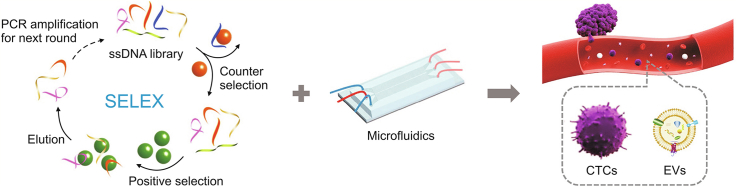
Abstract
Liquid biopsy is a technology that exhibits potential to detect cancer early, monitor therapies, and predict cancer prognosis due to its unique characteristics, including noninvasive sampling and real-time analysis. Circulating tumor cells (CTCs) and extracellular vesicles (EVs) are two important components of circulating targets, carrying substantial disease-related molecular information and playing a key role in liquid biopsy. Aptamers are single-stranded oligonucleotides with superior affinity and specificity, and they can bind to targets by folding into unique tertiary structures. Aptamer-based microfluidic platforms offer new ways to enhance the purity and capture efficiency of CTCs and EVs by combining the advantages of microfluidic chips as isolation platforms and aptamers as recognition tools. In this review, we first briefly introduce some new strategies for aptamer discovery based on traditional and aptamer-based microfluidic approaches. Then, we subsequently summarize the progress of aptamer-based microfluidics for CTC and EV detection. Finally, we offer an outlook on the future directional challenges of aptamer-based microfluidics for circulating targets in clinical applications.
Keywords: Aptamer, Microfluidic, Circulating tumor cells, Extracellular vesicles, Bioanalysis
Graphical abstract
Highlights
-
•
Liquid biopsy is important for cancer diagnosis and personalized therapy.
-
•
Aptamer is emerged as an ideal recognition tool for CTC and EV detection.
-
•
Microfluidic chip is considered as a powerful platform for CTC and EV detection.
-
•
Aptamer-based microfluidics for the detection of CTCs and EVs is summarized.
1. Introduction
Accurate diagnosis and personalized therapy remain a great challenge for precision cancer medicine [1]. As the gold standard, tissue biopsy can provide histological information and molecular information on cancer to assist in treating and managing cancer [2]. However, tissue biopsy is an invasive procedure, which limits its application in routine disease screening [3]. Moreover, a tissue biopsy is conducted only after a mass lesion is detected by medical imaging [4]. The assessments only involves the primary tumor tissues and do not reflect the real-time status of cancer and the efficacy of the targeted treatments [5,6]. To address these challenges, liquid biopsy, which is considered as a noninvasive technology, has become an acceptable alternative method to tissue biopsy for noninvasive diagnosis and real-time monitoring of cancer [[7], [8], [9]]. By analysing and detecting circulating targets in various biofluids, liquid biopsy with noninvasive sampling can realize dynamic and real-time monitoring of cancer therapy response and progression [10,11]. In addition, liquid biopsy provides all-sided dynamic information on both metastatic and primary tumors to assist cancer treatment due to the unique characteristics of circulating targets [12,13].
Circulating targets have the potential to detect tumors early because circulating targets are present at an early stage of cancer [14,15]. Circulating targets can also reflect the real-time pathological and physiological status of the original cancer tissue [16]. Circulating tumor cells (CTCs) and extracellular vesicles (EVs) are two important components of circulating targets. CTCs are secreted from primary tumor tissues and metastasize by blood circulation in the vasculature [17]. CTCs are associated with tumor progression and exhibit great prognostic and diagnostic value [18,19]. EVs are heterogeneous lipid-membrane vesicles derived from various parent cells in large quantities [20,21]. EVs convey cargoes, including lipids with superior stability, nucleic acids, and proteins, to carry out intercellular communications [22,23]. EVs have been used as prospective biomarkers for prognosis, therapy assessment, metastasis monitoring, and early detection of cancers because EVs contain abundant molecular information and exhibit unique stability [[24], [25], [26], [27]]. Thus, it is necessary to analyse CTCs/EVs at the single-cell/EV level due to their heterogeneity [28,29].
Microfluidic techniques with a high level of integration exhibit a superior capability of manipulating and controlling fluids accurately, providing unprecedented opportunities for liquid biopsy [21,30]. Based on unique hydrodynamic effects, such as inertial effects, viscoelastic effects, and dean flow, microfluidic chips combined with specially designed microstructures can be used to continuously isolate and rapidly detect circulating targets from body fluids [[31], [32], [33], [34], [35]]. Microfluidic platforms achieve highly specific and sensitive detection of CTCs and EVs by implementing various detection methods (e.g., visual methods, electrochemical methods, fluorescence methods, and surface-enhanced Raman scattering (SERS) methods), providing prognostic and diagnostic information for cancer research.
Microfluidics also offers a microstructured platform for anchoring recognition probes for capturing targets [16]. Affinity-based microfluidics can reduce nonspecific adsorbed interferents and enhance target-interface interactions to specifically and efficiently isolate circulating targets by manipulating fluids [36,37]. Recognition ligands play an important role in affinity-based liquid biopsy because of the particular interactions between circulating target biomarkers and recognition ligands [38,39]. As a recognition ligand, aptamers are single-stranded oligonucleotides used for specific binding to targets by unique tertiary structures [40,41]. Compared to antibodies, aptamers show many prominent characteristics for liquid biopsies, as aptamers are easily synthesized, evolved, tailored, and engineered. First, systematic evolution of ligands by exponential enrichment (SELEX) [42,43] can directly identify the aptamer. CTCs and EVs are used as evolutionary targets to identify aptamers and do not require prior knowledge owing to the high flexibility of SELEX [[44], [45], [46]]. Additionally, enhancing the affinity and specificity for aptamers does not require a complicated adjustment of the selection conditions [47]. Second, large quantities of aptamers are acquired by simple chemical synthesis with good reproducibility. Aptamers with specific sites could conveniently modify various functional moieties, providing favourable binding by the controllable and oriented immobilization of aptamers on recognition interfaces. Third, many strategies can be conducted by base-pairing rules for target release and signal amplification because aptamers are oligonucleotides [[48], [49], [50]]. Therefore, aptamers with these superior characteristics are considered as ideal recognition units for liquid biopsies.
Here, we summarize the recent advancements in aptamer-based microfluidics for the detection of CTCs and EVs. First, some strategies are briefly introduced for the evolution of aptamers of circulating targets. Then, aptamer-based microfluidic methods for the detection of CTCs and EVs are reviewed. Moreover, regarding the detection of CTCs and EVs, we also summarize some signal-probe-based amplification strategies. Finally, we provide prospects for future directions and challenges of aptamer-based microfluidics for the detection of CTCs and EVs in clinical applications.
2. Aptamer discovery via SELEX technology
As a gold-standard strategy, SELEX technology can be performed for the generation of nucleic acid aptamers. Very large advances in SELEX technology have occurred in recent decades to obtain aptamers, including recombinant proteins, CTC surface proteins, EVs, or whole cells [[51], [52], [53]]. Existing screening methods include cell SELEX [53], capillary electrophoresis SELEX [54,55], magnetic bead-based SELEX [56,57], microfluidic SELEX [[58], [59], [60]], capture SELEX [[61], [62], [63]], and some comprehensive methods [[64], [65], [66], [67]]. The advantages and disadvantages of commonly used SELEX methods are summarized in Table 1. Below, we mainly introduce conventional aptamer screening methods and screening methods based on microfluidics.
Table 1.
Summary of different methods of currently used systematic evolution of ligands by exponential enrichment (SELEX) for aptamer discovery.
| Method | Key aspects | Advantages | Disadvantages |
|---|---|---|---|
| Capillary electrophoresis SELEX | Electrophoretic mobility for the separation of ions | Fast; target immobilization not needed; and only few rounds of selection needed | Expensive equipment and not suitable for small molecules |
| Cell SELEX | Whole cells as targets | Protein purification not needed; knowledge of the target not needed; and many potential targets available on the cell surface | Costly; time consuming; high level of technical expertise; and suitable for cell surface targets |
| Microfluidic SELEX | SELEX with a microfluidic device | Rapid; automatable; efficient; and suitable for small molecules | Target immobilization needed and low recovery/purity |
| Capture SELEX | Oligonucleotide library is immobilized on a support instead of the targets to identify aptamers | Immobilization of the target not needed; structure-switching aptamers needed; and selection of aptamers for small molecules | Some oligonucleotides might be not selected/released |
2.1. Conventional SELEX
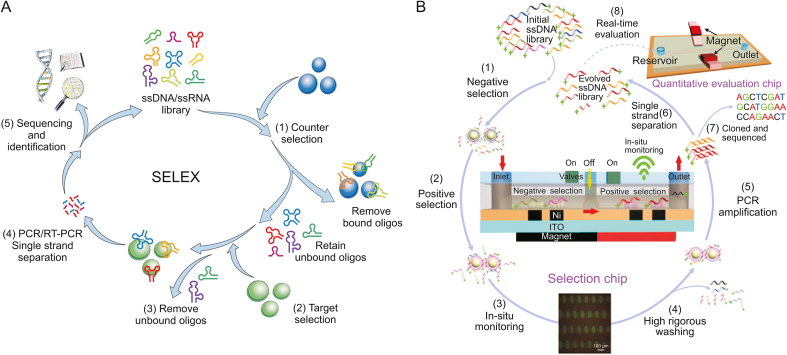
Overall, the traditional steps of aptamer screening, whether for cells, proteins or live animals, include the following (Fig. 1A) [68]: 1) A counterselection process to exclude interference from nontargets. 2) Incubation of the target with a gene library containing random sequences under certain selection conditions. 3) Isolation of oligonucleotides bound to the target. 4) Recovery and polymerase chain reaction (PCR) amplification of the bound oligonucleotides. The cycle of selection is then repeated until enrichment with the desired affinity sequence is achieved. 5) Sequencing and characterization of the obtained candidate aptamer sequences.
Fig. 1.
Aptamer discovery via systematic evolution of ligands by exponential enrichment (SELEX) technology. (A) Conventional and (B) chip-based microfluidic SELEX methods for aptamer discovery. ssDNA: single-stranded DNA; PCR: polymerase chain reaction; RT-PCR: reverse transcription PCR. Reprinted from Refs. [59,68] with permission.
2.2. Microfluidic SELEX
Notably, SELEX in general often encounters limitations, such as low separation efficiency, low success rate, and time-consuming problems. In contrast, microfluidic chips that integrate microchannels, PCR chambers, micropumps, microvalves, or reagents exhibit unique advantages, including high throughput, high automation, and low reagent consumption. The use of a series of ferromagnetic articles in the microfluidic device allows paramagnetic beads coated with the target to be stably and effectively captured, and the captured beads can be processed by a highly rigorous cleaning procedure achieved by the washing buffer. Most of the oligonucleotides that are unbound are washed off within minutes due to the automation advantage of the microfluidic device. By strictly and precisely controlling the washing conditions, aptamers that showed higher affinity could be screened after fewer rounds of selection using an automated microfluidic device compared to general SELEX [69].
