Abstract
To evaluate the composition of the microbial community of the middle nasal in paranasal sinus fungus ball (FB), chronic sinusitis with nasal polyps (CRSwNP) and healthy controls, providing new insights into the pathogenesis of FB and CRSwNP. Through 16 s rRNA gene high-throughput sequencing to determine the microbial characterization from patients with FB (n = 29) and CRSwNP (n = 10), and healthy controls (n = 4). The FB group had significantly lower αdiversity and significantly different β diversity compared to the other groups. All three groups mainly consisted of four bacterial phyla (Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria). In the FB group, the highest relative abundance was found in Proteobacteria (47.04%). However, pairwise comparisons resulted in statistically significant differences only for Firmicutes (CRSwNP, p = 0.003, Control, p = 0.008). The CRSwNP group was statistically different from the control group in TM7(p = 0.010), Chloroflexi(p = 0.018) and Bacteroidete(p = 0.027). At the genus level, the FB group had the highest relative abundance of Haemophilus (11.53%), followed by Neisseria (7.39%), and Neisseria abundance (p < 0.001) was significantly different from the remaining two groups. Ruminococcacea abundance (p < 0.001) and Comamonadaceae abundance (p < 0.001) were increased in the CRSwNP group. The relative abundance of Lactobacillus (p < 0.001), Bacteroides S24_7 (p < 0.001), and Desulfovibrio (p < 0.001) was significantly decreased in the FB and CRSwNP groups compared to the control group. The imbalance of the microbial community is related to the pathogenesis of sinusitis.
Keywords: 16S rRNA, Fungal ball, Chronic rhinosinusitis, Nasal polyps, Microbiota
Introduction
Paranasal sinus fungus ball(FB), described as a non-invasive conglomeration of fungal hyphae, usually found in a single sinus cavity [1]. With the widespread use of sinus CT and nasal endoscopy, the diagnosis of fungal rhinosinusitis is gradually increasing, and the variety of pathogenic microorganisms is also increasing [2]. Chronic sinusitis with nasal polyps (CRSwNP) is usually considered to be related to pathogenic factors such as bacterial infections, allergy, mucociliary damage, and anatomical variations of the nasal sinus [3]. The microbial perspective of human health/disease interrelationships has become increasingly topical in recent years. The human mucosal surface hosts a large microbiota that is functionally and quantitatively diverse. Metabolites of the microbiome interact to regulate the growth of harmful pathogens and the development of immune cells [4, 5]. The mucosal surface microenvironment, formed by the body's defense mechanisms, provides continuous selective pressure on epithelial microorganisms creating the ecological niche of the mucosal surface, while imbalances in the microbiota may contribute to the inflammatory process [6]. The interaction between the microbiota and the local immune system is considered as a potential etiology, but the mechanisms of their interactions are complex and still not sufficiently clarified [7]. A variety of diseases with intestinal [4], reproductive tract [8] and oral sites [9] have been shown to be closely related to microflora imbalances. However few studies have used 16S rRNA gene sequencing to evaluate the nasal microbiome, especially the FB mircobiota, and there are differences in the results of different category of sinusitis [10, 11]. Therefore, more sequencing results are needed to provide valuable analysis of the nasal microbiota.
Through this study, we hope to demonstrate the role of bacteria in the pathogenesis of fungal ball versus chronic sinusitis with nasal polyps, and also to investigate the differences in microbiome profiles through comparative analysis of microbial community diversity.
Method and Materials
Sample Collection
For this study, we collected 49 samples which were classified into three different types of groups:1) 29 samples from the affected side of FB, 2) 10 from CRSwNP with Lund-Mackay Scale ≥ 10 points, 3) Four from healthy participants. The exclusion criteria were patients using local or systemic antibiotics, hormones, antihistamines, and leukotriene receptor antagonists within the past month; nasal irrigation within the past two weeks or patients with pregnancy and lactation; combined with the severe respiratory system, immunodeficiency. All the samples were collected with nasal endoscopy (KARL STORZ, Germany, 0°, 70°) surgery under general anesthesia in the operating room. We use nasopharyngeal swabs (Copan, Italy) to collect the specimen before using the vasoconstrictors. Secretions from the middle meatus of maxillary sinus fungal ball of FB group and the remaining two groups and from the superior meatus of the sphenoid sinus fungal ball were put into the swabs and immersed into preservation solution completely. The swabs were all transferred to the laboratory for storage at -80˚C.
