Graphical abstract
Keywords: Chicken, Extreme environment, Whole-genome sequence, Genetic adaption
Highlights
-
•
Chickens from Southern China originated earlier than those from Northern China.
-
•
SLC33A1 and TSHR genes provided key evolutionary molecular adaption to tropical environment.
-
•
5 SNPs in SLC33A1 and TSHR appeared low mutant allele frequencies to adapt to tropical environment.
-
•
NDUFS4 gene helped chicken achieve rapid adaption to frigid environment.
-
•
SLC33A1, TSHR and NDUFS4 can be biomarkers to assess the adaptation to extreme environments.
Abstract
Introduction
Investigating the genetic footprints of historical temperature selection can get insights to the local adaptation and feasible influences of climate change on long-term population dynamics.
Object
Chicken is a significative species to study genetic adaptation on account of its similar domestication track related to human activity with the most diversified varieties. Yet, few studies have demonstrated the genetic signatures of its adaptation to naturally tropical and frigid environments.
Method
Here, we generated whole genome resequencing of 119 domesticated chickens in China including the following breeds which are in order of breeding environmental temperature from more tropical to more frigid: Wenchang chicken (WCC), green-shell chicken (GSC), Tibetan chicken (TBC), and Lindian chicken (LDC).
Results
Our results showed WCC branched off earlier than LDC with an evident genetic admixture between WCC and LDC, suggesting their closer genetic relationship. Further comparative genomic analyses solute carrier family 33 member 1 (SLC33A1) and thyroid stimulating hormone receptor (TSHR) genes exhibited stronger signatures for positive selection in the genome of the more tropical WCC. Furthermore, genotype data from about 3,000 African local ecotypes confirmed that allele frequencies of single nucleotide polymorphisms (SNPs) in these 2 genes appeared strongly associated with tropical environment adaptation. In addition, the NADH:ubiquinone oxidoreductase subunit S4 (NDUFS4) gene exhibited a strong signature for positive selection in the LDC genome, and SNPs with marked allele frequency differences indicated a significant relationship with frigid environment adaptation.
Conclusion
Our findings partially clarify how selection footprints from environmental temperature stress can lead to advantageous genomic adaptions to tropical and frigid environments in poultry and provide a valuable resource for selective breeding of chickens.
Introduction
Environment pressures have the feasibility to adversely affect the health of domestic animals, with consequences to reducing animal welfare and economic efficiency [1]. On the strength of differences of evolved genetic adaptations, resilience obtained from artificial and natural selection showed significant differences among varieties [2], which has resulted in a wide diversity of domestic animal breeds since they were domesticated. Hence, domestic animals are excellent models for genetic studies and identification of mutations underlying evolution adaption [3], which have contributed to basic and medical biology [4]. Due to the convenience and simple requirements of feeding conditions during migration, domestic chicken is closely related to human activity tracks and has become the most diversified domestication model among domestic animals [5]. According to 2007 FAO statistics, China owned 116 domestic chicken breeds, accounting for nearly 10% of the world’s breeds [6]. China situated in eastern Asia, and is located on the west coast of the Pacific Ocean, and it straddles the eastern and northern hemispheres with diversified geographical, climatic and ecological conditions. In particular, there is a great temperature difference (∼76 °C between hottest and coldest temperature) from North to South created the conditions for the formation of variety diversity and evolved genetic adaptations in chickens.
Considering their short breeding and growth periods, and a similar domestication track related to human activity with the most diversified domestic animals, chickens have served as model organisms for investigating genetic diversity and evolutionary selection mechanisms [7], [8]. Several studies have reported that the domestication of chicken started in multiple regions including South Asia, Southeast Asia, and Southwestern China thousands of years ago [9]. Subsequently, numerous indigenous local breeds were formed after a long time of natural and artificial selections, which have adapted to extensive eco-geographic conditions, especially those in tropical and frigid regions. In view of the current and future global influences of climate change, understanding the genetic adaptation of local chicken breeds to extreme environments and then developing applicable breeding plans to tackle climate change are of practical value for the modern chicken industry.
Extreme environment pressures have been considered as an important diver shaping the animal genome [10]. Identifying an adaptive selection signature driven by extreme environments has become a key focus of evolutionary biology. A series of genetic variations influenced by environmental adaptation have been reported in diverse species, such as Drosophila, chickens, honey bees, sheep, and horses [11], [12], [13], [14], [15]. Exploring the selective signature mediated by climate necessary for comprehending the genetic basis of native environmental adaption as well as any functionally vital variants [16]. For the past few years, whole genomic sequencing (WGS) has been widely used for genomic studies in domestic livestock. In fact, WGS can identify candidate genes of important traits and utilize SNPs to reveal detailed insights into biological questions such as evolutionary processes, environmental adaptation, and natural selection [17]. Previous genomic studies on poultry have mainly been conducted on the genetic diversity of breeds [18], evolutionary mechanisms [19], and genetic mechanisms of quality traits [20]. However, to our knowledge, there is a paucity of WGS studies characterizing poultry genetic adaptations to extreme environments, like tropical and frigid environments, although there has been some genetic work on adaptation of Tibetan chickens (TBC) to high altitudes [12] and tropical desert climate [7].
In this study, the whole genomes of 119 chickens from 4 Chinese local breeds in various temperature environments were sequenced (Fig. 1A and supplementary Fig. S2B, supplementary table S1, Supplementary Material online). Then we performed comprehensive analyses of the population structure, genomic diversity, and demographic history of those chicken breeds according to their genomic data. We also identified candidate genes and pathways that were potentially associated with rapid adaptation to tropical and frigid environments, since domestication of the chicken occurred in Southern Asia ∼ 6,000–8,000 years ago [21].
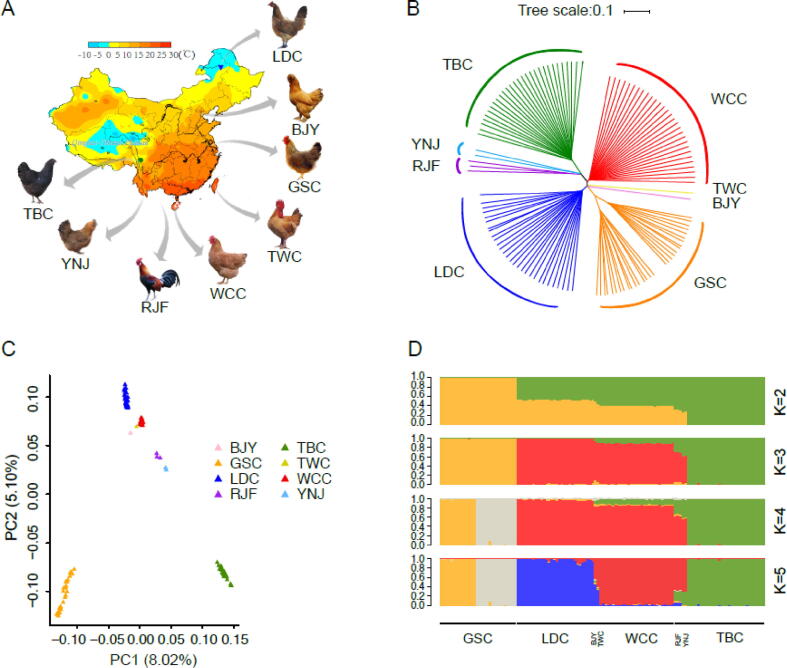
Fig. 1.
Population genetics. (A) Geographic locations of the 126 individuals. A total of 119 sampled Chinese chickens representing four breeds were included and 7 individuals of four breeds were used as outgroups. (B) The neighbor-joining tree constructed using p-distance between individuals. The 7 individuals of four breeds were used as outgroups. (C) Principal component plot of the 126 individuals. (D) Population genetic structure of the 126 individuals examined via the program FRAPPE v1.1. The number of assumed genetic clusters K ranged from 2 to 5 are shown.
Materials and methods
Ethics statement
Handling and sampling of 119 Chinese local chickens were conducted under the guidance of ethical regulations of the Poultry Institute, Chinese Academy of Agriculture Science, Jiangsu, China. All experiments involving chickens from Tanzani and Ghana were conducted according to the ethical policies and procedures of the Institutional Animal Care and Use Committee, the University of California, Davis (#:17853).
Sample collection
Blood was collected from 119 chickens for whole-genome sequencing, including 29 Wenchang chickens (female, 43 weeks, Wenchang, Hainan, 32 °C), 30 Green-shell laying hens (female, 42 weeks, Yangzhou, Jiangsu, 20 °C), 30 Lindian chickens (female, 42 weeks, Lindian County, Heilongjiang, −3 °C) and 30 Tibetan chickens (female, 40 weeks, Ganzi, Sichuan, 2 °C) from their national local poultry genetic resources conservation farm with different temperature zones (supplementary table S1, supplementary Fig S2B, Supplementary Material online). After blood collection of these 119 chickens, 16 liver tissues (4 chickens per breed) were sampled for RNA extraction and transcriptome analyses. Also, blood samples of 120 chickens originating from four local breeds (30 individuals per breed), including Minqing Mao Jiao chicken from Fujian (female, 43 weeks, 30 °C), Longsheng Feng chicken from Guangxi (female, 41 weeks, 29 °C), Jining Bairi chicken from Shangdong (female, 46 weeks, 17 °C), and Beijing You chicken from Beijing (female, 44 weeks, 13 °C), were collected for DNA extraction and genotyped by PCR. All these birds were reared in semi open chicken house and fed with traditional corn-soybean meal diet.
Blood samples of 1,399 indigenous chickens from the regions of Morogoro, Tanga, Shinyanga, Mwaza, and Singida in Tanzania (29 °C), as well as 1,439 indigenous chickens from Ghana (30 °C) were obtained for chicken Affymetrix 600 K SNP chip analysis. All the blood samples were collected from wing vein and stored in 5 ml EDTA sodium anticoagulant tube at −20 °C.
