Abstract
We generated a genetically heterogenous rat model by a 4-way cross strategy using 4 inbred strains (Brown Norway [BN], Fischer 344 [F344], Lewis [LEW], and Wistar Kyoto [KY]) to provide investigators with a highly genetically diverse rat model from commercially available inbred rats. We made reciprocal crosses between males and females from the 2 F1 hybrids to generate genetically heterogeneous rats with mitochondrial genomes from either the BN (OKC-HETB, a.k.a “B” genotype) or WKY (OKC-HETW a.k.a “W” genotype) parental strains. These two mitochondrial genomes differ at 94 nucleotides, more akin to human mitochondrial genome diversity than that available in classical laboratory mouse strains. Body weights of the B and W genotypes were similar. However, mitochondrial genotype antagonistically affected grip strength and treadmill endurance in females only. In addition, mitochondrial genotype significantly affected multiple responses to a high-fat diet (HFD) and treatment with 17α-estradiol. Contrary to findings in mice in which males only are affected by 17α-estradiol supplementation, female rats fed a HFD beneficially responded to 17α-estradiol treatment as evidenced by declines in body mass, adiposity, and liver mass. Male rats, by contrast, differed in a mitochondrial genotype-specific manner, with only B males responding to 17α-estradiol treatment. Mitochondrial genotype and sex differences were also observed in features of brain-specific antioxidant response to a HFD and 17α-estradiol as shown by hippocampal levels of Sod2 acetylation, JNK, and FoxO3a. These results emphasize the importance of mitochondrial genotype in assessing responses to putative interventions in aging processes.
Keywords: Aging, Estrogen, Genetic diversity, Mitochondria, Physical function, Sex differences
Largely because of its genetic tractability and short life span, the mouse has become the mainstay of mammalian biomedical research focused on elucidating mechanisms of aging and identifying putative anti-aging interventions. Because the translatability of effective pharmacological interventions from mice to humans has been weak (eg, cancer (1), Alzheimer’s disease (2), and amyotrophic lateral sclerosis (3)), it is critical to assess the generality of mouse findings with respect to disease-modifying interventions. One way to assess generality is to employ a second, experimentally tractable species to confirm or dispute the initial result, especially if the second species displays greater similarity to humans with a specific disease phenotype. Previously, we argued that the laboratory rat would be a useful animal model to test the translatability of observations in mice to humans (4). Rats diverged from a common ancestor at about the same time that humans diverged from African monkeys and differ from mice in multiple physiological parameters important in aging (eg, insulin sensitivity, muscle and bone biology, cognitive capacity, end-of-life pathology, and sex differences in longevity). In each of these cases, rats are more similar to humans than mice. Also, despite their larger size, which is advantageous for a variety of reasons, rats generally do not live longer than mice.
Because inbred rodents are genetically homozygous at every locus, they show strain-specific idiosyncrasies, some of which are known, others of which may not be. For example, it is well-established that multiple inbred mouse genotypes have lost the ability to synthesize melatonin in their pineal glands (5). Some strains also show idiosyncrasies in the primary pathologies of aging or cause of death (6). A major improvement in mouse models to study aging was the development of the UM-HET3 model, which is a controlled, genetically heterogeneous stock in which each mouse is genetically unique but the population can be replicated at will (7).
We describe below the development of a similar genetically heterogenous rat model (OKC-HETB/W), which allows investigators to test the translatability of data from mice to another species or to compare the translatability of results from a second rodent species to humans. Our rat model is generated using a 4-way cross strategy, which was initially used by Roderick (8) in producing a population of genetically heterogenous mice to identify mouse strains that show different sensitivities to X-irradiation. Miller and colleagues used a similar strategy to develop UM-HET3 mice, described above, which are now widely used in aging research, including the NIA Intervention Testing Program (9). The advantage of a 4-way cross is that anyone can generate a similar population by obtaining the parental strains from commercial vendors. Although the UM-HET3 mice are genetically heterogenous with respect to nuclear encoded alleles, the breeding scheme was designed in a manner that leads to all pups having the same BALB/c mitochondrial genome.
It is important to note that the interaction between mitochondrial and nuclear genomes is an underappreciated intracellular dynamic. Mitochondrial function depends on more than 1,200 proteins synthesized in the nucleus, but only thirteen produced from the mitochondria genome. These 13, however, are important in metabolism as they form critical components of 4 of the 5 electron transport chain complexes. Thus, coordination of transcription, translation, and intracellular transport of mito-nuclear proteins must be tightly regulated under a range of cellular conditions that can alter the dynamics of mitochondrial biogenesis or mitophagy (10). Illustratively, two mouse mitochondria that differ at only a handful of nucleotides can alter nuclear gene expression, bioenergetic and organismal metabolic efficiency, as well as cardiac function, and adiposity when paired with the same nuclear genome (11,12). Another unique feature of mitochondrial nuclear dynamics is that because mitochondria are transmitted intergenerationally only through the female lineage, selection will be stronger for optimizing female mito-nuclear coordination relative to males, in which any male-specific deleterious mutations will have minimal evolutionary consequence. This sex-specific mito-nuclear inheritance dynamic has been adduced to explain sex differences in longevity and later life diseases (13) and in Drosophila has been shown to affect sex-specific aging phenotypes (14).
The OKC-HETB/W rat model we describe below was generated by crossing 4 commercially available inbred strains deliberately selected to maximize genetic diversity (15,16). Traditional rat strains, because they represent multiple, disparate domestication events, are genetically far more diverse than traditional mouse strains used in research (4). Specifically, we used Brown Norway (BN), Fischer 344 (F344), Lewis (LEW), and Wistar Kyoto (WKY) inbred rats to produce 2 F1 lines [(BN/F344)F1 and (WKY/LEW)F1]. By performing reciprocal crosses between males and females from these 2 F1 hybrids, 2 F2 stocks (OKC-HETB and OKC-HETW) are created with all the advantages of typical 4-way cross populations but also with half of the progeny having the BN (henceforth “B”) mitochondrial genotype and the other half the WKY (henceforth “W”) mitochondrial genotype. These mitochondrial genomes, again because of the domestication of rats from multiple wild populations, are far more diverse than mitochondrial genomes from traditional inbred mouse strains, which have a single mitochondrial ancestor and thus differ at only a handful of nucleotides (17). Below, we describe an initial characterization of the OKC-HETB/W rats. In addition to assessing their baseline phenotypes, we also challenged them metabolically with a high-fat diet (HFD) to investigate whether such metabolic stress might result in different responses depending on mitochondrial genotype. We also investigated the generality of sex differences previously observed in mice by supplementing the rats’ diet with 17α-estradiol (17α-E2), an intervention demonstrated to have beneficial effects almost exclusively in male mice (18–29).