DNA Extraction and 16S rDNA Amplicon sequencing
Total bacterial genomic DNA samples were extracted using the Mag-Bind soil DNA kit(200)(M5635-02, OMEGA, USA), following the manufacturer’s instructions, and stored at − 20 °C prior to further analysis. PCR amplification of the 16S rRNA genes V3–V4 region was performed using the primer 338F-806R and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled in equal amounts, and pair-end 2 × 300 bp sequencing was performed using the Illlumina MiSeq platform with MiSeq Reagent Kit v3. The Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) pipeline was employed to process the sequencing data. Paired-end reads were assembled using FLASH. After chimera detection, the remaining high-quality sequences were clustered by UCLUST into operational taxonomic units (OTUs) with 97% sequence identity.
Bioinformatics and Statistical Analysis
Sequence data analyses were mainly performed using QIIME and R packages (v3.2.0). OTU-level alpha diversity indices, such as Chao1 richness estimator, ACE metric (Abundance-based Coverage Estimator), and Shannon diversity index, were calculated using the OTU table in QIIME. OTU-level ranked abundance curves were generated to compare the richness and evenness of OTUs among samples. Generate Venn diagrams using the R package "VennDiagram" to visualize shared and unique OTUs between samples or groups. Beta diversity analysis was performed using the UniFrac distance metric to study structural changes in microbial communities in different samples and visualized by non-metric multidimensional scaling (NMDS). Statistical comparisons of taxon abundance between samples or groups were performed by Metastats. LEfSe (Linear discriminant analysis effect size) was performed to detect differentially abundant taxa across groups using the default parameters. For normally distributed data, the microbiota richness index was examined by ANOVA, and for non-normally distributed data, the Kruskal–Wallis test was used. For normally and non-normally distributed data, pairwise comparisons were performed with Welch's t-test or Mann–Whitney U-test, respectively. Demographic and clinical characteristics were analyzed with chi-square test for binomial variables and t-test for continuous variables, with a P value < 0.05 indicating statistical significance.
Results
Study Population and Subject Characteristics