Whole genome sequencing library preparation
The DNA was extracted with 1.5 µg per sample. We formed sequencing libraries via Truseq Nano DNA HT Sample preparation Kit (Illumina USA) and then added index codes to each sample. In brief, every DNA sample was fragmented into 350 bp size, which were then end polished, A-tailed, and ligated. At last, we purified PCR products and analyzed these libraries by Agilent2100 Bioanalyzer as well as quantified using real-time PCR.
Genome sequencing
In the current research, we sequenced the whole genome of these samples with the Illumina Hiseq 2000 platform. In all, 3287.41 Gb raw data were sequenced. Firstly, low quality paired reads were removed (Reads with ≥ 10% unidentified nucleotides (N); > 10 nt aligned to the adaptor, allowing ≤ 10% mismatches; > 50% bases having phred quality < 5; and putative PCR duplicates generated in the library construction process). Consequently, we generated 3278.18 Gb (∼22.74-average fold per sample) high quality paired-end reads for 119 samples with the quality of 93.44% and 90.75% of the bases were ≥ Q20 and ≥ Q30, respectively.
Reads mapping
The rest of high quality reads were mapped to the Gallus_gallus-5.0 reference genome using BWA (Burrows-Wheeler Aligner) (Version: 0.7.8) with the command ‘mem -t 4 -k 32 –M’ [22]. In order to decrease mismatch generated by PCR amplificatio, duplicated reads were removed by Samtools v1.3.1 [23].
SNP and InDel calling
SNP and InDel were performed by a Bayesian approach which was carried out with the package Samtoolsv1.3.1 [24]. Then we calculated the genotype likelihoods from reads at each genomic location, and allele frequencies were calculated with a Bayesian approach. The ‘mpileup’ command was applied to authenticate SNPs with the parameters like ‘-q 1 -C 50 -t SP -t DP -m 2 -F 0.002′. Then, only high quality SNPs (coverage depth ≥ 5, RMS mapping quality ≥ 20, maf ≥ 0.05, miss ≤ 0.1) were reserved for pursuant analysis to eliminate SNP calling errors caused by incorrect mapping or InDels. Hence, 11,690,980 SNPs were reserved after filtering from 22,830,447 raw SNPs.
Variants of structure detection
Structure variants (SV) including insertions (INS), deletions (DEL), intra-chromosomal translocations (ITX), inter-chromosomal translocations (CTX) and inversions (INV) of all 119 samples and 4 groups was detected by lumpy-sv v 0.2.13 [25].
Functional annotation of genetic variants
We annotate those SNP and InDel according to the Gallus_gallus-5.0 genome using the package ANNOVAR [24] (Version: 2013–05-20). SNPs were grouped into intronic regions, exonic regions, upstream and downstream regions, splicing sites, and intergenic regions according to the genome annotation results. Then those SNPs in coding exons were moreover classed into synonymous SNPs or nonsynonymous SNPs.
Phylogenetic tree and population structure
We instituted a neighbour-joining (NJ) tree by the software TreeBest v1.9.2 to show the phylogenetic relationship based on the genome-wide SNPs. An expectation maximization algorithm was applied to inspect the population genetic structure in the program FRAPPE v1.1. We made the Principal component analysis (PCA) via the software GCTA v1.24.2. Then Tracey-Widom experiment was used to evaluate the eigenvector significance level.
Genomic diversity and linkage disequilibrium
The θπ was calculated by Vcftools v0.1.14, and θw was calculated by variscan 2.0. The ROH (regions of homozygosity) was identified by the runs of homozygosity tool in PLINK (v.1.07) with the parameters (--homozyg-density 10 --homozyg-window-het 1 --homozyg-window-kb 5000 --homozyg-window-snp 20 --homozyg-kb 100 --homozyg-snp 10). Then we conducted the squared correlation coefficient (r2) with the software Haploview [26] to estimate LD decay. The average r2 was counted in a 500-kb window as well as averaged through the whole genome.
Population Historical, differentiation time and migration events
Demographic history of 119 samples was reconstructed by a hidden Markov model (HMM) approach in pairwise sequentially Markovian coalescent (PSMC v 0.6.4-r49) [27]. Initially, genotype of each sample was called via the package Samtools v1.3.1. Then, the program ‘fq2psmcfa' was applied to switch the consensus sequence into a fasta-like format where the i-th character indicates whether there is at least one heterozygote in the bin [100i, 100i + 100], which the purpose of improving the accuracy of inferred historical recombination events. The parameters were set as follows: −N30, −t15, −r5 and − p ‘4 + 25*2 + 4 + 6′. The chicken neutral mutation rate per generation (μ) was 0.19 × 10−8 and generation time (g) was 1 years [12].
We estimated the divergent time by fastsimcoal2 v 2.6.0.3 [28], SNP located at intergenic regions were applied to transform SFS by easySFS (https://github.com/isaacovercast/easySFS#easysfs). Mutation ratio was set to 2E-9 per site per generation as well as generation time as 1 year. The fastsimcoal2 was conducted 100 times with changed starting points, and the fitting model was reserved based on the highest likelihood. Demographic estimates were obtained from 100,000 simulations (-n100,000) per parameter file, 40 Expectation/Conditional Maximization cycle (-L40). The population relationship and gene flow patterns were concluded with a maximum likelihood approach applied in TreeMix v 1.12 [29].
Genome-wide selective sweep test
To ascertain genome-wide selective sweeps concerned with temperature adaptation, the genome-wide assignment of fixation index (FST) values and θπ ratios were calculated for these defined group pairs (vcftools v0.1.14). Average SNP FST values were plotted in 40 kb genomic bins with a 20 kb step. Nucleotide diversity (π) was estimated for the same bins. The FST values were Z-transformed as follows: Z(FST) = (FST - µFST) / σFST, in which µFST is the mean FST and σFST is the standard deviation of FST. The θπ ratios were log2-transformed. Then we evaluated and arranged the empirical percentiles of Z(FST) and log2(θπ ratio). The windows with the top 5% Z(FST) and log2(θπ ratio) values contemporaneously considered as nominated outliers under strong selective sweeps. XP-EHH value of each SNP was calculated using the XP-EHH program (https://github.com/joepickrell/xpehh). The average XP-EHH values were computed with a 40 kb window size and a 20 kb step size. Windows with the top 5% XP-EHH values were selected.
Genotyping array analysis
The blood samples of chickens from Tanzania and Ghana were applied for DNA extraction and Array analysis. Genomic DNA was isolated and genotyping was analysed via the Aymetrix Axiom® 600 k Array at GeneSeek (Lincoln, NE, USA). Annotation files of Genotyping Array were referred to the Gallus gallus genome version 5 (Thermo Fisher Scientific Inc., Calsbad, CA, USA). We then performed the genotype data quality filtering by AxiomTM Analysis Suite 3.1 (Applied Biosystems, Thermo Fisher Scientific Inc., Calsbad, CA, USA) and these genotype data quality filtering included single nucleotide polymorphism (SNP) call rate ≥ 99% as well as minor allele frequency ≥ 0.05.
Transcriptome sequencing and analysis
Liver tissues from 16 chickens (4 biological replicates per condition) were collected and Sequenced for transcriptomic analysis. We generated sequencing libraries via NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA). Then we conducted the clustering of the index-coded samples on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia). Then the library preparations were sequenced via an Illumina Hiseq platform and 150 bp paired-end reads were created. Totally, 126.86 Gb RNA-seq reads were generated with 16 liver tissues. Index of the reference genome was built with Bowtie v2.2.3 and aligned the high-quality RNA-seq reads to the reference genome by the HISAT2 v2.0.4 program. HTSeq v0.6.1 was applied to cast the reads numbers of each gene. Then we calculated FPKM according to gene length and read count numbers.
Quantitative PCR analysis and Western blotting
qPCR mainly was conducted for five genes, SLC33A1, NFUFS4, TSHR and its downstream genes adenylate cyclase 1 (ADCY1), cAMP responsive element binding protein 3 like 3 (CREB3L3). The primer sequences of these genes were revealed in Supplementary Table S23, Supplementary Material online. Liver tissues randomly selected from 5 chickens per breed were used to verify the accuracy of the transcriptome data, and those heart and hypothalamus tissues were performed to reveal the temporal expression pattern. Total RNA was isolated and synthesized cDNA using FastKing Super Mix Kit (KR118, Tiangen Biotech Co., Ltd.). Then, we used SuperReal PreMix Plus (FP205, Tiangen Biotechnology Co., Ltd.) to perform Real-time PCR by StepOnePlus (Applied Biosystems). And the mRNA expression was calculated by 2−ΔΔCt method.
20 Liver proteins (5 chickens per breed) were extracted using RIPA Lysis Buffer. After complete lysis of samples, we collected the supernatant, and tested the protein concentrations with a BCA Protein Assay kit. Each sample was denatured at 100 °C for 5 min before being loaded into polyacrylamide gel and then moved to polyvinylidene fluoride membranes. The membranes were washed in Tween-Tris-bufferedsaline buffer and blocked for 2 h, and then cultivated by primary antibodies (SLC33A1 (LS-B11505, Lifespan) at 1:500, TSHR (PAB27584, abnova) at 1:500, NDUFS4 (DF9670, affinity) at 1:500, β-actin at 1:2000) at 4 °C overnight. After washes, we incubated the membranes via anti-rabbit IgG secondary antibody for 2 h. Then we visualized the immunoblots with enhanced chemiluminescence detection and quantified via densitometric scanning of the autoradiograph. The band densities of proteins were normalized with those of β-actin.
