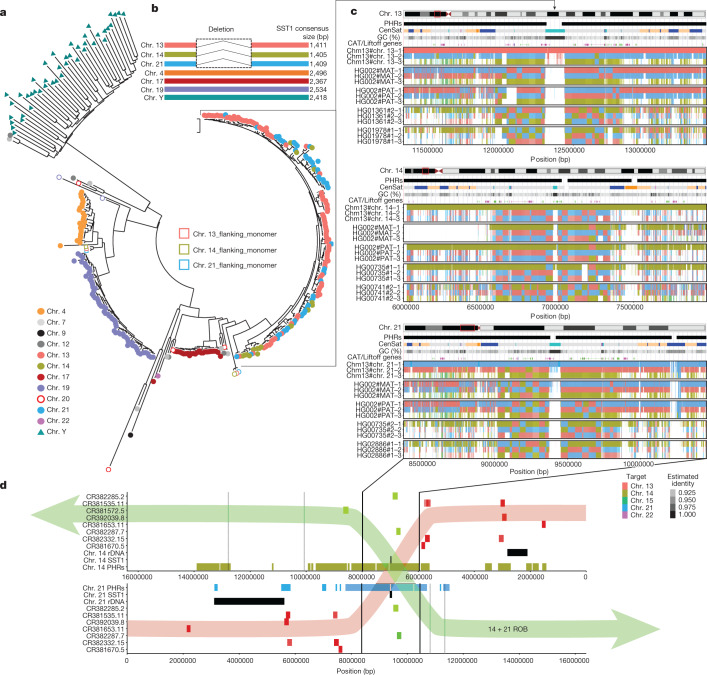
Fig. 4. PHRs of chr. 13, chr. 14 and chr. 21, centred on the SST1 array.
a, Maximum likelihood phylogenetic analysis of SST1 full-length elements indicates a recent homogenization process of acrocentric arrays. Coloured circles next to chromosome labels indicate individual monomers retrieved from the T2T-CHM13 assembly. Coloured triangles indicate SST1 full-length monomers retrieved from the HG002 chr. Y assembly. Partial or chimeric monomers flanking chr. 13, chr. 14 and chr. 21 arrays (located around 250 kb from the main array) are labelled as open circles or squares, respectively, coloured according to the corresponding chromosome. b, Schematic representation of SST1 consensus alignments, indicating a deletion that is present only in the SST1 unit from arrays on chr. 13, chr. 14 and chr. 21. c, Multiple untangling of T2T-CHM13, HG002-Verkko haplotypes and HPRCy1-acro contigs versus T2T-CHM13. Three of the five acrocentric chromosomes are represented. The degree of transparency indicates the estimated identity of the mappings. All mappings above 90% estimated pairwise identity are shown. To enable the display of simultaneous hits to all acrocentric regions, each grouping shows the first three best alternative mappings. SST1 arrays described in a are at the centre of a PHR that displays chequerboard patterns indicative of recombination between heterologous acrocentric chromosomes (black arrows link the SST1 arrays in all panels). These patterns are less common on chr. 15 and chr. 22 (Supplementary Figs. 20 and 22). d, The PHRs on T2T-CHM13 (yellow and light blue) in relation to BACs localized cytogenetically8 to recurrent chr. 14–chr. 21 ROB breakpoints. BACs shown in green are found in dicentric Robertsonian chromosomes, whereas those in red are not. Chr. 14 is shown in an inverted orientation aligned to chr. 21 at the breakpoint region suggested experimentally8. In a transparent overlay, we propose a retained dicentric chromosome (14+21 ROB, green) and lost (red) products of the studied recurrent translocations.