Method
Animals and Diets
Brown Norway (BN), Fischer 344 (F344), Lewis (LEW), and Wistar Kyoto (WKY) rats were obtained from Charles River at approximately 2 months of age. BN females were bred to F344 males to produce BN/F344F1 hybrids, and WKY females were bred to LEW males to produce WKY/LEWF1 hybrids. The female BN/F344F1’s were crossed with male WKY/LEWF1’s to generate the OKC-HETB stock, and male BN/F344F1’s were crossed with female WKY/LEWF1’s rats to generate the OKC-HETW stock. We found that litter size differed between the 2 stocks: WKY/LEWF1 females produced a mean litter size of 17 pups (range: 14–20), whereas BN/F344F1 females produced a mean litter size of 11 pups (range: 9–12). The rats were bred and maintained in the animal facilities at the Oklahoma City VA Medical Center and fed ad libitum a commercial rodent chow diet (Picolab Rodent Diet 5053, LabDiet, St. Louis, MO). Animals were group housed with the number per cage dependent on the size of the rats. Once the rats reached 15 months of age males were housed 1 or 2 per cage and the females were housed 2–3 per cage. Their environment was enriched with nesting materials, plastic lodges, and wooden chew sticks. In the HFD experiments, control rats were fed normal chow diet (58YP: 66.6% carbohydrates, 20.4% protein and 13% fat; TestDiet, IN) or a HFD (58V8: 35.5% carbohydrate, 18.3% protein, 45.7% fat; TestDiet, IN) with, or without, 14.4 ppm 17α-estradiol (Steraloids, Newport, RI). All experiments were performed in accordance with the National Institutes of Health’s guidelines and approved by the Institutional Animal Care and Use Committee at the Oklahoma City VA Medical Center.
Sequencing of Mitochondrial Genomes
DNA isolation
Total DNA was extracted from frozen quadriceps muscles using proteinase K digestion with SDS and EDTA, phenol/chloroform extraction, and ammonium acetate/ethanol precipitation as previously described (30).
Cas9 cleavage, library preparation, and nanopore sequencing
We used a tiling approach for nanopore re-sequencing of rat mtDNA. Custom guide RNA sequences targeting rat mitochondrial DNA (mtDNA) are listed in Table 1 and purchased from IDT-DNA.
Table 1.
RNA Primer Sequences Targeting Mitochondrial Genome
| Probe | Location of 5-prime End* | crRNA Sequence | Sense |
|---|---|---|---|
| Probe-1 | 469 | ACGATAGCTAAGACCCAAAC | Positive |
| Probe-2 | 4444 | TATTCATCAATTGCCCACAT | Positive |
| Probe-3 | 8917 | GTTCCTACCCACGACCTAGG | Positive |
| Probe-4 | 14370 | ATCCGATACCTACACGCCAA | Positive |
*Based on Brown-Norway mitochondrial DNA GenBank reference sequence AY172581.1.
Pre-complexed Alt-R CRISPR-Cas9 single guide RNA (sgRNA) (IDT) was diluted to a concentration of 10 µM and combined with HiFi Cas9 Nuclease V3 (IDT, cat 1081060) in CutSmart Buffer (NEB, cat B7204). Cas9 cleavage, library preparation, and nanopore sequencing was performed in accordance with previously described methods (31). Prepared libraries were run on a MinION flow cell (v9.4.1) using the MinION Mk1C sequencer. Sequencing and base-calling were done using the MinKNOW software (v21.10.8).
Sequence analysis
The analysis was done using python version 3.6.15 and R version 4.0.2. The reference mitochondrial genomes for the Brown-Norway strain was AY172581.1, and the WKY strain was DQ673907.1. Base calling was performed using MinKNOW (v4.5.4, release 4/20/21), and reads meeting a QC threshold of 5 were included in analysis. All fastqs passing quality control were aligned to the rotated mitochondrial genome using Minimap2, version 2.24. All nanopore sequencing data are available at BioProject PRJNA899912.
PCR-restriction length polymorphism to genotype BN and WKY DNA samples
To verify the mitochondrial DNA genotype in rat samples, we developed a polymerase chain reaction (PCR)-restriction length polymorphism screen. A 614-bp portion of the rat mtDNA minor arc from 4902 bp to 5515 bp flanking a polymorphism at position 5269 (BN G to WKY C numbering according to BN rat reference mtDNA AY172581.1) was PCR amplified using forward primer AAGTACCCTTACCCTACCGC (bp 4902-4922) and reverse primer sequence TTACGAATGCATGGGCTGTGA (bp 5515-5494; IDT DNA). Restriction enzyme digestion of the PCR amplicon was used to discriminate between the BN and WKY rat strains as shown in Supplementary Figure 1. Five microliters of each PCR reaction was prepared in 3 separate restriction digestion reactions with no restriction enzyme, BanII (Cat no.: R0119S to cut the BN sequence), or HaeIII (Cat no.: R0108S to cut the WKY sequence).
Grip Strength and Treadmill Performance
Rats from the Oklahoma VA Medical Center were sent to the University of Alabama at Birmingham at 2 to 3 months of age for performance-related outcomes. After habituation to their new environments, grip strength and treadmill performance was measured. Animals were also habituated and trained prior to the treadmill performance task using a Five Lane Touchscreen Convertible Treadmill (Panlab, Holliston, PA). On the first day of habituation, the rats were placed on the treadmill for 10 minutes. On the second through fifth day, the treadmill was set to 8 cm/s and then increased by 1 cm/s until reaching 12 cm/s (5 minutes) to train them to run a moderate speed. On the sixth day, the rats were placed on the treadmill apparatus at 12 cm/s for 2 minutes, after which the speed increased by 2 cm/s until 16 cm/s max speed was achieved and thereafter, animals were allowed to run at this speed. When rats stopped moving, they were prodded with a stylus 3 times. If they did not respond by the third prodding, they were considered to have reached their endurance limit.
Grip strength was tested using a Chatillon Grip Strength apparatus (Columbus Instruments; Columbus, OH). Briefly, rats were held by the tail and allowed to place their forelimbs on the mesh grid. Once they gripped the mesh, they were firmly pulled away from the machine until their grip was broken. This was repeated for a total of 3 trials. Maximum grip strength measurement was then corrected for body weight. The same experimenter performed all grip strength testing for all rats and was blinded with respect to the mitochondrial genotype.
Assays of Mitochondrial Function
Preparation of skeletal muscle fiber permeabilization was performed as described previously with minor modifications (32). A small piece (~3–5 mg) of red gastrocnemius muscle was dissected, and the separated fibers placed in ice-cold buffer X, containing (in mM): 7.23 K2ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid) (EGTA), 2.77 CaK2EGTA, 20 imidazole, 0.5 dithiothreitol (DTT), 20 taurine, 5.7 ATP, 14.3 PCr, 6.56 MgCl2-6H2O, 50 K-2-(N-morpholino)ethanesulfonic acid (MES) (pH 7.1). The muscle bundle was permeabilized in saponin-containing solution (30 µg/mL) for 30 minutes, followed by three 5-minute washes in ice-cold wash buffer Z containing (in mM): 105 K-MES, 30 KCl, 10 K2HPO4, 5 MgCl2-6H2O, 0.5 mg/mL bovine serum albumin, 0.1 EGTA (pH 7.1). Rates of mitochondrial respiration were determined using the Oxygraph-2k (O2k, OROBOROS Instruments, Innsbruck, Austria). We sequentially added substrates and inhibitors to measure complex-specific respirations as follows: glutamate (10 mM), malate (2 mM), ADP (5 mM), succinate (10 mM), rotenone (1 µM), antimycin A (1 µM), and TMPD (0.5 mM) immediately followed by ascorbate (5 mM). All respiration measurements were normalized to antimycin A to account for non-mitochondrial oxygen consumption. Data for OCR were normalized by milligrams of muscle bundle wet weight.