Results
Sequencing variants among different chicken breeds
We conducted whole-genome resequencing of 119 individuals from the 4 local chicken breeds which are in order of breeding environmental temperature from more tropical to more frigid: Wenchang chicken (WCC), Green-shell chicken (GSC), TBC, and Lindian chicken (LDC). We acquired a total of 3,278 Gb of high-quality genomic data, with an average of 183.61 M paired-end reads (150 bp) per individual. Alignment of the obtained sequences to the chicken reference genome (ftp://ftp.ensembl.org/pub/release-90/fasta/gallus_gallus/dna/Gallus_gallus.Gallus _gallus-5.0.dna.toplevel.fa.gz) resulted in an average of 92.52% sequencing coverage with 22.74 × effective depth of each individual (supplementary table S2, S3, Supplementary Material online). After accurate quality filtering of the identified SNPs, 11,690,980 high-quality SNPs were identified in the 119 individuals. Of these SNPs, 83.55% ∼ 88.81% were found in the chicken dbSNP database (supplementary Fig. S2A, supplementary table S4, Supplementary Material online), proving the high quality and validity of the SNP calling. Of these four populations, a majority of these SNPs identified in the chicken genome were lied in intronic and intergenic regions (11.11 million, 95.03%), while only 1.41% (0.17 million) of the SNPs were in exonic regions (supplementary table S5, Supplementary Material online). And 117,539 synonymous SNPs and 47,151 nonsynonymous SNPs were located in exons, resulting in a nonsynonymous/synonymous ratio of 0.40, which was consistent with previous studies in chickens [18]. In addition, we detected a total of 2,212,046 indels (<100 bp), with 1,942,963 for the WCC, 1,530,616 for the GSC, 1,757,682 for the TBC, and 1,780,716 for the LDC (supplementary table S6, Supplementary Material online). In total, 18,315 structural variations (SVs, 100 bp-chromosome level) were identified including deletion, inversion, and complex rearrangements with breakends (supplementary table S7, Supplementary Material online). Most of the indels and SVs were in intergenic regions (supplementary table S6, supplementary table S8, Supplementary Material online). And all the SNPs, indels and SVs distributed evenly on all chromosomes (supplementary Fig. S1, Supplementary Material online).
Phylogeny and population genetic structure as well as migration events
To infer genetic and evolutionary relationships of chicken breeds adapted to diverse temperature environments, we generated high quality whole genome sequencing data of 119 individuals from 4 populations, as previously mentioned. In addition, we also downloaded resequencing data from the NCBI database for the following four chicken breeds: Red jungle fowl (RJF, 3 individuals), Yunnan native chickens (YNC, 2 individuals), Taiwan chickens (TWC, 1 individual), and Beijing-You chickens (BJY, 1 individual) (Fig. 1A, supplementary table S9, Supplementary Material online). From these data we in all obtained 11,698,251 SNPs. Next, a phylogenetic tree from a total of eight chicken breeds with 126 chickens was constructed using the neighbor-joining (NJ) algorithm according to pairwise genetic distances between populations according to the polymorphic SNPs in the whole genome. The tree revealed that individuals from the same geographic region tended to cluster into a single group (Fig. 1B). RJF, as the ancestor of the domestic chicken, was in the root of the tree and clustered together with TBC and YNJ, which were 2 ancient native breeds raised in Southwest China near the geographic region of RJF. Those 3 breeds first branched off, then WCC, TWC, BJY, LDC and GSC successively branched off in order of their geographic position from south to north and environment from tropical to frigid. GSC branched off relatively late as it was a commercial breed selected by artificial selection. These findings were corroborated by both principal component analysis (PCA) and population structure analysis (Fig. 1C and 1D, supplementary Fig. S4, Supplementary Material online).
In the population structure analysis, when K ≤ 3, the overall individuals were genetically fallen into three clusters (WCC-LDC, GSC, or TBC populations) with WCC and LDC clustering together and the other 2 populations tending to cluster independently. When K = 4, two genetic clusters were formed within the population of GSC. This was in accordance with NJ tree and PCA analysis and revealed an obvious branch in the GSC population, suggesting that there are 2 different strains of GSC. As a commercial high-yield breed, plumage color change in GSC (black to hemp-colored feathers) appeared during the hybridization process. Also, we detected a number of nonsynonymous mutations in several plumage color related key genes including melanocortin 1 receptor (MC1R) and premelanosome protein (PMEL) [30], [31], with different SNP frequencies in the 2 GSC strains (supplementary Table S10, Supplementary Material online).
Furthermore, we observed a clear signature of genetic introgression or ancestral polymorphisms from TBC into WCC and LDC when K = 2. We also found WCC and LDC clustered more closely with each other from K = 2 to K = 4, and there was a slight gene flow between them (Fig. 1D). A division between WCC and LDC was found at K = 5, which was the optimal K, with slight genetic admixture between these two breeds, indicating a close genetic relationship and a similar domestication history between the two breeds, although the precise origins of WCC and LDC are unclear. Demographic modeling using Fastsimcoal analysis supported the idea that LDC originated from breeds of Southern China (Fig. 2B), further confirming the close relationship between WCC and LDC.
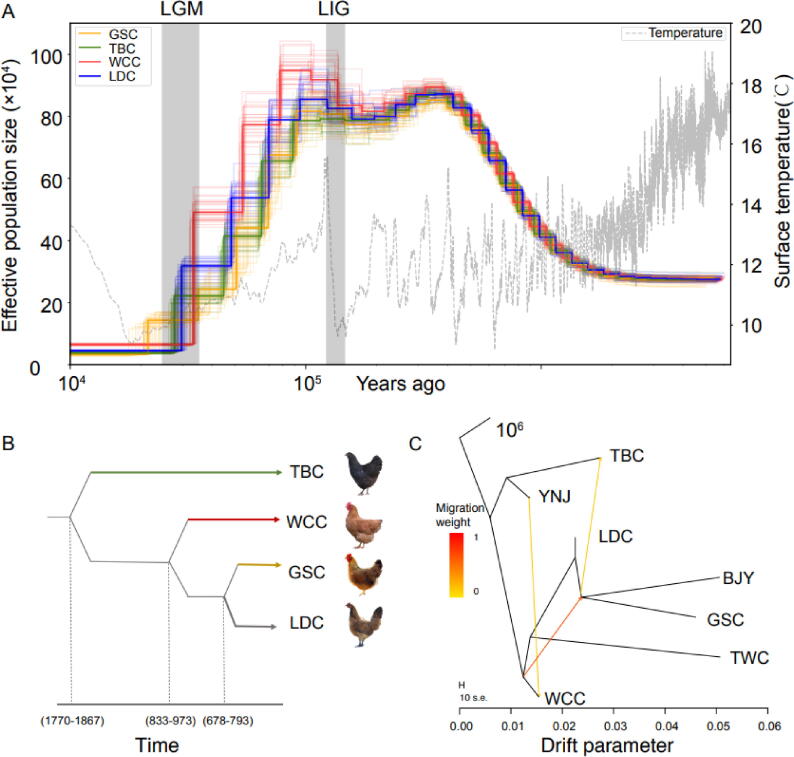
Fig. 2.
Demographic history of the chicken population. (A) Estimated effective population size of the 119 individuals with PSMC analysis based on SNP distribution. (B) The demographic history of the four populations from ∼ 2,000 years ago to present showed with fastsimcoal analysis. (C) Pattern of population splits and mixtures among the four populations and the four outgroups. The drift parameter is proportional to Ne generations, where Ne is the effective population size. Scale bar shows ten times the average standard error of the estimated entries in the sample covariance matrix. Arrows indicate migration events, and a spectrum of heat colors indicate the migration weights of the migration events.
We then rebuilt the maximum likelihood tree and residual matrix of these breeds and added sequential migration events to the tree by using TreeMix [29], to address population history relationships and to deduce migration patterns among those different breeds (Fig. 2C, supplementary Fig. S6, Supplementary Material online). We found that one supposed migration edge produced a tree with the smallest residuals and consequently best fit the data. We observed a statistically significant migration edge (p < 2.23e-308) with an estimated greatest weight of 62.65%. This edge provides evidence for the gene flow from WCC to the breeds in Northern China. In recent years, studies have provided a perspective on the pattern of gene flow between Southeastern and Northern breeds in China [32], though no direct evidence has been presented to demonstrate that WCC has been crossbred with Northern breeds. Similar to the previous TreeMix finding [33], our data found gene flow between LDC and TBC. Also consistent with previous results [34], current study revealed gene flow between YNC and WCC, which might be attributable to population migration from the Southwest to Hainan China [35]. These findings partly provided confidence to a single origin theory for Southern and Northern chicken breeds and frequent gene flows among them, further implying the multiple and close relationships of these breeds.
Genomic diversity and linkage disequilibrium
The genome-wide average θπ values of the four populations were estimated in this study, with the highest θπ value (2.99 × 10-3) being observed in WCC while the lowest θπ value (2.64 × 10-3) was found for GSC. The other genomic variation parameters (average number of regions of homozygosity (ROH) and mean ROH size) showed nearly congruent patterns (supplementary Fig. S2C - S2F, supplementary table S11, Supplementary Material online). In other words, WCC showed the lowest ROH number (3 7 7) and a relatively smaller average ROH size (347.87 kb), whereas the highest ROH number (5 8 1) and the largest average ROH size (511.75 kb) were observed in GSC. These data further indicate the greatest genomic diversity in WCC and extensive inbreeding in GSC, potentially due to artificial selection. Genome-wide linkage disequilibrium of the four populations also varied markedly (supplementary Fig. S3, Supplementary Material online). Specifically, GSC showed a slow decay rate and a high level of linkage disequilibrium, while WCC exhibited a rapid decay rate and a low level of linkage disequilibrium. TBC and LDC presented a similar decay rate and level of linkage disequilibrium in an intermediate status. Taken together, the patterns of linkage disequilibrium and genomic diversity may reflect the differences in population histories of genetic bases, and the low genetic diversity of GSC may have resulted from artificial selection and breeding in recent years.
