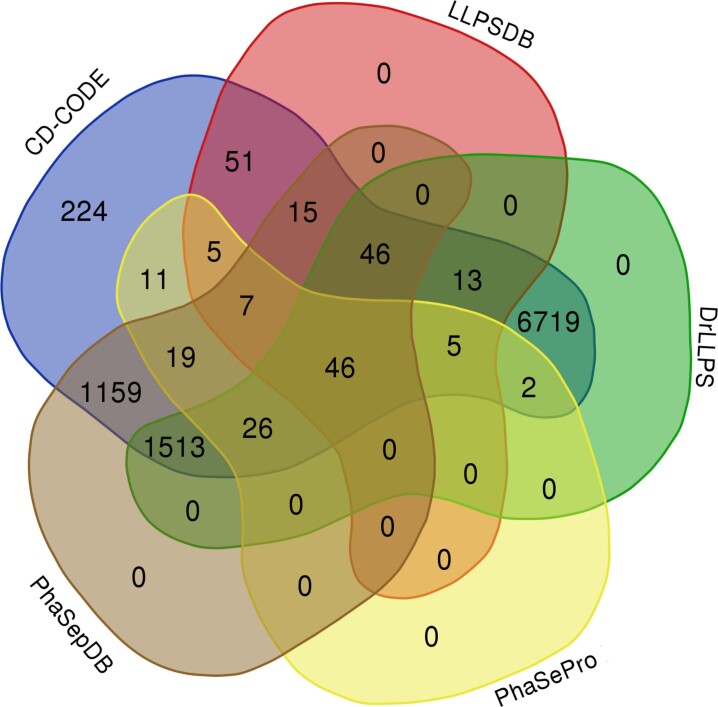
Extended Data Fig. 6. Venn diagram to compare protein entries in condensate/liquid-liquid phase separation (LLPS) databases.
The Venn diagram displays the overlap of the protein entries in CD-CODE and four other LLPS/condensate databases (LLPSD1, DrLLPS2, PhaSepDB3, PhaSePro4). CD-CODE incorporates data from all four databases and additionally, there are 224 new condensate-forming proteins in v1.01 that were added to CD-CODE from curating the literature.
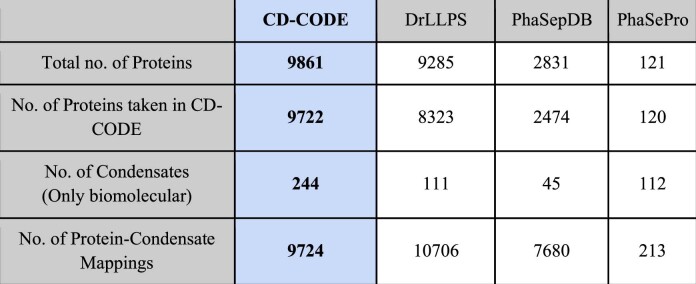
Extended Data Table 1.
Biomolecular condensates across databases
Comparison of total number of data points presented by CD-CODE in comparison to other databases in the context of biomolecular condensates or membrane-less organelles (MLOs). We count the condensates from different species as separate counts of biomolecular condensates. The other three databases here also report phase-separating proteins without any condensate or with condensates named ‘Droplets’ and ‘Others’. Such entries are removed from CD-CODE by manual curation.
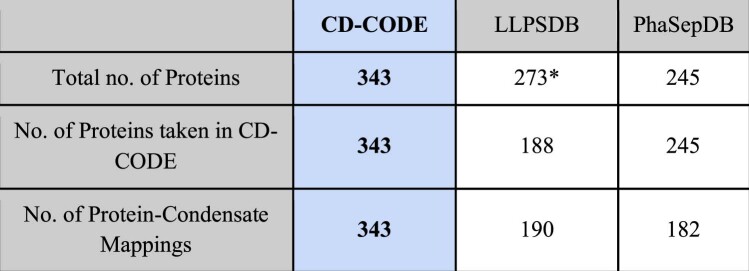
Extended Data Table 2.
Synthetic condensates across databases
Comparison of total number of proteins presented in CD-CODE to other databases in the context of synthetic condensates or in vitro experiments. Since LLPSDB only reports in vitro experiments, the condensate-related counts are not reported for that.
*Out of the 273 proteins reported by LLPSDB, 198 are natural proteins, and 75 are designed proteins (not all experiments result in phase separation).
