Abstract
Many group B Streptococcus agalactiae strains and other pathogenic streptococci express a cell-associated peptidase that inactivates C5a (C5a-ase), the major neutrophil chemoattractant produced by activation of the complement cascade. Type III group B streptococci (GBS) can be classified genotypically into three restriction digest pattern types. Functional C5a-ase activity of GBS correlates with this genetic typing; therefore, we sought to identify a genetic basis for this phenomenon. Southern hybridization confirms that all type III GBS contain scpB, the gene encoding GBS C5a-ase. GBS strains with high C5a-ase functional activity and those with no or very low activity both express immunoreactive C5a-ase. The scpB sequence of strain I30, which has high C5a-ase activity, is 98.2% homologous to the previously reported serotype II GBS scpB sequence. The scpB sequences of strains I25 and GW, which have low or no C5a-ase activity, are identical. The predicted I25 and GW C5a-ase proteins share a four-amino-acid deletion affecting the protease histidine active-site consensus motif. Recombinant I30 C5a-ase has good functional activity, whereas recombinant I25 C5a-ase has low activity. These data demonstrate that functional C5a-ase differences between type III GBS strains are attributable to a genetic polymorphism of scpB. The ubiquitous expression of C5a-ase, irrespective of functional activity, suggests that C5a-ase may have a second, as yet unidentified, function.
Streptococcus agalactiae, Lancefield group B beta-hemolytic streptococci (GBS), are the most common cause of serious bacterial infections in neonates and are major pathogens in parturient women and adults with underlying chronic diseases (3, 15). Infection in neonates is often fulminant and is characterized by poor host inflammatory responses (12).
C5a is produced by the cleavage of C5 following activation of the complement cascade. It is rapidly converted to C5adesarg by proteolytic cleavage in plasma (14). C5a and C5adesarg are potent chemoattractants for polymorphonuclear leukocytes (PMNs) (11). Many GBS produce a surface-associated peptidase, C5a-ase, that is able to inactivate C5a by cleavage between His67 and Lys68 (4, 6, 13). In animal models, C5a-ase-positive GBS strains fail to elicit the expected rapid neutrophil responses at sites of infection (5). C5a-ase, therefore, may be an important virulence factor. Serotype II GBS C5a-ase is encoded by a 3,450-bp open reading frame, scpB (8). Other pathogenic streptococci also exhibit C5a-ase activity, and genes homologous to scpB have been found in group A Streptococcus pyogenes (scpA) and group G streptococci (9).
Serotype III GBS are of considerable interest since they are responsible for the majority of disease in newborn infants (3). We previously described a system for classifying type III GBS based on restriction digest patterns (RDP) (20). By enumerating and quantitating restriction fragments produced by restriction endonuclease HindIII or Sse83871, it is possible to classify bacterial strains by genetic relatedness into three major groups, RDP III-1, III-2, and III-3. RDP III-3 can be further subdivided into subgroups RDP III-3a and RDP III-3b. Bacterial phenotype corresponds to this genotypic classification. Members of RDP type III-3a have high functional C5a-ase activity, whereas in RDP type III-3b strains activity is markedly reduced or absent, suggesting a genetic basis for this discrepancy. The importance of C5a-ase in the pathogenesis of GBS infections prompted us to identify the mechanism underlying the variability in C5a-ase expression.
MATERIALS AND METHODS
Bacterial strains.
S. agalactiae strains were isolated from infants hospitalized in Salt Lake City, Utah, or Tokyo, Japan. Strains I30, 630640, 560177, I31, and I05 are RDP type III-3a strains that have high C5a-ase functional activity (20). Strains I25, I12, I51, I53, I32, C39, C35, I10, and 861503 are RDP type III-3b strains that have little or no functional C5a-ase activity (20). Strain GW is a serotype III GBS that has no functional C5a-ase activity (13). Strain TOH-97, provided by C. Rubens and Theresa Harris, University of Washington, is an scpB isogenic mutant strain derived from strain COH-1. Strain TOH-87, also derived from serotype III strain COH-1, has a deletion in an unrelated gene, csp.
Southern hybridization of GBS DNA.
