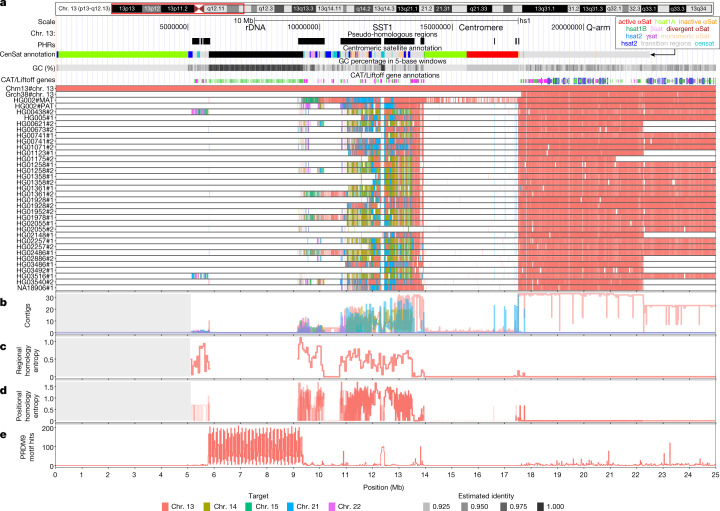
Fig. 3. Characteristics of the PHRs of acrocentric chromosomes.
a, We focus on the first 25 Mb of chr. 13, shown here as a red box over T2T-CHM13 cytobands. PHRs are highlighted relative to T2T-CHM13 genome annotations for centromere and satellite repeats (CenSat annotation), GC percentage and genes (CAT/Liftoff genes). Top, regions of interest described in the main text: rDNA, the SST1 array, the centromere and q-arm. Bottom, relative homology mosaics based on the T2T-CHM13 assembly for each chr. 13-matched contig from HPRCy1-acro, with colours indicating the most similar reference chromosome (target). b,c,d, Aggregated untangle results in the SAACs. b, The count of HPRCy1 q-arm-anchored contigs mapping to each acrocentric chromosome (Contigs) aggregated by target chromosome and (c) the regional (50 kb) untangle entropy metric (Regional homology entropy) computed over the contigs’ untangling relative to T2T-CHM13. d, By considering the multiple untangling of each HPRCy1-acro contig, we develop a point-wise metric that captures diversity in homology patterns relative to T2T-CHM13 (Positional homology entropy), leading to our definition of the PHRs. e, The patterns of homology mosaicism suggest ongoing recombination exchange in the SAACs. A scan over T2T-CHM13 reveals that the rDNA and SST1 array units are enriched for PRDM9 binding motifs, and thus may host frequent double-stranded breaks during meiosis. In b–d, the grey background indicates regions with missing data due to the lack of non-T2T-CHM13 contigs.