To accelerate the discovery of synergists, Hong et al. [59] designed a multifunctional microfluidic screening platform, which greatly improved the screening efficiency and yielded synergists with high specificity and affinity in two rounds of screening (Fig. 1B). This versatile screening platform allowed the magnetic beads to be precisely manipulated at the micron scale, further improving the performance of microfluidic continuous flow-based screening and enhancing the screening process control through in situ real-time monitoring and evaluation. Briefly, bovine serum albumin (BSA) was used as a negative control protein, and mucin 1 (MUC1) was used as a positive control protein. Magnetic nanospheres (MNs) coated with the two proteins were introduced into the selection chip. The single-stranded DNA (ssDNA) library was first pumped into the chip for negative selection, in which a portion of ssDNA bind the BSA protein, thereby reducing the nonspecific binding of some ssDNA to the target protein MUC1. After a first round of screening, the ssDNA library that did not bind then selectively bound to the target protein when flowed through the positive selection unit to obtain the candidate aptamer. The entire screening process occurred under an inverted fluorescence microscope, in which the fluorescence signal was observed before and after incubation with the library, allowing in situ detection to better guide the screening process. After the library was incubated with MUC1, the microchannel was washed strictly with washing buffer, the weakly bound nucleic acid was removed, and nucleosomes bound to target proteins on MNs were then collected and amplified by direct PCR. After PCR amplification, the library could be used for the next round of screening or sequencing. Later, using NaOH to denature double-stranded DNA as ssDNA, an evolved ssDNA pool consisting of fluorescein amidite (FAM)-labelled ssDNA strands was generated, which was isolated from biotinylated antisense DNA strands by strand avidin-coated MNs. A fraction of the evolved ssDNA library was transported into the chip, and the degree of enrichment for each screening round could then be quantitatively assessed by fluorescence intensity. The candidate aptamers of evolved ssDNA libraries could bind specifically to MUC1-coated MNs in microchannels. The selection was stopped if the fluorescence intensity that was enriched did not increase significantly, the PCR product was amplified and further sequenced, and the sequence was selected for further analysis and application. Compared with conventional methods, this method can inhibit approximately 50-fold nonspecifically bound nucleic acids and further remove weakly bound nucleic acids within minutes, with both negative and positive selection. The breeding effect was monitored on-site and in real time. The selected aptamer showed high affinity for MUC1, could specifically mark cancer cells, and could effectively capture exosomes with an efficiency of 64%. The results indicated that this multifunctional chip-based screening platform was an effective method for the rapid and economical preparation of high-quality aptamers.
Typically, microfluidic-based screening methods are very convenient for aptamer discovery, and Sanger sequencing is generally applicable to candidate aptamers that need to be screened with comparably few oligonucleotides. However, sometimes the number of sequences obtained is relatively few, and the oligonucleotides obtained may not be the best bound compound in the library. To solve the drawbacks of the low throughput of regular sequencing and analyse the obtained candidates better, high-throughput sequencing (HTS), together with microfluidic devices, is used in the selection cycle process. By monitoring the trajectory of enrichment in each selection cycle, HTS can discern the supreme fold enrichment in the early rounds of selection. Many researchers agree that compounds that bind most rapidly to their targets in the earliest cycles of selection usually exhibit the best affinity, and therefore, the combination of microfluidic SELEX and HTS has the potential to become a more general technique for aptamer discovery [69].
3. Aptamer-based microfluidics for CTC detection
CTCs, which detach from solid tumor tissue and circulate in blood vessels, are assumed to cause the haematogenous spread of cancer to distant areas [[70], [71], [72]]. With cellular structure, CTCs could deliver more integrated information on the origin of tumor tissue compared to that of other targets. The exceedingly rare CTCs are present in complicated whole blood containing millions of times more abundant blood cells, and the enrichment and detection of rare CTCs are hot spots in recent research. Various techniques and methods have been developed for the detection of CTCs, ranging from immunomagnetic beads [[73], [74], [75]] and size-based filtration systems [76,77] to microfluidic devices [11]. In these strategies, affinity interaction-based cell sorting methods generate higher efficiency and greater specificity and have been widely applied. In comparison, aptamers, as emerging recognition elements, have been shown to exhibit considerable affinities and greater specificities, comparable to that of antibodies. Controlled, directional, high-density immobilization on trapping substrates is achieved by the small size of aptamers and ease of modification by functional fragments. Aptamers can be chemically synthesized, labelled and immobilized at specific sites and have been selected for numerous cancers. The combination of magnetic beads [78], nanostructured interfaces, and microfluidic devices [[79], [80], [81]] can provide a good capturing and separation platform for CTCs.
Microfluidic devices consisting of microchannels allow liquids to be precisely controlled and manipulated, and microfluidic-based separation capabilities can be compatible with aptamer-based affinity strategies to achieve efficient separation and detection of CTCs [82]. The following section mainly concentrates on the latest advances in aptamer-based microfluidics for detecting CTCs.
3.1. Single-target aptamer-based microfluidics
3.1.1. Simple aptamer-based microfluidics
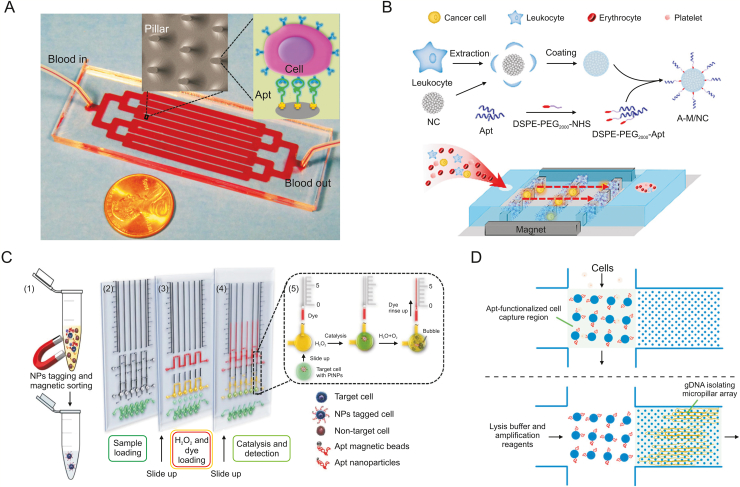
To realize quick analysis without preprocessing cells [83,84], Sheng et al. [85] reported an aptamer-functionalized, micropillar-based microfluidic device that can sort and analyse many types of cells (Fig. 2A). The device was the same size as the microscope slide and consisted of one inlet, one outlet, and eight channels connected by bifurcation. Avidin was first immobilized on the surface of the microchannel/microcolumn by physical adsorption, and then the aptamer was chemically immobilized by biotin-affinity interactions. Capture of target cancer cells was achieved by interaction between the receptor on the surface of the cell and the aptamer, and the large number of micropillars increased the probability of interaction between the cells and the inducer coated on the surface of the channel/column, leading to high capture efficiency. At the optimal flow rate, the device can obtain a capture efficiency of 95% and a purity of 81%. To improve cell release efficiency, Zhu et al. [86] presented a microfluidic device with a functionalized aptamer and temperature sensor on its surface to capture specific cells and release cells mediated by temperature. Microheaters and temperature sensors that generate appropriate temperature changes can reversibly disrupt selected chip regions, allowing the release and recovery of live target cells from these regions for downstream applications. Upon reversal of the temperature change, the inducer-functionalized surface regained binding affinity to the target cells, and this mild temperature change did not affect cell viability.
Fig. 2.
Simple aptamer-based microfluidics for the detection of circulating tumor cells (CTCs). (A) Multichannel microfluidic device for the detection of CTCs. (B) Combining biomimetic magnetosomes with a nickel pattern microfluidic device for the detection of CTCs. (C) Aptamer-conjugated Pt nanoparticles with volumetric bar-chart chip readout for visual quantifiable detection of CTCs. (D) A microfluidic device for the detection of CTCs and isolation of their genomic DNA for specific amplification and sequence analysis. NC: nanoclusters; DSPE: 1,2-distearoyl-sn-glycero-3-phosphoethanolamine; PEG2000: polyethyleneglycol-2000; NHS: N-hydroxysuccinimde; Apt: aptamer; A-M/NC: Apt-linked magnetic nanoclusters; NPs: nanoparticles; gDNA: genomic DNA. Reprinted from Refs. [85,88,91,92] with permission.
The combination of gold nanoparticles (AuNPs) and aptamers can significantly improve the sensitivity of aptamer sensors. In recent studies, Nguyen et al. [87] proposed a simple microfluidic platform. They conjugated sulfhydryl-labelled cell-specific aptamers to the AuNP layer within the channel to capture target cells. In the experiment, the impedance of the modified electrode was measured to show the successful fixation of the aptamer, and the highly selective capture performance of the device for CTC cells was demonstrated by the fluorescence imaging system. To achieve specific capture performance towards CTCs and eliminate the nonspecific adsorption interference of surrounding cells, Zhang et al. [88] proposed a novel and efficient CTC capture detection strategy that combines biomimetic magnetic nanoclusters with nickel-type microfluidic devices (Fig. 2B). Magnetic nanoclusters were disguised with white cell membrane fragments to modify the epithelial cell adhesion molecule (EpCAM) aptamer SYL3C in tumor cells. Next, it was loaded into the microfluidic chip with a magnet, and the aptamers loaded into the magnetron microfluidic device were arranged in a nickel square array, thereby increasing the chance of interaction with the target cells. The results showed that approximately 90% of tumor cells in the blood could be detected in less than 20 min without the need for a background of white blood cells.
The concentrations of circulating CTCs as a promising cancer biomarker must be measured not only at the initial diagnostic phase but also during therapy. The aptamer-based microfluidic devices proposed above provide an effective method to efficiently capture rare cancer cells, but it remains a great challenge to implement in situ detection in microfluidic devices. The main reasons are, on one hand, that most nucleic acid aptamer-cell interactions do not readily produce an output of the assay signal and, on the other hand, this process requires a complex washing procedure to improve the signal background ratio of the aptamer. To address these shortcomings, Cao et al. [89] proposed a miniature multiplexed microfluidic chip for the in situ detection of cancer cells using dye-labelled aptamers and graphene oxide as probes in combination with a Förster resonance energy transfer strategy. The probe underwent conformational changes and fluorescence activation upon binding to targeted cancer cells. Seven different cancer cell samples in this microfluidic chip could be measured simultaneously. The chip could achieve a detection limit of approximately 25 cells/mL, which was approximately 10 times lower than that of ordinary sensors. Chen et al. [90] integrated field-effect transistors (FETs) and chambers to develop a new microfluidic platform for capturing CTCs. The design enabled CTCs from whole blood samples to be captured and specific aptamer-CTC complexes to be counted by FET sensing. In brief, the cancer cell-specific aptamer was first immobilized on FETs to specifically identify cancer cells, the cell capture device allowed efficient and continuous capture of cancer cells, and the FET sensor array allowed cancer cells to be monitored and counted at physiological concentrations. The entire process can be automated on a microfluidic chip without additional sample preparation. Combining the microfluidic device and the FET sensor array enabled cell trapping in the detection region, and the results showed that each sensor can distinguish between zero and three cells and that the entire sensor array can capture up to 42 cancer cells. Moreover, those target cells were detected only when they bound to a specific aptamer on the FET sensor, so this design prevented unwanted interference from other blood cells.
For CTC detection, Yang and co-workers [91] designed a portable platform using a volumetric bar-chart chip (Fig. 2C). They first used nucleic acid aptamer-conjugated nanoparticles to label CTCs in buffer suspensions or artificial blood. Pt nanoparticles could be adsorbed on the sample to react with hydrogen peroxide to produce oxygen and quantify the number of cells by the distance the ink moves. This method was sufficiently sensitive to detect individual cells and enabled quantitative detection of CTCs even in the context of a high leukocyte count. This nanoparticle-based signal amplification method showed great potential for detecting rare cells. In addition, the quantitative results of the visual inspection can be read out by producing signals visible to the naked eye, allowing for disease diagnosis in resource-poor areas.