Population history and differentiation time
To explore the demographic history of the four chicken populations, the historical effective population sizes (Ne) of the chicken ancestors were estimated using PSMC method according to SNPs in the whole genome and the linkage disequilibrium among them. The results revealed that individuals from the same populations or the same geographic regions exhibited a highly similar pattern of demographic trajectories (Fig. 2A, supplementary Fig. S5, Supplementary Material online). The Ne of the four populations fluctuated over time with the earth’s climate, with two expansions and two bottlenecks in the demographic history, which is consistent with the findings of others [12]. We found that an increase in population sizes of the four chicken breeds occurred sometime around 1 Mya and reached a peak at approximately 100 kya, with a dramatic decline following that peak, which mirrors environmental changes in Southeast Asia where RJF are distributed. In time of the Quaternary ice ages (from ∼ 2.5 Ma to now), the rainforests in Southeast Asia that is known as one of the world’s biodiversity hotspots with a high concentration of local species, served as a refugia, empowering the persistence of forest biota[36]. Meanwhile a slight population size fluctuation occurred following that peak. This fluctuation was consistent with the Last Interglacial (LIG), during which increased seasonality in temperature extremes appeared [37]. An apparent reduction in population sizes of the four breeds was observed from the LIG until the Last Glacial Maximum (LGM), which might have resulted from a substantial decrease of the forested area that occurred in Southeast Asia during the Last Glacial (LG) period (∼125–10 kya) [38]. In addition, it was clear that similar Nes were observed in all 4 breeds, which implied none of the 4 breeds had branched off 10 kya ago. Fastsimcoal analysis showed TBC was likely domesticated more than 1,000 years, which was in accordance with a previous study [39]. WCC and LDC branched off ∼ 800 and ∼ 600 ya, respectively, indicating their time of domestication (Fig. 2B). Interestingly, we found the age estimate of LDC by fastsimcoal analysis coincided with the introduction time of WCC to Northern China. In fact, a historical record in “Lingnan omnivorous poem” [40], indicated that WCC was used as a tribute to the imperial court in the Ming Dynasty (CE 1368–1644). WCC was received by the emperor, and then WCC was introduced to Northern China and domesticated locally.
Genome-Wide selective sweep test
Adaptive mechanisms in tropical environments
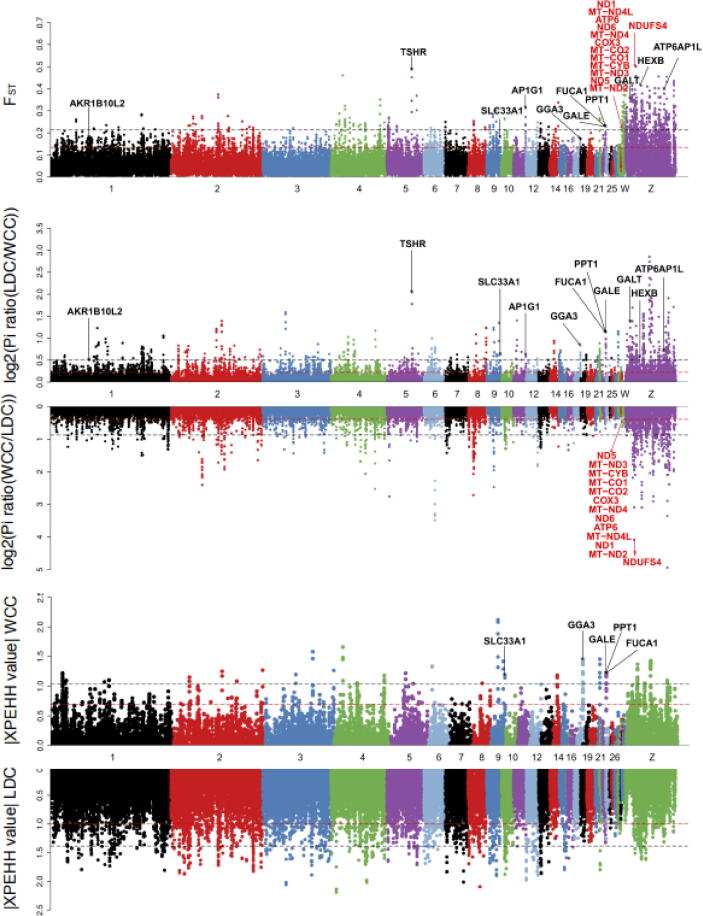
To study adaptive mechanisms of chickens in tropical environments, we used comparisons between WCC and LDC to screen WCC for a selective signal (supplementary Fig. S7, Supplementary Material online). Next, using the top 5% of both FST and θπ ratio cutoffs, we identified 17.92 Mb of 448 selective sweep regions encompassing 269 genes (Fig. 3, supplementary table S12, Supplementary Material online) and several highly enriched pathways (supplementary table S15, Supplementary Material online), which were mainly associated with glycometabolism and energy metabolism. In the negative control group GSC vs LDC and TBC vs LDC, we discovered a 36.96 Mb of 924 selective sweep region encompassing 455 genes with three highly enriched pathways (supplementary Fig. S8, supplementary table S13, Supplementary Material online) and a 22.60 Mb of 565 selective sweep region encompassing 278 genes with 4 highly enriched pathways (supplementary Fig. S9, supplementary table S14, Supplementary Material online). In total, 5 pathways with ten genes in WCC had no overlap with the other seven pathways, and 5 genes including solute carrier family 33 member 1 (SLC33A1); golgi associated, gamma adaptin ear containing, ARF binding protein 3 (GGA3); alpha-L-fucosidase 1 (FUCA1); UDP-galactose-4-epimerase (GALE); and palmitoyl-protein thioesterase 1 (PPT1) were also identified using the XP-EHH method (Fig. 3), which further speculated their significant role in tropical environmental adaption.
Fig. 3.
Positive selection scans for adaption to tropical and frigid environments. Chickens living in tropical environments (WCC) were compared with those living in frigid environments (LDC). The population genetic differentiation FST values, the nucleotide diversity θπ ratios (θπ-LDC/θπ-WCC) and θπ ratios (θπ-WCC/θπ-LDC) and the XP-EHH values are were plotted in 40 kb genomic bins with a 20 kb step. The significance threshold of the selection signature was arbitrarily assigned to the top 5% percentile outliers for each individual test and is indicated with red horizontal dashed lines. The black horizontal dashed lines display the top 1% quantile. The black font genes represent the WCC selective signal, and the red font genes represent the LDC selective signal.
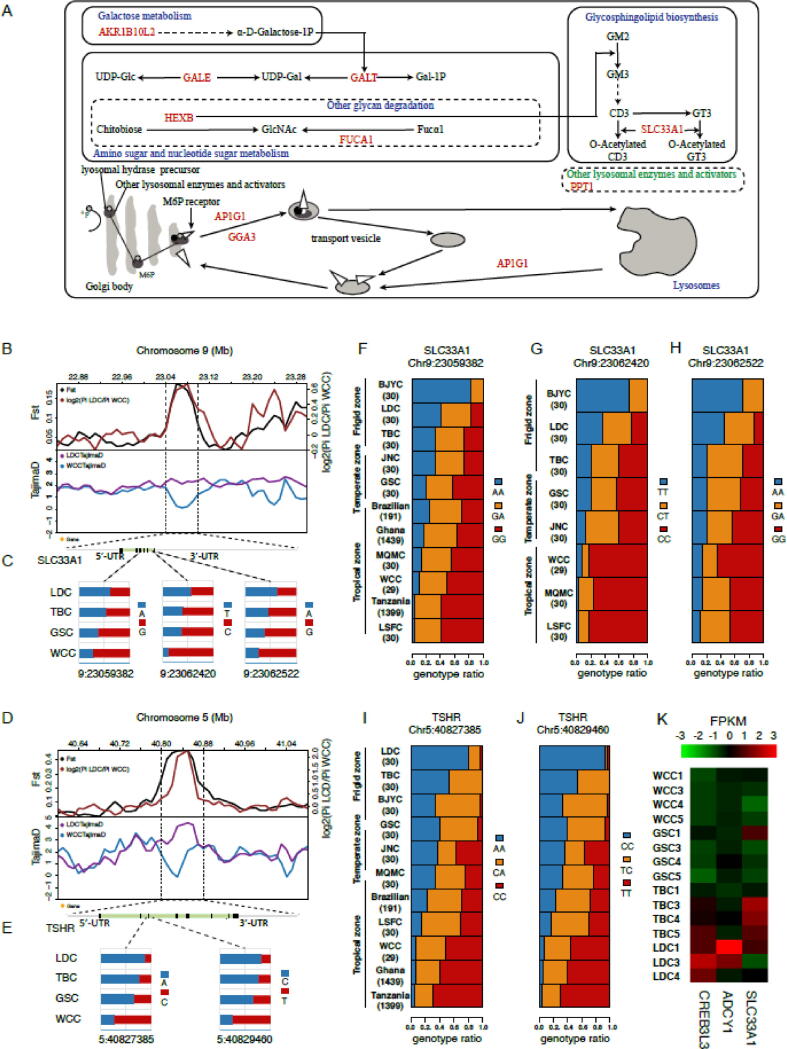
Among the 10 candidate genes for tropical environments, three genes [aldo–keto reductase family 1 member B10 (AKR1B10L2), GALE, and galactose-1-phosphate uridylyltransferase (GALT)] are in the galactose metabolism pathway, and three genes [GALE, hexosaminidase subunit beta (HEXB), and GALT] are in the amino sugar and nucleotide sugar metabolism pathway (supplementary table S15, Supplementary Material online). Both are found upstream of the pentose phosphate pathway which plays an important role in high temperature conditions because it generates NADPH to serve as a cofactor of GPX [41]. Meanwhile, UDP-Gla was found as an important metabolite in the amino sugar and nucleotide sugar metabolism pathway (Fig. 4A), which also participates in extreme environment adaption [42]. In addition, we discovered a positive metabolite, GlcNAc, which appears in the amino sugar and nucleotide sugar metabolism pathway and another glycan degradation pathway. It is a posttranslational sugar modification of nucleocytoplasmic and mitochondrial proteins which confers heat stress tolerance [43]. In the lysosome pathway, we also found 5 positively selected genes (ATP6AP1L, HEXB, AP1G1, PPT1 and GGA3), which directly affect substances for digestion via a series of processes including endocytosis, phagocytosis, and autophagy and which ultimately participate in heat stress-induced intestinal tissue and epithelial cell apoptosis [44]. 2 genes (HEXB and FUCA1) affect another glycan degradation pathway. 2 genes (HEXB and SLC33A1) influence the glycosphingolipid biosynthesis-ganglio series, and they are functionally related to cell proliferation, differentiation, and adhesion [45]. These 5 relevant pathways encompassing glycometabolism and energy metabolism, which are related to cellular metabolism, are involved in responses to high-temperature stress [46] and are, perhaps, the most important factors that allow chickens to adjust to tropical environmental pressures.
