Key Points
-
•
CH can be confined to specific bone marrow spaces and may be eliminated through surgical extraction.
-
•
Patients with osteoarthritis have a high prevalence of CH, involving genes encoding epigenetic modifiers and DNA damage repair proteins.
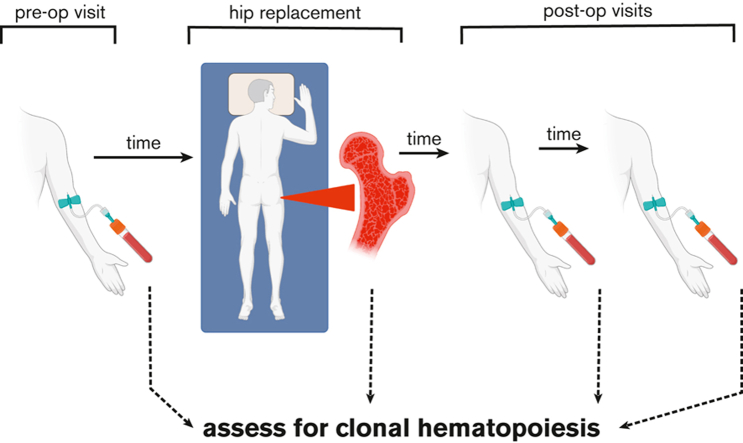
Visual Abstract
Abstract
Clonal hematopoiesis (CH) represents clonal expansion of mutated hematopoietic stem cells detectable in the peripheral blood or bone marrow through next generation sequencing. The current prevailing model posits that CH mutations detected in the peripheral blood mirror bone marrow mutations with clones widely disseminated across hematopoietic compartments. We sought to test the hypothesis that all clones are disseminated throughout hematopoietic tissues by comparing CH in hip vs peripheral blood specimens collected at the time of hip replacement surgery. Here, we show that patients with osteoarthritis have a high prevalence of CH, which involve genes encoding epigenetic modifiers and DNA damage repair pathway proteins. Importantly, we illustrate that CH, including clones with variant allele frequencies >10%, can be confined to specific bone marrow spaces and may be eliminated through surgical excision. Future work will define whether clones with somatic mutations in particular genes or clonal fractions of certain sizes are either more likely to be localized or are slower to disseminate into the peripheral blood and other bony sites.
Introduction
Next generation sequencing advancements have led to an increase in the identification of clonal hematopoiesis (CH) in older individuals having CH by the age of 70 years, by up to 10% to 25%.1, 2, 3, 4, 5, 6 CH is associated with hematologic malignancies,1, 2, 3,7 cardiovascular disease,8 and inflammatory conditions, including inflammatory bowel disease and vasculitis.9, 10, 11, 12 CH was recently added to the World Health Organization Classification of Haematolymphoid Tumours.6 CH mutations originate in hematopoietic stem and progenitor cells (HSPCs) in the bone marrow (BM), which differentiate into mature circulating blood cells.13 Accordingly, the prevailing model posits that mutations detected in the peripheral blood (PB) reflect mutations found in the BM, and clones are widely disseminated across BM compartments simultaneously with the PB.1, 2, 3,14 We sought to test the hypothesis that all clones are disseminated throughout hematopoietic tissues by comparing CH in hip specimens collected at hip replacement surgery with CH found in PB.
