Graphical abstract
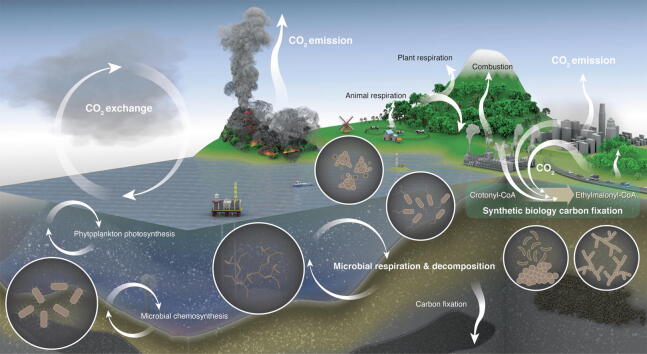
Carbon circulation on Earth. Anthropogenic and natural carbon emissions and carbon sequestration routes.
Keywords: Carbon fixation, Autotrophic, Thermophiles, Omics, Biochemistry, Synthetic pathway
Highlights
-
•
Carbon is often negatively associated with the anthropogenic emissions of greenhouse gases and, consequently, with environmental issues such as global warming. However, carbon is an element of paramount importance for life and ecological processes.
-
•
The knowledge of carbon fixation cycles, in addition to the ecological and evolutionary studies on the emergence of the first beings that inhabited Earth and metabolized carbon, provides a detailed understanding of the enzymes and important intermediaries involved in these cycles.
-
•
The central carbon metabolizing enzymes can be used for a variety of purposes; artificial CO2 fixation pathways can be designed and implemented in suitable host organisms; new solutions are possible for manipulating carbon fixation pathways and increasing their final yields.
-
•
The multidisciplinary integration of omics, synthetic biology, molecular biology, and biochemistry can enable the scientific community to describe new carbon fixation pathways and predict important routes and enzymes involved in these processes. This integration may further provide guidance for overcoming the limitations of natural carbon fixation pathways and allow the development of biotechnological applications.
-
•
Investigating thermophilic microorganisms is crucial for understanding many evolutionary events. These microorganisms are investigated extensively for developing biotechnological applications because of their metabolic diversity.
-
•
In addition to the six well-known natural carbon fixation pathways, the development of omics and systems biology has spearheaded the discovery of new natural and synthetic pathways for carbon metabolism.
Abstract
Background
Autotrophic carbon fixation is the primary route through which organic carbon enters the biosphere, and it is a key step in the biogeochemical carbon cycle. The Calvin–Benson–Bassham pathway, which is predominantly found in plants, algae, and some bacteria (mainly cyanobacteria), was previously considered to be the sole carbon-fixation pathway. However, the discovery of a new carbon-fixation pathway in sulfurous green bacteria almost two decades ago encouraged further research on previously overlooked ancient carbon-fixation pathways in taxonomically and phylogenetically distinct microorganisms.
Aim of Review
In this review, we summarize the six known natural carbon-fixation pathways and outline the newly proposed additions to this list. We also discuss the recent achievements in synthetic carbon fixation and the importance of the metabolism of thermophilic microorganisms in this field.
Key Scientific Concepts of Review
Currently, at least six carbon-fixation routes have been confirmed in Bacteria and Archaea. Other possible candidate routes have also been suggested on the basis of emerging “omics” data analyses, expanding our knowledge and stimulating discussions on the importance of these pathways in the way organisms acquire carbon. Notably, the currently known natural fixation routes cannot balance the excessive anthropogenic carbon emissions in a highly unbalanced global carbon cycle. Therefore, significant efforts have also been made to improve the existing carbon-fixation pathways and/or design new efficient in vitro and in vivo synthetic pathways.
Introduction
Carbon is the fourth-most abundant chemical element on Earth, after hydrogen, oxygen, and helium [1], [2], and it is considered as a base element for building organic compounds that are essential for life, such as proteins, carbohydrates, nucleic acids, and lipids. This element drives whole communities of living organisms and underpins the biogeochemical cycles on Earth [3].
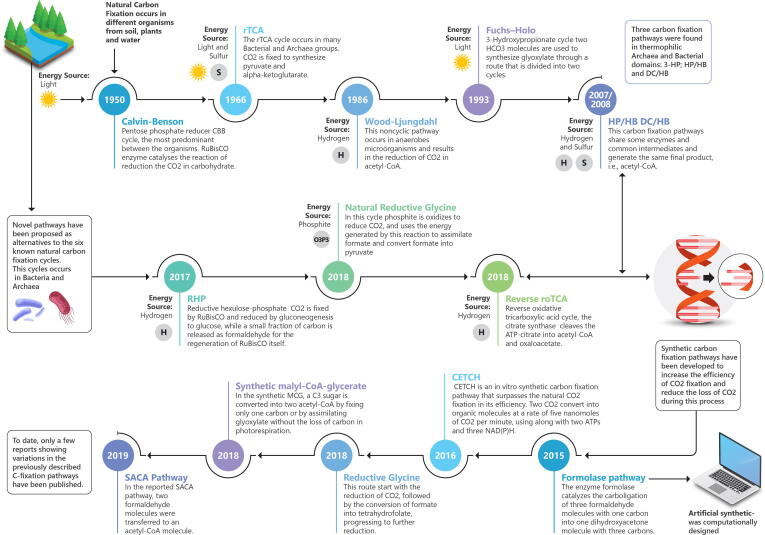
The entry of inorganic carbon into the biosphere is related to carbon fixation. Primary production of organic compounds is directly related to autotrophic carbon fixation, which converts inorganic carbon into biomass [4] (Fig. 1). At present, six natural autotrophic carbon-fixation pathways have been described and accepted, while other candidate routes have been recently proposed. With advances in synthetic biology, bioinformatics, and biochemical analyses, various synthetic carbon-fixation routes have also been developed (see Table 1), including the crotonyl-(CoA)/ethylmalonyl-CoA/hydroxybutyryl-CoA (CETCH; here, CoA represents coenzyme A) cycle, reductive glycine, synthetic malyl-CoA-glycerate pathway, synthetic acetyl-CoA (SACA) cycle, and formalase pathway [5], [6]. A timeline summary showing the key carbon fixation milestones discussed in our review is listed in Fig. 1.
Fig. 1.
A timeline summary showing key milestones and findings related to carbon fixation pathways discussed in our work.
Table 1.
Comparative description of the natural and synthetic carbon fixation pathways: microorganism examples, energy sources, necessary inputs, products of the cycle, reducing agents, main enzymes, and sensitivity of the enzymes to oxygen.
| Pathway | Status | Organisms | Energy Source | Input | Output | Reductants | Key Enzyme | O2 Sensitive | References |
|---|---|---|---|---|---|---|---|---|---|
| Calvin-Benson | Natural | Plants, Algae, Cyanobacteria, Aerobic Proteobacteria, Purple bacteria | Light | 3 CO2, 9 ATP,6 NAD(P)H | Glyceraldehyde-3-phosphate | NAD(P)H | RuBisCO | No | [23] |
| rTCA | Natural | Green sulfur bacteria, Proteobacteria, Aquificae, Nitrospirae | Light and Sulfur | 2 CO2, 2 ATP,4 NAD(P)H | Pyruvate | NAD(P)H and ferredoxin | 2-Oxoglutarate synthase, Isocitrate dehydrogenase | Yes | [54] |
| Wood–Ljungdahl | Natural | Acetogenic, Methanogenic Archaea, Planctomycetes, Sulfate, Archaeoglobales | Hydrogen | 2 CO2, 1 ATP, 4 NAD(P)H | Acetyl-CoA | Ferredoxin | NAD-independent formate dehydrogenase, Acetyl-CoA synthase-CO dehydrogenase | Yes | [9] |
| 3-HP | Natural | Chloroflexaceae | Light | 3 HCO3−, 5 ATP, 5 NAD(P)H | Pyruvate | NAD(P)H | Acetyl-CoA carboxylase, Propionyl-CoA carboxylase |
No |
[72], [187] |
| HP/HB | Natural | Aerobic Sulfolobales | Hydrogen and Sulfur | 2 HCO3−, 4 ATP, 4 NAD(P)H | Acetyl-CoA | NAD(P)H | Acetyl-CoA-Propionyl-CoA carboxylase | No | [78] |
| DC/HB | Natural | Anaerobic Thermoproteale, Desulfurococcales | Hydrogen and Sulfur | 1 CO2, 1 HCO3−, 3 ATP, 4 NAD(P)H | Acetyl-CoA | NAD(P)H and Ferredoxin | Pyruvate synthase, PEP carboxylase | Yes | [79] |
| RHP | Candidate Natural | Methanospirillum hungatei | Hydrogen | CO2, 3 ATP, 2 NAD(P)H | Gluconeogenesis and glycolysis | NAD(P)H | RuBisCO | No | [11] |
| Natural Reductive Glycine | Candidate Natural | Candidatus phosphitivorax anaerolimiDesulfovibrio desulfuricans | Phosphite | CO2, ATP, NAD(P)H |
Formate/ Pyruvate | NAD(P)H and ferredoxin |
CO2-reducing formate dehydrogenase (fdhAB) | – | [92] |
| Reverse oTCA | Candidate Natural | Desulfurella acetivorans | Hydrogen | CO2, ATP, NAD(P)H | Acetyl-CoA | Ferredoxin | Citrate synthase | – | [12] |
| CETCH | Synthetic | Theoretical | – | 2 CO2, 2 ATP, 3 NAD(P)H | Glyoxylate | NAD(P)H | CoA- dependent carboxylase | No | [104] |
| Reductive Glycine | Synthetic | Demonstrated in E. coli as host | – | CO2, NADH | Pyruvate | Ferredoxin | Glycine cleavage system | No | [93] |
| Synthetic malyl-CoA-glycerate | Synthetic | Demonstrated in E. coli and Synechococcus elongatus PCC7942 host | – | CO2, 3 ATP, 3 NADH | Acetyl-CoA | NAD(P)H | PEP-carboxylase, RuBisCO | No | [119] |
| SACA Pathway | Synthetic | Demonstrated in E. coli as host | – | CO2 | Acetyl-CoA | – | NAD-independent formate dehydrogenase | No | [6] |
| Formolase pathway | Synthetic | Theoretical | – | CO2, NADH, ATP | Dihydroxyacetone-phosphate | NADH | NAD-independent formate dehydrogenase | No | [5] |
The Calvin–Benson–Bassham (CBB) cycle is the most well-kwons mechanism of carbon assimilation and is found in all types of plants and algae and some prokaryotes [7]. However, new pathways of carbon fixation have also been elucidated in recent ecological, biochemical, and genomic studies [8]. In addition to the CBB cycle, autotrophic microorganisms can incorporate the carbon in biomass through five other natural carbon-fixation pathways: i) the reductive tricarboxylic acid (rTCA) cycle; ii) the reductive acetyl-CoA (also called the Wood–Ljungdahl [WL]) pathway; iii) the 3-hydroxypropionate [3-HP] bicycle; iv) the 3-hydroxypropionate/4-hydroxybutyrate (3-HP/4-HB) cycle; and v) the dicarboxylate/4-hydroxybutyrate (DC/4-HB) cycle [9], [10]. Three previously unknown natural carbon-fixation routes have also been described recently: i) the reductive hexulose-phosphate pathway (RHP), ii) the natural reductive glycine (nrGly) cycle, and iii) the reverse oxidative TCA cycle (roTCA) [11], [12].
Plants and photoautotrophic microorganisms are capable of fixing inorganic CO2 via the CBB cycle, which uses energy in the form of NAD(P)H and adenosine tri-phosphate (ATP) derived from photons. The global incident solar energy is 178,000 TW y-1, which is severalfold higher than that required by human society [13]. Oxygenic phototrophs, such as cyanobacteria, plants, and algae, have the ability to use this incident solar energy as a free source for photosynthesis-induced energy generation. Due to the release of oxygen in the process, these organisms are considered as the primary producers of the current biosphere [14]. Conversely, anoxygenic photosynthetic bacteria are also able to fix CO2 and grow autotrophically, typically using light as energy (ATP) and without producing oxygen in the process [15].
However, some autotrophic microorganisms are also capable of fixing inorganic carbon using other energy sources [16], [17]. Autotrophic organisms assimilate inorganic carbon into biomass using processes that are driven by light or chemical energy derived from organic molecule breakdown. Chemolithoautotrophic microbes use inorganic compounds (CO, CO2, H2, sulfate, phosphite, and Fe) as their energy sources, whereas chemoorganotrophic microorganisms rely on the energy produced from chemical transformations using organic compounds (sugars, organic acids, and amino acids) as carbon sources [17], [18].
Autotrophic organisms are crucial components of the global carbon cycle since they assimilate the organic carbon that is released by other organisms in the biosphere [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]. The fossil fuel reserves that are essential in the current linear global economy represent carbon stored from the primary autotrophic production of biomass and sequestered in geological deposits. However, the unsustainable rates of global anthropogenic carbon release have now surpassed the ability of natural carbon-fixation pathways to recapture and fix this carbon in the biosphere [19]. Thus, an understanding of the metabolic conversion of inorganic carbon into organic carbon is critical for developing potential alternative solutions for capturing and using these waste carbon sources.
The collective techniques that generate “omics” data on a massive scale are expected to facilitate the discovery of novel carbon-fixation pathways and efficient carbon-fixing enzymes [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. Future trends are focused on manipulating the known autotrophic carbon-fixation pathways to increase their efficiency and to understand their components, as well as the development of new and more efficient natural and synthetic fixation cycles [21], [22]. Moreover, thermophilic microorganisms are also valuable sources of metabolic diversity and are currently being investigated for alternative carbon-fixation pathways, because a chemolithoautotrophic thermophile is probably the most common precursor to life and provides the best model for investigating primordial metabolism [7]. This review summarizes and discusses the six known autotrophic pathways of carbon fixation, the newly identified and proposed natural pathways, as well as recent advances in designing new pathways for biological and synthetic carbon fixation (Fig. 2). We discuss the continued progress of “omics” tools and their role in unveiling new routes of carbon fixation and the efforts devoted to identifying the enzymes that participate in carboxylation reactions. Furthermore, efforts to develop engineered enzymes with activities that surpass those found in natural carbon-fixation cycles are also discussed. Subsequently, we focus on carbon fixation performed by thermophilic Bacteria and Archaea to understand the carbon-fixation pathways and obtain insights for future attempts at synthetic engineering. Finally, we highlight the knowledge gaps in the field as well as the challenges and new directions for future studies on carbon fixation.
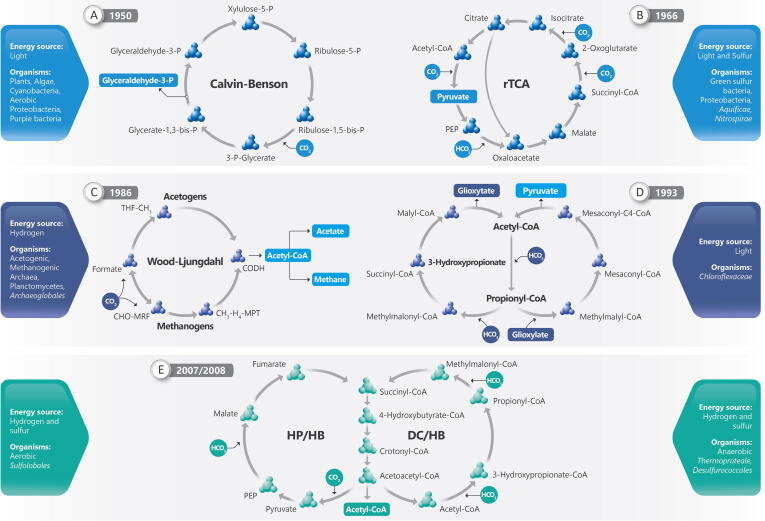
Fig. 2.
