Summary
Background
Rheumatoid arthritis (RA) shares genetic variants with other autoimmune conditions, but existing studies test the association between RA variants with a pre-defined set of phenotypes. The objective of this study was to perform a large-scale, systemic screen to determine phenotypes that share genetic architecture with RA to inform our understanding of shared pathways.
Methods
In the UK Biobank (UKB), we constructed RA genetic risk scores (GRS) incorporating human leukocyte antigen (HLA) and non-HLA risk alleles. Phenotypes were defined using groupings of International Classification of Diseases (ICD) codes. Patients with an RA code were excluded to mitigate the possibility of associations being driven by the diagnosis or management of RA. We performed a phenome-wide association study, testing the association between the RA GRS with phenotypes using multivariate generalized estimating equations that adjusted for age, sex, and first five principal components. Statistical significance was defined using Bonferroni correction. Results were replicated in an independent cohort and replicated phenotypes were validated using medical record review of patients.
Findings
We studied n = 316,166 subjects from UKB without evidence of RA and screened for association between the RA GRS and n = 1317 phenotypes. In the UKB, 20 phenotypes were significantly associated with the RA GRS, of which 13 (65%) were immune mediated conditions including polymyalgia rheumatica, granulomatosis with polyangiitis (GPA), type 1 diabetes, and multiple sclerosis. We further identified a novel association in Celiac disease where the HLA and non-HLA alleles had strong associations in opposite directions. Strikingly, we observed that the non-HLA GRS was exclusively associated with greater risk of the validated conditions, suggesting shared underlying pathways outside the HLA region.
Interpretation
This study replicated and identified novel autoimmune phenotypes verified by medical record review that share immune pathways with RA and may inform opportunities for shared treatment targets, as well as risk assessment for conditions with a paucity of genomic data, such as GPA.
Funding
This research was funded by the US National Institutes of Health (P30AR072577, R21AR078339, R35GM142879, T32AR007530) and the Harold and DuVal Bowen Fund.
Keywords: Rheumatoid arthritis, Genetic risk scores, Phenome-wide association study, Genetic architecture, Risk calibration, Vasculitis
Research in context.
Evidence before this study
Rheumatoid arthritis (RA) is the most common autoimmune inflammatory joint disease worldwide. Prior epidemiologic studies have identified associations between RA and other autoimmune conditions. Studies testing the relationship between RA risk alleles with these conditions confirmed that the associations between RA and other autoimmune conditions can in part be explained by shared genetic architecture.
Added value of this study
This phenome-wide association study (PheWAS) using a composite genetic risk score (GRS) for both HLA, not tested in prior studies, as well as the non-HLA RA risk alleles add several findings to our current body of knowledge. First, we performed a screen of association between the RA GRS and a broad range of conditions regardless of prior associations. Using this agnostic approach, we identified the previously observed associations between the RA GRS with other autoimmune conditions such as type 1 diabetes (T1D) and vasculitis. The medical record review performed as part of this study enabled more precise and accurate phenotyping, thus uncovering that the association between the RA GRS and TID was driven by those with poor outcomes such as retinopathy. Our study provides new information regarding differential associations between the HLA and non-HLA RA risk alleles associated with risk for other autoimmune conditions. For example, the RA HLA alleles were associated with reduced risk of Celiac, while non-HLA were associated with increased risk. In addition to understanding pathways for studies of etiology, these data may have potential applications in evaluating treatments. We further reviewed studies of therapeutic treatments that were tested in RA and other autoimmune conditions identified in this study. Among conditions where RA risk alleles were associated with increased risk for the condition, the conditions share common therapies. Among conditions where RA risk alleles were associated with reduced risk, RA treatments exacerbated the condition.
Implications of all the available evidence
The PheWAS approach using an RA GRS allowed testing across a broad range of phenotypes, and ultimately replicated findings from large-scale epidemiologic studies, demonstrating an alternate approach in using genetics to study relationships across conditions. RA shares genetic architecture mainly with other autoimmune conditions vs non-autoimmune conditions. However, differential associations can exist between the RA HLA and non-HLA alleles with other autoimmune conditions. For uncommon conditions such as vasculitis, with a paucity of data for genetic risk, the RA GRS could potentially be included in strategies for risk assessment. Knowledge of shared genetic architecture shared across conditions can inform studies of etiology, risk assessment, and potentially shared therapies.
Introduction
Rheumatoid arthritis (RA) is the most common autoimmune inflammatory joint disease worldwide and is often studied as a human model of inflammation.1, 2, 3, 4, 5, 6 The growth of large population-based biobanks, such as the UK Biobank, along with the development of methods for aggregating genetic variants into risk scores, afford novel approaches to examine genetic relationships and mechanisms across diseases, complementing existing epidemiologic studies.7, 8, 9, 10 Previous work has highlighted the possibility of leveraging biobanks and genome wide association study (GWAS) data to uncover novel genetic relationships that exist between complex diseases, demonstrating the utility of these datasets in furthering our understanding of the genetics of complex diseases.11,12 For example, a recent study provided novel insights into the shared genetics underlying circulatory and nervous system disorders.13 In particular, the RA genetic risk score (GRS), an aggregate representation of genetic susceptibility to RA, can explain up to 50–60% of risk for developing RA.14,15 Thus, one can consider the RA GRS as a proxy for individuals with elevated genetic susceptibility and risk of RA. Identifying the conditions significantly associated with the RA GRS can shed light on complex conditions which share genetic architecture with RA and inform novel ascertainment strategies of disease risk or help identify shared pathways early in the development of autoimmunity amenable for therapeutic targeting.
The Phenome Wide Association Study (PheWAS) approach is a method designed for biobanks with linked electronic health records and genomic data. Initially developed to screen for associations between a genomic variant of interest and multiple phenotypes, it can also be used to screen for association between an aggregate genetic risk score and a broad range of phenotypes. This bioinformatics approach identifies multiple EHR-based phenotypes associated with a genetic variant or risk score in an agnostic fashion and is less constrained by prior assumptions as previous genetic association studies are.16, 17, 18, 19
Existing PheWAS for RA risk alleles have focused on the non-HLA alleles, and thus may not capture differential associations between HLA and non-HLA alleles.17 This represents a current gap in the literature as the largest known genetic risk factor for RA is the shared epitope region in the human leukocyte antigen (HLA) region, which encodes a sequence of amino acids.20,21 The identification of five amino acids explaining the majority of the association with RA across 3 HLA proteins has enabled imputation of the shared epitope using GWAS data.20,22 Due to the large effect on disease risk, genomic studies in RA often stratify findings based on HLA or non-HLA regions.