This study involved 42 patients, including 29 FB patients, 10 CRSwNP patients and four controls. The demographics and clinical characteristics of the 3 groups revealed significant differences in the age of subjects at the time of sampling between the cohorts, with FB subjects being older. Meanwhile, there were significantly more females in the FB cohort. CRSwNP had significantly higher incidence of hyposmia symptom than FB group (Table 1).
Table 1.
The demographic and clinical data of the subjects
| FB | CRSwNP | Control | p value | |||||
|---|---|---|---|---|---|---|---|---|
| Age(years), mean ± SD | 59.72 ± 13.89 | 41.30 ± 15.97 | 38.00 ± 9.02 | 0.024 | ||||
| Female/Male | 23/6 | 3/7 | 2/2 | 0.008 | ||||
| Site, n | Middle meatus | Maxillary sinus | 25 | Middle meatus | 10 | Middle meatus | 4 | – |
| Superior meatus | Sphenoid sinus | 4 | – | |||||
| Symptom, n | Nasal obstruction | 16 | 9 | 0.064 | ||||
| Nasal discharge | 14 | 4 | 0.468 | |||||
| Headache | 11 | 1 | 0.131 | |||||
| Bloody secretions | 6 | 1 | 0.653 | |||||
| Postnasal drip | 5 | 0 | 0.302 | |||||
| Hyposmia | 0 | 6 | 0.000 | |||||
FB paranasal sinus fungal ball, CRSwNP chronic rhinosinusitis with nasal polyps
Bacterial Diversity
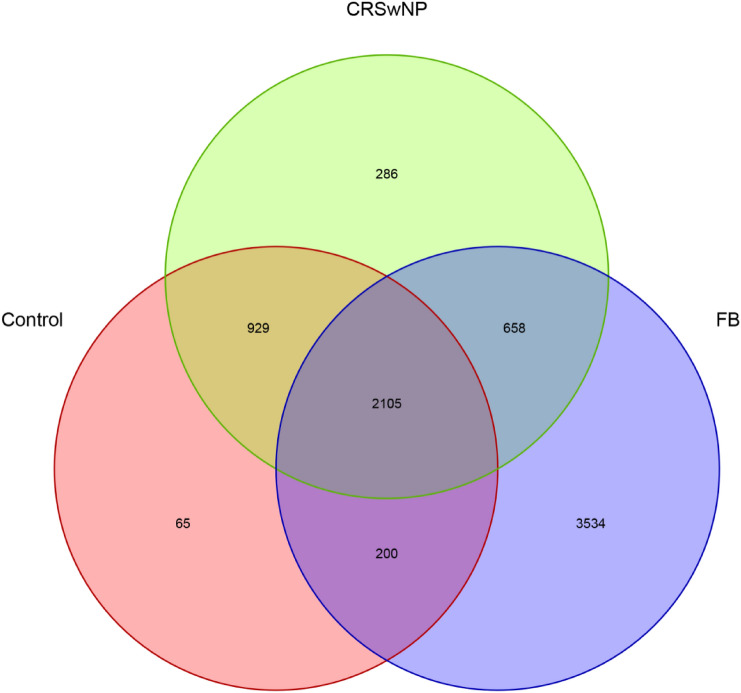
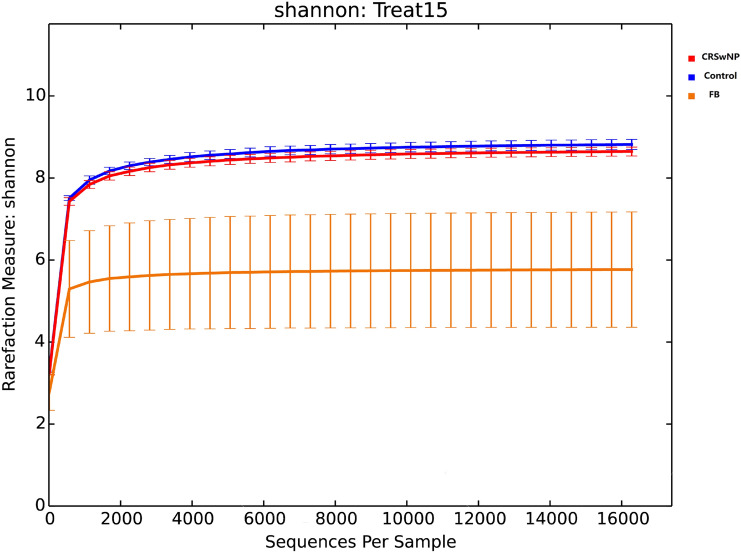
In total, 723,523 high-quality sequences were optimized using the NCBI against a library from the Greengenes database. 7777 OTUs were assigned to 29 bacteria phyla and 509 genera. 2105 OTUs were shared by the three groups. Measurements of unique OTUs of FB, CRSwNP and control groups were 3534, 286 and 65, respectively. Venn diagram was used to show the number of different OTUs common/unique to the group samples (Fig. 1). The Shannon index is used to construct a rarefaction curve. As the readings increase, the curve gradually flattens, which means that the current sequencing depth of each sample is sufficient to reflect the bacterial diversity of the community (Fig. 2).
Fig. 1.
The number of different OTUs common/unique to the group samples. FB stands for paranasal sinus fungal ball. CRSwNP indicates chronic sinusitis with nasal polyps. Control represents healthy control
Fig. 2.
The total number of sequences for each sample in the OTU abundance matrix is randomly sampled at different depths, and the rarefaction curve is constructed with the number of sequences drawn at each depth and their corresponding OTU numbers