Methods
Patients with planned hip replacement surgeries at the University of Chicago Medical Center from April 2018 to March 2021 gave written informed consent to participate in an institutional review board–approved study. Detailed methods are provided in the supplemental Materials. Briefly, PB samples were collected preoperatively. At surgery, femoral heads were bisected and hematopoietic cells were collected by rocking the bones in media for several hours. DNA was extracted from red blood cell–lysed cells, and libraries containing unique molecular identifiers allowing for error correction were prepared.15 DNA libraries were sequenced to ∼2000x mean overall depth of coverage and analyzed as described previously.4,15 Variants were filtered to eliminate those with low quality, recurrent artifacts, or those detected in NA12878 reference DNA. The resulting call set was enriched in somatic mutations using population allele frequency data (dbSNP v153 or ExAC adjusted population allele frequency ≤0.25%) to exclude likely inherited variants. Finally, variants that met all filtering criteria and those with variant allele fraction (VAF) >1% and <35% were reported.4 Variants with VAF >35% were included if reported previously on COSMIC v90 database or at least once in The Cancer Genome Atlas Acute Myeloid Leukemia study.16,17 To track variants in longitudinal or paired samples, variants with VAF <1% were included provided they were detected in at least 1 paired or longitudinal sample passing all filtering criteria and above VAF >1%. Manual revision of read piles was performed in cases showing discrepancies among compartments after force calling.
Results and discussion
Preoperative diagnoses were osteoarthritis (n = 52) and avascular necrosis after chemotherapy for acute lymphoblastic leukemia (n = 1). The median age was 65 years (range, 26-86 years), and 58% were females (supplemental Table 1). Preoperative PB samples of all patients were collected, but some hip samples were not collected because of logistical issues. Paired preoperative PB and BM samples were available for 26 patients (49%). At least 1 postoperative PB sample was collected from 14 patients (26%); of those, 4 had serial longitudinal samples collected between 1 and 17 months after surgery. Patient 2 had BM cells collected from both hips during surgeries that were performed 14 months apart (supplemental Figure 1). CH testing was performed on 102 pre- and postoperative PB and BM samples (supplemental Table 1).
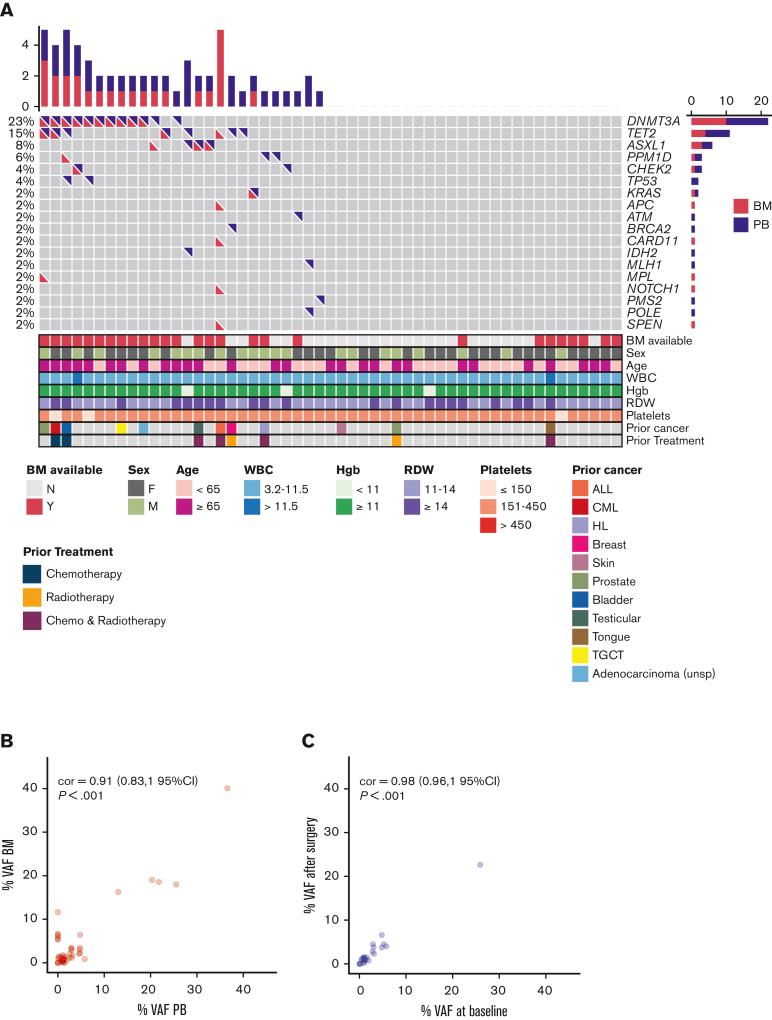