Western Blot Assays
Brains were rapidly extracted from the skull following decapitation, hippocampi dissected out, and the tissue flash frozen in liquid nitrogen. Lysates from hippocampi were prepared in RIPA buffer containing protease inhibitors (Roche cOmplete Protease Inhibitor Cocktail) and protein concentration determined using the Pierce BCA assay (Thermo Fisher, Waltham, MA). Equal amounts of protein (10 µg/lane) were subjected to SDS/PAGE and subsequently transferred to a nitrocellulose membrane. The membrane was blocked in a solution containing 5% bovine serine albumin in TBST (20 mM Tris, 0.9% NaCl, 0.1% Tween-20, pH 7.4) and probed with primary antibodies for Sod2 (Abcam, Waltham, MA, cat# ab13533, 1:5 000), Sod2-acetyl-K68 (Abcam, cat# ab137037, 1:5 000), Sod1 (Abcam, cat# ab51254, 1:1 000), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Cell Signaling, cat# 97166, 1:2 000), JNK (Cell Signaling, cat# 9252, 1:1 000), p-JNK (Cell Signaling, cat# 4668, 1:1 000), FoxO3a (Cell Signaling, cat# 2497, 1:1 000), and p-FoxO3a (Cell Signaling, cat# 9466, 1:1 000). Following secondary incubation with IRDye donkey anti-mouse and donkey anti-rabbit antibodies (LI-COR Biosciences, Lincoln, NE, 926-68072 and 926-32213, 1:5 000), images were captured using an Odyssey CLX imaging system (LI-COR Biosciences). Band densitometry was quantified using ImageStudio software (LI-COR Biosciences) with all signals normalized to GAPDH as a loading control.
Results
Generation of the OKC-HETB/W Rats and Mitochondrial DNA Resequencing of Both Genotypes
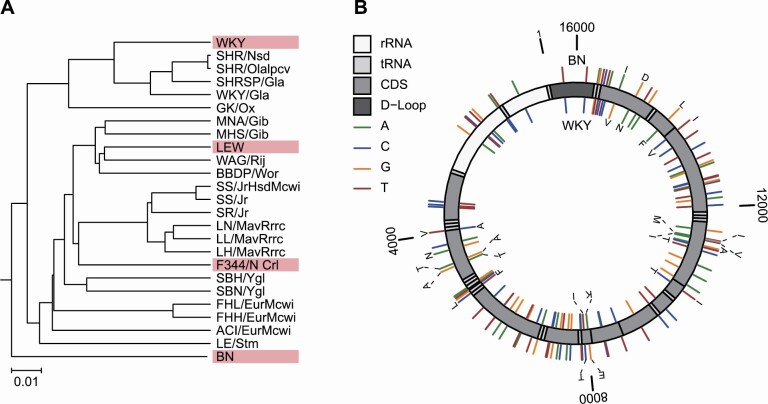
To produce rats with the greatest genetic heterogeneity, we selected 4 commercially available, but phylogenetically diverse, inbred strains of rats as shown in Figure 1A: BN, F344, LEW, and WKY. Using a 4-way cross of these strains, we generated 2 F2 populations of rats that have equal numbers of alleles from each of the 4 inbred strains but have mitochondrial genomes from either BN or WKY strains. We refer to these as OKC-HETB (abbreviated “B” genotype) and OKC-HETW (abbreviated “W” genotype) rats. The BN rat was the third complete mammalian genome, including the mitochondrial genome, to be sequenced after the human and mouse genomes (33) that we used as a reference sequence (GenBank Accession: AY172581.1). Similarly, the WKY rat mtDNA has been sequenced (GenBank Accession: DQ673907.1) (34). Using nanopore Cas9-targeted sequencing (31), we resequenced the mtDNA from our founder female BN and WKY animals as well as the female F1 hybrids. With greater than 100-fold coverage across the BN and WKY mtDNA, we found no differences in the BN mtDNA sequence from the AY172581.1 BN reference genome and two bases that differed from the DQ673907.1 WKY mtDNA reference genome: one at position 11339 (A to C synonymous mutation in ND4) and one at 15351 (C to A in tRNA-Pro). Comparing our updated WKY and BN mtDNA sequences, we identified 94 nucleotide differences between the two mitochondrial genomes with 88 base substitutions and 6 base insertions/deletions (Figure 1B). These substitutions were distributed throughout 7 of the 13 protein coding genes, 5 tRNAs, both rDNA subunits, and the D-loop. Of these substitutions, 16 are nonsynonymous substitutions in the genes for ND2, COI, ATPase8, ATPase6, ND4, ND6, and Cytb.
Figure 1.
(A) Phylogenetic tree of domesticated rats from Atanur et al. (16) for 28 inbred strains using 9.6 million single-nucleotide variants (SNVs). The phylogenetic distribution of the 4 inbred lines of rats used to generate the OKC-HETB/W rat are shown in red outline. (B) Nucleotide and amino acid differences between BN and WKY mitochondrial DNA. The rat mitochondrial genome is denoted by the gray scale circular diagram. The 88 nucleotide differences between the two genomes are denoted in the color of the corresponding divergent nucleotide. Nonsynonymous amino acid changes are denoted using the single letter amino acid code. Differences in the BN genome are on the outer track and differences in the WKY genome on the inner track. Numbering is according to the BN reference genome (AY172581.1).
General Phenotypic Characterization
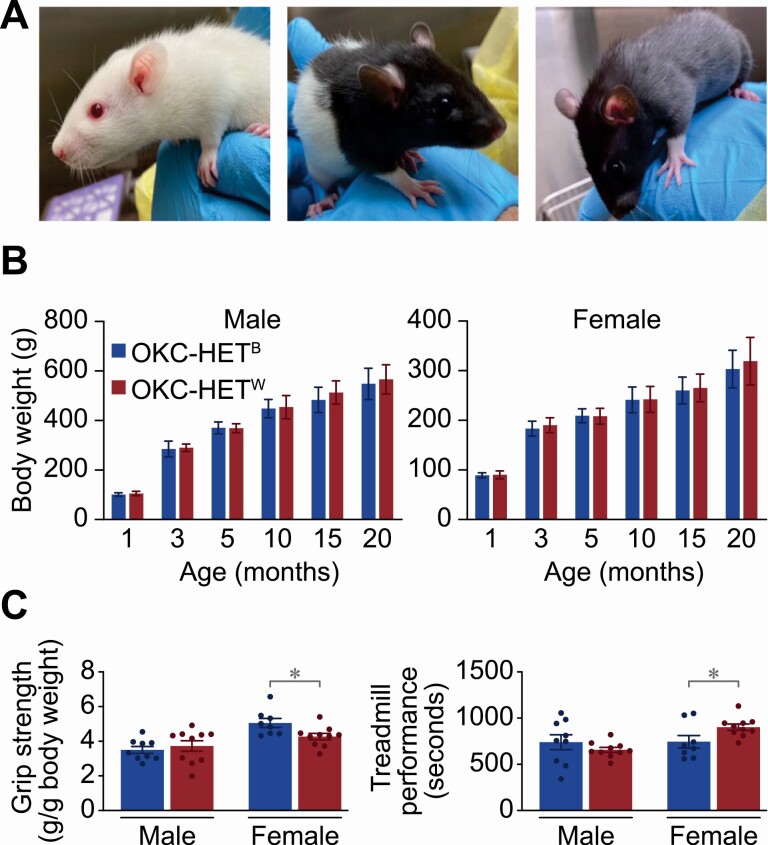
OKC-HETB/W rats come in 3 coat colors (Figure 2A). Approximately 50% of the OKC-HETB/W rats are white albino, as are the F344, LEW, and WKY parental strains. The remaining 50% are equally divided between charcoal gray, similar to the BN parental strain, or a combination of white and charcoal gray. We observed no significant differences among the two mitochondrial genotypes in the body weights of either sex up to 20 months of age (Figure 2B). At 20 months of age, the average size of males was ~550 ± ~60(SD) g compared to ~310 ± 40(SD) g for females. Overall, the animals appeared normal with no major observable pathology up to 24 months of age at which point approximately 85% of animals were still alive. The mean life span of F344, BN, and F344BNF1 rats in the NIA Biomarkers colony was 24–29, 29–30, and 31–32 (male–female) months, respectively, with females being the longer-lived sex in all 3 genotypes (35).