Fig. 4.
Analysis of the signatures of positive selection in the genome of chicken breeds and the adaptions to tropical Environments. (A) Schematic mechanisms of signaling pathways for genetic adaptions to tropical environments in chicken. The names of the KEGG pathways are shown in blue. The candidate genes positively selected in the two methods of FST and θπ ratio tests are shown in red. Dotted arrows indicate an indirect effect. (B) (D) log2 (θπ ratios), FST value and Tajima’s D values around the SLC33A1 locus, TSHR locus, respectively. The black and red line represent FST and log2 (θπ ratios) values, respectively. The blue and purple lines represent the WCC, LDC Tajima’s D values, respectively. (C) (E) Allele frequencies of two and three SNPs within the SLC33A1, TSHR gene across the four breeds, respectively. The red alleles are consistent with reference genome, and the blue alleles represent mutant alleles. (F)(G)(H) Genotype frequencies of three SNPs within the SLC33A1 gene were determined in our 119 individuals and chickens from Tanzania, Ghana, and Brazil using the Affymetrix 600 k SNP database, as well as in four Chinese breeds with PCR. For the abbreviations of the breeds, see supplementary table S16 and Supplementary Material online. (I)(J) Genotype frequencies of two SNPs within the TSHR gene were determined in our 119 individuals and chickens from Tanzania, Ghana, and Brazilian using the Affymetrix 600 k SNP database, as well as in four Chinese breeds with PCR. For the abbreviations of the breeds, see supplementary table S16 and Supplementary Material online. (E) Gene expression of SLC33A1, and the gene expression of the TSHR downstream genes, including ADCY1 and CREB3L3 in chicken liver of four breeds based on RNA-seq analysis. The FPKM (fragments per kilobase of transcript per million mapped reads) value is used to measure the expression level.
Notably, among 10 identified genes in WCC selective pathways, we observed significantly higher Z (FST) and log2 (θπ ratio) values but lower Tajima’s D values for the target gene SLC33A1 compared with those in the adjacent genomic regions (Fig. 4B), indicating that a strong selective sweep occurred in this gene. SLC33A1 encodes the acetyl-CoA (Ac-CoA) transporter (ACATN), which transports Ac-CoA from the cytoplasm to the golgi apparatus or the endoplasmic reticulum lumen that regulates protein acetylation [47]. SLC33A1 promotes GD3 acetylation that contributes to mitochondrial damage repair in the glycosphingolipid biosynthesis-ganglio series pathway [48], which plays a crucial role in high temperature reactions [49]. Additionally, we observed that the gene for thyroid stimulating hormone receptor (TSHR), though not a candidate gene in the 5 selective pathways, had the strongest selective sweep (Fig. 4D). Also, it had markedly higher Z (FST) and log2 (θπ ratio) values but lower Tajima’s D values in WCC compared with LDC in adjacent genomic regions. TSHR is the receptor for thyroid-stimulating hormone (TSH), the key regulator of thyroid function. TSHR has been reported to have a pivotal biological function in metabolic regulation during evolution [19], [50].
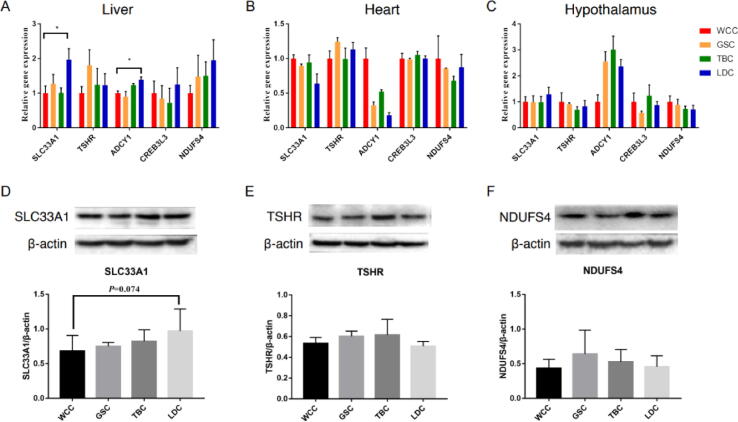
We found within SLC33A1, which is highly conserved with particular significance [51], that specific alleles of three SNPs in WCC had lower mutation frequency that gradually became higher from GSC to LDC (Fig. 4C, supplementary table S16, Supplementary Material online), which was similar in pattern to each breed environmental location. Similar to changes in the allele frequencies of the three SNPs, the liver expression levels of SLC33A1 were significantly higher in LDC than in WCC (Fig. 4K, Fig. 6). We also noticed that there were 2 SNPs within the most selective region of TSHR showed similar change law as the three SNPs of SLC33A1 (Fig. 4E). Meanwhile, the interspecies NJ tree of TSHR sequences exposed that an evolutionarily conserved nucleotide has mutated which induced a change of coded amino acid (G to R) in WCC but remained variant in LDC and other breeds (supplementary Fig. S10, Supplementary Material online). Previously, this mutation has been identified as contributing to chicken evolution [50]. We further examined the mRNA expression levels of TSHR in livers and its downstream genes ADCY1 and CREB3L3. The data revealed that liver but expression levels of ADCY1 and CREB3L3 genes decreased as environmental temperature surrounding WCC to LDC decreased (Fig. 4K, Fig. 6). Although there was no change in RNA expression levels in TSHR, the expression changes of ADCY1 and CREB3L3 suggested the related signal pathway of TSHR might be activated in liver in response to temperature.
Fig. 6.
Validation assays of the selective genes for adaption to tropical and frigid environments. (A)(B)(C) Temporal relative expression of five genes in liver, heart, hypothalamus across the four breeds measured by qRT-PCR, respectively. (D)(E)(F) Western-blotting analysis of SLC33A1, TSHR, NDUFS4.
To further explore the SNPs identified above, we utilized SNPs data (chicken Affymetrix 600 K SNP chip) from 1,399 indigenous chickens from Tanzania (Eastern Africa) [52], and 1,439 indigenous chickens from Ghana (Western Africa) [53], and from 156 Brazilian chickens [54]. The SNPs allele frequencies of the selected 11 candidate genes (including TSHR) in these populations were calculated and compared with allele frequencies in the breeds of the current study. 3 mutations [2 in the TSHR gene (at position 40,827,385 and 40829460) and 1 in the SLC33A1 gene (at position 23059382)] were found in the Affymetrix 600 k data, and their allele frequencies in these 2 large African populations (6 local ecotypes) supported their possible important roles in tropical environment adaption (Fig. 4F, 4I, 4 J, supplementary table S16, Supplementary Material online).
To further examine whether allele frequency patterns of the 5 mutations in SLC33A1 and TSHR were associated with tropical environments, 120 individuals originating from 4 Chinese breeds were genotyped by PCR (30 individuals per breed, supplementary fig. S11 and S12, supplementary table S17, Supplementary Material online). The collection test analyses were in support of a strong correlation between the frequencies of 3 variants and environment temperature (position 40827385, P-value = 1.05E-4; position 40829460, P-value = 1.11e-4, position 23059382, P-value = 0.03). WCC living at > 25 °C showed the lowest allelic frequencies of the three mutations (∼0.25), whereas LDC living in a low-temperature environment (<10 °C) showed the highest (∼0.60–0.90). The other chicken breeds presented corresponding frequencies (Fig. 4F-J, supplementary table S16, Supplementary Material online).
Adaptive mechanisms in frigid environments
For the purpose of understanding adaptive mechanisms in frigid environments, LDC, TBC, and GSC were compared to WCC, and the last 2 groups for comparison were set as the control group (supplementary Fig. S13, Supplementary Material online). Using the top 5% of both FST and θπ ratio cutoffs, we identified a 33.40 Mb of 835 selective sweep region encompassing 504 genes (Fig. 3, supplementary table S18, Supplementary Material online). Additionally, several highly enriched pathways were identified, including cardiac muscle contraction, oxidative phosphorylation, metabolic pathways, and a notch signaling pathway (supplementary table S21, Supplementary Material online). In the positive control group (TBC vs WCC) and the negative control group (GSC vs WCC), we discovered a 30.72 Mb of 768 selective sweep regions encompassing 407 genes with 5 highly enriched pathways (supplementary Fig. S14, supplementary table S19, Supplementary Material online) and a 44 Mb of 1100 selective sweep regions encompassing 564 genes with 7 highly enriched pathways (supplementary Fig. S15, supplementary table S20, Supplementary Material online). Interestingly, cardiac muscle contraction, oxidative phosphorylation and notch signaling pathways were selected pathways in both LDC and TBC but not in GSC, suggesting these three pathways’ crucial role in adaptation to frigid environments. In total, 13 common genes in cardiac muscle contraction and oxidative phosphorylation pathways were found in LDC and TBC (Fig. 3, supplementary table S21, Supplementary Material online), which suggests their key status. Meanwhile, we found no overlapping genes in notch signaling pathways between LDC and TBC, indicating their different genetic mechanisms of frigid environment adaption. In addition, the folate biosynthesis pathway was selected in TBC, which has been identified as being related to plateau adaptability [55].