In this study, we used an RA GRS which incorporates HLA and non-HLA risk loci to identify novel associations of conditions with RA and identified potential shared pathways across conditions using real-world EHR data from the UK Biobank in a PheWAS. All results were validated in an independent biobank, and phenotype accuracies were validated with manual medical record review. For select associated and validated phenotypes, we show effective genetic risk stratification via dose–response relationships using the RA GRS and identified potential shared genetic variants between associated conditions.
Methods
Study populations
The UK Biobank served as the main study cohort. The UK Biobank is a prospective study containing extensive genetic and phenotypic data of participants from the general population of the United Kingdom.7,8 Participants were between the ages of 40 and 69 at recruitment. Phenotype information available in the UK Biobank include individual inpatient International Statistical Classification of Diseases and Related Health Problems, Ninth and Tenth Revision (ICD-9, ICD-10) codes. ICD codes up to December 2020 were included in the phenotype data. Deidentified data from UK Biobank are available online following informed consent obtained from all participants.
The Mass General Brigham (MGB) Biobank is a cohort study from the MGB healthcare network and served as the replication cohort.23, 24, 25 The MGB healthcare network is a large healthcare network covering the Greater Boston area in the United States. The MGB Biobank contains EHR, genetic, and lifestyle data collected from community-based primary care facilities and tertiary care centers. Recruitment for the MGB Biobank is ongoing, and at the time of analysis clinical and genetic data were available for 34,195 participants. All recruited patients provided written informed consent upon enrollment. The study protocol was approved by the MGB Institutional Review Board.
Aggregate genetic-risk score
Study participants at the UK Biobank were genotyped centrally by the UK Biobank study staff and genetic data was obtained using array-based imputation procedures as previously published.7 Study participants of the MGB Biobank were genotyped on Illumina arrays such as the Infinium MEGA array. Standard quality control was performed for genotyped data, followed by imputation using the 1000 Genomes reference panel.24 Genotype data was used to calculate an RA GRS for study participants. Both HLA GRS and non-HLA RA GRS were calculated. The HLA and non-HLA GRS were then summed to estimate a composite RA GRS. Statistically imputed HLA haplotypes with an established association with RA were used to calculate the HLA GRS.20 Single nucleotide polymorphisms (SNPs) outside of the HLA region associated with RA from a meta-analysis of genome-wide association studies (GWAS) were used to calculate non-HLA GRS.4 The 95 genetic variants and their corresponding effect sizes used to construct the RA GRS are provided in Supplementary Table S1.
To construct the HLA-GRS, we utilized classical HLA-DRB1 allele data imputed using the HLA∗IMP:02 software and a merged reference panel representative of 8869 individuals of various ancestries.26,27 Alleles of the HLA-DRB1 gene associated with RA were included.20 The HLA GRS were subsequently calculated as:
where is the number of considered HLA-DRB1 alleles, is the assigned weight for each allele, and is the number of alleles identified (0, 1, or 2). Allele-specific weights were derived by taking a log transformation of odds ratios for RA among individuals of European descent as reported in Raychaudhuri et al.20 The non-HLA GRS was calculated as:
where is the number of RA risk SNPs considered, is the assigned weight for , and is the SNP frequency (0, 1, or 2). Weights for each SNP were calculated as the log transformation of trans-ethnic odds ratios derived from the Okada et al. RA GWAS meta-analysis.4
Statistical methods
Phenotypes were defined using published groupings of ICD-9 and ICD-10 codes into clinically relevant codes, termed PheWAS codes or PheCodes.28 As the original UKB data contained inpatient ICD codes, a participant was defined as having a phenotype if they had 1 or more PheCodes for a particular phenotype. PheCodes with a prevalence of 0.1% or less were excluded from the analysis.
The PheWAS study tested associations between the RA GRS and each phenotype defined by PheCodes. We constructed multivariate generalized estimating equation (GEE) methods using version 1.3.9 of the “geepack” package for each test with a logit link and an exchangeable working correlation structure to control for potential kinship relations between participants.29, 30, 31 The GEE approach using a working correlation matrix was designed to analyze potentially correlated or clustered data without requiring explicit specification or calculation of the correlation structure, i.e., kinship relations. In each model, RA GRS served as the independent variable, and age at enrollment, self-reported sex, first five principal components, and log-transformed number of hospital visits served as covariates. The number of hospital visits was used as a surrogate for healthcare utilization, was included in GEE methods to adjust for density of EHR data. To mitigate the possibility of phenotype associations with RA GRS being primarily driven by the diagnosis of RA, we performed after the primary analysis excluding patients with one or more PheWAS codes for RA (PheCode 714.1). We further conducted separate PheWAS using either HLA GRS or non-HLA GRS as independent variables to identify genomic risk contributions to phenotypes between these two regions.
To account for multiple testing, we defined statistical significance as a value less than a threshold controlling for Bonferroni correction at a familywise error rate of 5%.32 As a reference, we further reported the threshold controlling for a false discovery rate (FDR) of 5% using the Benjamini-Hochberg procedure.33
Next, we tested whether a higher burden of RA risk alleles was associated with a higher magnitude and odds for a phenotype by categorizing subjects by their decile of GRS. We then estimated the odds of developing the disease in the second through tenth deciles, using the first decile as the reference. We further used linear regression to test for dose–response relationships. To identify shared genetic variants among phenotypes associated with RA GRS, we fit multivariate adaptive LASSO regression frameworks to test for the association between individual RA risk loci and phenotypes associated with RA GRS. We used bootstrapping over 500 samples to estimate standard errors for significance testing.
All analyses were implemented in R, version 4.0.1 (the R Foundation).
Replication of findings in the mass general brigham biobank
We further studied UK Biobank phenotypes with significant associations with the RA GRS in the MGB Biobank. RA GRS were constructed for MGB Biobank patients in the same manner as was done in UK Biobank. Both ICD-9 and ICD-10 codes were mapped to PheWAS codes in the same manner. All replication studies used age at the last visit, sex, self-reported race, and log-transformed number of hospital visits as covariates. To define statistical significance in replication studies, a value less than 0.05 was considered significant.