Natural carbon fixation. (A) CBB cycle; enzymes: ribulose-1,5-bisphosphate carboxylase/oxygenase, 3-phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, ribulose-phosphate epimerase. (B) rTCA cycle; enzymes: ATP-citrate lyase, malate dehydrogenase, succinyl-CoA synthetase, ferredoxin (Fd)-dependent-2-oxoglutarate synthase, isocitrate dehydrogenase, PEP carboxylase. (C) Wood–Ljungdahl cycle, upstairs acetogens Archaea and downstairs methanogens Archaea; enzymes: MPT-methylene tetrahydromethopterin, MFR-methanofuran, THF, tetrahydrofolate. (D) 3HP cycle; enzymes: acetyl-CoA carboxylase, propionyl-CoA carboxylase, methylmalonyl-CoA epimerase, succinyl-CoA:(S)-malate-CoA transferase, trifunctional (S)-malyl-CoA, -methylmalyl-CoA, mesaconyl-CoA transferase, mesaconyl-C4-CoA hydratase. (E) HP/HP and DC/HP cycle; enzymes: pyruvate synthase, PEP-carboxylase, malate dehydrogenase, fumarate hydratase/reductase, acetyl-CoA/propionyl-CoA carboxylase, 3-hydroxypropionate-CoA ligase/dehydratase, methylmalonyl-CoA mutase, succinyl-CoA reductase, 4-hydroxybutyrate-CoA ligase, crotonyl-CoA hydratase, acetoacetyl-CoA-ketothiolase.
Autotrophic carbon-fixation pathways: The six known cycles
Calvin–Benson–Bassham cycle
The photosynthetic and pentose phosphate reducing pathway (CBB pathway) discovered by Melvin Calvin, Andrew Benson, and James Bassham in the 1940 s and 1950 s [23] was the first reported carbon-fixation pathway. The CBB cycle is present in algae, plants, cyanobacteria, phyla Proteobacteria and Firmicutes, and photoautotrophic and chemoautotrophic bacteria [24], [25]. In microorganisms, this cycle is the most important carbon dioxide fixation pathway [26]. The key enzymes in this pathway include ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and phosphoribulokinase (PRK) [27] (Fig. 2A). Another important enzyme in the CBB cycle is glyceraldehyde-3-phosphate dehydrogenase (GAPDH). GAPDH and PRK participate in the regeneration of RuBisCO and form an inactive complex with the regulatory protein CP12 when in a dark environment [28]. CP12 is formed by 80 amino acids and, after binding to GAPDH and PRK, becomes an important light/dark regulator [29]. In dark, the CP12 protein complex is deactivated, while under light conditions, this complex is dissociated, allowing the reactivation of GAPDH and PRK enzymes [30]. Most oxygenic photosynthetic organisms that use the CBB cycle to fix CO2 contain CP12, whereas nonoxygenic phototrophic bacteria do not have this protein [28], [29], [30]. In the CBB cycle, three molecules of CO2 are fixed by RuBisCO to form one molecule of glyceraldehyde 3-phosphate, in a pathway composed of a total of 29 reactions from 13 enzyme reactions [31]. RuBisCO drives the production of carbohydrates by catalyzing the removal of CO2 from the atmosphere and its conversion into carbohydrates for plants and other organisms [32].
Recently, a novel variant of the CBB cycle was described using computation of elementary flux modes or extreme paths to find all pathways leading to the formation of glyceraldehyde 3-phosphate from CO2 [31]. Named as the S7P-removing transaldolase variant, it involves transaldolase, and it bypasses the intermediate enzyme fructose 1,6-bisphosphate [31]. The author suggested that transaldolase can be an alternative for enhancing photosynthetic carbon metabolism. Previously, this theoretical path was proposed as the S7P-forming transaldolase variant because it requires an S7P-forming step [33], different from the variant described by Ohta [31]. The S7P-forming transaldolase variant works twice per three CO2 molecules fixed, in which transaldolase transfers three carbons from fructose 6-phosphate to erythrose 4-phosphate to make S7P and glyceraldehyde 3-phosphate [33], [34].
RuBisCO catalyzes both the carboxylation and oxygenation of its substrate [35]. CO2 and O2 compete for the same active site on RuBisCO when reacting with the same substrate. The carboxylation results in the formation of two molecules of a three-carbon-containing organic acid, 3-phosphoglycerate (3-PGA), whereas RuBisCO oxygenation produces one molecule of 3-PGA and another of 2-phosphoglycolate (2-PG) using a process called photorespiration [36]. The 3-PGA molecule resulting from RuBisCO carboxylation generates carbohydrates, whereas 2-PG is not metabolized in the CBB cycle and is instead used in respiration, wherein it consumes O2 and releases already fixed CO2 [37].
To increase the concentration of CO2 near RuBisCO and decrease the photorespiration rate, cyanobacteria and some chemoautotrophs contain an intracellular microcompartment–carboxysome–that functioning as an organelle [38]. The carboxysomes encapsulate RuBisCO and the carbonic anhydrases, and operate as semipermeable barriers that allow the passage of HCO3 and RuBisCO and exclude O2 that competes for the substrate. In the carboxysome, the carbonic anhydrases catalyze the conversion of HCO3 into CO2, which is fixed by RuBisCO to form 3-PGA [39]. The efficient CO2 fixation mechanism in the carboxysomes recently inspired their implementation in plants to increase crop productivity [40]. In some eukaryotic algae, carbon fixation is mediated by several mechanisms to enrich the CO2 concentration locally as the pyrenoid, which appears as a dynamic compartment of membranes and crystalline starch under a microscope [41]. A recent study provided the first glimpse of the cycle of a carboxysome inside living cells through a fluorescence imaging platform that allows the simultaneous measurement of the numbers, positions, and activities of carboxysomes in cyanobacteria [42].
Importance of RuBisCO
Four forms of RuBisCO, which has a structure formed by eight large subunits and eight small subunits, are known [43]. The I and II forms participate in the autotrophic assimilation of CO2. RuBisCO form I is more widely distributed, and is found in eukaryotes and some prokaryotes. Additionally, form I RuBisCO is roughly 2.4 million years old and emerged from the great oxygenation event that occurred owing to the transforming action of cyanobacteria, which began to introduce oxygen into the atmosphere driven by the abundant photosynthetic energy provided by the sunlight. Meanwhile, RuBisCO form II occurs in prokaryotes (mostly in Proteobacteria species) and microeukaryotes such as dinoflagellates (i.e., Symbiodinium) [44]; these are the only groups capable of processing RuBisCO form II, which is encoded by the nuclear DNA [45], [46].
Banda et al. [47] discovered an uncharacterized clade sister of RuBisCO form I, i.e., RuBisCO I’. Their study involved the metagenomic analysis of samples from environmental communities with a large amount of nonculturable bacteria. They reported that RuBisCO form I’ may have evolved under anaerobic conditions before the evolution of cyanobacteria. Unlike RuBisCO form I, RuBisCO form I’ consists of only eight large subunits without the small subunits. Twenty-four rbcL genes with gene products that share a high sequence homology (52 %–61 %) with the known RuBisCO form I were reported. However, a more detailed analysis of the genomes assembled from the metagenome indicated the absence of the rbcS genes [47]. These genes are always found in Bacteria and Archaea that use the CBB cycle to fix carbon [48]. On the basis of these results, the group suggested that RuBisCO I’ probably represents a distinct form of RuBisCO that presumably diverged from form I before the origin of life.
According to previously reported studies, form III of RuBisCO does not participate in autotrophic CO2 fixation; instead, it uses ribonucleotides via the pentose-bisphosphate pathway [49]. However, using “omics” and biochemical approaches, Frolov et al. [25] identified an operative form III RuBisCO in a transaldolase variant of the CBB cycle in Thermithiobacillus tepidarius and Thiobacillus thioparus. Form III was found exclusively in Archaea [50], and since then, it has been hypothesized to exist in bacterial candidates [51].
RuBisCO form IV is known to not perform carboxylase and oxygenase activities and is distributed in seven phylogenetically distinct subgroups, namely, IV-Photo (phototrophic bacteria), IV-Nonphoto (nonphototrophic bacteria), IV-AMC (microbial consortia found in acid mines lakes), IV-GOS (ocean sampling sequences), IV-DeepYkr (deep branch close to the YkrW clade), IV-Aful (Archaeoglobus fulgidus), IV-YkrW (Bacillus) [52], and a recently discovered form that acts as an oxygenase that converts 3-keto-d-ribitol-1,5-bisphosphate into two hydroxy acid phosphorylates: 3-d-phosphoglycerate and phosphoglycolate [53].
Reductive tricarboxylic acid cycle (rTCA)
The second carbon-fixation pathway was described in 1966 by Evans and collaborators [54]. This route was revealed in Chlorobium limicola, a green sulfur bacterium. Experiments with fermentative bacteria revealed a reaction that challenged the concepts of carbon fixation. Ferredoxin was found to serve directly as an electron donor for carbon fixation for the synthesis of pyruvate and alpha-ketoglutarate from CO2 and reduced ferredoxin and acetyl-CoA or succinyl-CoA, respectively, through an irreversible pathway [55].
This route was named rTCA or the Arnon–Buchanan cycle and is believed to be the most plausible candidate for the first autotrophic metabolism [56]. In the rTCA cycle, two cofactors mediate the carbon fixation: thiamine pyrophosphate, which converts acetyl-CoA to pyruvate, and succinyl-CoA to produce ketoglutarate [57]. This cycle behaves in a direction opposite to that of the oxidative citric acid cycle (oTCA; one of the stages of cellular respiration in heterotrophs, which is performed in the presence of O2 and releases fixed carbon) and forms acetyl-CoA from two CO2 molecules.
Some key enzymes of the Krebs cycle are considered to react irreversibly and are replaced in the rTAC cycle to reverse the cycle; for example, succinate dehydrogenase is substituted by fumarate reductase; NAD 2-oxoglutarate dehydrogenase is replaced by ferredoxin 2-oxoglutarate synthase; and citrate synthase is replaced by ATP-citrate lyase [54], [58], [59]. The main product of carbon fixation in the rTCA cycle is acetyl-CoA, which is later converted into other central intermediates of the carbon metabolism, including pyruvate/phosphoenolpyruvate (PEP), oxaloacetate, and 2-oxoglutarate [56]. Acetyl-CoA is carboxylated into pyruvate by ferredoxin-dependent pyruvate synthase, and pyruvate can also be converted into PEP; oxaloacetate is synthesized in the reactions of pyruvate or PEP carboxylase [54] (Fig. 2B).
The rTCA cycle occurs in many Bacteria and Archaea groups and includes enzymes that are sensitive to oxygen. For this reason, this cycle is observed in anaerobes or facultative anaerobes that can tolerate O2 concentrations lower than that found in the air [60]. Although discovered in photosynthetic green sulfur bacteria, the rTCA cycle has been shown to be present in plenty of chemoautotrophic microbes [55]. More recently, it was assumed that the complete TCA cycle can occur in Pandoravirus massiliensis, which showed genes with low similarity to all enzymes of the cellular TCA cycle and, most importantly, a functional isocitrate dehydrogenase, a key enzyme of this cycle [61]. The gene ORF132 that encodes isocitrate dehydrogenase was cloned and expressed in Escherichia coli, and was shown to encode an isocitrate dehydrogenase. The authors suggested the possible existence of an autonomous TCA pathway or another unknown metabolic pathway that may be involved in redirecting amoeba carbon metabolism where this virus was replicated [61].
Reductive acetyl-CoA pathway
In 1986, Ljungdahl and collaborators [9] described the third carbon-fixation route, i.e., the reductive acetyl-CoA or WL pathway. This noncyclic pathway occurs in anaerobic microorganisms and results in the reduction of CO2 in acetyl-CoA. According to the reported studies, these microbes may have been the first autotrophs to use inorganic compounds such as CO and H2 as energy sources and CO2 as an electron acceptor one billion years before the appearance of O2 [62].
In this cycle, two CO2 molecules are converted into acetyl-CoA through two different branches that operate in parallel: the eastern or methyl branch and the western or carbonyl branch [63]. In the eastern branch, one CO2 molecule undergoes six-electron reduction to form a methyl group, whereas the western branch involves the reduction of the other CO2 molecule to CO, which is condensed with the methyl group and CoA to form acetyl-CoA [64]. Acetyl-CoA is then incorporated into cellular carbon or converted to acetylphosphate, from which the phosphoryl group is transferred to adenosine diphosphate (ADP) to generate ATP and acetate, which are the main growth products of acetogenic bacteria [9], [65], [66].
The eastern branch begins with the reduction of CO2 in formate catalyzed by the formate dehydrogenase enzyme, which undergoes ATP-dependent condensation with tetrahydrofolate (H4F) to form 10-formyl-H4folate, which is converted to 5,10-methylenyl-H4folate. The next step is the reduction of 5,10-methylene-H4folate to 5-CH3-H4folate [63]. At the end of the branch, the methyltetrahydrofolate (corrinoid/iron–sulfur protein) methyltransferase enzyme catalyzes the relocation of the methyl group on N5 of (6S)–CH3-H4folate to the cobalt center of the corrinoid iron-sulfur protein (CfeSP). In contrast, the western branch involves one enzyme, i.e., the CO-dehydrogenase/acetyl-CoA synthase, which catalyzes the conversion of CO2 into CO [67].
When organisms are cultivated with CO, CO dehydrogenase generates CO2, which is converted into formate in the eastern branch, and CO is incorporated directly into the carbonyl group of acetyl-CoA. In the next step, the acetyl-CoA synthase catalyzes the condensation of CO, CoA, and the methyl group of a methylated CfeSP to generate acetyl-CoA; at this point, the western branch meets the eastern branch.
In the Archaea Methanosarcina acetivorans, the conversion of methanogen to acetogen has been shown to completely dispense with the conserved methanogenesis energy [68]. Methanosarcina acetivorans uses the reducing acetyl-CoA to conserve energy in the form of acetate [69]. The genes encoding Mtr, i.e., disruption from the methyl branch toward methane, were eliminated in M. acetivorans, and all carbon from the methanogenesis substrate was diverted to flow through acetyl-CoA [68]. Thus, M. acetivorans could be converted into an acetogenic organism, which suggests that methanogenesis may have evolved from the acetyl-CoA pathway [68].
The reductive acetyl-CoA route is found in prokaryotes that live at thermodynamic limits such as the acetogenic and methanogenic Archaea [23], which use this route not only for the reduction of CO2 in acetyl-CoA but also for the conservation of energy by generating an electrochemical gradient [70]. In the energy-conservation process occurring during autotrophic growth or in the fermentation process, the acetogens and methanogens generate acetate and methane, respectively [71] (Fig. 2C).