Genomic DNA from GBS strains was prepared by mutanolysin and proteinase K digestion as previously described (20). For genomic DNA dot blots, 5 μg of genomic DNA from each strain was denatured by incubation in denaturation buffer (2 M NaCl, 0.1 M NaOH, 5% ethanol) for 30 min at 50°C and applied to nylon membrane (Hybond-N+; Amersham Life Science Limited, Arlington Heights, Ill.), using a 96-chamber vacuum manifold. DNA was neutralized with 1 M NH4 acetate and cross-linked to the membrane by exposure to UV light. An scpB probe (provided by P. Cleary), which consists of the complete coding region of the gene, was labeled with fluorescein-dUTP (Gene Images labeling and detection system; Amersham) according to the manufacturer's protocol. Membranes were hybridized for 12 h with the heat-denatured scpB probe, washed at high stringency, and exposed to radiographic film according to the manufacturer's protocol.
Anti-C5a-ase monoclonal antibody (MAb).
An 8-week-old BALB/c mouse was immunized intraperitoneally once with 50 μg of recombinant C5a-ase (rC5a-ase; see below) in complete Freund's adjuvant, followed 4 weeks later by two weekly intraperitoneal immunizations of 25 μg of rC5a-ase in incomplete Freund's adjuvant and a final intravenous immunization with 25 μg of rC5a-ase. Splenocytes were isolated and fused to the nonsecreting murine myeloma cell line SP2 as previously described (1). Anti-C5a-ase hybridomas were identified by enzyme-linked immunosorbent assay. Ninety-six-well microtiter plates (Corning Costar Corporation, Oneonta, N.Y.) were coated with rC5a-ase (5 μg/ml in phosphate-buffered saline [PBS, pH 7.3]) for 2 h at 37°C. Plates were washed three times with PBS–0.05% Tween 20 and then incubated with undiluted hybridoma tissue culture supernatant for 16 h at 4°C. After washing, anti-C5a-ase antibody was detected by incubation with a 1:1,000 dilution of goat horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) antibody (Biosource International, Camarillo, Calif.) and addition of 2,2′-azino-di-[3-ethylbenzthiazoline sulfonate] substrate (Southern Biotechnology Associates, Birmingham, Ala.). Anti-C5a-ase IgG-secreting cell lines were cloned by limiting dilution, and the IgG2-secreting line F1 was selected for further studies.
Western blot detection of C5a-ase.
GBS strains were grown overnight at 37°C in Todd-Hewitt broth. Cultures were diluted 1:100 in fresh medium and grown at 37°C without shaking to an optical density at 600 nm of 0.500. After being washed in PBS (pH 7.4), 1010 cells were resuspended in 500 μl of mutanolysin (2 mg/ml in PBS–1 mM phenylmethylsulfonyl fluoride) and incubated for 30 min at 37°C. Cell debris was removed by centrifugation at 16,000 × g for 20 min at 4°C. Protein concentration was determined by spectrophotometry. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described, using a 4 to 15% gradient gel (6). Proteins were transferred to nitrocellulose membranes by electroblotting. Membranes were blocked by incubation in PBS–0.1%, Tween–3% nonfat dry milk overnight at 4°C, washed with PBS–0.1% Tween 20 (PBS-T), and then incubated with a 1:10 dilution of MAb F1 tissue culture supernatant in PBS-T for 1 h at 37°C. C5a-ase protein was detected by enhanced chemiluminescence (ECL) following incubation with a 1:2,500 dilution of horseradish peroxidase-conjugated mouse anti-IgG antibody in PBS-T (Sigma Chemical, St. Louis, Mo.) and ECL Western blotting system reagents (Amersham Pharmacia Biotech, Piscataway, N.J.) according to the manufacturer's protocol.
rC5a-ase production.