Figure 2.
Characterization of OKC-HETB and OKC-HETW rats. (A) Coat color for OKC-HETB/W rats. (B) Body weights for male and female B (blue bars) and W (red bars) genotypes. Data are the mean ± SD for 40–50 rats per group. (C) Grip strength (g/g body weight), and (D) Treadmill performance (seconds) for 4-month-old male and female rats as described in the Methods. The mean ± SEM are given for 8–10 rats per group, and the data were analyzed by two-way analysis of variance with Tukey posthoc comparisons. *p < .05.
Our small number of observations indicated antagonistic differences in grip strength and treadmill performance between females of the 2 mitochondrial genotypes at 4 months of age (Figure 3C and D). Specifically, females with the B mitochondrial genotype showed significantly greater grip strength, whereas W females displayed greater treadmill performance. We observed no significant differences in grip strength or treadmill performance among males.
Figure 3.
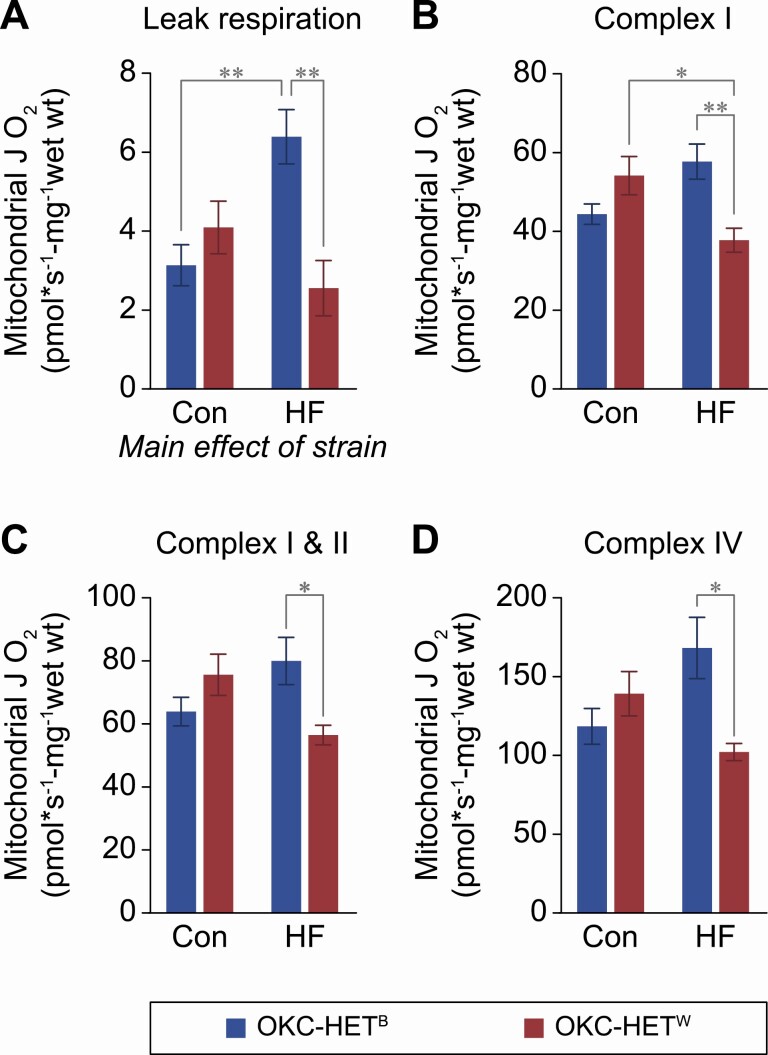
Mitochondrial bioenergetics in the gastrocnemius from OKC-HETB and OKC-HETW rats. Permeabilized fibers from red gastrocnemius of 13- to 14-month-old OKC-HETB (blue bars) and OKC-HETW (red bars) rats fed a normal chow (control) or high-fat diet (HFD) (45.7%) for 12 weeks were used to measure mitochondrial oxygen consumption rate. (A) Mitochondrial Leak respiration was measured after adding Complex I substrates in absence of ADP. Complex I (B), Complex I&II (C), and Complex IV (D) activities were measured by ADP-stimulated mitochondrial oxygen consumption rates with sequential additions of substrates and inhibitors. The mean ± SEM are given for 7–9 rats per group. Data were analyzed by two-way analysis of variance with Tukey posthoc comparisons. *p < .05.
Mitochondrial Function
We measured mitochondrial function under basal conditions and under metabolically challenging HFD conditions. Specifically, under both these conditions we assessed mitochondrial respiration in permeabilized fibers from the gastrocnemius of 13- to 14-month-old animals of both sexes (Figure 3A–D). To establish adequate power for assessment of whether respiration differed as a function of mitochondrial genotype, we combined sexes for the analysis because sex-specific effects were largely equivalent (Supplementary Figure 2). Leak respiration, which occurs when electrons passed down the respiratory chain exit prior to the reduction of oxygen to water at cytochrome C oxidase, had little response to the HFD in the W mitochondrial genotype, but was robustly higher for the B mitochondrial genotype (Figure 3A). These relative responses persisted for Complexes I, I&II, and IV. In general, it appears that the B genotype had compensatory increases in respiration to the HFD, whereas the W genotype did not. Further, it appeared that the W genotype may have some decline in respiratory rate to HFD as observed at Complex I (Figure 3B).