The ACE index, Chao1 index focus on the richness of the microbiota, while Shannon index takes into account the evenness of the microbiota at the same time. They all reflected the α-diversity of the microbiota. The indicators had statistically significant difference between the FB and other groups (Table 2). The α- diversity of FB showed a significant decrease. Control and CRSwNP had no significant difference in the three indicators. Although healthy controls tended to have higher α-diversity when compared to CRSwNP group this was not significant (p > 0.05).
Table 2.
The comparison of the α-diversity indexes of the three groups
| Estimators | p-value | ||
|---|---|---|---|
| Control- | Control- | CRSwNP- | |
| CRSwNP | FB | FB | |
| Chao1 | 0.267 | < 0.001 | < 0.001 |
| ACE | 0.283 | < 0.001 | < 0.001 |
| Shannon | 0.084 | < 0.001 | < 0.001 |
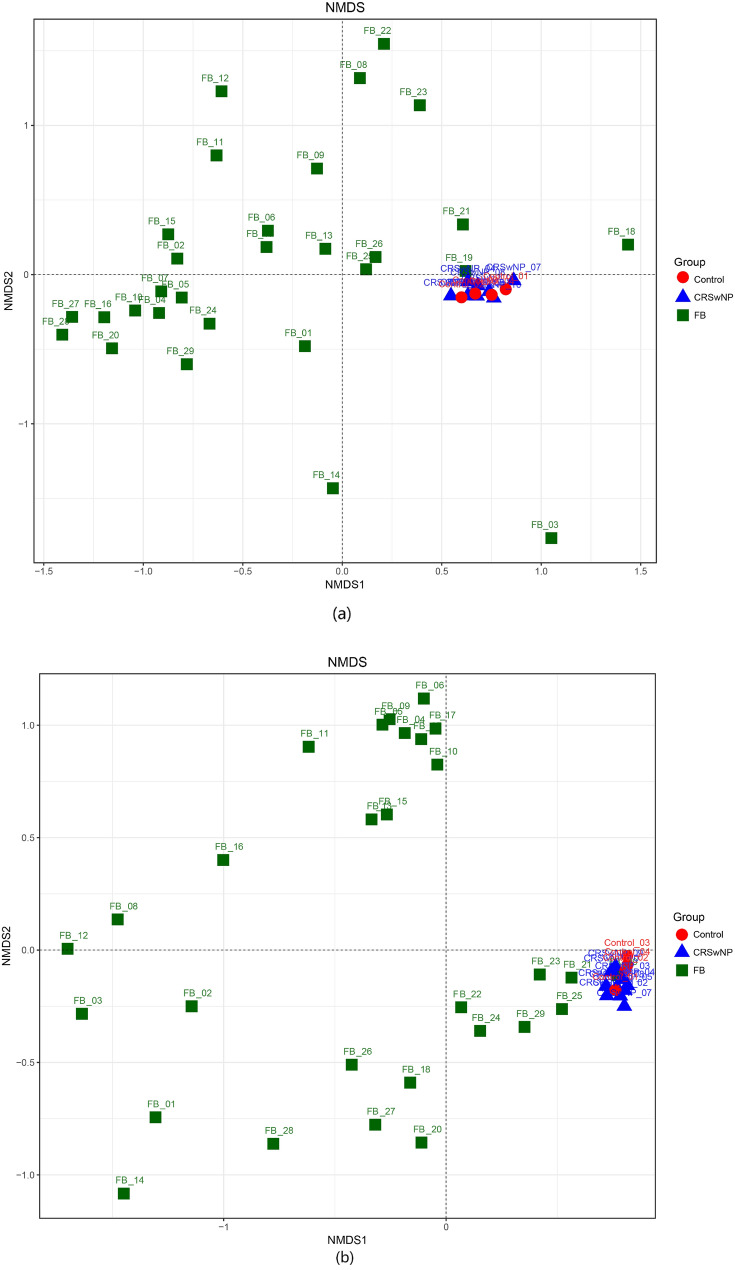
Nonmetric Multidimensional Scaling(NMDS) which based on the Unifrac distances reflect the β-diversity of the microbiota. (Fig. 3a, b) Healthy controls and CRSwNP groups were close in distance, indicating that the distribution of the microbiota of the two groups was relatively similar. While, significant different cluster had been observed between the FB and the other two groups. The distribution of samples within the FB group was relatively scattered.
Fig. 3.
NMDS of bacterial communities were conducted to describe the structural distribution of community samples by two-dimensional sorting diagrams based on the UniFrac distance metrics. Figure a was analyzed by Unweighted UniFrac NMDS, b was analyzed by Weighted UniFrac NMDS. The former considers only the presence of OTUs in the samples without considering their high or low abundance. The latter takes into account the phylogenetic relationships among the community members and their abundance in the respective samples
Bacterial Composition
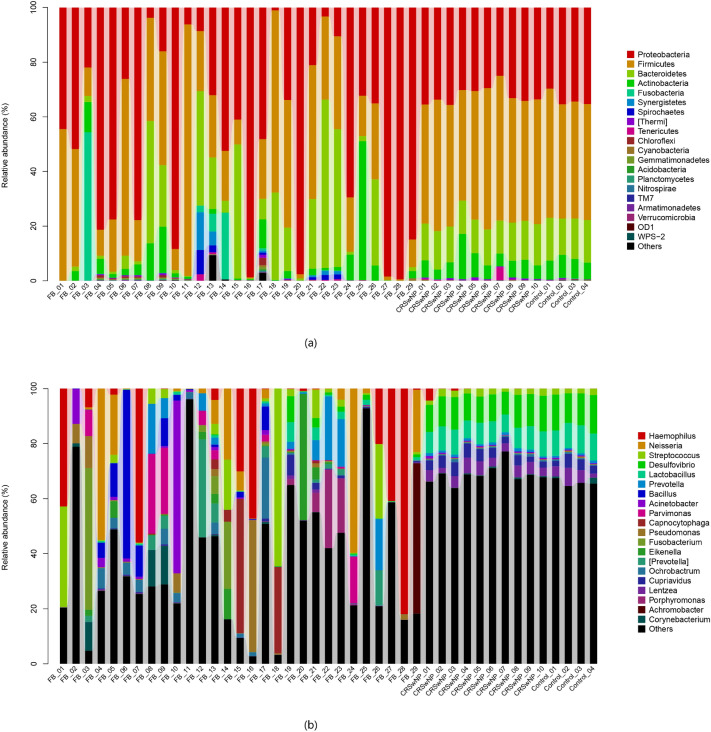
In the FB group, 491,244 high-quality sequences were obtained. OTU taxon analysis showed that 6497 OTUs referring to 22 phyla and 492 bacterial genera, and about 0.4% of all were no blast hit. Ranked by abundance, the predominant bacterial phyla were Proteobacteria (47.04%), Firmicutes (27.36%), Bacteroidetes (14.63%), Actinobacteria (5.18%). The predominant bacterial genera were Haemophilus (11.53%), Neisseria (7.39%), Staphylococcus (6.30%), Unclassified_Planococcaceae (5.13%), Bacillus (3.98%), Prevotella (3.63%). There was no statistical difference in the bacterial community between the maxillary and sphenoid sinuses(p = 0.901) (Fig. 4).