Vector pGEX-4T (Amersham Pharmacia Biotech), containing the complete serotype II scpB coding sequence, was provided by P. Cleary, University of Minnesota. rC5a-ase was expressed as a glutathione fusion protein and purified as previously described (8). The complete coding sequences of scpB from strains I30 and I25 were amplified from genomic DNA by PCR using 5′ sense (5′ AAGGACGACGGATCCCATAAA 3′) and 3′ antisense (5′ TTGAATTCCTTTTTGGCGTTT 3′) primers. Amplification reactions were performed in a 75-μl reaction mixture consisting of 300 ng of genomic DNA, 1.5 mM MgCl2, 2 mM deoxynucleotides, 50 pmol of each primer, 1× high-specificity additive, and 4 U of high-fidelity Bio-X-Act DNA polymerase (ISC Bioexpress, Kaysville, Utah) in the manufacturer's buffer. Reaction conditions consisted of denaturation at 95°C for 1 min, annealing at 42°C for 1.5 min, and extension at 72°C for 4 min. Thirty-five rounds of amplification were performed. Amplification products were cloned into pGEX-2T (Amersham Pharmacia Biotech), and sequences were confirmed. rC5a-ase was expressed as described above and quantitated by spectrophotometry. To engineer appropriate restriction sites, rI30 and rI25 C5a-ase share two amino acid substitutions from the native protein, Lys→Ser1 and Arg→His2, and both retain an additional Gly residue at the 5′ end following thrombin cleavage. These modifications would be expected to have similar effects on both recombinant proteins.
PMN adhesion assay for functional C5a-ase activity.
C5a-ase activity was determined by the ability of whole GBS, mutanolysin extracts, or rC5a-ase to inhibit the ability of C5a-stimulated adhesion of human PMNs to gelatin-coated tissue culture wells as previously described (6). Briefly, 10-fold dilutions ranging from 5 × 105 to 5 × 108 bacteria or 5-fold dilutions ranging from 1 to 25 μl of mutanolysin extracts (50 mg/ml) or rC5a-ase (0.01 to 1 μg/ml) were incubated with 500 μl of recombinant human C5a (1 μg/ml; Sigma, St. Louis, Mo.) in Hanks balanced salt solution–0.5% human serum albumin. If applicable, bacteria were removed by centrifugation and 25 μl of treated rC5a was added to 225 μl of 111In-labeled white blood cells at 37°C for 10 min. Nonadherent cells were removed, adherent cells were lysed with 1 M NH4OH, and cell lysates were counted in a gamma counter. Percentage adherence is expressed as counts of adherent cells/(adherent cells + nonadherent cells). Mean differences in PMN adhesion were compared by t test (Statview 4.01; SAS Institute, Inc., Cary, N.C.). A C5a-ase concentration curve was determined with each assay to ensure that percent adhesion was within the linear portion of this curve. For concentration curves, 225 μl of 111In-labeled PMNs was incubated with 25 μl of 0, 0.01, 0.10, or 1.00 μg of rC5a per ml for 10 min at 37°C, and adherent cells were quantified as described above. All assays were performed in triplicate or quadruplicate.
Cloning of GBS scpB.
scpB from representative GBS isolates was cloned by PCR. The following primers were designed to amplify full-length or overlapping segments of the scpB promoter and coding regions: S1, 5′ AAAGAATTCGGATAAGGAGGT 3′; AS1, 5′ CCTGCATCTCGAGGAGTTTG 3′; S2, 5′ AACAGCGAATTCTGAGGAAG 3′; AS2, 5′ CTGATAGCTCGAAGCGTAGTT 3′; S3, 5′ GTGTCGGAATTCTAAATGGA 3′; AS3, 5′ GACCGTCTTCTCGAGTGATA 3′; S4, 5′ GGACAAGGAATTCCCGATTG 3′; AS4, 5′ TGCTATTGGCTCGAGTTGTG 3′; S5, 5′ CCTAAAGAATTCTATGAGGCA 3′; AS5, 5′ GCGGACTCGAGAGGTGTAG 3′; S6, 5′ GATGAATTCGGCAAAGTTGT 3′; AS6, 5′ TAAGCTTCTTTTTGGCG 3′; spb2XF, 5′ ATGAAAAAGAAAATGATTCAATCG 3′; and spb2XR, 5′ AGAACGTAAACGACGACGAGC 3′.
Amplification reactions were performed in a 75-μl reaction mixture consisting of 500 ng of genomic DNA, 0.2 mM deoxynucleoside triphosphates, 3 mM MgCl2, 50 pmol of each primer, and 4 U of DNA polymerase (Bio-X-Act; ISC BioExpress) in the manufacturer's supplied buffer. Thirty-five cycles, consisting of denaturation for 1 min at 94°C, annealing for 1.5 min at 48 to 52°C, and extension for 2.5 to 4.0 min at 72°C, were performed. In most cases, amplification products were directly sequenced using an ABI-377 automated sequencer (Perkin-Elmer Corporation, Norwalk, Conn.) at the University of Utah Sequencing Core Facility or at the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital. In some cases, PCR products were cloned into pBSII KS+ (Stratagene, La Jolla, Calif.) or pBADtopo (Invitrogen, Carlsbad, Calif.) vectors and sequenced using vector-specific primers. All sequence differences between GBS strains were confirmed by a minimum of two independent amplification products from independent genomic DNA preparations.