Validation of significant outcomes with medical record review
For phenotypes in the UK Biobank which were significant after Bonferroni correction and replicated in the MGB Biobank, we validated the accuracy of each phenotype through manual medical record review. For each phenotype, 50 patients from the MGB Biobank were randomly selected among those who were defined to have the phenotype and reviewed for evidence of the phenotype in narrative notes or diagnostic reports. All reviewers were either clinically trained health professionals or supervised by clinically trained health professionals. We reported the positive predictive value (PPV) as defined by the number of confirmed phenotypes based on manual review divided by the number of participants with either 1 or more PheWAS codes. A PheWAS code with a PPV of 80% or greater was considered an accurate surrogate for the phenotype.
Since the majority of RA genetic risk alleles were identified from populations of majority European descent, we additionally performed a sensitivity analysis in the UK Biobank restricting the PheWAS to individuals of European ancestry. We followed an identical study design to the main PheWAS that was inclusive of all individuals regardless of their ancestry.
Ethics
Informed consent was obtained from all participants, and all ethical approvals for the study were obtained by the Institutional Review Boards of the MGB Healthcare System. Further, this project is under UK Biobank application ID 37072.
Role of funders
None of the funding sources played a role in the study design, data collection, data analyses, interpretation, or writing the manuscript.
Results
The RA GRS PheWAS consisted of 316,166 participants from the UK Biobank, of which 140,005 (44.3%) were male and 176,161 (55.7%) were female. The mean age (SD) of participants was 57.1 (8.1) years, and most participants self-reported European ancestry (94.2%) while fewer reported Asian (2.2%) or African ancestry (1.6%). The most common conditions present in the study population based on PheWAS codes were essential hypertension (28.2%), abdominal hernia (17.5%), osteoarthrosis (15.2%), hyperlipidemia (13.7%), and esophagitis and gastroesophageal reflux disease (13.6%). The mean age (SD) of participants in the MGB Biobank replication cohort was 58.8 (17.2), and 18,050 (52.8%) participants were male.
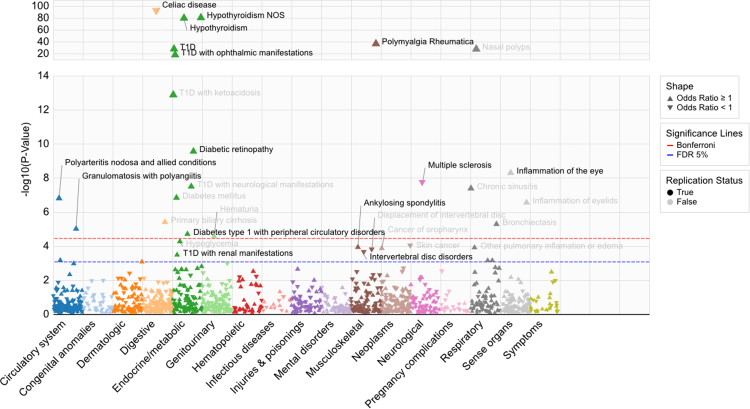
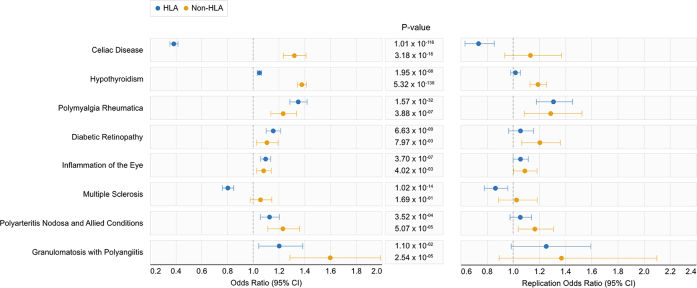
As a positive control, we first confirmed the association between the RA GRS for RA (OR, 1.46; 95% CI, 1.42–1.51; [Wald Test]). The PheWAS excluding participants with RA identified 20 phenotypes significantly associated with the RA GRS, of which 13 (65%) were immune mediated conditions (Fig. 1). The threshold for significance after Bonferroni correction at a familywise error rate of 5% was . Among 34,195 participants in the MGB Biobank, we replicated 11 of the 20 significant associations (Table 1). The phenotypes that replicated with the highest significance (lowest p-value) included Celiac disease (OR, 0.65; 95% CI, 0.63–0.68), hypothyroidism (OR, 1.14; 95% CI, 1.12–1.16), polymyalgia rheumatica (OR, 1.32; 95% CI, 1.26–1.38), type 1 diabetes (OR, 1.19; 95% CI, 1.15–1.23), and complications of type 1 diabetes, e.g. ophthalmic and circulatory manifestations (Table 1). Estimated effect sizes were highly concordant between the UK Biobank and MGB Biobank studies (Table 1 and Supplementary Table S2).
Fig. 1.
Phenome-Wide Association Manhattan Plot of Rheumatoid Arthritis Genetic-Risk Score in UK Biobank. Type 1 diabetes is abbreviated as “TID” in the figure. The Bonferroni threshold in red color denotes Bonferroni correction at a familywise error rate of 5%. The False Discovery Rate (FDR) 5% threshold in blue color was defined using the Benjamini-Hochberg procedure. values calculated using the Wald test.
Table 1.
Significant associations of rheumatoid arthritis genetic-risk score with HLA and non-HLA variants with Phenome-Wide Association Study (PheWAS) codes in the UK Biobank that were also replicated in the Mass General Brigham (MGB) Biobank.
| Phenotype description | UK Biobank |
MGB biobank (Replication cohort) |
|||
|---|---|---|---|---|---|
| Prevalence (%) | OR (95% CI) | value | OR (95% CI) | value | |
| Celiac disease | 0.66 | 0.65 (0.63–0.68) | 0.86 (0.76–0.96) | ||
| Hypothyroidism | 5.50 | 1.14 (1.12–1.16) | 1.07 (1.04–1.10) | ||
| Polymyalgia Rheumatica | 0.47 | 1.32 (1.26–1.38) | 1.31 (1.19–1.43) | ||
| Diabetes mellitus | |||||
| Type 1 diabetes | 1.10 | 1.19 (1.15–1.23) | 1.10 (1.03–1.16) | ||
| Type 1 diabetes with ophthalmic manifestations | 0.20 | 1.37 (1.28–1.47) | 1.40 (1.19–1.65) | ||
| Type 1 diabetes with peripheral circulatory disorders | 0.04 | 1.41 (1.20–1.64) | 1.30 (1.02–1.66) | ||
| Diabetic retinopathy | 0.60 | 1.14 (1.10–1.19) | 1.10 (1.02–1.19) | ||
| Inflammation of the eye | 1.11 | 1.09 (1.06–1.13) | 1.07 (1.02–1.12) | ||
| Multiple sclerosis | 0.46 | 0.88 (0.84–0.92) | 0.92 (0.84–1.00) | ||
| Vasculitides | |||||
| Polyarteritis nodosa and allied conditions | 0.31 | 1.16 (1.10–1.22) | 1.09 (1.02–1.16) | ||
| Granulomatosis with polyangiitis | 0.06 | 1.31 (1.16–1.47) | 1.29 (1.04–1.60) | ||
values calculated using the Wald test.