Response to High-fat Feeding and 17α-estradiol Supplementation
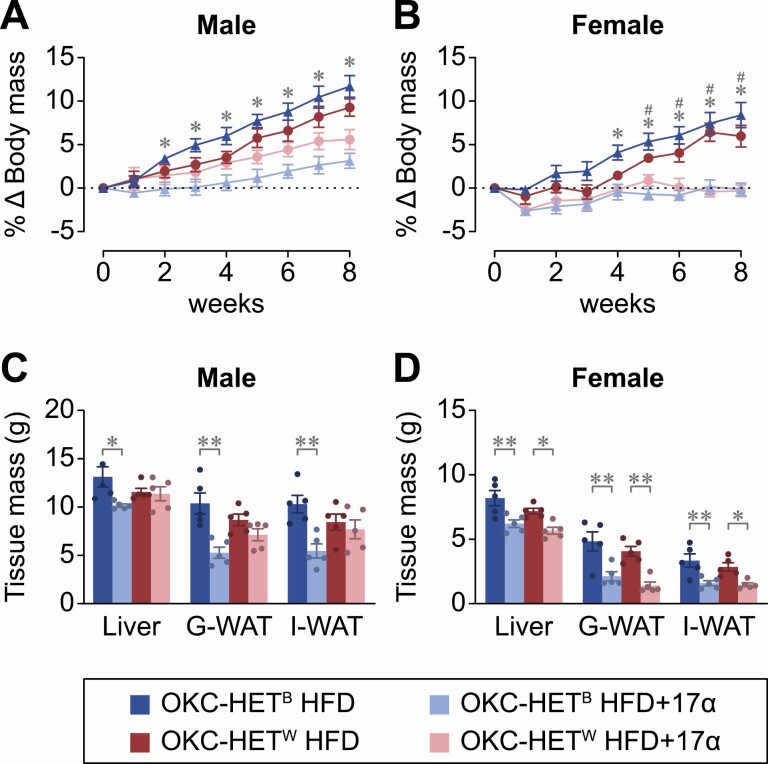
The NIA Intervention Testing Program found that long-term administration of 17α-estradiol (17α-E2) extended median life span of male, but not female, mice in a dose-dependent manner (18,20). Our group has been exploring potential mechanisms by which 17α-E2 may improve health span and extend life span in a male-specific manner. We reported that 17α-E2 supplementation reduces calorie intake, body mass, and regional adiposity in combination with significant improvements in a multitude of systemic metabolic parameters in both middle-aged obese and old male mice without inducing deleterious effects (19,25–29,36). Given that 17α-E2 elicits sex-specific effects in mice, which could be at least partially mediated through altered mitochondrial activity, we sought to determine if 17α-E2 would elicit similar sex-specific effects in 4-month-old OKC-HETB/W rats fed HFD ± 17α-E2 for 8 weeks. Interestingly, 17α-E2 prevented HFD-induced increases in body mass and organ mass (eg, liver and white adipose tissue [WAT]) in a mitochondrial genotype- and sex-specific manner. In males, 17α-E2 prevented gains in body mass, liver, gonadal WAT, and inguinal WAT mass in the B mitochondrial genotype only (Figure 4A and C). This was an unexpected finding because prior work with male mice showed that 17α-E2 treatment elicited benefits to health parameters in both UM-HET3 as well as inbred (C57BL/6J or C57BL/6N) mice, suggesting a conserved species pattern. We observed similar effect of 17α-E2 on body mass of 13- to 14-month-old OKC-HETB/W rats fed HFD ± 17α-E2 for 12 weeks (Supplementary Figure 3).
Figure 4.
Young OKC-HETB and OKC-HETW rats respond differently to high-fat feeding and 17α-E2 treatment. Male and female B (blue) and W (red) genotypes at 4 months of age received a 45.7% high-fat diet (HFD) ± 17α-E2 (14.4 ppm) over an 8-week intervention. (A, B) Longitudinal percent change in body mass in male and female rats. The data are the mean ± SEM for 5 animals per group and were analyzed by 2-way repeated measures analysis of variance (ANOVA) with Tukey post-hoc comparisons. *p < .05 between OKC-HETB HFD and OKC-HETB HFD+17α-E2; #p < .05 between OKC-HETW HFD and OKC-HETW HFD+17α-E2. (C,D) Liver, gonadal (G-WAT), and inguinal (I-WAT) white adipose tissue masses at necropsy. The data are mean ± SEM for 5 animals per group and were analyzed by 1-way ANOVA with Tukey posthoc comparisons within individual tissues and sexes. *p < .05, **p < .01.
Female OKC-HETB/W rats receiving 17α-E2 also displayed unexpected results. Both mitochondrial genotypes beneficially responded to 17α-E2 treatment as evidenced by a prevention of HFD-induced gains in body mass, liver, gonadal WAT, and inguinal WAT mass (Figure 4B and D). These observations are contrary to previous studies in mice, in which females are generally unresponsive to 17α-E2 treatment unless ovariectomized (21,37). These observations suggest that unlike in mice, 17α-E2 may have significant biological activity in female rats.
17α-E2 has also been shown to have neuroprotective effects by mitigating the toxic effects of oxidative stress in humans and ovariectomized rats (38). Therefore, we investigated whether markers of oxidative stress and response to a HFD and 17α-E2 are impacted in our rat model by mtDNA genotype. We previously reported that mitochondrial function and antioxidant expression were altered in a mouse model of brain aging with reduced circulating levels of IGF-1 (39). Additionally, a decline in levels of both Cu/Zn- and Mn-superoxide dismutase (Sod1 and 2, respectively) was associated with impaired hippocampal-dependent spatial learning (40,41). We therefore measured the levels of Sod1, Sod2, and acetylated Sod2, in the hippocampus of W and B genotypes under chow-fed (control) and HFD feeding with and without 17α-E2 supplementation. Acetylated Sod2 has been shown to lack its normal dismutation activity but a gain in peroxidase activity (42).
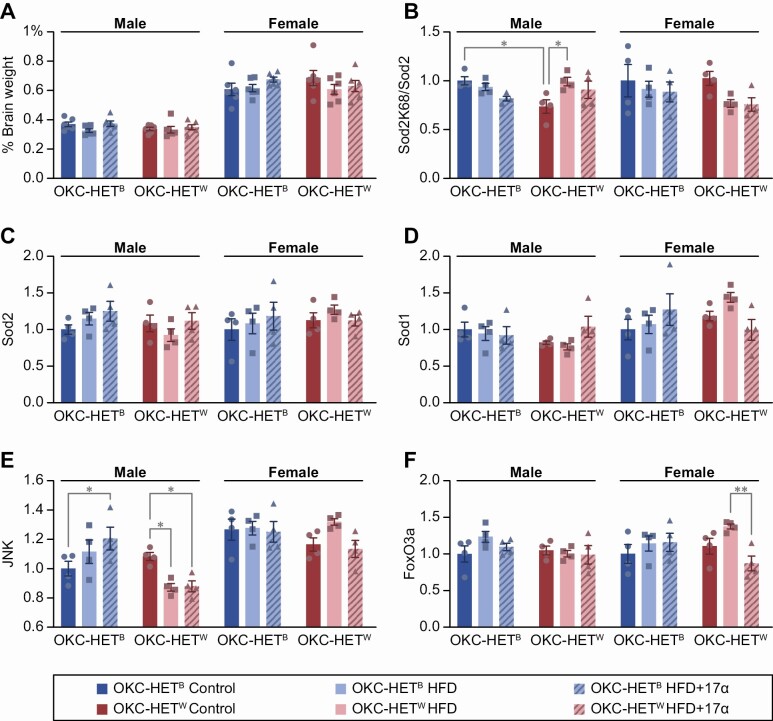
Brain weights (normalized to body weight) were comparable between mitochondrial genotypes within sex, albeit female percent brain weights were higher than males (Figure 5A). No significant difference was found between the genotypes with respect to the levels of Sod1 (Figure 5D) or Sod2 (Figure 5C) in either sex. However, we did observe that the levels of acetylated Sod2 (K68) were significantly lower in male W genotypes compared to B genotypes (Figure 5B). Conversely, when fed HFD, acetylated Sod2 levels were significantly increased in male W, but not B, genotypes. Acetylated Sod2 levels trended to be lower in female W, but not B genotype fed HFD with and without 17α-E2.
Figure 5.