Nucleotide sequence accession numbers.
The nucleotide sequences of the coding regions of I30, I25, and GW scpB are available from GenBank under accession numbers AF189004, AF189003, and AF189002, respectively.
RESULTS
C5a-ase binding by MAb F1.
In Western blots, MAb bound to rC5a-ase and a similarly sized protein in mutanolysin extracts of the C5a-ase+ strains COH-1 and TOH-85 (Fig. 1). SDS-PAGE typically overestimates the size of C5a-ase, which has an apparent molecular mass of 120 to 140 kDa on these gels (6, 7). The corresponding band is absent from strain TOH-97, a scpB deletion mutant of strain COH-1, confirming the specificity of the MAb for C5a-ase.
FIG. 1.
Western blot detection of C5a-ase by MAb F1. Shown is a Western blot of 0.1 μg of rC5a-ase (lane 1) and mutanolysin extracts of strains COH-1 (scpB+; lane 2), TOH-97 (scpB isogenic mutant of strain COH-1; lane 3), and TOH-85 (isogenic mutant of csp; lane 4). Positions of molecular mass standards are shown at the left in kilodaltons.
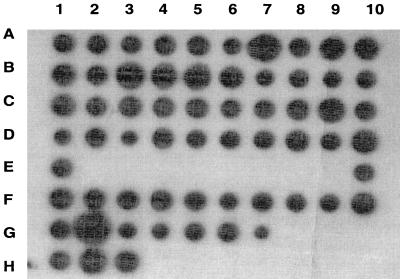
Southern dot blot analysis of genomic GBS DNA.
Genomic DNA from all tested type III GBS strains, including 17 strains with minimal or no functional C5a-ase activity, hybridized with the scpB probe, suggesting that scpB or a homologous gene is present in most type III GBS strains (Fig. 2).
FIG. 2.
Southern dot blot analysis of scpB. Five micrograms of genomic DNA from each of 62 type III GBS strains was hybridized with a full-length scpB probe. A gene homologous to scpB was present in all strains tested, including RDP type III-3b strains with markedly reduced or absent functional C5a-ase activity (wells C8 to D10).
Western blot analysis of C5a-ase expression.
Mutanolysin extracts of C5a-ase+ strain I30 and six C5a-ase− strains, including I25 and GW, were subjected to SDS-PAGE, and C5a-ase was detected with anti-C5a-ase MAb F1 (Fig. 3). This Western blot demonstrated a single band of similar size in mutanolysin extracts from each GBS strain, suggesting that a less functional protein is expressed by strains with limited C5a-ase activity.
FIG. 3.
Western blot of C5a-ase. Mutanolysin extracts from strain I30 (high functional C5a-ase activity; lane 1) and strains with absent or low C5a-ase activity (I25, GW, C39, 62059, I32, C35, and 830097; lanes 2 to 7, respectively) were subjected to SDS-PAGE on a 4 to 15% gel, electroblotted to nitrocellulose membranes, and detected with anti-C5a-ase MAb F1. A band corresponding to the predicted size of C5a-ase is detected in mutanolysin extracts from each GBS strain. Positions of molecular mass standards are indicated in kilodaltons.