Cancer cells are also subject to genetic mutations; thus cancer cells can evade regulatory processes necessary for the function of healthy tissues and organs. Assays that detect specific mutations have been developed, and all other mutations can be detected by sequencing. However, many bulk samples must be produced for these assays, and when genetic material is limited, this type of processing risks a loss of initial genetic material. Accordingly, an approach that incorporates sample preparation and enables the original gene template to be reused would be beneficial. To solve the above problems, Reinholt et al. [92] proposed a microfluidic device that uses aptamers to selectively capture cancer cells and isolate their genomic DNA for genetic analysis (Fig. 2D). First, cancer cells were captured by aptamers immobilized on the surface of the microchannel. The captured cells were lysed in situ, and their genomic DNA was isothermally amplified by a specific chip after isolation from the second smallest dimensional microcolumn array. The gene was then sequenced for any mutations. The device was designed so that cell capture and genomic DNA isolation were carried out in a single device, reducing pollution and sample loss. Theranostics has attracted much attention as a promising strategy in the fight against cancer, combining precise diagnosis and simultaneous treatment. With the goal of capturing cancer cells, Xu et al. [93] designed a liquid biopsy-based drug delivery system based on an aptamer microfluidic device that integrates cancer diagnosis and treatment. In brief, they designed two types of MNs, one to identify the target and the other to load the drug. When CTCs bind to the aptamer on the recognition MNs, the complementary chains hybridized with the aptamer are released to conjugate with the drug-loaded MNs, which in turn causes the release of anticancer drugs. The dosage of drug release could be controlled by the amounts of CTCs, and the diagnosis could provide the patient with the appropriate treatment.
3.1.2. Multivalent aptamer-based microfluidics
Due to the exceptional scarcity of CTCs in blood, capturing and detecting CTCs is a major technical challenge. The process of enriching CTCs with aptamers shows great promise, but the insufficient binding affinity has hindered its widespread application. Sheng et al. [94] reported a strategy combining multivalent DNA aptamer nanospheres and microfluidic devices to efficiently capture cancer cells from blood. Each AuNP could ligate up to 95 aptamers, thereby enhancing its molecular recognition capability. The binding of AuNP-aptamer conjugation resulted in a 39-fold increase in affinity compared with that of aptamer alone. To further achieve efficient cell capture, Zhao et al. [95] captured and isolated cells using a three-dimensional (3D) DNA network (Fig. 3A), which was synthesized from microfluidic surfaces amplified by rolling cycles and contained repeated adhesion aptamer domains. Aptamer-modified DNA strands could be effectively captured by synergistic binding of cell surface markers, thus improving the efficiency of cell capture. In addition, the extension of the DNA strand into three dimensions increases the frequency of interaction with the target and thus allows for the capture of cells at high flow rates. When the herringbone microfluidic device was included, the 3D DNA networks exhibited a higher capture efficiency than that of monovalent aptamers and antibodies and a better capture effect at a high flow rate. Finally, DNA strands could be cleaved by restriction enzymes, which allowed the captured cells to be easily released for further analysis.
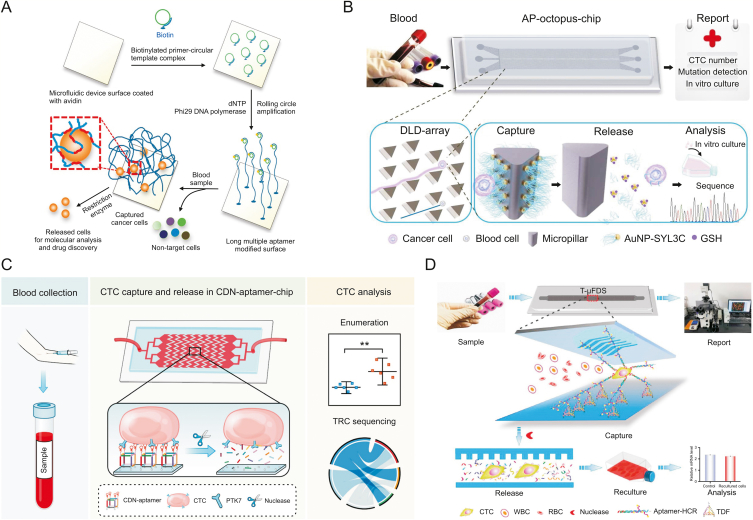
Fig. 3.
Multivalent aptamer (Apt)-based microfluidics for the detection of circulating tumor cells (CTCs). (A) Three-dimensional (3D) DNA network comprising repeating an adhesive aptamer-based microfluidic device for the detection of cancer cells in whole blood. (B) Aptamer-tailed octopus-chip for the detection of CTCs. (C) Cubic DNA nanostructure-aptamer-chip for the detection of CTCs. (D) Tetrahedral DNA framework-based microfluidic technology combined with hybridization chain reaction for the multivalent detection of CTCs. AP: aptamer-tailed; AuNPs: gold nanoparticles; DLD: deterministic lateral displacement; GSH: glutathione; CDN: cubic DNA nanostructure; TDFs: tetrahedral DNA framework; T-μFS: microfluidic system; WBC: white blood cells; RBC: red blood cells. Reprinted from Refs. [95,96,98,99] with permission.
To improve the binding affinity and stability of aptamers when enriched with CTCs in real samples, Yang and co-workers [96] combined a size-determined immune capture chip with a multivariable aptamer nanointerface (AuNPs-EpCAM) (Fig. 3B). The chip formed a biomimetic functional interface of nanometre size and a geometric microcolumn array of micron size. The chip was designed based on the principle of deterministic lateral displacement (DLD). On the one hand, the diameter of the microcolumn could be used to realize the effective separation of CTCs and blood cells. In addition, the rotating triangular microcolumn formed a smooth hydrodynamic gradient, which could reduce the flow rate and thus increase the time of contact between CTCs and the microcolumn. In the results, the capture efficiency of the geometric microfluidic chip was improved by more than 300%, and the adsorption of the background cell was reduced. Furthermore, because Au–S bonds were easily destroyed by thiol exchange reactions, glutathione was selected as the release agent, and enriched cancer cells could be released by thiol exchange reactions with up to 80% efficiency and 96% survival rate, which was fully compatible with downstream detection assays. Based on the very large application potential of nanoscale structure microfluidic chips, Zhang et al. [97] proposed DNA nanolithography in microfluidic chips based on the large application potential of nanoscale structure microfluidic chips. In their work, the nanoscale 3D DNA structure was used as the framework, and a vertical aptamer was added to the top. The nanoscale framework provided the aptamer with a highly ordered vertical orientation, avoiding the undesired orientation or crowding effects that occur during the fabrication of conventional microfluidic interfaces. Compared to the monoclonal aptamer-modified chip, the capture efficiency of the multivalent aptamer-based microfluidics was improved by nearly 60% due to its highly accurate size and rigid structure. In addition, the restricted tetrahedral nanostructured scaffold made the aptamer more accessible to deoxyribonuclease I, with a release efficiency of up to 83% and cell viability of up to 91%; thus, the aptamer was fully compatible with downstream molecular assays.
Although most previous studies have been conducted to improve the target capture efficiency by designing a single aptamer or a single multivalent aptamer, ignoring the orientation of the aptamer on the microfluidic interface, the accessibility and affinity of the aptamer in such cases still need further refinement. Due to programmability, aptamer-functionalized microfluidic interfaces have shown great potential for liquid biopsies. Thus, Peng et al. [98] reported a strategy for programming cubic DNA nanostructures to precisely control the orientation and valence of aptamers at the microfluidic interface, enhancing the efficiency of CTC capture and release (Fig. 3C). Aminated cubic DNA nanostructure aptamers, which bear four orientation-upright Sgc8 aptamers, could be obtained by DNA self-assembly. The aptamers were then decorated on N-hydroxysuccinimde (NHS) ester-functionalized herringbone chips for CTC-specific capture. The four amine groups on the cubic DNA nanostructure-aptamer can covalently react with the NHS ester-functionalized microfluidic interface, resulting in strong rigid immobilization of the cubic DNA nanostructure-aptamer. Cubic DNA nanostructure aptamers at the microfluidic interface benefit from the synergistic effect of well-ordered orientation and multivalent binding, showing favourable accessibility and affinity to CTCs, leading to high capture efficiency. Finally, the captured CTCs could be nondestructively released by nucleases for subsequent sequencing analysis.
In recent research, Wang et al. [99] constructed a microfluidic system for the first time by combining a tetrahedral DNA framework, a hybridization chain reaction (HCR), and a herringbone channel chip (Fig. 3D). Among them, tetrahedral DNA frameworks are immobilized on the chip as scaffolds and hybridized with multiple branched arms of the HCR product for multivalent binding to CTCs. With the help of the herringbone channel, more efficient contact between the CTCs and the capture probe was realized. Tetrahedral DNA frameworks could be specifically removed by benzonase nucleases due to their DNA nanostructures, thus resulting in the nondestructive release of CTCs for subsequent analysis. When the number of MCF-7 cells was 10–103, the capture efficiency was 83.3%–94.2%. The MCF-7 cell release efficiency was 96.2% in phosphate-buffered saline buffer, and the survival rate was 94.6%. The results showed that this chip could be used as an integrated and automated multivalent capture and release platform for CTCs, which has broad application prospects in tumor liquid biopsy.
3.2. Multiple-target aptamer-based microfluidics
Among CTC assay methods, many affinity-based methods rely on a single surface marker, EpCAM. However, highly metastatic CTCs may undergo epithelial to mesenchymal transformation, triggering a significant downregulation of EpCAM expression; thus, these methods focus only on the epithelial subtype of CTCs and neglect the mesenchymal subtype of CTCs, ultimately leading to the neglect of this fraction of CTCs. False-negative results may occur, indicating that the information reflecting disease progression will be lost to some extent. Therefore, there is an urgent need to develop a capture platform to isolate CTCs with different phenotypes. Overall, by considering the number of CTCs obtained by combining multiple aptamers, more comprehensive information on the disease is obtained.
Based on polydimethylsiloxane, Maremanda et al. [100] designed and fabricated a simple microfluidic device. To avoid the degradation of aptamers by nucleases, EpCAM and nucleolus expression were targeted by locking nucleic acid-modified aptamers for quick and efficient capture of CTCs and cancer cells. It was concluded that at the optimum flow rate (10 μL/min), the aptamer-modified device could be reused multiple times and still maintained the best capture efficiency (>90%) and specificity for the target cells. Zhao et al. [101] proposed a multiple aptamer strategy to synergistically increase cancer cell capture efficiency and accuracy. Composed of an aptamer-grafted silicon nanowire substrate and an overlaid polydimethylsiloxane chaotic mixer, the microfluidic chip achieved high affinity towards CTCs. It was demonstrated that using aptamer cocktails with synergistic effects could provide more comprehensive treatment monitoring information via CTC counts.