Expression of antioxidant and oxidative stress pathway proteins in hippocampus of OKC-HETB and OKC-HETW rats. (A) Percent brain weight (normalized to body weight). (B) Levels of acetylated SOD2(K68), (C) Sod2, (D) Sod1, (E) JNK, (F) FoxO3a from hippocampi of 13–14-month-old male and female rats fed either a control diet (dark blue or dark red bars), a 45.7% high-fat diet (HFD) (light blue or light red bars), or HFD + 17α-E2 (stripped blue or red bars) for 12 weeks, and levels were determined by Western blots, which are shown in Supplementary Figure 4. The data are the mean ± SEM for 4 animals and were analyzed using a 2-way analysis of variance with Tukey posthoc comparisons. *p < .05.
We also measured the levels of upstream regulators of the antioxidant pathway, Jun N-terminal kinase (JNK) and FoxO3a. It is well established that oxidative stress can activate the JNK pathway (43), and FoxO transcription factors, which are activated by the JNK pathway, regulate the transcription of genes coding for antioxidant proteins (44). Levels of JNK were significantly lower in W genotype male rats fed the HFD with or without 17α-E2, while JNK levels were significantly elevated in the OKC-HETB male rats fed the HFD with 17α-E2 only (Figure 5E). No significant differences were found in JNK levels in the 2 genotypes in female rats. FoxO3a levels were comparable in the hippocampus of the 2 mitochondrial genotypes for male rats, and feeding a HFD with or without 17αE2 had no detectable effect on FoxO3a levels (Figure 5F). However, female FoxO3a levels were significantly reduced in the W genotype when fed a HFD with 17α-E2 compared to rats fed HFD alone. These data suggest that adaptive changes in cellular response to stressful stimulus such as HFD, and responsiveness to 17α-E2 treatment, may be mediated through mtDNA variants in a sex-specific manner, potentially contributing to cognitive health.
Discussion
We have developed a new genetically heterogeneous 4-way cross rat model with 2 genetically divergent mitochondrial backgrounds. We have furthermore shown that mitochondrial genotype, often interacting with sex, affects multiple physiological parameters important to the biology of aging. Specifically, we observed that one mitochondrial genotype was associated with greater grip strength, whereas the other associated with greater exercise endurance in female rats only. The 2 mitochondrial genotypes also differentially affected gastrocnemius respiratory response to HFD. We also discovered that mitochondrial genotype interacting with sex had some dramatic effects on cellular and physiological response to HFD and supplementation with 17α-E2 and that these effects clearly differed from previous research done in mice. These findings emphasize the importance of this new model for understanding the role of mitochondrial genotype in multiple aspects of aging biology and also for distinguishing phenomena that may or may not be generalizable across species.
An important difference between laboratory mice and rats is the genetic diversity represented in commonly used domesticated stocks and strains. Virtually all traditional inbred strains of laboratory mice originated from a small number of “fancy mice,” so their diversity is highly constrained (45). Phylogenetic mtDNA studies show that 50 of 52 common inbred strains of mice descended from a single female Mus musculus domesticus mouse (17). In contrast, laboratory rats have come from multiple domestications and in fact were often bred back to wild-caught animals; hence, both nuclear and mitochondrial diversity are considerably greater in laboratory rats compared with laboratory mice.
A unique feature of the OKC-HETB/W rat that we have generated is that it gives investigators a rodent model where substantial differences in the mitochondrial genome can be studied in rats with the same genetically diverse nuclear backgrounds. Our BN-derived mitochondria differ from our WKY-derived mitochondria in 94 nucleotides with 88 base substitutions and 6 base insertions/deletions (Figure 2A). The importance of even considerably less divergent mitochondrial genomes on a variety of physiological outcomes was shown by Ballinger’s group when they generated mitochondrial nuclear exchange mouse models (MNX) in which the mtDNA from the C3H mouse was merged with the C57BL/6 nuclear background and vice versa (11). They showed that the mitochondrial genetic background modulated a variety of functions and pathways, e.g., bioenergetics and susceptibility to acute cardiac volume overload (11); macrosteatosis, inflammation, fibrosis, and mitochondrial function in liver induced by an atherogenic diet (46); metabolic efficiency, body composition, and gene expression in adipose tissue (12); and insulin sensitivity (47). Importantly, the mitochondrial genomes from C57BL/6 and C3H mice differ by only 5 nucleotides (17).
The normal response to relatively short-term HFD as seen in mice is a compensatory increase in mitochondrial biogenesis and respiration (48). Interestingly, our B genotype rats appeared to make this compensation with trends or significant increases as expected. However, the W genotype rats did not and, in fact, had significantly lower Complex I activity with HFD compared to control. Unfortunately, we did not assess ROS generation in this preliminary work to determine if the failure to respond was associated with increased ROS production, although we suspect it would (49). Finally, we found similar trends in the changes in respiration between sexes. It is worth noting that our preliminary data indicate the failure to adapt to HFD is most pronounced in females of the W genotype. This finding is interesting because rats (compared to mice) and female rats (compared to males) are generally more resistant to increases in fat deposition and declines in insulin sensitivity to a HFD (50).
Because 17α-E2 has been shown to modulate metabolic- and age-related outcomes in different strains of male, but not female mice, we studied the effects of 17α-E2 in both sexes and mitochondrial genotypes of rats undergoing HFD feeding. This is the first study to test the effect of feeding 17α-E2 to rats. Contrary to findings in mice (18,20–24,26,28), female rats of both genotypes beneficially responded to 17α-E2 treatment as evidenced by the prevention of HFD-induced gains in body mass and adiposity. Liver mass was also reduced by 17α-E2 treatment in female rats of both mitochondrial genotypes. Male rats in our study responded in a mitochondrial genotype-specific manner with only B genotype males showing a decline in body mass, adiposity, and liver mass, similar to previous studies in mice (19,21,22,25–29). Our findings in both sexes were unexpected and interesting because we anticipated that both male genotypes would beneficially respond to treatment and that female responsiveness would be mild or absent. The mechanisms underlying these observations remain unknown. It should be noted that the dose provided in these studies is equivalent to what mice are provided. As such, the dosing regimen has not yet been allometrically scaled downward to account for differences in size and surface area between mice and rats. Thus, in the current studies female rats are consuming a higher dose than female mice relative to size and surface area, which could contribute to female responsiveness in our studies. Future studies will be required to determine if lower dosing regimens will effectively mimic 17α-E2 blood levels observed in mice. We surmise that genotype-specific differences in male responsiveness to 17α-E2 arises from differences in mitochondrial activity, which almost certainly affects substrate utilization, and thus, attenuates effects on adiposity. Additional studies will be needed to confirm this possibility.
Sex and genotype-specific changes were also evident in the stress response in the hippocampus with HFD feeding, potentially ameliorated by 17α-E2. While Sod2 detoxifies superoxide in the mitochondrial matrix, acetylated Sod2 (K68) can generate hydrogen peroxide and have detrimental effects on tissue function (42). Thus, the increase in Sod2 acetylation we observed in OKC-HETW males could impact hippocampal mitochondrial and cognitive dysfunction with HFD feeding. Additional functional studies on brain function in these models would be pertinent to unraveling mitochondrial contribution to cognitive decline in aging and neurodegenerative disorders.
