Abstract
The global uptake of prostate cancer (PCa) active surveillance (AS) is steadily increasing. While prostate-specific antigen density (PSAD) is an important baseline predictor of PCa progression on AS, there is a scarcity of recommendations on its use in follow-up. In particular, the best way of measuring PSAD is unclear. One approach would be to use the baseline gland volume (BGV) as a denominator in all calculations throughout AS (nonadaptive PSAD, PSADNA), while another would be to remeasure gland volume at each new magnetic resonance imaging scan (adaptive PSAD, PSADA). In addition, little is known about the predictive value of serial PSAD in comparison to PSA. We applied a long short-term memory recurrent neural network to an AS cohort of 332 patients and found that serial PSADNA significantly outperformed both PSADA and PSA for follow-up prediction of PCa progression because of its high sensitivity. Importantly, while PSADNA was superior in patients with smaller glands (BGV ≤55 ml), serial PSA was better in men with larger prostates of >55 ml.
Patient summary
Repeat measurements of prostate-specific antigen (PSA) and PSA density (PSAD) are the mainstay of active surveillance in prostate cancer. Our study suggests that in patients with a prostate gland of 55 ml or smaller, PSAD measurements are a better predictor of tumour progression, whereas men with a larger gland may benefit more from PSA monitoring.
Keywords: Prostate cancer, Active surveillance, Prostate-specific antigen, Predictive modelling, Longitudinal data, Artificial intelligence, Recurrent neural networks
The past decade has witnessed a global increase in the uptake of active surveillance (AS) for management of patients with low-risk or favourable intermediate-risk prostate cancer (PCa) [1]. However, there is significant global variation in AS practices between centres and among guidelines [2], with the PRIAS study protocol most commonly used in Europe. Specifically, while some institutions favour protocol-driven biopsies to base their clinical decisions on histological ground truth, others argue for a more personalised approach in which the need for biopsy is guided by multiparametric magnetic resonance imaging (MRI) and prostate-specific antigen (PSA) kinetics [3]. While the latter strategy could indeed improve patient adherence to AS without compromising oncological outcomes, it requires the development of robust, dynamic, risk-adapted predictive models using high-quality multi-institutional data. Although highlighted as the current highest AS research priority [2], clinical translation of such models will require considerable time and resources. In parallel, application of longitudinal predictive modelling methods to existing MRI-driven AS cohorts can offer clinical insights that can shape future translational efforts.
We have encountered several clinical questions in our practice. First, while PSA density (PSAD) is an important baseline predictor of PCa progression on AS [4], [5], there is a scarcity of recommendations on its use during follow-up [2], and specifically on the best way of measuring MRI-derived PSAD. One approach would be to use baseline gland volume (BGV) as the denominator in all calculations throughout AS (nonadaptive PSAD, PSADNA), while another would be to remeasure gland volume whenever a new MRI scan is performed (adaptive PSAD, PSADA). Intuitively, PSADA is the preferred approach given its ability to provide more accurate values with dynamic increases in prostate volume in patients on AS [6]. However, PSADNA is easier to implement in routine clinical practice and there is no evidence regarding its comparative performance to either serial PSADA or PSA alone. In addition, the predictive performance of longitudinal PSA, PSADA, or PSADNA may vary for different BGVs. Specifically, in patients with smaller prostates, even a modest increase in volume may lead to a considerable decrease in PSAD, while this effect would be the opposite in men with larger glands. In this study we tested these hypotheses using machine learning for longitudinal predictive modelling of the risk of PCa progression in patients on AS using serial PSA, PSADA, and PSADNA.
We included 332 patients enrolled on our previously described AS programme [4] between March 2012 and August 2020 in this ethically approved, single-centre study (Health Research Authority and Health and Care Research Wales, IRAS project ID 288,185; Supplementary Fig. 1). Clinical and histopathological characteristics of the study cohort are presented in Table 1. Over median follow-up of 51 mo (interquartile range 35–75) we collected 4508 serial PSA measurements (median 12 per patient) and performed 1362 serial prostate MRI scans (median 4 per patient). BGV for PSADNA and follow-up gland volumes for PSADA were calculated from MRI scans according to Prostate Imaging-Reporting and Data System guidelines [7] using three-plane measurements by four consultant urogenital radiologists with 4–14 yr of prostate MRI reporting experience. A previously described [8] long short-term memory recurrent neural network with leave-one-out cross-validation was applied to the data to generate areas under the receiver operating characteristic curve (AUCs) for predicting PCa progression on AS. Progression was noted in 80/332 patients, defined as either histopathological (biopsy-confirmed International Society of Urological Pathology grade group upgrading) or clear radiological stage progression (PRECISE [9] score of 5). Notably, repeat biopsies were performed at protocol-driven time points or were triggered earlier by a rise in PSA or suspected MRI progression [4]. AUCs were compared using DeLong’s test.