Fig. 4.
The relative abundance of each sample of phyla a and genera b *In the FB group, samples 4,5,6,20 were sphenoid sinus fungal ball and the rest were from maxillary. There was no statistical difference between the two microbiota(p = 0.901)
From the ten CRSwNP samples, 165,594 high-quality sequences were assigned to 3978 OTUs. 18 different bacterial phyla and 244 bacterial genera were detected, while 0.01% of all were no blast hit. Compared with the FB group, Firmicutes (46.43%) was more predominant, followed with Proteobacteria (31.96%), Bacteroidetes (13.17%), Actinobacteria (7.07%). The predominant bacterial genera were Unclassified_Ruminococcaceae (17.04%), Unclassified_S24-7 (11.42%), Desulfovibrio (10.65%), Lactobacillus (7.9%), Unclassified_Comamonadaceae (4.97%), Unclassified_Clostridiales (4.45%).
The composition of the bacterial of the control group is similar to that of CRSwNP group. We obtained 66,685 high-quality sequences belonging to 3299 OTUs of 12 different bacterial phyla and 103 genera with 0.01% no blast hit in the control group. The major abundant phyla contained Firmicute (43.58%), Proteobacteria (33.73%), Bacteroidetes (14.85%), Actinobacteria (7.20%). Unclassified_Ruminococcaceae, Unclassified_S24-7, Desulfovibrio, Lactobacillus, Unclassified_Comamonadaceae, Unclassified_Clostridiales belong to the prevalent bacterial genera with relative abundances (%) of 16.98, 13.25, 12.38, 9.58, 4.93 and 4.23, respectively. The relative abundance of each group of phyla and genera was shown in Fig. 4a, b.
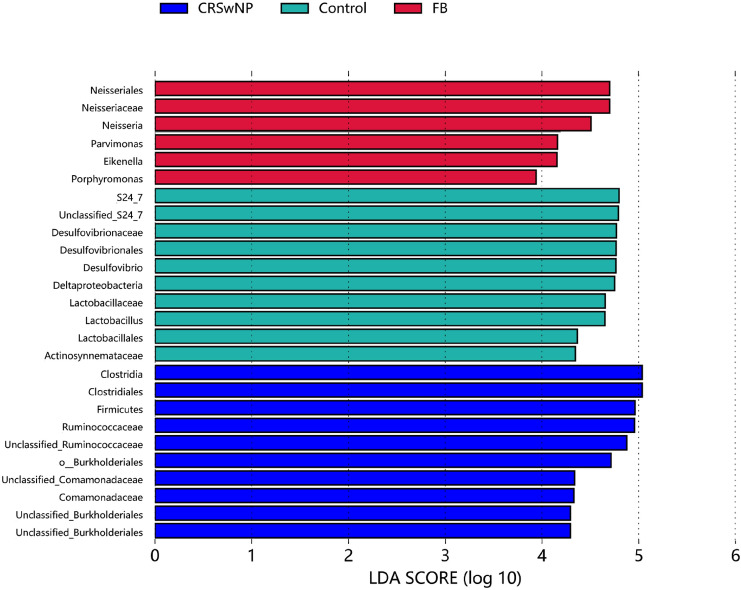
Based on the Metastats analysis, At the phylm level, the relative abundance of Firmicutes had significant difference between the FB group and the other two groups. (CRSwNP, p=0.003, Control, p=0.008). The abundance of TM7(p = 0.010), Chloroflexi (p = 0.018) and Bacteroidete (p = 0.027) were significantly different between the CRSwNP and control groups. At the genus level, CRSwNP(p = 0.003) and FB group(p < 0.001) revealed significant difference with control group in the relative abudunce of Lactobacillus. Haemophilus abundance is significantly different between the FB group and control group(p = 0.032), but not from the CRSwNP group. Further, based on the Linear discriminant analysis effect size(LEfSE), among the three groups, Neisseria (p < 0.001) has the highest relative abundance and the most significant difference in the FB group, followed with Parvimonas(p = 0.001) and Eikenella.(p = 0.017). In the CRSwNP group, The relative abundance of Ruminococcacea (p < 0.001) from the phylum of clostriadia and Comamonadaceae (p < 0.001) from the phylum of Burkholderiales was significantly higher than the two other groups. The control groups revealed siginificant difference with highest abundance of Bacteroides S24_7 (p < 0.001), and Desulfovibrio (p < 0,0.001). Other remaining taxa with statistical difference among the three groups were drawn into LDA diagrams based on the results of Lefse (Fig. 5).