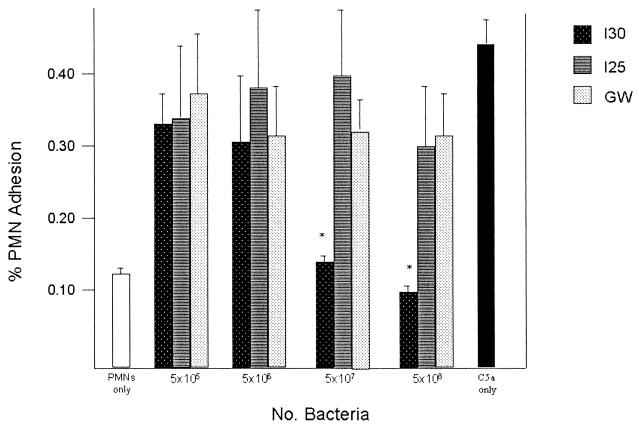
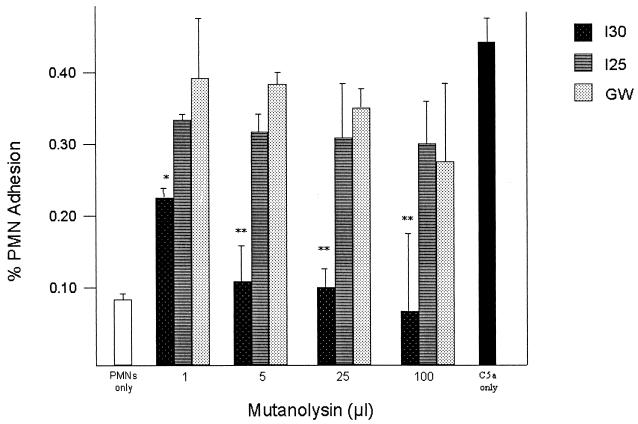
Functional activity of C5a-ase.
We examined the concentration-dependent C5a-ase activity of whole GBS and of cell-free C5a-ase in mutanolysin extracts to determine if the diminished functional C5a-ase activity noted in some type III strains is due to subtle differences in the amount of C5a-ase expressed by some strains of bacteria or limited accessibility of surface-bound C5a-ase. Incubation of rC5a with increasing numbers of whole I30 GBS resulted in decreasing PMN adherence, indicating functional C5a-ase activity, whereas no significant functional C5a-ase activity was detected at even the highest concentration of I25 and GW (Fig. 4). Treatment of rC5a with as little as 1 μl of mutanolysin extract of strain I30 significantly reduced PMN adhesion, whereas even 100 μl of mutanolysin extracts from strains I25 and GW did not significantly reduce C5a activity as measured by stimulation of PMN adhesion (Fig. 5). These data indicate that the deficient C5a-ase activity of strains I25 and GW is not attributable to quantitative differences in functional C5a-ase or differences in accessibility of this protease to C5a.
FIG. 4.
C5a-ase functional activity of GBS strains I30, I25, and GW. Shown is the percent adherence of PMNs in the presence of C5a pretreated with 5 × 106, 5 × 107, or 5 × 108 whole GBS. Asterisks indicate significant differences (P < 0.01) between the functional C5a-ase activity of strain I30 and that of strains I25 and GW. Also shown is the percent adhesion of untreated PMNs (left) and PMNs treated with 100 μg of rC5a per ml (right).
FIG. 5.
C5a-ase functional activity of mutanolysin extracts from GBS strains I30, I25, and GW. Shown is the percent adherence of PMNs exposed to human C5a pretreated with 1, 5, 25, or 100 μl of mutanolysin extract from strain I30, I25, or GW. Asterisks indicate significant differences between the functional C5a-ase activity of strain I30 and that of strains I25 and GW (∗, P = 0.01; ∗∗, P < 0.01). Also shown is the percent adhesion of untreated PMNs (left) and PMNs treated with 100 μg of rC5a per ml (right).
Purified rC5a-ase from I30 and I25 was tested for the ability to inactivate C5a in order to confirm that differences in the functional activity of I30 and I25 C5a-ase are attributable to differences in gene products. At concentrations of 0.1 to 1.0 μg/ml, rI30 C5a-ase had significantly greater C5a-ase functional activity than rI25 C5a-ase (Fig. 6).
FIG. 6.
Functional activity of recombinant I30 and I25 C5a-ase. Shown is the percent adherence of PMNs incubated with human C5a that has been pretreated with 0.01 to 1 μg of I30 or I25 rC5a-ase per ml. Asterisks indicate significant differences (P < 0.01) in functional C5a-ase activity. Also shown is the percent adhesion of untreated PMNs (left) and PMNs treated with 1 μg of rC5a per ml (right).
Nucleotide and predicted amino acid sequences of scpB.