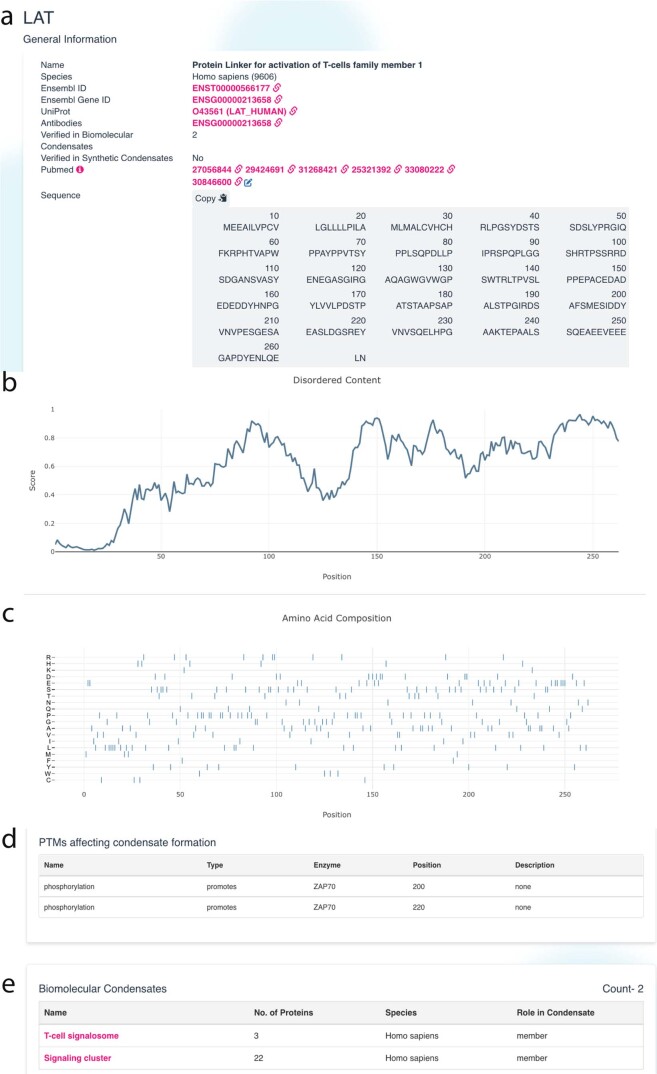
Extended Data Fig. 7. Example detail page to illustrate the general information and data points stored in CD-CODE.
a) General information about a protein is shown for the LAT Linker for activation of T-cells family member 1 protein (Uniprot ID: LAT_HUMAN). Links are provided to other popular protein-related databases (Uniprot, Ensembl, Human Protein Atlas). b) Disorder score computed by IUPred5; c) Barcode plot depicting amino acid composition bias; d) Table of PTMs affecting condensate behavior; e) List of biomolecular condensates of which the given protein is a member.
We standardized the names of condensates by creating an ontology from the literature and grouped condensates by functional categories (Supplementary Table 1) to reveal the evolutionary history of condensates. Most known condensates are found in mammals and many are clade-specific (Fig. 2a). Since our current knowledge is sparse and likely biased, the evolutionary origin of condensates remains an open future research direction that CD-CODE can facilitate.
Fig. 2. Selected features available at CD-CODE.