3-Hydroxypropionate bicycle
The 3-hydroxypropionate bicycle (or the Fuchs–Holo route) was found in Chloroflexus aurantiacus, which is a green sulfur thermophilic bacterium, by Helge Holo and Georg Fuchs in 1989 [58], [72]. In this pathway, two HCO3 molecules are used to synthesize glyoxylate through a route that is divided into two cycles. In the first cycle, glyoxylate is formed from the carboxylation of acetyl-CoA to produce malonyl-CoA, which is reduced to proprionyl-CoA via hydroxypropionate, after which the carboxylation of proprionyl-CoA produces succinyl-CoA. Subsequently, succinyl-CoA is converted to (S)-malyl-CoA, regenerating acetyl-CoA (starting molecule) and releasing glyoxylate as the first product of the carbon fixation. The second cycle begins with the assimilation of glyoxylate and its conversion to methylmalyl-CoA, which in turn is converted to form citramailyl-CoA by mesaconyl-CoA. Finally, the product citramailyl-CoA is converted into pyruvate and acetyl-CoA, thereby regenerating the precursors of the cycle [73] (Fig. 2D).
This pathway involves 19 steps and 13 enzymes; none of the steps are sensitive to oxygen and all the enzymes work under aerobic conditions [74]. This fourth carbon-fixation pathway appears to be exclusively restricted to a single clade of photosynthetic Chloroflexus [75]. This green sulfur bacterium uses the 3-hydroxypropionate bicycle in combination with the glyoxylate cycle to channel organic substrates such as glycolate, acetate, propionate, 3-hydroxypropionate, lactate, butyrate, or succinate for its central carbon metabolism under both autotrophic and heterotrophic growth conditions [76].
Hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle and dicarboxylate/4-hydroxybutyrate (DC/HB) cycle
The fifth and sixth carbon-fixation pathways share some enzymes and common intermediates and generate the same final product, i.e., acetyl-CoA [77] (Fig. 2E). The fifth carbon-fixation pathway—the 3-hydroxypropionate/4-hydroxybutyrate cycle (HP/HB) was described in Metallosphaera sedula, which is a thermophilic Archaea belonging to the Sulfolobales order [78]. The sixth pathway (dicarboxylate/4-hydroxybutyrate [DC/HB]) was found in Ignicoccus hospitalis, which is an Archaea belonging to the order Desulfurococcales [79]. Existing variants of both cycles, the orders Sulfolobales and Desulfurococcales, did not evolve from a common ancestor, and the representatives of these two orders have phylogenetically unrelated enzymes [80].
In these two cycles, two CO2 molecules are added to acetyl-CoA to produce succinyl-CoA and rearrange into acetoacetyl-CoA and cleave into two acetyl-CoA molecules [81]. In the HP/HB cycle, an acetyl-CoA and the propionyl-CoA carboxylase, fixes two HCO3 molecules, and in the DC/HB cycle, the pyruvate synthase and PEP carboxylase catalyze this reaction. These pathways vary mainly in relation to oxygen tolerance and the use of reduction cofactors: NAD(P)H is used in the HP/HB cycle, and ferredoxin/NAD(P)H is employed in the DC/HB cycle [23], [82]. The enzymes in the HP/HB cycle are oxygen-tolerant, whereas the 4-hydroxybutyryl-CoA dehydratase enzyme in the DC/HB cycle is inactivated in the presence of oxygen. Although the two cycles produce acetyl-CoA, their main difference lies in their link to the carbon metabolism. In the DC/HB pathway, pyruvate is synthesized from acetyl-CoA by using a pyruvate synthase, and in the HP/HB pathway, another half turn of the cycle is needed to produce succinyl-CoA, which is oxidized via succinate to produce pyruvate [83].
This cycle is also found in Nitrosopumilus maritimus, belonging to the order Sulfolobales. This species heterologously produces the protein Nmar_1028 that catalyzes the conversion of succinyl-CoA into two acetyl-CoA molecules. Nmar_1028 is homologous to the dehydrogenase domain of crotonyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase and seems to be the only (S)-3-hydroxybutyryl-CoA dehydrogenase in N. maritimus and is thus essential for the functioning of the 3-hydroxypropionate/4-hydroxybutyrate cycle [84]. In Metallosphaera sedula, Msed_2001 catalyzes the same reaction as Nmar_1028, and these two enzymes are homologous and have evolved independently from their respective bacterial homologs [15]. The existence of the HP/HB cycle in two distantly related archaeal groups with different physiologies is not evident in the course of evolution of autotrophic CO2 fixation, because this process would need to be adapted to the corresponding ecological niche [15].
New candidate pathways for natural carbon fixation
Recent studies have revealed that carbon fixation is not limited to these six cycles (Tab. 1). Genome analyses have shown that several autotrophs do not present genes from any of the already described autotrophic CO2-fixation pathways [23]. Therefore, investigations performed in this direction are of utmost importance for establishing novel ways of fixing inorganic carbon and exploring alternative routes for metabolic energy supply [85].
RHP pathway
To date, three novel pathways have been proposed as alternatives to the six known natural carbon-fixation routes. Kono et al. [11] fully described the RHP pathway in the methanogenic Archaea, Methanospirillum hungatei. The carbon metabolism has been shown to involve the enzymes RuBisCO and phosphoribulokinase (PRK), which are the same enzymes found in the CBB cycle. RuBisCO of M. hungatei seems to form a new clade with RuBisCO of another methanogenic Archaea that also has PRK; this clade is different from RuBisCO form III. The coexistence of the RuBisCO and PRK in this Archaea led the authors to propose that M. hungatei fixed carbon. However, the enzymes transketolase, ribulose-5-phosphate and sedoheptulose-1,7-bisphosphatase, which participate in the CBB cycle, are absent from the genome. Archaea are known to lack genes for a transketolase, which is essential in the CBB cycle [86].
The proposed RHP pathway is noncyclical and shares the majority of the reactions of the CBB cycle, except for those in the regeneration stage. In the RHP cycle, CO2 is fixed by RuBisCO and reduced by gluconeogenesis to glucose, while a small fraction of carbon is released as formaldehyde for the regeneration of RuBisCO itself [11]. Because the RHP cycle differs from the CBB cycle in some steps, the reported results provide insights into the evolutionary and functional associations between carbon metabolism and the pathways involving the RuBisCO enzyme present in photosynthetic organisms and methanogenic Archaea. The authors speculate that the CBB cycle and RHP originated from a primitive carbon metabolic pathway that used RuBisCO, but with the replacement of a few steps and without carbon release. Although the energy requirements of the RHP pathway are significantly lower than those of the CBB cycle in plants and cyanobacteria, it does not proliferate as in the case of the CBB.
Natural reductive glycine (nrGly) pathway
Next-generation sequencing can provide a holistic approach to understand metabolisms through multiple “omics” datasets [87], [88]. The data generated by genomics, proteomics, transcriptomics, and metabolomics have significantly increased the understanding of cell physiology and provided answers to important questions about the metabolisms of microorganisms [88], [89]. Studies on the (meta)genome and (meta)transcriptome have also made it possible to identify the key genes involved in carbon fixation, allowing the prediction of potential carbon-fixation routes in (unculturable) environmental microbes as well as the discovery and design of new pathways for carbon fixation [90], [91].
Figueroa et al. [92] proposed another candidate carbon-fixation route, nrGly, using 16S rRNA region sequencing. The group proposed that inorganic carbon is assimilated into the glycine-reducing pathway found in the uncultivated bacterium Candidatus Phosphitivorax anaerolimi strain Phox-21, belonging to the class Deltaproteobacteria. This pathway was previously described as a synthetic route for carbon fixation [93]. In this proposed route, Phox-21 oxidizes phosphite to reduce CO2, and uses the generated energy to assimilate formate through the glycine-reducing pathway. Due to the lack of alternative electron acceptors, Phox-21 has been suggested to couple phosphite oxidation to CO2 reduction; in addition to the presence of CO2 reductase and formate dehydrogenase (FdhAB) in its genome, this assumption could explain the mechanism by which this metabolism occurs [92]. The bacteria reduce CO2 to formate by using FdhAB and convert formate into pyruvate via the glycine-reducing pathway. All the necessary precursors (acetyl-CoA, oxaloacetate, 2-oxoglutarate, and succinyl-CoA) can subsequently be generated from the pyruvate route in the partial rTCA cycle. The results of Phox-21 genomic analysis suggest that it may be possible for microorganisms to exploit the energy derived from phosphite oxidation, enabling autotrophic growth through the glycine-reducing pathway with CO2 as the only electron acceptor [92].
Because of its very low redox potential, phosphite might be the only biological electron donor that can drive the fixation of CO2 in chemotrophs through the reductive glycine pathway (an ATP-consuming network) in the absence of an additional energy source or electron acceptor. Furthermore, according to other reported studies, phosphite oxidation may be coupled with CO2 reduction through the WL pathway [94]. Although genomic analysis has indicated a new carbon-fixation pathway, experimental evidence is required to verify the validity of this hypothesis.
Sánchez-Andrea et al. [95] described the reductive glycine pathway in Desulfovibrio desulfuricans. In addition to performing genomic, transcriptomic, proteomic, and metabolomic analyses, they also performed in vivo evaluation of autotrophic growth of bacteria. Unlike C. Phosphitivorax anaerolimi, the results of the study on D. desulfuricans demonstrated the enrichment of the culture medium with an organic carbon compound (cysteine and ruminal fluid) via autotrophic growth. The studies performed with labeled CO2 and functional “omics” experiments proved the fixation of carbon through the reductive glycine pathway. The results of this study demonstrate that the reductive glycine pathway is the only carbon-fixation pathway that allows autotrophic growth of D. desulfuricans. In the reductive glycine pathway, firstly, CO2 is first reduced to formate, which is further reduced and condensed with a second CO2 molecule to generate glycine. The resulting product is then reduced by the glycine reductase to acetyl-P and then to acetyl-CoA, which is further condensed with another CO2 molecule to form pyruvate [95]. Thus far, it seems that this is the first reported study to prove the operation of the entire reductive glycine pathway in a microorganism.
Recently, Hao et al. [96] identified two C. Phosphitivorax strains (F81 and R76) as butyrate oxidizers. Transcriptome analyses were performed, and the entire ptx-ptd gene cluster for phosphite oxidation was reconstructed in strain F81; interestingly, this cluster was not found in strain R76. Genes involved in the glycine-reduction pathway were found in both the strains, whereas the essential genes for other autotrophic CO2 pathways were absent. However, according to the authors, although genes for the glycine-reduction pathway were observed, they were not expressed. This observation indicates that these microorganisms, unlike the Phox-21 strain that was externally enriched with CO2 and phosphate, live as heterotrophic syntrophs besides autotrophs. Other studies demonstrated that the glycine-cleavage system can support glycine and serine biosynthesis from formate in an engineered E. coli strain at increased CO2 concentration [97], [98]. Nonetheless, it is necessary to verify the growth of the bacterium on formate (and also CO2), which remains an unresolved challenge [99].
Reverse oxidative tricarboxylic acid cycle (roTCA)
Although metagenomic analyses have gained considerable acceptance in the studies of organisms and microbial communities, researchers have reported the need for joint efforts involving both “omics” and biochemistry [12]. In studying Desulfurella acetivorans, which is a sulfur-reducing and thermophilic Deltaproteobacteria (optimal growth at 52 °C to 57 °C), the authors elucidated a new version of the rTCA cycle, which was named the reverse oTCA cycle (roTCA) (this report also mentions the oTCA cycle that provides energy for aerobic organisms and has citrate synthase as the key enzyme). The genome analysis of D. acetivorans revealed the absence of key genes and enzymes involved in the known pathways of carbon fixation normally found in organisms exhibiting autotrophic growth. On the basis of this information, the authors investigated D. acetivorans in detail using classical biochemical techniques. D. acetivorans can grow heterotrophically using acetate as an electron donor and carbon source [100] or autotrophically with H2 [101]. As previously described, the key enzyme in the rTCA cycle is the ATP-dependent citrate lyase enzyme; however, D. acetivorans do not have the genes that encode that enzyme.
In the roTCA pathway, citrate synthase is the key enzyme that cleaves ATP-citrate into acetyl-CoA and oxaloacetate. Using ultra-performance liquid chromatography, the cleavage of citrate into acetyl-CoA and oxaloacetate was observed in bacterial cell extracts, with the latter being further reduced to malate by malate dehydrogenase. The only enzyme in D. acetivorans that could possibly catalyze this ATP-independent cleavage is citrate synthase. In the cell extracts of D. acetivorans, fumarate reductase activity, which is another enzyme characteristic of the rTCA cycle, was also detected, and the regulation of carbon flow was proposed to occur as follows: the presence of H2 changes the flow toward the roTCA cycle or drives the exogenous acetate from the TCA cycle in the oxidative direction [12].
Zhang et al. [102] also identified the roTCA pathway in Geobacter sulfurreducens. Until then, it was believed that this group of bacteria was unable to fix carbon. However, the identification of the roTCA cycle in this bacterium suggested that it may grow chemolithoautotrophically. The authors used genomic analysis to investigate whether G. sulfurreducens could fix inorganic carbon, and serially transferred the strain to a chemolithoautotrophic culture medium containing formate as an electron donor and carbon source, as well as iron as an electron acceptor. Furthermore, enzymatic assays also corroborated the carbon fixation, showing that citrate synthase can achieve citrate cleavage, which is necessary for the function of the roTCA cycle. These findings showed that G. sulfurreducens can fix CO2 through the roTCA cycle after adaptation; this previously unexplored metabolic pathway can be used for biotechnology and may elucidate previously unclear ecological functions for Geobacter [102].
Development of synthetic pathways for carbon fixation
The global energy crisis and greenhouse effect have impeded the sustainable development of society [4]. Thus, reducing the emission of CO2, which causes the greenhouse effect, is an urgent task. However, increasingly efficient carbon-fixation processes are also required because the rate of natural carbon fixation is inadequate to balance the CO2 released from industries. Carbon fixation performed by photosynthetic organisms allows the recycling of greenhouse gas emissions into high value-added products. However, the dependence on light to drive carbon fixation can be limiting for industrial chemical synthesis [103]. Although natural carbon-fixing processes are not feasible for use in industrial production, they offer a variety of carbon-fixing enzymes and corresponding pathways, opening options for artificial engineering [4]. Thus, in recent years, synthetic carbon-fixation pathways have been developed to increase the efficiency of CO2 fixation and reduce the loss of CO2 during this process.
Crotonyl-(CoA)/ethylmalonyl-CoA/hydroxybutyryl-CoA (CETCH) cycle
The CBB cycle is the most prevalent CO2 assimilation mechanism in the biosphere and is found in all plants, algae, and some prokaryotes. However, its efficiency is limited for natural carbon fixation, rendering the productivity of this path extremely low despite its widespread ecological success owing to the availability of free energy through sunlight.
Schwander et al. [104] described an in vitro synthetic carbon-fixation pathway called the CETCH cycle, which surpasses natural CO2 fixation in its efficiency. This pathway involves 17 enzymes that convert CO2 into organic molecules at a rate of five nanomoles of CO2 per minute, using two CO2 molecules along with two ATPs and three NAD(P)H cofactors, which are the same energy carriers as for the oxygenic photoautotrophs.
The CETCH cycle was realized through a combinatorial approach involving enzyme engineering and metabolic review that was described as a radical and reductionist approach. This CO2-fixation cycle was assembled from its main components in an ascending manner. First, a comparison was made between the kinetic and biochemical properties of all the known classes of carboxylases, and based on these analyses, the enzymes CoA-dependent carboxylases and enoyl-CoA carboxylases/reductases (ECRs) were chosen. ECRs are found in Alphaproteobacteria and Streptomyces, whose carboxylation activities surpass that of the propionyl-CoA carboxylase involved in the 3-HP bicycle, HP-HB cycle, and RuBisCO carboxylation [105], [106]. In comparison with other carboxylases, ECRs work with a broad spectrum of substrates, are insensitive to oxygen, do not accept oxygen as a substrate, and catalyze CO2 fixation with a high catalytic efficiency [107]. The CETCH pathway converts two CO2 molecules from proprionyl-CoA to form a glyoxylate molecule through 13 main reactions catalyzed by 17 enzymes belonging to nine different organisms from four domains of life, including plants, humans, and microbes (Bacteria and Archaea), thereby indicating the potential for natural diversity.