Alterations in the oxidative phosphorylation efficiency of mitochondria could play a crucial role in increased heat production of animals and plants exposed to cold [56], [57], because this efficiency is necessary to maintain energy homeostasis. Along with increased oxidative phosphorylation, oxygen consumption increases, which modulates cardiac muscle contractility and adjusts cardiac output and pulmonary artery blood flow to increase the internal and external gas exchange rate [58]. In total, 13 genes notably gathered in the oxidative phosphorylation pathway as well as cardiac muscle contractility pathway are mainly oxidation–reduction enzyme genes in the mitochondrial genome, and they are considered important in adaptation to cold environments [59].
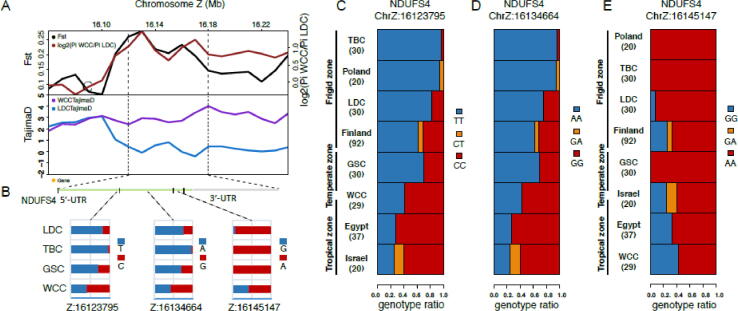
The target gene, NADH dehydrogenase ubiquinone Fe-S protein 4 (NDUFS4), had the largest Z (FST) value among these 13 genes, with much higher Z (FST) and log2 (θπ ratio) values but lower Tajima’s D values in LDC (Fig. 5A), suggesting a strong selective sweep. NDUFS4 belongs to a group of karyogenes of mitochondrial complex Ⅰ (CⅠ), which is responsible for regulation of CI activity and mitochondrial respiration [60]. Within the NDUFS4 genes, specific alleles of two SNPs (Z:16123795, Z:16134664) in the LDC as well as in the TBC populations owned notably higher frequencies than those observed in WCC. The other SNP (Z: 16145147) exhibited an opposite trend simultaneously (Fig. 5B).
Fig. 5.
Analysis of the signatures of positive selection in the genome of chicken breeds and adaptions to frigid environments. (A) log2 (θπ ratios), FST value and Tajima’s D values around the NDUFS4 locus. The black and red line represent FST and log2 (θπ ratios) values, respectively. The blue and purple lines represent the WCC, LDC Tajima’s D values, respectively. (B) Allele frequencies of three SNPs within the NDUFS4 gene across the four breeds, respectively. The red alleles are consistent with reference genome, and the blue alleles represent mutant alleles. (C)(D)(E) Genotype frequencies of three SNPs within the NDUFS4 gene were identified in our 119 individuals and in chickens from Poland, Finland, Israel and Egypt. For the abbreviations of the breeds, see supplementary table S22 and Supplementary Material online.
Next, we sought to further investigate the potential functional consequences of SNPs from the gene NDUFS4. We studied whether they indicated any connection with frigid environments, stretching our analysis to other populations including 20 individuals from Poland, 92 individuals from Finland, 20 individuals from Israel and 37 individuals from Egypt [61], [62]. We found the allele frequencies of three mutations of NDUFS4 in Poland, Finland, Israel and Egypt population displayed similar variation tendency as the populations of WCC, GSC, TBC and LDC have suggested, based on their respective ambient temperatures. These results suggest strong correlation between the variants frequencies and environmental temperature (Fig. 5C-E, supplementary table S22, Supplementary Material online).
Discussion
In this research, we sequenced the genomes of 119 chickens from four Chinese local breeds spanning a wide range of environmental temperatures, from tropical to frigid. A total of ∼ 11.69 million SNPs (∼2.21 million indels and SVs) were identified based on our vast genome data, which helped uncover the population structure of these Chinese local chickens. The location and time of chicken domestication in China has remained controversial even in recent years [63]. However, in line with previous work based on mitochondrial genomes of the chicken [9], our population genetic diversity analysis revealed some chickens from Southern China originated earlier than those from Northern China.
Sweeping the genome of the WCC breed for positive selection signatures to tropical environments, we uncovered a series of genes involved in a few major signaling pathways that were related to glycometabolism and energy metabolism and that played central roles in regulating cellular responses to tropical environments [46]. Within the top-candidate genes, SLC33A1 is a key member of the endoplasmic reticulum (ER) acetylation machinery that belongs to the SLC family [64]. The SLC family facilitates transfer of various molecules, including sugars, nucleotides, and amino acids to maintain a constant internal environment [65]. Several SLC members (SLC13A1, SLC5A11) have been mined and described in connection with tropical environment adaption based on microarray analyses [66]. Additionally, some other members including SLC7A7, SLC2A3, SLC16A1 and SLC16A7 were proposed as having a high potential as heat stress biomarkers for different organisms [67], [68]. Remarkably, SLC33A1 is located in the glycosphingolipid biosynthesis-ganglio series pathway, one of the enriched pathways in our analysis, which regulates ganglioside GD3 and GT3, both are acidic glycosphingolipids that are important lipid and glycolipid mediators of the adaptive responses to various stressors [48], and it was also selected in Hainan pigs (Ding’an and Tunchang pigs) [69], which are exposed to warm environmental conditions; and, therefore, it is reasonable to assume that SLC33A1 is important for adaption to tropical environments. The 3 SNPs within SLC33A1 were found at lower mutation frequencies in chicken breeds from tropical zones than from temperate and frigid zones. This data was collected across a large quantity and different varieties of chickens, that were derived initially from Southeast Asia and that have a high adaptability to tropical environments [9]. Meanwhile, the expression levels of SLC33A1 in the WCC breed were lower than in other breeds, because of its downregulation results in widespread cell death and induction of features characteristic of autophagy to maintain ER homeostasis [64].
It's also worth noting that TSHR, a critical gene with a strong selective sweep, has been described as a “domestication locus” in the chicken [19], [70]. Functional consequences of mutations in the TSHR gene remain to be explored, though previous studies have confirmed a pivotal mediating role for TSHR in reducing aggression to conspecifics, decreasing ascites, controlling reproductive machinery, and influencing broodiness [19], [70], [71]. Interestingly, in our study TSHR appears to be involved in high temperature adaptation, one of the prevailing environmental stressors for which natural selection footprints are often associated [72]. The TSHR SNP variant exhibited a glycine-to-arginine substitution that was unique to the WCC breed, and it was almost completely conserved among the other chicken breeds (supplementary Fig. S10, Supplementary Material online). Currently, this identified missense substitution has not revealed any indication that it is a deleterious mutation. It is interesting to note that lower mutation frequencies of the other two SNPs (5: 40827385, 5: 40829460) within TSHR were discovered in chicken breeds from tropical zones, and these 2 TSHR SNPs exhibited a similar variation tendency to that of the 3 SNP mutations of SLC33A1. We speculate that the TSHR allele exhibits an advantage of increasing metabolic activity and growth. We intend to follow up on the discovery of the TSHR sweep with functional receptor studies.
Genetic variation in the genome of the LDC breed, which is characterized by adaptations to frigid environments, particularly evolved genes associated with mitochondria oxidative phosphorylation (OXPHOS) in the heart that will ultimately generate heat to make an organism more resistant to low temperatures [73]. Several selected genes, including ND2, ND3, ND4, ND5, MT-CYB and ATP6, are known to be important for adaptation to frigid environments in many organisms [74], [75], [76]. Variations in these genes are speculated to influence the mitochondrial coupling efficiency and coordination of the ATP synthesis efficiency ratio to thermogenesis efficiency, which assists in climate adaptation [77]. In addition, a NADH-ubiquinone oxidoreductase gene, NDUFS4, was notably found to exhibit a strong selective sweep in our results. A previous study revealed mutations in NDUFS4 can enhance cold, salt, and osmotic stress tolerance in Arabidopsis [78]. Moreover, NDUFS4 can catalyze electron transport coupled with proton translocation across the inner mitochondrial membrane, thus providing the driving force for ATP synthesis and antioxidant defense maintenance [79], which are strongly related to the cold stress response [80]. In our studies, higher frequencies of 3 SNP variants in the conserved region of NDUFS4 were observed in chickens from frigid environments (LDC and TBC), as well as chickens from Poland and Finland, but not in the more tropical WCC breed, and chickens from Israel and Egypt. These results suggest the value of these detailed advantageous alleles for adaption of chickens to the frigid environments.
Conclusion
In conclusion, the present study provided a whole-genome sequence analysis of Chinese native chicken breeds. The analysis included 119 individuals of 4 breeds from tropical environments to frigid environments. Patterns of genetic diversity revealed differentiation as well as close relationships among these breeds following domestication. Genomic regions exhibited a signature of positive selection revealing unique difference due to selection pressure of the environment. The more tropical WCC revealed a stronger selection toward a variety of novel genes located in 5 pathways that mainly participate in glycometabolism and energy metabolism, which are potentially involved in adaptation to tropical environments. While the LDC from frigid regions exhibited more selection pressure toward cardiac muscle contraction, oxidative phosphorylation and notch signaling pathways related to energy homeostasis in response to frigid environments. The SLC33A1 gene displayed a stronger signature for positive selection in WCC, with lower mutation frequency in 3 selected SNPs and lower expression in the liver than in the other breeds. Another gene, TSHR, exhibited the same nonsynonymous mutation as has been found in previous studies, and it also exhibited 2 other SNPs with lower mutation frequency in WCC. 6 SNPs were confirmed in 1395 Tanzania and 1439 Ghana local African chickens, 156 chickens from Brazil, and 120 individuals of 4 Chinese breeds; and a lower mutation frequency was obtained in these chickens from tropical environments. Additionally, the NDUFS4 gene presented a higher frequency of SNPs in breeds from LDC and TBC as well as other chickens from the colder environments of Poland and Finland. Overall, these results indicate that the genetic differences seen in these very distinct populations likely contribute to their tolerance of and survival in extreme environments. Our genomic data formed in this research will be of value for genetic breeding of chickens that will face extreme climate change.