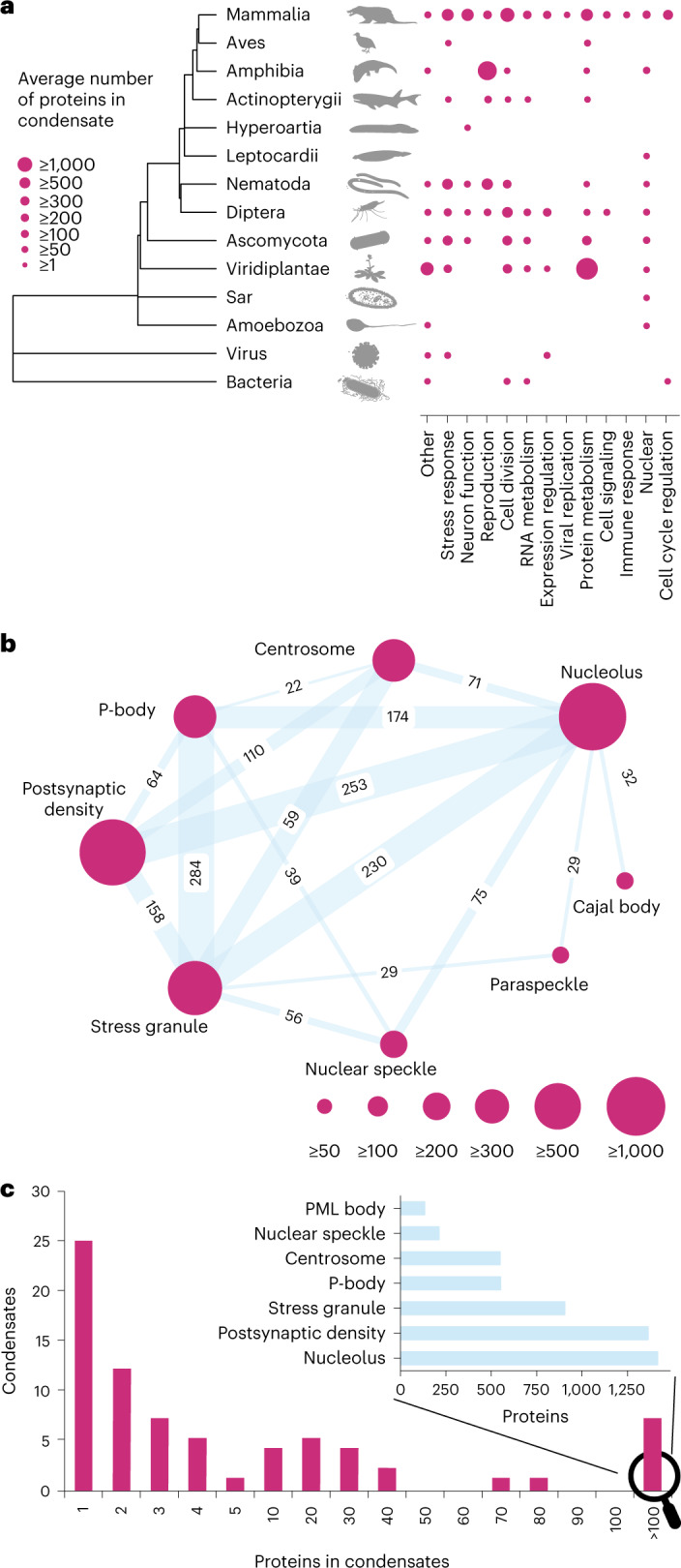
a, Biomolecular condensates across the tree of life. CD-CODE contains information about 244 condensates across 49 species. Here, only major clades are shown for clarity and condensates were grouped into functional categories. b, Many proteins localize to multiple condensates. There is a large overlap between the proteomes of different biomolecular condensates in humans. The largest condensates in humans are represented as circles and the shared proteins between every two condensates are shown (only condensates with >20 connections are shown). c, The distribution of condensate proteome sizes in humans. Most biomolecular condensates have a few known protein members. The largest condensates contain >1,000 different proteins (inset).
While many proteins undergo liquid–liquid phase separation in vitro, it is unclear which proteins form condensates in cells and which condensates they partition into. To facilitate our understanding of condensate-specificity of proteins, we collected all known condensates a given protein was found in, and we curated the experimental evidence for association of each protein with a given condensate (confidence score, corresponding to zero to five stars: 1 star: literature evidence, PubMed identifier (ID); 2 stars, high-throughput; 3 stars, in vitro; 4 stars, in cellulo; and 5 stars, in vivo evidence). Condensates and proteins that have zero or one star rating have not been manually curated yet.
As expected, for dynamic cellular compartments, many proteins partition into different condensates and the overlap between condensate proteomes is substantial (Fig. 2b). While proteins may localize to multiple condensates (members), a few are obligate and essential components (drivers). We annotated 205 driver proteins in specific condensates, providing the corresponding experimental evidences. Our database revealed that several proteins that are drivers in one condensate are nonessential members of another (for example, G3BP1, a driver of stress granules, is also present in processing bodies (P-bodies) and neuronal ribonucleoprotein particle granules). CD-CODE will aid our understanding of the determinants of condensate-specific driver behavior, and whether a driver protein can be used as a ‘marker’ of a condensate in experiments.