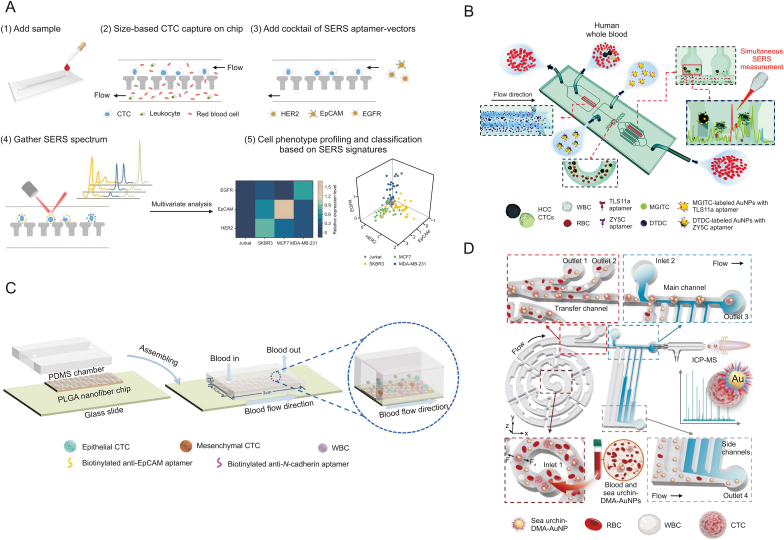
Isolating and in situ analysis of heterogeneous molecular phenotypes of CTCs are significant methods for clinical cancer diagnostics and individualized therapy. SERS is a sensitive optical analytical technique, and Raman scattering can be enhanced by factors up to 106 and even larger. Zhang et al. [102] used a microfluidic chip to profile the CTC phenotype (Fig. 4A). In their work, size-based microfluidic cell separation combined with multispectral orthogonal SERS analysis was used to analyse cell membrane proteins in situ and identify cancer subpopulations. The designed microfluidic chip can screen tumor cells from blood based on size differences. They chose three major subtypes of breast cancer for examples in their experiment. First, based on the size difference between CTCs and blood cells, the microfluidic filter efficiently separated CTCs from blood, and then multiple SERS aptamer carriers could concurrently target distinct biomarkers on the cell surface. The revised classic least squares algorithm could be used to decompose complex SERS fingerprints and obtain 3D phenotype information at single-cell resolution. Finally, partial least squares discriminant analysis was used to accurately classify the different subtypes of cells.
Fig. 4.
Multiple-target aptamer-based microfluidics for the detection of circulating tumor cells (CTCs). (A) Combining multiplex surface-enhanced Raman scattering (SERS) nanovectors and multivariate analysis for in situ profiling of CTC phenotypes using a microfluidic chip. (B) Simultaneous single-cell phenotype analysis of hepatocellular carcinoma CTCs using a SERS-aptamer-based microfluidic chip. (C) A poly(lactic-co-glycolic acid) (PLGA) nanofiber microfluidic device for highly efficient isolation and release of CTCs with different phenotypes based on dual aptamers. (D) Dual-multivalent-aptamer-conjugated nanoprobes for superefficient discerning of single CTCs in a microfluidic chip with inductively coupled plasma-mass spectrometry (ICP-MS) detection. EGFR: epidermal growth factor receptor; EpCAM: epithelial cell adhesion molecule; HER2: human epidermal growth factor receptor-2; HCC: hepatocellular carcinoma; WBC: white blood cells; RBC: red blood cells; MGITC: malachite green isothiocyanate; DTDC: 3,3′-diethylthiadicarbocyanine iodide; AuNPs: gold nanoparticles; PDMS: polydimethylsiloxane; PLGA: poly(lactic-co-glycolic acid); Fi: inertial lift forces; Fd: dean drag force; DMA: dual-multivalent-aptamers. Reprinted from Refs. [[102], [103], [104], [105]] with permission.
SERS has shown outstanding advantages in the multiplex detection of CTCs due to the unique fingerprinting characteristics of different SERS probes. To achieve high specificity and excellent affinity for rare CTCs, variable antigenic phenotypes, such as EpCAM and epidermal growth factor receptor, should be discussed. For example, based on the SERS aptamer, Gao et al. [103] developed a microfluidic chip for in situ single-cell phenotyping by characterizing the expression levels of surface biomarkers of CTCs. (Fig. 4B). Novel microfluidic chips with lantern-like bypass structures were developed to capture large CTCs from whole blood. Through two types of SERS-aptamer nanotags, the expression of surface membrane proteins in individual CTCs could be identified spectrally. This purpose-designed microfluidic channel provided the following functions: isolation, capture, and in situ analysis. In addition, multivalent SERS-aptamer nanotags were prepared to avoid DNA degradation and improve the stability of nanoparticles, thereby simultaneously identifying different biomarkers on cell membranes. Finally, synchronous SERS in situ analysis of CTCs can be performed on microfluidic chips. The results showed that the device could accurately identify hepatocellular carcinoma-related CTCs in clinical specimens. At the same time, the expression of specific biomarkers on CTCs could be quantitatively analysed by SERS intensity.
Based on dual aptamers, Wu et al. [104] designed a poly(lactic-co-glycolic acid) (PLGA) nanofiber microfluidic device for highly efficient and specific detection of different CTCs with different phenotypes (Fig. 4C). They modified both EpCAM and N-calmodulin aptamers on PLGA fibrillated substrates. While ensuring the capture of normal EpCAM-expressing CTCs, they were also able to reduce the number of missed EpCAM-low-expressing CTCs, improving the overall capture rate and reducing the number of false-negative results. According to the level of expression of N-calmodulin, the amount of EpCAM varied in different cells, such as ovarian cancer A2780 cells with high expression of N-calmodulin and OVCAR-3 cells with high expression of EpCAM. Taking ovarian cancer A2780 cells and OVCAR-3 cells as examples, the capture efficiency of this device was 91% and 89%, respectively, and the sensitivity was 92% and 88%. Finally, the microfluidic device achieved the capture of 1–13 CTCs in clinical blood samples from ovarian cancer patients with the help of immunofluorescence staining identification, providing guidance for clinical application-diagnostic, therapeutic and prognostic potential.
To overcome the deficiency of weak binding affinity and poor stability of aptamers in real samples, Zhang et al. [105] integrated dual-multivalent-aptamer-conjugated nanoprobes, microfluidic, inductively coupled plasma-mass spectrometry (ICP-MS) detection to efficiently identify and detect CTCs (Fig. 4D). They coupled two aptamers, Sgc8 and SYL3C, to AuNPs to form nanoprobes, which could specifically bind to the biomarker proteins protein tyrosine kinase 7 and EpCAM, which were overexpressed on the surface of CTCs, to achieve high capture efficiency. The sorting of CTCs was achieved on a specially designed multifunctional microfluidic configuration, which integrated an enhanced helical separation unit and a hydrodynamic filtration purification unit without damaging the cells. ICP-MS resulted in background-free, efficient and sensitive detection and analysis of proteins in individual CTCs. The binding efficiency of the dual aptamer nanoprobe was improved by more than 200% compared to that of the single aptamer nanoprobe. The separation rate of this microfluidic device was 93.6% at the optimal flow rate, and its measurement efficiency for a single cell was 73.8% ± 5.0%. The results indicate that this device can provide a new method for the isolation assay of rare cells.
Liu et al. [106] used multiple aptamers to construct a multimarker microfluidic chip based on a DNA nanoframework for the enrichment of heterogeneous CTCs from peripheral blood samples of breast cancer patients. As a scaffold, their precisely tuneable triangular prism DNA nanostructure allowed three distinct aptamers to be arranged in a highly ordered and vertical pattern in a controlled manner, facilitating the interaction of multilabelled aptamers with heterogeneous CTC surfaces. A DLD micropillar array was used as a foothold to fix the three vertices at the bottom of the triangular prism on a microfluidic chip, thereby forming a nanointerface for interaction with the target. Based on the principle of DLD clutches, a cylindrical microcolumn array geometry design was applied to a microfluidic chip to reduce the collision between CTCs and microcolumn efficiency while reducing the blood cells and other collisions between the microcolumn. Simultaneously, the rigidity of DNA nanostructures on the interface provided a reasonable space distance, making it easier for nuclease degradation of the adapter body and achieving effective release of captured cells. Compared to microarrays modified with monovalent inducers, the multiple aptamer-labelled microfluidic microarrays displayed higher identifying efficiency for both high and low EpCAM-expressing cell lines, and the DNA nanostructure-functionalized microarrays were able to accurately capture CTCs with different phenotypes.
4. Aptamer-based microfluidics for EV detection
EVs are a specific type of lipid bilayer-enclosed heterogeneous microvesicle secreted from most cell types [16,107,108]. In the 1980s, Harding et al. [109] and Pan et al. [110] reported a complicated endocytic pathway in which the endosomal multivesicular body (MVB) membrane secretes EVs by inwards budding. EVs carry unique molecular cargo, including nucleic acids, proteins, and lipids, from their original cells to recipient cells, playing a critical role in intercellular information communication.
According to their size and biogenesis, EVs are mainly classified into exosomes, microvesicles, and apoptotic bodies [111]. Microvesicles originated from the plasma membrane with a density of 1.02–1.22 g/mL and a heterogeneous diameter of approximately 100−1000 nm. In contrast, exosomes have a density of 1.10–1.18 g/mL and diameters of approximately 30−150 nm and are related to the endocytic pathway involving double invagination of the plasma membrane. First, early endosomes are formed during endocytosis by invaginating the plasma membrane and contain soluble proteins and membrane proteins of the extracellular space. As early endosomes evolve into late endosomes, their shape changes to spherical, and they migrate to the domain. Then, intraluminal vesicles (future exosomes) are provided inside the lumen by invaginating the endosomal limiting membrane of early endosomes. These late endosomes are named MVBs and have two potential fates. MVBs are degraded by fusing with autophagosomes or lysosomes or secreting intraluminal vesicles into the extracellular milieu as exosomes after migrating to the cell membrane surface and fusing with the plasma membrane [16].
Exosomes carry various molecular cargoes from the original cells, including some special lipids, nucleic acids, and proteins, which are associated with the biological properties, functions, and structures of exosomes [24]. Exosomal proteins are derived from the membranes of endosomes, plasma membranes, or cytosol. Exosomes mainly carry many common sets of proteins, including proteins participating in membrane fusion and exosome release, MVB biogenesis-related proteins, transmembrane proteins from tetraspanins, cytoskeletal proteins, adhesion proteins, heat-shock proteins, metabolic enzymes, and major histocompatibility complex class II molecules [112,113]. Aside from common protein sets, some specific proteins are carried by exosomes depending on different original cell types. For example, tumor-derived exosome surfaces contain some overexpressed tumor markers that assist in the detection and treatment of cancer. Therefore, exosomal proteins have great potential in the therapeutic monitoring and diagnosis of diseases.
In addition to protein, nucleic acids are also important cargoes carried by exosomes [114]. Exosomes have been confirmed to transport functional RNA from one cell to another both in vivo and in vitro. Exosomes may play a crucial role in protein translation and the regulation of gene expression. Exosome-shuttle nucleic acids show excellent stability compared to that of cell-free nucleic acids because of the protection of the lipid bilayer. Some specific lipid components are included in exosomes, such as phosphatidylethanolamine, glycerophospholipids, phosphatidylcholine, ceramide, sphingolipids, and cholesterol. Thus, although exosomes and the plasma membrane have a similar lipid bilayer, the lipid composition of exosomes is different from that of the original cells.
These excellent characteristics have received extensive attention in EV-based liquid biopsy [24]. Therefore, tumor-derived EVs containing molecular information, such as specific tumor marker miRNAs and proteins, can be used for prognosis, therapy evaluation, metastasis monitoring, and the early detection of cancers. With some merits, such as stability and abundance in circulation, EVs are promising biomarkers for the classification [27], metastasis detection [26], and treatment monitoring of cancer [25].