Fig. 5.
The vertical coordinates are the taxonomic units with significant differences between groups, and the horizontal coordinates show the logarithmic LDA difference analysis scores, ranked by the size of the scores, to describe the magnitude of their differences in different subgroups of samples. The longer lengths indicate the more significant differences of the taxonomic units, and the different colors of the bars indicate the sample group with higher abundance for that taxonomic unit. (The graph only shows taxonomic unit with the most significant differences in the top 20% of each group)
Discussion
Patients with FB tend to have a mixture of fungal and bacterial infections. It is often diagnosed as secondary to a long history of recurrent sinusitis, and about 3.7% of patients with chronic sinusitis in the late twentieth century were diagnosed with fungal sinusitis [12]. Stammberger believed that the purulent secretions produced by bacteria were an ideal medium for fungi to grow that the infection induces an environment conducive to fungal growth [13]. Fungi have a role in the development of CRSwNP, but are difficult to determine [14]. Recent studies of the sinus microbiota have demonstrated that the sinuses of patients with chronic sinusitis had a loss of bacterial diversity and a simultaneous enrichment of pathogens (resident microorganisms with pathogenic potential), with an imbalance in the microbiota community [15]. Patients with CRS always receive multiple antibiotic treatments, leading to adverse factors such as microbiota disruption and multi-drug resistant bacteria [16].
In this study, the mean age of the FB group (59.72 ± 13.89) was significantly different from the remaining two groups with an older age (p = 0.024). Meanwhile, the FB group possess higher incidence in female patients (p = 0.008). In the other studies, the FB mostly appeared in elder individuals. Females over 50 years of age are susceptible to FB infections possibly related to decreased estrogen levels and mucosal dysfunction [17].
Among the three groups, though there were no significance in α-diversity index between the control and CRSwNP groups, the mean α-diversity index was lower in the CRSwNP group. In the FB group the index was significantly lower than that of the two groups. Based on the β- diversity analysis, the samples of the CRSwNP group and the control group were mixed with each other. This result is accordance to the previous study [18]. The FB group shows significant difference in β-diversity with other groups, but at the same time the differences within the FB group were also more obvious than the other two groups.
The four bacterial phyla (Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria) mainly constituted the nasal microbiota. Firmicutes was dominant in the control and the CRSwNP group. The relative abundance of Ruminococcaceae and Lactobacillaceae which are the families of Firmicutes decreased significantly in the FB group. Rumenococcaceae confirmed to be associated with allergic rhinitis and positively associated with the levels of Th2-related factor [19]. Th2 differentiated cytokines IL-4, IL-5, IL-8, IL-10 and IL-13 can be involved in the development and progression of nasal polyps [20]. Rumenococcaceae was also a major bacterial colonies of the CRSwNP group in this study.
Lactobacillaceae are regarded as promising probiotics which have been safely used daily as probiotics in food supplements for over a century [21]. However, their potential as upper respiratory tract probiotics has not been widely considered. Some studies have described them as part of the normal upper respiratory microbiota in healthy adults and/or children. Lacticaseibacillus casei AMBR2 has been demonstrated to have the antimicrobial effects and the ability to repair the respiratory epithelium in patients with CRSwNP [22]. Lactobacillaceae are expected to be further investigated in clinical trials for their potential as upper respiratory tract probiotics.
The relative abundance of Bacteroidete is decreased in CRSwNP compared to controls, especially Bacteroidete_S247 which are called Muribaculaceae now. The members of this family have been identified as potential mucus degraders [23]. In the previous study, through a murine model, C. tuberculostearicum is able to cause mucosal inflammation with goblet cell proliferation and excessive mucus secretion in a decreased microbiota abundance condition [15]. This is similar to the pathophysiological mechanism of sinusitis [24]. Muribaculaceae have the potential to reduce Clostridiodes difficile colonization by using mucin-derived sugars in competition with Clostridium in situ [25]. CRSwNP is always associated with asthma and allergic rhinitis. The increasing abundance of Comamonadaceae is highly correlated with the degree of bronchial hyperresponsiveness [26].