Medical record review in the MGB Biobank estimated PPV of PheWAS codes ranging from 32% to 98% (Table 2), and phenotypes with PPV>0.8 were reported in the downstream analyses (Fig. 2, Fig. 3). The PheWAS codes with highest PPV were those for granulomatous polyangiitis (GPA), polymyalgia rheumatica, multiple sclerosis (MS), hypothyroidism, and Celiac disease. The phenotype group for inflammation of the eye included inflammation of the eyelids, conjunctivitis, and, uveitis (Supplementary Table S3).
Table 2.
Positive Predictive Value (PPV) based on Medical Record Review of Phenome-Wide Association Study (PheWAS) codes significantly associated with rheumatoid arthritis genetic-risk score.
| Characteristic | Positive predictive value |
|---|---|
| Celiac disease | 0.86 |
| Hypothyroidism | 0.90 |
| Polymyalgia Rheumatica | 0.98 |
| Type 1 diabetes | 0.32 |
| Type 1 diabetes with ophthalmic manifestations | 0.70 |
| Type 1 diabetes with peripheral circulatory disorders | 0.60 |
| Diabetic retinopathy | 0.84 |
| Inflammation of the eye | 0.84 |
| Multiple sclerosis | 0.94 |
| Polyarteritis nodosa and allied conditions | 0.86 |
| Granulomatosis with polyangiitis | 0.98 |
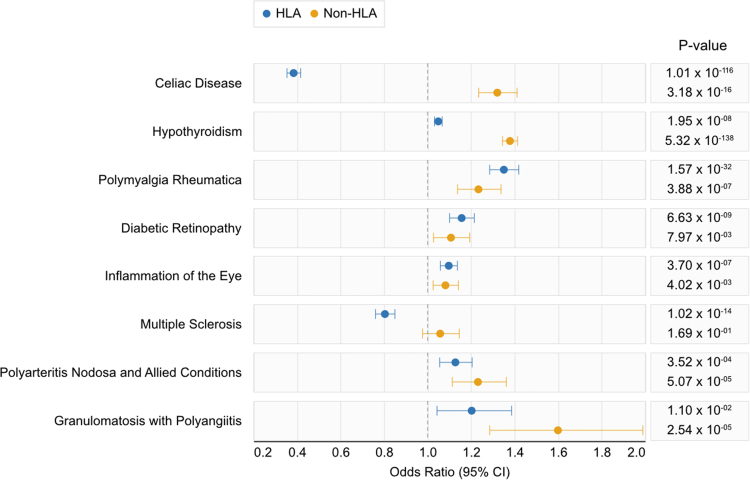
Fig. 2.
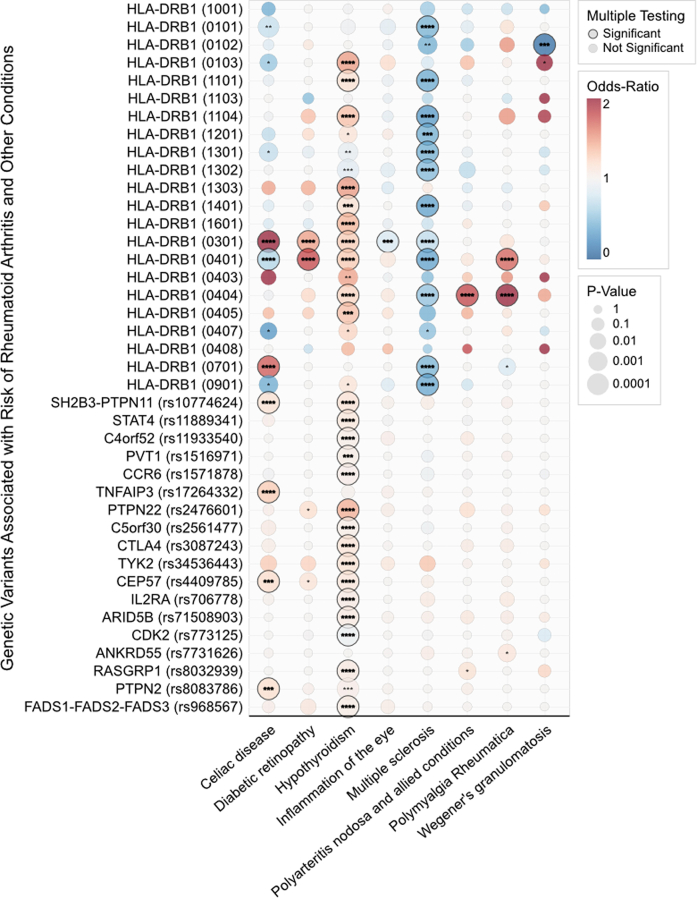
Effect sizes from the UK Biobank of human leukocyte antigen (HLA) GRS and non-HLA GRS among significantly associated phenotypes that were successfully replicated and confirmed using medical record review. Effect sizes were estimated by regressing the GRS on the phenotype while adjusting for age at enrollment, sex, healthcare utilization, and first five principal components. values calculated using the Wald test.