The CETCH cycle shares four reactions and five intermediates with the HP-HB cycle [108]. At the time of writing this manuscript, the CETCH cycle was not implemented in a living host (neither a heterotrophic host, nor a photoautotrophic host). Indeed, balancing the expression of 17 separate enzymes in a heterologous system is challenging even for the most genetically tractable organisms. Thus, the development of artificial carbon-fixation pathways may be limited by the gap between theoretical predictions and their experimental realization in synthetic biology. Moreover, attempts to synthesize new metabolic pathways in living organisms are challenging because of the limited understanding of the interactions between enzymes in heterologous systems [104]. Although the CETCH cycle presents attractive features for implementation in vivo, its synthetic engineering in microorganisms remains undemonstrated [109].
Synthetic reductive glycine pathway (rGlyP).
Carbon fixation can be implemented through assimilation routes based on natural or synthetic formats [110]. In recent synthetic biology studies, a formate-assimilation pathway, called the rGlyP pathway, that supports carbon fixation if coupled with CO2 reduction has been demonstrated [93], [110], [111]. The rGlyP cycle is structurally similar to the most efficient carbon-fixation cycle known, i.e., the WL route. Both routes start with the reduction of CO2, followed by the conversion of formate into tetrahydrofolate, and progressing to further reduction. This cycle uses an enzymatic complex to assimilate carbon and generate CO2 compounds and convert them to pyruvate by condensation with another unit of carbon [110]. Formate is the product of CO2 reduction by an electron pair and is considered to be the simplest organic compound that can supply cells with carbon and reducing power [110]. An efficient carbon-fixation pathway is one that combines CO2 reduction with carboxylation, as shown by the rGlyP cycle [93]. Thus, the synthetic rGlyP pathway represents an efficient route because of the combination of ATP-free CO2 reduction, carboxylation, and versatility.
The rGlyP route seems be a promising pathway for the assimilation of formate and other sustainable C1-feedstocks and could be applicable in future biotechnology-based attempts [112]. Modular engineering was used to implement the rGlyP for supporting synthetic formatotrophic growth as well as for growth on methanol in Escherichia coli [99]. The rGlyP was divided into four metabolic modules: 1) the C1 module (C1M), which consisted of the enzymes formate-THF ligase, methenyl-THF cyclohydrolase, and methylene-THF dehydrogenase from Methylobacterium extorquens that together converted formate into methylene-THF; 2) the C2 module (C2M), which consisted of endogenous enzymes of the GCS (GcvT, GcvH and GcvP) that converted methylene-THF with CO2 and ammonia to form glycine; 3) the C3 module (C3M), which utilized serine hydroxymethyltransferase and serine deaminase that condenses glycine and methylene-THF to generate serine and pyruvate; and 4) the energy module, which used formate dehydrogenase from Pseudomonas sp. [99]. This approach was used to redesign the central carbon metabolism of the model in E. coli that supported the growth of the bacteria with one carbon using rGlyP, and the growth in methanol and CO2 was achieved by the additional expression of a methanol dehydrogenase. Biologically adapted microbial growth using formate and methanol has been investigated by the synthetic biology community in recent years [113]. Thus, this synthetic route appears to be finally becoming possible.
Formaldehyde fixation 1 - the formolase pathway
Even though formaldehyde is considered an organic carbon, sections 4.3 and 4.4 were included to discuss the complementary synthesis mechanisms of carbon fixation. To shorten the process of using carbon and accelerate growth, an artificial synthetic route the formolase pathway—was computationally designed [17], and the computationally designed enzyme, called formolase, performs a reaction that catalyzes carboligation by directly fixing the units of one carbon in units of three carbons that feed the central metabolism. By integrating formolase with various naturally occurring enzymes, a new pathway for carbon fixation has been created that assimilates units of a carbon via formate. The enzyme formolase catalyzes the carboligation of three formaldehyde molecules with one carbon into one dihydroxyacetone molecule with three carbons [5].
Formaldehyde fixation 2 - synthetic acetyl-CoA (SACA)
The efficiency of carbon fixation can also be improved during the synthesis of acetyl-CoA, which is one of the central precursors involved in the biosynthesis of various products [114]. Recently, a synthetic route for acetyl-CoA (SACA) was designed using a glycolaldehyde synthase and an acetyl phosphate synthase and was introduced in E. coli [6]. In the reported SACA pathway, two formaldehyde molecules were transferred to an acetyl-CoA molecule in just three steps by using two ATP and two NADH molecules. First, the formaldehyde was condensed into glycolaldehyde by an glycolaldehyde synthase. Subsequently, the glycolaldehyde was converted to acetyl phosphate by an acetyl phosphate synthase. The glycolaldehyde synthase and acetyl phosphate synthase were selected and designed to increase their catalytic efficiency with their new substrates. At the end of the cycle, the acetyl phosphate group was replaced by CoA in a process catalyzed by a phosphate acetyltransferase, and both in vitro and in vivo production of acetyl-CoA was achieved. The pathway exhibited high carbon-fixation capabilities [115], [116]. The SACA pathway works with five reaction steps and enables acetyl-CoA production from formaldehyde; however, employing this route in E. coli can be challenging due to the low enzyme activity of glycolaldehyde synthase engineered, but glycolaldehyde synthase seems to show improved affinity to substrate formaldehyde [117], [118]. Another question is whether the SACA pathway requires high intracellular concentration of formaldehyde, which can be toxic for the cells [118].
To overcome the deficiency of the CBB cycle for the efficient synthesis of acetyl-CoA, Yu and collaborators [119] designed the synthetic malyl-CoA-glycerate (MCG) pathway. The CBB cycle has not evolved for optimal production of acetyl-CoA; when 3-phosphoglycerate, which is the product of the CBB cycle, is converted to acetyl-CoA, one fixed carbon is lost as CO2 [119]. In the synthetic MCG, a C3 sugar is converted into two acetyl-CoA by fixing only one carbon or by assimilating glyoxylate without the loss of carbon in photorespiration. In this synthetic carbon-fixation pathway, two oxaloacetate molecules are reduced to malate, which is then converted to malyl-CoA and later splits into two acetyl-CoAs and two glyoxylates. These two acetyl-CoAs are the final products of the pathway, and the two glyoxylates are used to regenerate PEP. The MCG pathway converts a PEP and HCO3 into an acetyl-CoA using three ATPs and three NADHs. The functionality of the MCG pathway was first demonstrated in vitro and in E. coli. Later, the pathway was implemented in the photosynthetic strain Synechococcus elongatus PCC7942, which showed an increase in the intracellular pool of acetyl-CoA as well as an improvement in the assimilation of HCO3 [119]. Recently, a system of CO2 fixation oxygen insensitive, self-replenishing with opto-sensing was demonstrated through the junction of the MCG pathway and synthetic reductive glyoxylate and pyruvate synthesis in vitro. This system produced acetyl-CoA, pyruvate, and malate from CO2, and its self-replenishing feature allowed every intermediate of system to be produced from CO2 [120].
Engineering RuBisCO for enhanced CO2 fixation
The low efficiency of carbon fixation in autotrophs, limited by the enzyme RuBisCO, is a biotechnological shortcoming that has led to numerous studies focused on genetic engineering with different strategies to optimize the carboxylase function of this enzyme. RuBisCO is considered the most abundant enzyme on Earth, consituting 50 % of the total soluble protein in the chloroplasts of a C3 plant or in bacteria that use this cycle [121], [122]. In addition, RuBisCO also catalyzes a competitive reaction with oxygen and initiates the photorespiration process, which leads to a loss of fixed carbon and involves significant consumption of ATP to fix CO2. However, in comparison with other enzymes in the CBB cycle, RuBisCO has a low turnover rate, implying that large amounts of this enzyme are needed to sustain the efficiency of CO2 assimilation [123].
Improving RuBisCO by using bioengineering is a complex challenge due to two key aspects: 1) identification of the structural changes that promote performance and 2) identification of the ways to efficiently transplant these changes into RuBisCO within a target organism. Additionally, this task requires a satisfactory understanding of the regulatory pathways of the chloroplast gene, as well as the complex nature of catalysis and biogenesis promoted by this enzyme [32], [124].
Despite the challenges in production, substantial progress has been made in bioengineering RuBisCO. In a previous study, the construction of a mutant RuBisCO was adopted as a strategy to improve the rate of carbon fixation in plants. Substitutions were made in the small subunit of the RuBisCO enzyme, thereby increasing the carboxylation activity by 85 % and enhancing the catalytic efficiency toward CO2 by 45 % [125]. According to that study, the RuBisCO mutant could still be transplanted into higher plants if they shared the same hexadecameric L8S8 structure.
Another strategy already used to improve RuBisCO performance is the replacement of native RuBisCO by an exogenous homolog. Lin et al. [126] worked on replacing the RuBisCO of tobacco plants with the functional RuBisCO of the cyanobacterium Synechococcus elongatus PCC7942 (Se7942). The native gene encoding the large RuBisCO subunit was knocked out, and the genes for the large and small subunits of Se7942 corresponding to a chaperone, RbcX, or an internal carboxysomal protein were inserted. The transformed plants were photosynthetically competent and supported autotrophic growth, and the respective forms of RuBisCO exhibited greater rates of CO2 fixation per unit of enzyme than the control plants [126].
The discovery of the new RuBisCO assembly factors (C3 and C4 plants) has provided next steps for improving this enzyme for higher plants [127], [128]. For example, improving CO2 assimilation now includes equipping C3 plants with a CO2 concentration mechanism and generating alternative metabolic pathways to bypass the oxygenation [129], [130].
Gene editing tools, such as chemical and physical mutagenic agents, that operate on nuclear DNA can be used to modify the family of multiple rbcS genes or insert new rbcS copies and are commonly applicable on various cultures [131]. Because the transformation of the chloroplast is viable in only a few species, modifying the rbcL gene in the chloroplast genome is excessively complicated [132]. Thus, the implementation of RuBisCO in practical applications has necessitated the establishment of chloroplast transformation in several varieties of plants [133].
Mixotrophic-mediated inorganic carbon uptake enhancement
Many photosynthetic eukaryotic microorganisms are capable of consuming organic carbon sources in addition to CO2. Higher plants arose from the first photosynthetic flagellated protists, which acquired a cyanobacterial endosymbiont that led to the chloroplast in photosynthetic eukaryotes. As these organisms are found in every environment, they possess the ability to consume organic carbon in the absence of light and CO2, exhibiting an advantageous metabolic capacity. Within the green algae, the genus of Chlorella is ubiquitous around the world, and C. vulgaris has been generally regarded to show a safe status as an alternative protein-rich biomass, nutraceutical, and source of bio-compounds.
This organism can be fermented like yeasts on simple sugars as a carbon source [134], [135]. Chlamydomonas is another genus of green algae, known for only being able to metabolize organic acetic acid via the glyoxylate cycle [136], whereas in the red algae, the extremophile Galdieria sulphuraria is known for its ability to metabolize over 50 different carbon sources [137]. The consumption of glucose in algae has been observed to correlate with a reduction in photosynthetic activity, in which the heterotrophic growth is favored over the autotrophic growth [138]. However, glycerol addition to mixotrophic cultures of the diatom Phaeodactylum tricornutum or G. sulpheraria increases their overall growth rates, resulting in higher growth rates in the presence of light and CO2. Chlamydomonas reinhardtii demonstrates marked improvements in growth behavior in photo-mixotrophic conditions of high-CO2 cultures fed with acetic acid [139], [140]. Each of these organic carbon sources is metabolized through a different pathway and clearly stimulates different cellular responses. For example, although acetic acid mixotrophy reduces the chlorophyll content in exponential-phase C. reinhardtii cultures, the overall photosynthetic rates and biomass formation are improved [140]. We postulate that large amounts of organic carbon precursors can increase the amounts of CBB intermediates and, concomitantly, improve their ability to sequester CO2 [140].
Algal cultivation is generally aimed at maximizing biomass production for a range of different applications, and now, with the advent of synthetic biological techniques, the engineering possibilities in these hosts have increased [141], [142]. Examples of engineered algae as sources of polyamines, isoprenoids, and recombinant proteins have been demonstrated using mixotrophic cultivation strategies [143], [144], [145]. Algae offer a unique platform for waste carbon conversion, and the study of mixotrophic carbon uptake enhancement can increase the yield of CO2 and provide desired products via waste-stream conversions.
Carbon fixation in thermophilic microorganisms
Thermophiles are microorganisms that can survive above the typical thermal limits of life, in inhospitable environments [146]. Although the temperature range for these microbes varies between 45 °C and 80 °C, the hyperthermophiles can survive even at temperatures above 80 °C [147], [148]. They are found in volcanic areas, hydrothermal vents, solfatara fields, soils heated by steam, geothermal plants, and deserts [20]. Archaea, Bacteria, and Eukarya are the three domains of life in which extremophiles are found [149]. These microorganisms have genetic and physiological complexes that assist them in tolerating the environmental changes and adapting and repairing the damage caused by extreme environmental disturbances [150], [151].
Thermophiles have attracted interest for biotechnological applications requiring whole cells, pure cultures, or consortia, and for applications wherein their macromolecules, metabolites and extreme enzymes are used [152], [153]. Proteins and cell membranes with thermostable capabilities are not denatured at high temperatures; some can also resist proteolysis, making them highly desirable for industrial applications [20], [154]. The discovery of the polymerase enzyme in the thermophiles Thermus aquaticus and Pyrococcus furiosus significantly influenced and revolutionized the use of the polymerase chain reaction owing to its stability and reasonable cost [62], [155], [156].
Life is believed to have originated in a hydrothermal environment, and chemoevolution began in volcanoes through a transition metal-catalyzed autocatalytic carbon-fixation cycle [77]. Three of the six carbon-fixation pathways were found in thermophilic microorganisms, in the Archaea and Bacteria domains. For example, approximately 50 % of the bacterial cellular carbon originates from pyruvate/PEP (a gluconeogenic substrate that forms cell wall components and nucleotides), followed by acetyl-CoA (approximately 30 %), oxaloacetate (approximately 13 %), and alpha-ketoglutarate (approximately 7 %) [157]. The 3-HP cycle was found in the bacterium Chloroflexus aurantiacus, known as a thermophilic filamentous anoxygenic photoheterotrophic bacterium, that was initially isolated in hot springs and found to develop at temperatures between 45 °C and 75 °C [158], [159].
The carbon-fixation pathways HP/HB and DC/HB have been found in thermophilic Archaea. The HP/HB cycle was found in the Archaea Metallosphaera sedula, which grows autotrophically at 73 °C [160], and the DC/HB cycle was reported for the hyperthermophilic Archaea Ignicoccus hospitalis [79], [161]. Ignicoccus was isolated from the submarine hydrothermal ventilation systems, which are anaerobic, hyperthermophilic, always chemolithoautotrophic, and exhibit an optimal growth temperature of 90 °C [162].