Marker proteins are used to define the identity of the condensates and inform designing of condensate-targeting drug screening pipelines3. They are thought to be uniquely associated with a given condensate, and are commonly used to visualize condensates using microscopy, for example, in colocalization experiments to prove the localization of proteins into condensates. Our database revealed that several known marker proteins are not specific to a condensate. For example, whilst DCP1A is used as a marker for P-bodies, it also localizes to stress granules and nucleoli. Knowing specific protein components will facilitate the experimental design for accurate, specific identification of condensates.
CD-CODE enables us to answer the questions posed at the beginning: (1) there are currently 136 unique biomolecular condensates documented in the literature; (2) as an example, P-granules, which are the germ granules of Caenorhabditis elegans, have 190 documented protein components: one of them, pgl-3 (PGL3_CAEEL), is a driver for P-granule formation, and its presence within P-granules is supported by in vivo experimental evidence (5 star). Pgl-3 is exclusively reported to be associated with P-granules; thus, it is a P-granule-specific marker protein.
Databases that curate proteins undergoing liquid–liquid phase separation have facilitated the development of machine learning algorithms to predict phase separation13–15 and the discovery of what protein properties drive phase separation15. The next open question is which biomolecular condensate does a specific protein belong to. Our database contains a curated list of condensate proteomes (Fig. 2c), which can facilitate investigations of protein recruitment into specific condensates. Our resource can provide high-quality benchmarking data for machine learning algorithms aimed at predicting the protein components of condensates.
Furthermore, our comprehensive curation of condensate types and their respective composition in multiple species, and the level of experimental support, provides a valuable resource for drug hunters, which can inform the design of assays and screening pipelines. For example, in high-content imaging phenotypic screens, it is desired that the protein or protein combination chosen to be monitored is/are selective for the target condensate3. Additionally, through regular updating of the database by the community and via curation of new publications, CD-CODE supports and accelerates nomination of new condensate-associated drug targets.
The field of biomolecular condensates is highly transdisciplinary and ever-developing, where definitions and terms keep changing, creating a need for constant updates that require consensus within the community. The encyclopedia, as a standalone wiki, serves as a platform to aggregate knowledge about condensate research. In the future, we are planning weekly updates to integrate new data from the users, and yearly updates with new features and data points that become relevant to store, as the research field develops.
The main feature of CD-CODE is that it contains experimentally validated entries. However, caution should be exercised by users when interpreting lack of data on a particular protein, condensate or species, as this may simply reflect the biased interest of the community towards particular model systems and biological pathways. Any missing information could mean that (1) the protein or condensate has not been studied yet; (2) there is a research paper but the information has not been added to the database yet; (3) the condensate truly does not exist; or (4) the protein truly does not belong to a given condensate. As such, CD-CODE aims to highlight the unknowns in the field to guide future research questions to fill the gaps. These gaps in experimental evidence can be bridged by computational predictions16,17, which are beyond the scope of CD-CODE. Evolution of the CD-CODE database through ongoing curation of new experimental evidence will lead to a progressive increase in high-scoring condensate entries.
In summary, we present CD-CODE, a semi-manually curated condensate database, and a community-editable web application. The crowdsourcing platform allows the community to further scrutinize definitions and evidence as the field evolves. This will ensure that the ever-growing knowledge on condensate research is integrated into the database and into the encyclopedia in a timely manner.
Methods
Technical architecture of the web application
The web application is internally divided into four distinct components (Extended Data Fig. 3).
The main database contains the condensates, proteins and other related datasets. The database service used for this component is MongoDB, where the interlinking of resources is done at the logical layer. The Application Programming Interface (API) layer exposes consumable data for the frontend to visualize data. This can also be used for programmatic access to the data by statisticians and bioinformaticians. It also facilitates filter, search, sort and some basic aggregate functionalities. We used Flask, a simple lightweight Python framework, for this.
The frontend allows visualization of data—list and detail pages for condensates and proteins. It provides user-interactable controls for the addition/modification of data. The frontend is built using a Javascript framework called Vue.js. A subcomponent embedded within the frontend is the Content Management System (CMS), which facilitates user management and workflow of each update item submitted by users. The CMS also helps to maintain a history of contribution for each data record.
The contribution database stores the history of data from the crowdsourcing effort and helps versioning the database. All data edits submitted by contributors and review actions performed by maintainers are stored here to aid transparency.
The Sync script is a scheduled process that runs at regular time intervals (daily/weekly) to copy the update items safely to the main database. This python program interacts with both PostgreSQL from CMS and MongoDB from the main database in a producer–consumer paradigm.