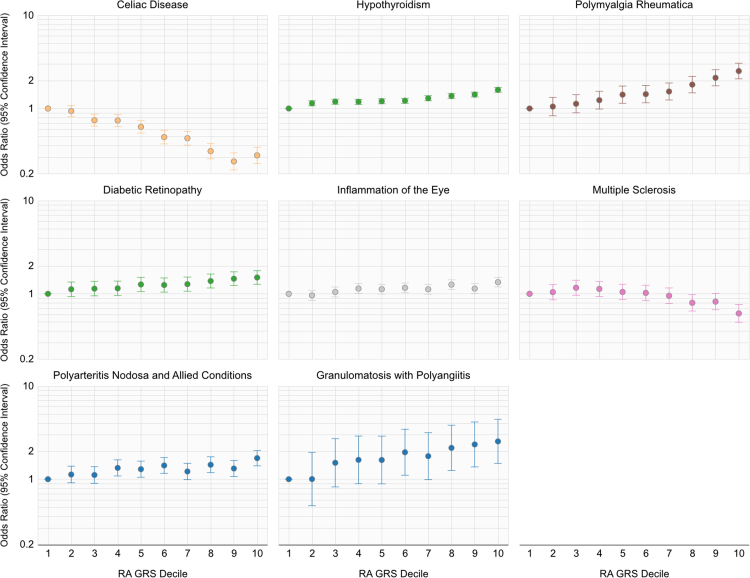
Fig. 3.
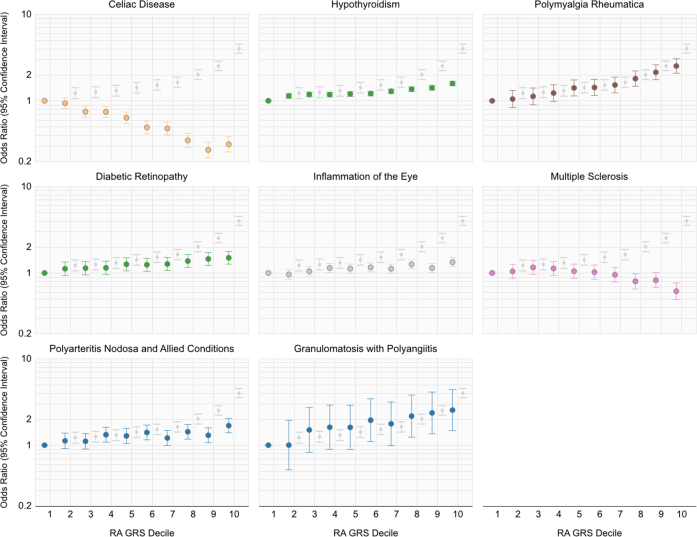
Odds-ratio (OR) for a phenotype in a specified decile relative to individuals in the first decile of the composite rheumatoid arthritis genetic-risk score incorporating HLA and non-HLA variants.
Eight phenotypes were statistically significant after Bonferroni correction, replicated in MGB Biobank cohort, and confirmed to have a PPV>0.8 for the accuracy of the phenotype based on medical record review. The relative contribution of the genetic effect between HLA and non-HLA alleles for these phenotypes is shown in Fig. 2. The ORs between HLA GRS and non-HLA GRS largely shared the same direction of association, with the exception of Celiac disease and multiple sclerosis. The RA HLA GRS was associated with reduced risk for Celiac disease, while the non-HLA GRS was associated with significantly increased risk for Celiac disease (Fig. 2). In MS, the HLA GRS was associated with reduced odds, while the non-HLA GRS had no association (Fig. 2). Strikingly, the non-HLA GRS was strongly associated with greater odds of disease across all eight phenotypes, seven of which are statistically significant effects after Bonferroni correction (Fig. 2). Supplementary Fig. S1 shows a side-by-side comparison of the results in the UK Biobank with the MGB Biobank, demonstrating consistent directions of effect.
The odds of developing a phenotype among participants in each decile relative to participants with lowest the GRS in the first decile is reported in Fig. 3, and significant dose–response relationships were observed for all depicted curves. We further plot the odds of developing RA among participants in each decile relative to participants in the first decile as a reference in Supplementary Fig. S2. A higher composite RA GRS decile was associated with a higher odds of having complications of diabetes, i.e., retinopathy, with values of the trends of the OR's across GRS deciles ranging from to [Wald Test]. For Celiac disease and MS, each increasing decile for the RA GRS was associated with a reduced odds of disease, with values of the trends of the ORs across GRS deciles being to [Wald Test].
The individual risk allele analysis showed that the majority of the shared associations across phenotypes were driven by the HLA-DRB1, 03:01, 04:01, and 04:04 alleles, however in varying directional effects between phenotypes (Supplementary Fig. S3).
The results of the sensitivity analyses in the UK Biobank restricting to individuals of European ancestry were highly concordant to the results using the entire cohort adjusting for population stratification (Supplementary Table S4).
Discussion
In this large-scale population-based study validated in 2 independent cohorts, we identified conditions with increased or reduced risk in association with an increasing burden of RA risk alleles. A higher RA GRS was associated with increased risk for GPA, polyarteritis nodosa, inflammatory conditions of the eye, PMR and T1D complications. In contrast, the RA GRS was inversely associated with celiac and multiple sclerosis. We also provide novel findings demonstrating the opposite effects of HLA and non-HLA alleles particularly for celiac disease risk. Additionally, the HLA GRS was associated with reduced odds for MS while the non-HLA GRS had no association. Further, we found that the non-HLA GRS was associated with greater odds of all eight validated conditions, in contrast to disease-specific HLA GRS directions of associations. These shared genetic risk factors suggest robustly shared pathways for pathogenesis with RA especially outside of the HLA region and provide intriguing hypothesis generating data with implications for screening or for targeted treatment either for the development of the condition or disease-related complications.
In this study, we built upon a few studies identifying the association between RA risk alleles with GPA by replicating across multiple large independent cohorts for this uncommon condition with a poor prognosis.34,35 The association between RA risk factors and GPA were driven by non-HLA alleles. In this study we demonstrated a significant dose–response relationship between RA GRS and risk for GPA ( [Wald Test]), suggesting that the RA that the GRS may provide data to assist with classifying high- and low-risk individuals in real-world settings. To our knowledge, a GRS for GPA or polyarteritis nodosa has not yet been developed. As concerted efforts are underway to develop methods using genetics to screen for at-risk patients prior to the presentation of the condition, future studies can consider evaluating subjects with a high RA GRS for RA and GPA.