Although EV-based liquid biopsy has attracted widespread research interest, some elusive technological challenges remain in the detection of EVs. Aptamers have been extensively developed for efficiently isolating and sensitively detecting EVs because they feature convenient signal amplification and affinity regulation. Compared with other conventional methods, microfluidic detection technologies are beneficial, as they involve less contamination, low reagent consumption, higher throughput, and shorter analysis times, as well as easy automatization and integration. Microfluidic technologies are particularly suitable for the selective, sensitive, and rapid analysis and detection of EVs. Thus, aptamer-based microfluidics offers improved purity and isolation efficiency of EVs by combining the advantages of aptamers as recognition tools and microfluidic chips as isolation platforms [16]. Aptamer-based microfluidics offers a high-performance platform for CTCs and EVs in integration with the advantages of both micro/nanostructured microdevices and aptamers (Table 2) [85,88,91,[96], [97], [98],[103], [104], [105],115,116].
Table 2.
Aptamer-based microfluidic for the detection of circulating tumor cells (CTCs) and extracellular vesicles (EVs).
| Aptamer | Target | Cell line | Detection type | Microdevice characteristics | Refs. |
|---|---|---|---|---|---|
| KCHA10 | Cell | HCT116 | CTCs and single-target simple aptamer-based microfluidics | Aptamer-modified micropillars; unprocessed peripheral blood; and fluorescence analysis | [85] |
| SYL3C | EpCAM | MCF-7 | CTCs and single-target simple aptamer-based microfluidics | Aptamer-modified magnetic nanoclusters; biomimetic microfluidic device; and fluorescence analysis | [88] |
| Sgc8 | PTK7 | CCRF-CEM | CTCs and single-target simple aptamer-based microfluidics | Volumetric bar chart chip and visual quantitative detection | [91] |
| SYL3C | EpCAM | SW480 | CTCs and single-target multivalent aptamer-based microfluidics | DLD-based microfluidic chip; aptamer-functionalized AuNPs; micropillars; capture and release of CTCs; and fluorescence analysis | [96] |
| SYL3C | EpCAM | SW480 | CTCs and single-target multivalent aptamer-based microfluidics | DNA nanolithography in microfluidic chip; tetrahedral DNA structures as frameworks; and fluorescence analysis | [97] |
| Sgc8 | PTK7 | CCRF-CEM | CTCs and single-target multivalent aptamer-based microfluidics | Herringbone chip; cubic DNA nanostructures as frameworks; and fluorescence analysis | [98] |
| TLS11a | Cell | HepG2 | CTCs and multiple-target aptamer-based microfluidics | Heterogeneous phenotype analysis of CTCs and SERS analysis | [103] |
| ZY5C | Vimentin | SK-HEP-1 | |||
| NC3S | Cadherin | A2780 | CTCs and multiple-target aptamer-based microfluidics | Dual aptamer-modified PLGA nanofiber; different phenotypic CTC capture; and fluorescence analysis | [104] |
| EpCAM-5-1 | EpCAM | OVCAR-3 | |||
| Sgc8 | PTK7 | MCF-7, HepG2 | CTCs and multiple-target aptamer-based microfluidics | Dual multivalent-aptamers modified AuNPs and ICP-MS analysis | [105] |
| SYL3C | EpCAM | ||||
| CD63 aptamer | CD63 | / | EVs and single-target aptamer-based microfluidics | Two-stage microfluidic platform and on-chip isolation and in situ electrochemical detection | [115] |
| HER2 aptamer | HER2 | / | EVs and multiple-target aptamer-based microfluidics | Viscoelastic microfluidics; EV subpopulations at the single-EV level; and fluorescence detection | [116] |
| EpCAM aptamer | EpCAM |
EpCAM: epithelial cell adhesion molecule; DLD: deterministic lateral displacement; AuNPs: gold nanoparticles; PTK7: protein tyrosine kinase 7; SERS: surface-enhanced Raman scattering; PLGA: poly(lactic-co-glycolic acid); ICP-MS: inductively coupled plasma mass spectrometry; HER2: human epidermal growth factor receptor-2.
4.1. Single-target aptamer-based microfluidics
Aptamers with unique tertiary structures combine with the surface protein of EVs by specific recognition. In addition, aptamers as mediators are applied to signal transduction and amplification due to their versatile engineering and structural design. Electrochemical detection has attracted widespread attention in liquid biopsy due to its outstanding advantages, including easy operation, excellent portability, high sensitivity, and low cost. Aptamers can be used as capture probes or signal probes on the electrode surface with convenient immobilization for EV detection. For example, Zhou et al. [117] designed an electrochemical aptasensor chip for the quantitative detection of exosomes (Fig. 5A). Redox moieties labelled with probing strands were designed to hybridize with aptamer molecules anchored on the electrode surface of a microfluidic system. In the presence of exosomes, the aptamer recognized CD63, a transmembrane protein of exosomes. Then, probing strands combined with redox reporters were released, resulting in a drop in the electrochemical signal. These biosensors could be applied in detecting exosomes with a limit of detection of 1 × 106 particles/mL.
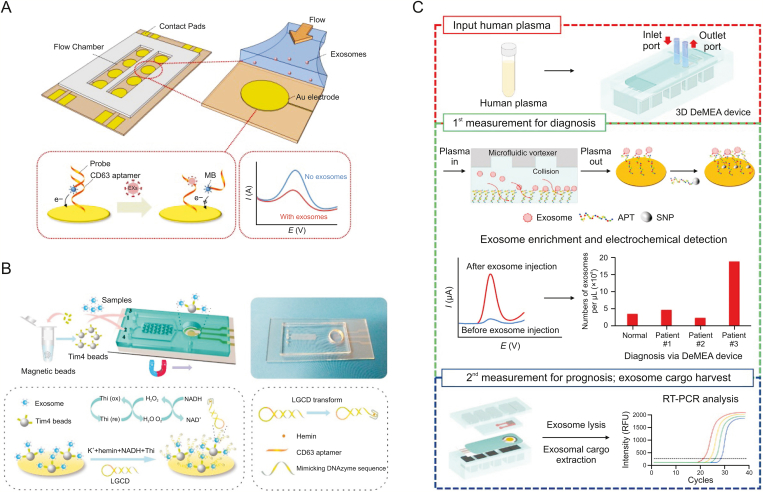
Fig. 5.
Single-target aptamer-based microfluidics for electrochemical detection of extracellular vesicles (EVs). (A) An electrochemical microfluidic aptasensor with a CD63 aptamer hybridized with signal strands. (B) A two-stage microfluidic platform using a hairpin structure with a CD63-aptamer sequence to continuously catalyse hydrogen peroxide. (C) A detachable microfluidic electrochemical aptasensor with an epithelial cell adhesion molecule aptamer specifically capturing cancer-derived exosomes. MB: methylene blue; Thi: thionine; LGCD: a label-free and immobilization-free electrochemical aptasensor; DNAzyme: deoxyribozyme; DeMEA: a detachable microfluidic device implemented with an electrochemical aptasensor; APT: aptamer; SNP: silver nanoparticles; RT-PCR: reverse transcription polymerase chain reaction; RFU: relative fluorescence units. Reprinted from Refs. [115,117,118] with permission.
Xu et al. [115] proposed a two-stage microfluidic platform to on-chip isolate and analyse exosomes from serum in situ electrochemically by combining magnetic enrichment and a new signal transduction strategy (Fig. 5B). Magnetic enrichment was realized by specific phosphatidylserine-Tim4 protein recognition. Tim4 immobilized with magnetic beads capturing exosomes is Ca2+ dependent, and intact exosomes can be eluted easily with a chelating agent. In the detection stage, a hairpin structure with a G-quadruplex and CD63 aptamer sequence was designed to continuously catalyse the newly generated H2O2 by nicotinamide adenine dinucleotide oxidation, and obvious signal enhancement was observed. The reported platform provided a linear range spanning five orders of magnitude for CD63-positive exosomes with a highly sensitive detection of 4.39 × 103 particles/mL.
Kashefi-Kheyrabadi et al. [118] introduced an electrochemical aptasensor implemented with a detachable microfluidic device for in situ quantification that was highly sensitive to cancerous exosomes (Fig. 5C). They constructed a sensing system with a microfluidic vortexer to enhance the collision between the sensing surface and exosomes by using hydrodynamically generated transverse flow. To improve the physiochemical properties of the aptasensor, they designed a nanocomposite dropped on the electrode surface, including MoS2 nanosheets, graphene nanoplatelets, and chitosan. Subsequently, an aptamer labelled with silver nanoparticles was applied on the electrode surface for detecting cancer-derived exosomes. This chip exhibits high specificity and sensitivity with a wide detection range of 1 × 102 to 1 × 109 exosomes/μL and an ultralow detection limit of 17 exosomes/μL.
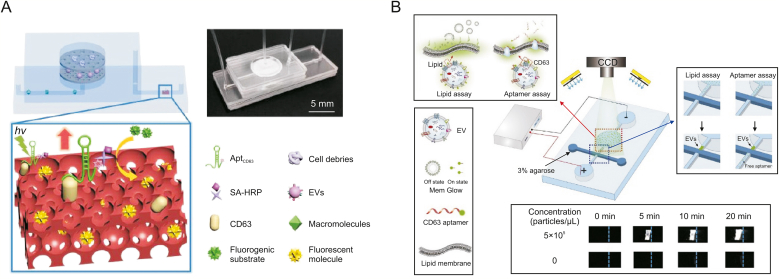
Due to the fluorescence enhancement effect of photonic crystal nanostructures, EVs can be sensitively quantified by analysing the concentration of excess aptamers. Dong et al. [119] reported a microfluidic chip with a double-filtration unit for isolating and detecting EVs by photonic crystal nanostructures (Fig. 6A). The device is integrated with two nanofiltration membranes and can enrich and isolate 20–200 nm EV by size exclusion. Two nanofiltration membranes with pore sizes of 20 and 200 nm filtrate particles smaller than 20 nm or larger than 200 nm to minimize interference, such as cell debris, nucleic acids, and free proteins. After EV enrichment, excessive aptamers targeting CD63 were introduced into the chip to recognize the CD63 surface protein of EVs. The excess CD63 aptamers are filtered out by the filtration membrane and captured by the CD63 protein immobilized on the PC membrane. Then, the injected streptavidin-horseradish peroxidase solution was combined with the biotinylated CD63 aptamer. After adding the H2O2 substrate and the fluorogenic solution, the fluorescent product of resorufin was produced, and fluorescence images were obtained by using a fluorescence microscope. This device only consumes 20 μL of sample and has a limit of detection of 8.9 × 103 EV/mL.
Fig. 6.
Single-target aptamer-based microfluidics for fluorescent detection of extracellular vesicles (EVs). (A) A microfluidic chip with a double-filtration unit filtering excess CD63 aptamers to quantify EVs. (B) A microfluidic platform detecting EVs by two label strategies with fluorescein amidite (FAM)-labelled CD63 aptamer and Mem Glow-labelled phospholipid bilayers of EVs. AptCD63: aptamers targeting CD63; SA-HRP: streptavidin-horseradish peroxidase; CDD: charge coupled device. Reprinted from Refs. [119,120] with permission.