Table 1.
Baseline clinicopathological characteristics of the study cohort a
| Variable | Overall cohort (n = 332) |
Progressors (n = 80) |
Nonprogressors (n = 252) |
p value |
|---|---|---|---|---|
| Median age, yr (IQR) | 66 (61–69) | 66 (62–69) | 66 (61–69) | 0.57 |
| Median PSA, ng/ml (IQR) | 5.6 (4.1–7.8) | 5.8 (4.1–7.7) | 5.5 (4.1–7.9) | 0.59 |
| Median BGV, ml (IQR) | 45.8 (35.4–64.2) | 42.0 (29.8–53.6) | 49.6 (37.0–67.2) | 0.007 |
| Median PSAD, ng/ml/ml (IQR) | 0.12 (0.08–0.17) | 0.14 (0.09–0.22) | 0.11 (0.08–0.16) | 0.003 |
| Median AS follow-up, mo (IQR) | 51.0 (35.0–75.8) | 43.5 (28.5–59.0) | 56.5 (38.0–79.8) | 0.0002 |
| Biopsy ISUP grade 1, n (%) | 220 (66) | 48 (60) | 172 (68) | 0.18 |
| Biopsy ISUP grade 2, n (%) | 112 (34) | 32 (40) | 80 (32) |
BGV = baseline gland volume; IQR = interquartile range; ISUP = International Society of Urological Pathology; PSA = prostate-specific antigen; PSAD = PSA density.
Intergroup comparisons of patient characteristics were performed using the Mann-Whitney U test and Fisher’s exact test, as appropriate.
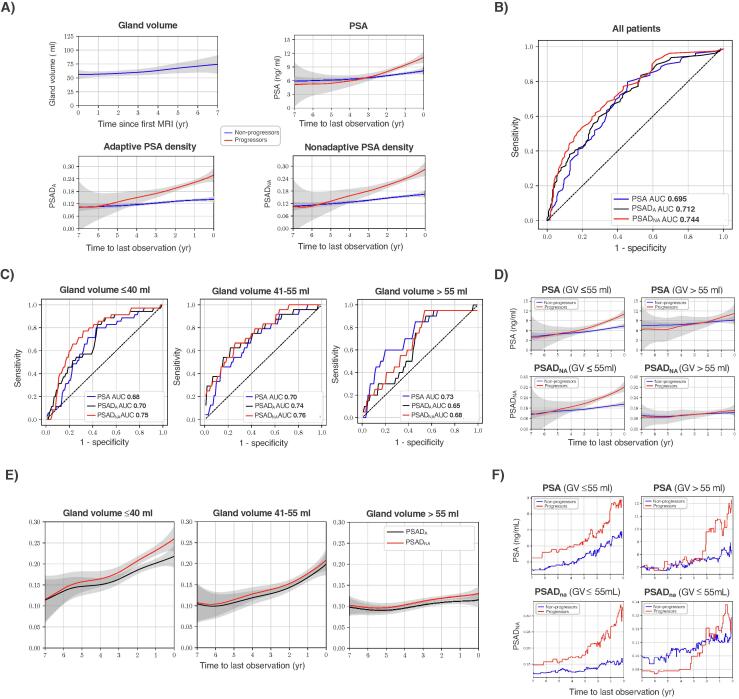
Prostate volume increased over time (Fig. 1A), consistent with previous results [6]. At the cohort level, serial PSADNA significantly outperformed both PSADA and PSA for prediction of PCa progression (p < 0.0001 for all; Fig. 1A,B and Supplementary Table 1). To assess the impact of BGV on biomarker performance, we a priori defined three BGV cutoffs (group A, ≤40 ml; group B, 41–55 ml; group C, >55 ml) to divide the cohort into three groups of similar sample size and distribution of progressors and nonprogressors (Supplementary Table 2). In groups A and B, PSADNA showed significantly better performance in comparison to both PSADA and PSA (p < 0.0001 for all; Fig. 1C and Supplementary Table 3). This can be explained by the higher sensitivity of PSADNA (Fig. 1B,C), which effectively overestimates the “true” PSAD by maximising the impact of increasing PSA with a stable denominator of BGV. Conversely, in group C, PSA significantly outperformed both PSADA and PSADNA (p < 0.0001 for all; Fig. 1C and Supplementary Table 3). This probably reflects the need for a much higher relative increase in PSA to change PSAD values sufficiently to match the more rapidly increasing gland volume in patients with BGV >55 ml (Supplementary Fig. 2). Importantly, the diagnostically superior PSADNA and PSA are easier to use clinically given the inconsistent reporting of follow-up gland volumes as required for calculating PSADA.