In contrast, the CBB cycle occurs in several thermophiles, but never in hyperthermophiles, showing a temperature limit between approximately 70 °C and 75 °C [8]. This temperature limit of the cycle can be explained by the heat-induced instabilities of some intermediates in the cycle. Such instabilities are mainly observed in glyceraldehyde-3-phosphate, which produces toxic methylglyoxal at high temperatures [163]. The thermophilic bacteria Thermosulfidibacter takaii ABI70S6T showed a greatly efficient and reversible citrate synthase that needs reduced ferredoxin in the pathway [12], [164]. Until this discovery, it was believed that the CBB cycle can be reversed only to cleave citrate and fix CO2 autotrophically, however, this can be achieved only with alternative enzymes (e.g., citrate lyase). According to the authors, this question cannot be answered by metagenomics approach, but classical biochemistry can fill the gaps between genome sequences and organism phenotypes.
Although most species that use the rTCA cycle are mesophilic, the representatives of Aquificae are thermophiles that grow best at ≥ 70 °C, whereas Aquifex aeolicus grows best at temperatures up to 95 °C [23]. Hydrogenobacter thermophilus, a member of the phylum Aquificae, invests additional ATP in the conversion of 2-oxoglutarate to form isocitrate by the combination of an irreversible biotin-dependent 2-oxoglutarate carboxylase and a noncarboxylating isocitrate dehydrogenase [165]. This process possibly becomes effective and irreversible at high temperatures [23]. A recent fixation cycle describing the roTCA cycle was discovered in a thermophilic Deltaproteobacterium, Desulfurella acetivorans (a sulfur-reducing thermophile showing optimum growth at 52 °C-57 °C), through metagenomic analysis. In addition to the carbon-fixation pathways, the key enzymes involved in these routes have also been described in these thermophiles. Aoshima et al. [166] reported a new key enzyme, called the citryl-CoA synthetase, which was isolated from Hydrogenobacter thermophilus. This enzyme, which is involved in the rTCA cycle, could reportedly catalyze the first citrate cleavage step.
Carboxydotrophic bacteria are also found in geothermal environments, suggesting that CO-oxidizing Bacteria and Archaea perform an important role in thermophilic environments [167]. For example, acetogens and methanogens can survive in thermophilic systems and use CO as a carbon source [168]. In the WL pathway, CO is a key intermediate in the fixation of CO2 into acetyl-CoA in acetogens, methanogens, and sulfate-reducing bacteria [169], [170].
Recently, a novel Archaea phylum from terrestrial hot spring and deep-sea hydrothermal vent sediments that presented unique and versatile carbon cycling pathways was described [171]. Metagenome-assembled genomes were reconstructed from 15 Archaea genomes, and a new phylum classified as Brockarcheota was proposed. The Brockarchaeota are capable of mediating nonmethanogenic anaerobic methylotrophs via the tetrahydrofolate methyl branch of the WL pathway and the reductive glycine pathway. The members of this unique phylum exhibit a type of anaerobic methylotrophic metabolism that has not been described previously in Archaea. The methyltransferase process is composed of two key steps: first, specific methyltransferases break the C-R bonds in several substrates (e.g., methanol and trimethylamine) and transfer the methyl moieties to the subunit MtaC; then, the methyltransferase (MtaA for methanol and MtbA for methylamines) transfers the methyl group from the corrinoid protein to coenzyme M in the methanogens or to tetrahydrofolate in the acetogens [171], [172].
Brockarchaeota lack the key enzymes from the methyl and carbonyl branches of the WL pathway, which are involved in the transfer and reduction of the C1 fraction for methane production. Furthermore, Brockarchaeota do not encode the pyrroloquinoline quinone-linked methanol dehydrogenase pathway for aerobic methylotrophic reactions. This lack of methylotrophic pathways suggests that the new phylum metabolizes methanol and trimethylamine through the convergent action of the tetrahydrofolate (H4F) methyl branches of the WL and glycine-reduction pathways [171].
The phylum Brockarchaeota also presents an unknown pathway for butanol metabolism, since key enzymes involved in the fermentation of pyruvate to butanol are absent in their genomes [171]. Nevertheless, most of the 15 genomes encode a putative aldehyde dehydrogenase that can convert butyraldehyde to butyric acid. In addition, a putative enoyl-CoA hydratase/isomerase protein has been found to be involved in the further conversion of butyric acid to acetyl-CoA, which suggests an alternative pathway for the oxidation of butanol. Members of this phylum can also perform the following activities: (i) degrade complex carbon compounds such as xylan, (ii) assimilate formaldehyde and formate, (iii) use arsenate as an electron acceptor, (iv) possibly reduce sulfur during fermentative growth and produce H2S, and (v) possibly use group 3b [NiFe]-hydrogenases for the oxidation of H2 with NADP or NAD(P) as the electron acceptors [171], [172], [173].
In the early 1990 s, Gadkari et al. [174] described a new species of Streptomyces, which was a thermophilic-carboxidotrophic-chemolithoautotrophic microbe isolated from soil covering a burning charcoal pile, and named it Streptomyces thermoautrotophicus UBT1. In contrast to the other carboxidotropics, this strain is unique in its exclusive use of a lithotrophic substrate with a new CO dehydrogenase. Several decades later, MacKellar et al. [175] described a new strain of Streptomyces thermoautrotophicus (strain H1), which was isolated from another burning charcoal pile; furthermore, multiple CO dehydrogenase gene clusters were identified in the genomes of the strains UBT1 and H1 [176].
Recently, Volpiano et al. [176] proposed a new reclassification of the species and genus Streptomyces thermoautrotophicus, which was modified to Carbonactinospora thermoautotrophica based on its phylogenetic placement and distinctive phenotypes among Actinomycetes. Multiple genes related to carbon metabolism were found in this species, including ribulose bisphosphate carboxylase (rbcL), phosphoenolpyruvate carboxykinase, glucose/mannose-6-phosphate isomerase and PFK 6-phosphofructokinase. Thus, more investigations are necessary to identify the carbon path used by these thermophilic strains [176].
These findings indicate that thermophilic microorganisms are important for identifying new ways of fixing carbon. Thus, it is crucial to identify microorganisms that can participate in the global carbon cycle and provide alternatives for applications in various sectors of biotechnology.
Challenges and perspectives
The main challenge underlying the manipulation and modification of carbon-fixation routes is the insufficient understanding of how the pathways perform together within the studied organism. The use of fossil fuel resources is one of the main causes of global warming, being strictly related to the increase of atmospheric CO2 concentration [110]. Natural photosynthesis can convert approximately 100 billion tons of CO2 into biomass annually; however, RuBisCO, which is the most abundant enzyme on Earth, itself has a limited carboxylation rate, indicating an extremely low productivity of this pathway [126]. Synthetic biologists have made numerous efforts to overcome these challenges and improve the specificity of RuBisCO. Natural carbon-fixation processes are often slow, increasing the difficulty in designing genetically modified autotrophic plants and organisms and improving native CO2 fixation [4], [110].
Since the discovery of other natural pathways of carbon fixation besides the well-known CBB cycle, studies have attempted to identify the existence of other pathways in organisms that were not previously thought to fix carbon. This has led to a better understanding of the functioning of the pathways and the catalytic power of key enzymes related to carbon fixation. In addition, these findings have raised the question of increasing the efficiency of already known pathways as well as redesigning new pathways of carbon fixation to naturally reduce the amount of anthropogenically emitted CO2.
The global objective is to achieve liquid zero carbon emissions by the year 2050 [177]. This emitted carbon can be fixed, stored, and converted into sustainable feedstock. The present review aimed to elaborate the natural carbon pathways, as well as highlight the synthetic pathways that have been developed over the last decades. Given the need for shedding light on these two subjects, we collected and centralized the available data on the recent research in this field. In recent years, different pathways have emerged to improve carbon-fixation efficiency, demonstrating the importance of this discussion. Furthermore, the most important step will be to apply these efficient pathways in hosts and make them functional.
Based on the recent progress made in this field, certain problems related to biological carbon fixation have been identified [4]. Due to the low activities of the key carbon-fixing enzymes, there are limitations on natural carbon-fixation pathways. However, the rapid advancements in microbiome studies and their related “omics” analyses have generated large amounts of data, from which novel and more efficient carbon-fixing enzymes can be identified. Natural carbon-fixation pathways have several shortcomings, with many reaction steps and low efficiencies. However, the development of synthetic biology permits the exploration of new natural carbon-fixation pathways and the design of novel synthetic carbon-fixation techniques. Another barrier is the low solar energy-absorption efficiency of carbon fixation, which significantly reduces the economic benefits. These limitations can be addressed by converting light to other sources such as electric energy and hydrogen for direct carbon fixation. Chemoautotrophs provide an alternative platform because they can be driven chemically by renewable H2 instead of light [103]. Electrical energy can also be applied to produce energy carriers like formate, hydrogen, carbon monoxide, methanol, and methane [178]. Li et al. [179] described Ralstonia eutropha H16, an autotrophic genetically engineered microorganism, that produced high levels of alcohol in an electric bioreactor using CO2 as the sole carbon source and electricity as the sole energy input. Another approach could be to improve light-utilization efficiency. Recently, Nürnberg et al. [180] discovered specific photosystems I and II in Chroococcidiopsis thermalis that can use energy from far-infrared radiation.
Due to the low efficiencies and limited scope for performing genetic modifications of the natural carbon-fixation systems, various researchers are exploring these systems using different ways; for example, introducing partial or total carbon-fixation pathways in heterotrophic chassis cells. Generally, heterotrophic microorganisms exhibit superior growth and production yields than autotrophic microorganisms [17]. The significant improvements in synthetic biology permit the engineering of novel functions and metabolic networks in heterotrophic microorganisms for innumerous biotechnological applications [178], [179], [180], [181], [182]. Novel genes mediating the CO2 fixation pathway in autotrophic microorganisms may apply negative influences on the regulatory networks in cells, such as carbon/nitrogen metabolism; however, they have been widely used as chassis to express natural CO2-fixation pathways in the model heterotrophic microbes Escherichia coli and Saccharomyces cerevisiae [183], [184].
In the near future, novel enzymes and entire pathways of carbon fixation are expected to be identified with rapid development of “omics” tools and microbiome projects [4]. These advancements will facilitate the development of feasible methods for changing and/or increasing the production of the desired product. Moreover, the use and availability of “omics” tools has progressed rapidly, highlighting the limitations of single techniques in dealing with the complexities of biological systems [185].
One of the future possibilities is the integration of data obtained using the “omics” tools with studies on metabolism and kinetics, classical biochemistry, and molecular biological techniques. In addition, the development of mathematical models to analyze biological processes on a micro-scale will allow us to predict their effects on a macro scale. In the near future, these suggested methods may facilitate the exploration, modeling, manipulation, and engineering of metabolic pathways in natural and synthetic microbial systems. According to Bar-Even et al. [115] and Fuchs [7], the number of known autotrophic pathways is expected to increase with the aid of genome searching and the application of synthetic biological tools. Notably, “omics” analyses have revolutionized the research on carbon fixation and unveiled the genomic black box. Nevertheless, for better results, these analyses must be linked to bench tests and classical biochemical analyses to obtain detailed and robust results because some resources may not be identified solely with bioinformatics tools.
Although carbon-fixation pathways have not yet been used at the industry level owing to the low catalytic efficiencies of the enzymes and the need for a significant energy input, studies of these routes are anticipated to provide a plethora of possibilities for biotechnological advances. The major challenge, i.e., the joining of carbon-fixation pathways with synthetic pathways due to the differences between the metabolism of autotrophs and heterotrophs that are used as chassis, can be resolved. Carbon-fixation pathways based on synthetic biology have already been described, and the energy supply and more efficient enzymes can be redesigned in these pathways [186].
Conclusion (Remarks)
Carbon is often negatively associated with the anthropogenic emissions of greenhouse gasses; however, it is also an element of paramount importance for life and ecological processes. The knowledge of carbon-fixation cycles, in addition to ecological and evolutionary studies on the emergence of the first beings that inhabited Earth and metabolized carbon, can provide a detailed understanding of the enzymes and important intermediaries involved in these cycles.
The central carbon-metabolizing enzymes can be used for a variety of purposes; for example, artificial CO2 fixation pathways can be designed and implemented in suitable host organisms. Additionally, new solutions are possible for manipulating carbon-fixation pathways and increasing their final yields. The multidisciplinary integration of “omics”, synthetic biology, molecular biology, and biochemistry can enable the scientific community to describe new carbon-fixation pathways and predict important routes and enzymes involved in these processes. This integration may provide further guidance for overcoming the limitations of natural carbon-fixation pathways and allow the development of biotechnological applications.
Nevertheless, investigation of thermophilic microorganisms is crucial for understanding many evolutionary events. Overall, these microorganisms have been extensively investigated for developing biotechnological applications because of their metabolic capabilities; however, thermophiles remain underexplored with regard to the identification of novel natural routes of carbon fixation and the development of efficient synthetic pathways.
Funding
This study was funded by KAUST (BAS/1/1096-01-01). SSC was awarded scholarships from the Foundation for Research Support of the State of Rio de Janeiro (FAPERJ) and Coordination for the Improvement of Higher Education Personnel (CAPES).
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Sulamita Santos Correa: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. Junia Schultz: Data curation, Writing – original draft, Writing – review & editing. Kyle J. Lauersen: Validation, Writing – review & editing. Alexandre Soares Rosado: Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Graduate Program of Plant Biotechnology and Bioprocesses (PBV) at the Federal University of Rio de Janeiro for supporting this work and Prof. Patricia Moura, Ricardo Chaloub and Alex Prast for their valuable suggestions to improve the manuscript. The graphical abstract was created by Heno Hwang, scientific illustrator at King Abdullah University of Science and Technology (KAUST).
Biographies

Sulamita Correa MSc. Sulamita Correa is currently doing a PhD in Plant Biotechnology and Bioprocesses at the Federal University of Rio de Janeiro and is developing her project at the Molecular Microbial Ecology Laboratory (UFRJ), where she works with the nitrogen and carbon fixation pathway in thermophilic actinobacteria. She holds a master’s degree in Plant Biotechnology and Bioprocesses from the Federal University of Rio de Janeiro, developed her project in the genetics and biochemistry laboratory at Embrapa Agrobiology, in the lines of research: plant microorganism interaction; abiotic stress in plants. She has a degree in Agricultural Sciences from the Federal Rural University of Rio de Janeiro. MSc. Correa has experience on microbial ecology, biotechnology, and molecular biology.

Junia Schultz Dr. Junia Schultz is biologist and holds a PhD in Biotechnology and bioprocesses, pursued in Federal University of Rio de Janeiro, Brazil, with PhD internships in the University of Queensland (Australia), University of Western Brittany (France) and Helmholtz Institute (Germany). Dr. Schultz has experience on microbial ecology and biotechnology, especially with extreme environments and extremophiles. Currently she is a postdoctoral researcher at the King Abdullah University of Science and Technology (Saudi Arabia), and her research interests are focused on unraveling the culturable and yet-to-be-cultured microorganisms of extreme Saudi Arabian environments and exploit those extremophiles for biotechnological and astrobiological applications.