Data aggregation
The four most popular protein phase-separation databases are PhaSePro6, PhaSepDB8, LLPSDB7 and data resource of liquid–liquid phase separation (DrLLPS)5, which provide an excellent resource for phase-separating proteins along with their possible localization (membraneless organelles) and also synthetic condensates (in vitro experiments). However, all these databases are protein-centric and not condensate-centric, that is, a protein may or may not be part of a biomolecular condensate. To fill this gap and augment our current knowledge, we have built a condensate-centric database. At CD-CODE, we aggregated data from the four databases to create a seed dataset of condensates and their protein members (Extended Data Fig. 6). This was followed by a manual curation of the data and annotation of up-to-date evidences (PubMed references, experimental evidences, confidence scores explained below).
Literature curation
All the 674 condensate literature-related PubMed references which were published after the release of the last four databases were checked manually, and new condensates and proteins involved in condensates were identified and added to CD-CODE along with the relevant PubMed IDs. In total, 26 new biomolecular condensates and 224 new proteins were added to CD-CODE which were not part of the previous databases (Extended Data Fig. 6 and Extended Data Tables 1 and 2).
Data curation
Condensate names were standardized and condensates were merged based on synonyms that were manually assembled from the literature (Supplementary Table 1). In total, 9,916 protein–condensate relationships are supported by a PubMed ID as evidence.
Annotation of driver proteins
We defined the ‘Role in condensate’ of a protein as the role it plays in a condensate formation and integrity. Based on their role in the formation of the condensate, proteins can be categorized into one of two classes—‘driver’ and ‘member’.
Drivers are defined based on the following criteria:
They induce the formation of a condensate.
They are essential for the integrity of a condensate.
Driver proteins are also called ‘scaffolds’ since they bind many other proteins. It is important to note that these annotations are valid in the context of a specific condensate. A protein acting as a driver for one condensate can be a member in another condensate.
Confidence scores
We designed the confidence score criteria and started manually adding the experimental evidences. 1 star, PubMed reference-annotated; 2 stars, high-throughput experiment (for example, mass spectrometry); 3 stars, in vitro evidence; 4 stars, in cellulo evidence; 5 stars, in vivo evidence. We also collected the experimental methodology, for example, fluorescence recovery after photobleaching or microscopy colocalization.
Crowdsourcing functionalities
Registered users can contribute to data curation, including any kind of attribute data update for any protein/condensate we already have at CD-CODE, such as marker addition, name change, description update, add/remove PubMed evidence and so on (Extended Data Fig. 4). Additionally, users can add new proteins to existing condensates and create new condensates by filling in a form.
Encyclopedia
The encyclopedia (wiki.cd-code.org), another crowdsourcing element of CD-CODE, contains descriptive data related to definitions, synonyms and terminologies in the world of biomolecular condensates and liquid–liquid phase separation in biology. Contributors have the necessary credentials to create and edit content at the encyclopedia. It is powered by Wiki.js and provides easy content management functionalities to users who are not equipped with programming or HTML skills.
Data analysis
Condensate counts for species were taken from CD-CODE v.1.01 and summarized by functional group annotation (Supplementary Table 1). The tree was downloaded from timetree.org (ref. 18) and scaled with the chronos function in the ape (5.6–2) R (v.4.2.0) package for better visualization. The tree was plotted using ggtree (v.3.4.4) and ggplot (v.3.4.0). Figure 2b,c was created using custom Python scripts (pandas: 1.2.4; matplotlib: 3.3.4; networkx: 2.5).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41592-023-01831-0.
Supplementary information
Documentation.
Condensate nomenclature, synonyms and functional groups.
Source data
Source data for Fig. 2.
Source data for Extended Data Fig. 6.
Acknowledgements
This project was funded by the Max Planck Gesellschaft (A.T.-P., A.A.H. and A.H.) and Dewpoint Therapeutics (A.T.-P.). C.F.W.C. was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC-2068 grant no. 390729961—Cluster of Excellence Physics of Life of TU Dresden. A.H. was supported by the ELBE postdoctoral fellowship. We thank F. Friedrich for logo design and MPI-CBG Computing Services, especially M. Boes, for setting up the server.