Fig. 1.
Use of serial PSA and PSAD for predicting prostate cancer progression on active surveillance. (A) LOWESS curves demonstrating serial changes in prostate GV, PSA, PSADA, and PSADNA for patients with and without progression. (B) ROC curves for serial PSA, PSADA, and PSADNA applied to the whole cohort to assess the ability to predict prostate cancer progression in patients on active surveillance. (C) ROC curves for serial PSA, PSADA, and PSADNA for patients with differing BGV. (D) LOWESS curves demonstrating changes in serial PSA and PSADNA for patients with smaller (≤55 ml) and larger (>55 ml) BGV. (E) LOWESS curves demonstrating the difference between PSADA and PSADNA by BGV. (F) Serial changes in median PSA and PSADNA for patients with smaller (≤55 ml) and larger (>55 ml) BGV. AUC = area under the ROC curve; BGV = baseline GV; GV = gland volume; LOWESS = locally weighted scatterplot smoothing; PSA = prostate-specific antigen; PSAD = PSA density (in ng/ml/ml); PSADA = adaptive PSAD; PSADNA = nonadaptive PSAD; ROC = receiver operating characteristic.
These findings can be visualised in locally weighted scatterplot smoothing curves that show more prominent differences in longitudinal trends for PSA and PSADNA between progressors and nonprogressors in patients with smaller (≤55 ml) and larger (>55 ml) BGV (Fig. 1D). The same trend is evident from plots of dynamic changes in median PSA and PSADNA values (Fig. 1F). For patients with smaller glands, PSADNA grew steadily in progressors and plateaued in nonprogressors, while PSA showed a proportionate increase in both groups until the last year before progression/censorship. This trend was reversed for patients with larger glands: median PSA showed a much clearer relative increase in progressors in comparison to PSADNA. Interestingly, in group A the difference between serial PSADNA and PSADA was considerably larger than in groups B and C, for which the two methods produced similar values (Fig. 1E).
Our study has several limitations, including its single-centre nature, retrospective design, limited sample size, and lack of assessment of inter-reader variability for MRI-derived gland volume measurement (which is generally >0.90 for expert readers [10]). While identifying specific serial PSADNA and PSA cutoffs sufficient to trigger unscheduled MRI or biopsy was beyond the scope of this study, our data provide several observations to be tested in future work. First, regardless of BGV, a consistent increase in PSADNA beyond the median value of 0.18 ng/ml/ml was a characteristic feature of progressors that could be first noted 3 yr before their clinical reclassification (Fig. 1A,D,F). Second, albeit less pronounced, a similar trend was observed for the median PSA value of 9 ng/ml; this was breached approximately 2 yr before clinical progression in patients with BGV of >55 ml (Fig. 1F).
Overall, this study offers three main observations:
-
•
Dynamic monitoring of PSADNA consistently outperformed PSADA in predicting PCa progression on AS both for the whole AS population and in particular for patients with BGV ≤55 ml.
-
•
Patients with BGV >55 ml benefit more from serial PSA monitoring, since PSAD is more stable and less predictive as the volume has a higher denominator value.
-
•
If clinicians prefer more accurate PSAD, they should prioritise measurement of PSADA in patients with BGV of ≤40 ml, for whom the discrepancy with PSADNA is more pronounced.
These results may help in informing both current clinical practice and future multicentre studies to develop personalised AS algorithms using dynamic risk-adapted predictive modelling and incorporating all available clinical data, including serial MRI and biopsy results.