Prior studies have identified an association between RA risk alleles with increased risk of type 1 diabetes.36, 37, 38 The present study observed that RA risk alleles may be informative not only for predicting risk of type 1 diabetes, but also for poor outcomes among those with type 1 diabetes, such as retinopathy.39 Both the HLA and non-HLA GRS were associated with increased risk of type 1 diabetes, demonstrating that the genetic effects of the two regions were in the same direction. Of note, a variant in PTPN22 is a known genetic risk factor for both RA and type 1 diabetes and is likely contributing to the shared association of the two phenotypes with the non-HLA GRS.40
This study also highlights pleiotropy resulting in opposite directions of effect observed, with the largest differences in the HLA region for RA and celiac. Previous studies have implicated variants in the HLA class II region as risk alleles for Celiac and RA, suggesting a similar molecular pathogenesis in both diseases through autoantigen specific CD4+ T cell immune responses.20,21,41, 42, 43, 44, 45 However, genetic predisposition for Celiac disease has been linked to HLA-DQ2 and HLA-DQ8 haplotypes, while genetic predisposition for RA is instead attributed to HLA-DRB1 haplotypes.20,43,44 The observed shared but opposite associations in the HLA-DRB1 region may be a result of differing functionality across cell types, or differences in the host environment resulting in a differing gene–environment interaction.46 At the population level, small-scale studies have also reported decreased prevalence of RA among patients with Celiac disease.47, 48, 49, 50
A decreased risk for MS was associated with the RA GRS, a finding which has been similarly observed in other genetic and epidemiologic studies.17,51, 52, 53 When separating the RA GRS into HLA and non-HLA components, we observed that the HLA GRS drove the observed signal for reduced odds while the effect size of the non-HLA GRS was minimal (OR: 1.06; 95%: 0.98–1.15; [Wald Test]). It has been established that the HLA-DRB1 15:01 variant confers the most risk for MS and further studies have found this variant to be associated with a reduced odds for RA.20,54,55 While the HLA-DRB1 15:01 variant was not specifically examined in this study, the observations of a similar and reciprocal negative relationship, where RA risk variants in the HLA-DRB1 gene were associated with reduced odds for MS is in line with the literature. Previous genetic studies have also reported that RA and MS shared non-HLA risk alleles which increased risk of both diseases, however, the current study did not find any significant association between non-HLA GRS and MS, suggesting that previously reported associations may have been attenuated when considering additive effects of many risk alleles in the GRS.56
When linking our knowledge from current clinical care, we observed that the genetic relationships somewhat mirror treatments that mitigate or exacerbate the conditions, i.e., phenotypes where RA GRS associated with increased risk share similar therapies. Methotrexate is the first-line therapy for RA and is also an option for PMR refractory to steroid therapy, and specific manifestations of small vessel vasculitis, e.g., inflammatory joint disease and upper airway disease. Two trials tested MTX in inducing T1D remission had mixed results. In one study remission was achieved in combination with another immunomodulator, while the other had no effect.57,58 However, MTX has not been tested in preventing complications of T1D such as diabetic retinopathy observed in this study. We did not identify studies of MTX for treatment or prevention of autoimmune thyroiditis.
The RA GRS was associated with a reduced odds of celiac and MS and there are no common therapies to date. However, the experience of targeting effective pathways for RA exacerbating MS correlates with the genomic findings. The tumor necrosis factor (TNF) pathway is a well-established effective target for the treatment of RA. This targeting this pathway was tested in MS using lenercept and was observed to increase MS exacerbations and neurologic deficits.59,60 The mainstay of treatment for celiac is avoidance of gluten.
Limitations
This study has limitations. PheWAS codes are based on ICD codes whose accuracy is known to vary. We therefore performed chart review in the MGB Biobank where we had access to the medical records and reported associations with PPV>0.8. This approach has the potential to miss true associations for phenotypes that are less well defined by ICD codes due to smaller numbers of cases defined and corresponding insufficient statistical power. Further, the standard mapping of ICD codes to PheWAS codes defines phenotypes that are not independent of each other. We sought to account for correlated outcomes using Bonferroni and Benjamini-Hochberg FDR control procedures.
Second, the majority of study participants were of European ancestry, and thus our findings may not generalize well to other populations, and further work is needed to robustly study these outcomes in non-European populations. In line with this point, the previously published effect sizes of genetic variants to construct the RA GRS were estimated from populations that were predominantly of European ancestry. Thus, we performed a sensitivity analysis restricting the population to individuals of individual ancestry. The sensitivity analysis’ results were nearly identical with results from the main analysis, suggesting that the reported signals were not due to differences in populations. Overall, however, more data are needed regarding RA risk factors among individuals of non-European ancestry, specifically African ancestry. Further, when constructing the HLA GRS, we were unable to use amino acid polymorphism data as it was not centrally available at the UK Biobank. Lastly, further replication studies in independent cohorts are needed to confirm and generalize the observed associations in this study.
Conclusion
In this study, we provide a roadmap approach to study the relationships across conditions using RA genetics as an anchor point. Our conclusions were supported by existing studies, highlighting the advantages of using a data-driven, phenome-wide approach over genetic studies with one or two outcomes. Overall, we identified novel associations demonstrating differential risk between HLA and non-HLA alleles, and their associations with other autoimmune conditions, particularly in Celiac disease where HLA and non-HLA alleles had opposite directions of effect. As the biomedical field moves toward incorporating genetics into clinical practice, it will become important to thoroughly understand how RA risk loci may predispose patients to other inflammatory conditions. Finally, we observed that the patterns of shared genetics mirror effects of treatments. Specifically, the genetic variants associated with RA were associated with a reduced risk for MS. The most common targeted therapy in RA inhibits the TNF pathway resulting in control of RA symptoms, and in a trial of TNF in MS, inhibiting the pathway had an opposite effect. Future directions include further elucidating the specific pathways that these conditions share in their pathogenesis as well as developing novel therapeutics that target these pathways. Data from this study can serve as hypothesis generating when considering therapies for other autoimmune conditions or their complications.
Contributors
Conceptualization: HGZ, TC, KPL; Data Curation: HGZ, SH, KD, CLB, JC; Formal Analysis: HGZ; Funding Acquisition: TC, KPL; Investigation: HGZ, GM, TS, SH, SL, DW, PM, SR, TRC, JC, CH, TC, KPL; Methodology: HGZ, TC, KPL; Project Administration: JS, DW; Resources: KHY, TC, KPL; Supervision: TC, KPL; Validation: HGZ, GM, TS, SH, PM, SR, TRC, JC, CH, TC, KPL; Visualization: SL; Writing-Original Draft: HGZ, KPL; Writing-review & editing: HGZ, GM, KY, SR, TC, KPL. All authors read and approved the final version of the manuscript. HGZ, KD, CLB, TC, KPL verified the underlying data.
Data sharing statement
All data will be available to approved users of the UK Biobank upon application. Researchers must obtain proper Institutional Review Board ethical approval to access data of the MGB healthcare system.