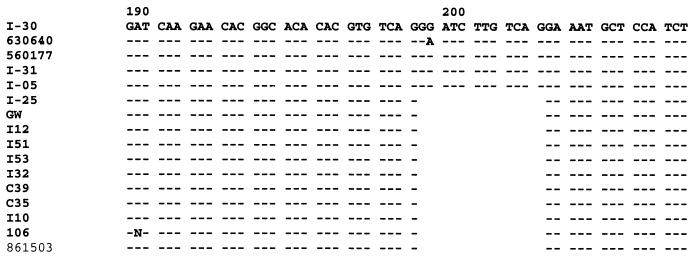
The nucleotide sequences of the coding region of I30 scpB differs from the previously reported serotype II GBS scpB sequence by 62 bases, 59 of which are shared by all three serotype III scpB genes. The predicted amino acid sequences of serotype III scpB genes differ from the serotype II sequence by 34 to 38 amino acid residues (Fig. 7). I25 and GW scpB share a 12-bp in-frame deletion from the I30 sequence that results in a four-amino-acid deletion (Ile200 to Gly203, inclusive). The region flanking this deletion was amplified from an additional four RDP type III-3a strains and nine RDP type III-3b strains. Each of the strains with high C5a-ase functional activity lacked this deletion, whereas the deletion was present in scpB of all strains with low or absent C5a-ase functional activity (Fig. 8).
FIG. 7.
Comparison of the predicted amino acid sequences of the strain I30, I25, and GW scpB to the previously reported GBS C5a-ase amino acid sequence (top row) and the translated sequences of group A streptococcal scpA12 and scpA49 (7, 8). The predicted active sites of the enzyme are indicated by asterisks. A slash indicates the 51-bp deletion present in scpB relative to scpA.
FIG. 8.
Nucleic acid sequences of codons 190 to 207 of the scpB genes from RDP type III-3a (I30, 630640, 560177, I05, and I31) and III-3b (I25, GW, I12, I51, I53, I32, C39, C35, I10, I06, and 861503) strains.
I30 C5a-ase is also closely homologous to the M12 C5a peptidase (95.6% amino acid homology) and, to a lesser degree, M49 C5a peptidase (93.7% amino acid homology) (Fig. 7). A number of differences between the I30 and the type II scpB sequences are shared by the scpA genes, supporting the previous suggestion that it is possible that scpB genes originated from a common scpA12-like precursor (8). All serotype III scpB have the 51-bp deletion previously described for serotype II scpB relative to scpA.
DISCUSSION
We previously described a system for the classification of type III GBS by RDP (20). This classification divides type III GBS into three major genotypic groups on the basis of the similarity of restriction fragment patterns produced by digestion with the restriction enzyme HindIII or Sse83871. RDP type III-3 strains can be further divided into subgroups III-3a and III-3b. This genetic classification correlates with a number of phenotypic features, including capsular sialic acid content and expression of R protein, as well as the geographic origin of isolates (20). Members of RDP type III-3a have high functional C5a-ase activity, whereas members of the RDP type III-3b subgroup have little or no activity. Here we show that in some RDP type III-3b strains a deletion mutation markedly diminishes C5a-ase activity. This finding supports the genetic basis for this functional difference and the validity of our RDP genetic typing system.
Group A S. pyogenes expresses a cell wall-associated peptidase, C5a peptidase, which also inactivates C5a/C5adesarg by cleavage at His67-Lys68 (21). Nucleotide sequences of genes encoding C5a-ase, scpA, and GBS C5a-ase, scpB, that have been previously reported are ≥97% homologous to one other, and a homologous gene is also present in group G streptococci (8, 9). The remarkable homology of these genes is attributed to horizontal transmission between these two species or, alternatively, to evolution from a common ancestor (8). The three type III scpB genes reported here are more closely homologous to one another than to type II scpB and share a number of nucleotide differences with the serotype II sequence, suggesting a clonal origin that parallels that of other virulence factors such as genes responsible for capsular polysaccharide synthesis. The deletion mutation observed in RDP type III-3b scpB is likely to have resulted from an error in replication of the short tandem repeat AGGAA that flanks the deletion and may have been facilitated by the ability of intervening single-stranded DNA to assume a hairpin conformation (Fig. 8). The presence of this deletion in the majority of III-3b strains further supports the clonal origin of these genetically related strains.