These observations on 17α-E2 provide support for our concept that studying anti-aging compounds in both mice and rats will provide additional insights, particularly related to the mechanisms that underlie sex-specific aging. Although our findings should be considered preliminary, they suggest that the sex differences in the impact of 17α-E2 on life span might be different in rats and that mitochondrial genotype may play a role in male-responsiveness to 17α-E2. However, more studies are needed to unravel the interaction between mitochondrial genotypes and sex-specific aging and disease burden. Notably though, this short interventional study provides proof-of-concept that mice and rats may age differently in a sex-specific fashion and these differences, depending on mitochondrial genotype could impact the effect of aging-interventions. These findings raise the prospect that the OKC-HETB/W rat may be applied to study dietary interventions that purport to improve cognitive and performance phenotypes associated with improvements in mitochondrial function.
We note that tissues and OKC-HETB/W rats are available to the scientific community and can be obtained by contacting Arlan Richardson (Arlan-Richardson@ouhsc.edu), Michael Stout (Michael-Stout@omrf.org), or Steven Austad (austad@uab.edu).
Supplementary Material
Funding
The efforts of authors were supported by the following National Institutes of Health (NIH) grants: R21AG072137 (A.R., S.N.A., M.B.S.), R21 AG058811 & R01 AG057434 (S.N.A.), R00 AG064143 (B.A.), R00 AG51661 (M.B.S.), R01 AG070035 (M.B.S.), R00 AG056662 (S.L.), R01AG055518 (J.W.), R01AG069924 (J.W.), K02AG059847 (J.W.) and grants from the Department of Veterans Affairs: 1IK6BX005238 (A.R.) and I01BX004538 (A.R.). Scientific illustration assistance was provided by Kate Baldwin, PhD (K8Baldwin.com).
Contributor Information
Roshini Sathiaseelan, Department of Nutritional Sciences, University of Oklahoma Health Sciences Center, Oklahoma City, USA.
Bumsoo Ahn, Gerontology and Geriatric Medicine, Wake Forest University School of Medicine, Winston-Salem, North Carolina, USA.
Michael B Stout, Aging & Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA; Oklahoma City VA Medical Center, Oklahoma City, Oklahoma, USA.
Sreemathi Logan, Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Jonathan Wanagat, Divisions of Geriatrics and Dermatology, Department of Medicine, University of California Los Angeles and Veterans Administration Greater Los Angeles Healthcare System, Los Angeles, California, USA.
Hoang Van M Nguyen, Department of Nutritional Sciences, University of Oklahoma Health Sciences Center, Oklahoma City, USA.
Norman G Hord, Department of Nutritional Sciences, University of Oklahoma Health Sciences Center, Oklahoma City, USA.
Amy R Vandiver, Divisions of Geriatrics and Dermatology, Department of Medicine, University of California Los Angeles and Veterans Administration Greater Los Angeles Healthcare System, Los Angeles, California, USA.
Ramasamy Selvarani, Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Rojina Ranjit, Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Hannah Yarbrough, Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Anthony Masingale, Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Benjamin F Miller, Aging & Metabolism Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA; Oklahoma City VA Medical Center, Oklahoma City, Oklahoma, USA.
Roman F Wolf, Oklahoma City VA Medical Center, Oklahoma City, Oklahoma, USA.
Steven N Austad, Department of Biology, University of Alabama at Birmingham, Birmingham, Alabama, USA.
Arlan Richardson, Oklahoma City VA Medical Center, Oklahoma City, Oklahoma, USA; Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA.
Conflict of Interest
None declared.
References
- 1. Ireson CR, Alavijeh MS, Palmer AM, Fowler ER, Jones HJ. The role of mouse tumour models in the discovery and development of anticancer drugs. Br J Cancer. 2019;121:101–108. doi: 10.1038/s41416-019-0495-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res Ther. 2014;6:37. doi: 10.1186/alzrt269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benatar M. Lost in translation: treatment trials in the SOD1 mouse and in human ALS. Neurobiol Dis. 2007;26:1–13. doi: 10.1016/j.nbd.2006.12.015 [DOI] [PubMed] [Google Scholar]
- 4. Carter CS, Richardson A, Huffman DM, Austad S. Bring Back the Rat!. J Gerontol A Biol Sci Med Sci. 2020;75:405–415. doi: 10.1093/gerona/glz298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kasahara T, Abe K, Mekada K, Yoshiki A, Kato T. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc Natl Acad Sci USA. 2010;107:6412–6417. doi: 10.1073/pnas.0914399107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller RA, Nadon NL. Principles of animal use for gerontological research. J Gerontol A Biol Sci Med Sci. 2000;55:B117–B123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nadon NL, Strong R, Miller RA, et al. Design of aging intervention studies: the NIA interventions testing program. AGE. 2008;30:187–199. doi: 10.1007/s11357-008-9048-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roderick TH. Selection for radiation resistance in mice. Genetics. 1963;48:205–216. doi: 10.1093/genetics/48.2.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller RA, Austad S, Burke D, et al. Exotic mice as models for aging research: polemic and prospectus. Neurobiol Aging. 1999;20:217–231. doi: 10.1016/s0197-4580(99)00038-x [DOI] [PubMed] [Google Scholar]
- 10. Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23 [DOI] [PubMed] [Google Scholar]
- 11. Fetterman JL, Zelickson BR, Johnson LW, et al. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J. 2013;455:157–167. doi: 10.1042/BJ20130029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunham-Snary KJ, Sandel MW, Sammy MJ, et al. Mitochondrial - nuclear genetic interaction modulates whole body metabolism, adiposity and gene expression in vivo. EBioMedicine. 2018;36:316–328. doi: 10.1016/j.ebiom.2018.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frank SA. Evolution: mitochondrial burden on male health. Curr Biol. 2012;22:R797–R799. doi: 10.1016/j.cub.2012.07.066 [DOI] [PubMed] [Google Scholar]
- 14. Camus MF, Clancy DJ, Dowling DK. Mitochondria, maternal inheritance, and male aging. Curr Biol. 2012;22:1717–1721. doi: 10.1016/j.cub.2012.07.018 [DOI] [PubMed] [Google Scholar]
- 15. Mashimo T, Voigt B, Tsurumi T, et al. A set of highly informative rat simple sequence length polymorphism (SSLP) markers and genetically defined rat strains. BMC Genet. 2006;7:19. doi: 10.1186/1471-2156-7-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atanur SS, Diaz AG, Maratou K, et al. Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell. 2013;154:691–703. doi: 10.1016/j.cell.2013.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu X, Gimsa U, Wester-Rosenlof L, et al. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res. 2009;19:159–165. doi: 10.1101/gr.078865.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison DE, Strong R, Allison DB, et al. Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell. 2014;13:273–282. doi: 10.1111/acel.