CH was detected in either the preoperative PB or the BM sample in 47% (25/53) of our patients. (Figure 1A). The top mutated genes were DNMT3A, TET2, and ASXL1 (76%, 19/25).1, 2, 3 Mutations in genes involved in DNA damage repair, including ATM, CHEK2, TP53, and PPM1D were found more frequently (25%) than expected when compared with both healthy (∼5%) and cancer populations (∼12.5%)4,5,18, 19, 20 (Figure 1A). Moreover, in contrast to earlier studies, 44% (11/25) of our patients with CH had multiple comutations in the same sample.1, 2, 3 The mean VAF in the preoperative PB or BM was 6.76% (range, 1-40), with no differences in VAF between paired PB and BM (Figure 1B). The mean VAF for DNMT3A, TET2, and ASXL1 genes were higher than DNA damage repair or other genes, 8.2% vs 1.5% vs 5.4%, respectively (P = .0035). The mean corpuscular volume was higher in patients with CH (P = .037) (supplemental Figure 2). Patients with CH appeared to be more likely to have a history of prior malignancy (P = .053) (supplemental Figure 3).
Figure 1.
Spectrum of mutations in the pre- and postoperative blood and BM sample of patients with CH. (A) Oncoprint for all study participants based on preoperative PB and BM findings. Each row represents a gene, and each column corresponds to a study participant. Bar plots indicate the number of mutations per patient (top bar plot) and the number of patients with mutations in each gene (side bar plot). For each patient, bottom panels indicate age, sex, prior cancer history, prior exposure to chemotherapy and/or radiation, and blood parameters. (B) VAFs of variants found in the BM are plotted against VAFs of variants found in the corresponding preoperative PB. Most patients showed concordance between VAFs in the BM and VAFs in the preoperative PB. (C) VAFs of variants found in the postoperative PB are plotted against VAFs found on the preoperative baseline PB sample. VAFs in the baseline sample correlated well with postoperative VAFs in follow-up PB samples. Pearson correlation is shown (95% confidence interval) for panels B and C. ALL, acute lymphoblastic leukemia; CML, chronic myeloid leukemia; cor, correlation; Hgb, hemoglobin; HL, Hodgkin lymphoma; RDW, red cell distribution width; TGCT, tenosynovial giant cell tumor; WBC, white blood cells.
Although we observed CH concurrently in preoperative PB and BM for most patients (n = 15/18) (Figure 1B), we noted highly discordant results in 4 patients (Table 1; Figure 1A). Importantly, 6 clones from 2 patients were detected in the BM that were not identified in the corresponding preoperative PB samples (Table 1). If clones were widely disseminated, we would have expected to see these variants in the PB based on their presence in the BM. Our observation indicates that some clones are secluded within the BM space and fail to disseminate into the PB. In another patient, an ATM variant, found with a VAF of 1.54% in a preoperative PB sample, was not detected in the corresponding BM sample (Table 1).
Table 1.