We found that Neisseriaceae had the most significant difference abundance in the FB group in our cohort. Neisseria is a genus of Proteobacteria, including pathogenic and commensal species that mainly colonize the human oro-nasopharynx. Neisseria have also been found to be associated with human infections, including valve endocarditis, periodontitis, and otitis media [27]. One study found that sex hormone levels drop related to Neisseria mucosa abundance increased [28]. In our study, the majority within the FB group were middle-aged and menopausal women with decreased estrogen levels which may leading to the increased abundance of Neisseria. Eikenella was first isolated from human abscesses. The previous study has reported that empyema could be secondary to Eikenella induced sinusitis. The increase in the abundance may correlate with the severity of the infection [29].
The relative abundance of Haemophilus was increased in both the CRSwNP and fungal groups, while only the FB group was statistically different from the control group. The current studies have observed that in vitro Haemophilus inhibits the function of the mucociliary clearance system. This strategy can benefit fungus in the sinuses [30]. Many species of Haemophilus and Neisseria are naturally transformative, able to take up DNA fragments from their environment and integrate them into chromosomes. This behavior may also explain the prevalence of Haemophilus species in the Neisseria meningitidis environment [31].
In our study, there are some limitations. Firstly, the small sample size of healthy control may cause contingency of statistical results. Secondly, the large variation within the fungal group may be caused by the different microbiota brought about by different species of fungi. Further classification of the samples according to fungal species is needed in the future. For the CRSwNP group, the group should be better to be distinguished by eosinophilic and non-eosinophilic. Finally, further studies are needed to confirm whether the microbiota difference is the cause or the result of sinusitis formation, such as animal tests with significantly different genera to confirm whether the microbiota can induce fungal ball formation, or changes in microbiota before and after sinus implantation in animal models to elucidate the causal relationship between microbiota changes in fungal ball formation.
Conclusion
Among the three groups, the FB group differed from the polyp group, and the control group in terms of microbial diversity. Neisseria abundance revealed the most significance difference in the FB group compared with other groups. The possible role of microbial dysbiosis as a pathogenetic mechanism of nasal mucosal inflammation was further confirmed in this study. In contrast, the increase relative abundance of S247 as well as Lactobacillus in the healthy group may have an inhibitory effect on inflammation in the nasal mucosa, but the interaction between these bacterial microbiota requires further experiments to verify.
Acknowledgements
The research was funded by the National Natural Science Foundation of China (grant award number 81070768) and Wu Jieping Clinical Research Special Assistance Fund (grant award number 320.6750); National Key Research & Development Program of China (2017YFC0112500), Beijing Municipal Administration of Hospitals’ Mission Plan (SML20150201).
Ethical Approval
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pengyu Shi and Hongzheng Wei have contributed equally to this research.
References
- 1.Ferreiro JA, Carlson BA, Cody DT., 3rd Paranasal sinus fungus balls. Head Neck. 1997;19:481–486. doi: 10.1002/(sici)1097-0347(199709)19:6<481::aid-hed4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 2.Durrani Z, Naeem M, Khan MA. Assessment of bacterial infection in patients operated for complications of chronic fungal rhinosinusitis. Pak J Med Health Scis. 2018;12:153–156. [Google Scholar]
- 3.Lou H, Meng Y, Piao Y, et al. Cellular phenotyping of chronic rhinosinusitis with nasal polyps. Rhinology. 2016;54:150–159. doi: 10.4193/Rhino15.271. [DOI] [PubMed] [Google Scholar]
- 4.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 5.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DW, Kim YM, Min JY, et al. Clinicopathologic characteristics of paranasal sinus fungus ball: retrospective, multicenter study in Korea. Eur Arch Otorhinolaryngol. 2020;277:761–765. doi: 10.1002/lary.27726. [DOI] [PubMed] [Google Scholar]