Some researchers use fluorescent-labelled aptamers as recognition probes to obtain detection signals. Ren et al. [120] designed a microfluidic platform to enrich EVs by using electrophoresis technology and to detect by two label strategies with FAM-labelled CD63 aptamer and Mem Glow-labelled phospholipid bilayers of EVs (Fig. 6B). Due to the electromigration characteristics of EVs, the platform removed free dye molecules and achieved effective enrichment of EVs. A customized fluorescence imaging system was used to record the increased fluorescent signal of EVs, and this platform achieved over a 400-fold enhancement of fluorescent intensity in 20 min. However, the amount of EVs cannot be calculated by quantitative analysis based on aptamers targeting membrane proteins because heterogeneous EVs derived from different cells express various proteins, such as the marker proteins CD81, CD63, and CD9. In contrast, the signal with Mem Glow-labelled phospholipid bilayers of EVs reflected the amount and size of EVs. Thus, the expression level of CD63 was compared before and after normalization based on the EV amount. Due to the different protein expression levels in different EV samples, the deviation can be minimized by lipid assay.
4.2. Multiple-target aptamer-based microfluidics
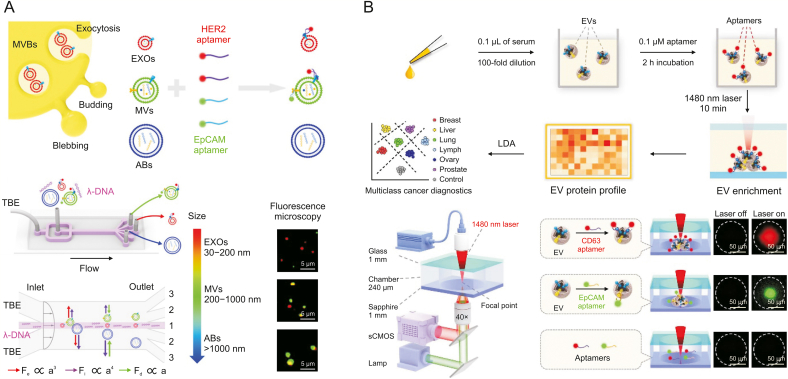
EVs are derived from molecular information related to tumor cell cargo, such as tumor surface marker proteins. Due to the specific recognition, fluorescent-labelled aptamer as a probe can be applied to label various styles of EVs, and different aptamers against the respective surface protein of EVs can be integrated to classify cancer types. Liu et al. [116] designed a microfluidic coflow system containing Newtonian sheath fluid and viscoelastic sample fluid to isolate EVs and visualize the separation process by using two labelled aptamers with FAM-conjugated EpCAM aptamer and Cy5-conjugated human epidermal growth factor receptor-2 (HER2) aptamer (Fig. 7A). Under different interaction forces, including the viscous drag force, inertial lift force, and elastic lift force, EVs of different sizes were immersed in distinct lateral positions downstream. Small labelled aptamers (2 to 3 nm) were used to efficiently recognize target surface proteins of the EV surface, and aptamer-based single EV analysis with high signal-to-noise ratios was allowed. The expression levels of EpCAM/HER2 were determined by fluorescence intensity measurements of individual exosomes, apoptotic bodies, and microvesicles, which were significantly different among individual EVs. Even if EpCAM and HER2 on EV subpopulations originate from the same cell line, the mean expression levels also varied, which was demonstrated by dot blotting. The heterogeneous marker expression of single EV subpopulations was revealed by this multiple-target aptamer-based microfluidic chip.
Fig. 7.
Multiple-target aptamer-based microfluidics for fluorescent detection of extracellular vesicles (EVs). (A) A microfluidic coflow system using two labelled aptamers with Cy5-conjugated human epidermal growth factor receptor-2 (HER2) aptamer and fluorescein amidite (FAM)-conjugated epithelial cell adhesion molecule (EpCAM) aptamer to visualize the separation process. (B) A thermophoretic aptasensor using a panel of seven fluorescent Cy5-conjugated aptamers to label serum EVs. MVB: multivesicular body; EXO: exosomes; MVs: microvesicles; ABs: apoptotic bodies; TBE: tris-boric acid-ethylenediamine tetraacetic acid; Fe: the centerline-directed elastic lift force; Fi: inertial lift force; Fd: viscous drag force; a: diameter of EV; LDA: linear discriminant analysis; sCMOS: scientific complementary metal-oxide semiconductor. Reprinted from Refs. [27,116] with permission.
Liu et al. [27] presented a thermophoretic aptasensor by using a panel of seven fluorescent Cy5-conjugated aptamers to label serum EVs (Fig. 7B). To rapidly accumulate EVs in the laser spot centre, a thermophoretic aptasensor was used to locally heat the diluted serum sample (10 μL) in the microchamber through applying a 1480 nm laser for 10 min. Due to local laser heating, which leads to the interplay of convection, diffusion, and thermophoresis, EVs accumulated in a size-dependent manner. Moreover, laser heating could not enrich free proteins and aptamers of small sizes. Then, an amplified fluorescence signal was produced upon the accumulation of the aptamer-bound EVs, and the expression level of the target surface protein of the EVs was correlated with the fluorescence signal intensity. By using this method, the researchers obtained profiles for EV surface proteins in 232 serum samples. The design can classify cancer automatically across six cancer types by a linear discriminant analysis algorithm, discriminate prostate cancer from benign disease, and monitor biochemical recurrence after radical prostatectomy.
5. Conclusions and future perspectives
Liquid biopsy of CTCs and EVs offers a promising technique for cancer diagnostics in a noninvasive, sensitive, and dynamic manner. This review described recent developments in aptamer-based microfluidic platforms for the detection of CTCs and EVs. Although promising, there are few aptamer-based microfluidics with routine clinical utility. Some obstacles must be addressed before aptamer-based microfluidics can be implemented as a real-world tool.
First, aptamers exhibit unique properties, such as facile modification, convenient synthesis, and affinity regulation; thus, they are superior to antibodies as recognition ligands. However, aptamers have not been widely applied clinically for CTCs and EVs, as expected, due to their compromised stabilities and binding affinities in complex body fluids. The formation of tertiary structures and binding to targets for aptamers can be interfered with by high-concentration proteins, nucleases, and blood cells of the whole blood. Moreover, some standards with aptamer-based liquid biopsy for clinical applications have not been established, and almost all reported approaches do not discuss these topics, such as preserved conditions, sampling, enrichment, detection, and result readout. Thus, more advanced SELEX techniques are needed to improve the properties of aptamers (variety, affinity, stability, etc.). Moreover, SELEX-integrated microdevices can simplify processing procedures for aptamer discovery. This device provides the possibility to monitor the aptamer evolution process in real-time.
Second, aptamer-based microfluidic platforms are emerging as powerful tools for circulating target studies, especially CTCs and EVs. The elaborate micro/nanostructure design and precise liquid manipulation in microdevices enable increased collision opportunities between circulating targets and capture substrates. The application of multivalent aptamer units further enhances the capturing capability of substrates in microdevices, reducing nonspecific interactions in the sample and improving the purity and capture efficiency of circulating targets. Integrated microdevices should be developed to achieve circulating target isolation and analysis in one step, minimizing contamination and improving automation. Aptamers against different CTC and EV subpopulations should be identified and immobilized on different microchannels to solve the heterogeneity of circulating targets. Multiplex profiling of the phenotype and genotype of CTCs and EVs can offer comprehensive and accurate molecular information for guiding personalized antitumor therapy and understanding the mechanism of metastasis and tumorigenesis. More importantly, clinical samples should be analysed to determine the reliability, sensitivity, and feasibility of aptamer-based microfluidics techniques for liquid biopsy before routine clinical application.
Finally, integrated devices and standardized processing procedures are important factors for clinical application. With mature protocols, commercial instruments, and reagent kits, aptamer-based microfluidic methods can achieve a smooth transition from bench to bedside. Furthermore, portable devices can achieve point-of-care testing without requiring highly trained personnel in a resource-limited area, which together with low-cost and convenient sampling will make it possible for disease screening. Additionally, commercial reagent kits with standardized operation procedures and quality control will ensure the reproducibility and reliability of aptamer-based microfluidics methods. With the concerted efforts of clinicians and researchers, aptamer-based microfluidics for liquid biopsies could become a powerful analytical approach in clinical applications for tumor diagnosis and personalized therapy.
CRediT author statement
Duanping Sun: Formal analysis, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing - Original draft preparation, Reviewing and Editing; Ying Ma and Maoqiang Wu: Data curation, Software, Writing - Original draft preparation; Zuanguang Chen: Methodology, Conceptualization, Investigation; Luyong Zhang: Funding acquisition, Supervision, Project administration; Jing Lu: Funding acquisition, Resources, Supervision, Project administration, Writing - Reviewing and Editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos.: 82003710 and 82173808), the Natural Science Foundation of Guangdong Province (Grant Nos.: 2020A1515010075 and 2021B1515020100), the Project of Educational Commission of Guangdong Province (Grant No.: 2021ZDZX2012), the Guangzhou Basic and Applied Basic Research Project (Grant No.: 2023A04J1163), the National Key Clinical Specialty Construction Project (Clinical Pharmacy), and High-Level Clinical Key Specialty (Clinical Pharmacy) in Guangdong Province, China.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Contributor Information
Duanping Sun, Email: sundp@gdpu.edu.cn.
Luyong Zhang, Email: lyzhang@gdpu.edu.cn.
Jing Lu, Email: lujing28@mail.sysu.edu.cn.