Discordant clones in the PB vs BM
| Patient | Variant | VAF in PB (%) | VAF in BM (%) | VAF in PB 6-12 mo (%) |
VAF in PB 12-17 mo (%) |
|---|---|---|---|---|---|
| 31 | CHEK2 Ile459AsnfsTer7 |
3.04 | 1.15 | 3.79 | 3.62 |
| 31 | DNMT3A Phe731Ile |
Undetectable | 0.07 | Undetectable | Undetectable |
| 31 | DNMT3A Cys517Ter |
3.11 | 1.78 | 2.29 | 3.95 |
| 31 | TET2 His1717IlefsTer3 |
0.88 | 0.51 | 0.19 | 0.12 |
| 31 | TP53 Arg282Trp |
1.97 | 0.62 | 0.75 | 1.49 |
| 35 | ASXL1 Cys193Arg |
0.13 | 1.38 | Undetectable | No sample |
| 35 | ASXL1 Ser207Gly |
0.10 | 1.05 | Undetectable | No sample |
| 35 | DNMT3A splice site | 5.78 | 0.87 | 3.99 | No sample |
| 42 | ATM Thr306Arg |
1.54 | Undetectable | No sample | No sample |
| 43 | APC Ser1126Arg |
Undetectable | 1.16 | No sample | No sample |
| 43 | CARD11 Thr532Met |
Undetectable | 5.51 | No sample | No sample |
| 43 | NOTCH1 Arg1263His |
Undetectable | 6.38 | No sample | No sample |
| 43 | SPEN Arg2305_Ala2306delinsSer |
Undetectable | 5.99 | No sample | No sample |
| 43 | TET2 Gly1606Ala |
Undetectable | 5.30 | No sample | No sample |
Our postoperative longitudinal PB samples addressed several additional questions. First, in patient 31, having observed that 1 DNMT3A-mutated clone [Phe731Ile] was present in the BM but was not widely disseminated into the PB at the time of surgery (Table 1), we asked whether the hip replacement was sufficient to remove the mutated HSPCs. Although most clones found concurrently in the BM and PB (eg, CHEK2, TET2, TP53, and DNMT3A Cys517Ter) persisted in the PB after surgery, the DNMT3A Phe731Ile clone (present in the BM) was not detected as distantly as 17 months after surgery, indicating that this clone was most likely confined spatially to the resected hip. Second, in patient 35, 2 ASXL1 variants (Cys193Arg and Ser207Gly) were present in both the preoperative PB and BM. However, both disappeared from the PB at 8 months after surgery, suggesting that these clones were likely confined to the resected hip (Table 1). Despite high sequencing depth, PB clones could be missed if they are below the technical detection limit, whereas expanded BM clones could have microenvironment advantages.
Remarkably, clone dissemination did not appear to be related to the size of the clone, with large clones with VAFs of up to 11% remaining confined locally to BM (Table 1). In patient 2 who underwent bilateral hip replacements 14 months apart, a small (VAF 1%) KRAS-mutated clone (Gly12Ser) was detected in both hips. Our cohort showed a surprisingly high prevalence of CH (47%) consistent with a prior osteoarthritis report.21 Whether CH clones directly contribute to or result from chronic inflammation in osteoarthritis, is unknown.22
Leukemias are disseminated throughout the marrow spaces at diagnosis. 14,23,24 Our data question the prevailing model that CH, a leukemia precursor state, is disseminated throughout the marrow spaces and into the PB. We propose that CH in its earliest stage could be confined to specific spatial spaces in the BM. In our patients, the size of the clone did not seem to affect the likelihood of dissemination. Furthermore, patient 43 had undergone an allogeneic stem cell transplant years before our study for treatment of acute lymphoblastic leukemia. It remains unclear whether this contributed to the confinement of the patient’s CH to the BM. The nondissemination of some clones could result from spatial restriction to BM compartments because of differential microenvironment, competing clones, or regression owing to reduced inflammation after surgery. Future work will define what determines dissemination and rates of dissemination, whether clonal fractions make dissemination more/less likely, whether certain mutations/conditions in HSPCs do not contribute to terminal hematopoiesis, and whether surgical excision can eliminate clones.
Conflict-of-interest disclosure: P.D. and D.C.H. have a patent for CH testing “Methods for early detection of AML”. The remaining authors declare no competing financial interests.
Acknowledgments
The authors thank the patients for their participation in this research. This work was supported by a Team Science award from the University of Chicago Comprehensive Cancer Center (B.J. and L.A.G.), R21AG066552 (M.L.G.), and R01 CA248747-01A1 (P.D.).