Declaration of interests
SR is a founder of Mestag, a scientific advisor for Rheos Medicines, serves on the advisory boards for Janssen and Pfizer, and is a consultant for Sanofi. The remaining authors have no declarations of interests.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104581.
Appendix A. Supplementary data
Supplementary Fig. S1.
Supplementary Fig. S2.
Supplementary Fig. S3
References
- 1.Sparks J.A. Rheumatoid arthritis. Ann Intern Med. 2019 Jan 1;170(1):ITC1–16. doi: 10.7326/AITC201901010. [DOI] [PubMed] [Google Scholar]
- 2.Mian A., Ibrahim F., Scott D.L. A systematic review of guidelines for managing rheumatoid arthritis. BMC Rheumatol. 2019 Oct 22;3(1):42. doi: 10.1186/s41927-019-0090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janke K., Biester K., Krause D., et al. Comparative effectiveness of biological medicines in rheumatoid arthritis: systematic review and network meta-analysis including aggregate results from reanalysed individual patient data. BMJ. 2020 Jul 7;370:m2288. doi: 10.1136/bmj.m2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada Y., Wu D., Trynka G., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014 Feb 20;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smolen J.S., Aletaha D., Barton A., et al. Rheumatoid arthritis. Nat Rev Dis Prim. 2018 Feb 8;4 doi: 10.1038/nrdp.2018.1. [DOI] [PubMed] [Google Scholar]
- 6.Smolen J.S., Aletaha D., McInnes I.B. Rheumatoid arthritis. Lancet. 2016 Oct 22;388(10055):2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 7.Bycroft C., Freeman C., Petkova D., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018 Oct;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudlow C., Gallacher J., Allen N., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015 Mar 31;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igo R.P., Kinzy T.G., Bailey J.N.C. Genetic risk scores. Curr Protoc Hum Genet. 2019 Dec;104(1):e95. doi: 10.1002/cphg.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan L., Shen H., Gelaye B., et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019 Jul 25;10(1):3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ombrello M.J., Sikora K.A., Kastner D.L. Genetics, genomics and their relevance to pathology and therapy. Best Pract Res Clin Rheumatol. 2014 Apr;28(2):175–189. doi: 10.1016/j.berh.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gratten J., Visscher P.M. Genetic pleiotropy in complex traits and diseases: implications for genomic medicine. Genome Med. 2016 Jul 19;8(1):78. doi: 10.1186/s13073-016-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X., Lucas A.M., Veturi Y., et al. Large-scale genomic analyses reveal insights into pleiotropy across circulatory system diseases and nervous system disorders. Nat Commun. 2022 Jun 14;13(1):3428. doi: 10.1038/s41467-022-30678-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarwood A., Huizinga T.W.J., Worthington J. The genetics of rheumatoid arthritis: risk and protection in different stages of the evolution of RA. Rheumatology. 2016 Feb;55(2):199–209. doi: 10.1093/rheumatology/keu323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X.H., Bo L., Cao R.R., et al. Systematic evaluation of rheumatoid arthritis risk by integrating lifestyle factors and genetic risk scores. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.901223. https://www.frontiersin.org/articles/10.3389/fimmu.2022.901223 [cited 2022 Aug 23]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritsche L.G., Gruber S.B., Wu Z., et al. Association of polygenic risk scores for multiple cancers in a phenome-wide study: results from the Michigan genomics initiative. Am J Hum Genet. 2018 Jun 7;102(6):1048–1061. doi: 10.1016/j.ajhg.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai V.K., Shi M., Feng Q., et al. Pleiotropy in the genetic predisposition to rheumatoid arthritis: a phenome-wide association study and inverse variance–weighted meta-analysis. Arthritis Rheumatol. 2020;72(9):1483–1492. doi: 10.1002/art.41291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen X., Howard D.M., Adams M.J., et al. A phenome-wide association and Mendelian Randomisation study of polygenic risk for depression in UK Biobank. Nat Commun. 2020 May 8;11(1):2301. doi: 10.1038/s41467-020-16022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai T., Zhang Y., Ho Y.L., et al. Association of interleukin 6 receptor variant with cardiovascular disease effects of interleukin 6 receptor blocking therapy. JAMA Cardiol. 2018 Sep;3(9):849–857. doi: 10.1001/jamacardio.2018.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raychaudhuri S., Sandor C., Stahl E.A., et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44(3):291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregersen P.K., Silver J., Winchester R.J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987 Nov;30(11):1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 22.Jia X., Han B., Onengut-Gumuscu S., et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013 Jun 6;8(6) doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gainer V.S., Cagan A., Castro V.M., et al. The biobank portal for partners personalized medicine: a query tool for working with consented biobank samples, genotypes, and phenotypes using i2b2. J Personalized Med. 2016 Feb 26;6(1):11. doi: 10.3390/jpm6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlson E.W., Boutin N.T., Hoffnagle A.G., Allen N.L. Building the partners HealthCare biobank at partners personalized medicine: informed consent, return of research results, recruitment lessons and operational considerations. J Personalized Med. 2016 Jan 14;6(1):2. doi: 10.3390/jpm6010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro V.M., Gainer V., Wattanasin N., et al. The Mass General Brigham Biobank Portal: an i2b2-based data repository linking disparate and high-dimensional patient data to support multimodal analytics. J Am Med Inf Assoc. 2021 Nov 28:ocab264. doi: 10.1093/jamia/ocab264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dilthey A., Leslie S., Moutsianas L., et al. Multi-population classical HLA type imputation. PLoS Comput Biol. 2013 Feb 14;9(2) doi: 10.1371/journal.pcbi.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.https://biobank.ndph.ox.ac.uk/showcase/field.cgi?id=22182 Data-Field 22182 (HLA Imputation values) [Internet]. [cited 2023 Mar 14]. Available from:
- 28.Wu P., Gifford A., Meng X., et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform. 2019 Nov 29;7(4) doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Højsgaard S., Halekoh U., Yan J. The R package geepack for generalized estimating equations. J Stat Software. 2006;15:1–11. [Google Scholar]