Compliance with ethics requirements
All Institutional and National Guidelines for the care and use of animals were followed.
Data availability
The accession numbers for genome re-sequencing data and transcriptome data reported in this paper are NCBI SRA: PRJNA800119. Other data that supported the discovers could been obtained from the corresponding authors.
CRediT authorship contribution statement
Shourong Shi: Conceptualization, Methodology, Visualization, Writing – review & editing. Dan Shao: Visualization, Writing – review & editing. Lingyun Yang: Software, Data curation. Qiqi Liang: Software, Data curation. Wei Han: Investigation. Qian Xue: Software, Data curation. Liang Qu: Investigation. Li Leng: Investigation. Yishu Li: Investigation. Xiaogang Zhao: Investigation. Ping Dong: Investigation. Muhammed Walugembe: Data curation. Boniface B. Kayang: Data curation. Amandus P. Muhairwa: Data curation. Huaijun Zhou: Methodology, Writing – review & editing. Haibing Tong: Conceptualization, Methodology, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Genome Sequencing Technology Platform at Novogene for performing the sequencing and suppling computer resources. We thank F.Cheng. (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences), G.Chang., H.Bai. and Q. Guo. (Yangzhou University) for helpful discussion and comments. This work was supported by grants from the Supported by the Jiangsu Provincial Key Laboratory of Poultry Genetics & Breeding (JQLAB-ZZ-202006).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.07.005.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Miraglia M., Marvin H.J.P., Kleter G.A., Battilani P., Brera C., Coni E., et al. Climate change and food safety: an emerging issue with special focus on Europe. Food Chem Toxicol. 2009;47(5):1009–1021. doi: 10.1016/j.fct.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Thornton P.K., Steeg J.V.D., Notenbaert A., Herrero M. The impacts of climate change on livestock and livestock systems in developing countries: a review of what we know and what we need to know. Agr Syst. 2009;101(3):113–127. [Google Scholar]
- 3.Sayyab S. Swedish University of Agricultural Sciences; 2014. Bioinformatic screening for candidate mutations underlying phenotypic traits in domestic animals. PHD Thesis: [Google Scholar]
- 4.Andersson L., Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5(3):202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- 5.Boivin N., Fuller D.Q. Shell middens, ships and seeds: exploring coastal subsistence, maritime trade and the dispersal of domesticates in and around the Ancient Arabian Peninsula. J World Prehist. 2009;22(2):113–180. [Google Scholar]
- 6.Rischkowsky B., Pilling D. The state of the world's animal genetic resources for food and agriculture. Food Agric Org. 2007:102. [Google Scholar]
- 7.Tian S., Zhou X., Phuntsok T., Zhao N., Zhang D., Ning C., et al. Genomic Analyses Reveal Genetic Adaptations to Tropical Climates in Chickens. iScience. 2020;23(11):101644. doi: 10.1016/j.isci.2020.101644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burt D.W. Emergence of the chicken as a model organism: implications for agriculture and biology. Poult Sci. 2007;86(7):1460–1471. doi: 10.1093/ps/86.7.1460. [DOI] [PubMed] [Google Scholar]
- 9.Wang M.-S., Thakur M., Peng M.-S., Jiang Y.u., Frantz L.A.F., Li M., et al. 863 genomes reveal the origin and domestication of chicken. Cell Res. 2020;30(8):693–701. doi: 10.1038/s41422-020-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ai H., Fang X., Yang B., Huang Z., Chen H., Mao L., et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 2015;47(3):217–225. doi: 10.1038/ng.3199. [DOI] [PubMed] [Google Scholar]
- 11.Hancock A.M., Brachi B., Faure N., Horton M.W., Jarymowycz L.B., Sperone F.G., et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334(6052):83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- 12.Wang M.-S., Li Y., Peng M.-S., Zhong L.i., Wang Z.-J., Li Q.-Y., et al. Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol Biol Evol. 2015;32(7):1880–1889. doi: 10.1093/molbev/msv071. [DOI] [PubMed] [Google Scholar]
- 13.Chen C., Liu Z., Pan Q.i., Chen X., Wang H., Guo H., et al. Genomic analyses reveal demographic history and temperate adaptation of the newly discovered Honey Bee Subspecies Apis mellifera sinisxinyuan n. ssp. Mol Biol Evol. 2016;33(5):1337–1348. doi: 10.1093/molbev/msw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J., Li W.R., Lv F.H., He S.G., Tian S.L., Peng W.F., et al. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol Biol Evol. 2016;33(10):2576–2592. doi: 10.1093/molbev/msw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X., Zhang Y., Li Y., Pan J., Wang D., Chen W., et al. EPAS1 gain-of-function mutation contributes to high-altitude adaptation in Tibetan horses. Mol Biol Evol. 2019;36(11):2591–2603. doi: 10.1093/molbev/msz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen R., Hellmann I., Hubisz M., Bustamante C., Clark A.G. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8(11):857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stratton M. Genome resequencing and genetic variation. Nat Biotechnol. 2008;26(1):65–66. doi: 10.1038/nbt0108-65. [DOI] [PubMed] [Google Scholar]
- 18.Fan W.L., Ng C.S., Chen C.F., Lu M.Y., Chen Y.H., Liu C.J., et al. Genome-wide patterns of genetic variation in two domestic chickens. Genome Biol Evol. 2013;5(7):1376–1392. doi: 10.1093/gbe/evt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin C.J., Zody M.C., Eriksson J., Meadows J.R., Sherwood E., Webster M.T., et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 20.Feng C., Gao Y.u., Dorshorst B., Song C., Gu X., Li Q., et al. A cis-regulatory mutation of PDSS2 causes silky-feather in chickens. PLoS Genet. 2014;10(8):e1004576. doi: 10.1371/journal.pgen.1004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mwacharo J.M., Nomura K., Hanada H., Han J.L., Amano T., Hanotte O. Reconstructing the origin and dispersal patterns of village chickens across East Africa: insights from autosomal markers. Mol Ecol. 2013;22(10):2683–2697. doi: 10.1111/mec.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15(6):R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Excoffier L., Dupanloup I., Huerta-Sánchez E., Sousa V.C., Foll M., Akey J.M. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9(10):e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickrell J.K., Pritchard J.K., Tang H. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Zhou R., Peng Y., Zhang C., Li L., Lu C., et al. Feather follicles transcriptome profiles in Bashang long-tailed chickens with different plumage colors. Genes Genomics. 2019;41(11):1357–1367. doi: 10.1007/s13258-018-0740-y. [DOI] [PubMed] [Google Scholar]
- 31.Yang C.-w., Ran J.-S., Yu C.-L., Qiu M.-H., Zhang Z.-R., Du H.-R., et al. Polymorphism in MC1R, TYR and ASIP genes in different colored feather chickens. 3 Biotech. 2019;9(5) doi: 10.1007/s13205-019-1710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W.Y., Zhao C.J. Comprehensive genetic analysis with mitochondrial DNA data reveals the population evolution relationship between Chinese gamecocks and their neighboring native chicken breeds. Asian J Anim Vet Adv. 2010;5(6):388–401. [Google Scholar]
- 33.Hui-Fang L.I., Chen K.W., Han W., Zhang X.Y., Gao Y.S., Chen G.H., et al. Comparison of analyses of genetic structure among Chinese indigenous chicken breeds using distance-based and model-based methods. Chinese J Anim Vet Sci. 2009;S1:5. [Google Scholar]
- 34.Chun-Hong S., Hong-Ju C., Yue-Hui M.A., Hui T., Ding-Guo C., Xin-Zhong F., et al. Maternal origins of six indigenous chicken breeds in China. Acta Veterinaria Et Zootechnica Sinica. 2007;38(7):735–740. [Google Scholar]
- 35.Michaud J. From Southwest China into Upper Indochina: an overview of Hmong (Miao) migrations. Asia Pacific Viewpoint. 1997;38(2):119–130. [Google Scholar]
- 36.Sodhi N.S., Lian P.K., Brook B.W., Ng P. Southeast Asian biodiversity: an impending disaster. Trends Ecol Evol. 2004;19(12):654–660. doi: 10.1016/j.tree.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Kim S.-J., Lü J.M., Yi S., Choi T., Kim B.-M., Lee B.Y., et al. Climate response over Asia/Arctic to change in orbital parameters for the last interglacial maximum. Geosci J. 2010;14(2):173–190. [Google Scholar]
- 38.Wurster C.M., Bird M.I., Bull I.D., Creed F., Bryant C., Dungait J.A.J., et al. Forest contraction in north equatorial Southeast Asia during the Last Glacial Period. Proc Natl Acad Sci U S A. 2010;107(35):15508–15511. doi: 10.1073/pnas.1005507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H., Wu C.X., Chamba Y., Ling Y. Blood characteristics for high altitude adaptation in Tibetan chickens. Poult Sci. 2007;86(7):1384–1389. doi: 10.1093/ps/86.7.1384. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z. Lingnan omnivorous poem.Beijing. Zhonghua Book Company. 1985 [Google Scholar]
- 41.Habashy W.S., Milfort M.C., Rekaya R., Aggrey S.E. Expression of genes that encode cellular oxidant/antioxidant systems are affected by heat stress. Mol Biol Rep. 2018;45(3):389–394. doi: 10.1007/s11033-018-4173-0. [DOI] [PubMed] [Google Scholar]