Author contributions: Nikita Sushentsev had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Sushentsev, Zaikin, Blyuss, Barrett.
Acquisition of data: Sushentsev, Colarieti, Sanmugalingam, Stanzione, Zawaideh, Caglic.
Analysis and interpretation of data: Sushentsev, Abrego, Caglic, Zaikin, Blyuss, Barrett.
Drafting of the manuscript: Sushentsev.
Critical revision of the manuscript for important intellectual content: Abrego, Caglic, Zaikin, Blyuss, Barrett.
Statistical analysis: Abrego, Zaikin, Blyuss.
Obtaining funding: Sushentsev, Zaikin, Blyuss, Barrett.
Administrative, technical, or material support: Zaikin, Blyuss, Barrett
Supervision: Zaikin, Blyuss, Barrett.
Other: None.
Financial disclosures: Nikita Sushentsev certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: This research was supported by the National Institute for Health and Care Research (NIHR) Cambridge Biomedical Research Centre (NIHR203312). The study sponsor did not play a role in data collection and analysis. The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care. The authors also acknowledge support from Cancer Research UK (CRUK; Cambridge Imaging Centre grant number C197/A16465; ACED Pilot Award A095792/EICEDAAP\100009), the CRUK National Cancer Imaging Translational Accelerator (C42780/A27066), and the Engineering and Physical Sciences Research Council Imaging Centre in Cambridge and Manchester. Nikita Sushentsev acknowledges support from the Gates Cambridge Trust. Oleg Blyuss acknowledges support from Cancer Research UK and EPSRC joint award EDDCPJT/100022.
Associate Editor: Guillaume Ploussard
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.euros.2023.04.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fletcher S.A., von Landenberg N., Cole A.P., et al. Contemporary national trends in prostate cancer risk profile at diagnosis. Prostate Cancer Prostat Dis. 2020;23:81–87. doi: 10.1038/S41391-019-0157-Y. [DOI] [PubMed] [Google Scholar]
- 2.Moore C.M., King L.E., Withington J., et al. Best current practice and research priorities in active surveillance for prostate cancer—a report of a Movember international consensus meeting. Eur Urol Oncol. 2023 doi: 10.1016/j.euo.2023.01.003. In press. [DOI] [PubMed] [Google Scholar]
- 3.Bangma C.H., Schoots I.G. Magnetic resonance imaging-based monitoring in active surveillance: are we ready to jump on the bandwagon? Eur Urol Open Sci. 2022;38:49. doi: 10.1016/j.euros.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caglic I., Sushentsev N., Gnanapragasam V., et al. MRI-derived PRECISE scores for predicting pathologically-confirmed radiological progression in prostate cancer patients on active surveillance. Eur Radiol. 2020;31:2696–2705. doi: 10.1007/s00330-020-07336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giganti F., Stabile A., Stavrinides V., et al. Natural history of prostate cancer on active surveillance: stratification by MRI using the PRECISE recommendations in a UK cohort. Eur Radiol. 2020;31:1644–1655. doi: 10.1007/s00330-020-07256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stavrinides V., Papageorgiou G., Danks D., et al. Mapping PSA density to outcome of MRI-based active surveillance for prostate cancer through joint longitudinal-survival models. Prostate Cancer Prostat Dis. 2021;24:1028–1031. doi: 10.1038/S41391-021-00373-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turkbey B., Rosenkrantz A.B., Haider M.A., et al. Prostate Imaging Reporting and Data System version 2.1: 2019 update of Prostate Imaging Reporting and Data System version 2. Eur Urol. 2019;76:340–351. doi: 10.1016/j.eururo.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 8.Sushentsev N., Rundo L., Abrego L., et al. Time series radiomics for the prediction of prostate cancer progression in patients on active surveillance. Eur Radiol. 2023 doi: 10.1007/S00330-023-09438-X. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore C.M., Giganti F., Albertsen P., et al. Reporting magnetic resonance imaging in men on active surveillance for prostate cancer: the PRECISE recommendations—a report of a European School of Oncology task force. Eur Urol. 2017;71:648–655. doi: 10.1016/j.eururo.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Wasserman N.F., Niendorf E., Spilseth B. Measurement of prostate volume with MRI (a guide for the perplexed): biproximate method with analysis of precision and accuracy. Sci Rep. 2020;10:575. doi: 10.1038/s41598-019-57046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.