Kyle J. Lauersen Dr. Kyle J. Lauersen is originally from Kingston, Ontario, Canada, where he completed his Bachelor of Science with honours, Bachelor of Education, and Master of Science degrees at Queen’s University. Dr. Lauersen’s research background is in molecular biology, green biotechnology, and genetic control of gene expression. In 2011, he was awarded a full PhD scholarship funded by the Cluster Industrial Biotechnology Graduate Cluster (CLIB-GC) at the Center for Biotechnology (CeBiTec), Bielefeld University, Germany. During his doctoral research, Dr. Lauersen discovered that long-observed recalcitrance of eukaryotic microalgae to genetic engineering could be overcome by synthetically adapting transgene expression constructs to mimic host regulation machinery. This innovation enabled reliable transgene expression from eukaryotic green algal nuclear genomes for the first time and opened new possibilities for engineering these hosts. Dr. Lauersen was nominated by Bielefeld University for the German Research Foundation 2019 Heinz Maier-Leibnitz award. In 2019 he became Assistant Professor at King Abdullah University of Science & Technology (KAUST) in the Biological and Environmental Sciences and Engineering Division (BESE). Currently, he leads the Sustainable & Synthetic Biotechnology research group at KAUST. His team works towards increasing fundamental understanding of algal metabolism and the development of sustainable, light-driven, bio-processes with these hosts.

Alexandre Soares Rosado Professor of Bioscience at King Abdullah University of Science and Technology (KAUST). Former Professor at Federal University of Rio de Janeiro (UFRJ) and Visiting Professor at the Department of Land, Air and Water Resources, University of California - Davis, USA. Former Director of the Institute of Microbiology and Vice-President of the Brazilian Society of Microbiology (SBM); holds a BA and BSc in Biological Sciences, MSc (Microbiology), PhD in Microbiology from UFRJ and Wageningen University & Research (WUR), The Netherlands. Professor Rosado is an environmental microbiologist specialized in Microbiome Science, biotechnology, and Microbiology of Extreme Environments (source of new biotechnological products). His main research interests are centered around the study of microbial diversity, evolution, and new microbial metabolisms, using conventional tools associated to multi-omics approaches to understand the evolutionary history, ecological roles, and physiological capacity and capabilities of free-living and symbiotic microorganisms, including those found at the extremes of where life exists.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Aversa R., Petrescu R.V., Apicella A., Petrescu F.I. The basic elements of life's. American J E and A S. 2016;9(4):1189–1197. [Google Scholar]
- 2.Loeve, S., & Vincent, B. B. The multiple signatures of carbon. In Res. Obj. in their Tech. Setting (pp. 185-200). (2017).
- 3.Anthony M.A., et al. Distinct assembly processes and microbial communities constrain soil organic carbon formation. One earth. 2020;2(4):349–360. [Google Scholar]
- 4.Gong F., Zhu H., Zhang Y., Li Y. Biological carbon fixation: from natural to synthetic. J of CO2 Uti. 2018;28:221–227. [Google Scholar]
- 5.Siegel J.B., Smith A.L., Poust S., Wargacki A.J., Bar-Even A., Louw C., et al. Computational protein design enables a novel one-carbon assimilation pathway. Proc of the Nat Aca of Scie. 2015;112(12):3704–3709. doi: 10.1073/pnas.1500545112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu X., Liu Y., Yang Y., Wang S., Wang Q., Wang X., et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Natu Comm. 2019;10(1) doi: 10.1038/s41467-019-09095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annual Rev of Microb. 2011;65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 8.Hugler M., Sievert S.M. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev of Mar Scie. 2011;3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 9.Ljungdhal L.G. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Ann Rev of Microb. 1986;40(1):415–450. doi: 10.1146/annurev.mi.40.100186.002215. [DOI] [PubMed] [Google Scholar]
- 10.Bar-Even A., Noor E., Milo R. A survey of carbon fixation pathways through a quantitative lens. J of Expe Bot. 2012;63(6):2325–2342. doi: 10.1093/jxb/err417. [DOI] [PubMed] [Google Scholar]
- 11.Kono T., Mehrotra S., Endo C., Kizu N., Matusda M., Kimura H., et al. A RuBisCO-mediated carbon metabolic pathway in methanogenic Archaea. Natu comm. 2017;8(1):1–12. doi: 10.1038/ncomms14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mall A., et al. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Scien. 2018;359(6375):563–567. doi: 10.1126/science.aao2410. [DOI] [PubMed] [Google Scholar]
- 13.Kruse O., Rupprecht J., Mussgnug J.H., Dismukes G.C., Hankamer B. Photosynthesis: a blueprint for solar energy capture and biohydrogen production technologies. Photo & Photobi Scien. 2005;4(12):957–970. doi: 10.1039/b506923h. [DOI] [PubMed] [Google Scholar]
- 14.Sciuto K., Moro I. Cyanobacteria: the bright and dark sides of a charming group. Biodi and Conse. 2015;24(4):711–738. [Google Scholar]
- 15.Liu L., Brown P.C., Könneke M., Huber H., König S., Berg I.A. Convergent evolution of a promiscuous 3-hydroxypropionyl-CoA dehydratase/crotonyl-CoA hydratase in crenarchaeota and thaumarchaeota. Msphere. 2021;6(1):e01079–e10120. doi: 10.1128/mSphere.01079-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Souza Y.P.A., Rosado A.S. Microbial Diversity in the Genomic Era. Acad. Press; 2019. Opening the Black Box of Thermophilic Autotrophic Bacterial Diversity; pp. 333–343. [Google Scholar]
- 17.Liang B., Zhao Y., Yang J. Recent Advances in Developing Artificial Autotrophic Microorganism for Reinforcing CO2 Fixation. Front in Microb. 2020;11:2848. doi: 10.3389/fmicb.2020.592631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel, A. S. Chemolithoautotrophy. In Encyclopedia of Caves, Acad. Press, 267-276. (2019).
- 19.Irfan M., Bai Y., Zhou L., Kazmi M., Yuan S., Mbadinga S.M., et al. Direct microbial transformation of carbon dioxide to value-added chemicals: a comprehensive analysis and application potentials. Bior Tech. 2019;288 doi: 10.1016/j.biortech.2019.121401. [DOI] [PubMed] [Google Scholar]
- 20.Sysoev M., Grötzinger S.W., Renn D., Eppinger J., Rueping M., Karan R. Bioprospecting of novel extremozymes from prokaryotes - The advent of culture-independent methods. Front in Microb. 2021;12 doi: 10.3389/fmicb.2021.630013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atsumi S., Higashide W., Liao J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Natu Biotec. 2009;27(12):1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 22.Durall C., Lindblad P. Mechanisms of carbon fixation and engineering for increased carbon fixation in cyanobacteria. Algal Res. 2015;11:263–270. [Google Scholar]
- 23.Berg I.A. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl and Envir Micro. 2011;77(6):1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjornsson L., Hugenholtz P., Tyson G.W., Blackall L.L. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Micro. 2002;148(8):2309–2318. doi: 10.1099/00221287-148-8-2309. [DOI] [PubMed] [Google Scholar]
- 25.Frolov E.N., Kublanov I.V., Toshchakov S.V., Lunev E.A., Pimenov N.V., Bonch-Osmolovskaya E.A., et al. Form III RuBisCO-mediated transaldolase variant of the Calvin cycle in a chemolithoautotrophic bacterium. Proc of the Nat Aca of Scien. 2019;116(37):18638–18646. doi: 10.1073/pnas.1904225116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Li W., Xiao Y., Cheng A., Shen T., Zhu M., et al. Abundance and diversity of carbon-fixing bacterial communities in karst wetland soil ecosystems. Catena. 2021;204 [Google Scholar]
- 27.Kusian B., Bowien B. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Micro Rev. 1997;21(2):135–155. doi: 10.1111/j.1574-6976.1997.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 28.Gurrieri L., Fermani S., Zaffagnini M., Sparla F., Trost P. Calvin-Benson cycle regulation is getting complex. Trends Plant Sci. 2021;26(9):898–912. doi: 10.1016/j.tplants.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Launay H., Shao H., Bornet O., Cantrelle F.X., Lebrun R., Receveur-Brechot V., et al. Flexibility of Oxidized and Reduced States of the Chloroplast Regulatory Protein CP12 in Isolation and in Cell Extracts. Biomolecules. 2021;11(5):701. doi: 10.3390/biom11050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao H., Huang W., Avilan L., Receveur-Bréchot V., Puppo C., Puppo R., et al. A new type of flexible CP12 protein in the marine diatom Thalassiosira pseudonana. Cell Communication and Signaling. 2021;19(1):1–13. doi: 10.1186/s12964-021-00718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta J. A novel variant of the Calvin-Benson cycle bypassing fructose bisphosphate. Sci Rep. 2022;12(1):1–8. doi: 10.1038/s41598-022-07836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharwood R.E. Engineering chloroplasts to improve RuBisCO catalysis: prospects for translating improvements into food and fiber crops. New Phyto. 2017;213(2):494–510. doi: 10.1111/nph.14351. [DOI] [PubMed] [Google Scholar]
- 33.Sharkey T.D., Weise S.E. The glucose 6-phosphate shunt around the Calvin-Benson cycle. J Exp Bot. 2016;67(14):4067–4077. doi: 10.1093/jxb/erv484. [DOI] [PubMed] [Google Scholar]
- 34.Sharkey T.D. Pentose phosphate pathway reactions in photosynthesizing cells. Cells. 2021;10(6):1547. doi: 10.3390/cells10061547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cleland W.W., Andrews T.J., Gutteridge S., Hartman F.C., Lorimer G.H. Mechanism of RuBisCO: the carbamate as general base. Chem Rev. 1998;98(2):549–562. doi: 10.1021/cr970010r. [DOI] [PubMed] [Google Scholar]
- 36.Ducat D.C., Silver P.A. Improving carbon fixation pathways. Cur Opi in Chem Biol. 2012;16(3–4):337–344. doi: 10.1016/j.cbpa.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caldwell P.E., MacLean M.R., Norris P.R. Ribulose bisphosphate carboxylase activity and a Calvin cycle gene cluster in Sulfobacillus species. Micro. 2007;153(7):2231–2240. doi: 10.1099/mic.0.2007/006262-0. [DOI] [PubMed] [Google Scholar]
- 38.Yeates T.O., Kerfeld C.A., Heinhorst S., Cannon G.C., Shively J.M. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Natu Rev Micro. 2008;6(9):681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 39.Savage D.F., Afonso B., Chen A.H., Silver P.A. Spatially ordered dynamics of the bacterial carbon fixation machinery. Scien. 2010;327(5970):1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 40.Long B.M., Hee W.Y., Sharwood R.E., Rae B.D., Kaines S., Lim Y.-L., et al. Carboxysome encapsulation of the CO 2-fixing enzyme RuBisCO in tobacco chloroplasts. Natu Comm. 2018;9(1) doi: 10.1038/s41467-018-06044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackinder L.C.M., Chen C., Leib R.D., Patena W., Blum S.R., Rodman M., et al. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell. 2017;171(1):133–147.e14. doi: 10.1016/j.cell.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill N.C., Tay J.W., Altus S., Bortz D.M., Cameron J.C. Life cycle of a cyanobacterial carboxysome. Scien Adv. 2020;6(19):eaba1269. doi: 10.1126/sciadv.aba1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabita F.R., Satagopan S., Hanson T.E., Kreel N.E., Scott S.S. Distinct form I, II, III, and IV RuBisCO proteins from the three kingdoms of life provide clues about RuBisCO evolution and structure/function relationships. J of Expe Bot. 2008;59(7):1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- 44.Wilkes E.B., Carter S.J., Pearson A. CO2-dependent carbon isotope fractionation in the dinoflagellate Alexandrium tamarense. Geoch et Cosm Acta. 2017;212:48–61. [Google Scholar]
- 45.Morse D.P., Salois P., Markovic J.W.H. A nuclear-encoded form II RuBisCO in dinoflagellates. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 46.Rowan R.S.M., Whitney A., Fowler D.Y. Rubisco in marine symbiotic dinoflagellates: form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell. 1996;8:539–553. doi: 10.1105/tpc.8.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banda D.M., Pereira J.H., Liu A.K., Orr D.J., Hammel M., He C., et al. Novel bacterial clade reveals origin of form I RuBisCO. Natu Plan. 2020;6(9):1158–1166. doi: 10.1038/s41477-020-00762-4. [DOI] [PubMed] [Google Scholar]
- 48.Whitney S.M., Andrews T.J. The gene for the ribulose-1, 5-bisphosphate carboxylase/oxygenase (RuBisCO) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into RuBisCO. Plant Cell. 2001;13(1):193–205. doi: 10.1105/tpc.13.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida S., Atomi H., Imanaka T. Engineering of a type III RuBisCO from a hyperthermophilic archaeon in order to enhance catalytic performance in mesophilic host cells. Appl and Enviro Micro. 2007;73(19):6254–6261. doi: 10.1128/AEM.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aono R., Sato T., Imanaka T., Atomi H. A pentose bisphosphate pathway for nucleoside degradation in Archaea. Natu Chem Bio. 2015;11(5):355–360. doi: 10.1038/nchembio.1786. [DOI] [PubMed] [Google Scholar]
- 51.Wrighton K.C., et al. RuBisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. The ISME J. 2016;10(11):2702–2714. doi: 10.1038/ismej.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tabita F.R., Hanson T.E., Li H., Satagopan S., Singh J., Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71(4):576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S.M., Lim H.S., Lee S.B. Discovery of a RuBisCO -like Protein that Functions as an Oxygenase in the Novel D-Hamamelose Pathway. Biotec and Biopr Eng. 2018;23(5):490–499. [Google Scholar]
- 54.Evans M.C., Buchanan B.B., Arnon D.I. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proce. of the Nat. Aca of Scien of the Unit Sta of Amer. 1996;55(4):928. doi: 10.1073/pnas.55.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchanan B.B., Sirevåg R., Fuchs G., Ivanovsky R.N., Igarashi Y., Ishii M., et al. The Arnon-Buchanan cycle: a retrospective, 1966–2016. Photo Res. 2017;134(2):117–131. doi: 10.1007/s11120-017-0429-0. [DOI] [PubMed] [Google Scholar]
- 56.Kitadai N., Kameya M., Fujishima K. Origin of the reductive tricarboxylic acid (rTCA) cycle-type CO2 fixation: a perspective. Life. 2017;7(4):39. [Google Scholar]
- 57.Kluger R., Tittmann K. Thiamin diphosphate catalysis: enzymic and nonenzymic covalent intermediates. Chem Rev. 2008;108(6):1797–1833. doi: 10.1021/cr068444m. [DOI] [PubMed] [Google Scholar]
- 58.Fuchs G. Alternative pathways of autotrophic carbon dioxide fixation in autotrophic bacteria. In Bio. of Autotr. Bac., ed. HG Schlegel, 365-82. (1989).