- 42.Liu H.L., Dai X.Y., Xu Y.Y., Chong K. Over-expression of OsUGE-1 altered raffinose level and tolerance to abiotic stress but not morphology in Arabidopsis. J Plant Physiol. 2007;164(10):1384–1390. doi: 10.1016/j.jplph.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Kazemi Z., Chang H., Haserodt S., McKen C., Zachara N.E. O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner. J Biol Chem. 2010;285(50):39096–39107. doi: 10.1074/jbc.M110.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Česen M.H., Pegan K., Spes A., Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp Cell Res. 2012;318(11):1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Ge J., Jin Y., Lv X., Liao Q., Luo C., Ye G., et al. Expression profiles of circular RNAs in human colorectal cancer based on RNA deep sequencing. J Clin Lab Anal. 2019;33(7) doi: 10.1002/jcla.22952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao J.L., Zhang H.Y., Liu J.B., Zhong P.A., Huang Y.J. Identification of candidate genes related to rice grain weight under high-temperature stress. Plant Sci. 2012;196:32–43. doi: 10.1016/j.plantsci.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Lin P., Li J., Liu Q., Mao F., Li J., Qiu R., et al. A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42) Am J Hum Genet. 2008;83(6):752–759. doi: 10.1016/j.ajhg.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malisan F., Testi R. GD3 ganglioside and apoptosis. Biochim Biophys Acta. 2002;1585(2–3):179–187. doi: 10.1016/s1388-1981(02)00339-6. [DOI] [PubMed] [Google Scholar]
- 49.Balogh G., Péter M., Glatz A., Gombos I., Török Z., Horváth I., et al. Key role of lipids in heat stress management. FEBS Lett. 2013;587(13):1970–1980. doi: 10.1016/j.febslet.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Qanbari S., Rubin C.-J., Maqbool K., Weigend S., Weigend A., Geibel J., et al. Genetics of adaptation in modern chicken. PLoS Genet. 2019;15(4):e1007989. doi: 10.1371/journal.pgen.1007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirabayashi Y., Nomura K.H., Nomura K. The acetyl-CoA transporter family SLC33. Mol Aspects Med. 2013;34(2–3):586–589. doi: 10.1016/j.mam.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Walugembe M., Mushi J.R., Amuzu-Aweh E.N., Chiwanga G.H., Msoffe P.L., Wang Y., et al. Genetic analyses of Tanzanian local chicken ecotypes challenged with newcastle disease virus. Genes (Basel) 2019;10(7):546. doi: 10.3390/genes10070546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.da Silva A.P., Aston E.J., Chiwanga G.H., Birakos A., Muhairwa A.P., Kayang B.B., et al. Molecular characterization of newcastle disease viruses isolated from chickens in Tanzania and Ghana. Viruses. 2020;12(9):916. doi: 10.3390/v12090916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walugembe M., Bertolini F., Dematawewa C.M.B., Reis M.P., Elbeltagy A.R., Schmidt C.J., et al. Detection of selection signatures among Brazilian, Sri Lankan, and Egyptian chicken populations under different environmental conditions. Front Genet. 2019;9 doi: 10.3389/fgene.2018.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge R.-L., Cai Q., Shen Y.-Y., San A., Ma L., Zhang Y., et al. Draft genome sequence of the Tibetan antelope. Nat Commun. 2013;4(1) doi: 10.1038/ncomms2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panagos S., Beyer R.E., Masoro E.J. Oxidative phosphorylation in liver mitochondria prepared from cold-exposed rats. Biochim Biophys Acta. 1958;29(1):204–205. doi: 10.1016/0006-3002(58)90164-1. [DOI] [PubMed] [Google Scholar]
- 57.Liu J., Xu Y., Zhang L., Li W., Cai Z., Li F., et al. Correction: De novo assembly and analysis of the transcriptome of Rumex patientia L. during cold stress. PLoS One. 2017;12(12):e0190154. doi: 10.1371/journal.pone.0190154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mann D.L. Targeting myocardial energetics in the failing heart: are we there yet? Circ Heart Fail. 2017;10(12) doi: 10.1161/CIRCHEARTFAILURE.117.004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price E.R., Sirsat T.S., Sirsat S.K.G., Kang G., Keereetaweep J., Aziz M., et al. Thermal acclimation in American alligators: effects of temperature regime on growth rate, mitochondrial function, and membrane composition. J Therm Biol. 2017;68(Pt A):45–54. doi: 10.1016/j.jtherbio.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 60.Papa S., Scacco S., Sardanelli A.M., Vergari R., Papa F., Budde S., et al. Mutation in the NDUFS4 gene of complex I abolishes cAMP-dependent activation of the complex in a child with fatal neurological syndrome. FEBS Lett. 2001;489(2–3):259–262. doi: 10.1016/s0014-5793(00)02334-6. [DOI] [PubMed] [Google Scholar]
- 61.Fleming DS, Weigend S, Simianer H, Weigend A, Rothschild M, Schmidt C, et al. Genomic comparison of indigenous African and Northern European chickens reveals putative mechanisms of stress tolerance related to environmental selection pressure. G3 (Bethesda) 2017;7(5):1525-37. [DOI] [PMC free article] [PubMed]
- 62.Malomane D.K., Simianer H., Weigend A., Reimer C., Schmitt A.O., Weigend S. The SYNBREED chicken diversity panel: a global resource to assess chicken diversity at high genomic resolution. BMC Genomics. 2019;20(1):1–15. doi: 10.1186/s12864-019-5727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiang H., Gao J., Yu B., Zhou H., Cai D., Zhang Y., et al. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci U S A. 2014;111(49):17564–17569. doi: 10.1073/pnas.1411882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jonas M.C., Pehar M., Puglielli L. AT-1 is the ER membrane acetyl-CoA transporter and is essential for cell viability. J Cell Sci. 2010;123(19):3378–3388. doi: 10.1242/jcs.068841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueckler M., Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34(2–3):121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao H., Wang H., Jia D., Yan T., Liu F. Cluster analysis of differentially expressed heat stress genes in Rat Jejunal Mucosal. Agr Sci Tech. 2014;15(7):1082–1085. [Google Scholar]
- 67.Bao Z.Q., Liao T.T., Yang W.R., Wang Y., Luo H.Y., Wang X.Z. Heat stress-induced autophagy promotes lactate secretion in cultured immature boar Sertoli cells by inhibiting apoptosis and driving SLC2A3, LDHA, and SLC16A1 expression. Theriogenology. 2017;87:339–348. doi: 10.1016/j.theriogenology.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 68.Yadav S.K., Pandey A., Kumar L., Devi A., Kushwaha B., Vishvkarma R., et al. The thermo-sensitive gene expression signatures of spermatogenesis. Reprod Biol Endocrinol. 2018;16(1):56. doi: 10.1186/s12958-018-0372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diao S., Huang S., Chen Z., Teng J., Zhang Z. Genome-Wide Signatures of Selection Detection in Three South China Indigenous Pigs. Genes. 2019;10(5):346–360. doi: 10.3390/genes10050346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qanbari S., Seidel M., Strom T.M., Mayer K.F., Preisinger R., Simianer H. Parallel selection revealed by population sequencing in chicken. Genome Biol Evol. 2015;7(12):3299–3306. doi: 10.1093/gbe/evv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liisa L., Thomas M.G., Ross B., Richard A., Naomi S., Paxinos P.D., et al. Inferring allele frequency trajectories from ancient DNA indicates that selection on a chicken gene coincided with changes in medieval husbandry practices. Mol Biol Evol. 2017;34(8):1981–1990. doi: 10.1093/molbev/msx142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elbeltagy A.R., Bertolini F., Fleming D.S., Goor A.V., Ashwell C.M., Schmidt C.J., et al. Natural selection footprints among African chicken breeds and village ecotypes. Front Genet. 2013;10 doi: 10.3389/fgene.2019.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brand M.D. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35(6–7):811–820. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 74.Balloux F., Handley L.J., Jombart T., Liu H., Manica A. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc Biol Sci. 2009;276(1672):3447–3455. doi: 10.1098/rspb.2009.0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morales H.E., Pavlova A., Joseph L., Sunnucks P. Positive and purifying selection in mitochondrial genomes of a bird with mitonuclear discordance. Mol Ecol. 2015;24(11):2820–2837. doi: 10.1111/mec.13203. [DOI] [PubMed] [Google Scholar]
- 76.Sun J.T., Duan X.Z., Hoffmann A.A., Liu Y., Garvin M.R., Chen L., et al. Mitochondrial variation in small brown planthoppers linked to multiple traits and probably reflecting a complex evolutionary trajectory. Mol Ecol. 2019;28(14):3306–3323. doi: 10.1111/mec.15148. [DOI] [PubMed] [Google Scholar]
- 77.Gershoni M., Levin L., Ovadia O., Toiw Y., Shani N., Dadon S., et al. Disrupting mitochondrial-nuclear coevolution affects OXPHOS complex I integrity and impacts human health. Genome Biol Evol. 2014;6(10):2665–2680. doi: 10.1093/gbe/evu208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Delannoy E., Pogson B.J. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 2009;151(4):2187. doi: 10.1104/pp.109.141770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Q., Zeng Y., Wang H., Yang L., Yang Y., Zhu H., et al. Molecular characterization and expression analysis of NDUFS4 gene in m. longissimus dorsi of Laiwu pig (Sus scrofa) Mol Biol Rep. 2013;40(2):1599–1608. doi: 10.1007/s11033-012-2208-5. [DOI] [PubMed] [Google Scholar]
- 80.Colinet H. Disruption of ATP homeostasis during chronic cold stress and recovery in the chill susceptible beetle (Alphitobius diaperinus) Comp Biochem Physiol A Mol Integr Physiol. 2011;160(1):63–67. doi: 10.1016/j.cbpa.2011.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for genome re-sequencing data and transcriptome data reported in this paper are NCBI SRA: PRJNA800119. Other data that supported the discovers could been obtained from the corresponding authors.