C5a-ase is a member of the subtilisin family of serine proteases (7, 18). Catalytic activity of subtilisins results from a charge relay system comprised of Asp, Ser, and His residues. The three residues of C5a peptidase and C5a-ase hypothesized to be critical to serine protease activity include Asp130, His193, and Ser512 (7). Differences in the functional C5a-ase activity of type III GBS strains reported here can be attributed to a mutation altering the His consensus pattern of the triad, H - G - [STM] - x - [VIC] - [STAGC] - [GS] - x - [LIVMA]-[STAGCLV]-[SAGM]. Comparison of proteins included in the PROSITE database demonstrate the His motif is present in 87 of 89 subtilisins reported to date (2). Pasteurella haemolytica A1 Ssa1 and Aeromonas salmonicida AspA both lack the His active site (17, 22). AspA, interestingly, retains proteolytic activity, but whether or not Ssa1 is a protease has not been confirmed.
In addition to the six strains demonstrated here (Fig. 3), we have noted that an additional five GBS strains with minimal or no functional activity also express immunoreactive C5a-ase, suggesting that, in addition to possessing an scpB gene, most or all GBS produce its gene product. It is possible that polymorphisms of scpB are a relatively recent genetic event and that, with time, protein expression will be lost as consequence of additional mutations. A more intriguing alternative possibility, however, is that the ubiquitous expression of GBS C5a-ase is related to a second, as yet unidentified important function of this protein. Many bacterial subtilisins have broad substrate specificity. In contrast, C5a peptidase is highly specific for C5a (10). This is, as previously suggested, consistent with the evolution of a highly specialized mechanism by which some streptococci may avoid prompt detection by phagocytic defenses (10). However, C5a-ase might have, independent of its antichemotactic function, other effects resulting in, for example, a competitive advantage against other bacteria in a particular ecological niche. In Lactococcus lactis, for example, the subtilisin NisP is responsible for cleavage of leader peptide from a specific substrate, the lantibiotic nisin (19).
The ubiquitous expression of GBS C5a-ase and evidence of the importance of C5a peptidase in the pathogenesis of both group A and group B streptococcal infections suggest that C5a-ase may be a good candidate for a vaccine antigen. Immunization of rabbits with a truncated S. pyogenes C5a peptidase elicits neutralizing antibody, and intranasal immunization of mice with this candidate vaccine reduced numbers of bacteria and the duration of nasopharyngeal colonization with wild-type group A S. pyogenes strains in a mouse model (16). The homology between C5a peptidase and GBS C5a-ase suggests that the latter protein may also be immunogenic and potentially protective against colonization or GBS disease. The absence of functional C5a-ase activity would not necessarily limit the efficacy of such a vaccine if C5a-ase is a target for opsonizing antibody or if it has other functions in addition to its established role in limiting neutrophil chemotaxis.
ACKNOWLEDGMENTS
This work was supported by grants AI40918 from the National Institutes of Health, the Primary Children's Medical Center Research Foundation, the University of Utah Undergraduate Research Program (D.V.M.), Cancer Center Support CORE grant P30 CA 21765, and the American Lebanese Syrian Associated Charities (ALSAC). E.E.A. is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Adderson E E, Shackelford P G, Quinn A, Carroll W L. Restricted IgH chain V gene usage in the human antibody response to Haemophilus influenzae type b capsular polysaccharide. J Immunol. 1991;147:1667–1674. [PubMed] [Google Scholar]
- 2.Bairoch A. Prosite. Geneva, Switzerland: Swiss Institute of Bioinformatics; 1999. [Google Scholar]
- 3.Baker C J, Edwards M S. Group B streptococcal infections. In: Remington J, Klein J O, editors. Infectious diseases of the fetus and newborn infant. 4th ed. Philadelphia, Pa: W. B. Saunders; 1995. pp. 980–1054. [Google Scholar]