Extended data
Author contributions
A.T.-P., A.A.H. and D.M.M. conceived the project with input from C.F.W.C., C.L. and S.G. S.G., H.M. and L.H. conceived the code concept and design. Backend coding development and implementation was done by S.G. Frontend coding development and implementation was done by H.M. and R.G. N.R. and C.F.W.C. created the content for the encyclopedia. N.R. curated the data manually and served as the maintainer of the data. S.G. collected data and designed the data structure. D.M.M. and I.A.K. provided resources. S.G., A.H. and C.L. analyzed the data. A.H., C.L., A.T.-P., S.G., H.M. and N.R. visualized data and prepared the figures. A.T.-P., L.H. and S.G. supervised the project. A.T.-P. wrote the main manuscript with input from all the coauthors. S.G. prepared supplementary information and documentation of the code. A.T.-P. acquired the funding and administered the project.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Arunima Singh, in collaboration with the Nature Methods team.
Funding
Open access funding provided by Max Planck Society.
Data availability
Our database is available at cd-code.org and the encyclopedia at wiki.cd-code.org. Rolling feature update releases and data dumps will be noted here: https://cd-code.org/release. Source data are provided with this paper.
Code availability
Software and documentation: https://git.mpi-cbg.de/dd-code-team/dd-code-docs.
Competing interests
A.A.H. is a founder and shareholder of Dewpoint Therapeutics. I.A.K. and D.M.M. are employees and shareholders of Dewpoint Therapeutics. A.T.-P. is a recipient of research support from Dewpoint Therapeutics. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nadia Rostam, Soumyadeep Ghosh.
Extended data
is available for this paper at 10.1038/s41592-023-01831-0.
Supplementary information
The online version contains supplementary material available at 10.1038/s41592-023-01831-0.
References
- 1.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti S, Dormann D. Liquid-liquid phase separation in disease. Annu. Rev. Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 3.Mitrea DM, Mittasch M, Gomes BF, Klein IA, Murcko MA. Modulating biomolecular condensates: a novel approach to drug discovery. Nat. Rev. Drug Discov. 2022;21:841–862. doi: 10.1038/s41573-022-00505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conti BA, Oppikofer M. Biomolecular condensates: new opportunities for drug discovery and RNA therapeutics. Trends Pharmacol. Sci. 2022;43:820–837. doi: 10.1016/j.tips.2022.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Ning W, et al. DrLLPS: a data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res. 2020;48:D288–D295. doi: 10.1093/nar/gkz1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mészáros B, et al. PhaSePro: the database of proteins driving liquid-liquid phase separation. Nucleic Acids Res. 2020;48:D360–D367. doi: 10.1093/nar/gkz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, et al. LLPSDB: a database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res. 2020;48:D320–D327. doi: 10.1093/nar/gkz778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You K, et al. PhaSepDB: a database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 2020;48:D354–D359. doi: 10.1093/nar/gkz847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham F, et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 12.Mészáros B, Erdos G, Dosztányi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saar KL, et al. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl Acad. Sci. USA. 2021;118:e2019053118. doi: 10.1073/pnas.2019053118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Mierlo G, et al. Predicting protein condensate formation using machine learning. Cell Rep. 2021;34:108705. doi: 10.1016/j.celrep.2021.108705. [DOI] [PubMed] [Google Scholar]
- 15.Hardenberg M, Horvath A, Ambrus V, Fuxreiter M, Vendruscolo M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl Acad. Sci. USA. 2021;117:33254–33262. doi: 10.1073/pnas.2007670117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatos A, Tosatto SCE, Vendruscolo M, Fuxreiter M. FuzDrop on AlphaFold: visualizing the sequence-dependent propensity of liquid-liquid phase separation and aggregation of proteins. Nucleic Acids Res. 2022;50:W337–W344. doi: 10.1093/nar/gkac386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, et al. Screening membraneless organelle participants with machine-learning models that integrate multimodal features. Proc. Natl Acad. Sci. USA. 2022;119:e2115369119. doi: 10.1073/pnas.2115369119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, et al. TimeTree 5: an expanded resource for species divergence times. Mol. Biol. Evol. 2022;39:msac174. doi: 10.1093/molbev/msac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Documentation.
Condensate nomenclature, synonyms and functional groups.
Source data for Fig. 2.
Source data for Extended Data Fig. 6.
Data Availability Statement
Our database is available at cd-code.org and the encyclopedia at wiki.cd-code.org. Rolling feature update releases and data dumps will be noted here: https://cd-code.org/release. Source data are provided with this paper.
Software and documentation: https://git.mpi-cbg.de/dd-code-team/dd-code-docs.