References
- 1.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sumida Y., Nakajima A., Itoh Y. Limitations of liver biopsy and non-invasive diagnostic tests for the diagnosis of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J. Gastroenterol. 2014;20:475–485. doi: 10.3748/wjg.v20.i2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W., Wang H., Zhao Z., et al. Emerging nanotechnologies for liquid biopsy: The detection of circulating tumor cells and extracellular vesicles. Adv. Mater. 2019;31 doi: 10.1002/adma.201805344. [DOI] [PubMed] [Google Scholar]
- 4.Frable W.J. Fine-needle aspiration biopsy: A review. Hum. Pathol. 1983;14:9–28. doi: 10.1016/s0046-8177(83)80042-2. [DOI] [PubMed] [Google Scholar]
- 5.Munzone E., Nolé F., Goldhirsch A., et al. Changes of HER2 status in circulating tumor cells compared with the primary tumor during treatment for advanced breast cancer. Clin. Breast Cancer. 2010;10:392–397. doi: 10.3816/CBC.2010.n.052. [DOI] [PubMed] [Google Scholar]
- 6.Guarneri V., Giovannelli S., Ficarra G., et al. Comparison of HER-2 and hormone receptor expression in primary breast cancers and asynchronous paired metastases: Impact on patient management. Oncologist. 2008;13:838–844. doi: 10.1634/theoncologist.2008-0048. [DOI] [PubMed] [Google Scholar]
- 7.Crowley E., Di Nicolantonio F., Loupakis F., et al. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 8.Di Meo A., Bartlett J., Cheng Y., et al. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer. 2017;16 doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L., Wang Y., Zhu L., et al. Aptamer-based liquid biopsy. ACS Appl. Bio Mater. 2020;3:2743–2764. doi: 10.1021/acsabm.9b01194. [DOI] [PubMed] [Google Scholar]
- 10.Chen G., Huang A.C., Zhang W., et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagrath S., Sequist L.V., Maheswaran S., et al. Isolation of rare circulating tumor cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chemi F., Rothwell D.G., McGranahan N., et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 2019;25:1534–1539. doi: 10.1038/s41591-019-0593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maheswaran S., Sequist L.V., Nagrath S., et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellby L.D., Nyberg A.P., Johansen J.S., et al. Serum biomarker signature-based liquid biopsy for diagnosis of early-stage pancreatic cancer. J. Clin. Oncol. 2018;36:2887–2894. doi: 10.1200/JCO.2017.77.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W., Xia W., Lv Z., et al. Liquid biopsy for cancer: Circulating tumor cells, circulating free DNA or exosomes? Cell. Physiol. Biochem. 2017;41:755–768. doi: 10.1159/000458736. [DOI] [PubMed] [Google Scholar]
- 16.Wu L., Wang Y., Xu X., et al. Aptamer-based detection of circulating targets for precision medicine. Chem. Rev. 2021;121:12035–12105. doi: 10.1021/acs.chemrev.0c01140. [DOI] [PubMed] [Google Scholar]
- 17.Tian F., Liu C., Lin L., et al. Microfluidic analysis of circulating tumor cells and tumor-derived extracellular vesicles. Trends Analyt. Chem. 2019;117:128–145. [Google Scholar]
- 18.Alix-Panabières C., Pantel K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 19.Baccelli I., Schneeweiss A., Riethdorf S., et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 20.Boriachek K., Islam M.N., Möller A., et al. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 2018;14 doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- 21.Wang W., Luo J., Wang S. Recent progress in isolation and detection of extracellular vesicles for cancer diagnostics. Adv. Healthcare Mater. 2018;7 doi: 10.1002/adhm.201800484. [DOI] [PubMed] [Google Scholar]
- 22.Kim K.M., Abdelmohsen K., Mustapic M., et al. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA. 2017;8 doi: 10.1002/wrna.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thind A., Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles. 2016;5 doi: 10.3402/jev.v5.31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.B. Lin, Y. Lei, J. Wang, et al., Microfluidic-based exosome analysis for liquid biopsy, Small Methods 5 (2021), 2001131. [DOI] [PubMed]
- 25.Shao H., Chung J., Balaj L., et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C., Zhao J., Tian F., et al. Low-cost thermophoretic profiling of extracellular-vesicle surface proteins for the early detection and classification of cancers. Nat. Biomed. Eng. 2019;3:183–193. doi: 10.1038/s41551-018-0343-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhang R., Le B., Xu W., et al. Magnetic “squashing” of circulating tumor cells on plasmonic substrates for ultrasensitive NIR fluorescence detection. Small Methods. 2019;3 [Google Scholar]
- 29.Sun D., Lu J., Zhang L., et al. Aptamer-based electrochemical cytosensors for tumor cell detection in cancer diagnosis: A review. Anal. Chim. Acta. 2019;1082:1–17. doi: 10.1016/j.aca.2019.07.054. [DOI] [PubMed] [Google Scholar]
- 30.Liu C., Feng Q., Sun J. Lipid nanovesicles by microfluidics: Manipulation, synthesis, and drug delivery. Adv. Mater. 2019;31 doi: 10.1002/adma.201804788. [DOI] [PubMed] [Google Scholar]
- 31.Lin S., Yu Z., Chen D., et al. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small. 2020;16 doi: 10.1002/smll.201903916. [DOI] [PubMed] [Google Scholar]
- 32.Li G., Tang W., Yang F. Cancer liquid biopsy using integrated microfluidic exosome analysis platforms. Biotechnol. J. 2020;15 doi: 10.1002/biot.201900225. [DOI] [PubMed] [Google Scholar]
- 33.Jackson J.M., Witek M.A., Kamande J.W., et al. Materials and microfluidics: Enabling the efficient isolation and analysis of circulating tumour cells. Chem. Soc. Rev. 2017;46:4245–4280. doi: 10.1039/c7cs00016b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian W., Zhang Y., Chen W. Capturing cancer: Emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small. 2015;11:3850–3872. doi: 10.1002/smll.201403658. [DOI] [PubMed] [Google Scholar]
- 35.Myung J.H., Hong S. Microfluidic devices to enrich and isolate circulating tumor cells. Lab Chip. 2015;15:4500–4511. doi: 10.1039/c5lc00947b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Xu D., Tan W. Aptamer-functionalized nano/micro-materials for clinical diagnosis: Isolation, release and bioanalysis of circulating tumor cells. Integr. Biol. 2017;9:188–205. doi: 10.1039/c6ib00239k. [DOI] [PubMed] [Google Scholar]
- 37.Yu Z.T.F., Aw Yong K.M., Fu J. Microfluidic blood cell sorting: Now and beyond. Small. 2014;10:1687–1703. doi: 10.1002/smll.201302907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng N., Du D., Wang X., et al. Recent advances in biosensors for detecting cancer-derived exosomes. Trends Biotechnol. 2019;37:1236–1254. doi: 10.1016/j.tibtech.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Song Y., Tian T., Shi Y., et al. Enrichment and single-cell analysis of circulating tumor cells. Chem. Sci. 2017;8:1736–1751. doi: 10.1039/c6sc04671a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma H., Liu J., Ali M.M., et al. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem. Soc. Rev. 2015;44:1240–1256. doi: 10.1039/c4cs00357h. [DOI] [PubMed] [Google Scholar]
- 41.Fang X., Tan W. Aptamers generated from cell-SELEX for molecular medicine: A chemical biology approach. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 43.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 44.Tang Z., Shangguan D., Wang K., et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 45.Mallikaratchy P., Tang Z., Kwame S., et al. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol. Cell. Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Shangguan D., Li Y., Tang Z., et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang M., Song J., Huang P., et al. Molecular crowding evolution for enabling discovery of enthalpy-driven aptamers for robust biomedical applications. Anal. Chem. 2019;91:10879–10886. doi: 10.1021/acs.analchem.9b02697. [DOI] [PubMed] [Google Scholar]
- 48.Shen C., Liu S., Li X., et al. Electrochemical detection of circulating tumor cells based on DNA generated electrochemical current and rolling circle amplification. Anal. Chem. 2019;91:11614–11619. doi: 10.1021/acs.analchem.9b01897. [DOI] [PubMed] [Google Scholar]
- 49.Shen W., Guo K., Adkins G.B., et al. A single extracellular vesicle (EV) flow cytometry approach to reveal EV heterogeneity. Angew. Chem. Int. Ed. Engl. 2018;57:15675–15680. doi: 10.1002/anie.201806901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu L., Zhu L., Huang M., et al. Aptamer-based microfluidics for isolation, release and analysis of circulating tumor cells. Trends Analyt. Chem. 2019;117:69–77. [Google Scholar]
- 51.Dickey D.D., Giangrande P.H. Oligonucleotide aptamers: A next-generation technology for the capture and detection of circulating tumor cells. Methods. 2016;97:94–103. doi: 10.1016/j.ymeth.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassan E.M., Willmore W.G., DeRosa M.C. Aptamers: Promising tools for the detection of circulating tumor cells. Nucleic Acid Ther. 2016;26:335–347. doi: 10.1089/nat.2016.0632. [DOI] [PubMed] [Google Scholar]
- 53.Duan Y., Zhang C., Wang Y., et al. Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Mol. Biol. Rep. 2022;49:7979–7993. doi: 10.1007/s11033-022-07317-0. [DOI] [PubMed] [Google Scholar]
- 54.Yang J., Bowser M.T. Capillary electrophoresis-SELEX selection of catalytic DNA aptamers for a small-molecule porphyrin target. Anal. Chem. 2013;85:1525–1530. doi: 10.1021/ac302721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo Z., Zhou H., Jiang H., et al. Development of a fraction collection approach in capillary electrophoresis SELEX for aptamer selection. Analyst. 2015;140:2664–2670. doi: 10.1039/c5an00183h. [DOI] [PubMed] [Google Scholar]
- 56.Song K.-M., Jeong E., Jeon W., et al. Aptasensor for ampicillin using gold nanoparticle based dual fluorescence-colorimetric methods. Anal. Bioanal. Chem. 2012;402:2153–2161. doi: 10.1007/s00216-011-5662-3. [DOI] [PubMed] [Google Scholar]
- 57.Chen J., Liu J., Wang J., et al. Fluorescent biosensor based on FRET and catalytic hairpin assembly for sensitive detection of polysialic acid by using a new screened DNA aptamer. Talanta. 2022;242 doi: 10.1016/j.talanta.2022.123282. 123282. [DOI] [PubMed] [Google Scholar]
- 58.Hung L.Y., Wang C.H., Hsu K.F., et al. An on-chip Cell-SELEX process for automatic selection of high-affinity aptamers specific to different histologically classified ovarian cancer cells. Lab Chip. 2014;14:4017–4028. doi: 10.1039/c4lc00587b. [DOI] [PubMed] [Google Scholar]
- 59.Hong S., Wan Y., Tang M., et al. Multifunctional screening platform for the highly efficient discovery of aptamers with high affinity and specificity. Anal. Chem. 2017;89:6535–6542. doi: 10.1021/acs.analchem.7b00684. [DOI] [PubMed] [Google Scholar]
- 60.Ni S., Zhuo Z., Pan Y., et al. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl. Mater. Interfaces. 2021;13:9500–9519. doi: 10.1021/acsami.0c05750. [DOI] [PubMed] [Google Scholar]
- 61.Aminova O., Disney M.D. A microarray-based method to perform nucleic acid selections. Methods Mol. Biol. 2010;669:209–224. doi: 10.1007/978-1-60761-845-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y., Chandra P., Ban C., et al. Electrochemical evaluation of binding affinity for aptamer selection using the microarray chip. Electroanalysis. 2012;24:1057–1064. [Google Scholar]
- 63.Liu X., Li H., Jia W., et al. Selection of aptamers based on a protein microarray integrated with a microfluidic chip. Lab Chip. 2017;17:178–185. doi: 10.1039/c6lc01208f. [DOI] [PubMed] [Google Scholar]
- 64.Glökler J., Schütze T., Konthur Z. Automation in the high-throughput selection of random combinatorial libraries — Different approaches for select applications. Molecules. 2010;15:2478–2490. doi: 10.3390/molecules15042478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogawa N., Biggin M.D. High-throughput SELEX determination of DNA sequences bound by transcription factors in vitro. Methods Mol. Biol. 2012;786:51–63. doi: 10.1007/978-1-61779-292-2_3. [DOI] [PubMed] [Google Scholar]