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stout MB, Steyn FJ, Jurczak MJ, et al. 17alpha-estradiol alleviates age-related metabolic and inflammatory dysfunction in male mice without inducing feminization. J Gerontol A Biol Sci Med Sci. 2017;72:3–15. doi: 10.1093/gerona/glv309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strong R, Miller RA, Antebi A, et al. Longer lifespan in male mice treated with a weakly estrogenic agonist, an antioxidant, an alpha-glucosidase inhibitor or a Nrf2-inducer. Aging Cell. 2016;15:872–884. doi: 10.1111/acel.12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garratt M, Bower B, Garcia GG, Miller RA. Sex differences in lifespan extension with acarbose and 17-alpha estradiol: gonadal hormones underlie male-specific improvements in glucose tolerance and mTORC2 signaling. Aging Cell. 2017;16:1256–1266. doi: 10.1111/acel.12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garratt M, Lagerborg KA, Tsai YM, Galecki A, Jain M, Miller RA. Male lifespan extension with 17-alpha estradiol is linked to a sex-specific metabolomic response modulated by gonadal hormones in mice. Aging Cell. 2018;17:e12786. doi: 10.1111/acel.12786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Debarba LK, Jayarathne HSM, Miller RA, Garratt M, Sadagurski M. 17-alpha-estradiol has sex-specific effects on neuroinflammation that are partly reversed by gonadectomy. J Gerontol A Biol Sci Med Sci. 2022;77:66–74. doi: 10.1093/gerona/glab216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garratt M, Leander D, Pifer K, et al. 17-alpha estradiol ameliorates age-associated sarcopenia and improves late-life physical function in male mice but not in females or castrated males. Aging Cell. 2019;18:e12920. doi: 10.1111/acel.12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steyn FJ, Ngo ST, Chen VP, et al. 17alpha-estradiol acts through hypothalamic pro-opiomelanocortin expressing neurons to reduce feeding behavior. Aging Cell. 2018;17:e12703. doi: 10.1111/acel.12703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mann SN, Hadad N, Nelson Holte M, et al. Health benefits attributed to 17alpha-estradiol, a lifespan-extending compound, are mediated through estrogen receptor alpha. Elife. 2020;9:e59616. doi: 10.7554/eLife.59616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller BF, Pharaoh GA, Hamilton KL, et al. Short-term calorie restriction and 17alpha-estradiol administration elicit divergent effects on proteostatic processes and protein content in metabolically active tissues. J Gerontol A Biol Sci Med Sci. 2020;75:849–857. doi: 10.1093/gerona/glz113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sidhom S, Schneider A, Fang Y, et al. 17alpha-Estradiol modulates IGF1 and hepatic gene expression in a sex-specific manner. J Gerontol A Biol Sci Med Sci. 2021;76:778–785. doi: 10.1093/gerona/glaa215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ali Mondal S, Sathiaseelan R, Mann SN, et al. 17alpha-estradiol, a lifespan-extending compound, attenuates liver fibrosis by modulating collagen turnover rates in male mice. Am J Physiol Endocrinol Metab. 2022;324(2):E120-E134. doi: 10.1152/ajpendo.00256.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Herbst A, Choi S, Hoang AN, et al. Remdesivir does not affect mitochondrial DNA copy number or deletion mutation frequency in aged male rats: a short report. PLoS One. 2022;17:e0271850. doi: 10.1371/journal.pone.0271850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vandiver AR, Pielstick B, Gilpatrick T, et al. Long read mitochondrial genome sequencing using Cas9-guided adaptor ligation. Mitochondrion. 2022;65:176–183. doi: 10.1016/j.mito.2022.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahn B, Ranjit R, Kneis P, et al. Scavenging mitochondrial hydrogen peroxide by peroxiredoxin 3 overexpression attenuates contractile dysfunction and muscle atrophy in a murine model of accelerated sarcopenia. Aging Cell. 2022;21:e13569. doi: 10.1111/acel.13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gibbs RA, Weinstock GM, Metzker ML, et al. ; Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426 [DOI] [PubMed] [Google Scholar]
- 34. Schlick NE, Jensen-Seaman MI, Orlebeke K, Kwitek AE, Jacob HJ, Lazar J. Sequence analysis of the complete mitochondrial DNA in 10 commonly used inbred rat strains. Am J Physiol Cell Physiol. 2006;291:C1183–C1192. doi: 10.1152/ajpcell.00234.2006 [DOI] [PubMed] [Google Scholar]
- 35. Sprott RL, Austad SN. Animal models for aging research. In: Schneider EL, Rowe JW, eds. Handbook of the Biology of Aging. 4th ed. San Diego: Academic Press; 1996:3–23. [Google Scholar]
- 36. Isola JVV, Veiga GB, de Brito CRC, et al. 17alpha-estradiol does not adversely affect sperm parameters or fertility in male mice: implications for reproduction-longevity trade-offs. Geroscience. 2022. doi: 10.1007/s11357-022-00601-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mann SN, Pitel KS, Nelson-Holte MH, et al. 17alpha-Estradiol prevents ovariectomy-mediated obesity and bone loss. Exp Gerontol. 2020;142:111113. doi: 10.1016/j.exger.2020.111113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yi KD, Perez E, Yang S, Liu R, Covey DF, Simpkins JW. The assessment of non-feminizing estrogens for use in neuroprotection. Brain Res. 2011;1379:61–70. doi: 10.1016/j.brainres.2010.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pharaoh G, Owen D, Yeganeh A, et al. Disparate central and peripheral effects of circulating IGF-1 deficiency on tissue mitochondrial function. Mol Neurobiol. 2020;57:1317–1331. doi: 10.1007/s12035-019-01821-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baier MP, Nagaraja RY, Yarbrough HP, et al. Selective ablation of Sod2 in astrocytes induces sex-specific effects on cognitive function, d-serine availability, and astrogliosis. J Neurosci. 2022;42:5992–6006. doi: 10.1523/JNEUROSCI.2543-21.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Logan S, Royce GH, Owen D, et al. Accelerated decline in cognition in a mouse model of increased oxidative stress. Geroscience. 2019;41:591–607. doi: 10.1007/s11357-019-00105-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hjelmeland AB, Patel RP. SOD2 acetylation and deacetylation: another tale of Jekyll and Hyde in cancer. Proc Natl Acad Sci USA. 2019;116:23376–23378. doi: 10.1073/pnas.1916214116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shen HM, Liu ZG. JNK signaling pathway is a key modulator in cell death mediated by reactive oxygen and nitrogen species. Free Radic Biol Med. 2006;40:928–939. doi: 10.1016/j.freeradbiomed.2005.10.056 [DOI] [PubMed] [Google Scholar]
- 44. Klotz LO, Sanchez-Ramos C, Prieto-Arroyo I, Urbanek P, Steinbrenner H, Monsalve M. Redox regulation of FoxO transcription factors. Redox Biol. 2015;6:51–72. doi: 10.1016/j.redox.2015.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Silver LM. Mouse Genetics: Concepts and Applications. New York: Oxford University Press; 1995. [Google Scholar]
- 46. Betancourt AM, King AL, Fetterman JL, et al. Mitochondrial-nuclear genome interactions in non-alcoholic fatty liver disease in mice. Biochem J. 2014;461:223–232. doi: 10.1042/BJ20131433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sammy MJ, Connelly AW, Brown JA, Holleman C, Habegger KM, Ballinger SW. Mito-Mendelian interactions alter in vivo glucose metabolism and insulin sensitivity in healthy mice. Am J Physiol Endocrinol Metab. 2021;321:E521–E529. doi: 10.1152/ajpendo.00069.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Newsom SA, Miller BF, Hamilton KL, Ehrlicher SE, Stierwalt HD, Robinson MM. Long-term rates of mitochondrial protein synthesis are increased in mouse skeletal muscle with high-fat feeding regardless of insulin-sensitizing treatment. Am J Physiol Endocrinol Metab. 2017;313:E552–E562. doi: 10.1152/ajpendo.00144.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anderson EJ, Lustig ME, Boyle KE, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. 2009;119:573–581. doi: 10.1172/JCI37048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maric I, Krieger JP, van der Velden P, et al. Sex and species differences in the development of diet-induced obesity and metabolic disturbances in rodents. Front Nutr. 2022;9:828522. doi: 10.3389/fnut.2022.828522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.