- 8.Agostinis C, Mangogna A, Bossi F, Ricci G, Kishore U, Bulla R. Uterine Immunity and Microbiota: A Shifting Paradigm. Front Immunol. 2019;17:2387. doi: 10.3389/fimmu.2019.02387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He XS, Shi WY. Oral microbiology: past, present and future. Int J Oral Sci. 2009;1:47–58. doi: 10.4248/ijos.09029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan W, Zhang H, Yang F, et al. The influence of nasal bacterial microbiome diversity on the pathogenesis and prognosis of chronic rhinosinusitis patients with polyps. Eur Arch Otorhinolaryngol. 2021;278:1075–1088. doi: 10.1007/s00405-020-06370-4. [DOI] [PubMed] [Google Scholar]
- 11.Rom D, Bassiouni A, Eykman E, et al. The association between disease severity and microbiome in chronic Rhinosinusitis. Laryngoscope. 2019;129:1265–1273. doi: 10.1002/lary.27726. [DOI] [PubMed] [Google Scholar]
- 12.Wagner Mackenzie B, Waite DW, Hoggard M, et al. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ Microbiol. 2017;19:381–392. doi: 10.1111/1462-2920.13632. [DOI] [PubMed] [Google Scholar]
- 13.Stammberger H, Jakse R, Beaufort F. Aspergillosis of the paranasal sinuses x-ray diagnosis, histopathology, and clinical aspects. Ann Otol Rhinol Laryngol. 1984;93:251–256. doi: 10.1177/000348948409300313. [DOI] [PubMed] [Google Scholar]
- 14.Qaisar Sajjad SM, Suhail Z, Ahmed R. Prevalence of fungal infection in nasal polyposis - A cross-sectional study, conducted at a tertiary care hospital in Karachi. J Pak Med Assoc. 2020;70:48–52. doi: 10.5455/JPMA.296507. [DOI] [PubMed] [Google Scholar]
- 15.Abreu NA, Nagalingam NA, Song Y, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2020;4:151ra124. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner Mackenzie B, West AG, Waite DW, et al. A novel description of the human sinus archaeome during health and chronic Rhinosinusitis. Front Cell Infect Microbiol. 2020;10:398. doi: 10.3389/fcimb.2020.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai JC, Lee HS, Chen MK, et al. Patient satisfaction and treatment outcome of fungus ball rhinosinusitis treated by functional endoscopic sinus surgery. Eur Arch Otorhinolaryngol. 2011;268:227–230. doi: 10.1007/s00405-010-1299-7. [DOI] [PubMed] [Google Scholar]
- 18.Gan W, Yang F, Meng J, et al. Comparing the nasal bacterial microbiome diversity of allergic rhinitis, chronic rhinosinusitis and control subjects. Eur Arch Otorhinolaryngol. 2021;278:711–718. doi: 10.1007/s00405-020-06311-1. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y, Chen Y, Li Q, et al. Tetrahydrocurcumin alleviates allergic airway inflammation in asthmatic mice by modulating the gut microbiota. Food Funct. 2021;12:6830–6840. doi: 10.1039/d1fo00194a. [DOI] [PubMed] [Google Scholar]
- 20.Zhong Y, Li Y, Xiao X. The relationship of the expression of thymic stromal lymphopoietin in nasal polyps tissues and Th2 inflammatory response. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2014;28:817–820. [PubMed] [Google Scholar]
- 21.Salvetti E, O’Toole PW. When regulation challenges innovation: the case of the genus Lactobacillus. Trends Food Sci Technol. 2017;66:S0924224416300759. doi: 10.1016/j.tifs.2017.05.009. [DOI] [Google Scholar]
- 22.De Boeck I, van den Broek MFL, Allonsius CN, et al. Lactobacilli Have a Niche in the Human Nose. Cell Rep. 2020;31:107674. doi: 10.1016/j.celrep.2020.107674. [DOI] [PubMed] [Google Scholar]
- 23.Lagkouvardos I, Lesker TR, Hitch TCA, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. 2019;7:28. doi: 10.1186/s40168-019-0637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraniuk JN, Petrie KN, Le U, et al. Neuropathology in rhinosinusitis. Am J Respir Crit Care Med. 2005;171:5–11. doi: 10.1164/rccm.200403-357OC. [DOI] [PubMed] [Google Scholar]
- 25.Pereira FC, Wasmund K, Cobankovic I, et al. Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat Commun. 2020;11:5104. doi: 10.1038/s41467-020-18928-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castillo Vizuete JA, Sastre J, Del Cuvillo BA, et al. Asthma, rhinitis, and nasal polyp multimorbidities. Arch Bronconeumol (Engl Ed) 2019;55:146–155. doi: 10.1016/j.arbres.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Weyand NJ. Neisseria models of infection and persistence in the upper respiratory tract. Pathog Dis. 2017;75:ftx031. doi: 10.1093/femspd/ftx031. [DOI] [PubMed] [Google Scholar]
- 28.Adriaens LM, Alessandri R, Spörri S, et al. Does pregnancy have an impact on the subgingival microbiota? J Periodontol. 2009;80:72–81. doi: 10.1902/jop.2009.080012. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez-Rojas L, Suarez-López A, Cantón R, et al. Eikenella corrodens causing deep-seated infections Six-year experience in a University Hospital in Madrid. Enferm Infecc Microbiol Clin (Engl Ed) 2020;38(2):76–78. doi: 10.1016/j.eimc.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 31.Kroll JS, Wilks KE, Farrant JL, et al. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc Natl Acad Sci U S A. 1998;95:12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]