- 4.Bohnsack J F, Mollison K W, Buko A M, Ashworth J W, Hill H R. Group B streptococci inactivate complement component C5a by enzymic cleavage at the C-terminus. Biochem J. 1991;273:635–640. doi: 10.1042/bj2730635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohnsack J F, Widjaja K, Ghazizadeh S, Rubens C E, Hillyard D, Parker C J, Albertine K H, Hill H R. A role for C5 in the acute neutrophil response to group B streptococcal infections. J Infect Dis. 1997;175:847–855. doi: 10.1086/513981. [DOI] [PubMed] [Google Scholar]
- 6.Bohnsack J F, Zhou X, Williams P A, Cleary P P, Parker C J, Hill H R. Purification of the protease from group B streptococci that inactivates human C5a. Biochim Biophys Acta. 1991;1079:222–228. doi: 10.1016/0167-4838(91)90129-n. [DOI] [PubMed] [Google Scholar]
- 7.Chen C C, Cleary P P. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J Biol Chem. 1989;265:3161–3167. [PubMed] [Google Scholar]
- 8.Chmouryguina I, Surorov A, Ferrieri P, Cleary P P. Conservation of the C5a peptidase genes in group A and B streptococci. Infect Immun. 1996;64:2387–2390. doi: 10.1128/iai.64.7.2387-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary P P, Peterson J, Chen C, Nelson C. Virulent human strains of group G streptococci express a C5a peptidase enzyme similar to that produced by group A streptococci. Infect Immun. 1991;59:2305–2310. doi: 10.1128/iai.59.7.2305-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary P P, Prahbu U, Dale J B, Wexler D E, Handley J. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect Immun. 1992;60:5219–5223. doi: 10.1128/iai.60.12.5219-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez H N, Henson P M, Otani A, Hugli T E. Chemotactic response to human C3a and C5a anaphylatoxins. J Immunol. 1978;120:109–115. [PubMed] [Google Scholar]
- 12.Hemming V G, Hall R T, Rhodes P G, Shigeoka A O, Hill H R. Assessment of group B streptococcal opsonins in human and rabbit serum by neutrophil chemiluminescence. J Clin Investig. 1976;58:1379–1387. doi: 10.1172/JCI108593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill H R, Bohnsack J F, Morris E Z, Augustine N H, Parker C J, Cleary P P, Wu J T. Group B streptococci inhibit the chemotactic activity of the fifth component of complement. J Immunol. 1988;141:3551–3556. [PubMed] [Google Scholar]
- 14.Hugli T E. Structure and function of the anaphylatoxins. Spring Semin Immunopathol. 1984;7:193–219. doi: 10.1007/BF01893020. [DOI] [PubMed] [Google Scholar]
- 15.Jackson L A, Hilsdon R, Farley M M, Harrison L H, Reingold A L, Plikaytis B D, Wenger J D, Schuchat A. Risk factors for group B streptococcal disease in adults. Ann Intern Med. 1995;123:415–420. doi: 10.7326/0003-4819-123-6-199509150-00003. [DOI] [PubMed] [Google Scholar]
- 16.Ji Y, McLandsborough L, Kondagunta A, Cleary P P. C5a-peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64:503–509. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo R Y C, Strathdee C A, Shewen P E, Cooney B J. Molecular studies of Ssa1, a serotype-specific antigen of Pasteurella haemolytica A1. Infect Immun. 1991;59:3398–3406. doi: 10.1128/iai.59.10.3398-3406.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siezen R J, de Vos W M, Leunissen J A, Dijkstra B W. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991;4:719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- 19.Siezen R J, Rollema H S, Kuipers O P, de Vos W M. Homology modelling of the Lactococcus lactis leader peptidase NisP and its interaction with the precursor of the lantibiotic nisin. Protein Eng. 1995;8:117–125. doi: 10.1093/protein/8.2.117. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi S, Adderson E E, Nagano Y, Nagano N, Briesacher M R, Bohnsack J F. Identification of a highly encapsulated, genetically related group of invasive type III group B streptococci. J Infect Dis. 1998;177:1116–1119. doi: 10.1086/517408. [DOI] [PubMed] [Google Scholar]
- 21.Wexler D E, Nelson R C, Cleary P P. Human neutrophil chemotactic response to group A streptococci: bacteria-mediated interference with complement-derived chemotactic factors. Infect Immun. 1983;39:239–246. doi: 10.1128/iai.39.1.239-246.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitby P W, Landon M, Coleman G. The cloning and nucleotide sequence of the serine protease gene (aspA) of Aeromonas salmonicida. FEMS Microbiol Lett. 1992;78:65–71. doi: 10.1016/0378-1097(92)90289-z. [DOI] [PubMed] [Google Scholar]