- 59.Ivanovsky R.N., Fal Y.I., Berg I.A., Ugolkova N.V., Krasilnikova E.N., Keppen O.I., et al. Evidence for the presence of the reductive pentose phosphate cycle in a filamentous anoxygenic photosynthetic bacterium, Oscillochloris trichoides strain DG-6. Micro. 1999;145(7):1743–1748. doi: 10.1099/13500872-145-7-1743. [DOI] [PubMed] [Google Scholar]
- 60.Thauer R.K. A fifth pathway of carbon fixation. Scien. 2007;318(5857):1732–1733. doi: 10.1126/science.1152209. [DOI] [PubMed] [Google Scholar]
- 61.Aherfi S., Brahim Belhaouari D., Pinault L., Baudoin J.P., Decloquement P., Abrahao J., et al. Incomplete tricarboxylic acid cycle and proton gradient in Pandoravirus massiliensis: is it still a virus? The ISME journal. 2022;16(3):695–704. doi: 10.1038/s41396-021-01117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brock T.D. The value of basic research: discovery of Thermus aquaticus and other extreme thermophiles. Genet. 1997;146(4):1207. doi: 10.1093/genetics/146.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adam P.S., Borrel G., Gribaldo S. An Archaeal origin of the Wood-Ljungdahl H 4 MPT branch and the emergence of bacterial methylotrophy. Natu Micro. 2019;4(12):2155–2163. doi: 10.1038/s41564-019-0534-2. [DOI] [PubMed] [Google Scholar]
- 64.Youssef N.H., Farag I.F., Rudy S., Mulliner A., Walker K., Caldwell F., et al. The Wood-Ljungdahl pathway as a key component of metabolic versatility in candidate phylum Bipolaricaulota (Acetothermia, OP1) Enviro Micro Rep. 2019;11(4):538–547. doi: 10.1111/1758-2229.12753. [DOI] [PubMed] [Google Scholar]
- 65.Drake H.L., Daniel S.L., Küsel K., Matthies C., Kuhner C., Braus-Stromeyer S. Acetogenic bacteria: what are the in situ consequences of their diverse metabolic versatilities? Biofac. 1997;6(1):13–24. doi: 10.1002/biof.5520060103. [DOI] [PubMed] [Google Scholar]
- 66.Shafiee S., Topal E. When will fossil fuel reserves be diminished? Ener Poli. 2009;37(1):181–189. [Google Scholar]
- 67.Roberts J.R., Lu W.P., Ragsdale S.W. Acetyl-coenzyme A synthesis from methyltetrahydrofolate, CO, and coenzyme A by enzymes purified from Clostridium thermoaceticum: attainment of in vivo rates and identification of rate-limiting steps. J of Bac. 1992;174(14):4667–4676. doi: 10.1128/jb.174.14.4667-4676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schöne, C., Poehlein, A., Jehmlich, N., Adlung, N., Daniel, R., von Bergen, M., ... & Rother, M. Deconstructing Methanosarcina acetivorans into an acetogenic archaeon. Proceedings of the National Academy of Sciences, 119(2), e2113853119. (2022). [DOI] [PMC free article] [PubMed]
- 69.Orsi W.D., Vuillemin A., Rodriguez P., Coskun Ö.K., Gomez-Saez G.V., Lavik G., et al. Metabolic activity analyses demonstrate that Lokiarchaeon exhibits homoacetogenesis in sulfidic marine sediments. Nat Microbiol. 2020;5(2):248–255. doi: 10.1038/s41564-019-0630-3. [DOI] [PubMed] [Google Scholar]
- 70.Biegel E., Müller V. Bacterial Na+-translocating ferredoxin: NAD+ oxidoreductase. Proce. of the Nat. Aca of Scien. 2010;107(42):18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borrel G., Adam P.S., Gribaldo S. Methanogenesis and the Wood-Ljungdahl pathway: an ancient, versatile, and fragile association. Geno Bio and Evo. 2016;8(6):1706–1711. doi: 10.1093/gbe/evw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holo H. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch of Micro. 1989;151(3):252–256. [Google Scholar]
- 73.Zarzycki J., Brecht V., Müller M., Fuchs G. Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proce of the Nat Aca of Scie. 2009;106(50):21317–21322. doi: 10.1073/pnas.0908356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shih P.M., Ward L.M., Fischer W.W. Evolution of the 3-hydroxypropionate bicycle and recent transfer of anoxygenic photosynthesis into the Chloroflexi. Proce of the Nat Aca of Scien. 2017;114(40):10749–10754. doi: 10.1073/pnas.1710798114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ward L.M., Shih P.M. The evolution and productivity of carbon fixation pathways in response to changes in oxygen concentration over geological time. Free Rad Bio and Med. 2019;140:188–199. doi: 10.1016/j.freeradbiomed.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 76.Zarzycki J., Fuchs G. Coassimilation of organic substrates via the autotrophic 3-hydroxypropionate bi-cycle in Chloroflexus aurantiacus. Appl Environ Microbiol. 2011;77(17):6181–6188. doi: 10.1128/AEM.00705-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berg I.A., Ramos-Vera W.H., Petri A., Huber H., Fuchs G. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Micro. 2010;156(1):256–269. doi: 10.1099/mic.0.034298-0. [DOI] [PubMed] [Google Scholar]
- 78.Berg I.A., Kockelkorn D., Buckel W., Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Scien. 2007;318(5857):1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 79.Huber H., Gallenberger M., Jahn U., Eylert E., Berg I.A., Kockelkorn D., et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc of the Nat Acad of Scien. 2008;105(22):7851–7856. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flechsler, J., Heimerl, T., Huber, H., Rachel, R., & Berg, I. A. Functional compartmentalization and metabolic separation in a prokaryotic cell. Proceedings of the National Academy of Sciences, 118(25), e2022114118. (2021). [DOI] [PMC free article] [PubMed]
- 81.Hawkins A.S., Han Y., Bennett R.K., Adams M.W., Kelly R.M. Role of 4-hydroxybutyrate-CoA synthetase in the CO2 fixation cycle in thermoacidophilic Archaea. J of Bio Chem. 2013;288(6):4012–4022. doi: 10.1074/jbc.M112.413195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Auernik K.S., Kelly R.M. Physiological versatility of the extremely thermoacidophilic archaeon Metallosphaera sedula supported by transcriptomic analysis of heterotrophic, autotrophic, and mixotrophic growth. Appl and Enviro Micro. 2010;76(3):931–935. doi: 10.1128/AEM.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Estelmann S., Hügler M., Eisenreich W., Werner K., Berg I.A., Ramos-Vera W.H., et al. Labeling and enzyme studies of the central carbon metabolism in Metallosphaera sedula. J of Bac. 2011;193(5):1191–1200. doi: 10.1128/JB.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berg I.A., Liu L., Schubert D., Koenneke M. (S)-3-Hydroxybutyryl-CoA dehydrogenase from the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Nitrosopumilus maritimus. Front Microbiol. 2021;12:1846. doi: 10.3389/fmicb.2021.712030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rosgaard L., de Porcellinis A.J., Jacobsen J.H., Frigaard N.U., Sakuragi Y. Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J of Biot. 2012;162(1):134–147. doi: 10.1016/j.jbiotec.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 86.Soderberg T. Biosynthesis of ribose-5-phosphate and erythrose-4-phosphate in archaea: a phylogenetic analysis of archaeal genomes. Arch. 2005;1(5):347–352. doi: 10.1155/2005/314760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.White R.A., III, Rivas-Ubach A., Borkum M.I., Köberl M., Bilbao A., Colby S.M., et al. The state of rhizospheric science in the era of multi-omics: A practical guide to omics technologies. Rhizo. 2017;3:212–221. [Google Scholar]
- 88.Palazzotto E., Weber T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Cur Opi in Micro. 2018;45:109–116. doi: 10.1016/j.mib.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 89.Murugadas V., Prasad M.M. ICAR-Cen. Inst. of Fish; Tec: 2018. Bioinformatics in Microbial systematics. [Google Scholar]
- 90.Christel S., Herold M., Bellenberg S., El Hajjami M., Buetti-Dinh A., Pivkin I.V., et al. Multi-omics reveals the lifestyle of the acidophilic, mineral-oxidizing model species Leptospirillum ferriphilum T. Appl and Enviro Micro. 2017;84(3):e02091–e10117. doi: 10.1128/AEM.02091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McAllister S.M., Polson S.W., Butterfield D.A., Glazer B.T., Sylvan J.B., Chan C.S. Validating the Cyc2 neutrophilic iron oxidation pathway using meta-omics of Zetaproteobacteria iron mats at marine hydrothermal vents. Msys. 2020;5(1):e00553–e619. doi: 10.1128/mSystems.00553-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Figueroa I.A., Barnum T.P., Somasekhar P.Y., Carlström C.I., Engelbrektson A.L., Coates J.D. Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc of the Nat Acad of Scien. 2018;115(1):92–101. doi: 10.1073/pnas.1715549114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cotton C.A., Edlich-Muth C., Bar-Even A. Reinforcing carbon fixation: CO2 reduction replacing and supporting carboxylation. Curr Opi in Biote. 2018;49:49–56. doi: 10.1016/j.copbio.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 94.Schink B., Thiemann V., Laue H., Friedrich M.W. Desulfotignum phosphitoxidans sp. nov., a new marine sulfate reducer that oxidizes phosphite to phosphate. Arch of Micro. 2002;177(5):381–391. doi: 10.1007/s00203-002-0402-x. [DOI] [PubMed] [Google Scholar]
- 95.Sánchez-Andrea I., Guedes I.A., Hornung B., Boeren S., Lawson C.E., Sousa D.Z., et al. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Natu Comm. 2020;11(1):1–12. doi: 10.1038/s41467-020-18906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hao L., Michaelsen T.Y., Singleton C.M., Dottorini G., Kirkegaard R.H., Albertsen M., et al. Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics. The. ISME J. 2020;14(4):906–918. doi: 10.1038/s41396-019-0571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yishai O., Bouzon M., Döring V., Bar-Even A. In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli. ACS Synth Biol. 2018;7(9):2023–2028. doi: 10.1021/acssynbio.8b00131. [DOI] [PubMed] [Google Scholar]
- 98.Tashiro Y., Hirano S., Matson M.M., Atsumi S., Kondo A. Electrical-biological hybrid system for CO2 reduction. Metab Eng. 2018;47:211–218. doi: 10.1016/j.ymben.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 99.Kim S., Lindner S.N., Aslan S., Yishai O., Wenk S., Schann K., et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol. 2020;16(5):538–545. doi: 10.1038/s41589-020-0473-5. [DOI] [PubMed] [Google Scholar]
- 100.Bonch-Osmolovskaya E.A., Sokolova T.G., Kostrikina N.A., Zavarzin G.A. Desulfurella acetivorans gen. nov. and sp. nov. a new thermophilic sulfur-reducing eubacterium. Arch of Micro. 1990;153(2):151–155. [Google Scholar]
- 101.Pradella S., Hippe H., Stackebrandt E. Macrorestriction analysis of Desulfurella acetivorans and Desulfurella multipotens. FEMS Micro Lett. 1998;159(1):137–144. doi: 10.1111/j.1574-6968.1998.tb12852.x. [DOI] [PubMed] [Google Scholar]
- 102.Zhang T., Shi X.C., Ding R., Xu K., Tremblay P.L. The hidden chemolithoautotrophic metabolism of Geobacter sulfurreducens uncovered by adaptation to formate. The ISME J. 2020;14(8):2078–2089. doi: 10.1038/s41396-020-0673-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thevasundaram, K., Gallagher, J. J., Cherng, F., & Chang, M. C. Engineering nonphotosynthetic carbon fixation for production of bioplastics by methanogenic archaea. Proceedings of the National Academy of Sciences, 119(23), e2118638119. (2022). [DOI] [PMC free article] [PubMed]
- 104.Schwander T., von Borzyskowski L.S., Burgener S., Cortina N.S., Erb T.J. A synthetic pathway for the fixation of carbon dioxide in vitro. Scie. 2016;354(6314):900–904. doi: 10.1126/science.aah5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erb T.J., Berg I.A., Brecht V., Müller M., Fuchs G., Alber B.E. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proce of the Nat Acad of Scien. 2007;104(25):10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peter D.M., Schada von Borzyskowski L., Kiefer P., Christen P., Vorholt J.A., Erb T.J. Screening and engineering the synthetic potential of carboxylating reductases from central metabolism and polyketide biosynthesis. Ange Chem Inter Ed. 2015;54(45):13457–13461. doi: 10.1002/anie.201505282. [DOI] [PubMed] [Google Scholar]
- 107.Erb T.J., Brecht V., Fuchs G., Müller M., Alber B.E. Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoyl-thioester reductase. Proce of the Nat Acad of Scien. 2009;106(22):8871–8876. doi: 10.1073/pnas.0903939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gong F., Li Y. Fixing carbon, unnaturally. Scien. 2016;354(6314):830–831. doi: 10.1126/science.aal1559. [DOI] [PubMed] [Google Scholar]
- 109.Naduthodi M.I.S., Claassens N.J., D’Adamo S., van der Oost J., Barbosa M.J. Synthetic biology approaches to enhance microalgal productivity. Trends Biotechnol. 2021;39(10):1019–1036. doi: 10.1016/j.tibtech.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 110.Bar-Even A. Formate assimilation: the metabolic architecture of natural and synthetic pathways. Bioch. 2016;55(28):3851–3863. doi: 10.1021/acs.biochem.6b00495. [DOI] [PubMed] [Google Scholar]
- 111.Bar-Even A., Noor E., Flamholz A., Milo R. Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes. Bioch et Bioph Acta (BBA)-Bioene. 2013;1827(8–9):1039–1047. doi: 10.1016/j.bbabio.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 112.Claassens N.J. Reductive glycine pathway: a versatile route for one-carbon biotech. Trends Biotechnol. 2021;39(4):327–329. doi: 10.1016/j.tibtech.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 113.Wang X., Wang Y., Liu J., Li Q., Zhang Z., Zheng P., et al. Biological conversion of methanol by evolved Escherichia coli carrying a linear methanol assimilation pathway. Bioresources and Bioprocessing. 2017;4(1):1–6. [Google Scholar]
- 114.Lan E.I., Liao J.C. Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bior Tec. 2013;135:339–349. doi: 10.1016/j.biortech.2012.09.104. [DOI] [PubMed] [Google Scholar]
- 115.Bar-Even A., Noor E., Lewis N.E., Milo R. Design and analysis of synthetic carbon fixation pathways. Proce of the Nat Acad of Scien. 2010;107(19):8889–8894. doi: 10.1073/pnas.0907176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li D., Huang L., Liu T., Liu J., Zhen L., Wu J., et al. Electrochemical reduction of carbon dioxide to formate via nano-prism assembled CuO microspheres. Chemo. 2019;237 doi: 10.1016/j.chemosphere.2019.124527. [DOI] [PubMed] [Google Scholar]
- 117.Bang J., Ahn J.H., Lee J.A., Hwang C.H., Kim G.B., Lee J., et al. Synthetic formatotrophs for one-carbon biorefinery. Adv Sci. 2021;8(12):2100199. doi: 10.1002/advs.202100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao T., Li Y., Zhang Y. Biological carbon fixation: a thermodynamic perspective. Green Chem. 2021;23(20):7852–7864. [Google Scholar]
- 119.Yu H., Li X., Duchoud F., Chuang D.S., Liao J.C. Augmenting the Calvin–Benson-Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway. Natu Comm. 2018;9(1):1–10. doi: 10.1038/s41467-018-04417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luo S., Lin P.P., Nieh L.Y., Liao G.B., Tang P.W., Chen C., et al. A cell-free self-replenishing CO2-fixing system. Nat Catal. 2022;5(2):154–162. [Google Scholar]