- 66.Fraser L.A., Kinghorn A.B., Tang M.S.L., et al. Oligonucleotide functionalised microbeads: Indispensable tools for high-throughput aptamer selection. Molecules. 2015;20:21298–21312. doi: 10.3390/molecules201219766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dunn M.R., Jimenez R.M., Chaput J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017;1 [Google Scholar]
- 68.Zhang J., Huang Y., Sun M., et al. Recent advances in aptamer-based liquid biopsy. ACS Appl. Bio Mater. 2022;5:1954–1979. doi: 10.1021/acsabm.1c01202. [DOI] [PubMed] [Google Scholar]
- 69.Gotrik M.R., Feagin T.A., Csordas A.T., et al. Advancements in aptamer discovery technologies. Acc. Chem. Res. 2016;49:1903–1910. doi: 10.1021/acs.accounts.6b00283. [DOI] [PubMed] [Google Scholar]
- 70.Ludwig J.A., Weinstein J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 71.Yu M., Stott S., Toner M., et al. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alix-Panabières C., Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 73.Hardingham J.E., Hewett P.J., Sage R.E., et al. Molecular detection of blood-borne epithelial cells in colorectal cancer patients and in patients with benign bowel disease. Int. J. Cancer. 2000;89:8–13. [PubMed] [Google Scholar]
- 74.Cristofanilli M., Broglio K.R., Guarneri V., et al. Circulating tumor cells in metastatic breast cancer: Biologic staging beyond tumor burden, Clin. Breast Cancer. 2007;7:34–42. [PubMed] [Google Scholar]
- 75.Nadal R., Fernandez A., Sanchez-Rovira P., et al. Biomarkers characterization of circulating tumour cells in breast cancer patients. Breast Cancer Res. 2012;14:R71. doi: 10.1186/bcr3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim S.H., Ito H., Kozuka M., et al. Cancer marker-free enrichment and direct mutation detection in rare cancer cells by combining multi-property isolation and microfluidic concentration. Lab Chip. 2019;19:757–766. doi: 10.1039/c8lc00772a. [DOI] [PubMed] [Google Scholar]
- 77.Zhang L., Zou L., Ma Y., et al. Multifaceted modifications for a cell size-based circulating tumor cell scope technique hold the prospect for large-scale application in general populations. Cell Biol. Int. 2021;45:345–357. doi: 10.1002/cbin.11491. [DOI] [PubMed] [Google Scholar]
- 78.Dong Z., Tang C., Zhao L., et al. A microwell-assisted multiaptamer immunomagnetic platform for capture and genetic analysis of circulating tumor cells. Adv. Healthcare Mater. 2018;7 doi: 10.1002/adhm.201801231. [DOI] [PubMed] [Google Scholar]
- 79.Sun N., Wang J., Ji L., et al. A cellular compatible chitosan nanoparticle surface for isolation and in situ culture of rare number CTCs. Small. 2015;11:5444–5451. doi: 10.1002/smll.201501718. [DOI] [PubMed] [Google Scholar]
- 80.Dou B., Xu L., Jiang B., et al. Aptamer-functionalized and gold nanoparticle array-decorated magnetic graphene nanosheets enable multiplexed and sensitive electrochemical detection of rare circulating tumor cells in whole blood. Anal. Chem. 2019;91:10792–10799. doi: 10.1021/acs.analchem.9b02403. [DOI] [PubMed] [Google Scholar]
- 81.Xiao Y., Lin L., Shen M., et al. Design of DNA aptamer-functionalized magnetic short nanofibers for efficient capture and release of circulating tumor cells. Bioconjug. Chem. 2020;31:130–138. doi: 10.1021/acs.bioconjchem.9b00816. [DOI] [PubMed] [Google Scholar]
- 82.Song Y., Lin B., Tian T., et al. Recent progress in microfluidics-based biosensing. Anal. Chem. 2019;91:388–404. doi: 10.1021/acs.analchem.8b05007. [DOI] [PubMed] [Google Scholar]
- 83.Phillips J.A., Xu Y., Xia Z., et al. Enrichment of cancer cells using aptamers immobilized on a microfluidic channel. Anal. Chem. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Y., Phillips J.A., Yan J., et al. Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal. Chem. 2009;81:7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheng W., Chen T., Kamath R., et al. Aptamer-enabled efficient isolation of cancer cells from whole blood using a microfluidic device. Anal. Chem. 2012;84:4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu J., Shang J., Jia Y., et al. Spatially selective release of aptamer-captured cells by temperature mediation. IET Nanobiotechnol. 2014;8:2–9. doi: 10.1049/iet-nbt.2013.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nguyen N.V., Jen C.-P. Selective detection of human lung adenocarcinoma cells based on the aptamer-conjugated self-assembled monolayer of gold nanoparticles. Micromachines (Basel) 2019;10:195. doi: 10.3390/mi10030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang F., Wu L., Nie W., et al. Biomimetic microfluidic system for fast and specific detection of circulating tumor cells. Anal. Chem. 2019;91:15726–15731. doi: 10.1021/acs.analchem.9b03920. [DOI] [PubMed] [Google Scholar]
- 89.Cao L., Cheng L., Zhang Z., et al. Visual and high-throughput detection of cancer cells using a graphene oxide-based FRET aptasensing microfluidic chip. Lab Chip. 2012;12:4864–4869. doi: 10.1039/c2lc40564d. [DOI] [PubMed] [Google Scholar]
- 90.Chen Y.-H., Pulikkathodi A.K., Ma Y.-D., et al. A microfluidic platform integrated with field-effect transistors for enumeration of circulating tumor cells. Lab Chip. 2019;19:618–625. doi: 10.1039/c8lc01072b. [DOI] [PubMed] [Google Scholar]
- 91.Abate M.F., Jia S., Ahmed M.G., et al. Visual quantitative detection of circulating tumor cells with single-cell sensitivity using a portable microfluidic device. Small. 2019;15 doi: 10.1002/smll.201804890. [DOI] [PubMed] [Google Scholar]
- 92.Reinholt S.J., Craighead H.G. Microfluidic device for aptamer-based cancer cell capture and genetic mutation detection. Anal. Chem. 2018;90:2601–2608. doi: 10.1021/acs.analchem.7b04120. [DOI] [PubMed] [Google Scholar]
- 93.Xu C., Tang M., Feng J., et al. A liquid biopsy-guided drug release system for cancer theranostics: Integrating rapid circulating tumor cell detection and precision tumor therapy. Lab Chip. 2020;20:1418–1425. doi: 10.1039/d0lc00149j. [DOI] [PubMed] [Google Scholar]
- 94.Sheng W., Chen T., Tan W., et al. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano. 2013;7:7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao W., Cui C.H., Bose S., et al. Bioinspired multivalent DNA network for capture and release of cells. Proc. Natl. Acad. Sci. U S A. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Song Y., Shi Y., Huang M., et al. Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angew. Chem. Int. Ed. Engl. 2019;58:2236–2240. doi: 10.1002/anie.201809337. [DOI] [PubMed] [Google Scholar]
- 97.Zhang J., Lin B., Wu L., et al. DNA nanolithography enables a highly ordered recognition interface in a microfluidic chip for the efficient capture and release of circulating tumor cells. Angew. Chem. Int. Ed. Engl. 2020;59:14115–14119. doi: 10.1002/anie.202005974. [DOI] [PubMed] [Google Scholar]
- 98.Peng J., Liu Y., Su R., et al. DNA-programmed orientation-ordered multivalent microfluidic interface for liquid biopsy. Anal. Chem. 2022;94:8766–8773. doi: 10.1021/acs.analchem.2c01359. [DOI] [PubMed] [Google Scholar]
- 99.C. Wang, Y. Xu, S. Li, et al., Designer tetrahedral DNA framework-based microfluidic technology for multivalent capture and release of circulating tumor cells, Mater. Today Bio 16 (2022), 100346. [DOI] [PMC free article] [PubMed]
- 100.Maremanda N.G., Roy K., Kanwar R.K., et al. Quick chip assay using locked nucleic acid modified epithelial cell adhesion molecule and nucleolin aptamers for the capture of circulating tumor cells. Biomicrofluidics. 2015;9 doi: 10.1063/1.4930983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao L., Tang C., Xu L., et al. Enhanced and differential capture of circulating tumor cells from lung cancer patients by microfluidic assays using aptamer cocktail. Small. 2016;12:1072–1081. doi: 10.1002/smll.201503188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y., Wang Z., Wu L., et al. Combining multiplex SERS nanovectors and multivariate analysis for in situ profiling of circulating tumor cell phenotype using a microfluidic chip. Small. 2018;14 doi: 10.1002/smll.201704433. [DOI] [PubMed] [Google Scholar]
- 103.Gao R., Zhan C., Wu C., et al. Simultaneous single-cell phenotype analysis of hepatocellular carcinoma CTCs using a SERS-aptamer based microfluidic chip. Lab Chip. 2021;21:3888–3898. doi: 10.1039/d1lc00516b. [DOI] [PubMed] [Google Scholar]
- 104.Wu Z., Pan Y., Wang Z., et al. A PLGA nanofiber microfluidic device for highly efficient isolation and release of different phenotypic circulating tumor cells based on dual aptamers. J. Mater. Chem. B. 2021;9:2212–2220. doi: 10.1039/d0tb02988b. [DOI] [PubMed] [Google Scholar]
- 105.Zhang X., Wei X., Men X., et al. Dual-multivalent-aptamer-conjugated nanoprobes for superefficient discerning of single circulating tumor cells in a microfluidic chip with inductively coupled plasma mass spectrometry detection. ACS Appl. Mater. Interfaces. 2021;13:43668–43675. doi: 10.1021/acsami.1c11953. [DOI] [PubMed] [Google Scholar]
- 106.Liu Y., Lin Z., Zheng Z., et al. Accurate isolation of circulating tumor cells via a heterovalent DNA framework recognition element-functionalized microfluidic chip. ACS Sens. 2022;7:666–673. doi: 10.1021/acssensors.1c02692. [DOI] [PubMed] [Google Scholar]
- 107.Z. Han, F. Wan, J. Deng, et al., Ultrasensitive detection of mRNA in extracellular vesicles using DNA tetrahedron-based thermophoretic assay, Nano Today 38 (2021), 101203.
- 108.Tian F., Liu C., Deng J., et al. Microfluidic separation, detection, and engineering of extracellular vesicles for cancer diagnostics and drug delivery. Acc. Mater. Res. 2022;3:498–510. [Google Scholar]
- 109.Harding C., Heuser J., Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pan B.T., Teng K., Wu C., et al. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Belotti Y., Lim C.T. Microfluidics for liquid biopsies: Recent advances, current challenges, and future directions. Anal. Chem. 2021;93:4727–4738. doi: 10.1021/acs.analchem.1c00410. [DOI] [PubMed] [Google Scholar]
- 112.Patil A.A., Rhee W.J. Exosomes: Biogenesis, composition, functions, and their role in pre-metastatic niche formation. Biotechnol. Bioprocess Eng. 2019;24:689–701. [Google Scholar]
- 113.Tschuschke M., Kocherova I., Bryja A., et al. Inclusion biogenesis, methods of isolation and clinical application of human cellular exosomes. J. Clin. Med. 2020;9:436. doi: 10.3390/jcm9020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sato-Kuwabara Y., Melo S.A., Soares F.A., et al. The fusion of two worlds: Non-coding RNAs and extracellular vesicles - diagnostic and therapeutic implications (Review) Int. J. Oncol. 2015;46:17–27. doi: 10.3892/ijo.2014.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu H., Liao C., Zuo P., et al. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes. Anal. Chem. 2018;90:13451–13458. doi: 10.1021/acs.analchem.8b03272. [DOI] [PubMed] [Google Scholar]
- 116.Liu C., Zhao J., Tian F., et al. λ-DNA- and aptamer-mediated sorting and analysis of extracellular vesicles. J. Am. Chem. Soc. 2019;141:3817–3821. doi: 10.1021/jacs.9b00007. [DOI] [PubMed] [Google Scholar]
- 117.Zhou Q., Rahimian A., Son K., et al. Development of an aptasensor for electrochemical detection of exosomes. Methods. 2016;97:88–93. doi: 10.1016/j.ymeth.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 118.Kashefi-Kheyrabadi L., Kim J., Chakravarty S., et al. Detachable microfluidic device implemented with electrochemical aptasensor (DeMEA) for sequential analysis of cancerous exosomes. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112622. [DOI] [PubMed] [Google Scholar]
- 119.Dong X., Chi J., Zheng L., et al. Efficient isolation and sensitive quantification of extracellular vesicles based on an integrated ExoID-Chip using photonic crystals. Lab Chip. 2019;19:2897–2904. doi: 10.1039/c9lc00445a. [DOI] [PubMed] [Google Scholar]
- 120.Ren Y., Ge K., Sun D., et al. Rapid enrichment and sensitive detection of extracellular vesicles through measuring the phospholipids and transmembrane protein in a microfluidic chip. Biosens. Bioelectron. 2022;199 doi: 10.1016/j.bios.2021.113870. 113870. [DOI] [PubMed] [Google Scholar]