- 30.Liang K.Y., Zeger S.L. Longitudinal data analysis using generalized linear models. Biometrika. 1986 Apr 1;73(1):13–22. [Google Scholar]
- 31.Chen M.H., Liu X., Wei F., et al. A comparison of strategies for analyzing dichotomous outcomes in genome-wide association studies with general pedigrees. Genet Epidemiol. 2011 Nov;35(7):650–657. doi: 10.1002/gepi.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bland J.M., Altman D.G. Multiple significance tests: the Bonferroni method. BMJ. 1995 Jan 21;310(6973):170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 34.Chung S.A., Xie G., Roshandel D., et al. Meta-analysis of genetic polymorphisms in granulomatosis with polyangiitis (Wegener's) reveals shared susceptibility loci with rheumatoid arthritis. Arthritis Rheum. 2012 Oct;64(10):3463–3471. doi: 10.1002/art.34496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemminki K., Li X., Sundquist J., Sundquist K. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 2009 Mar;60(3):661–668. doi: 10.1002/art.24328. [DOI] [PubMed] [Google Scholar]
- 36.Liao K.P., Gunnarsson M., Källberg H., et al. A specific association exists between type 1 diabetes and anti-CCP positive rheumatoid arthritis. Arthritis Rheum. 2009 Mar;60(3):653–660. doi: 10.1002/art.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiani A.K., Jahangir S., Jahngir S., et al. Genetic link of type 1 diabetes susceptibility loci with rheumatoid arthritis in Pakistani patients. Immunogenetics. 2015 Jun;67(5–6):277–282. doi: 10.1007/s00251-015-0839-0. [DOI] [PubMed] [Google Scholar]
- 38.Onengut-Gumuscu S., Chen W.M., Burren O., et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015 Apr;47(4):381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noble J.A., Valdes A.M. Genetics of the HLA region in the prediction of type 1 diabetes. Curr Diabetes Rep. 2011 Dec;11(6):533–542. doi: 10.1007/s11892-011-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maziarz M., Janer M., Roach J.C., et al. The association between the PTPN22 1858C>T variant and type 1 diabetes depends on HLA risk and GAD65 autoantibodies. Genes Immun. 2010 Jul;11(5):406–415. doi: 10.1038/gene.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koning F., Thomas R., Rossjohn J., Toes R.E. Coeliac disease and rheumatoid arthritis: similar mechanisms, different antigens. Nat Rev Rheumatol. 2015 Aug;11(8):450–461. doi: 10.1038/nrrheum.2015.59. [DOI] [PubMed] [Google Scholar]
- 42.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978 Apr 20;298(16):869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 43.Tjon J.M.L., van Bergen J., Koning F. Celiac disease: how complicated can it get? Immunogenetics. 2010 Oct;62(10):641–651. doi: 10.1007/s00251-010-0465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abadie V., Sollid L.M., Barreiro L.B., Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol. 2011;29:493–525. doi: 10.1146/annurev-immunol-040210-092915. [DOI] [PubMed] [Google Scholar]
- 45.Vader W., Stepniak D., Kooy Y., et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12390–12395. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ota M., Nagafuchi Y., Hatano H., et al. Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases. Cell. 2021 May 27;184(11):3006–3021.e17. doi: 10.1016/j.cell.2021.03.056. [DOI] [PubMed] [Google Scholar]
- 47.Lauret E., Rodrigo L. Celiac disease and autoimmune-associated conditions. BioMed Res Int. 2013;2013 doi: 10.1155/2013/127589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iqbal T., Zaidi M.A., Wells G.A., Karsh J. Celiac disease arthropathy and autoimmunity study. J Gastroenterol Hepatol. 2013 Jan;28(1):99–105. doi: 10.1111/j.1440-1746.2012.07272.x. [DOI] [PubMed] [Google Scholar]
- 49.Francis J., Carty J.E., Scott B.B. The prevalence of coeliac disease in rheumatoid arthritis. Eur J Gastroenterol Hepatol. 2002 Dec;14(12):1355–1356. doi: 10.1097/00042737-200212000-00011. [DOI] [PubMed] [Google Scholar]
- 50.Neuhausen S.L., Steele L., Ryan S., et al. Co-occurrence of celiac disease and other autoimmune diseases in celiacs and their first-degree relatives. J Autoimmun. 2008 Sep;31(2):160–165. doi: 10.1016/j.jaut.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sirota M., Schaub M.A., Batzoglou S., Robinson W.H., Butte A.J. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009 Dec;5(12) doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Somers E.C., Thomas S.L., Smeeth L., Hall A.J. Autoimmune diseases co-occurring within individuals and within families: a systematic review. Epidemiology. 2006 Mar;17(2):202–217. doi: 10.1097/01.ede.0000193605.93416.df. [DOI] [PubMed] [Google Scholar]
- 53.Restrepo N.A., Butkiewicz M., McGrath J.A., Crawford D.C. Shared genetic etiology of autoimmune diseases in patients from a biorepository linked to de-identified electronic health records. Front Genet. 2016 Oct 20;7:185. doi: 10.3389/fgene.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caillier S.J., Briggs F., Cree B.A.C., et al. Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J Immunol. 2008 Oct 15;181(8):5473–5480. doi: 10.4049/jimmunol.181.8.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olerup O., Hillert J. HLA class II-associated genetic susceptibility in multiple sclerosis: a critical evaluation. Tissue Antigens. 1991 Jul;38(1):1–15. doi: 10.1111/j.1399-0039.1991.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki A., Kochi Y., Okada Y., Yamamoto K. Insight from genome-wide association studies in rheumatoid arthritis and multiple sclerosis. FEBS Lett. 2011 Dec 1;585(23):3627–3632. doi: 10.1016/j.febslet.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 57.Buckingham B.A., Sandborg C.I. A randomized trial of methotrexate in newly diagnosed patients with type 1 diabetes mellitus. Clin Immunol. 2000 Aug;96(2):86–90. doi: 10.1006/clim.2000.4882. [DOI] [PubMed] [Google Scholar]
- 58.Sobel D.O., Henzke A., Abbassi V. Cyclosporin and methotrexate therapy induces remission in type 1 diabetes mellitus. Acta Diabetol. 2010 Sep;47(3):243–250. doi: 10.1007/s00592-010-0188-2. [DOI] [PubMed] [Google Scholar]
- 59.TNF neutralization in MS: results of a randomized, placebo-controlled multicenter study. The lenercept multiple sclerosis study group and the university of British columbia MS/MRI analysis group. Neurology. 1999 Aug 11;53(3):457–465. [PubMed] [Google Scholar]
- 60.Huang H., Fang M., Jostins L., et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017 Jul 13;547(7662):173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.