- 121.Sarles L.S., Tabita F.R. Derepression of the synthesis of D-ribulose 1, 5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum. J of Bac. 1983;153(1):458–464. doi: 10.1128/jb.153.1.458-464.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wildman S.G. Along the trail from Fraction I protein to Rubisco (ribulose bisphosphate carboxylase-o xygenase) Photo Res. 2002;73(1):243–250. doi: 10.1023/A:1020467601966. [DOI] [PubMed] [Google Scholar]
- 123.Parry M.A., Keys A.J., Madgwick P.J., Carmo-Silva A.E., Andralojc P.J. RuBisCO regulation: a role for inhibitors. J of Expe Bot. 2008;59(7):1569–1580. doi: 10.1093/jxb/ern084. [DOI] [PubMed] [Google Scholar]
- 124.Bock R. Genetic engineering of the chloroplast: novel tools and new applications. Cur Opi in Biotec. 2014;26:7–13. doi: 10.1016/j.copbio.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 125.Cai Z., Liu G., Zhang J., Li Y. Development of an activity-directed selection system enabled significant improvement of the carboxylation efficiency of RuBisCO. Prote & cell. 2014;5(7):552–562. doi: 10.1007/s13238-014-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin M.T., Occhialini A., Andralojc P.J., Parry M.A., Hanson M.R. A faster RuBisCO with potential to increase photosynthesis in crops. Natu. 2014;513(7519):547–550. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hauser T., Popilka L., Hartl F.U., Hayer-Hartl M. Role of auxiliary proteins in RuBisCO biogenesis and function. Natu Plan. 2015;1(6):1–11. doi: 10.1038/nplants.2015.65. [DOI] [PubMed] [Google Scholar]
- 128.Whitney S.M., Birch R., Kelso C., Beck J.L., Kapralov M.V. Improving recombinant RuBisCO biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proce of the Nat Acad of Scien. 2015;112(11):3564–3569. doi: 10.1073/pnas.1420536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hanson M.R., Lin M.T., Carmo-Silva A.E., Parry M.A. Towards engineering carboxysomes into C3 plants. The Plant J. 2016;87(1):38–50. doi: 10.1111/tpj.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Long B.M., Rae B.D., Rolland V., Förster B., Price G.D. Cyanobacterial CO2-concentrating mechanism components: function and prospects for plant metabolic engineering. Cur Opi in Plant Bio. 2016;31:1–8. doi: 10.1016/j.pbi.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 131.Songstad D.D., Petolino J.F., Voytas D.F., Reichert N.A. Genome editing of plants. Crit Rev in Plant Scien. 2017;36(1):1–23. [Google Scholar]
- 132.Bock R. Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Ann Rev of Plant Bio. 2015;66:211–241. doi: 10.1146/annurev-arplant-050213-040212. [DOI] [PubMed] [Google Scholar]
- 133.Conlan B., Whitney S. Preparing RuBisCO for a tune up. Natu Plant. 2018;4(1):12–13. doi: 10.1038/s41477-017-0089-2. [DOI] [PubMed] [Google Scholar]
- 134.Hu J., Nagarajan D., Zhang Q., Chang J.S., Lee D.J. Heterotrophic cultivation of microalgae for pigment production: A review. Biotec Adva. 2018;36(1):54–67. doi: 10.1016/j.biotechadv.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 135.Canelli G., Neutsch L., Carpine R., Tevere S., Giuffrida F., Rohfritsch Z., et al. Chlorella vulgaris in a heterotrophic bioprocess: study of the lipid bioaccessibility and oxidative stability. Algal Res. 2020;45 [Google Scholar]
- 136.Lauersen K.J., Willamme R., Coosemans N., Joris M., Kruse O., Remacle C. Peroxisomal microbodies are at the crossroads of acetate assimilation in the green microalga Chlamydomonas reinhardtii. Algal Res. 2016;16:266–274. [Google Scholar]
- 137.Barbier G., Oesterhelt C., Larson M.D., Halgren R.G., Wilkerson C., Garavito R.M., et al. Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physi. 2005;137(2):460–474. doi: 10.1104/pp.104.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sloth J.K., Wiebe M.G., Eriksen N.T. Accumulation of phycocyanin in heterotrophic and mixotrophic cultures of the acidophilic red alga Galdieria sulphuraria. Enzy and Micro Tech. 2006;38(1–2):168–175. [Google Scholar]
- 139.Fields F.J., Ostrand J.T., Mayfield S.P. Fed-batch mixotrophic cultivation of Chlamydomonas reinhardtii for high-density cultures. Algal Res. 2018;33:109–117. [Google Scholar]
- 140.Puzanskiy R., Shavarda A., Romanyuk D., Shishova M. The role of trophic conditions in the regulation of physiology and metabolism of Chlamydomonas reinhardtii during batch culturing. J of App Phyco. 2021;33(5):2897–2908. [Google Scholar]
- 141.Hallmann A. Algae biotechnology green cell-factories on the rise. Cur Biotec. 2015;4(4):389–415. [Google Scholar]
- 142.Südfeld C., et al. High-throughput insertional mutagenesis reveals novel targets for enhancing lipid accumulation in Nannochloropsis oceanica. Meta Eng. 2021;66:239–258. doi: 10.1016/j.ymben.2021.04.012. [DOI] [PubMed] [Google Scholar]
- 143.Lauersen K.J. Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Plant. 2019;249(1):155–180. doi: 10.1007/s00425-018-3048-x. [DOI] [PubMed] [Google Scholar]
- 144.Specht E., Miyake-Stoner S., Mayfield S. Micro-algae come of age as a platform for recombinant protein production. Biotec Lett. 2010;32(10):1373–1383. doi: 10.1007/s10529-010-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Freudenberg R.A., Baier T., Einhaus A., Wobbe L., Kruse O. High cell density cultivation enables efficient and sustainable recombinant polyamine production in the microalga Chlamydomonas reinhardtii. Bior Tech. 2021;323 doi: 10.1016/j.biortech.2020.124542. [DOI] [PubMed] [Google Scholar]
- 146.Donato P., Finore I., Poli A., Nicolaus B., Lama L. The production of second generation bioethanol: The biotechnology potential of thermophilic bacteria. J of Clea Prod. 2019;233:1410–1417. [Google Scholar]
- 147.Holden, J. F. Hot Environments. In: Schaechter M (ed) Schmidt TM. Top. in Eco. and Enviro. Micro. Elsevier Inc, 313-332. (2012).
- 148.Straub C.T., Zeldes B.M., Schut G.J., Adams M.W., Kelly R.M. Extremely thermophilic energy metabolisms: biotechnological prospects. Cur Opi in Biotec. 2017;45:104–112. doi: 10.1016/j.copbio.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 149.Robb F., Antranikian G., Grogan D., Driessen A., editors. Thermophiles: biology and technology at high temperatures. CRC Press; 2007. [Google Scholar]
- 150.Ranawat P., Rawat S. Stress response physiology of thermophiles. Arch of Micro. 2017;199(3):391–414. doi: 10.1007/s00203-016-1331-4. [DOI] [PubMed] [Google Scholar]
- 151.Schultz J., Rosado A.S. Extreme environments: a source of biosurfactants for biotechnological applications. Extremo. 2020;24(2):189–206. doi: 10.1007/s00792-019-01151-2. [DOI] [PubMed] [Google Scholar]
- 152.Urbieta M.S., Donati E.R., Chan K.G., Shahar S., Sin L.L., Goh K.M. Thermophiles in the genomic era: biodiversity, science, and applications. Biotech Adv. 2015;33(6):633–647. doi: 10.1016/j.biotechadv.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 153.Jorquera M.A., Graether S.P., Maruyama F. Bioprospecting and biotechnology of extremophiles. Front in Bioen and Biotec. 2019;7:204. doi: 10.3389/fbioe.2019.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sarmiento F., Peralta R., Blamey J.M. Cold and hot extremozymes: industrial relevance and current trends. Front in Bioen and Biotec. 2015;3:148. doi: 10.3389/fbioe.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bruins M.E., Janssen A.E., Boom R.M. Thermozymes and their applications. Appl Bioch and biotechno. 2001;90(2):155–186. doi: 10.1385/abab:90:2:155. [DOI] [PubMed] [Google Scholar]
- 156.Irwin J.A., Baird A.W. Extremophiles and their application to veterinary medicine. Irish Vet J. 2004;57(6):1–7. doi: 10.1186/2046-0481-57-6-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Alber B.E. Biotechnological potential of the ethylmalonyl-CoA pathway. Appl Micro and Biotec. 2011;89(1):17–25. doi: 10.1007/s00253-010-2873-z. [DOI] [PubMed] [Google Scholar]
- 158.Pierson B.K., Castenholz R.W. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch of Micro. 1974;100(1):5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- 159.Kawai S., Nishihara A., Matsuura K., Haruta S. Hydrogen-dependent autotrophic growth in phototrophic and chemolithotrophic cultures of thermophilic bacteria, Chloroflexus aggregans and Chloroflexus aurantiacus, isolated from Nakabusa hot springs. FEMS Micro Lett. 2019;366(10):fnz122. doi: 10.1093/femsle/fnz122. [DOI] [PubMed] [Google Scholar]
- 160.Keller M.W., et al. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proce of the Nat Acad of Scien. 2013;110(15):5840–5845. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jahn U., Huber H., Eisenreich W., Hügler M., Fuchs G. Insights into the autotrophic CO2 fixation pathway of the archaeon Ignicoccus hospitalis: comprehensive analysis of the central carbon metabolism. J of Bac. 2007;189(11):4108–4119. doi: 10.1128/JB.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Heimerl T., et al. A complex endomembrane system in the archaeon Ignicoccus hospitalis tapped by Nanoarchaeum equitans. Front in Micro. 2017;8:1072. doi: 10.3389/fmicb.2017.01072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Imanaka H., Fukui T., Atomi H., Imanaka T. Gene cloning and characterization of fructose-1, 6-bisphosphate aldolase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J of Biosc and Bioeng. 2002;94(3):237–243. doi: 10.1263/jbb.94.237. [DOI] [PubMed] [Google Scholar]
- 164.Nunoura T., Chikaraishi Y., Izaki R., Suwa T., Sato T., Harada T., et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Scien. 2018;359(6375):559–563. doi: 10.1126/science.aao3407. [DOI] [PubMed] [Google Scholar]
- 165.Aoshima M., Igarashi Y. Non decarboxylating and decarboxylating isocitrate dehydrogenases: oxalosuccinate reductase as an ancestral form of isocitrate dehydrogenase. J of Bac. 2008;190(6):2050–2055. doi: 10.1128/JB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Aoshima M., Ishii M., Igarashi Y. A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mole Micro. 2004;52(3):751–761. doi: 10.1111/j.1365-2958.2004.04009.x. [DOI] [PubMed] [Google Scholar]
- 167.Robb, F. T., & Techtmann, S. M. Life on the fringe: microbial adaptation to growth on carbon monoxide. F1000 Res., 7. (2018). [DOI] [PMC free article] [PubMed]
- 168.Liu Y., Whitman W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic Archaea. Annals of the New York Acad. of Scien. 2008;1125(1):171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- 169.Seravalli J., Kumar M., Lu W.P., Ragsdale S.W. Mechanism of carbon monoxide oxidation by the carbon monoxide dehydrogenase/acetyl-CoA synthase from Clostridium thermoaceticum: kinetic characterization of the intermediates. Bioche. 1997;36:11241–11251. doi: 10.1021/bi970590m. [DOI] [PubMed] [Google Scholar]
- 170.Techtmann S.M., Colman A.S., Robb F.T. ‘That which does not kill us only makes us stronger’: the role of carbon monoxide in thermophilic microbial consortia. Enviro Micro. 2009;11(5):1027–1037. doi: 10.1111/j.1462-2920.2009.01865.x. [DOI] [PubMed] [Google Scholar]
- 171.De Anda V., Chen L.-X., Dombrowski N., Hua Z.-S., Jiang H.-C., Banfield J.F., et al. Brockarchaeota, a novel Archaeal phylum with unique and versatile carbon cycling pathways. Natu Comm. 2021;12(1) doi: 10.1038/s41467-021-22736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sousa D.Z., Visser M., van Gelder A.H., Boeren S., Pieterse M.M., Pinkse M.W.H., et al. The deep-subsurface sulfate reducer Desulfotomaculum kuznetsovii employs two methanol-degrading pathways. Natu Comm. 2018;9(1) doi: 10.1038/s41467-017-02518-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Van Haaster D.J., Silva P.J., Hagedoorn P.L., Jongejan J.A., Hagen W.R. Reinvestigation of the steady-state kinetics and physiological function of the soluble NiFe-hydrogenase I of Pyrococcus furiosus. J of Bac. 2008;190(5):1584–1587. doi: 10.1128/JB.01562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gadkari D., Schricker K., Acker G., Kroppenstedt R.M., Meyer O. Streptomyces thermoautotrophicus sp. nov., a thermophilic CO-and H2-oxidizing obligate chemolithoautotroph. Appl and Enviro Micro. 1990;56(12):3727–3734. doi: 10.1128/aem.56.12.3727-3734.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.MacKellar D., et al. Streptomyces thermoautotrophicus does not fix nitrogen. Sci Rep. 2016;6:20086. doi: 10.1038/srep20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Volpiano, C. G., Sant'Anna, F. H., da Mota, F. F., Sangal, V., Sutcliffe, I., Munusamy, M., ... & Rosado, A. S. Proposal of Carbonactinosporaceae fam. nov. within the class Actinomycetia. Reclassification of Streptomyces thermoautotrophicus as Carbonactinospora thermoautotrophica gen. nov., comb. nov. Syst. and Appl. Micro., 44(4), 126223. (2021). [DOI] [PubMed]
- 177.Guterres, A. United Nations Secretary-General. Carbon Neutrality by 2050: The World’s Most Urgent Mission (United Nations, 2020);Carbon neutrality by 2050: the world’s most urgent mission | United Nations Secretary-General.
- 178.Chen S.C., Sun G.X., Yan Y., Konstantinidis K.T., Zhang S.Y., Deng Y., et al. The Great Oxidation Event expanded the genetic repertoire of arsenic metabolism and cycling. Proce of the Nat Acad of Scien. 2020;117(19):10414–10421. doi: 10.1073/pnas.2001063117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li H., Opgenorth P.H., Wernick D.G., Rogers S., Wu T.Y., Higashide W., et al. Integrated electromicrobial conversion of CO2 to higher alcohols. Scien. 2012;335(6076):1596. doi: 10.1126/science.1217643. [DOI] [PubMed] [Google Scholar]
- 180.Nürnberg D.J., Morton J., Santabarbara S., Telfer A., Joliot P., Antonaru L.A., et al. Photochemistry beyond the red limit in chlorophyll f-containing photosystems. Scien. 2018;360(6394):1210–1213. doi: 10.1126/science.aar8313. [DOI] [PubMed] [Google Scholar]
- 181.Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164(6):1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 182.Clarke L., Kitney R. Developing synthetic biology for industrial biotechnology applications. Bioch Society Trans. 2020;48(1):113–122. doi: 10.1042/BST20190349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.from heterologous RuBisCO expression to the Calvin-Benson-Bassham cycle Antonovsky, N., Gleizer, S., Milo, R. Engineering carbon fixation in E. coli. Cur Opi in Biotec. 2017;47:83–91. doi: 10.1016/j.copbio.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 184.Song S., Timm S., Lindner S.N., Reimann V., Hess W.R., Hagemann M., et al. Expression of formate-tetrahydrofolate ligase did not improve growth but interferes with nitrogen and carbon metabolism of Synechocystis sp. PCC 6803. Front. Micro. 2020;11:1650. doi: 10.3389/fmicb.2020.01650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bathke J., Konzer A., Remes B., McIntosh M., Klug G. Comparative analyses of the variation of the transcriptome and proteome of Rhodobacter sphaeroides throughout growth. BMC Geno. 2019;20(1):1–13. doi: 10.1186/s12864-019-5749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fuchs G., Berg I.A. Unfamiliar metabolic links in the central carbon metabolism. J of Biotec. 2014;192:314–322. doi: 10.1016/j.jbiotec.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 187.Strauss, G., & fuchs, G. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3‐hydroxypropionate cycle. European Journal of Biochemistry, 215(3), 633-643. (1993). [DOI] [PubMed]