Authorship
Contribution: A.E.G.O., M.L.G., P.D., and L.A.G. designed the study; A.E.G.O., C.S., L.M., A.K., G.H., M.P., K.S., N.D., and B.J. contributed to sample collection; N.M.-T. supervised DNA sequencing and performed data analyses; A.E.G.O. and N.M.-T. generated all tables and figures; A.E.G.O., N.M.-T., M.L.G., P.D., L.A.G. wrote and edited the manuscript; and all authors approved the final submission.
Footnotes
∗A.E.G.O. and N.M.-T. contributed equally to this work.
Data are available on request from the corresponding author, Lucy A. Godley (lgodley@medicine.bsd.uchicago.edu).
The full-text version of this article contains a data supplement.
Contributor Information
Monica L. Guzman, Email: mlg2007@med.cornell.edu.
Pinkal Desai, Email: pid9006@med.cornell.edu.
Lucy A. Godley, Email: lgodley@medicine.bsd.uchicago.edu.
Supplementary Material
References
- 1.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai P, Mencia-Trinchant N, Savenkov O, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24(7):1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoury JD, Solary E, Abla O, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–1719. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niroula A, Sekar A, Murakami MA, et al. Distinction of lymphoid and myeloid clonal hematopoiesis. Nat Med. 2021;27(11):1921–1927. doi: 10.1038/s41591-021-01521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bick AG, Popadin K, Thorball CW, et al. Increased CHIP prevalence amongst people living with HIV. medRxiv. 2020;12(1):577. doi: 10.1038/s41598-021-04308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young NS, Ogawa S. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373(17):1675–1676. doi: 10.1056/NEJMc1509703. [DOI] [PubMed] [Google Scholar]
- 11.Arends CM, Weiss M, Christen F, et al. Clonal hematopoiesis in patients with anti-neutrophil cytoplasmic antibody-associated vasculitis. Haematologica. 2020;105(6):e264–e267. doi: 10.3324/haematol.2019.223305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang CRC, Nix D, Gregory M, et al. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp Hematol. 2019;80:36–41.e33. doi: 10.1016/j.exphem.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arends CM, Galan-Sousa J, Hoyer K, et al. Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia. 2018;32(9):1908–1919. doi: 10.1038/s41375-018-0047-7. [DOI] [PubMed] [Google Scholar]
- 14.Whiteley AE, Price TT, Cantelli G, Sipkins DA. Leukaemia: a model metastatic disease. Nat Rev Cancer. 2021;21(7):461–475. doi: 10.1038/s41568-021-00355-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A, Mencia-Trinchant N, Griffiths EA, et al. Mutant PPM1D- and TP53-Driven hematopoiesis populates the hematopoietic compartment in response to peptide receptor radionuclide therapy. JCO Precis Oncol. 2022;6:e2100309. doi: 10.1200/PO.21.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52(11):1219–1226. doi: 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabre MA, de Almeida JG, Fiorillo E, et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature. 2022;606(7913):335–342. doi: 10.1038/s41586-022-04785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecker JS, Hartmann L, Riviere J, et al. CHIP & HIPs: clonal hematopoiesis is common in hip arthroplasty patients and associates with autoimmune disease. Blood. 2021 doi: 10.1182/blood.2020010163. [DOI] [PubMed] [Google Scholar]
- 22.Robinson WH, Lepus CM, Wang Q, et al. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2016;12(10):580–592. doi: 10.1038/nrrheum.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas F, Michaels PD, Wang D, Kim AS. Mutational analysis of hematologic neoplasms in 164 paired peripheral blood and bone marrow samples by next-generation sequencing. Blood Adv. 2020;4(18):4362–4365. doi: 10.1182/bloodadvances.2020002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohamedali AM, Gaken J, Ahmed M, et al. High concordance of genomic and cytogenetic aberrations between peripheral blood and bone marrow in myelodysplastic syndrome (MDS) Leukemia. 2015;29(9):1928–1938. doi: 10.1038/leu.2015.110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.